Abstract
In-solution conjugation is the most commonly used strategy to label peptides and proteins with fluorophores. However, lack of site-specific control and high costs of fluorophores are recognised limitations of this approach. Here, we established facile access to grams of Cy5-COOH via a two-step synthetic route, demonstrated that Cy5 is stable to HF treatment and therefore compatible with Boc-SPPS, and coupled Cy5 to the N-terminus of α-conotoxin RgIA while still attached to the resin. Folding of the two-disulfide containing Cy5-RgIA benefitted from the hydrophobic nature of Cy5 resulting in only the globular disulfide bond isomer. In contrast, wild-type α-RgIA folded into the inactive ribbon and bioactive globular isomer under the same conditions. Labelled α-RgIA retained its ability to inhibit acetylcholine(100 μM)-evoked current reversibly with an IC50 of 5.0 nM (Hill coefficient = 1.7) for α-RgIA and an IC50 of 1.6 (Hill coefficient = 1.2) for Cy5-RgIA at the α9α10 nicotinic acetylcholine receptors (nAChRs) heterologeously expressed in Xenopus oocytes. Cy5-RgIA was then used to successfully visualise nAChRs in RAW264.7 mouse macrophage cell line. This work introduced not only a new and valuable nAChR probe, but also a new versatile synthetic strategy that facilitates production of milligram to gram quantities of fluorophore-labelled peptides at low cost, which is often required for in vivo experiments. The strategy is compatible with Boc- and Fmoc-chemistry, allows for site-specific labelling of free amines anywhere in the peptide sequence, and can also be used for the introduction of Cy3/Cy5 FRET pairs.
Keywords: α-conotoxin α-RgIA; cyanine (Cy5) dye; α9α10 nicotinic acetylcholine receptor (nAChR); fluorescence imaging, molecular probe; on-resin solid phase peptide synthesis (SPPS)
Introduction
Nicotinic acetylcholine receptors (nAChRs) are ligand-gated ion channels that mediate fast synaptic transmission within the peripheral and central nervous systems.[1] They play an important role in many physiological processes including muscle contraction, cholinergic synaptic transmission, cognitive functions such as learning and memory, participation in the pain signal transduction, neuroprotection and regulation of the immune responses.[1] nAChRs are also implicated in a number of disease states including Parkinson’s and Alzheimer’s disease, pain, Tourette’s syndrome, lung cancer, nicotinic addiction, schizophrenia, attention-deficit hyperactivity disorder, epilepsy, autism, depression and anxiety.[1a, 1d, 1e, 1g, 2] nAChRs therefore attract substantial interest from the scientific community interested in fundamental problems of neurobiology, pharmacology and drug design. Structurally, they are pentameric assemblies in which five transmembrane spanning subunits are symmetrically arranged around a central pore and channel opening is induced by the endogenous neurotransmitter acetylcholine, or by exogenous agonists such as nicotine. Seventeen nAChR subunits have been cloned encoding the α1−α10, β1−β4, δ, ε, and γ subunits, which assemble in various combinations to form heteromeric or homomeric receptors, which display distinct pharmacological and biophysical properties.[1d, 1e, 1g]
Three-finger toxins α-bungarotoxin (74 amino acids (AA) long, 5 disulfide bonds), isolated from the venom of the Taiwanese banded krait Bungarus multicinctus, and α-cobratoxin (71 AA long, 5 disulfide bonds), isolated from the venom of the Naja cobras, bind to the α subunits of the nAChRs and have been valuable tools for the last several decades to study nAChR expression, structure and function.[3] Conjugation of fluorophores, biotin, and radioactive tags to these snake toxins extend their use from electrophysiology experiments to receptor visualisation and quantification experiments. Their large three-finger structure however is optimised to bind to more than just a single nAChR subtype, hence they are not able to discriminate between individual subtypes. This selectivity issue was resolved by the discovery and characterisation of another venom peptide class, namely the α-conotoxins, isolated from the venom of the marine predatory cone snail (Genus Conus).[3c, 4] These fairly short yet structurally well-defined peptides (12-20 AA long, 2 disulfide CC-C-C framework with a globular Cys1-3 and Cys 2-4 connectivity) bind competitively to the orthosteric sites of the interface of the individual subunits of the nAChRs thereby not only inhibiting their function but also differentiating between combinations of homo and heteromeric subunits with high potency (low nanomolar) and selectivity (often >1,000 fold differences). Some of these α-conotoxins are even able to discriminate different subunit stoichiometry within a subtype (e.g., α-AuIB at α3β4),[5] making them valuable probes to study the individual nAChR subtypes).[3c, 4] Even though α-conotoxins have been widely used to study nAChRs over the last 30 years and some have even entered clinical trials,[4c] they have not reached the same status as α-bungarotoxin or α-cobratoxin in terms of commercial availability and modifications for other applications (e.g., receptor visualisation, pull-down experiments, autoradiography, etc.). Only a few reports successfully carried out labelling of α-conotoxins: α-MII was labelled with different fluorophores (tetramethylrhodamine, Bodipy FL, Alexa Fluor 488, and terbium chelates)[6] and 125I-Tyr for autoradiography at its N-terminus,[7] and α-[V11L;V16A]-ArIB was labelled with Cy3[8] and Alexa Fluor546[9] at its N-terminus and with 125I at its single histidine residue at position 15.[10] In all cases, binding and selectivity was retained thereby making them valuable probes to visualise nAChR subtypes. Nevertheless, commercial availability of such probes is still not offered, which is likely due to the labelling complexity, the low yields and small scales involved, and due to the lack of a general method that would apply for the majority of conotoxins (e.g., also for conotoxins that have lysine residues in their sequences or where the N-terminus is important for binding).
In an effort to expand the chemical repertoire of peptide labelling and to provide a method that could facilitate larger-scale synthesis of labelled (venom) peptides, we explored in this work an on-resin labelling strategy using the cyanine 5 (Cy5) fluorophore and α-conotoxin RgIA.
Materials and Methods
Reagents
Nα-Boc-L-amino acids and reagents used during chain assembly were of peptide synthesis grade purchased from Novabiochem (Merck Pty., Kilsyth, Vic., Australia) and Bachem (Bubendorf, Switzerland). Nα-Boc-L-amino acid-phenylacetamidomethyl (Pam)-resin was purchased from Peptides International (Louisville, Kentucky, USA). [2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] (HBTU) was purchased from Fluka (Buchs, Switzerland), 2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyl uronium hexafluorophosphate (HATU) from GenScript Corporation (Piscataway, NJ, USA), N,N’-diisopropylethylamine (DIEA), trifluoroacetic acid (TFA), dichloromethane (DCM) and N,N’-dimethylformamide (DMF) from Auspep (Melbourne, Vic., Australia). Anhydrous hydrogen fluoride (HF) was imported from BOC Gases (Sydney, NSW, Australia) and the scavengers, p-cresol and p-thiocresol, as well as all other organic reagents and solvents, unless stated otherwise, were purchased in their highest purity from Sigma-Aldrich (Sydney, NSW, Australia). All solvents for solid phase peptide synthesis (SPPS) were of peptide synthesis grade and used without further purification. HPLC grade acetonitrile (Lab Scan, Bangkok, Thailand) and water measuring 18.2 MΩ (ELGA, Melbourne, Vic., Australia) were used for the preparation of all solvents for liquid chromatography.
Reversed-phase high performance liquid chromatography (RP-HPLC)
Analytical RP-HPLC was performed on a Shimadzu LC-10ATVP solvent delivery system equipped with a SIL-10ADVP auto-injector and a SPD-M10AVP photodiode array detector with deuterium light source using a Vydac C18 column (300 Å, 5 μm, 250 mm x 4.6 mm). Data were recorded and processed with Shimadzu Class VP software. 1% /min linear gradient of 0-40% solvent B (solvent A = H2O/0.05% TFA, solvent B = 90% CH3CN/10% H2O/0.043% TFA) were employed at a flow rate of 1 mL/min over 40 min. The eluent was monitored by a UV detector between 200 and 700 nm.
Purification of the peptides was achieved by preparative RP-HPLC on a Vydac C18 column (300 Å, 10μm, 250 mm x 21.2 mm) using a Waters 600E solvent delivery system at a flow rate of 8 mL/min and a 0.5%/min linear gradient of 0-50% B over 100 min. For the isolation and purification of smaller quantities, a semi-preparative Vydac C18 column (300 Å, 1μm, 250 mm x 10 mm) was used at a flow rate of 4 mL/min and a 0.5% /min linear gradient of 0-50% B over 100 min. The absorbance was monitored at 222 nm with a Waters 484 UV absorbance detector. Final purification of the Cy5-COOH was also achieved by preparative RP-HPLC on a Vydac C18 column (300 Å, 10μm, 250 mm x 21.2 mm) using the same solvent delivery system and a flow rate of 8 mL/min and a 1%/min linear gradient of 30-80% B over 60 min at a wavelength of 635 nm. Fractions taken during RP-HPLC purification were analysed by mass spectrometry and fractions containing the desired mass were further analysed by analytical RP-HPLC. Pure fractions with correct mass were gathered and lyophilised.
Peptide concentrations were determined based on peak area detected at 214 nm by analytical RP-HPLC (0–90% B in 10 min; n=3; Phenomenex – Prodigy 100 Å, 5 μm, 4.6 x 50 mm, 2 mL/min) against a peptide standard (χ-MrIA) with known peptide content established by amino acid analysis. Using Beer-Lambert Law, the peptide concentrations were calculated based on absorptions of standards and samples using calculated extinction coefficients.[11]
Electro-spray ionisation mass spectrometry (ESI-MS) and liquid chromatography mass spectrometry (LCMS)
Mass spectra were obtained using a LCT-TOF mass spectrometer (MicroMass, Manchester, UK) equipped with an ESI source. Samples (5-15 μL) were injected into solvent flowing at 100 μL/min (75% CH3CN / 0.1% formic acid (aq)). The orifice potential was set to 90 V and spectra were acquired over the mass range 400 to 2000 Da.
LCMS was performed using a Phenomenex Jupiter LC-MS C18 column (90 Å, 4 μm, 250 mm x 2 mm) column with 0.1% formic acid (solvent A) and 90% CH3CN in 0.1% formic acid (solvent B) with linear gradients (1% solvent B /min over 50 min) at a flow rate of 200 μL/min. Solvent delivery and gradient formation was achieved using an Agilent 1100 binary solvent delivery system. ESI mass spectra were acquired on a SCIEX QSTAR Pulsar QqTOF mass spectrometer equipped with an atmospheric pressure ionisation source. Samples were analysed in the positive ion mode at a sprayer potential of 5kV. Full scan data were acquired at a declustering and focusing potential of 60V and 260V respectively over the mass range of m/z 400-2000 and a 1 second scan time. Data acquisition and processing were carried out using Analyst QS software (SCIEX, Concord, ON, Canada).
Cyanine 5 (Cy5) dye synthesis
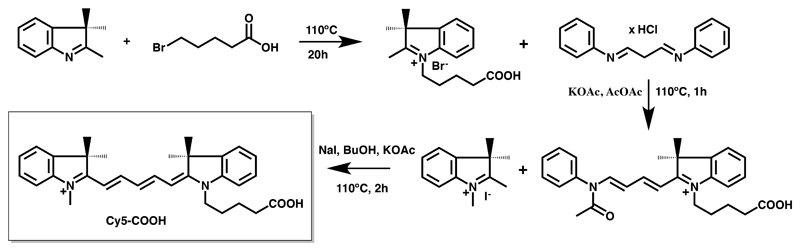
Cy5 with a carboxylic acid as its functional conjugation group (Cy5-COOH) was synthesised on gram-scale using an amended protocol of Korbel et al..[12]
1-[(4'-Carboxybutyl)]-2,3,3-trimethyl-3H-indolium bromide (1)
15.8 g of 2,3,3-trimethylindolenine (100 mmol, 1.2 eq) was added to 15 g of 5-bromovaleric acid (83 mmol, 1 eq) and the reaction was heated, with vigorous stirring, for 20 h at 110°C. The burgundy solid was broken up and washed with 6x 80 mL portions of refluxing ethyl acetate, 2x 80 mL portions of refluxing acetone, and 2x 240 mL portions of refluxing acetone. Filtration of the solid and drying under vacuum afforded 15.4 g (55% yield) of 1 as a pale pink powder. MS[m/z]: MS[calc.]: 260.1645; MS[observed]: 260.1638.
1-[4''-(1''-Carboxybutyl)]-1',2,2,2',2'-pentamethyl-indodicarbocyanide iodide, Cy5-COOH
288 mL of acetic anhydride was added to 4 g of 1 (11.8 mmol, 1 eq) and 2.4 g potassium acetate (24.7 mmol, 2.1 eq). The mixture was heated to 110°C and 6.1 g malonaldehydebis(phenylimine) monohydrochloride (23.5 mmol, 2 eq) was added slowly over a period of 45 min (6x 1 g; pink to blue colour change observed). The reaction was maintained at 110°C for an additional hour and then cooled to room temperature. Acetic acid and acetic anhydride were removed by vacuum distillation on the rotary evaporator as their azeotropes with heptane. The crude product was dried over vacuum overnight and used without further purification. To this intermediate 7.79 g of 1,2,3,3-tetramethyl-3H-indolium iodide (25.9 mmol, 2.2 eq), 4.6 g of potassium acetate (47 mmol, 4 eq), 35.2 g of sodium iodide (235.1 mmol, 20 eq) and 288 mL 1-butanol were added and the reaction was kept at 110°C for 2 h. The reaction was then cooled slightly and 1-butanol was removed on the rotary evaporator. The remaining solid was dissolved in CH2Cl2 and washed with water. The organic phase was dried over anhydrous sodium sulfate, filtered, and concentrated in vacuo to a blue solid. The crude product was purified by flash chromatography using 8% MeOH in CHCl3 as eluent. The fractions were analysed by ESI-MS and TLC, and appropriate fractions were combined and the solvent removed by rotary evaporation. The solid product was then purified once more by preparative C18-RP-HPLC affording 1.4 g of pure Cy5-COOH (20% yield, >95 % pure). MS[m/z]: MS[calc.]: 469.2855; MS[observed]: 469.8422, 1H-NMR, (400 MHz, CDCl3), δ [ppm]: 7.80 (t, 1H, J=13.2 Hz, CH=CH-CH); 7.78 (t, 1H, J=12.8 Hz, CH=CH-CH); 7.40-7.34 (m, 4H, aromatic); 7.25-7.20 (m, 2H, aromatic); 7.10-7.07 (dd, 2H, J1= 8.0 Hz, J2= 3.6 Hz, aromatic); 6.76 (t, 1H, J=12.4 Hz, CH-CH=CH); 6.32-6.29 (d, 2H, J=13.6 Hz); 4.04 (t, 2H, J=7.0 Hz, N-CH2-CH2); 3.60 (s, 3H, N-CH3); 2.59 (t, 2H, J=6.8 Hz, CH2-CH2-COOH); 1.9-1.82 (m, 4H, CH2-CH2-CH2-CH2); 1.68 (s, 6H, 2xCH3); 1.67 (s, 6H, 2xCH3)
α-RgIA and Cy5-RgIA synthesis
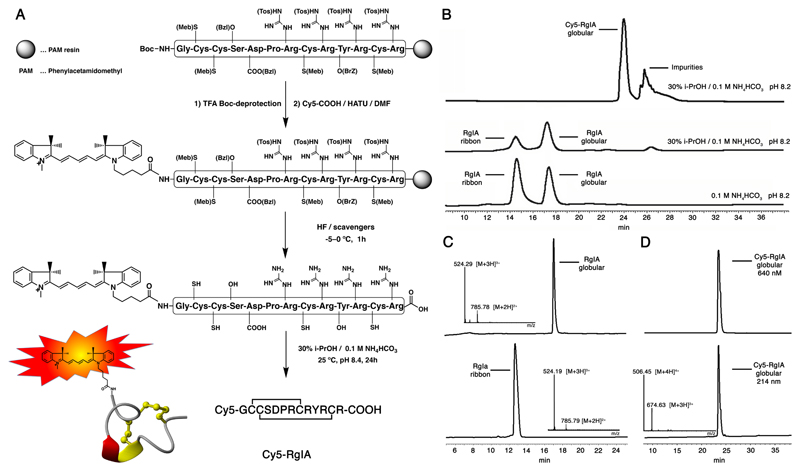
α-RgIA (GCCSDPRCRYRCR-COOH; 0.25 mmol scale) was assembled on a PAM-resin by manual Boc-SPPS using the HBTU-mediated in situ neutralisation protocol with DMF as solvent.[13] The individual SPPS steps were performed in screw-cap glass reaction vessels, fitted with a sintered glass frit. The resin was washed and allowed to swell for at least 1 h in a reaction vessel with DMF prior to any chemistry was performed. After complete assembly of α-RgIA, the N-terminal Boc protecting group was removed via a 1x 1 min followed by a 1x 5 min TFA deprotection step, a 1x 1 min 10% DIEA/DMF neutralisation step and a 1 min DMF flow-wash. Half of the α-RgIA on resin was used to couple Cy5-COOH (2 eq) to the deprotected N-terminus (2x for 4 h using HATU and DIEA to activate the carboxylic acid). Side chain deprotection and peptide cleavage from the resin was performed by treatment of the dried peptide resin (100-500 mg) with 10 mL HF/p-cresol/p-thio-cresol (18/1/1, v/v/v) for 1 h at 0°C.[14] Following evaporation of HF, the peptides were precipitated and washed with cold ether, filtered, redissolved in 50% CH3CN / 1% TFA and lyophilised. The peptides were then purified by RP-HPLC.
Peptide folding
The reduced linear peptides were folded in a solution of 30% i-PrOH/ 0.1 M NH4HCO3 buffer at pH 8.4 for 24 h. The peptide concentrations were kept between 0.1–0.5 mg/mL (60-300 μM) and the oxidation was monitored by ESI-MS and analytical RP-HPLC. After complete oxidation, the solution was quenched with neat TFA to pH 2 and the peptides were purified by C18 RP-HPLC to >95% purity.
Electrophysiology – Xenopus Oocytes
RNA preparation, oocyte preparation and expression of nAChRs in Xenopus oocytes were performed as described previously.[15] Plasmids with cDNA encoding the rat α9 and α10 nAChR subunits were kindly provided by Dr. A.B. Elgoyhen (Universidad de Buenos Aires, Argentina). Oocytes were injected with a total of 5 ng of cRNA and then kept at 18°C in ND96 buffer (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2 and 5 mM HEPES at pH 7.4) supplemented with 50 mg/L gentamycin and 5 mM pyruvic acid and 5% Horse serum 2–5 days before recording. Membrane currents were recorded from Xenopus oocytes using a two-electrode voltage virtual ground circuit on a GeneClamp 500B amplifier (Molecular Devices, Union City, CA, USA) or an automated OpusXpress™ 6000A workstation (Molecular Devices). Electrodes were pulled from borosilicate glass (Harvard Apparatus Ltd., Edenbridge, UK) and filled with 3M KCl with resistances of 0.3–1.5 MΩ. All recordings were conducted at room temperature (20–23°C). During recordings, the oocytes were perfused continuously at a rate of ~2 mL/min. Acetylcholine (ACh; 100 μM) was applied for 2 s at 5 mL/min, with 300 s washout periods between applications with conotoxins bath applied for 300 s. Peak current amplitude was measured before and following (co-applied with conotoxin) incubation of the conotoxin. Cy5-azide (Sigma-Aldrich #777323) was used as a control for Cy5-RgIA. Oocytes were voltage clamped at a holding potential of –80 mV. Data were sampled at 500 Hz and filtered at 200 Hz. All data were pooled (n=3–7 for each data point) and represent as arithmetic means ± S.E.M. Concentration-response curves for antagonists were fitted by unweighted non-linear regression to the logistic equation: Ex = EmaxXn/(Xn + IC50n), where Ex is the response; X the antagonist concentration; Emax the maximal response; nH is the slope factor (Hill coefficient); and IC50, the concentration of antagonist that gives 50% inhibition of the agonist response. Computation was carried out using SigmaPlot 8.0 (Jandel Corporation, San Rafael, CA).
Cell culture and confocal microscopy
RAW264.7 cells were a kind gift from Prof. Matthew Sweet and were maintained in Sterilin petri dishes (VWR International, Australia) using RPMI1640 medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2. Cells were split every 3–4 days at a 1:10 dilution as they approached 80–90% confluency using fresh media forcefully applied to surface of the dish using a syringe with an 18 g needle attached.
For Cy5-RgIA labelling experiments 50 x 103 cells were seeded on a sterile coverslip and incubated overnight. The following day growth media was removed and the cells were washed 3x with Dulbecco’s phosphate buffered saline (DPBS) (Life Technologies, Australia). 10 μM Cy5-RgIA and the nuclear stain 4',6-diamidino-2-phenylindole (DAPI) were applied to cells and incubated for 1 h at room temperature. Cells were then washed 3x with DPBS to remove unbound Cy5-RgIA and excess DAPI stain and fixed with 4% paraformaldehyde at room temperature for 15 min. Cells were again washed 3x with DPBS and mounted onto a glass slide with n-propyl gallate. Cy5-RgIA binding was observed using a Zeiss LSM 510 meta confocal microscope.
Results
Cy5-COOH synthesis and on-resin labelling to produce Cy5-RgIA
On-resin labelling requires significantly more amounts of fluorophores than in-solution labelling due to peptide losses during the cleavage and purification steps. Cy5 was chosen as a suitable fluorophore due to its well-established properties, its wide applicability for fluorescent imaging techniques (e.g., FRET, near-infrared imaging, flow cytometry, screening platforms, bioassays, etc.), its compatibility with standard filters, its low autofluorescence, and its scale-able two-step synthesis (Scheme 1).[12] α-RgIA was selected as a representative α-conotoxin due to its well-established synthesis and three-dimensional structure, and its ability to potently block nAChRs, which can be tested by electrophysiology.[16] We scaled up the Cy5-COOH synthesis reported by Korbel et al.[12] to gram-scale, which provided enough material to carry out the on-resin strategy (Figure 1A). α-RgIA assembly was carried out using Boc-SPPS and the HBTU-mediated in situ neutralization protocol[13] and Cy5-COOH was coupled on half of the resin split to the deprotected N-terminus using HATU and a two-fold excess (2x, 4h). 1h anhydrous hydrogen fluoride (HF) treatment of the RgIA-resin and Cy5-RgIA-resin yielded reduced α-RgIA and Cy5-RgIA in good yields. Importantly, HF treatment did not affect the Cy5 fluorophore, in contrast to another fluorophore, BODIPY, that we tried as an alternative (data not shown). Oxidative folding of α-RgIA in 0.1 M NH4HCO3 (peptide concentration 60–300 μM, pH 8.4) yielded the inactive ribbon disulfide bond isomer as well as the desired bioactive globular isomer in nearly equivalent amounts (Figure 1B), which aligned well with literature.[16a] Addition of 30% i-PrOH to the folding buffer shifted the ribbon/globular-folding ratio towards the more hydrophobic, bioactive globular isomer. This folding buffer (30% i-PrOH/ 0.1 M NH4HCO3, pH=8.2) was then used for Cy5-RgIA, which only folded into a single bioactive globular isomer. The hydrophobic nature of Cy5 produced a significant right shift in retention time of Cy5-RgIA on the analytical RP-HPLC (7 min; using a 1% linear gradient) compared to the more hydrophilic α-RgIA (Figure 1B). The purified RgIA analogues and their corresponding analytical HPLC traces and MS data are presented in Figure 1C & D.
Scheme 1. Synthetic scheme of Cy5-COOH.
Figure 1. On-resin labelling strategy of Cy5-RgIA and analytical RP-HPLC and MS analysis of folding and purification steps of wild-type α-RgIA and Cy5-RgIA.
A RgIA was assembled on a PAM resin using Boc-SPPS. The N-terminal Boc protecting group was removed upon TFA treatment and Cy5-COOH coupled to the N-terminal amine. HF treatment deprotected the side chain protecting groups and released the labelled peptide from the resin. Oxidative folding in 30% i-PrOH/ 0.1 M NH4HCO3 buffer at pH 8.4 for 24 h yielded the correctly folded, globular Cy5-RgIA. B Analytical HPLC analysis of different folding conditions of α-RgIA and Cy5-RgIA. The more hydrophobic Cy5-RgIA folds only into the single, bioactive, globular isomer, while unlabelled, less hydrophobic α-RgIA folds also into the inactive ribbon disulfide bond isomer in addition to the bioactive globular isomer. Addition of 30% i-PrOH to 0.1 M NH4HCO3 (peptide concentration 60–300 μM, pH 8.4) shifts the ribbon/globular-folding ratio towards the more hydrophobic, bioactive globular isomer. C Analytical RP-HPLC and MS analysis of folded and purified ribbon (inactive) and globular (active) α-RgIA. D Analytical RP-HPLC and MS analysis of folded and purified globular (active) Cy5-RgIA displaying its characteristic peptide bond absorbance at 214 nm and its 640 nm absorbance of Cy5.
Effect of Cy5 label on the ability of Cy5-RgIA to bind to nAChRs
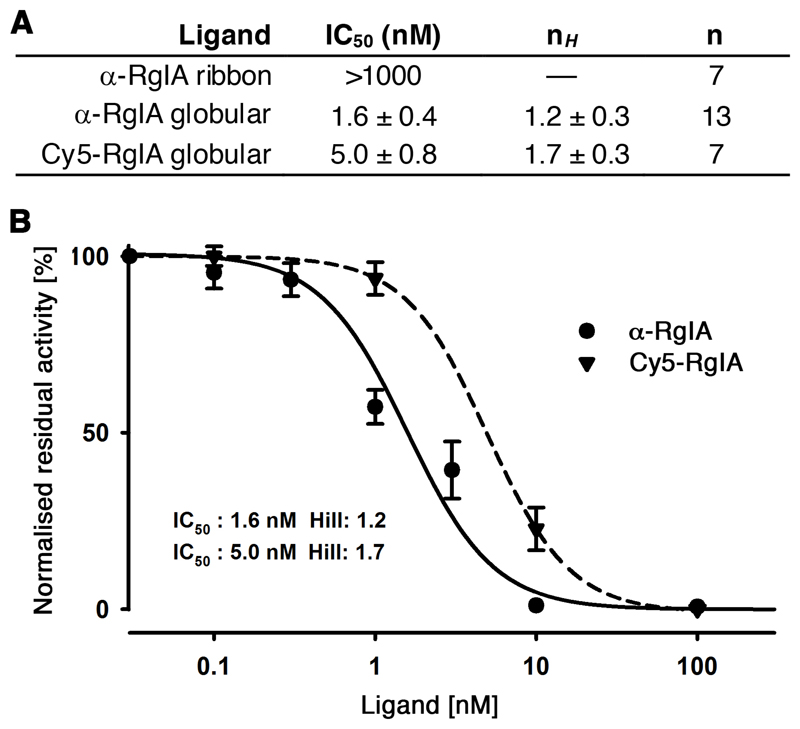
RgIA (globular and ribbon) and Cy5-RgIA were tested in parallel on their capability to block the rat α9α10 nAChRs heterologously expressed in Xenopus oocytes (Figure 2). Both α-RgIA and Cy5-RgIA inhibited the ACh (100 uM)-evoked current reversibly with an IC50 of 1.6 ± 0.4 nM (nH = 1.2 ± 0.3) for α-RgIA and an IC50 of 5.0 ± 0.8 nM (nH = 1.7 ± 0.3) for Cy5-RgIA. Ribbon RgIA was inactive at α9α10 nAChRs up to 1 μM. Bath application of 10 μM Cy5 alone had no effect on ACh-evoked current amplitude or time course mediated by rα9α10 nAChRs (n = 4). Complete function and potency of Cy5-RgIA was maintained, demonstrating that the Cy5 fluorophore attached to the N-terminus did not interfere with its ability to bind to the α9α10 nAChRs.
Figure 2. Inhibition of rat α9α10 nAChRs expressed in Xenopus oocytes by α-RgIA (globular and ribbon) and globular Cy5-RgIA.
A Table listing half-maximal inhibitory concentration (IC50), Hill coefficient (nH,), and number of observations (n) of α-RgIA (globular and ribbon) and globular Cy5-RgIA. B Representative concentration-response curves obtained for bioactive α-RgIA and Cy5-RgIA inhibition of rat α9α10 nAChRs.
Confocal imaging of Cy5-RgIA targeting α9α10 nAChRs in macrophages
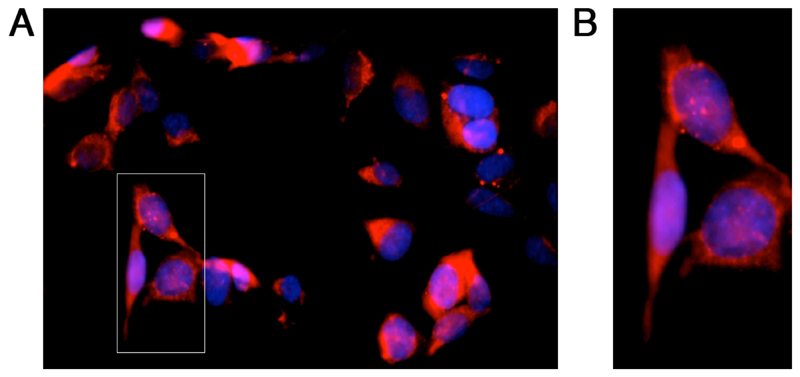
α-RgIA reduces immune cell activity in animal models of neuropathic pain.[17] We therefore assessed the functionality of Cy5-RgIA as a fluorescent probe on a mouse macrophage cell line (RAW264.7) natively expressing nAChRs.[18] Following the application of 10 μM Cy5-RgIA confocal images were acquired. Application of Cy5-RgIA resulted in bright and punctate labelling of cells indicating that Cy5-RgIA bound to the membrane and vesicular structures of the nAChR expressing cell line (Figure 3).
Figure 3. Confocal image of Cy5-RgIA targeting α9α10 nAChRs in a mouse macrophage cell line (RAW264.7).
Cells were labelled with Cy5-RgIA (red) and the nuclear DAPI stain (blue). A Representative image after 1 h incubation with 10 μM Cy5-RgIA and DAPI, washes with DPBS, 4% paraformaldehyde staining, and again washes with DPBS. A Zoomed in section showing bright and punctate labelling of the membrane and vesicular structures of the nAChR expressing cells.
Discussion
There exist two main strategies to produce fluorescent-labelled peptide probes: the first and most commonly used strategy entails in-solution labelling of purified and folded peptides with fluorophores that possess a functional group able to undergo (chemo-selective) reaction with the peptide. Such functional groups include isothiocyanate, amine-reactive succinimidyl/pentafluorophenyl esters and sulfhydryl-reactive maleimides. The advantage of this labelling strategy is commercial availability (of peptides and state-of-the-art fluorophores) and well-established protocols/kits with near quantitative labelling yields that only require minute amounts of (generally very expensive) fluorophores. Disadvantages include lack of site-specific control, which often restricts labelling to the N-terminus and complicates matters when other lysine groups are present, and reaction scale, which is a problem if larger quantities for in vivo experiments are needed.
The second strategy, on-resin labelling, was explored in this work and entails labelling of the peptide while it is still attached to the resin. This strategy offers better flexibility in terms of orthogonality and site-specific introduction of one or multiple fluorophores during the assembly phase (e.g., important for lysine-containing sequences or for the production of FRET probes). At the same time, it requires fluorophores that are chemically stable to standard cycles of SPPS (piperidine, TFA, HF) and that can be synthesised in large quantities.
Hence we compared the properties of a range of fluorophores that fall into this category such as fluorescein, BODIPY, 5-dimethylamino-1-naphthalenesulfonyl (dansyl), 7-nitrobenz-2-oxa-1,3-diazole (NBD), sulforhodamine B and the cyanine dye series, particularly in regards to brightness, quantum yield, commercial/synthetic availability, quantum yield, chemical stability, wavelength, and fluorescence half-life. Sulforhodamine B, BODIPY analogues and the cyanine series came out on top, particularly regarding brightness and synthetic/commercial availability. Our final choice fell on Cy5 due to its superior chemical stability to piperidine, TFA and HF that makes it compatible with Boc- and Fmoc-SPPS (e.g., BODIPY is not stable to HF, therefore incompatible with Boc-SPPS), its wide applicability and compatibility with available filters, its large molar extinction coefficient (250,000), and its emission maximum in the red region (649 nm excitation, 670 nm emission), where many detectors have maximum sensitivity with minimal background quenching/autofluorescence. Furthermore, Cy3/Cy5 is a known FRET pair; incorporation of Cy3 and Cy5 sequentially into the peptide sequence could be of high value when it comes to developing bioassays.
Cy5-COOH was produced on gram-scale and used to successfully label α-RgIA. Cy5-RgIA was significantly more hydrophobic compared to native α-RgIA, which turned out to be beneficial during folding as it only resulted in the desired globular isomer (Figure 1). This is in agreement with observations made in the folding of other α-conotoxins, where a more hydrophobic folding environment appears to favour the encoded structural information within the amino acid sequence to generate predominantly the bioactive native fold.[19] Although this did not cause any problems for the folding of Cy5-RgIA, one could anticipate problems with different and maybe more hydrophobic venom-derived peptides (e.g., the class of δ-conotoxins; known for their hydrophobicity to target membrane ion channels). There we recommend taking advantage of the development of a series of water-soluble cyanine dyes that have SO3 groups incorporated in their structure.[20] Cy5-RgIA was nearly equipotent (IC50=5.0 nM) to native α-RgIA (IC50=1.6 nM) in inhibiting ACh-evoked currents, confirming that the N-terminus is not significantly involved in binding to the nAChR.
Finally, Cy5-RgIA was assessed with confocal microscopy as a fluorescent marker on a mouse macrophage cell line natively expressing nAChRs (determined by mRNA) (Figure 3).[18] Considering the controversial role of the α9α10 nAChR subtype in inflammation caused by nerve injury and of RgIA as a potent analgesic,[16b, 17, 21] probes such as Cy5-RgIA will become highly valuable probes to assist in elucidating the precise molecular target mechanisms involved. They will also lead the way in validating and quantifying the presence of specific nAChR subtypes on the protein level, a task that is currently challenging due to a lack of subtype-specific nAChR antibodies and imaging probes. We would like to point out that the confocal work presented here represents only a rudimentary data set and more extensive work (that is outside of the scope of this work) is currently being carried out to further define the application of such imaging probes and to expand our knowledge on the presence, distribution and function of nAChRs in macrophages.
In conclusion, we have expanded our chemical repertoire to label peptides on-resin and produced a potent and valuable new probe targeting the α9α10 nAChRs. Access to grams of Cy5-COOH through a simple two-step synthesis in combination with Cy5’s chemical stability to piperidine, TFA and HF makes this strategy compatible with Fmoc- and Boc-SPPS, and enables production of large amounts of labelled peptide probes for in vivo experiments. Our strategy furthermore enables straightforward site-specific introduction of Cy5 via orthogonally protected lysine residues (e.g., via Lys(Fmoc) for Boc-SPPS or Lys(ivDde) for Fmoc-SPPS) as well as introduction of FRET pairs (e.g., Cy3 at the N-terminus and Cy5 within the sequence). This forms an important alternative to in-solution labelling and can be applied to a wide range of bioactive peptide probes.
Acknowledgements
We thank Dr. Debby Feytens for some of the Cy5 precursor synthesis work and Dr. Han-Shen Tae for the Cy5 control experiments. MM was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (714366), by the Australian Research Council (DE150100784, DP190101667), and by the Vienna Science and Technology Fund (WWTF; LS18-053). PFA and DJA were supported by Program Grant APP1072113 from the National Health & Medical Research Council (NHMRC) of Australia and a NHMRC Principal Research Fellowship to PFA (APP1080593). Finally, we want to recognise Professor Paul Alewood’s outstanding contributions to the field of peptide science and thank him for his long-lasting collegiality, mentorship and friendship.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
References
- [1].(a) Hurst R, Rollema H, Bertrand D. Pharmacol Ther. 2013;137:22. doi: 10.1016/j.pharmthera.2012.08.012. [DOI] [PubMed] [Google Scholar]; (b) Millar Neil S, Gotti C. Neuropharmacology. 2009;56:237. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]; (c) Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Physiol Rev. 2009;89:73. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gotti C, Zoli M, Clementi F. Trends Pharmacol Sci. 2006;27:482. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]; (e) Gotti C, Clementi F. Prog Neurobiol. 2004;74:363. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]; (f) Papke RL. Biochem Pharmacol. 2014;89:1. doi: 10.1016/j.bcp.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Romanelli MN, Gratteri P, Guandalini L, Martini E, Bonaccini C, Gualtieri F. ChemMedChem. 2007;2:746. doi: 10.1002/cmdc.200600207. [DOI] [PubMed] [Google Scholar]
- [2].Dineley KT, Pandya AA, Yakel JL. Trends Pharmacol Sci. 2015;36:96. doi: 10.1016/j.tips.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].(a) Kudryavtsev D, Shelukhina I, Vulfius C, Makarieva T, Stonik V, Zhmak M, et al. Toxins. 2015;7:1683. doi: 10.3390/toxins7051683. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Barber CM, Isbister GK, Hodgson WC. Toxicon. 2013;66:47. doi: 10.1016/j.toxicon.2013.01.019. [DOI] [PubMed] [Google Scholar]; (c) Dutertre S, Nicke A, Tsetlin VI. Neuropharmacology. 2017;127:196. doi: 10.1016/j.neuropharm.2017.06.011. [DOI] [PubMed] [Google Scholar]
- [4].(a) Lebbe EKM, Peigneur S, Wijesekara I, Tytgat J. Mar Drugs. 2014;12:2970. doi: 10.3390/md12052970. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muttenthaler M, Akondi KB, Alewood PF. Curr Pharm Des. 2011;17:4226. doi: 10.2174/138161211798999384. [DOI] [PubMed] [Google Scholar]; (c) Akondi KB, Muttenthaler M, Dutertre S, Kaas Q, Craik DJ, Lewis RJ, et al. Chem Rev. 2014;114:5815. doi: 10.1021/cr400401e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Giribaldi J, Dutertre S. Neurosci Lett. 2018;679:24. doi: 10.1016/j.neulet.2017.11.063. [DOI] [PubMed] [Google Scholar]
- [5].Grishin AA, Wang C-IA, Muttenthaler M, Alewood PF, Lewis RJ, Adams DJ. J Biol Chem. 2010;285 doi: 10.1074/jbc.M110.111880. 22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vishwanath VA, McIntosh JM. Bioconjugate Chem. 2006;17:1612. doi: 10.1021/bc060163y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Whiteaker P, McIntosh JM, Luo S, Collins AC, Marks MJ. Mol Pharmacol. 2000;57:913. [PubMed] [Google Scholar]
- [8].Hone AJ, Whiteaker P, Christensen S, Xiao Y, Meyer EL, McIntosh JM. J Neurochem. 2009;111:80. doi: 10.1111/j.1471-4159.2009.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hone AJ, Whiteaker P, Mohn JL, Jacob MH, McIntosh JM. J Neurochem. 2010;114:994. doi: 10.1111/j.1471-4159.2010.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Whiteaker P, Marks MJ, Christensen S, Dowell C, Collins AC, McIntosh JM. J Pharmacol Exp Ther. 2008;325:910. doi: 10.1124/jpet.108.136895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].(a) Conibear AC, Daly NL, Craik DJ. Biopolymers. 2012;98:518. doi: 10.1002/bip.22121. [DOI] [PubMed] [Google Scholar]; (b) Buck MA, Olah TA, Weitzmann CJ, Cooperman BS. Anal Biochem. 1989;182:295. doi: 10.1016/0003-2697(89)90597-6. [DOI] [PubMed] [Google Scholar]; (c) Moffatt F, Senkans P, Ricketts D. J Chromatogr A. 2000;891:235. doi: 10.1016/s0021-9673(00)00620-8. [DOI] [PubMed] [Google Scholar]
- [12].Korbel GA, Lalic G, Shair MD. J Am Chem Soc. 2001;123:361. doi: 10.1021/ja0034747. [DOI] [PubMed] [Google Scholar]
- [13].Schnölzer M, Alewood PF, Jones A, Alewood D, Kent SBH. Int J Pept Protein Res. 1992;40:180. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- [14].Muttenthaler M, Albericio F, Dawson PE. Nat Protoc. 2015;10:1067. doi: 10.1038/nprot.2015.061. [DOI] [PubMed] [Google Scholar]
- [15].Clark RJ, Fischer H, Nevin ST, Adams DJ, Craik DJ. J Biol Chem. 2006;281 doi: 10.1074/jbc.M604550200. 23254. [DOI] [PubMed] [Google Scholar]
- [16].(a) Clark RJ, Daly NL, Halai R, Nevin ST, Adams DJ, Craik DJ. FEBS Lett. 2008;582:597. doi: 10.1016/j.febslet.2008.01.027. [DOI] [PubMed] [Google Scholar]; (b) Ellison M, Haberlandt C, Gomez-Casati ME, Watkins M, Elgoyhen AB, McIntosh JM, et al. Biochemistry. 2006;45:1511. doi: 10.1021/bi0520129. [DOI] [PubMed] [Google Scholar]
- [17].Vincler M, Wittenauer S, Parker R, Ellison M, Olivera BM, McIntosh JM. Proc Natl Acad Sci USA. 2006;103 doi: 10.1073/pnas.0608715103. 17880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].(a) Wang H, Yu M, Ochani M, Amella Carol A, Tanovic M, Susarla S, et al. Nature. 2003;421 doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]; (b) Wang YY, Liu Y, Ni XY, Bai ZH, Chen QY, Zhang Y, et al. Oncol Rep. 2014;31:1480. doi: 10.3892/or.2013.2962. [DOI] [PubMed] [Google Scholar]; (c) Yi L, Luo J-f, Xie B-b, Liu J-x, Wang J-y, Liu L, et al. Shock. 2015;44:188. doi: 10.1097/SHK.0000000000000389. [DOI] [PubMed] [Google Scholar]
- [19].(a) Moroder L, Musiol H-J, Götz M, Renner C. Biopolymers. 2005;80:85. doi: 10.1002/bip.20174. [DOI] [PubMed] [Google Scholar]; (b) Bulaj G, Olivera BM. Antioxid Redox Signal. 2008;10:141. doi: 10.1089/ars.2007.1856. [DOI] [PubMed] [Google Scholar]
- [21].(a) Bouteiller C, Clave G, Bernardin A, Chipon B, Massonneau M, Renard P-Y, et al. Bioconjugate Chem. 2007;18:1303. doi: 10.1021/bc0700281. [DOI] [PubMed] [Google Scholar]; (b) Li X, Gao X, Shi W, Ma H. Chem Rev. 2014;114:590. doi: 10.1021/cr300508p. [DOI] [PubMed] [Google Scholar]
- [21].(a) Adams DJ, Callaghan B, Berecki G. Br J Pharmacol. 2012;166:486. doi: 10.1111/j.1476-5381.2011.01781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Callaghan B, Adams DJ. Channels. 2010;4:51. doi: 10.4161/chan.4.1.10281. [DOI] [PubMed] [Google Scholar]; (c) Di Cesare Mannelli L, Cinci L, Micheli L, Zanardelli M, Pacini A, McIntosh JM, et al. Pain. 2014;155:1986. doi: 10.1016/j.pain.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nevin ST, Clark RJ, Klimis H, Christie MJ, Craik DJ, Adams DJ. Mol Pharmacol. 2007;72:1406. doi: 10.1124/mol.107.040568. [DOI] [PubMed] [Google Scholar]