Abstract
Lissencephaly-1 (Lis1) is a key cofactor for dynein-mediated intracellular transport towards the minus-ends of microtubules (MTs). It remains unclear whether Lis1 serves as an inhibitor or an activator of mammalian dynein motility. Here we use single-molecule imaging and optical trapping to show that Lis1 does not directly alter the stepping and force production of individual dynein motors assembled with dynactin and a cargo adaptor. Instead, Lis1 promotes the formation of an active complex with dynactin. Lis1 also favors the recruitment of two dyneins to dynactin, resulting in increased velocity, higher force production and more effective competition against kinesin in a tug-of-war. Lis1 dissociates from motile complexes, indicating that its primary role is to orchestrate the assembly of the transport machinery. We propose that Lis1 binding releases dynein from its auto-inhibited state, which provides a mechanistic explanation for why Lis1 is required for efficient transport of many dynein-associated cargoes in cells.
Cytoplasmic dynein (dynein hereafter) is an AAA+ motor responsible for nearly all motility and force generation towards the MT minus-end1–3. Dynein is involved in a wide variety of cellular functions, such as positioning of intracellular organelles, breakdown of the nuclear envelope and assembly of the mitotic spindle4–6. The partial loss of dynein function has been implicated in a range of neurogenerative and neurodevelopmental conditions, including spinal muscular atrophy, amyotrophic lateral sclerosis, Alzheimer’s disease, and schizophrenia7–9.
The core of the dynein complex (1.4 MDa) is a homodimer of two heavy chains10. The C-terminal motor domain of the heavy chain is a catalytic ring of six AAA modules (AAA1-6). Unlike kinesin, dynein’s MT binding domain is separated from the catalytic domain by a coiled-coil stalk11. Nucleotide-dependent conformational changes of the linker drive the motility towards the MT minus-end12, 13. The N-terminal tail dimerizes the heavy chains14–16 and binds smaller polypeptides1, 17, 18. When dynein is not bound to its cargo, it forms two distinct conformations, the phi-particle, and open conformation, both of which move poorly along MTs19, 20. In the phi conformation, two motor domains self-dimerize through interactions between their linker, AAA+ ring and stalk regions and dynein weakly interacts with MTs. In the open conformation, the motor domains are more flexible and point towards each other, which is unfavorable for processive motility19, 21. Transitions between the phi and open conformation are proposed to be an important part of dynein regulation19, 21, but the molecular cues that govern this transition remain unclear.
Recent studies suggested that dynein and its cofactor dynactin are recruited to cargos through coiled-coil adaptor proteins in a mutually dependent manner (Fig. 1a)22–24. Formation of a dynein-dynactin-cargo adaptor complex aligns the dynein motor domains in a parallel conformation and activates processive motility along MTs25, 26. These adaptors recruit dynein to a specific set of cargos24, 27, enabling a single dynein gene to be responsible for nearly all minus-end directed functions along MTs. Members of the BicD family, BicD2 and BicDR1, are well-characterized coiled-coil adaptors that link dynein to Golgi-derived Rab6 vesicles, as well as nuclear pore complexes and viruses26, 28, 29. In vitro reconstitution studies showed that BicDR1 recruits two dyneins to dynactin, while the N-terminal coiled-coil domain of BicD2 (BiCD2N) mostly recruits a single dynein30, 31. Recruitment of two dyneins per dynactin results in complexes assembled with BicDR1 (DDR) moving faster and producing more force than complexes formed with BicD2N (DDB)30, 32. The differences elicited by BicD2 and BicDR1 in dynein motility may play a critical role in the sorting of Rab6 vesicles during neuronal differentiation23, 28.
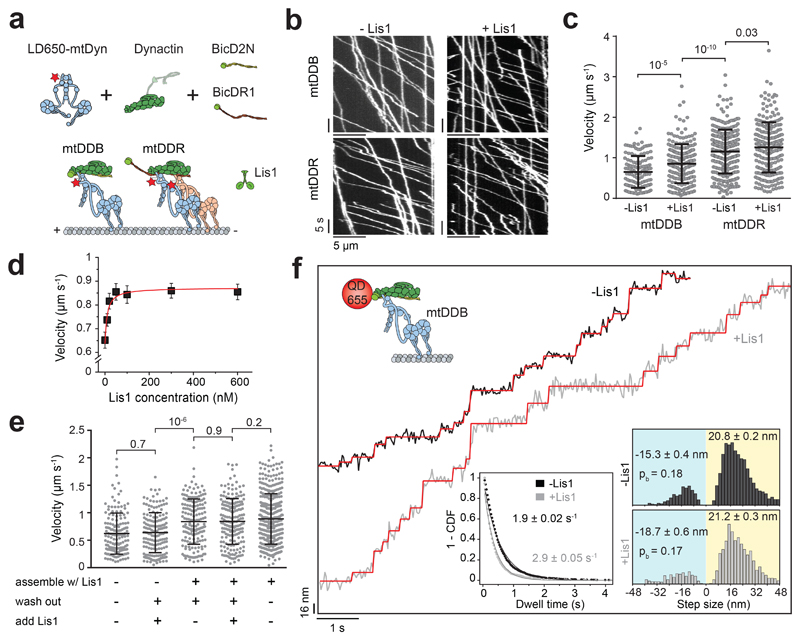
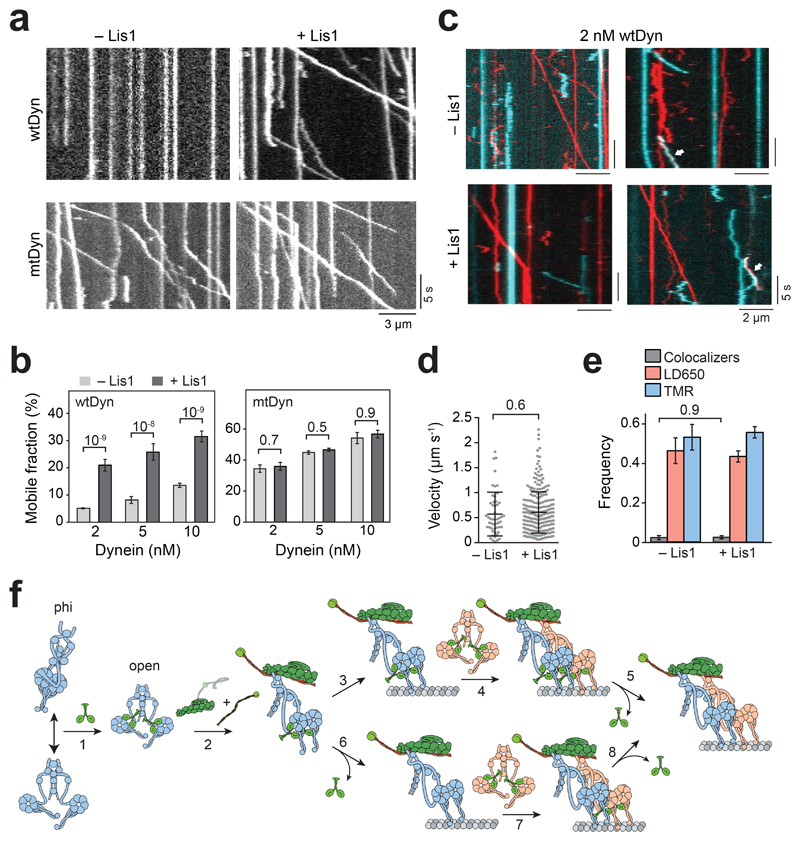
Figure 1. Lis1 increases the stepping rate of dynein-dynactin.
(a) Schematic depiction of the mammalian dynein-dynactin-cargo adaptor complexes. BicD2N primarily recruits single dynein to dynactin (DDB), whereas BicDR1 recruits two dyneins (DDR). Lis1 binds to the dynein motor domain. (b) Kymographs show the motility of mtDDB and mtDDR on MTs. (c) Velocity distribution of mtDDB and mtDDR with and without 600 nM Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 132, 217, 307, and 241, and mean values are 652, 854, 1155, and 1259 nm s-1. In a-c, four independent experiments were performed per condition. (d) The velocity of mtDDB under different Lis1 concentrations (mean ± SEM). mtDDB complex was assembled in the presence of Lis1, followed by removing excess protein and introducing Lis1 into the flow chamber. The red curve represents a fit to a Hill equation with n = 1. From left to right, n = 132, 216, 177, 204, 156, 179, and 217 (three independent experiments). (e) Velocity distribution of mtDDB assembled in the absence and presence of 600 nM Lis1 under different assembly conditions. The line and whiskers represent the mean and SD, respectively. From left to right, n = 152, 161, 170, 183, and 387 and mean values are 622, 638, 838, 842, and 888 nm s-1 (three independent experiments). (f) Step analysis of QD-labeled mtDDB (top insert) at 2 μM ATP in the presence and absence of 600 nM Lis1. Red staircases represent a fit to a step finding algorithm. (Bottom, left) Inverse cumulative distribution of dwell times between consecutive steps along the longitudinal axis. Solid curves represent fitting to an exponential decay (decay rate ± SE, n = 2138 for -Lis1 and 1441 for +Lis1). (Bottom, right) Normalized histograms of step sizes (n = 2,076 steps for -Lis1 and 1,374 for +Lis1, six independent experiments). Average forward and backward step sizes and the probability of backward stepping (pb) are shown (± SEM). In c and e, p values are calculated from a two-tailed t-test.
Dynein motility is also regulated by Lis1, which directly interacts with the dynein motor domain33. Lis1 inhibition reduces the transport of a wide variety of cargoes in eukaryotic cells, including endosomes, lysosomes, mRNAs, centrosomes and nuclei34–41. The critical role of Lis1 is underscored by the discovery that haploinsufficiency of the Lis1 gene causes a smooth brain disorder (lissencephaly) in humans, which is associated with a failure of nuclear migration42. Lis1 forms a homodimer, with each monomer comprising of an N-terminal dimerization domain and a C-terminal β-propeller domain that binds dynein at the interface between AAA3/4 and the coiled-coil stalk (Fig. 1a)33, 43.
The mechanism by which Lis1 regulates dynein motility is controversial. In vitro studies on yeast dynein revealed that a Lis1 homolog, Pac1 increases MT affinity, blocks nucleotide-dependent remodeling of the linker domain, and significantly reduces dynein velocity33, 44. However, the view of Lis1 as a dynein inhibitor is difficult to reconcile with the ability of Lis1 to promote dynein-mediated cargo transport in vivo34–41. Studies on isolated mammalian dynein proposed that Lis1 transiently interacts with dynein, enhances dynein’s affinity to MTs on high-load cargos by inducing a persistent-force generation state37, 45. However, Lis1’s function is not restricted to high-load cargos, and it is also required for the transport of smaller cargos36, 38, 39, 41. These studies were performed before it was understood that isolated dynein motors are autoinhibited in the absence of dynactin and a cargo adaptor, and may not reflect the force generation mechanism of active dynein-dynactin complexes24, 27, 46. In vivo studies gave rise to models that Lis1 is only required for targeting dynein to the MTs, with dissociation of Lis1 triggering the initiation of transport34, 38, 47–49 and that Lis1 promotes the interaction of dynein and dynactin39, 40. Consistent with these models, recent in vitro studies showed that mammalian Lis1 can increase the frequency and velocity of DDB motility49–51, but the underlying mechanism remained unknown.
In this study, we determined the effect of Lis1 binding on the motility, stepping, and force generation of DDB and DDR using single-molecule imaging and optical trapping in vitro. We found that Lis1 has no significant effect on the stepping and force generation of single dyneins after they have associated with dynactin and the cargo adaptor. Instead, Lis1 promotes the assembly of dynein with dynactin. Lis1 also favors the association of two dyneins to dynactin, and this accounts for the increase in both velocity and force generation of the complex. The presence of Lis1 shifts the force balance towards dynein’s direction during a tug-of-war with a plus-end directed kinesin. Our work reveals how Lis1 activates the motility of mammalian dynein-dynactin and is thereby required for efficient transport of cargos in cells.
Results
Lis1 increases the stepping rate of dynein-dynactin
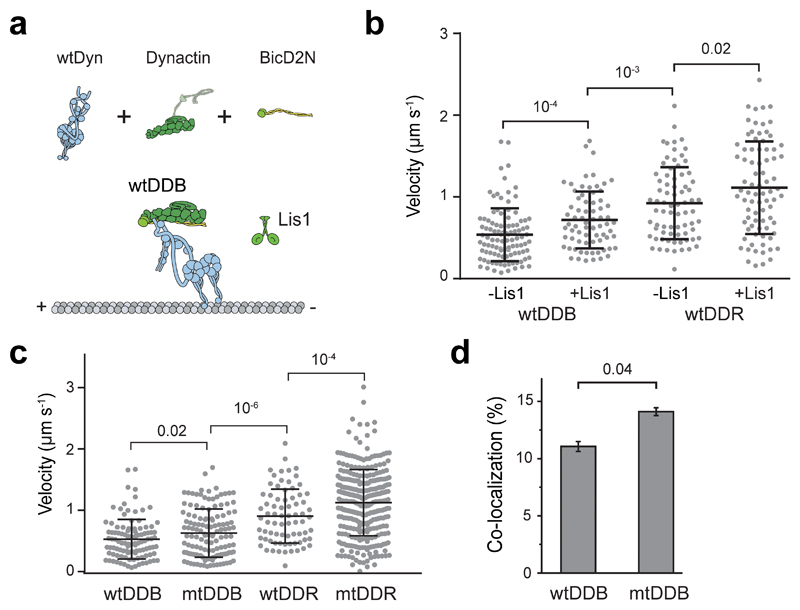
We first tested the effect of human Lis1 on the velocity of DDB and DDR complexes assembled with wild-type human dynein (wtDyn). In agreement with previous measurements50, 51, wtDDB moved 30% faster in 600 nM Lis1 (two-tailed t-test, p = 10-4). We also observed a modest (10%) increase in wtDDR velocity by Lis1 addition (Extended Data Fig. 1). Similar results were obtained using a dynein mutant (mtDyn) that disfavors the autoinhibited phi conformation21 (Fig. 1a-c, and Supplementary Movies 1 and 2). By titrating the Lis1 concentration, we found that 20 nM Lis1 is sufficient for the increased velocity of mtDDB (Fig. 1d). Because mtDyn favors the assembly of active dynein-dynactin complexes21, we used this construct to study the effect of Lis1 on dynein motility and force generation.
We altered the order of dynein-dynactin assembly and Lis1 addition to determining whether Lis1 is needed before or after the assembly of mtDDB for its faster movement. First, mtDDB was assembled in the presence of 75 nM Lis1. Removal of excess Lis1 from the flow chamber as the complexes moved along MTs did not lead to the slowdown of motility. Second, we assembled mtDDB without Lis1 and removed excess dynein, dynactin, and BicD2N from the flow chamber before adding Lis1. We observed that the addition of Lis1 after the complexes were being formed had no positive effect on mtDDB speed (Fig. 1e). These results show that Lis1 must be present during the assembly of dynein-dynactin to increase velocity, and it is dispensable after the complexes walk along the MT.
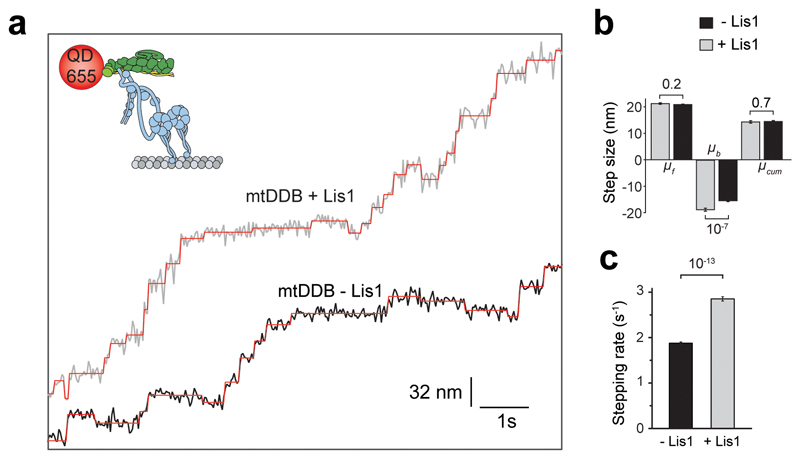
To distinguish whether Lis1 addition increases dynein step size or stepping rate for faster movement, we determined the stepping behavior of dynein-dynactin with and without 600 nM Lis1 at limiting (2 μM) ATP concentrations. We labeled dynein with a bright quantum dot (QD) at its N-terminus and tracked the motility of single mtDDB complexes at nanometer precision. In the absence of Lis1, mtDDB has a highly variable step size, frequently taking backward steps (Fig. 1f)32. Lis1 addition did not alter the size and direction of steps taken by dynein. Instead, mtDDB stepped more frequently in the presence of Lis1 (2.9 ± 0.05 vs. 1.9 ± 0.02 s-1, ±SE; two-tailed t-test, p = 10-13; Fig. 1f and Extended Data Fig. 2). We concluded that Lis1 increases the dynein stepping rate, not the mean step size, which accounts for the faster movement.
Lis1 increases the force production of dynein-dynactin
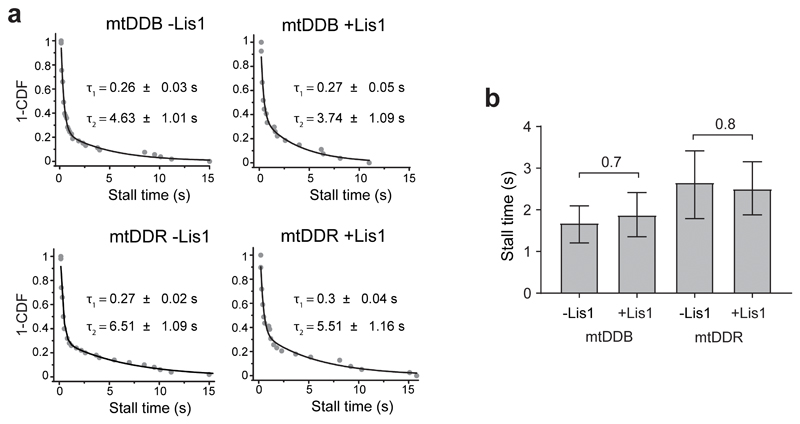
We tested whether Lis1 addition affects the force generation of dynein when this motor forms an active complex with dynactin and a cargo adaptor using an optical trap. We sparsely coated polystyrene beads with BicD2N and BicDR1 adaptors and assembled mtDDB and mtDDR complexes on beads32. Using a fixed trap, we observed that the stall force of mtDDB increases by 22% in 600 nM Lis1 (4.1 ± 0.1 vs. 5.4 ± 0.1 pN, mean ± SEM; two-tailed t-test, p = 10-11; Fig. 2a-c). Lis1 addition also resulted in a modest increase in mtDDR stall force (5.4 ± 0.1 vs. 6.1 ± 0.1 pN; two-tailed t-test, p = 10-4; Fig. 2c). Unlike isolated dynein45, we did not observe an increase in the stall duration of mtDDB and mtDDR in the presence of Lis1 (Fig. 2d and Extended Data Fig. 3).
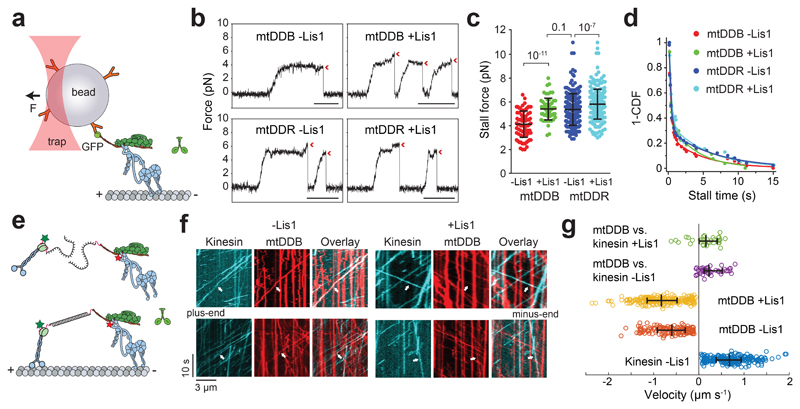
Figure 2. Lis1 increases the force production of dynein-dynactin.
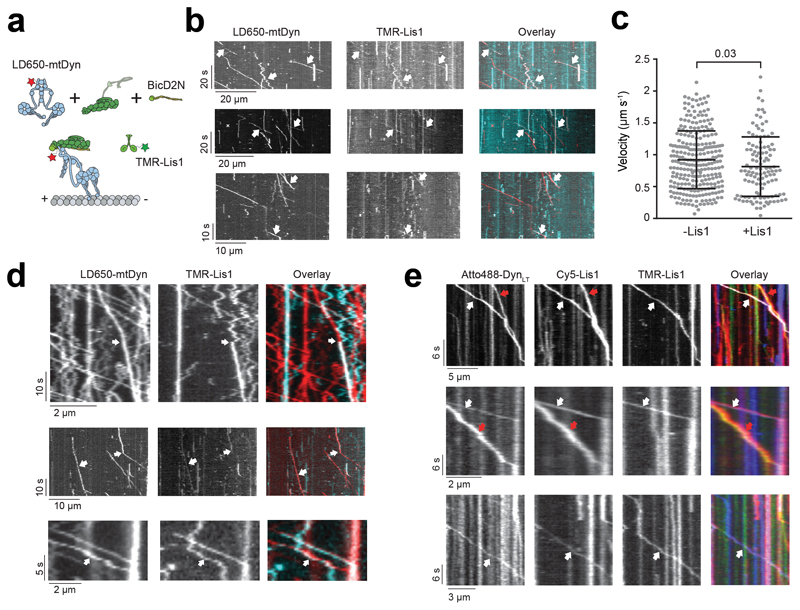
(a) Schematic of a fixed optical trapping assay for measuring the dynein stall force. (b) Typical stalls of beads driven by a single mtDDB or mtDDR. Red arrowheads denote the detachment of the motor from the MT after the stall event. Scale bars are 1 s. (c) Distribution of motor stall forces in the absence and presence of 600 nM Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 80 from 19 beads, 61 from 15 beads, 212 from 38 beads, and 152 from 32 beads, and mean values, are 4.1, 5.4, 5.4, and 6.1 pN. p-values are calculated from a two-tailed t-test. In b and c, four independent experiments were performed per condition. (d) Inverse cumulative distribution of stall durations of mtDDB and mtDDR in the presence and absence of Lis1. Solid curves represent fitting to a two-exponential decay (decay time ± SE, n = 53, 27, 50, and 39 from left to right). (e) Schematic depiction of the in vitro tug-of-war assay. DDB and kinesin were labeled with different-colored fluorescent dyes and tethered using a DNA scaffold. (f) Representative kymographs show the motility of LD650-dynein (red) and TMR-kinesin (cyan) in the absence and presence of 600 nM Lis1. White arrows show DDB-kinesin colocalizers. (g) Velocity distribution of mtDDB, kinesin, and mtDDB-kinesin assemblies in the absence and presence of Lis1. The centerline and whiskers represent the median and 65% CI, respectively. From top to bottom, n = 33, 45, 217, 132, and 210, and median values are 233, 185, −836, −604, and 670 nm s-1. In f and g, three independent experiments were performed per condition. Negative velocities represent movement towards the MT minus-end.
We then tested whether the increase in mtDDB force production by Lis1 also increases the likelihood of DDB to win a tug-of-war against a plus-end-directed kinesin-1. We labeled mtDyn and tail-truncated wild-type human kinesin-1 with different fluorescent dyes and pitted one mtDDB against one kinesin-1 using a DNA tether. Consistent with our previous measurements46, the majority (87%) of kinesin-DDB assemblies moved towards the plus-end in the absence of Lis1. The median velocity (185 nm s-1 towards the plus-end) was noticeably higher than previous measurements (26 nm s-1) that used a cysteine-light mutant of kinesin-146, likely because the cysteine-light kinesin mutant, but not native kinesin, can be forced to move backward under resistive forces52. The addition of Lis1 increased the percentage of complexes moving towards the minus-end from 13% to 22% and increased the mean velocity of minus-end-moving assemblies by 6-fold (353 ± 68 vs. 55 ± 8 nm s-1; two-tailed t-test, p = 0.02; Fig 2e-g and Supplementary Movie 3). Collectively, these results demonstrate that Lis1 addition increases the likelihood of DDB winning against kinesin in a tug-of-war.
Lis1 does not affect force generation of complexes with single dynein
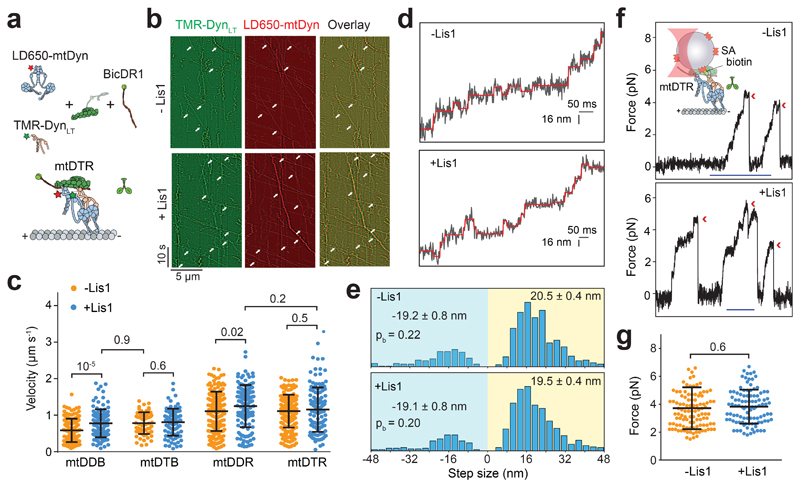
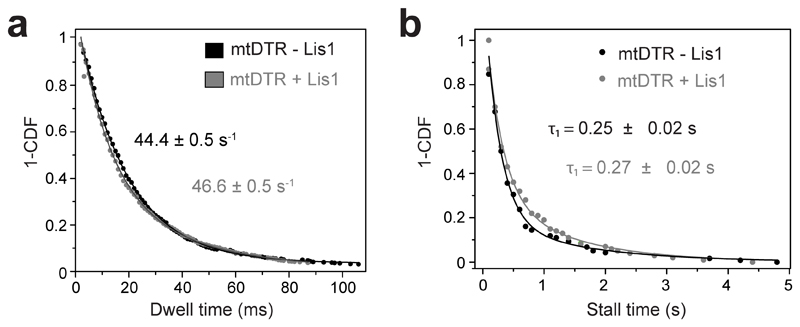
We next turned our attention to understanding how Lis1 increases the velocity and force production of mtDDB and mtDDR. To test whether Lis1 alters mechanochemical properties of single dynein assembled with dynactin, we mixed LD650-labeled full-length mtDyn and a TMR-labeled dynein tail construct (DynLT, containing residues 1–1,074 of the heavy chain and associated chains)30. Because DynLT lacks the motor domain, processive runs of this construct can only be achieved through its side-by-side recruitment with single full-length mtDyn to dynactin. We measured the velocity of dual-labeled complexes that contain both mtDyn and DynLT in the presence of BicD2N (mtDTB) and BicDR1 (mtDTR). Remarkably, Lis1 addition did not affect the mean velocity of complexes containing single dynein (Fig. 3a-c and Supplementary Movie 4). To test whether Lis1 alters the stepping properties of single dynein bound to dynactin, we tracked beads driven by single mtDTR under constant 1 pN hindering force exerted by the trap. Unlike mtDDB, Lis1 addition did not alter the stepping rate of mtDTR in 1 mM ATP (two-tailed t-test, p = 0.83; Fig. 3d-e and Extended Data Fig. 4a). In addition, Lis1 addition did not affect the stall force and duration (two-tailed t-test, p = 0.6; Fig. 3f-g and Extended Data Fig. 4b). We concluded that Lis1 does not directly affect the mechanical properties of single dynein bound to dynactin.
Figure 3. Lis1 does not affect the force generation and velocity of single dynein complexed to dynactin and a cargo adaptor.
(a) Schematic depiction of the mtDTR complex. Full-length dynein and DynLT are labeled with LD650 and TMR dyes, respectively. (b) Representative kymographs show the motility of mtDyn and DynLT. Arrows represent the colocalization of TMR and LD650. (c) Velocity distribution of mtDDB, mtDTB, mtDDR and mtDTR in the presence and absence of 600 nM Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 144, 117, 65, 88, 209, 134, 213, and 126 and mean values are 584, 778, 783, 809, 1108, 1248, 1111, and 1154 nm s-1. In b and c, three independent experiments were performed per condition. (d) Example traces of beads driven by mtDTR in the presence and absence of 600 nM Lis1 against 1 pN hindering force. The raw stepping data are shown in black and the steps fitting are in red. (e) Normalized histograms of mtDTR steps taken in the longitudinal direction. In d and e, n = 734 for -Lis1 and 724 for +Lis1 (three independent experiments per condition). Average sizes of steps taken in forward and backward directions (± SEM) and the probability of backward stepping in the presence and absence of Lis1 are indistinguishable (p = 0.6, two-tailed t-test). (f) (Top insert) Streptavidin (SA)-coated beads are sparsely decorated with biotin-DynLT in the presence of mtDyn, dynactin, and BicDR1, and trapped with a focused laser beam. Traces represent typical stalls of beads driven by mtDTR in the absence and presence of 600 nM Lis1. Red arrowheads denote the detachment of the motor from the MT after the stall event. Scale bar is 1 s. (g) Distribution of mtDTR stall force. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 111 stalls from 23 beads and 101 stalls from 21 beads, and mean values are 3.7 and 3.8 pN. In f and g, three independent experiments were performed per condition. In c and g, p-values are calculated from a two-tailed t-test.
Lis1 promotes the recruitment of two dyneins to dynactin
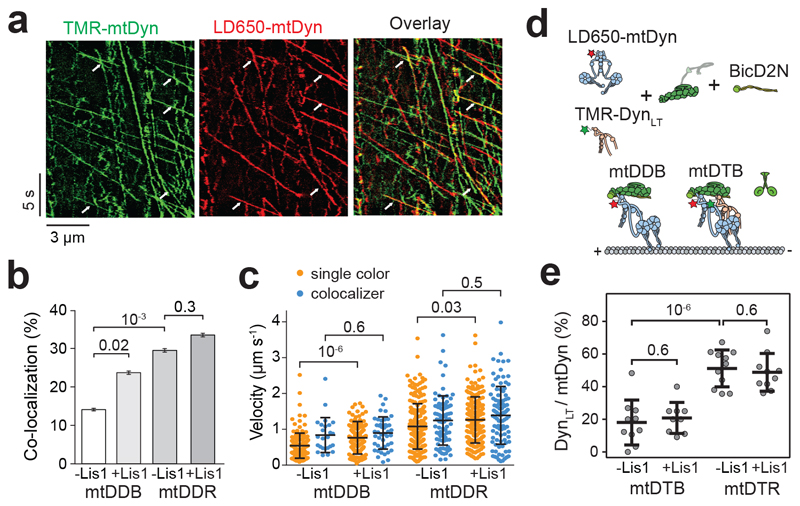
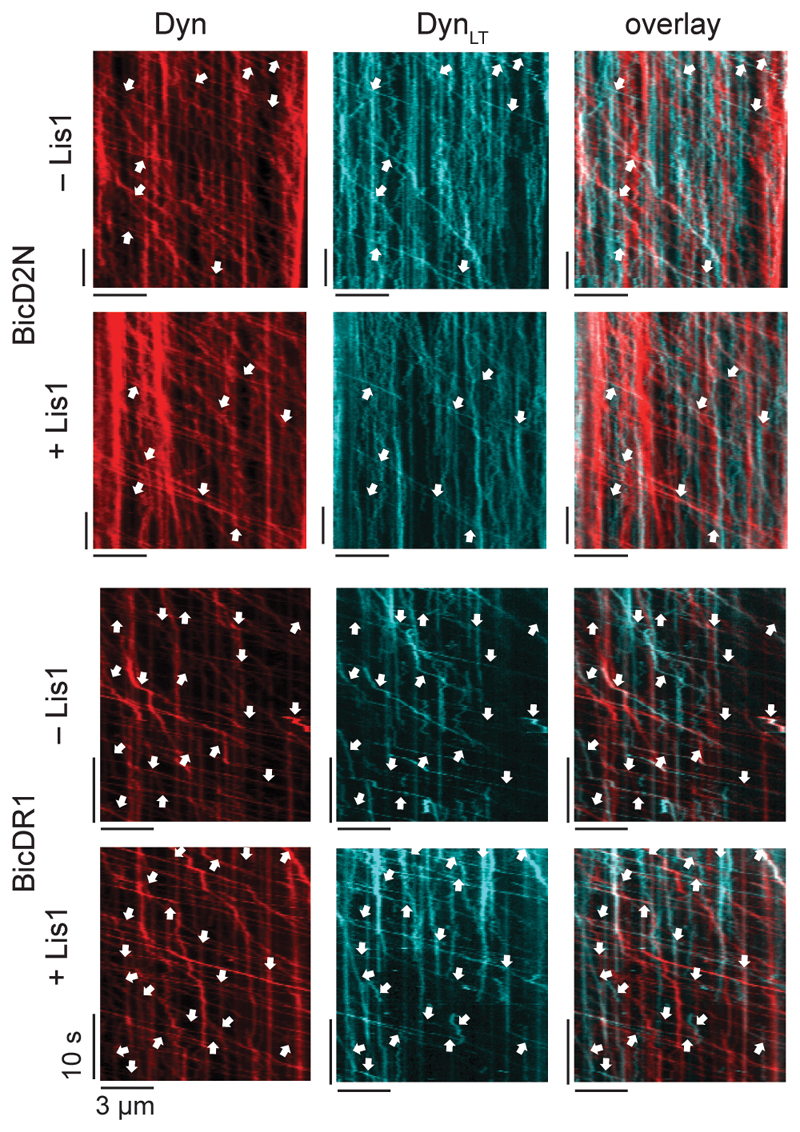
The effect of Lis1 on DDB velocity and force production is strikingly similar to the recruitment of second dynein to dynactin30, 32, leading us to hypothesize that Lis1 regulates the stoichiometry of dynein per dynactin. To test this possibility, we mixed TMR- and LD650-labeled dynein with dynactin and BicD2N. Dual-colored complexes contain two dyneins, while single-colored complexes contain either one or two dyneins. Lis1 addition increased the percentage of dual-colored complexes from 14% to 24% (p = 0.018, two-tailed t-test, Fig. 4a-b and Supplementary Movie 5). After correction for labeling efficiency and complexes dual-labeled with the same color30, we estimated that Lis1 addition increases the percentage of complexes containing two dyneins from 22% to 42%. Consistent with our hypothesis, Lis1 addition did not increase the velocity of dual-colored complexes that contain two dyneins (Fig. 4c). Similar results were obtained when BicDR1 was used as a cargo adaptor, but the effect of Lis1 addition was modest, presumably because DDR complexes are already predisposed to contain two dyneins30. Collectively, these results demonstrate that Lis1 favors the recruitment of two dyneins to dynactin, which accounts for the faster velocity of these complexes30, 32, 50, 51. We did not observe an increase in the recruitment of DynLT side-by-side with mtDyn to dynactin in the presence of Lis1 (Fig. 4d-e and Extended Data Fig. 5), suggesting that Lis1 favors the recruitment of two dyneins to dynactin through its interactions with the motor domain, not with the tail domain53, 54 of dynein.
Figure 4. Lis1 favors the recruitment of two dyneins to dynactin.
(a) Representative kymographs show the motility of LD650- and TMR-labeled dynein in the presence of dynactin, BicD2N, and 600 nM Lis1. Arrows represent TMR and LD650 colocalization. (b) The percentage of processive complexes that contain both TMR and LD650 signals (mean ± SEM, n = 178, 190, 289, 290 from left to right). Error bars represent SE calculated from multinomial distribution and p-values are calculated from a two-tailed z-test. (c) Velocity distribution of single-colored and dual-colored complexes of DDB and DDR in the presence and absence of Lis1. The line and whiskers represent the mean and SD, respectively. From left to right, n = 153, 25, 145, 45, 204, 85, 193, and 97 and mean values are 544, 840, 766, 899, 1082, 1248, 1263, and 1390 nm s-1. p-values are calculated from a two-tailed t-test. In a-c, four independent experiments were performed per condition. (d) Schematic shows the assembly of mtDDB and mtDTB complexes using TMR-DynLT, LD650-mtDyn, dynactin, and BicD2N. (e) The ratio of processive runs by TMR-DynLT to LD650-mtDyn on individual MTs in the presence and absence of Lis1. The line and whiskers represent the mean and SD, respectively (n = 10, 9, 11, and 10 MTs from left to right, three independent experiments). p values are calculated from a two-tailed t-test.
Lis1 dissociates from motile complexes
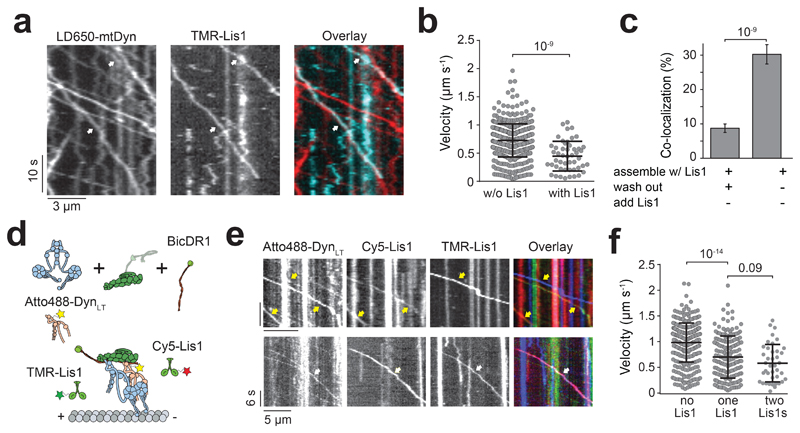
To determine whether Lis1 remains stably bound when dynein moves along MTs49–51, we mixed LD650-mtDyn and 50 nM TMR-Lis1 in the presence of dynactin and BicD2N. 30% of the motile mtDDB complexes contained Lis1. Removal of free Lis1 in assay solution after the initiation of processive motility reduced Lis1-dynein colocalization to 9%, suggesting that Lis1 can dynamically interact with dynein-dynactin as it moves along MT. In both cases, Lis1-bound complexes had a lower velocity than other mtDDB complexes (Fig. 5a-c, Extended Data Fig. 6a-c). On rare occasions, Lis1 diffused on an MT, hopped onto a motile DDB complex on the same MT (Extended Data Fig. 6d) and slowed down the motility33, 50. Therefore, Lis1 typically dissociates from motile complexes but reduces velocity if it remains bound to dynein. These results are not fully consistent with previous reports that Lis1 remains bound to nearly all motile complexes51 and Lis1-bound complexes have the same velocity50 or move faster than complexes that move without Lis151. The disparities may be related to differences in assay conditions and in vitro reconstitution methods.
Figure 5. Lis1 binding decreases the velocity of dynein/dynactin.
(a) Representative kymographs show the motility of mtDDB and Lis1 on MTs. White arrows represent the colocalization of LD650-Dyn (red) and TMR-Lis1 (cyan). (b) Velocity distribution of mtDDB and mtDDB-Lis1 assemblies. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 512, and 49 from left to right and mean values are 726 and 447 nm s-1. The p-value is calculated from a two-tailed t-test. In a and b, four independent experiments were performed per condition. (c) The percentage of processive complexes that contain both LD650-mtDyn and Lis1-TMR signals using different assembly conditions (see Methods; mean ± SEM, n = 561 and 387 from left to right). Error bars represent SE calculated from multinomial distribution and p-values are calculated from a two-tailed z-test. (d) Schematic depiction of mtDTR complex assembled in the presence of 50 nM TMR- and Cy5-Lis1. (e) Representative kymographs show the motility of mtDTR and Lis1 on MTs. Yellow arrows represent the colocalization of DynLT (green) and one color of Lis1. White arrows represent the colocalization of DynLT with Cy5-Lis1 (red), and TMR-Lis1 (cyan). (f) Velocity distribution of mtDTR that colocalizes with zero, one and two colors of Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 357, 172, and 40 and mean values are 985, 701, and 582 nm s-1. In e and f, three independent experiments were performed per condition. p-values are calculated from a two-tailed t-test.
Stoichiometry of Lis1 binding to dynein is also not well understood. DDB can concurrently recruit two Lis1 dimers50. However, it remains unclear whether each of the two dyneins in DDB binds to Lis1 or single dynein can simultaneously bind to two Lis1 dimers. To address this, we tested if two Lis1 molecules could bind to single full-length dynein recruited side-by-side with Atto488-DynLT (Fig. 5d). By quantifying DTR complexes colocalized with both TMR- and Cy5-Lis1, we estimated that 33% of DTR complexes with at least one Lis1 bound contain two Lis1s (Fig. 5e, Extended Data Fig. 6e and Supplementary Movie 6). Recruitment of two Lis1s leads to a further slowdown of the motility in comparison to single Lis1 (Fig. 5f). These results showed that single dynein can simultaneously recruit two Lis1 dimers.
Lis1 promotes the assembly of active dynein-dynactin complexes
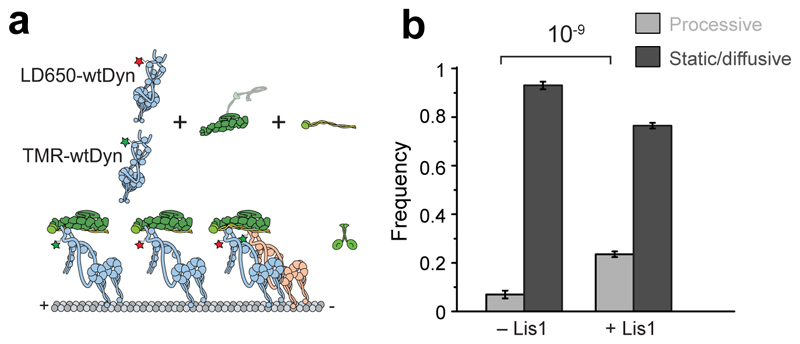
Lis1 stimulates the frequency of minus-end-directed transport under conditions insufficient to induce motility, such as when BicD2N concentration is low51. To determine how Lis1 favors initiation of dynein motility when the complex formation is strongly limiting, we quantified wtDDB motility while we lowered the wtDyn concentration 10-, 20-, and 50-fold compared to our standard assay condition (see Methods). In the absence of Lis1, the percentage of complexes exhibiting motility was decreased at lower dynein concentrations, and motility was almost fully abolished with the 50-fold dilution. Lis1 addition increased the percentage of motility by ~5-fold (Fig. 6a-b and Supplementary Movie 7), which is consistent with Lis1 favoring association of dynactin with dynein and the BicD2 orthologue in Drosophila cell extracts39. However, when we used mtDyn that does not form the phi conformation, we observed robust motility even in the 50-fold dilution condition and no significant increase in the percentage of motile complexes with the addition of Lis1 (Fig. 6a-b). We also mixed equal amounts of TMR- and LD650-wtDyn with dynactin and BicD2N and quantified the percentage of colocalizing complexes moving along the MTs under limiting dynein conditions (Extended Data Fig. 7). Only ~ 5% of these complexes were assembled with two dyneins and we did not observe an increase in wtDDB velocity by Lis1 addition (Fig 6c-e), suggesting that Lis1 promotes motility by recruiting single dynein to dynactin under these conditions. Therefore, Lis1 is also required for the assembly of the first dynein to dynactin and recruitment of the second dynein does not have to occur together with the first dynein.
Figure 6. Lis1 promotes assembly of the dynein transport machinery.
(a) Representative kymographs show the motility of DDB at 5 nM concentration of dynein in the absence and presence of 600 nM Lis1. (b) Ratio comparison of the number of processive runs by DDB to the total number of landed motors on MT (mean ± SEM). From left to right, n = 508, 355, 491, 262, 1244, and 392 for wtDyn, and 234, 459, 426, 1352, 457, and 859 for mtDyn. In a and b, three independent experiments were performed per condition. Error bars represent SE calculated from multinomial distribution and p values are calculated from a two-tailed z-test. (c) Representative kymographs show the motility of LD650- (red) and TMR- (cyan) wtDDB at 2 nM concentration of dynein in the absence and presence of 600 nM Lis1. Left kymographs show single-colored runs and right kymographs show rare events of TMR-LD650 colocalization (white arrows). (d) Velocity distribution of wtDDB motility assembled at 5 nM dynein concentration in the absence and presence of 600 nM Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 51 and 257, and mean values are 572 and 604 nm s-1. In c and d, three independent experiments were performed per condition. (e) Fraction of processive complexes that contain TMR, LD650, and TMR-LD650 colocalizers (mean ± SEM, n = 59 for -Lis1 and 303 for + Lis1). The p values are calculated from a two-tailed t-test in d and a two-tailed z test in e. (f) A model for Lis1-mediated assembly of the dynein-dynactin complex. (1) Lis1 binds to the open-conformation of dynein with one Lis1 dimer for each dynein motor domain. (2) Lis1 binding prevents transitions of the open conformation to the phi conformation, which increases the affinity of dynein to dynactin. This mechanism also favors the recruitment of second dynein to the complex resulting in higher force production and faster movement. Lis1 dissociates from active dynein-dynactin-cargo adaptor motors, either after pairing of two dyneins with dynactin (3-5) or during the assembly of the complex (6-8).
Discussion
Our results challenge previous views on how Lis1 binding regulates dynein motility. A previous study on isolated dynein suggested that Lis1 binding induces pausing of dynein motility and enhances MT affinity when dynein is subjected to force45. Studies on yeast dynein also suggested that Lis1 binding interferes with the powerstroke of the linker domain44, suggesting that Lis1 binding reduces dynein force generation. Our results with mammalian dynein-dynactin suggest a different mechanism. We found that the presence of Lis1 has no major effect on force generation of single dynein motors bound to dynactin and does not increase the time dynein stalls before dissociating from MTs under resistive loads. Therefore, our results do not support the view that the primary function of Lis1 is to regulate the tenacity of isolated dynein complexes to MTs33, 44. Instead, Lis1 favors the recruitment of dynein to dynactin, thereby promoting the assembly of an active complex for dynein motility. This finding explains the requirement for Lis1 for transport initiation in vivo34, 38. Consistent with two recent reports that studied the role of Lis1 in mammalian and yeast dynein-dynactin motility55, 56, we show that Lis1’s ability to promote the association of dynein with dynactin also favors the adoption of the two-motor state, which accounts for more frequent stepping and higher force generation per complex. The increased probability of recruiting two dyneins to dynactin also means that Lis1 induces more effective competition against kinesin in a tug-of-war, a result consistent with an increase in anterograde velocity observed when Lis1 is inhibited in cells37, 39. Remarkably, Lis1 does not have to be part of the complex to exert its effects on motility. Lis1 dissociates from most complexes before initiation of movement (Fig. 5), revealing that its primary role occurs during complex assembly. These results also provide an explanation for the recruitment of two dyneins to dynactin when the complex was pulled down by BicD2N from the brain lysate in which Lis1 is present31, compared to the recruitment of mostly a single dynein when the complex is assembled from purified components in the absence of Lis130.
Our results provide insights into how Lis1 enhances the affinity of dynein to dynactin. We show that this function is not dependent on reported interactions between Lis1 and the dynein tail, pointing instead to a mechanism that involves Lis1’s binding to the motor domain. Structural studies on yeast dynein showed that Lis1 binds to the motor domain at the interface between AAA3 and AAA4, and the coiled-coil stalk33, 43. Assuming that mammalian Lis1 binds dynein in a similar orientation, Lis1 binding sites are positioned close to the dimerization interface on the AAA+ ring and stalk in the phi particle21. We propose that Lis1 binds to the open conformation of dynein and prevents switching back to the phi conformation (Fig. 6f), thereby reducing dynein autoinhibition48. Because the open conformation has a higher affinity to dynactin than the phi conformation21, Lis1 promotes the assembly of dynein with dynactin and the cargo adaptor and favors the recruitment of two dyneins to each dynactin. The model explains why mtDDB is more likely to have two dynein motors and moves faster than wtDDB in the absence of Lis1 (Extended Data Fig. 1c-d). The model is also consistent with a recent report that the requirement of Lis1and NudE in HookA-mediated dynein activation in A. nidulans can be bypassed by expressing a mtDyn57. Since we obtained a similar increase in the velocity of mtDDB and wtDDB in the presence of Lis1, it is possible that Lis1-bound dynein has additional structural features not present in the open conformation and further stimulates the assembly of dynein and dynactin after the opening of the phi conformation. High-resolution structural studies will be needed to determine whether Lis1 binding induces conformational changes on the dynein heavy chain.
Our results are not consistent with a model in which a Lis1 dimer forms a bridge between two dyneins and recruits them simultaneously to dynactin, as we find that Lis1 is required for the assembly of complexes with single dynein at limiting dynein concentrations (Fig. 6 and Extended Data Fig. 7). Moreover, monomeric Lis1, which cannot crosslink two dyneins, also stimulates the assembly of dynein-dynactin55. Therefore, Lis1 favors assembly of the first dynein to dynactin, which does not have to occur simultaneously with the second dynein. It remains to be studied whether Lis1 remains bound to dynein motors for the assembly of the fully activated complex, or early dissociation of Lis1 from the first dynein could occur before the recruitment of the second Lis1-bound dynein (Fig. 6f).
Mutagenesis studies have indicated that the transition between the phi-particle and open conformation is a tightly regulated process in cells21. Future studies are required to test whether Lis1-mediated opening of the phi conformation is also regulated by other dynein-associated proteins, such as the Lis1 binding proteins NudE and NudEL47, 58–60.
Methods
Protein expression, labeling, and purification
Human SNAPf-wtDyn, SNAPf-mtDyn, SNAPf-DynLT (containing residues 1-1,074 of the heavy chain), BicD2N-GFP (containing residues 1-400), BicDR1-GFP, and Lis1-SNAPf were expressed in Sf9 cells and purified using IgG affinity chromatography (using a cleavable ZZ tag), as described previously27, 30. Sf9 cells were regularly tested for mycoplasma infection and no positive results were found. SNAP-tagged proteins were labeled with BG-functionalized biotin, TMR, Atto488 (NEB) or LD650 probes, and purified as described previously30. Dynactin was purified from pig brains using the large-scale SP-sepharose and MiniQ protocol26. Western blotting and mass spectrometry show no detectable levels of Lis1 in dynactin preps (V. Madan and S.L.B., in preparation). Human Kinesin-1(1-560)-SNAPf-GFP was expressed in BL21DE3 cells and purified using Ni-NTA affinity chromatography, as described previously61. Concentration of isolated proteins was quantified using the Bradford colorimetric assay.
Motility assays
Biotinylated MTs were prepared by mixing 190 μM of 2% biotin-labeled biotin with 0.9 mM of unlabeled pig brain tubulin in BRB80 buffer (80 mM PIPES pH 6.8, 1mM MgCl2, 1 mM EGTA), followed by the addition of equal volume of polymerization buffer (2x BRB80 supplemented with 2 mM GTP and 20% anhydrous Dimethyl sulfoxide (DMSO)). Tubulin was allowed to polymerize by incubation for 40 mins at 37°C, followed by the addition of 10 nM taxol and incubation for another 40 mins. Taxol-stabilized MTs were then pelleted at 20,000 g for 12 min and resuspended in BRB80 buffer containing 10 nM taxol and 1 mM Dithiothreitol (DTT).
The glass surface of motility chambers was first passivated by BSA and functionalized with biotin by flowing 1 mg/ml BSA-biotin (Sigma), followed by washing the chamber with 40 μl dynein buffer (DB: 30 mM HEPES pH 7.0, 5 mM MgSO4, 1 mM EGTA, 1 mM TCEP (tris(2- carboxyethyl)phosphine) supplemented with 1.25 mg/ml casein (DB-C) . To immobilize biotinylated MTs on the functionalized surface, the chamber was incubated with 1 mg/ml SA (NEB) and washed with DB-C.
For DDB, DDR, and DTR motility, 70 nM LD650-dynein was mixed with 150 nM dynactin, and 700 nM cargo adaptor (BicD2N-GFP or BicDR1-GFP) in 10 μl DB supplemented with 1 mg/ml Bovine Serum Albumin (BSA). For DTR experiments, 300 nM TMR-dynein tail was added to the mixture. For dynein co-localization experiments, 70 nM TMR-dynein was additionally included in the motility mix. The complexes were incubated on ice for 10 mins, diluted in DB-C, and flowed into the chamber. The motility mix was kept for 2 mins and then washed with 40 μl DB-C. To record motility, 20 μl dynein motility buffer (DMB: DB-C supplemented with 1 mM Mg.ATP, 2.5 mM PCA (protocatechuic acid), 35 μg/ml PCD (protocatechuate-3,4-dioxygenase)) was introduced into the chamber and the sample immediately imaged for 3 mins at 23°C. Unless otherwise indicated, experiments with unlabeled Lis1 were performed by mixing Lis1-SNAPf with DDB and introducing the reaction mixture into the chamber. The chamber was then washed with 40 μl DB-C, and Lis1-SNAPf was then reintroduced at the same concentration into the chamber with DMB before recording the motility. In Figure 5a-b, 50 nM TMR-Lis1 was added to the diluted motility mix (~ 1 nM LD650-mtDDB) and introduced into the motility chamber. The chamber was then washed with 40 μl DB-C and motility was recorded in the absence of free Lis1 in DMB. In Figure 5e-f, 50 nM TMR-Lis1 and 70 nM Cy5-Lis1 were incubated with 1 nM Atto488-mtDTR before introducing the mixture into the flow chamber. After washing out unbound proteins from the chamber with 40 μl DB-C, motility was recorded in the absence of free Lis1 in DMB. In Fig 1e, the role of Lis1 in DDB assembly and motility was tested by introducing and removing Lis1 in different stages of the sample preparation. In Extended Data Figure 6b-c 1 nM LD650-mtDDB and 75 nM TMR-Lis1 were introduced into the chamber in DMB containing 75 mM KOAC and motility was recorded without washing excess TMR-Lis1 in solution. Although Lis1 has a weak affinity to MTs, this has not affected the velocity of dynein-dynactin motility (Fig. 1d).
Single-molecule imaging was performed using a custom-built TIRF microscope equipped with a 100x 1.49 N.A. apochromat oil-immersion objective (Nikon) and a perfect focusing system on an inverted microscopy body (Nikon Ti-Eclipse). The fluorescence signal was detected using an electron-multiplied charge-coupled device (EM-CCD) camera (Andor, Ixon EM+). The sample that contained LD650 was excited with a 0.05 kW cm-2 633 laser beam (Coherent), and the emission signal was filtered using a 655/40 nm bandpass emission filter (Semrock). Movies were recorded using an effective pixel size of 160 nm at 300 ms per frame. For two- and three-color fluorescence assays, imaging was performed on a multi-color TIRF microscope (Nikon) using alternating excitation and time-sharing mode of emission collection. Atto488-, TMR- and LD650-labeled samples were excited using 0.05 kW cm-2 488, 532 and 633 nm laser beams (Coherent) and fluorescence signals were detected on an ImagEM X2 EM-CCD camera (Hamamatsu). Movies were recorded at 150 ms per frame per color for two-color and 100 ms per frame per color for three-color fluorescence assays. The effective pixel size was 108 nm.
Kymographs were generated from movies in ImageJ. Processive movement and velocity were then defined and measured, as described previously46. Briefly, kymographs were created by plotting segmented lines along each MT using ImageJ and individual runs were manually scored. Complexes that were static, diffusive or run shorter than 5 pixels (750 nm) along the MT were excluded from the analysis. In trajectories that exhibit pauses longer than 30 s, only the segments with unidirectional motility were analyzed. Pauses shorter than 30 s were included in the analysis. For two-color imaging, the two channels were overlaid using the merge function. Resulting kymographs were then manually scored for processive events that show co-localization between the two channels. In Figures 3b and 4a, kymographs were post-processed using the fast Fourier transform function of ImageJ for clarity. Labeling efficiency of dynein and Lis1 with at least one dye was 96% and 95% respectively, as determined by spectrophotometry. The fractions of the complexes containing two dyneins or Lis1s were calculated using the colocalization measurements, after accounting for unlabeled complexes, and complexes assembled with two dyneins that are labeled with the same color30.
High-resolution fluorescence-tracking assays
QDs were functionalized with benzyl guanine by mixing 8 μM amino (PEG) QDs emitting at 655 nm (ThermoFisher) with 20 mM BG-GLA-NHS (NEB) in 100 mM sodium borate buffer, pH 8.0 for 40 min at room temperature. To remove excess BG-GLA-NHS, functionalized QDs were concentrated through five consecutive spins through 100,000 MWCO centrifugal filter units (Amicon). Finally, spin-concentrated QDs were suspended in 100 μl DB and stored at 4°C.
For tracking the motility of individual dynein complexes, 70 nM SNAPf-dynein was mixed with 150 nM dynactin, and 700 nM BicD2N-GFP, and 600 nM Lis1-SNAPf in DB supplemented with 1 mg/ml BSA. The complex was incubated on ice for 10 mins followed by the addition of 50 nM BG-functionalized QDs for another 10 mins in ice. The mixture was then diluted in DB-C and flowed into the motility chamber for 2 mins, followed by washing with 80 μl DB-C. Movies were recorded immediately after washing the chamber with 20 μl DMB containing 2 μM Mg.ATP. For tracking the stepping of dynein in the presence of Lis1, 600 nM Lis1-SNAPf was included in DMB. The sample was excited with a 1 kW cm-2 488 nm beam (Coherent) and movies were recorded at 30 ms per frame on Ixon EM+ EM-CCD camera (Andor). For stepping analysis, fluorescence spots of QDs were localized using a 2-dimensional Gaussian fitting and the resulting trajectories fitted into steps using a custom-written algorithm based on Schwartz Information Criterion 62, 63.
Tug-of-war assays
To prepare a DNA tether between DDB and kinesin, two complementary DNA strands were first functionalized with benzyl guanine as described previously. Briefly, 25 μM DNA oligos containing an amino group modification at their 5’ends were mixed with 5 mM BG-GLA-NHS (NEB) in 50 mM HEPES buffer containing 50% anhydrous DMSO, pH 8.5. The reaction was kept overnight at room temperature. The excess unreacted ligand was removed and the BG-functionalized oligos were purified by ethanol precipitation of DNA. Finally, isolated DNA was dissolved in DB and stored at 4°C. The concentration of BG-DNA was estimated from the absorbance at 260 nm.
BicD2N-SNAPf and kinesin-SNAPf-GFP were labeled with BG-functionalized oligos by mixing protein with DNA in DB for 1 hr at 4°C. DNA and protein concentrations were optimized to yield ~ 30% efficiency of protein labeling to ensure that the likelihood of dual-labeling of a single dimeric protein with two DNA oligos was minimized (μ9%). The labeling efficiency was quantified by comparing the intensities of labeled to unlabeled bands on 4-12% Bis-Tris SDS-PAGE (Invitrogen). Excess unreacted DNA was removed from BicD2N-SNAPf using a TSKgel G4000SWXL size exclusion column (Tosoh). In the case of kinesin, a 10-fold molar excess of BG-GLA-TMR (NEB) was added to the motor-DNA mixture and the sample was incubated for an additional 30 min at 4°C. Excess DNA and dye were removed by an MT bind and release assay.
In tug-of-war experiments, 200 nM DNA-labeled BicD2N-SNAPf was mixed with 70 nM LD650-labeled dynein and 150 nM dynactin in 10 μl DB supplemented with 1 mg/ml BSA. For experiments with Lis1, 600 nM Lis1-SNAPf was added to the mixture. The mixture was incubated on ice for 10 mins, followed by the addition of 150 mM NaCl and 200 nM DNA- and TMR- labeled kinesin-SNAPf-GFP, and incubation on ice for a further 20 mins. Proteins were then diluted in DB-C and flowed into the chamber, followed by washing with DB-C and imaging in DMB. The buffer was supplemented with 600 nM Lis1-SNAPf in case of Lis1 experiments.
Optical trapping assays
For DDB and DDR experiments, complexes assembled with SNAPf-dynein, dynactin, and BicD2N-GFP or BicDR1-GFP were mixed with 860 nm diameter anti-GFP coated latex beads in ice for 10 mins. This assay geometry ensures that beads are driven by active dynein motors assembled to cargo adaptors and eliminates the possibility of cargo adaptor multimerization. Carboxyl latex beads (860 nm-diameter, Life Technologies) were washed and resuspended in activation buffer (10 mM MES, 100 mM NaCl, pH 6.0). The beads were coated by mixing with ~ 2 mg of custom-made polyclonal rabbit anti-GFP antibodies (BioLegend, previously Covance, catalog# MMS-118P) in activation buffer supplemented with 1 mg each of N-hydroxysulfosuccinimide (Sulfo-NHS) and 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) crosslinkers (Pierce) dissolved in dimethylformamide (DMF)64. The beads were passivated with BSA, washed and stored in phosphate-buffered saline (PBS) supplemented with 0.5 mg/ml BSA and 0.1% sodium azide at 4 °C. For DTR experiments, SNAPf-DynLT was labeled with biotin; and biotin-DynLT, mtDyn, dynactin, and BicDR1 were incubated with 800 nm diameter SA-coated beads (Spherotech) in ice for 10 mins. The protein-coated beads were then diluted in DB-C and flowed into the motility chamber in DMB. The protein concentration in the mixture was gradually reduced in assays until less than 30% of the tested beads exhibited motility activity in contact with Cy5-labeled axonemes to ensure that >95% of the beads are driven by a single complex. For force measurements in the presence of Lis1, 600 nM Lis1-SNAPf was added to the bead-protein mixture and also later added to DMB.
Force measurements were performed on a custom-built optical trap on a Nikon Ti-Eclipse microscope body consisting of a 2 W 1,064 nm continuous wave laser beam (Coherent) and a 100 x 1.49 NA Plan-Apo objective (Nikon), as described previously 64. Beads were trapped by the laser beam steered by two computer-controlled perpendicular acousto-optical deflectors (AA Electronics). The sample was excited with a 633 nm laser (Coherent) and Cy5-labeled axonemes were imaged using a monochrome camera (The Imaging Source). To detect the bead position relative to the center of the trap, a position-sensitive detector (First Sensor Inc.) was placed at the back focal plane of a 1.4 N.A. oil-immersion condenser (Nikon). Trap stiffness was derived by fitting the power spectrum of a trapped bead that was rapidly raster-scanned in both x and y directions using the acousto-optical deflectors to a Lorentzian spectrum. The typical spring constant used in these experiments was ~ 0.04 pN/nm to allow motors to travel 100-150 nm before stalling. The PSD data were recorded at 20 kHz during calibration and the resulting curve was fit to a cubic polynomial to calibrate the response of the PSD in each sample. For fixed trap assays, PSD data were collected at 5 kHz and downsampled to 500 Hz for ease of visualization. To qualify as a stall event, the bead position should remain stationary for at least 100 ms before rapid (μ2 ms) jumping towards the trap center, implicating release of the motor from the MT. Stall force histograms are then generated from individual stall events that were manually scored. For force-clamps assays, the PSD signal was acquired at 5 kHz and position feedback was performed at 100 Hz. Beads that walked for at least 100 nm were subjected to force feedback and resulting runs were downsampled to 500 Hz and fit to a step-finding algorithm as described previously64. Force-clamp runs that are shorter than 200 nm or included instant jumps larger than 50 nm were excluded from the analysis.
Statistics and reproducibility
At least three independent experiments were performed. Independent experiments showed similar results. The exact number of replicates (n) of every dataset is given at the corresponding figure legends. Statistical analysis methods are stated in the main text or the figure legend.
Extended Data
Extended Data Fig. 1. Lis1 increases the velocity of complexes assembled with wtDyn.
(a) Assembly of wtDDB and wtDDR. (b) Velocity distribution of wtDDB and wtDDR complexes assembled in the presence and absence of 600 nM Lis1. The line and whiskers represent the mean and SD, respectively. From left to right, n = 106, 72, 75, and 81, and mean values are 538, 718, 924, 1113 nm s-1 (three independent experiments). p-values are calculated from a two-tailed t-test. (c) Velocity distribution of complexes assembled with wtDyn and mtDyn in the absence of Lis1. The line and whiskers represent the mean and SD, respectively. From left to right, n = 106, 132, 75, and 307, and mean values are 538, 652, 924, 1155 nm s-1 (three independent experiments). p-values are calculated from a two-tailed t-test. (d) The percentage of processive wtDDB complexes that are dual-labeled when an equimolar mixture of TMR- and LD650-dynein motors were assembled with dynactin and BicD2N in the absence of Lis1 (mean ± SEM, n = 246 and 178 from left to right). Error bars represent SE calculated from multinomial distribution and the p-value is calculated from the two-tailed z-test.
Extended Data Fig. 2. Step analysis of mtDDB in the presence and absence of Lis1.
(a) Additional examples of mtDDB stepping in the presence and absence of 600 nM Lis1. (b) The average size of steps taken in forward (μf), backward, (μb), and both (μcum) directions along the longitudinal axis of the MT. Error bars are SEM. In a and b, six independent experiments were performed per condition. (c) Stepping rates estimated from the exponential fit in Figure 1f. Error bars are SE of the fit. In b and c, p values are calculated from a two-tailed t-test; sample size (n) distribution of data are provided in Fig. 1f.
Extended Data Fig. 3. Lis1 does not increase the stall duration of dynein bound to dynactin and a cargo adaptor.
(a) Inverse cumulative distribution of stall durations in the absence and presence of 600 nM Lis1. Solid curves represent fitting to a two-exponential decay (decay time ± SE). (b) Mean stall times of mtDDB and mtDDR in absence and presence of 600 nM Lis1 (± SEM). p values are calculated from a two-tailed t-test. In a and b, n = 53, 27, 50, and 39 from left to right, four independent experiments per condition.
Extended Data Fig. 4. Lis1 does not affect stall time and stepping rate of single dynein bound to dynactin.
(a) Distribution of dwell times between consecutive steps along the longitudinal axis of the MT. A fit to an exponential decay reveals the decay rate (rate ± SE, n = 734 for mtDTR-Lis1 and 724 for mtDTR+Lis1). (b) Inverse cumulative distribution of stall durations of mtDTR in the presence and absence of 600 nM Lis1. Solid curves represent fitting to a two-exponential decay (decay time ± SE, n = 118 for mtDTR-Lis1 and 100 for mtDTR+Lis1, three independent experiments).
Extended Data Fig. 5. Lis1 does not stimulate the recruitment of dynein tail to dynactin.
Representative kymographs show the motility of LD650-Dyn and TMR-DynLT assembled with BicD2N or BicDR1 in the presence and absence of 600 nM Lis1. White arrows point to complexes that contain both LD650-mtDyn and TMR-DynLT (three independent experiments were performed per condition).
Extended Data Fig. 6. Additional examples of binding events of Lis1 to mtDDB and mtDTR during processive movement.
(a) Schematic depiction of mtDDB complex assembled in the presence of TMR-Lis1. (b) Representative kymographs show binding of Lis1 to motile mtDDB complexes assembled by mixing 1 nM LD650-mtDDB and 75 nM TMR-Lis1 and immediately recording motility with free proteins in solution (see methods). White arrows represent colocalization of LD650-Dyn (red) and Lis1-TMR (cyan). (c) Velocity distribution of mtDDB complexes not bound to Lis1 moves faster than complexes that are bound to Lis1 during single-molecule motility. The line and whiskers represent the mean and SD, respectively. From left to right, n = 270 and 117 and mean values are 921 and 813 nm s-1. In b and c, three independent experiments were performed per condition. The p-value is calculated from a two-tailed t-test. (d) Rare events of dynamic binding of Lis1 to dynein as mtDDB walks along an MT assembled in the presence of 50 nM Lis1. White arrows represent the colocalization of LD650-Dyn (red) and TMR-Lis1 (cyan). In the top kymograph, Lis1 initially diffuses on an MT and then binds to mtDDB during processive movement. Lis1 binding reduces the velocity of the complex. In the middle kymograph, dissociation of Lis1 during mtDDB motility increases the velocity. In the bottom kymograph, a diffusing Lis1 initially binds and later dissociates from mtDDB, without significantly affecting the velocity of the complex (four independent experiments). (e) Additional kymographs show single- and dual Lis1 binding to motile mtDTR complexes assembled in the presence of 50 nM Lis1. Red arrows represent the colocalization of Atto488-DynLT (green) and Cy5-Lis1 (red). White arrows represent the colocalization of Atto488-DynLT (green) with both Cy5-Lis1 (red), and TMR-Lis1 (cyan). Three independent experiments were performed per condition.
Extended Data Fig. 7. At limiting dynein concentration, Lis1 recruits single dynein to dynactin and BicD2N.
(a) Schematic depiction of wtDDB assembly using 5 nM LD650-wtDyn and TMR-wtDyn in the absence and presence of 600 nM Lis1. (b) Fraction of processive and static/diffusive wtDDB complexes on MTs (mean ± SEM, n = 59, 788, 303 and 984 from left to right, three independent experiments).
Supplementary Material
Motility of single DDBs along MTs at limiting dynein concentration. 5 nM of LD650-mtDyn or LD650-wtDyn was mixed with dynactin and BicD2N in the absence and presence of 600 nM Lis1. Motility along surface-attached MTs was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Recruitment of two dyneins to dynactin by BicD2N and BicDR1. LD650-mtDyn (red) and TMR-mtDyn (cyan) were mixed with dynactin and a cargo adaptor in the absence and presence of 600 nM Lis1. Motility of DDB and DDR complexes along surface-attached MTs were imaged with two-color TIRF illumination. Colocalizers along a single MT are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.
Colocalization of two Lis1 dimers to DTR. Atto488-DynLT (green), TMR-Lis1 (blue), and LD650-Lis1 (red) were mixed with dynactin and BicDR1. Motility along surface-attached MTs was imaged with three-color TIRF illumination. Atto488, TMR, LD650, and overlaid frames are separately shown for ease of visualization. Colocalizers are denoted by white arrows. Scale bar is 3 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of single DTRs along MTs at 1 mM ATP. LD650-mtDyn (red) and TMR-DynLT (cyan) were mixed in the presence of dynactin and BicDR1. Motility along surface-attached MTs in the absence and presence of 600 nM Lis1 was imaged with two-color TIRF illumination. Colocalizers that move along a single MT are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of DDB-kinesin co-localizers. LD650-mtDyn was assembled with dynactin and BicD2N. LD650-DDB (red) and TMR-kinesin (cyan) were tethered using a DNA scaffold and motility along surface-attached MTs in the absence and presence of 600 nM Lis1 was imaged with two-color TIRF illumination. Colocalizers are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of single DDBs along MTs at 1 mM ATP. LD650-mtDyn was assembled with dynactin and BicD2N and motility along surface-attached MTs in the absence and presence of Lis1 was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.
Motility of single DDRs along MTs at 1 mM ATP. LD650-mtDyn was assembled with dynactin and BicDR1 and motility along surface-attached MTs in the absence and presence of Lis1 was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.
Acknowledgments
We are grateful to the members of the Yildiz laboratory for helpful discussions and to V. Madan (MRC) for sharing unpublished results. This work was funded by grants from the NIH (GM094522), and NSF (MCB-1055017, MCB-1617028) to A.Y., the MRC (MC_U105178790) to S.L.B., and a DFG research fellowship (BA5802/1-1) to J.B.
Footnotes
Code availability
Codes used in this paper are available from the corresponding author upon request.
Data availability
All data that support the conclusions are available from the authors on request.
Authors contribution
M.M.E., J.B., S.L.B. and A.Y. conceived the study and designed the experiments. M.M.E. purified dynein, dynactin and cargo adaptors. J.B. purified Lis1 proteins. M.M.E. and E.K. labeled the proteins with DNA and fluorescent dyes and performed the single-molecule motility experiments. M.M.E. and E.K. performed fluorescent tracking assays. M.M.E., S.V., and E.K. performed optical trapping assays. M.M.E., S.L.B., and A.Y. wrote the manuscript, and all authors read and edited the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nature reviews Molecular cell biology. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. Mechanism and regulation of cytoplasmic dynein. Annual review of cell and developmental biology. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroer TA, Steuer ER, Sheetz MP. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 4.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter AP, Diamant AG, Urnavicius L. How dynein and dynactin transport cargos: a structural perspective. Current opinion in structural biology. 2016;37:62–70. doi: 10.1016/j.sbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Hafezparast M, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes JM, Katsanis N. Microtubule transport defects in neurological and ciliary disease. Cell Mol Life Sci. 2005;62:1556–1570. doi: 10.1007/s00018-005-5007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiavo G, Greensmith L, Hafezparast M, Fisher EM. Cytoplasmic dynein heavy chain: the servant of many masters. Trends in neurosciences. 2013;36:641–651. doi: 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrone CA, et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and Mammalian cells. Molecular biology of the cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 12.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Can S, Lacey S, Gur M, Carter AP, Yildiz A. Directionality of dynein is controlled by the angle and length of its stalk. Nature. 2019 doi: 10.1038/s41586-019-0914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kon T, et al. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nature structural & molecular biology. 2012;19:492–497. doi: 10.1038/nsmb.2272. S491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. Journal of neurobiology. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 18.Bingham JB, King SJ, Schroer TA. Purification of dynactin and dynein from brain tissue. Methods in enzymology. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- 19.Torisawa T, et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nature cell biology. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- 20.Walter WJ, Brenner B, Steffen W. Cytoplasmic dynein is not a conventional processive motor. J Struct Biol. 2010;170:266–269. doi: 10.1016/j.jsb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell. 2017;169:1303–1314 e1318. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Splinter D, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Molecular biology of the cell. 2012;23:4226–4241. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlager MA, et al. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell reports. 2014;8:1248–1256. doi: 10.1016/j.celrep.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 24.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayloo S, et al. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nature communications. 2014;5 doi: 10.1038/ncomms5807. 4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. The EMBO journal. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlager MA, et al. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. The EMBO journal. 2010;29:1637–1651. doi: 10.1038/emboj.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carnes SK, Zhou J, Aiken C. HIV-1 Engages a Dynein-Dynactin-BICD2 Complex for Infection and Transport to the Nucleus. Journal of virology. 2018;92 doi: 10.1128/JVI.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urnavicius L, et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 2018;554:202–206. doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grotjahn DA, et al. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nature structural & molecular biology. 2018;25:203–207. doi: 10.1038/s41594-018-0027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elshenawy MM, et al. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat Chem Biol. 2019 doi: 10.1038/s41589-019-0352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a "clutch" between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. The EMBO journal. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi JY, et al. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dix CI, et al. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol. 2013;202:479–494. doi: 10.1083/jcb.201211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, et al. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Molecular biology of the cell. 2013;24:3522–3533. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur EL. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon HM, Wynshaw-Boris A. Cytoskeleton in action: lissencephaly, a neuronal migration disorder. Wiley interdisciplinary reviews. Developmental biology. 2013;2:229–245. doi: 10.1002/wdev.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeSantis ME, et al. Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein. Cell. 2017;170:1197–1208 e1112. doi: 10.1016/j.cell.2017.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toropova K, et al. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife. 2014;3 doi: 10.7554/eLife.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belyy V, et al. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nature cell biology. 2016;18:1018–1024. doi: 10.1038/ncb3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lammers LG, Markus SM. The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J Cell Biol. 2015;211:309–322. doi: 10.1083/jcb.201506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markus SM, Lee WL. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Developmental cell. 2011;20:639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jha R, Roostalu J, Cade NI, Trokter M, Surrey T. Combinatorial regulation of the balance between dynein microtubule end accumulation and initiation of directed motility. The EMBO journal. 2017;36:3387–3404. doi: 10.15252/embj.201797077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. The Journal of biological chemistry. 2017;292:12245–12255. doi: 10.1074/jbc.M117.790048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumbach J, et al. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife. 2017;6 doi: 10.7554/eLife.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreasson JO, et al. Examining kinesin processivity within a general gating framework. eLife. 2015;4 doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith DS, et al. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nature cell biology. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 54.Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Htet ZM, et al. Lis1 promotes the formation of maximally activated cytoplasmic dynein-1 complexes. biorXiv. 2019 doi: 10.1038/s41556-020-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marzo MG, Griswold JM, Markus SM. Pac1/LIS1 promotes an uninhibited conformation of dynein that coordinates its localization and activity. biorXiv. 2019 doi: 10.1038/s41556-020-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu R, Zhang J, Xiang X. LIS1 regulates cargo-adapter-mediated activation of dynein by overcoming its autoinhibition in vivo. J Cell Biol. 2019 doi: 10.1083/jcb.201905178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki SO, et al. Expression patterns of LIS1, dynein and their interaction partners dynactin, NudE, NudEL and NudC in human gliomas suggest roles in invasion and proliferation. Acta neuropathologica. 2007;113:591–599. doi: 10.1007/s00401-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 59.McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. The Journal of biological chemistry. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy BJ, et al. Load-induced enhancement of Dynein force production by LIS1-NudE in vivo and in vitro. Nature communications. 2016;7 doi: 10.1038/ncomms12259. 12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dogan MY, Can S, Cleary FB, Purde V, Yildiz A. Kinesin's front head is gated by the backward orientation of its neck linker. Cell reports. 2015;10:1967–1973. doi: 10.1016/j.celrep.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cleary FB, et al. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nature communications. 2014;5 doi: 10.1038/ncomms5587. 4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belyy V, Hendel NL, Chien A, Yildiz A. Cytoplasmic dynein transports cargos via load-sharing between the heads. Nature communications. 2014;5 doi: 10.1038/ncomms6544. 5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Motility of single DDBs along MTs at limiting dynein concentration. 5 nM of LD650-mtDyn or LD650-wtDyn was mixed with dynactin and BicD2N in the absence and presence of 600 nM Lis1. Motility along surface-attached MTs was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Recruitment of two dyneins to dynactin by BicD2N and BicDR1. LD650-mtDyn (red) and TMR-mtDyn (cyan) were mixed with dynactin and a cargo adaptor in the absence and presence of 600 nM Lis1. Motility of DDB and DDR complexes along surface-attached MTs were imaged with two-color TIRF illumination. Colocalizers along a single MT are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.
Colocalization of two Lis1 dimers to DTR. Atto488-DynLT (green), TMR-Lis1 (blue), and LD650-Lis1 (red) were mixed with dynactin and BicDR1. Motility along surface-attached MTs was imaged with three-color TIRF illumination. Atto488, TMR, LD650, and overlaid frames are separately shown for ease of visualization. Colocalizers are denoted by white arrows. Scale bar is 3 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of single DTRs along MTs at 1 mM ATP. LD650-mtDyn (red) and TMR-DynLT (cyan) were mixed in the presence of dynactin and BicDR1. Motility along surface-attached MTs in the absence and presence of 600 nM Lis1 was imaged with two-color TIRF illumination. Colocalizers that move along a single MT are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of DDB-kinesin co-localizers. LD650-mtDyn was assembled with dynactin and BicD2N. LD650-DDB (red) and TMR-kinesin (cyan) were tethered using a DNA scaffold and motility along surface-attached MTs in the absence and presence of 600 nM Lis1 was imaged with two-color TIRF illumination. Colocalizers are denoted by white arrows. Scale bar is 5 μm. Stopwatch shows time in seconds. Three independent experiments were performed for each condition.
Motility of single DDBs along MTs at 1 mM ATP. LD650-mtDyn was assembled with dynactin and BicD2N and motility along surface-attached MTs in the absence and presence of Lis1 was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.
Motility of single DDRs along MTs at 1 mM ATP. LD650-mtDyn was assembled with dynactin and BicDR1 and motility along surface-attached MTs in the absence and presence of Lis1 was imaged under TIRF illumination. Scale bar is 5 μm. Stopwatch shows time in seconds. Four independent experiments were performed for each condition.