Abstract
Malignant gliomas have a very poor prognosis. The current standard of care for these cancers consists of extended adjuvant treatment with the alkylating agent temozolomide after surgical resection and radiotherapy. Although a statistically significant increase in survival has been reported with this regimen, nearly all gliomas recur and become insensitive to further treatment with this class of agents. We sequenced 500 kb of genomic DNA corresponding to the kinase domains of 518 protein kinases in each of nine gliomas. Large numbers of somatic mutations were observed in two gliomas recurrent after alkylating agent treatment. The pattern of mutations in these cases showed strong similarity to that induced by alkylating agents in experimental systems. Further investigation revealed inactivating somatic mutations of the mismatch repair gene MSH6 in each case. We propose that inactivating somatic mutations of MSH6 confer resistance to alkylating agents in gliomas in vivo and concurrently unleash accelerated mutagenesis in resistant clones as a consequence of continued exposure to alkylating agents in the presence of defective mismatch repair. The evidence therefore suggests that when MSH6 is inactivated in gliomas, alkylating agents convert from induction of tumor cell death to promotion of neoplastic progression. These observations highlight the potential of large scale sequencing for revealing and elucidating mutagenic processes operative in individual human cancers.
Introduction
Gliomas account for approximately half of primary brain tumors diagnosed each year and are responsible for the majority of mortality due to central nervous system neoplasms.6 Malignant gliomas, such as glioblastoma, have exceedingly poor prognoses with 5-year survival rates of <5%. Recent studies have shown a modest, statistically significant increase in overall and disease-free survival of patients with glioblastoma when treatment with the alkylating agent temozolomide is added to the standard regimen of radiation therapy (1). Nevertheless, essentially all glioblastomas recur after alkylating agent chemotherapy and become refractory to further treatment (2). The mechanism of resistance in such cases is not well understood. To investigate the mutational profiles of and to identify genes causally implicated in human cancer, we have undertaken large-scale systematic sequencing for somatic mutations in several classes of human cancer (3–5). We have now extended these studies by sequencing the kinase domains of 518 protein kinases in human gliomas. The observed mutational patterns have revealed a likely mechanism for glioma recurrence after temozolomide treatment.
Materials and Methods
Human malignant glioma and blood samples were obtained with institutional review board approval. Pathologic diagnoses were confirmed by one of the authors (D.N. Louis). For tumor to be established in culture, samples were enzymatically dissociated and plated in defined DMEM cell culture medium (Life Technologies, Gaithersburg, MD) with 10% fetal bovine serum (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). Cells were split 1:2 upon confluence. After expansion through 15 to 20 passages, DNA was extracted. PCR primers amplifying the coding exons of all kinases domains and a subset of juxtamembrane exons are available online.7 The total length of coding sequence targeted was ∼554 kb, of which 532 kb (98%) was successfully covered. Amplification of sequencing templates from the tumors, bidirectional sequencing, and confirmations were done as previously described (3). Greater than 90% of designed exons were successfully analyzed in >90% of samples. DNA from pretreatment tumor material within the H&E formalin-fixed paraffin-embedded slide from case PD1487a was prepared using the Gentra Puregene DNA Purification kit and sequenced using USB P-33 Thermosequenase kit. MGMT methylation analysis was done as detailed elsewhere (6, 7). In addition to the protein kinase domains, TP53 and PTEN were sequenced in the set (Supplementary Data). Genome-wide analysis of copy number was done on paired normal/tumor samples as previously described (8). To assess the presence of biological selection on the observed somatic mutations, analyses of the ratio of nonsynonymous to synonymous mutations were done as previously described (4). Additional statistical methods applied can be found in Supplementary Data.
Results
Approximately 500 kb DNA corresponding to the kinase domains of 518 protein kinases was sequenced in each of nine human malignant gliomas. Sixty-five somatic mutations were detected including 39 missense, 6 nonsense, 3 conserved splice-site junctions, and 17 synonymous mutations (silent; Supplementary Data).8 Assuming that all the synonymous mutations are biologically neutral (i.e., ‘‘passenger’’ mutations) and that the expected ratio of nonsynonymous to synonymous neutral mutations is 2:1 (4), there was a slight, nonsignificant excess of nonsynonymous mutations (n = 45) compared with that expected by chance in the series (n = 36). Overall, however, the large majority of observed somatic mutations are likely to be passenger mutations.
The 48 nonsynonymous and splice site mutations were distributed among 44 genes. Three nonsynonymous mutations were found in BRD2 (one in PD1487a and two in PD1489a) and two were found in TRIM33 (in PD1487a and PD1489a). Two nonsynonymous mutations were also found in ERN1 (both in PD1487a). The remainder of the genes each had one or zero nonsynonymous mutations. Mutations in FGFR1, recently reported in a screen of 20 kinase domains (9), were not observed.
No frequently mutated protein kinase was identified in this screen. Nevertheless, there were individual mutations of potential interest. These include a mutation in the G-loop that mediates ATP binding (MLCK), mutations within the activation segment (MAPK8, PRKCB1, PTK2, and TLK1), a mutation of the canonical proline within the PSTAIRE motif of CDK2 important in interactions with cyclins/CDK inhibitor proteins (10), and a mutation in PDGFRA, amplification of which has previously been associated with glioma (11). To evaluate the significance of mutations detected, a follow-up screen was conducted of 21 glioblastomas through the kinase domain exons of 40 genes in which mutations had been found. Only two silent mutations were detected in the follow-up screen (Supplementary Data).
Six gliomas had no somatic mutations and one had a single mutation. In contrast to this generally low prevalence, two gliomas had numerous mutations, PD1487a with 34 and PD1489a with 30. Both PD1487a and PD1489a are early cultures derived from recurrences of glioblastomas. Analysis of a subset of these mutations in archival formalin-fixed, paraffin-embedded tissues showed that the mutations were present in the recurrences from which the cultures were derived (Supplementary Data).
The mutational patterns in PD1487a and PD1489a are shown in Tables 1 and 2. Because they exhibit similar patterns, the data from these two tumors have been combined and their features are summarized together. All the somatic mutations detected were single base substitutions. In both PD1487a and PD1489a, most substitutions were C:G>T:A transitions (63 of 64, 98%). These C:G>T:A mutations occurred in a particular sequence context (P = 0.0006; Table 2). Considering the strand on which each C:G>T:A mutation is a C>T change, there was an excess of mutations at CpC dinucleotides where the 5′ cytosine is mutated (50%) compared with that expected by chance (28%). There was also a reduction in guanine immediately 5′ to the mutated cytosine (11% compared with 25%) and a weak strand bias for mutation of guanine on the nontranscribed strand (61% G>A to 39% C>T).
Table 1. Details of cases studied and abnormalities found through mutation screening.
| Sample | Diagnosis | Sex | Age (y) | Treatment before sample removal | Sample used | Somatic mutations in this study | C:G>TA transitions | C:G>T:A transitions at CpG | MSH6 mutation |
|---|---|---|---|---|---|---|---|---|---|
| PD1487a/Gli 56 T | Glioblastoma | M | 54 | Radiotherapy, chemotherapy | Recurrence | 34 | 34 | 0 | Het C1453T, Q485X; Het G3907A, A1303T |
| PD1489a/Gli 60 T | Glioblastoma | M | 47 | Radiotherapy, chemotherapy | Recurrence | 30 | 29 | 2 | Hom delG2425, V809X |
| PD1493a/Gli 70 T | Anaplastic oligoastrocytoma (grade 3) | F | 48 | None | Primary | 1 | 1 | 1 | Absent |
| PD1494a/Gli 72 T | Anaplastic oligoastrocytoma (grade 3) | F | 50 | None | Primary | 0 | 0 | 0 | Absent |
| PD1536a/xT-155 TD | Glioblastoma | M | 51 | Radiotherapy | Recurrence | 0 | 0 | 0 | Absent |
| PD1537a/xT-924 TD | Glioblastoma | M | 71 | None | Primary | 0 | 0 | 0 | Absent |
| PD1538a/xT-2560 TD | Glioblastoma | M | 71 | None | Primary | 0 | 0 | 0 | Absent |
| PD1540a/xT-3058 TD | Glioblastoma | F | 69 | None | Primary | 0 | 0 | 0 | Absent |
| D542MG | Glioblastoma | F | 45 | Not known | Not known | 0 | 0 | 0 | Absent |
Table 2. Sequence context of C:G>T:A mutations observed in PD1487a and PD1489a compared with the expected sequence context.
| Sequence context of mutation | ACA | CCA | GCA | TCA | ACC | CCC | GCC | TCC | ACG | CCG | GCG | TCG | ACT | CCT | GCT | TCT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Percent observed mutations (%) | 0 | 8 | 5 | 6 | 11 | 22 | 3 | 14 | 0 | 2 | 0 | 2 | 9 | 8 | 3 | 7 |
| Percent expected mutations (%) | 7 | 10 | 7 | 8 | 6 | 7 | 7 | 8 | 2 | 4 | 3 | 3 | 5 | 8 | 8 | 8 |
NOTE: The expected sequence context was derived from the entire set of trinucleotides centered around all cytosines in the coding sequences of the protein kinases. Underlined C is the mutated base.
Due to the high rate of deamination of 5-methyl cytosine, C:G>T:A transitions generally occur at elevated frequency at CpG dinucleotides, accounting for the high proportion of single nucleotide polymorphisms at CpGs. C:G>T:A transitions at CpG dinucleotides also account for a high proportion of somatic mutations in human cancer genes such as TP53.9 However, only 2 of 64 (3%) C:G>T:A mutations in PD1487a and PD1489a were at CpG dinucleotides, less than the expected frequency (11%) based on the prevalence of CpG dinucleotides in the coding sequences of the protein kinases.
To identify the underlying causes of the mutator phenotype, several known DNA repair genes were evaluated. Inactivation of MGMT (O-6-methylguanine-DNA methyltransferase) by promoter methylation has been implicated in the response of gliomas to alkylating agent chemotherapy (7). MGMT status was evaluated by bothsequencing and promoter methylation analysis. No mutations were observed in the tumors under study. Methylation of the MGMT promoter was observed in both the primary tumor recurrence and early culture of PD1489a and in one additional glioblastoma (PD1537), but not in PD1487a. Thus, MGMT status was not concordant with the mutator phenotype.
Somatic mutations were found in the MSH6 gene in both PD1487a and PD1489a. MSH6 heterodimerizes with MSH2 recognizing double-stranded DNA mismatches (12). Homozygous-inactivating germ line alleles of MSH6 have recently been reported in two children with glioma (13, 14) but somatic mutations of MSH6 have not previously been reported in gliomas. In PD1489a, a single base pair deletion, c.2425delG/p.809V>X, resulting in a translational frameshift was present. This protein truncating mutation was homozygous due to complete loss of the second copy of chromosome 2 (Supplementary Data). A somatic heterozygous nonsense mutation, c1453C>T/p.485Q>X, was detected in PD1487a. Loss of heterozygosity on chromosome 2 was not observed in this sample. However, a second somatic missense mutation, c.3907G>A/p.1303A>T, was detected. Alanine 1303 is conserved in nearly all MutS orthologues through prokaryotes and is within the ATPase domain (Supplementary Data), critical for the mismatch repair function of this family of proteins. Therefore, p.1303A>T is likely to abrogate MSH6 function. These somatic mutations were confirmed in DNA from the recurrent gliomas from which the early cultures were derived. Evaluation of the NIHNational Cancer Institute consensus markers and an additional seven dinucleotide repeats revealed no evidence of microsatellite instability in PD1487a, PD1489a, or in any other glioma (Supplementary Data).
A single H&E-stained section of homogeneous, densely cellular tumor was available from the pretreatment glioblastoma specimen of PD1487a. DNA was extracted from this section and PCR amplified to investigate the presence of the two MSH6 mutations found in the recurrent glioma. Neither of the two mutations was detected (Fig. 1). This result suggests that the MSH6 mutations were selected as a subclone of the primary glioma that subsequently expanded to constitute most of the posttreatment recurrence. No pretreatment material was available for PD1489a. MSH6 mutations were not found in the six primary malignant gliomas that had not previously been subject to therapy or in the glioma recurrent after radiotherapy alone (PD1536a). Screening of an additional series of 21 pretreatment malignant gliomas also failed to reveal any further MSH6 mutations.
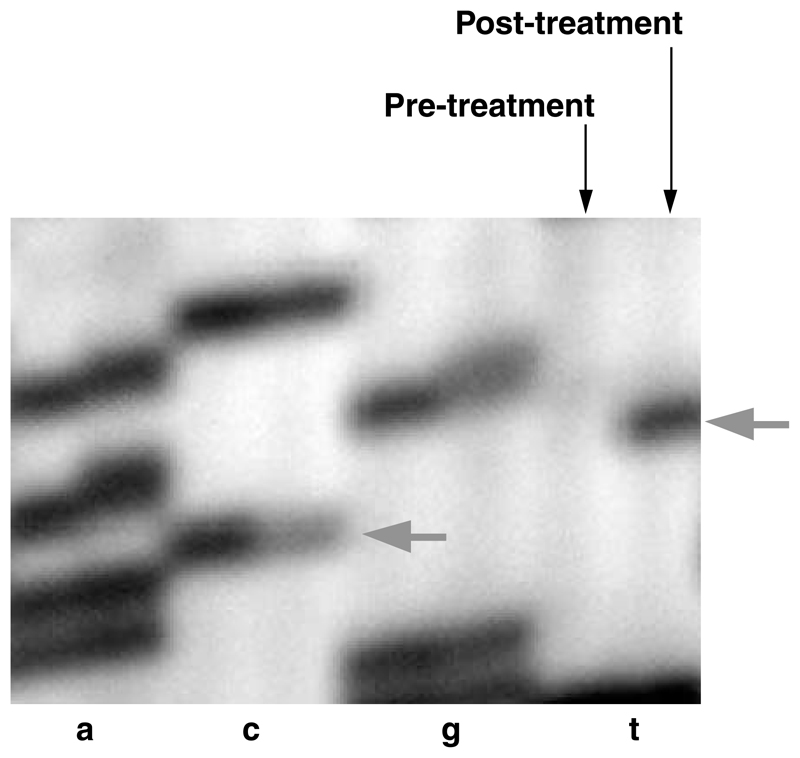
Figure 1.
Sequence analysis of the MSH6 gene in pretreatment and posttreatment samples of PD1487a. Arrows, positions of the wild-type (C) and mutated (T) bases of the heterozygous c.1453C>T mutation found in the posttreatment tumor.
Despite the presence of inactivating MSH6 mutations, the mutational signature of PD1487a and PD1489a clearly differs from that found in Msh6−/− mice, in other mice null for mismatch repair genes and in human cells with MSH6 mutations (Table 3; refs. 15–18). In these models, the proportion of C:G>T:A transitions is lower. Moreover, most of the observed C:G>T:A transitions are at CpG dinucleotides.
Table 3. Mutational signatures of wild-type and mismatch repair defective cells with and without exposure to alkylating agents.
| Mutation | Species | Reporter sequence | Tissue | Inducing agent | Percent C:G>T:A (%) | Percent C:G>T:A at CpG (%) | Percent C:G>T:A at CpC (%) | Percent T:A>C:G (%) | Percent C:G>A:T (%) | Percent ins/del (%) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Msh6−/− | Mouse | lacI | Small intestine | Spontaneous | 73 | 58 | — | 11 | 5 | 9 | (15) |
| Wild type | Mouse | lacI | Small intestine, thymus | Spontaneous | 50 | 80 | 0 | 13 | — | 9 | (16) |
| Wild type | Mouse | lacI | Small intestine, thymus | N -methyl-Nnitrosourea | 74 | 7 | 59 | 0 | — | 8 | (16) |
| Msh2−/− | Mouse | lacI | Small intestine, thymus | Spontaneous | 42 | 60 | 20 | 25 | — | 17 | (16) |
| Msh2−/− | Mouse | lacI | Small intestine, thymus | N-methyl-Nnitrosourea | 82 | 15 | 65 | 0 | — | 16 | (16) |
| hMSH6 3510del5 868delC | Human | HPRT | HCT15 colorectal cancer cell line | Spontaneous | 25 | 41 | 19 | 6 | 46 | 5 | (17) |
| hMSH6 D1145V V1192I | Human | HPRT | MT1 cells | Spontaneous | 0 | 0 | 0 | 60 | 20 | 20 | (18) |
| hMSH6 D1145V V1192I | Human | HPRT | MT1 cells | N′-Methyl-N ’-nitro-N -nitrosoguanidine | 94 | 0 | 43 | 0 | 1 | 0 | (18) |
| hMSH6 Het C1453T, Q485X; Het G3907A, A1303T | Human | 500 kb (exons of protein kinases) | PD1487a | Temozolomide | 100 | 0 | 47 | 0 | 0 | 0 | This study |
| hMSH6 Hom delG2425, V809X | Human | 500 kb (exons of protein kinases) | PD1489a | Temozolomide and BCNU | 97 | 7 | 55 | 0 | 3 | 0 | This study |
NOTE: The 5′ (underlined) cytosine in the CpC dinucleotide is the one mutated.
Abbreviations: HPRT, hypoxanthine phosphoribosyltransferase; BCNU, 1,3-bis(2-chloroethyl)-1-nitrosourea.
PD1487a and PD1489a are derived from glioblastomas recurrent after radiation and chemotherapy. Both had received temozolomide, which has become the standard therapy for such high-grade lesions (2), and PD1489 had also been treated with the alkylating agent 1,3-bis(2-chloroethyl)-1-nitrosourea. The mutational signature of PD1489a and PD1487a is entirely consistent withpreviously published signatures associated with alkylating agent exposure in various model systems (Table 3). Both sets are characterized by a monotonous pattern of C:G>T:A changes, not enriched at CpG dinucleotides, enriched at CpC (GpG) dinucleotides (Table 3) and with a tendency to mutation at guanine on the nontranscribed strand.
Discussion
Through sequencing 500 kb of DNA in a series of malignant gliomas, we have shown that glioblastomas recurrent after temozolomide treatment carry large numbers of somatic mutations. The remarkable clarity of the mutational patterns observed in these clinical samples and the striking similarity to results obtained with alkylating agents in experimental systems strongly suggest that most of the mutations detected were caused by alkylating agent mutagenesis.
Exposure of a population of cells to a mutagen will result in a different set of mutations in each cell within the population. Such scattered mutations would not be detectable by sequencing bulk DNA made from the whole cell population. The fact that we can detect these somatic mutations therefore indicates that expansion of a clone of tumor cells underlies the emergence of these recurrences after alkylating agent chemotherapy. The observation of MSH6 mutations in the PD1487a recurrence but not in the pretreatment glioblastoma sample provides direct evidence in favor of this hypothesis. Therefore, the results show for the first time that, at least in some cases, recurrence after temozolomide therapy in patients with glioblastoma is through clonal evolution.
Alkylating agents induce adducts on DNA, notably O6-methylguanine. O6-methylguanine can mispair with thymine in double-stranded DNA (12). This mismatch is recognized by the MSH2/MSH6 dimer component of the mismatch repair system. Overwhelming numbers of adducts and G/T mismatches cause cells to enter apoptosis. This cell killing effect is dependent on the integrity of the mismatch repair system and specifically of MSH2/MSH6 dimers. Several studies in experimental models indicate that if the mismatch recognition mechanism is not functional, cells become resistant to the killing effects of alkylating agents (19, 20). Malignant gliomas that recur following alkylator chemotherapy typically do not respond to further alkylating agents (2). Given the observed MSH6 mutations in recurrent gliomas, we propose that somatic MSH6 mutations confer resistance to alkylating agents on clones of tumor cells that subsequently expand and manifest as clinical recurrence.
The etiology of the somatic MSH6 mutations is unclear. They could be due to endogenous mutagenic processes or could have themselves been the result of mutagenesis due to alkylating agents. In PD1489, the single base pair deletion in MSH6 together with loss of the other chromosome 2 copy are not typical of alkylating agent exposure. Conversely, both mutations in PD1487 were C:G>T:A changes, neither of which were at CpG dinucleotides, a pattern more compatible with the mutagenic effects of alkylating agents.
Mutational inactivation of MSH6 and the mismatch repair system seems to have had further consequences. In experimental models, cells with mismatch repair deficiency exposed to alkylating agents have an elevated mutation rate because G/T mismatches are no longer recognized and repaired (12). PD1487a and PD1489a exhibit the highest prevalence of somatic mutations per nucleotide screened of any primary cancers or cancer cell lines we have examined thus far by extensive sequencing (3–5). On the assumption that most of the somatic mutations observed are passenger mutations and hence that the prevalence of mutations is similar in the genome as a whole, these two cancers each have at least 200,000 somatic point mutations in their genomes, ∼1 per 15 kb. If distributed randomly, these would be expected to generate ∼1,500 amino acid changes in each cancer, affecting ∼8% protein coding genes. Thus, in addition to conferring resistance, inactivating mutations of MSH6 in the context of alkylating agent exposure foster a hypermutational process in gliomas, which is very likely to facilitate rapid evolution of clones with growth advantage and contribute to progression.
It has been shown in cell culture systems that alkylator chemoresistance emerges from mismatch repair deficiency (21). Our findings represent the first demonstration of this paradigm in vivo. They also reveal the development of rampant mutagenesis as a consequence of continuing alkylator exposure that is likely contributory to disease progression. Given the inherent difficulties in obtaining posttreatment brain tumor samples for molecular evaluation, a coordinated multicenter study is now indicated to explore in greater detail the role of MSH6 and potentially other mismatch repair defects in recurrent, chemoresistant malignant gliomas.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
Grant support: NIH, American Association of Neurosurgeons Neurosurgery Research and Education Foundation Fellowship, Institute of Cancer Research, and Wellcome Trust.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank the patients who participated in this research, Wendy Haynes for administrative support, Nazneen Rahman for comments on the manuscript, and Darrell Bigner (Department of Pathology, Duke University Medical Center, Durham, NC) for the D-542MG cell line.
Footnotes
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Lonardi S, Tosoni A, Brandes AA. Adjuvant chemotherapy in the treatment of high grade gliomas. Cancer Treat Rev. 2005;31:79–89. doi: 10.1016/j.ctrv.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–5. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 5.Bignell G, Smith R, Hunter C, et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumours of adolescents and adults. Genes Chromosomes Cancer. 2006;45:42–6. doi: 10.1002/gcc.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7. [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens A-C, Gorlia T, et al. Mgmt gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–95. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rand V, Huang J, Stockwell T, et al. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–9. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherr CJ, Roberts JM. Cdk inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 11.Collins V. Amplified genes in human gliomas. Semin Cancer Biol. 1993;4:27–32. [PubMed] [Google Scholar]
- 12.Karran P, Offman J, Bignami M. Human mismatch repair, drug-induced DNA damage, and secondary cancer. Biochimie. 2003;85:1149–60. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Menko FH, Kaspers GL, Meijer GA, et al. A homozygous msh6 mutation in a child with cafe-aulait spots, oligodendroglioma and rectal cancer. Fam Cancer. 2004;3:123–7. doi: 10.1023/B:FAME.0000039893.19289.18. [DOI] [PubMed] [Google Scholar]
- 14.Hegde MR, Chong B, Blazo ME, et al. A homozygous mutation in msh6 causes Turcot syndrome. Clin Cancer Res. 2005;11:4689–93. doi: 10.1158/1078-0432.CCR-04-2025. [DOI] [PubMed] [Google Scholar]
- 15.Mark S, Sandercock L, Luchman H, et al. Elevated mutant frequencies and predominance of g:C to a:T transition mutations 6 (−/−) small intestinal epithelium. Oncogene. 2002;21:7126–30. doi: 10.1038/sj.onc.1205861. [DOI] [PubMed] [Google Scholar]
- 16.Andrew SE, McKinnon M, Cheng BS, et al. Tissues of msh2-deficient mice demonstrate hypermutability on exposure to a DNA methylating agent. Proc Natl Acad Sci U S A. 1998;95:1126–30. doi: 10.1073/pnas.95.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohzeki S, Tachibana A, Tatsumi K, Kato T. Spectra of spontaneous mutations at the hprt locus in colorectal carcinoma cell lines defective in mismatch repair. Carcinogenesis. 1997;18:1127–33. doi: 10.1093/carcin/18.6.1127. [DOI] [PubMed] [Google Scholar]
- 18.Tomita-Mitchell A, Kat AG, Marcelino LA, et al. Mismatch repair deficient human cells: spontaneous and MNNG-induced mutational spectra in the hprt gene. Mutat Res. 2000;450:125–38. doi: 10.1016/s0027-5107(00)00020-8. [DOI] [PubMed] [Google Scholar]
- 19.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci U S A. 1999;96:10764–9. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papouli E, Cejka P, Jiricny J. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 2004;64:3391–4. doi: 10.1158/0008-5472.CAN-04-0513. [DOI] [PubMed] [Google Scholar]
- 21.Fink D, Aebi S, Howell S. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.