Abstract
Frameshift mutations in the DMD gene, encoding dystrophin, cause Duchenne muscular dystrophy (DMD), leading to terminal muscle and heart failure in patients. Somatic gene editing by sequence-specific nucleases offers new options for restoring the DMD reading frame, resulting in expression of a shortened, but largely functional dystrophin protein. Here, we validated this approach in a pig model of DMD lacking exon 52 of DMD (DMDΔ52), as well as in a corresponding patient-derived induced pluripotent stem cell (iPSC) model. In DMDΔ52 pigs1, intramuscular injection of adeno-associated viral vectors of serotype 9 carrying an intein-split Cas92 and a pair of guide RNAs targeting sequences flanking exon 51 (AAV9-Cas9-gE51) induced expression of a shortened dystrophin (DMDΔ51-52) and improved skeletal muscle function. Moreover, systemic application of AAV9-Cas9-gE51 led to widespread dystrophin expression in muscle, including diaphragm and heart, prolonging survival and reducing arrhythmogenic vulnerability. Similarly, AAV6-Cas9-gE51-mediated excision of exon 51 in DMDΔ52-patient iPSC-derived myoblasts and cardiomyocytes restored dystrophin expression and ameliorated skeletal myotube formation as well as cardiac abnormal cardiomyocyte Ca2+ handling and arrhythmogenic susceptibility. The ability of Cas9-mediated exon excision to improve DMD pathology in these translational models paves the way for new treatment approaches in patients with this devastating disease.
Duchenne muscular dystrophy represents the most frequent hereditary childhood myopathy, leading to progressive muscle degeneration and weakness, and to premature death due to respiratory and cardiac failure. The vast majority of patients carry frameshift mutations in the DMD gene encoding dystrophin, which are mainly exon deletions3,4. The X-chromosomal location of DMD renders 1 in 3,500 to 5,000 male newborns affected5. Antisense oligonucleotide (AON)-mediated exon skipping aimed at reframing DMD transcripts6 has already been translated into clinical trials7,8. However, AONs - though initially efficient in a dose-dependent manner6 - offer only temporary and limited efficacy of DMD expression9. Endonuclease-based gene editing strategies provide a more efficient and permanent genomic correction, as demonstrated in mdx mouse models10–14. Recently, intravenous (i.v.) application of AAV9 delivering CRISPR/Cas9 components in a beagle model of DMD (exon 50 deficiency) proved successful in restoring expression of a shortened dystrophin in various muscles, including the heart15. However, functional data have not been reported as of yet.
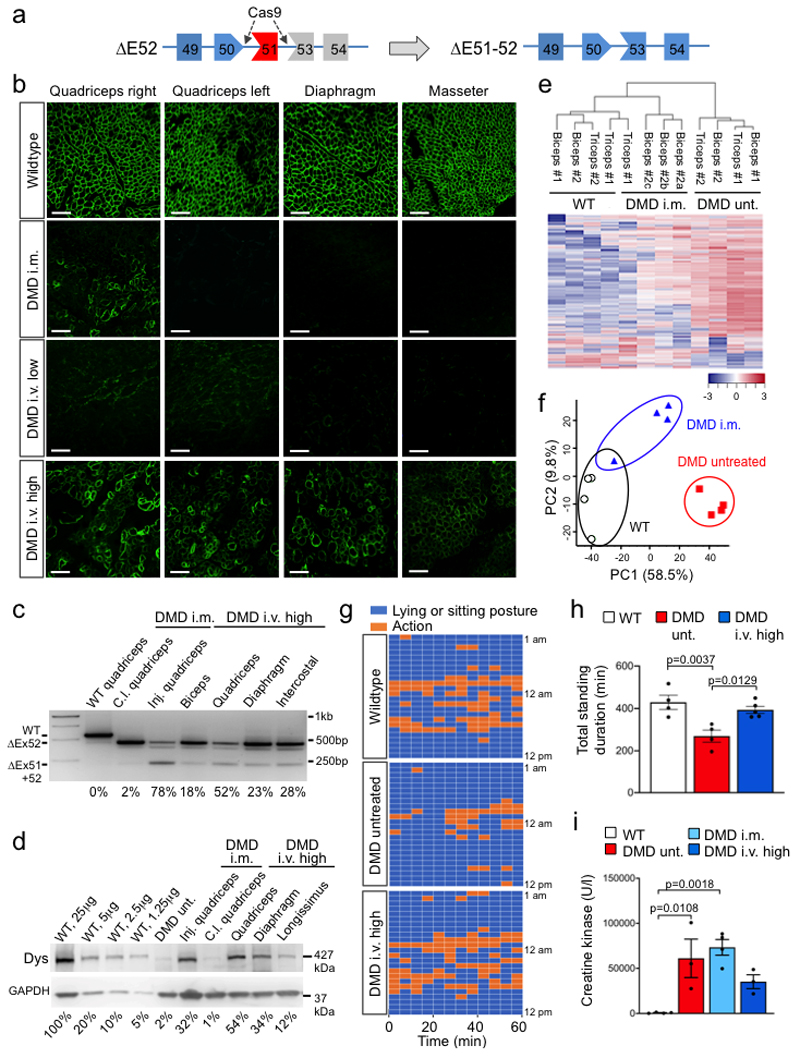
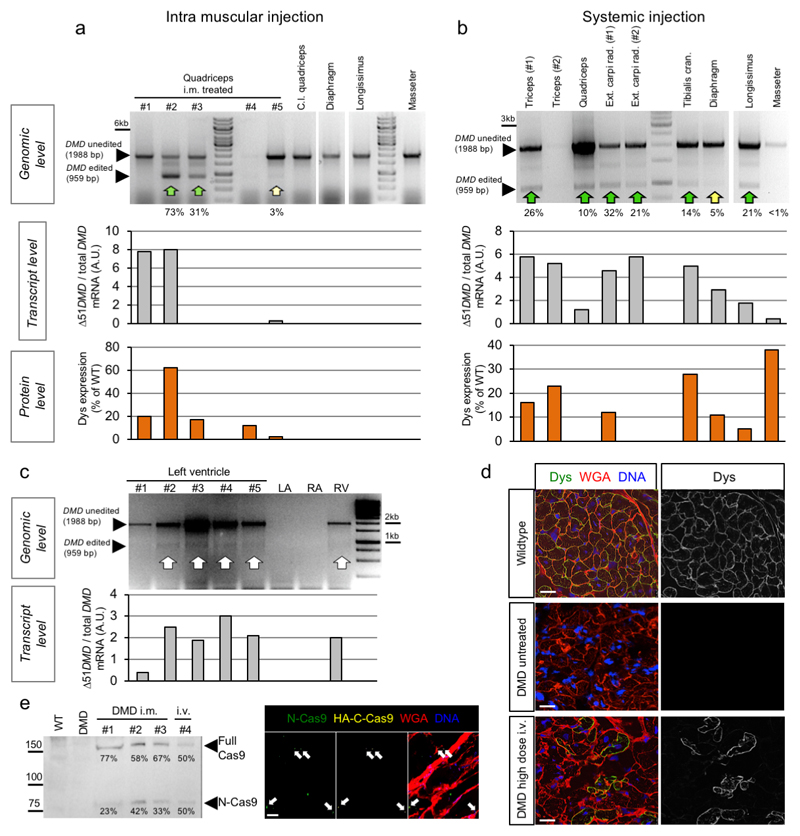
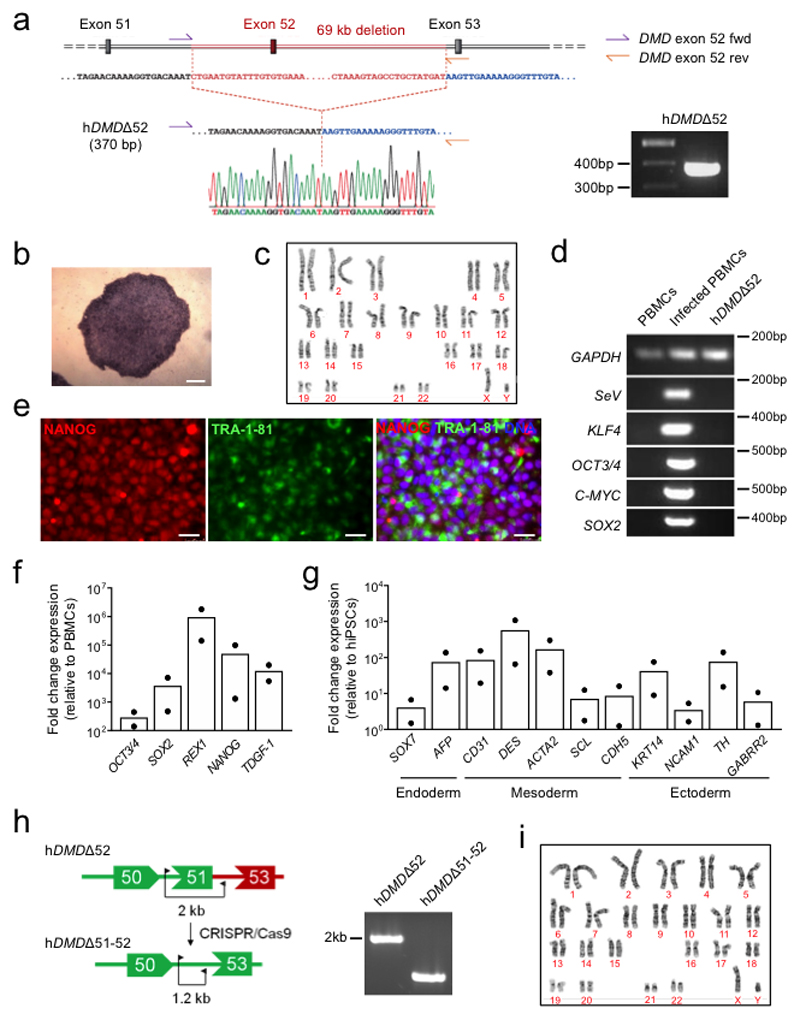
We have generated a DMD pig model lacking DMD exon 521, resulting in a complete loss of dystrophin expression (Fig. 1a, Methods). First, we assessed whether local application of Cas9 and selected gRNAs targeting exon 51 (Extended Data Fig. 1a-c, Source Data Extended Data Fig. 1c) induces expression of a shortened, but stable dystrophin in vivo (Fig. 1a). Ten to fourteen-day-old piglets were subjected to unilateral fore- and hindlimb intramuscular (i.m.) injection of a pair of intein-split Streptococcus pyogenes (Sp)-Cas92 and gRNA-encoding virus particles (AAV9-Cas9-gE51, 2x1013 vp/kg each) (Extended Data Fig. 1d-f, Source Data Extended Data Fig. 1f). After six weeks, histological analysis revealed restitution of membrane-localized dystrophin in the treated areas, and - due to leakage of the vector - in low levels at the contralateral limb. Successful elimination of exon 51 and expression of DMDΔ51-52 was confirmed at genomic, transcript, and protein levels (Fig. 1b-d and Extended Data Fig. 2a, Source Data Extended Data Fig. 2a), though full congruence could not be reached due to sample variability. Mass spectrometry analysis (Supplementary Fig. 1a-c) of treated muscle tissue indicated partial normalization of proteins dysregulated in DMD (Fig.1e), with several fibrosis-related proteins significantly reduced (Supplementary Fig. 2a). Principal component analysis of the proteome confirmed that the global protein profile of AAV9-Cas9-gE51-injected muscles resided closer to healthy than DMD animals (Fig. 1f).
Figure 1. Genome editing of DMDΔ52 pigs by Cas9-mediated exon 51 excision.
a, Schematic diagram of gene editing in DMDΔ52 pigs. Loss of exon 52 of the DMD gene (ΔE52) leads to an out-of-frame mutation with a premature stop codon, preventing protein translation. Cas9-mediated excision of exon 51 (ΔE51-52) corrects the reading frame, resulting in translation of an internally truncated but functional protein. Out of frame exons are illustrated in gray. b, Immunofluorescent staining for dystrophin expression in the indicated muscles (Quadriceps = Musculus quadriceps) in wildtype and DMDΔ52 (DMD) pigs, either at the injection sites after intramuscular (i.m.) AAV9-Cas9-gE51 treatment or after intravenous (i.v.) treatment with 2x1013 virus particles/kg (low dose) or 2x1014 virus particles/kg (high dose), representative of 9 images collected in n=3 independent experiments per group. Scale bars, 200 μm. c, Levels of wildtype (WT), mutated DMD (ΔE52) and edited DMD (ΔE51-52) in WT limb muscle or in the indicated muscles of DMDΔ52 pigs treated with i.m. or high dose i.v. G2-AAV9-Cas9-gE51, as assessed by RT-PCR, representative of 3 technical. Percentages indicate expression level of the corrected transcript (ΔE51-52) relative to total transcript (Δ52 + ΔE51-52). Inj. = injected, c.l. = contralateral (cf. Source Data Fig. 1). d, Immunoblotting for dystrophin (Dys) in extracts from WT quadriceps at the indicated concentrations, DMD untreated (unt.) quadriceps, and indicated muscles of DMD pigs after either i.m. or i.v. AAV9-Cas9-gE51 treatment. Percentages indicate expression of dystrophin protein relative to WT. Dystrophin levels were normalized to GAPDH, used as a loading control, representative of 3 independent experiments (cf. Source Data Fig.1). e, Unsupervised hierarchical clustering of normalized label free quantification intensity values for skeletal muscle samples of WT, i.m. injected DMD, and untreated (unt.) DMD pigs. The color code indicates z-score normalized expression values. f, Principal component analysis (PCA) of proteins isolated from biceps and triceps muscles from WT, untreated and i.m. treated DMD pigs (n=2 animals per group) presented in (e). Each symbol represents an individual sample (n=4 per group). Percentage of variance explained by each component is indicated. g, h, Representative data from 24h behavioral observation of a single wildtype, a DMD untreated, and a DMD high dose i.v. treated pig (g) and quantification of the total standing time per pig for WT (n=5), DMD (n=4) and DMD high dose i.v. treated (n=5), indicated as mean±SEM with p values from a one-way ANOVA (with Bonferroni’s multiple comparison test, F=9.819; df=10) (h, cf. Source Data Fig. 1). i, Serum creatine kinase levels of WT (n=4) and DMD pigs, the latter untreated (=unt., n=3) or after i.m. injection of AAV9-Cas9-gE51 (n=4) or after high dose i.v. (n=3) treatment with G2-AAV9-Cas9-gE51, indicated as mean±SEM (cf. Source Data Fig. 1) with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=9.986, df=10)
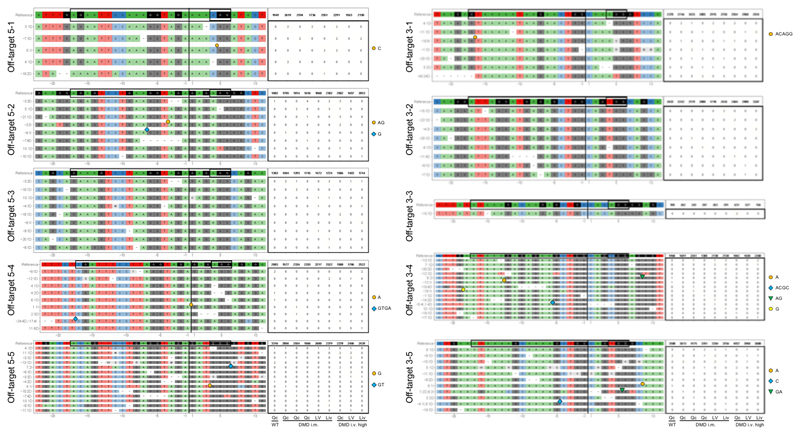
Of note, the diaphragm and the heart – two muscles contributing substantially to the mortality of Duchenne patients16,17 – were not affected by i.m. limb injection (Fig. 1b). We sought to transduce these organs by intravenous (i.v.) application of AAV9-Cas9-gE51, which was coated with PAMAM-G2 nanoparticles to increase the myotropism of the vector without enhancing toxicity (Supplementary Fig. 3a-c and 4, Supplementary Table 1)18,19. In contrast to a low dose (1-2x1013 coated-vp/kg G2-AAV9-Cas9-gE51), which sporadically transduced skeletal muscle specimen (Fig. 1b), a high dose (2x1014 vp/kg each) of G2-AAV9-Cas9-gE51 enabled dystrophin protein expression in skeletal muscles, diaphragm and heart (Fig. 1b,d, Extended Data Fig. 2b-d, Source Data Extended Data Fig. 2b,c) after self-assembly of both intein-carrying Cas9-halves (Extended Data Fig. 2e, Source Data Extended Data Fig. 2e). Accordingly, high-dose i.v. treatment prolonged the time that animals spent in upright position (Fig. 1g,h) and tended to attenuate highly elevated creatine kinase levels (indicating less muscle decay, Fig. 1i). No mutations were detected in the 5 most likely predicted off-target sites by deep sequencing (Extended Data Fig. 3). The INDELs in the target regions showed no concordant alterations in the treated samples. The mean observed alteration frequency was 0.01% (SD: 0.03), which laid in the background error levels of the DNA sequencer with a maximum sequencing error rate of 0.1%. Due to the limited number of animals available, we have not studied AAV9-Cas9 without gRNAs. We also cannot exclude that Cas9 presence in myocytes20,21 may potentially induce more off-target effects and immune reactions22,23 beyond the 3-4 months observation period in our study.
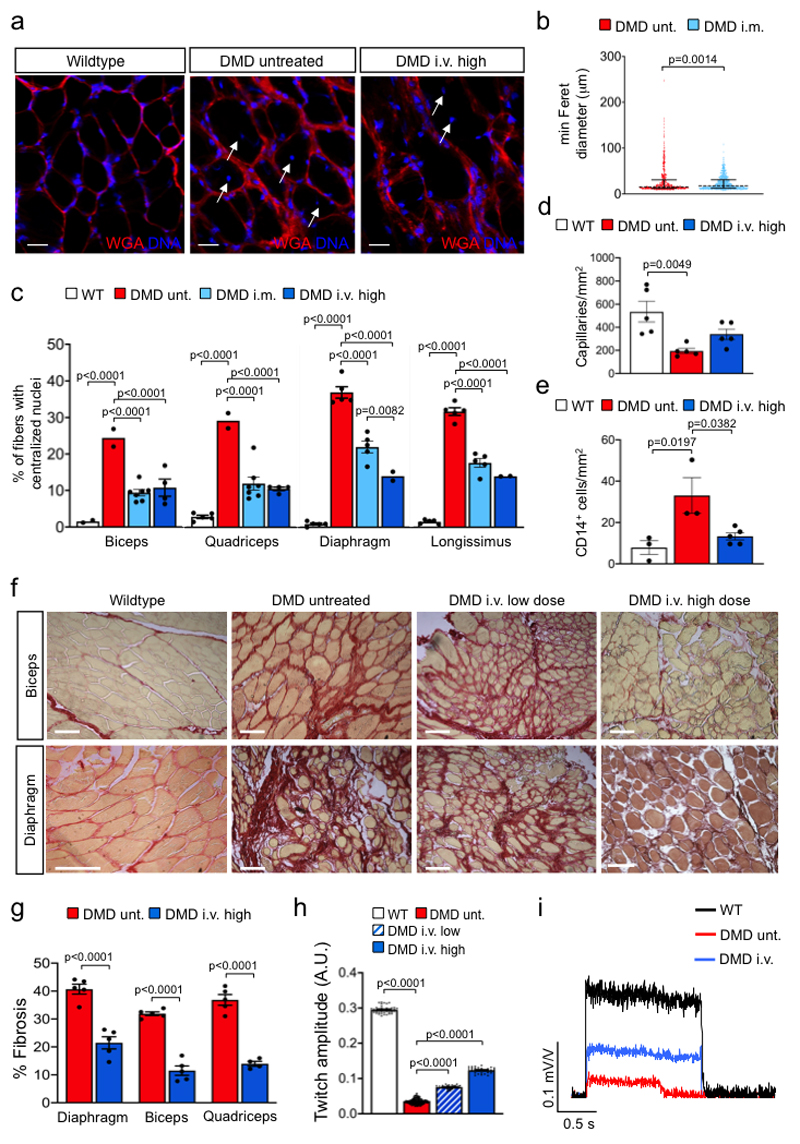
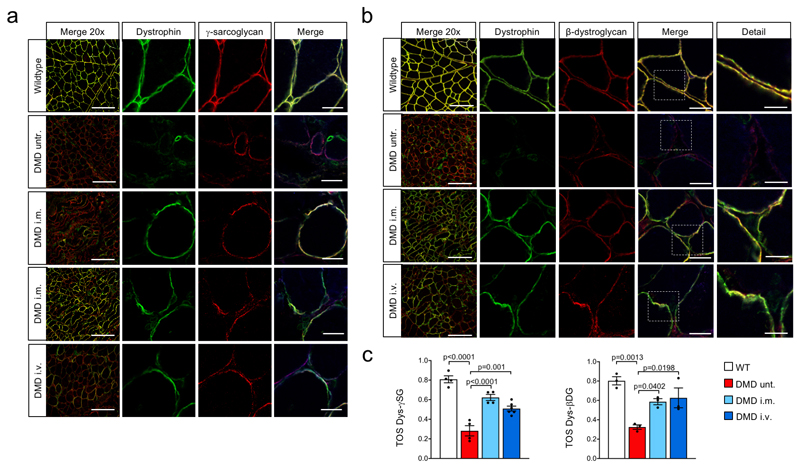
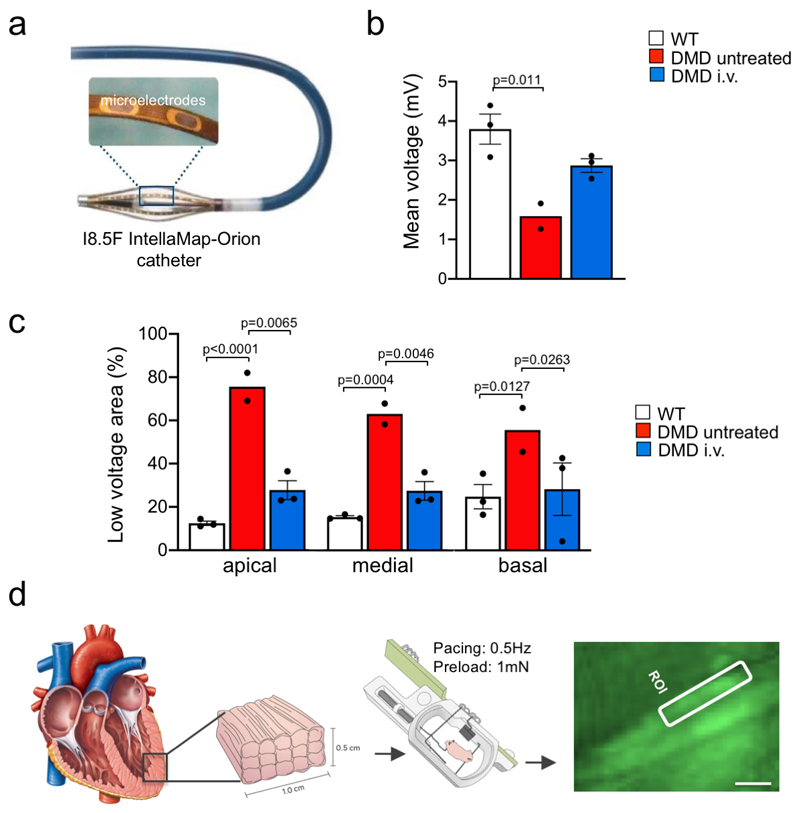
Lack of dystrophin in DMDΔ52 muscles induces the collapse of the dystrophin-associated glycoprotein complex (DGC)24. Local i.m. or high-dose i.v. treatment resulted in increased levels of membrane-bound γ-sarcoglycan and β-dystrogylcan and restoration of the DGC (Extended Data Fig. 4a-c). Further structural evaluation of skeletal muscles revealed that expression of membrane-bound DMDΔ51-52 significantly reduced the occurrence of rounded myofibers with centralized nuclei (Fig. 2a-c). Upon G2-AAV9-Cas9-gE51 application, an improved skeletal muscle structure was indicated by an attenuation of capillary loss as well as decreased mononuclear cell infiltration and interstitial fibrosis (Fig. 2d-g, Supplementary Fig. 2a,b). In accordance, augmented muscle twitch amplitude and tetanic contraction force indicated functional improvement after high-dose G2-AAV9-Cas9-gE51 treatment (Fig. 2h,i). The porcine DMDΔ52 model displayed a high mortality: 61.6% (45/73) of the affected males died within the first week after birth and none survived longer than 105 days. G2-AAV9-Cas9-gE51 treatment increased the maximum survival to 136 days (Fig. 3a). Video analysis revealed the occurrence of sudden cardiac death in untreated DMDΔ52 pigs (Supplementary Video 1). Thus, we investigated the global function and arrhythmogenicity of DMD hearts. Left ventricular (LV) angiography showed a reduction of ejection fraction in DMDΔ52 animals compared to wildtype controls, which was not significantly attenuated by systemic G2-AAV9-Cas9-gE51 treatment (p=0.08, Fig. 3b). Contraction and relaxation velocities (dP/dtmax/min) did not yield significant differences between groups (Fig. 3b), potentially due to modest expression levels of dystrophin (Fig. 3c, Extended Data Fig. 2d). No overt signs of heart failure (e.g. pleural effusion, pulmonary edema) were observed in pathologic investigations of DMDΔ52 animals, suggesting malignant arrhythmias as primary cause of death. Indeed, detailed electrophysiological mapping analysis (Extended Data Fig. 5a) demonstrated a decrease of voltage amplitude (Fig. 3d, Extended Data Fig. 5b) in ventricles of DMDΔ52 pigs compared to wildtype hearts, corresponding to an increased area of low voltage (<1.3mV) (Extended Data Fig. 5c). Since low voltage may indicate cardiac fibrosis25, we further investigated the left ventricles histologically and found extensive fibrotic areas in untreated DMDΔ52 hearts, whereas high dose i.v. G2-AAV9-Cas9-gE51 treatment restricted low-voltage area (Extended Data Fig. 5b,c) as well as fibrotic degeneration (Fig. 3e and Supplementary Fig. 2c).
Figure 2. Genome editing of DMDΔ52 restores the structure and function of diseased skeletal muscle.
a, Wheat germ agglutinin (WGA) staining (red) of cell borders and DNA labeling (blue) in quadriceps muscles of wildytpe pigs and untreated or high dose intravenous (i.v.) G2-AAV9-Cas9-gE51-injected DMD pigs, representative of 9 images collected in 3 pigs per group. Scale bars, 25 μm. b, Minimum diameter of cross-sectional fibers in untreated (unt.) DMD (n=406 of 3 pigs) and intramuscularly treated (i.m.) DMD muscle fibers (n=641 of 2 pigs), represented as dot plots showing all data points as well as the median (dashed line) and the quartiles (solid lines) (cf. Source Data Fig. 2), with the p value from an unpaired two-tailed t-test (t=3.212, df=1045). c, Percentage of fibers with centralized nuclei in skeletal muscle samples from biceps (B), quadriceps (Q) muscles, diaphragm (D) and longissimus dorsi (LD) of WT (B n=2; Q, D, LD n=5), untreated DMD (B, Q n=2; D, LD n=5) and i.m. (B, Q, n=7; D, LD n=5) or high dose i.v. treated DMD (B, Q n=4; D, LD n=2) of 2 animals per group (cf. Source Data Fig. 2), mean±SEM with p values from a two-way ANOVA with Tukey’s multiple comparison test (F=219.3, df=51). d, Density of CD31+ capillaries in quadriceps muscles of WT, untreated DMD and high dose i.v. treated DMD pigs (n=5 Q sections from n=3 animals, cf. Source Data Fig. 2), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=8.228, df=12). e, Quantification of CD14+ cells in skeletal muscle tissue of WT, untreated DMD and high dose i.v. treated DMD pigs (n=3 Q sections for WT and untreated DMD, 5 sections for high dose i.v. treated DMD from n=3 animals per group, cf. Source Data Fig.2), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=7.610, df=8). f, g, Sirius red staining showing the level of interstitial fibrosis in the peripheral muscle and diaphragm of WT, untreated DMD and low dose or high dose i.v. treated DMD pigs, representative of 10 sections collected in 3 independent experiments (f) and percentage of fibrosis in D and Q of untreated DMD (n=3) and high dose i.v. treated DMD pigs (n=3) (Source Data Fig. 2), indicated as mean±SEM with p values from a two-way ANOVA with Bonferroni’s multiple comparison test (F=238, df=23) (g). Scale bars in (f), 50 μm. h, Twitch amplitude after common peroneal nerve stimulation (cf. Methods) in WT (n=41 traces in n=3 pigs), untreated DMD (n=138 traces in n=3 pigs) and low dose (n=30 traces in n=2 pigs) or high dose (n=29 traces in n=3 pigs) i.v. treated DMD pigs (cf. Source Data Fig.2), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=11241, df=234). i, Tetanic contraction after 1.5 s common peroneal nerve stimulation at 50 Hz in WT, untreated DMD and i.v. treated DMD pigs, representative of 3 independent experiments for WT, DMD untreated (= unt.) and DMD high dose i.v..
Figure 3. Genome editing of DMDΔ52 improves survival and reduces cardiac arrhythmogenic vulnerability.
a, Kaplan-Meier curve of the survival time of untreated DMDΔ52 (DMD) (n=13) and high dose intravenously (i.v.) G2-AAV9-Cas9-gE51 treated DMD (n=7) pigs (Log Rank (Mantel Cox) p=0.032). b, Left, ejection fraction (EF) of wildtype (WT) (n=3), untreated DMD (n=3) and i.v. treated DMD (n=4) pigs, indicated as mean±SEM with p value from a one-way ANOVA with Bonferroni’s multiple comparison test (F=10.53, df=7). Middle and right, dP/dtmax and dPt/dtmin, respectively, of WT (n=3), untreated DMD (n=3) and high dose i.v. treated DMD (n=4) pigs, indicated as mean±SEM (cf. Source Data Fig. 3). c, Immunoblotting for dystrophin (Dys) in heart extracts (A=atrium, LV=left ventricle) from a WT pig at the indicated extract concentration), an untreated DMD pig, and four different high dose i.v. treated DMD pigs, representative of 3 replicates. Percentages indicate the level of dystrophin protein relative to WT based on the standard curve obtained by loading the indicated amounts of WT extract. Dystrophin levels were normalized to GAPDH, used as a loading control (cf. Source Data Fig. 3). d, High-resolution electrophysiological mapping indicating areas of normal excitation amplitude (> 1.3 mV, violet), low excitation amplitude (< 1.3 mV, yellow) or scar (< 0.3 mV, red) in WT (n=3), untreated DMD (n=2) and i.v. treated DMD pigs (n=3, see also Extended Data Figure 5a-c). Insets indicate angulation of the detection plane (front and rear) in DMD untreated and DMD high dose i.v., 45° right and left angulation in wildtype. e, Top, Sirius red staining showing the level of interstitial fibrosis in equivalent apical regions of WT, untreated DMD and i.v. treated DMD hearts, representative of 10 sections collected in 3 pigs per group. Scale bar, 200 μm. Bottom, corresponding quantification of the percentage of fibrosis (all n=5, cf. Source Data Fig. 3), indicated as mean±SEM with p value from an unpaired two-tailed t-test (t=7.619, df=8). f, Exemplary single-cell calcium transients in myocardial slices (loaded with Fluo-4 AM) from n=3 WT, n=2 untreated DMD and n=2 i.v. treated DMD pig hearts. g, Time to peak (top) and amplitude (bottom) of single-cell calcium transients in WT (n=10 cells from 2 animals), untreated DMD (unt., n=12 cells from 2 animals) and high dose i.v. treated DMD (n=8 cells from 3 animals) myocardial slices (cf. Source Data Fig. 3), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=63.24, df=27 and F=8.094, df=27, respectively). h, Left, exemplary images of Fluo-4 fluorescence in single cardiomyocytes within untreated DMD and high dose i.v. treated DMD myocardial slices with 4 regions of interest (ROI). Right, traces of the averaged fluorescence intensity within the 4 ROIs showing calcium transient propagation within single cells. The data are representative of 5 samples from n=2 animals in each group. Scale bars, 10 μm.
In addition, intrinsic arrhythmogenic potentials of DMD cardiomyocytes might contribute to malignant ventricular arrhythmias. Therefore, we performed ex vivo intracellular Ca2+ analysis of single cardiomyocytes within 300 μm-thick heart slices maintained in biomimetic chambers (Methods, Extended Data Fig. 5d)26. Compared to wildtype heart samples, cells from untreated DMDΔ52 cardiac tissue displayed abnormal Ca2+ transients (Fig. 3f,g) and unsynchronized intracellular Ca2+ waves (Fig. 3h). These pathological features were almost completely abolished by systemic G2-AAV9-Cas9-gE51 application (Fig. 3f-h), supporting the notion that an inherent arrhythmogenic susceptibility of DMDΔ52-cardiomyocytes can be ameliorated by genomic snipping of exon 51.
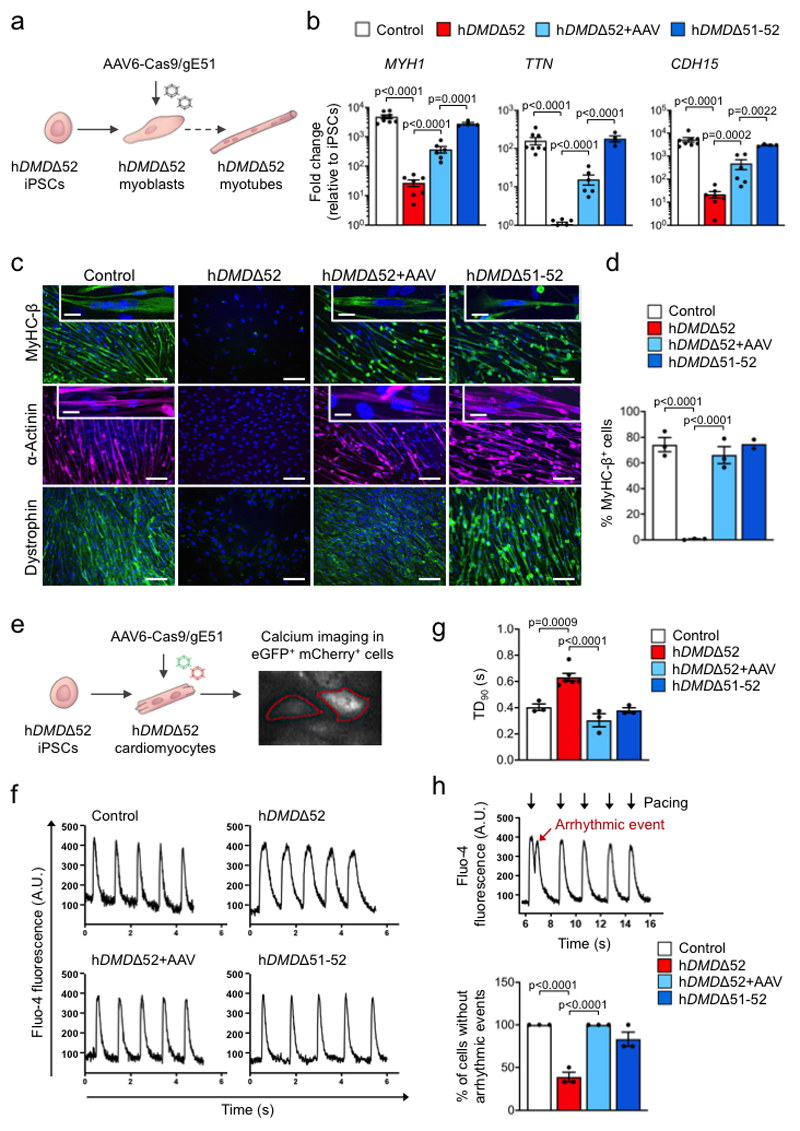
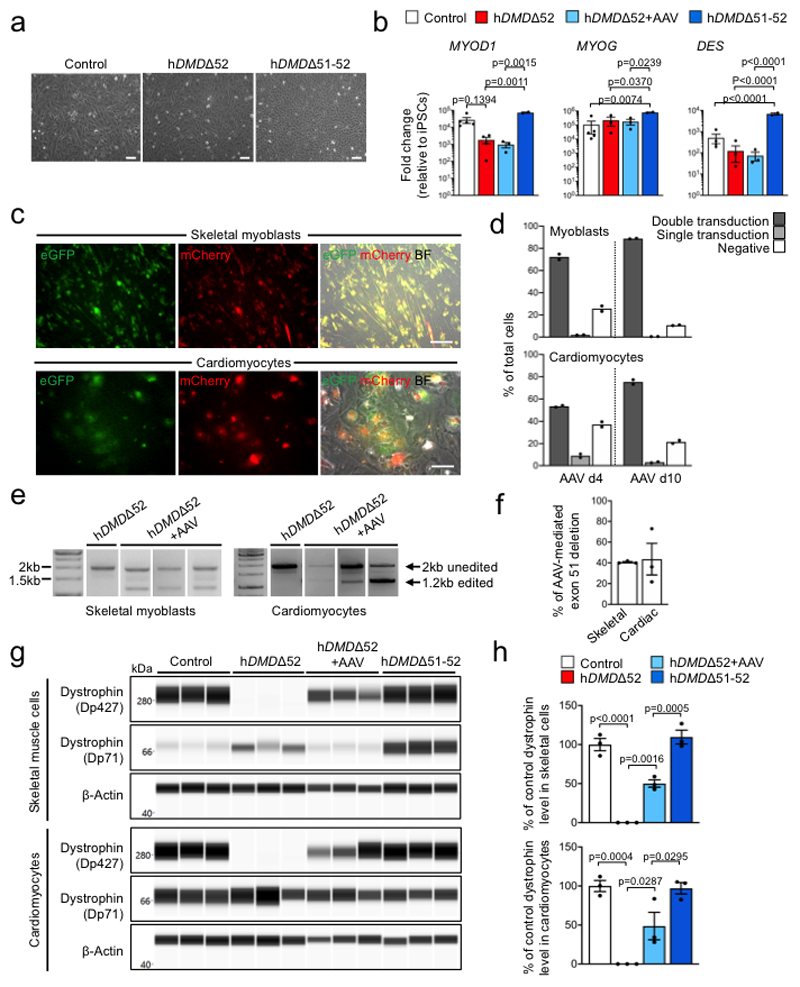
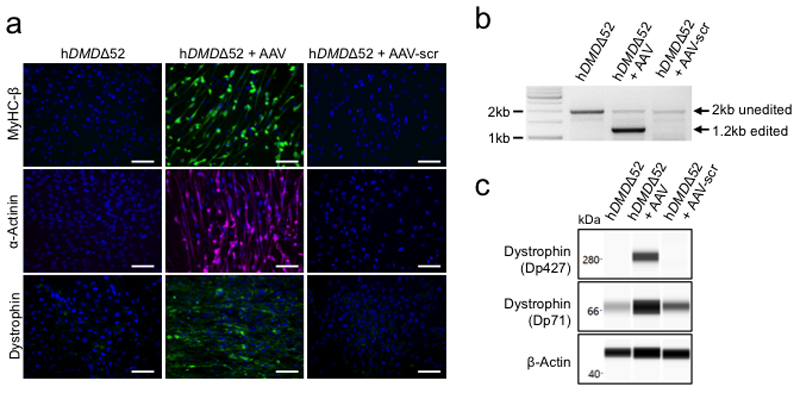
We finally investigated the muscle-specific targeting efficacy of the intein-split Cas9 AAV-mediated approach in human cells (Fig. 4a,e, Supplementary Fig. 5a-c). We generated iPSCs27 from a DMD patient carrying likewise a deletion of DMD exon 52 (hDMDΔ52), leading to appearance of a premature stop codon in exon 53 (Extended Data Fig. 6a-g). As controls, we used isogenic hDMDΔ51-52 iPSCs that were obtained by CRISPR/Cas9-mediated excision of exon 51 in the undifferentiated hDMDΔ52 iPSCs (Extended Data Fig. 6h-i) as well as iPSCs from a healthy, young male (Extended Data Fig. 7a-f). When directed to differentiate into skeletal muscle by a growth factor-based differentiation approach28, hDMDΔ52 cells expressed significantly lower levels of skeletal muscle genes (Fig. 4b) compared to controls and failed to generate multinucleated and spontaneously contractile myotubes (Fig. 4c,d and Supplementary Videos 2-4). This is in line with other studies having shown entirely defective myotube formation29 when DMD hiPSCs were subjected to directed skeletal muscle differentiation without overexpression of the skeletal master regulator gene myogenic differentiation 1 (MYOD1), although one study showed myotube formation without MYOD130. Double transduction of hDMDΔ52 iPSC-derived myoblasts with a pair of AAV6-Cas9-gE51 vectors (106 vp/cell) (Extended Data Fig. 8a-f, Source Data Extended Fig. 8b, d-f) induced expression of the human DMDΔ51-52 protein and rescued myotube formation (Fig. 4b-d, Extended Data Fig. 8g,h), while AAVs containing scrambled guide RNAs were ineffective (Extended Data Fig. 9a-c).
Figure 4. Somatic genome editing of human DMDΔ52 rescues disease phenotypes of skeletal and cardiac muscle cells from patient-specific iPSCs.
a, Schematic indicating strategy to rescue defective skeletal myotube formation in myoblasts differentiated from hDMDΔ52 hiPSCs by transduction with two AAV6 vectors containing an intein-split Cas9 and gRNAs designed to induce DMD exon 51 excision. b, RT-qPCR analysis of skeletal myotube markers 7-14 days after myotube induction in control (n=8 independent differentiations), hDMDΔ52 (n=7), hDMDΔ52+AAV (n=6) and hDMDΔ51-52 (n=4) myoblasts (cf. Source Data Fig. 4), indicated as mean fold change±SEM with p values from a one-way ANOVA of the logarithmized values with Bonferroni’s multiple comparison test (MYH1 F=26.21, df=3; TTN F=14.32, df=21; CDH15 F=10.84, df= 21). c, Immunofluorescence analysis of myosin heavy chain β (MyHC-β), α-actinin and dystrophin 14 days after myotube induction in myoblasts of all groups, representative of >30 images collected in 3 independent differentiations except hDMDΔ51-52 n=2. Scale bars, 100 μm. Insets show multinucleation (top) and sarcomeric striations (bottom). Scale bars, 25 μm. d, Percentage of MyHC-β+ cells 7-14 days after myotube induction of myoblasts of each of the indicated groups (cf. Source Data Fig. 4), represented as mean fold change±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=53.74, df=7), n=3 independent experiments in which hiPSCs were differentiated to skeletal muscles except hDMDΔ51-52 n=2. e, Schematic of the experimental design in which cardiomyocytes differentiated from DMDΔ52 hiPSCs were transduced with two AAV6 vectors containing an intein-split Cas9 and gRNAs designed to induce DMD exon 51 excision (as well as either eGFP or mCherry), followed by calcium imaging. f, Exemplary single-cell Ca2+ traces of hiPSC-derived cardiomyocytes of each of the indicated (1 Hz pacing) measured by Fluo-4 fluorescence. Data are representative of 3 independent experiments in which hiPSCs were differentiated to cardiomyocytes. g, Ca2+ transient durations at 90% peak decay (TD90) in hiPSC-derived cardiomyocytes of each of the indicated groups (1 Hz pacing), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=21.64, df=11), n=3 independent experiments in which hiPSCs were differentiated to cardiomyocytes, except hDMDΔ52 n=6. h, Top, single-cell Ca2+ trace showing an arrhythmic event in a hDMDΔ52 cardiomyocyte, representative of 3 independent experiments in which hiPSCs were differentiated to cardiomyocytes; bottom, percentage of cells measured without an arrhythmic event occurring in hiPSC-derived cardiomyocytes of each of the indicated groups (cf. Source Data Fig. 4), indicated as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (F=33.23, df=8), n=3 independent differentiations experiments in which hiPSCs were differentiated to cardiomyocytes.
Co-transduction yield was around 90%, as assessed using viruses carrying the CRISPR/Cas9 gene editing components together with a pair of fluorescence reporters (Extended Data Fig. 8c,d). AAV6-Cas9-mediated exon 51 excision and expression of re-framed DMDΔ51-52 (Extended Data Fig. 8e-h, Source Data Extended Fig. 8g,h) abolished Ca2+ handling defects and arrhythmogenic susceptibility in hDMDΔ52 hiPSC-derived cardiomyocytes (Fig. 4e-h), corroborating the in vivo results. Of note, we detected some dystrophin expression in hDMDΔ52 cells (Fig. 4c), which corresponded to the ubiquitous, short isoform Dp71 (Extended Data Fig. 8g), whose promoter/first exon is located in intron 6231,32 and is thus not affected by the DMD exon 52 deletion. Whole genome sequencing of hDMDΔ52 and hDMDΔ51-52 isogenic iPSCs revealed several nucleotide alterations that were further investigated for their potential to serve as a binding site for either of the two gRNAs. The analysis showed that none of the observed de novo mutations could be caused by the administered gRNAs, since we found at least 7 mismatches between the gRNAs and any of the mutated sites (Extended Data Fig. 10).
Taken together, we found that a large animal model of Duchenne muscular dystrophy, displaying disease hallmarks such as muscle weakness, cardiomyopathy and premature death, can be treated by somatic genome editing of the mutated DMDΔ52 gene via AAV9-Cas9-gRNA. Intramuscular therapy provided a robust expression of the internally truncated, but partially functional DMDΔ51-52 in the injected skeletal muscles, with minimal editing of other muscles (such as contralateral muscles, diaphragm and heart). In the case of systemic application, 2x1014 vp/kg of each of the two AAV9 vectors induced efficient and broad muscle transduction, including diaphragm and heart. To achieve this effect, dendrimer viral coating with G2-PAMAMs was required to overcome the need for even higher virus titers, potentially exceeding a recently reported level of toxicity in non-human primates and piglets33. No intracellular off-target effects were detected at this systemic dose in highly transduced peripheral muscle tissue. In the liver and kidney, gene editing was a rare event, although transduction of the liver was detected readily by PCR against virus DNA, and liver gene editing was seen in mdx mice treated by uncoated AAV8-SaCas21. Although transduction of satellite cells has been demonstrated before in mdx mice expressing a reporter gene under the control of the Pax7 promoter14, we got no evidence for such events in pigs with our viral system.
Using iPSCs, it was possible to model the effects of the human DMDΔ52 mutation and evaluate the efficacy of AAV-Cas9-mediated somatic excision of exon 51 in human muscle cells, which could theoretically reframe about 14% of all DMD mutations34,35. For the first time, we demonstrate that restoration of dystrophin by genome editing can be achieved in differentiated myoblasts as well as cardiomyocytes. Of note, the beneficial functional outcomes of that approach are comparable to direct correction of undifferentiated iPSCs.
In summary, we have demonstrated that AAV9-Cas9-gE51 application is capable of snipping exon 51 in vitro and in vivo and restoring the DMD reading frame. In vivo, intramuscular application yielded high local rates of gene editing, at the expense of distribution to vital muscles such as diaphragm and heart, which was instead achieved by intravenous application of G2-armed AAV9-Cas9-gE51 transducing a wide range of muscles. The shortened DMDΔ51-52 sufficed to improve muscle function and mobility and to prevent malignant arrhythmias, prolonging lifespan in this clinically severe DMD pig model. Further developments pending, the intravenous AAV9-Cas9 genome editing approach may prove clinically useful for the treatment of DMD patients in the future.
Methods
All animal experiments were approved by the Bavarian Animal Care and Use Committee and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996, Approved Institution #A5637-01)
DMDΔ52 animal generation
Animals were produced by breeding from a herd comprising heterozygous female DMD+/- pigs. The breeding herd was established from a single sow with a heterozygous DMD exon 52 deletion (DMDΔ52). Exon 52 of the porcine DMD gene was deleted according to1. In brief, BAC CH242-9G11 was modified to carry a neomycin selection cassette in place of exon 52; the BAC was nucleofected into a female primary kidney cell line (PKCf) and single cell clones were generated36. Genomic DNA was isolated from a batch of each cell clone and the copy number of the DMD exon 52 was compared to the copy numbers of two reference loci within the NANOG and the POU5F1 genes for identifying cell clones with a heterozygous DMD exon 52 deletion1. A total of 258 cell clones was screened and 9 of them had one modified allele as well as an intact one (3.49% efficacy). These cell clones were used for somatic cell nuclear transfer (SCNT) to produce heterozygous DMDΔ52 carrier sows37. One animal was raised and inseminated with wild-type sperm. Male DMDY/- offspring were used as experimental animals and female DMD+/- animals were used to expand the breeding capacity. Genotyping of the offspring was performed by PCR using the primer pair 5´-tgcacaatgctggagaacctca / 5´-gttctggcttcttgattgctgg for detecting the wild-type DMD allele and primer pair 5´-cagctgtgctcgacgttgtc / 5´-gaagaactcgtcaagaaggcgatag for the DMDΔ52 allele.
gRNA design
Specific gRNAs in intron 50 and 51 have been predicted using the CRISPOR web tool38, cloned into the N-Cas9_N-Intein_v2 and C-Intein_C-Cas9_v2 vectors respectively2 and transfected into a porcine or human cell line for testing editing efficacy. The gRNA combination showing highest activity has been chosen. Sequences of chosen gRNAs are listed in Supplementary Table 2.
Virus preparation and G2-PAMAM coating
Recombinant adeno-associated viruses of the serotype 9 and 6 were produced with the triple transfection method as described previously39. Briefly, the packaging cell line HEK 293T was transfected with the vector Cbh-N-Cas9/CRISPR 5-1 or Cbh-C-Cas9/CRISPR 3-1, a plasmid encoding the cap sequences of AAV9 (pigs) and AAV6 (hiPSCs) and rep AAV2 sequences and the helper plasmid delta F6 (Puresyn, Pennsylvania) using PEI Max (Polysciences). After 72 hours, cells were harvested and virus was purified by iodixanol-gradient centrifugation. The virus was further purified by a gravity flow size exclusion purification using Sepharose G100 SF resin (Sigma-Aldrich) in Econopac colums (Biorad). Virus was concentrated in PBS using Amicon Ultra-15 Centrifugal Filter Units (Merck) and stored at 4°C. Viral titer was quantified by ITR-Probe qPCR. For G2-PAMAM coating, 2x1014 virus particle (vgs)/kg were mixed with 18μg of G2 PAMAM-nanoparticles in Opti-MEM (Invitrogen, Germany) by gentle pipetting and allowed to incubate for at 30 min at RT before further use.
AAV9-Cas9-gE51 transduction and follow up
For intramuscular transduction, the right side of the animal was treated by injections of 200μl each (100μl of each virus subsequently) in 15 injection sites (9x thigh and 6x upper arm) at day 14 after birth. Of each Cas9-intein-half (N-Cas9 and C-Cas9), 2.5x1013 vg /kg bodyweight were injected in total in 5 animals. At the end of the experiment, tissue was recovered from injected and contralateral muscles and remote organs and analyzed for dystrophin expression.
For systemic application, the complex of AAV9 and G2 PAMAM-nanoparticles was formed by diluting indicated amounts of AAVs in Opti-MEM (Invitrogen, Germany). Viral particles (vp) were added to the PAMAM diluted solution, immediately mixed by gentle pipetting and allowed to incubate for at 30 min at RT before further use. 4 weeks old piglets were injected 2x1013 vp/kg for each, N-Cas9 and C-Cas9 into the ear vein (low dose) or 2x1014 vp/kg for each strain, totaling e.g. 2x1015 vp for a 5 kg piglet.
Mortality and termination of experiments
In total, 73 affected DMDY/- piglets could be produced in our DMD pig breeding herd, of which 45 newborn pigs died within the first week despite intense nursing and adjuvant feeding and were excluded from further studies. We equally distributed birthweights prior to gene therapy, trying to establish matched pairs (siblings with or without treatment), whenever possible. The birthweights were 1118±155g (DMD control), 1567±213g (DMD i.m.), 1444±305g (DMD i.v. low) and 1222±80g (DMD i.v. high, p=0.294). Of 28 male DMDY/- pigs used in this study, 2 animals (1 i.m., 1 i.v. animal) were observed for a pre-specified period (69 and 77 days, respectively) to investigate protein expression. Other DMD pigs were followed until physical deterioration (tachypnea, exhaustion, stridor), triggering termination of the experiment due to animal protection regulation. Alternately, DMD animals succumbed to sudden cardiac death, which occurred at rest (with video documentation in place for all i.v. treated animals) or during individual and veterinarian-accompanied transportation (2 i.m. treated DMD animals and 1 high-dose treated DMD animal), such that transportation of the last 7 DMD animals was performed after intubation under anesthesia with only 1 animal lost before measurements once intubated.
Finally, we were able to investigate 4 high dose i.v. treated animals, 3 DMD animals with untreated hearts (2 i.m. and 1 untreated heart) and 3 wildtype siblings of DMD pigs by cardiac catheterization (cf. Fig. 3b-d). Electrophysiological mapping was performed in 3 high dose treated DMD animals, 2 untreated DMD hearts and 3 wildtype animals.
Video surveillance
Digital night and day vision infrared cameras, type VTC-E220IRP (Santec, Ahrensburg), provided a 24/7 video surveillance of the pigs. Each box contained one camera giving an overview over the whole compartment. A control room on site was used for monitoring all cameras. Camera signals were digitalized by IndigoVision 9000 encoders (IndigoVision Inc., Edinburgh, U.K.). Via an AT-FS708 switch (Allied Telesis Inc., USA) the encoders were connected to a computer. Videos were evaluated with the IndigoVision control center software.
Functional behavior measurement
Behavioral observation methods were used according to 40. Instantaneous sampling records the state of an individual animal at predetermined time intervals. Animals were observed at every 5th minute for 24 hours. In order to ensure a reliable measurement, the second before and after was involved in the evaluation but only the centisecond of the instant was graded. Blue color indicates a lying or sitting posture, orange color indicates action.
Continuous recording enabled the observation of total activity and resting periods in 24 hours. Two states (upright and lying posture) were recorded to the second. Frequency and duration of standing and lying postures were evaluated.
Cardiac catheterization and high-resolution 3D mapping
Pigs were anesthetized and instrumented as previously described41,42. Briefly, global myocardial function was assessed by pressure-tip catheter placement in the left ventricle (for LV enddiastolic and systolic pressures, dP/dtmax, dP/dtmin) at rest and rapid atrial pacing (150/min), whereas analysis of ejection fraction was performed after LV angiography in anterior-posterior position (yielding slightly smaller control values than a right anterior oblique view).
The Rhythmia mapping system was used for high-resolution 3D-mapping (Boston Scientific, Natick, Massachusetts), as described before43. Bipolar activation maps were created in 3 wildtype hearts, an untreated DMD heart, and 3 high-dose treated hearts (Fig.3e). The I8.5F IntellaMap-Orion catheter (Extended Data Fig. 5a) contains 64 flat microelectrodes (0.8 mm diameter) in a basket configuration with 8 splines. The basket is steerable in 2 directions and can be opened and closed to provide appropriate wall contact for detection of electrophysiological signals. Cardiac beats were automatically selected by the mapping system based on standard beat acceptance criteria: cycle length stability, 12-lead electrocardiogram morphology match, electrode location stability, and respiratory gating. If the signals did not satisfy the above criteria, the information was not included in the map. The LV surface geometry was generated by including all points recorded within 2 mm from the outermost surface of the map (defined by outermost reach of any of the electrodes in 3D space). The voltage for bipolar electrograms was derived measuring from peak to peak. The low-voltage area and endocardial scar are were defined on the bipolar voltage map as <1.3mV and <0.3 mV, respectively.
For quantitative analysis of the electroanatomical maps, maximal, minimal and mean bipolar electrogram voltage was calculated for each LV-map. Quantification of low voltage scar areas - defined as bipolar voltage <1.3mV – was done using the paraView open-source, multi-platform data analysis and visualization application (Kitware, Clifton Park, New York, USA) (Supplementary Fig. 9b,c). In displayed voltage maps, the angulation of the detection plane with respect to the frontal plane is indicated by a pig cartoon (rendered from model provided by www.cadnav.com).
Tibiotarsal joint force measurements
The physiology rig was set up as described by Childers et al.44 using a bridge interface and load cell obtained from Phidgets Inc., Calgary, Canada. Anesthetized pigs were placed on the rig in dorsal recumbent position and hoof was strapped to foot pedal with maintaining a 90° angle for the coxofemoral, knee and tibiotarsal joints. Needle electrodes were placed on either side of the common peroneal nerve to stimulate tibiotarsal flexion. Isometric twitches were triggered with individual 150 V, 100 μsec pulses, tetanic contraction was obtained with 1½ sec train of pulses at 50Hz.
Histochemistry and quantification
Fibrosis was detected by Sirius red staining of paraffin-embedded tissues. Pictures were taken at a 20-fold magnification. Fibrosis quantity was determined from 10 independent images each with Image J Software.
Dystrophin was detected in frozen tissues with antibodies directed against the C-terminus (Novocastra NCL-DYS2, Wetzlar, Germany). An anti-CD14 antibody (Biorad MCA1218F, Munich, Germany) was used for detection of immune cells (Fig.2).
For DGC analysis, all images were recorded from 7 μm sections of muscle tissue frozen in isopentane chilled in liquid nitrogen using identical confocal imaging parameters (Olympus FluoView F1000). Primary antibodies are listed in Supplementary Table 3. To quantify colocalization of dystrophin with either γ-sarcoglycan or β-dystroglycan, monochromatic TIFF 32-bit images were analyzed with the ‘EzColocalization’ ImageJ plugin, according to the protocol described previously45. Briefly, due to the lack of significant non-specific background staining, automatic selection of the thresholds was chosen (Costes’ method) to decrease the potential for user bias. The threshold overlap score (TOS) was calculated to give a value between 0 and 1, reflecting the degree of co-occurrence of signals between dystrophin and γ-sarcoglycan or dystrophin and β-dystroglycan (0=no colocalisation, 1=full colocalisation, without unit).
Hydroxyproline assay
Hydroxyproline in porcine tissue samples was quantified via Colorimetric Hydroxyproline Assay Kit (ab222941, Abcam, Cambridge, UK). 100 mg of each sample was homogenized in 300 μl and subjected to alkaline lysis at 121°C for 2 hours. 10 μl of the lysates was assayed as per manufacturer’s protocol. 6 samples of two independent animals were investigated per group.
Gel electrophoresis, immunoblotting and preparation of bands for mass spectrometry analysis
Muscle tissue samples were homogenized in lysis buffer (125 mm Tris pH 8.8, 40% glycerol, 4% SDS, 0.5 mm PMSF, 100 mm DTT) using an ultrasonic device (46 kJ, Sonoplus GM3200 with BR30 cup booster, Bandelin, Berlin, Germany). Protein concentration was determined using the Pierce 660 nm Protein Assay (Thermo Fisher Scientific, Rockford, IL, USA). SDS gel electrophoresis was performed using a 4–20% Mini-PROTEAN® TGX™ precast gel (Bio-Rad, Hercules, CA, USA). After separation the gel was Coomassie stained using Roti-Blue (Carl Roth, Karlsruhe, Germany). Immunoblotting was performed with an antibody binding the C-terminus of dystrophin (Abcam ab15277, Cambridge, UK).
Mass spectrometry-based identification of dystrophin from gel bands
The Coomassie stained gel slice was excised and de-stained using 50% acetonitrile (ACN) in 50 mM NH4HCO3. Proteins were subjected to in-gel digestion. For reduction the gel piece was incubated in 45 mM DTT/50 mM NH4HCO3 for 30 min at 55°C. Alkylation of sulfhydryl (-SH) groups was done by incubation of the gel slice in 100 mM iodoacetamide/50 mM NH4HCO3 at RT in the dark for 30 min. Digestion was carried out using 70 ng LysC (FUJIFILM Wako Chemicals Europe, Neuss, Germany) for 4 h at 37°C followed by a second digestion step using 70 ng porcine trypsin (Promega, Fitchburg, WI, USA) overnight. Peptides were extracted using 70% ACN. Prior to mass spectrometry analysis the samples were dried using a SpeedVac vacuum concentrator. The tryptic peptides were separated on an Ultimate 3000 nano-LC system (Thermo Fisher Scientific, MA, USA) and identified on an online coupled Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). For separation, a 50 cm column was used (Column: PepMap RSLC C18, 75 μm x 50 cm, 2 μm particles, Thermo Scientific) and a 160 min gradient from 5 % solvent A (0.1 % formic acid in water) to 25 % solvent B (0.1% formic acid in acetonitrile) followed by a 10 min gradient from 25 % to 40 % solvent B. For MS measurement a top 15 data dependent CID method was used. MS data were searched using MASCOT V2.6.1 (Matrix Science, London, UK) against the porcine subset of the NCBI refseq database and filtered for an FDR < 1%. Data were further validated using Scaffold V4 (Proteome Software, Portland, OR).
Selected reaction monitoring (SRM) analysis
Sample preparation for selected reaction monitoring was carried out as for the mass spectrometry-based identification of dystrophin described in the chapters before, with the difference that 50 fmol of the synthetic heavy peptides (JPT, Berlin, Germany) were spiked in prior to digestion. SRM runs were performed on a nanoACQUITY UPLC system (Waters, Milford, MA, USA) coupled to a triple-quadrupole linear ion trap mass spectrometer (QTRAP 5500, AB SCIEX, Framingham, MA, USA). Tryptic peptides were transferred to a trap column (PepMap100 C18, 5 μm, 300 μm i.d. ×5 mm, Thermo Scientific) at a flow rate of 10 μl/min and separated at 280 nl/min on a reversed-phase C18 nano-LC column (ReproSil-Pur 120 C18-AQ, 2.4 μm, 75 μm i.d. ×15 cm, Dr. Maisch, Ammerbuch-Entringen, Germany). The following consecutive linear gradients were used: 1–5% B (0.1% formic acid in acetonitrile) in 1 min, 5–35% B in 45 min and 35–85% B in 5 min. For every peptide, three transitions were measured and chromatograms were evaluated using Analyst V 1.5.1. (AB SCIEX, Framingham, MA, USA).
Holistic proteome analysis of skeletal muscle samples
Protein concentration of the lysates was adjusted to a concentration of 2.3 μg/μl using 8 M Urea/0.4 M NH4HCO3. 250 μg of total protein was reduced using DTE at a final concentration of 5 mM for 30 min at 37°C. Cysteins were alkylated at room temperature for 30 min in the dark with iodoacetamide (final concentration 15 mM). Proteins were digested for 4 h at 37°C using 2.5 μg LysC (FUJIFILM Wako Pure Chemicals, Osaka, Japan). The samples were diluted with water to 1 M urea and digested overnight with 5 μg porcine trypsin (Promega, Madison, WI, USA) at 37°C. 1.5 μg of tryptic peptides were subjected to LC-MS/MS analysis as described above. For protein identification (FDR < 1%) and label free quantification, the acquired spectra were analyzed using the MaxQuant software platform (V1.6.1) in combination with the porcine subset of the NCBI refseq database. Hierarchical clustering, principal component analysis and Student's t-test were calculated with Perseus (V 1.5.3.2) part of the MaxQuant proteomics pipeline46. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE47 partner repository with the dataset identifier PXD014893.
hiPSC reprogramming and culture
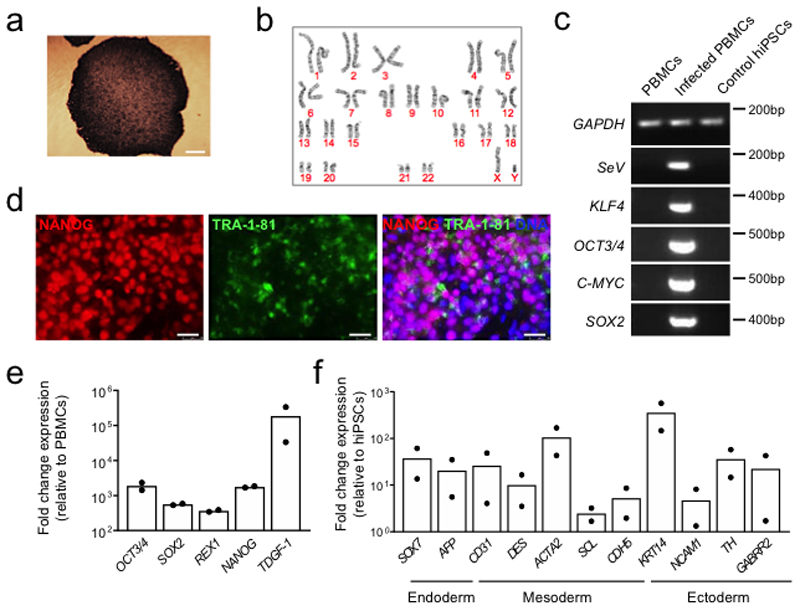
The DMDΔ52 human iPSC line was reprogrammed with the CytoTune-iPS 2.0 Sendai Reprogramming kit (Invitrogen A16517), as previously described48, using the peripheral blood mononuclear cells (PBMCs) of a male Duchenne muscular dystrophy patient carrying a deletion of DMD exon 52 leading to a premature stop codon. The healthy hiPSC line was reprogrammed from the PBMCs of a young, male volunteer following the same protocol. All recruitment and consenting procedures were done under institutional review board-approved protocols of both the Klinikum rechts der Isar, Technical University of Munich, and the Klinikum of the Ludwig-Maximilian University, Munich. Written informed consent was obtained from the affected patient and healthy volunteer.
Pluripotency was assessed after reprogramming via alkaline phosphatase staining (Roche 11681451001), immunofluorescence analysis of the pluripotency markers NANOG and TRA-1-81 (all antibodies listed in Supplementary Table 3) and qPCR analysis of the pluripotency markers OCT4, SOX2, NANOG, REX1 and TDGF-1 (all primers listed in Supplementary Table 4), as previously described22. Germ-layer differentiation potential was tested via spontaneous embryoid body differentiation in DMEM/F12 medium containing 20% FBS, 50 μg/mL L-ascorbic acid (Sigma-Aldrich A5960), 1% L-glutamine, 1% non-essential amino acids and 0.5% Penicillin-Streptomycin for 21 days followed by qPCR analysis of markers of endoderm (SOX7, AFP), mesoderm (CD31, DES, ACTA2, SCL, CDH5) and ectoderm (KRT14, NCAM1, TH, GABRR2) using GAPDH as an endogenous control. Loss of Sendai virus was confirmed after 13 passages via immunofluorescence analysis and RT-PCR of the Sendai vector and viral transgenes OCT4, SOX2, KLF4 and c-MYC using GAPDH as an endogenous control. Karyotyping was performed by the Institute of Human Genetics of the Klinikum rechts der Isar, Technical University of Munich. hiPSCs were maintained in mTeSR1 medium (Stemcell Technologies 85850) on Matrigel-coated plates (Corning 354277).
RNP-mediated CRISPR/Cas9 deletion of DMD exon 51 in DMDΔ52 hiPSCs (DMDΔ51-52 line)
For CRISPR/Cas9-mediated deletion of DMD exon 51 in DMDΔ52 hiPSCs, the Alt-R CRISPR-Cas9 system (IDT) was used according to the manufacturer’s instructions. Briefly, crRNA oligonucleotides targeting the human DMD exon 51 locus (5’ TAATTTGAAGCTGGACCCTA and 5’ GTCTAGGAGAGTAAAGTGAT) were purchased from IDT and duplexed with fluorescently labeled tracrRNA (IDT 1075927). The obtained gRNAs (Supplementary Table 2) were then each used to generate equimolar ribonucleoprotein (RNP) complexes with the S. pyogenes Cas9 protein (IDT 1074181) in Opti-MEM medium (Gibco 31985062). The RNP complexes were then reverse transfected into DMDΔ52 hiPSCs dissociated with TrypLE Express (Gibco 12604013) using Lipofectamine Stem Transfection reagent (Invitrogen STEM00003). A final RNP concentration of 10 nM was applied for 4x105 cells per well of a Matrigel-coated 96 well plate. Transfected cells were dissociated into single cells 24 hours after transfection with a 10-minute Accutase treatment (Gibco A1110501) and 1000 cells were seeded into a Matrigel-coated 10 cm plate in mTeSR1 containing 10 μM Y27632 (Calbiochem 688000). mTeSR1 was replaced every day until colonies were large enough to cut in half for clone screening and passaging. Deletion of exon 51 was verified by PCR and Sanger sequencing by Eurofins Genomics. The generated DMDΔ51-52 line was confirmed to have a normal karyotype by the Institute of Human Genetics of the Klinikum rechts der Isar, Technical University of Munich.
hiPSC muscle differentiation
Skeletal muscle differentiation of hiPSCs was induced using a commercially available kit (Amsbio SKM-KITM). Briefly, hiPSCs were dissociated with Accutase on day 0 and seeded into plates coated with 5 μg/cm2 type I collagen (Cell applications 122-20) at a density of 5000 cells/cm2 in Skeletal Muscle Induction Medium (Amsbio SKM01). Myogenic precursors were obtained within 6-8 days, at which point the cells were dissociated with TrypLE Express (Gibco 12604013) and replated into type I collagen coated plates at a density of 5000 cells/cm2 in Skeletal Myoblast Medium (Amsbio SKM02). The cells reached the myoblast stage within 6-8 days, after which the medium was replaced with Myotube Medium (Amsbio SKM03) to induce the formation of skeletal muscle myotubes. After 5 days, the Myotube Medium was replaced with Skeletal Muscle Cell Differentiation Medium containing 2% horse serum (Promocell C-23061; C-39366). Myotubes were maintained in culture a total of 7 or 14 days from the switch to Myotube Medium.
Differentiation into cardiomyocytes was induced by modulation of Wnt/β-catenin signaling, following a protocol described by Lian and colleagues, with some modifications49. Briefly, hiPSCs were seeded onto 12-well plates coated with 2 μg/cm2 fibronectin (Sigma-Aldrich F1141) at a density of 2x105 cells/well. Upon reaching 90% confluence after 3-4 days, cardiac differentiation was induced on day 0 by changing to RPMI1640 (Gibco 21875091) with B27 minus insulin (Gibco A1895601) (defined as basal cardiac differentiation medium) supplemented with 6 μM CHIR99021 (Axon Medchem 1386). On day 2, medium was replaced with basal cardiac differentiation medium supplemented with 5 μM IWR1 (Tocris 3132). After maintaining the cells in basal cardiac differentiation medium another 14 days, beating areas were mechanically transferred to fibronectin-coated plates and cultured in DMEM/F-12 (Gibco 11320033) with 2% FBS, 1% non-essential amino acids (Gibco 11140050), 1% Penicillin-Streptomycin-Glutamine (Gibco 10378016) and 0.1 mM β-mercaptoethanol.
AAV-mediated CRISPR/Cas9 deletion of DMD exon 51 in patient-derived muscle cells
Skeletal myoblasts and cardiomyocytes derived from DMDΔ52 patient hiPSCs were transduced with a dual AAV system (AAV2/6-Cas9/gE51) carrying the sequences for the split-intein Cas9 protein used in the pig model and two gRNAs targeting the human DMD exon 51 locus (Supplementary Table 2 and Supplementary Fig. 5a-c). Both AAVs were added to the cells at a concentration of 106 virus particles per cell and removed after 20 hours. Transduced myoblasts were maintained in Skeletal Myoblast Medium for 6 days before inducing skeletal muscle myotube formation as described above.
Immunofluorescence analysis
Immunofluorescence staining was performed as previously described50 using the primary antibodies listed in Supplementary Table 3. Sample imaging was performed with an inverted or confocal laser scanning microscope (DMI6000B and TCS SP8, Leica Microsystems, Wetzlar, Germany).
Capillary Western immunoassay
Cells were lysed in RIPA buffer (Sigma-Aldrich R0278) containing proteinase inhibitor (Roche 11836170001) and total protein concentration was determined via Pierce BCA assay (Thermo Fisher 23225). Dystrophin levels were analyzed with the size-based Wes system (ProteinSimple) using an antibody targeting the C-terminus of human dystrophin and an antibody targeting α-actin as a loading control (Supplementary Table 3). Samples were loaded at a protein concentration of 0.05 mg/mL or 0.1 mg/mL for hiPSC-derived skeletal myotubes and cardiomyocytes, respectively.
Ex vivo myocardial cultivation
For ex vivo heart slice cultivation, porcine myocardial tissue was obtained from left mid-ventricular transmural sections and immediately placed in a 30 mM 2,3-butadione-2-monoxime solution (BDM, Sigma-Aldrich B0753) at 4 °C. The sections were embedded in 5% agarose and further processed to 300 μm thick tissue slices by vibratome cutting (VT1200S, Leica Biosystems, Germany). Slices were anchored in biomimetic culture chambers via small plastic triangles attached to the slices with tissue adhesive (Histoacryl, B. Braun 69390) according to the fiber direction and immediately subjected to physiological preload of 1 mN and stimulation at 0.5 Hz (50 mA pulse current, 1 ms pulse duration). Before calcium imaging, the slices were maintained for 24 hours in M199 medium (Sigma-Aldrich M4530) supplemented with 1% Penicillin-Streptomycin, 0.5% insulin/transferrin/selene and 50 μM β-mercaptoethanol on a rocker plate (60 rpm, 15°C tilt angle) placed in an incubator set at 37 °C, 5 % CO2, 20% O2 and 80 % humidity. A continuous readout of contraction force was obtained via the biomimetic chamber21.
Calcium imaging
Myocardial tissue slices were incubated in culture medium containing 3 μM Fluo-4-AM (Thermo Fisher F14201), 0.75% Kolliphor EL (Sigma-Aldrich C5135) and 30 mM 2,3-butadione-2-monoxime (BDM, Sigma-Aldrich B0753) for 60 min at 37°C, then washed and incubated in Tyrode’s solution supplemented with Ca2+ (135 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2, and 10 mM HEPES; pH 7.35) containing 30 mM BDM for another 30 min at 37°C. Spontaneous calcium signals from the tissue slices were subsequently imaged using an upright epifluorescence microscope (Zeiss Axio Examiner) equipped with a 40x objective, a GFP filter set, and a Rolera em-c2 EMCCD camera.
For calcium imaging in hiPSC-derived cardiomyocytes, two months-old cells were dissociated to single cells using a papain-based protocol described previously51, Cells were then seeded onto 3.5 cm glass bottom cell culture microdishes (MatTek Corporation P35G-1.5-14-C) coated with 2 μg/cm2 fibronectin (Sigma-Aldrich F1141) at a density of 5x103 cells/cm2. Ten days after seeding, loading with 2 μM Fluo-4-AM (Thermo Fisher F14201) for 30 min at 37 °C, de-esterification of the dye for 30 min at 37 °C and imaging were all performed in Tyrode’s solution supplemented with Ca2+ (135 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2, and 10 mM HEPES; pH 7.35). The glass bottom microdishes were placed on the stage of an inverted epifluorescence microscope (DMI6000B, Leica Microsystems, Wetzlar, Germany), equipped with GFP filter sets, a HCX PL APO 63X/1.4-0.6 oil immersion objective (Leica Microsystems) and a Zyla V sCMOS camera (Andor Technology, Belfast, UK). Field stimulation electrodes (RC-37FS, Warner Instruments, Hamden, CT, USA) were connected to a stimulus generator (HSE Stimulator P, Hugo Sachs Elektronik, March-Hugstetten, Germany) providing depolarizing pulses (50 V, 5 ms duration) at 1 Hz as indicated. Imaging settings (illumination intensity, camera gain, binning) were adjusted to achieve an optimal signal-to-noise ratio while avoiding pixel saturation. Imaging rates were 14 Hz in the tissue slices and 100 Hz in the iPSC-derived cardiomyocytes. ImageJ (National Institutes of Health, Bethesda, MD) was used to quantify fluorescence over single cells and over background regions. Subsequent analysis was performed in RStudio (RStudio Team (2015). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA) using custom-written scripts. After subtraction of background fluorescence, the time course of Fluo-4 fluorescence was expressed either in arbitrary units or normalized to the initial value (F/F0). After manual selection of the starting points and the peaks of the calcium transients, the transient duration at 90% decay (TD90), the rise time and the monoexponential decay time constant τ were automatically determined by the script.
PCR, RT-PCR and quantitative Real-time PCR
PCR analysis to study genomic editing in DMD pigs was performed on genomic DNA extract from various tissues using the Wizard® Genomic DNA Purification Kit (Promega). PCR was performed using Q5 polymerase (NEB) and the following program: an initial step of 95°C for 2min and then 40 cycles of 95°C for 15s and 62°C for 20s and 72°C for 20s.
RT-PCR was performed on Trizol (Invitrogen, #15596-026) or RNeasy Mini Kit (Quiagen) extracted RNA samples from snap frozen tissue. Reverse transcription was performed using random hexamers and SuperScript®-VILO (Invitrogen, #11904-018) according to the manufacturer's instructions.
Quantitative DMD analysis was performed using genomic DNA from different regions and the ABI PRISM 7900 Sequence Detection System (Applied Biosystems) and TaqMan® reaction mixes for detecting unedited (APKA34W, Applied Biosystems) versus exon 51 deleted DMD (APMFXPU, Applied Biosystems). All samples were measured in triplicates in a 20-μl reaction contained 10μl of TaqMan® Universal PCR Master Mix (Applied Biosystems), 60 ng of HindIII fragmented gDNA template, 300 nmol/L of each primer and 200 nmol/L of the specific FAM-labelled probe. The fluorescent signal intensities were recorded and analyzed during PCR amplification using the Sequence Detection Software (SDS, Applied Biosystems) software. Following, the ratio of unedited versus edited DMD was determined (2-[ΔCT_del- ΔCT_WT]). Sequences of primers and probes can be found in Supplementary Tables 4 and 5.
Data was analyzed using Sigma Plot 12.0 (Systat Software, Inc, Chicago, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, USA).
gRNA design and off-target analysis
Specific gRNAs in intron 50 and 51 have been predicted using the CRISPOR web tool38, cloned into the N-Cas9_N-Intein_v2 and C-Intein_C-Cas9_v2 vector respectively2 and transfected into a porcine cell line for testing editing efficacy. The gRNA combination showing highest activity has been chosen (Supplementary Table 2).
Potential off-targets have been predicted using the CRISPOR and the CHOPCHOP web tool52 and ranked according to their CFD and MIT score. The top five predicted off-targets (see Supplementary Table 6) for both the intron 50 and intron 51 gRNA were amplified by PCR using Q5 polymerase (NEB) with standard conditions on genomic DNA from one control pig, two intramuscular treated animals and one systemically injected pig. A total of 400 ng of genomic DNA has been used per reaction; sequences of primers can be found in Supplementary Table 7. PCR products subsequently were analyzed by deep sequencing.
Sequencing for off-target analysis
Libraries were sequenced in paired end mode with 100 bases read length on an Illumina HiSeq 1500. Data analysis was performed with the Bioconductor package CrispRVariants [1]. For each off target region, the reference sequence shows the gRNAs and PAM sequence marked by a black rectangle and additional 5 nucleotide up- and downstream.
Position referred to the cut site and type of detected INDELs or INDEL combinations found in a dedicated read were listed at the left. Deletions were marked with '-' and distinct colored symbols indicate the position of insertions. The tables show in the first line the number of sequence reads matching the reference sequence and in the following lines the number of INDELS found in each sample.
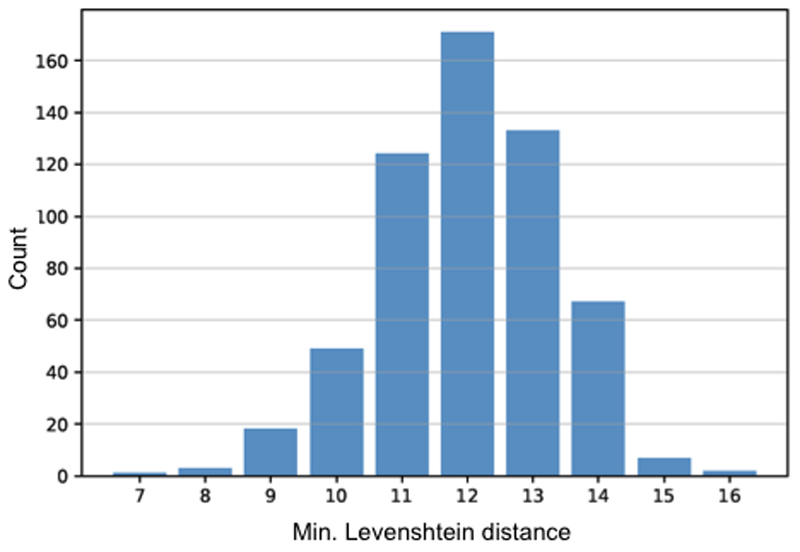
Levenshtein distance of gRNA around genomic SNP/INDELs
Genome-wide sequencing of human DMDΔ52 iPSCs and an isogenic edited DMDΔ51-52 iPSC clone was achieved using an Illumina HiSeq 1500 sequencer. The reads were sequenced in paired-end mode with a length of 100 nt. SNPs and INDELs in each sample were called using the GATK somatic SNV + INDEL pipeline53 and filtered for SNVs/INDELs specific to the edited DMDΔ51-52 iPSC clone. In total, 769 SNVs and 88 INDELs were identified. To clarify if these variants represent off-target effects or occurred randomly during the clonal expansion of the edited DMDΔ51-52 iPSCs, a minimal Levenshtein distance analysis was performed. The two guide RNAs were aligned in a sliding window starting 25 bp upstream and ending 25 bp downstream of each variant position and the alignment with the smallest distance was determined.
Statistical methods
The results are given as mean ± SEM, if not indicated otherwise. Two experimental groups were compared by Student´s t-test. Statistical analysis of results between >2 experimental groups was performed with one-way analysis of variance ANOVA. Whenever a significant effect was obtained with ANOVA, we performed multiple comparison tests between the groups using the Bonferroni procedure (parametric) or Kruskal-Wallis test (non-parametric). For non-parametric testing of two groups, the Mann Whitney U test was performed (Fig. 2b). Kaplan-Meier analysis was used in Fig. 3a. All procedures were performed with an SPSS statistical program (Version 25). Differences between groups were considered significant for p<0.05
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE database with the dataset identifier PXD014893. Unprocessed full scans of agarose gels and Western blots for Figs. 1 and 3, Extended Data Figs. 1, 2, 6, 7, 8 and 9 and Supplementary Figs. 3 and 5 are available online. Source data for Figs. 1-4, Extended Data Figs. 4-8 and Supplementary Figs. 2 and 3 are available online. A “Life Sciences Reporting Summary” with detailed information on experimental design and reagents is accompanying this publication.
Extended Data
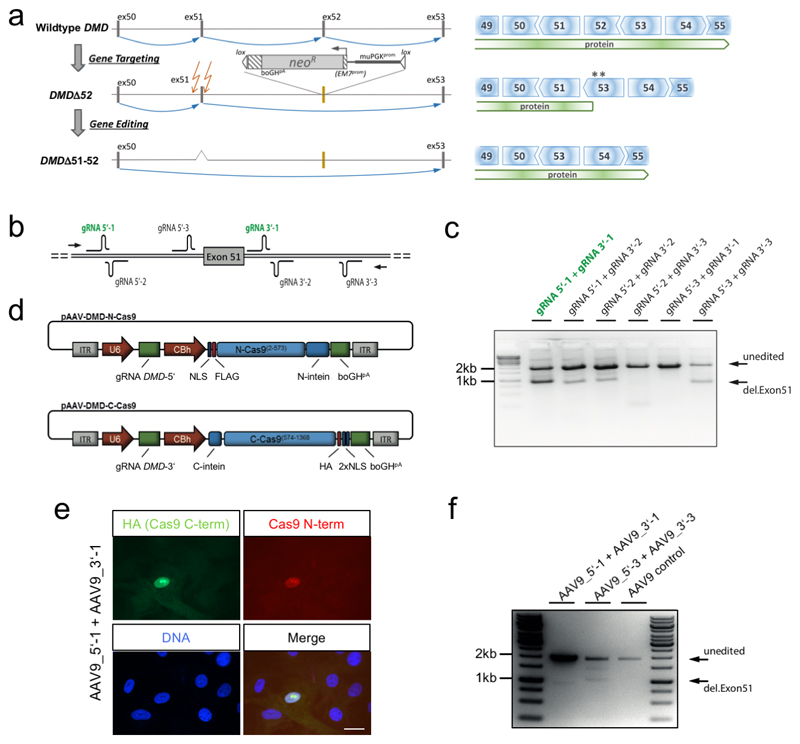
Extended Data Figure 1. DMDΔ52 pig model and in-vitro testing of gene editing strategy.
a, Scheme of the generation and gene editing strategy of the DMDΔ52 pig model (left) and consequences of the genetic alterations at the protein level (right). DMD Exon 52 was replaced with a neomycin selection cassette (neoR), flanked by a murine PGK (muPGKprom) and an EM7 promoter (EM7prom), a bovine growth hormone polyadenylation signal (boGHpA) and loxP sites (lox). Blue arrows indicate splice events. Asterisks indicate stop codons occurring in exon 53 due to reading frame incompatibility. Intended cutting sites for therapeutic gene editing are indicated by orange arrows. b, Scheme of gRNAs cloned into pAAV-N-Cas9 and pAAV-C-Cas9 vectors and tested in different combinations by transfection of porcine cells and PCR amplification (arrows: primer locations) of genomic DNA. c, Gel showing results of this test. d, Schematic representation of the intein-split-Cas9 system, consisting of two AAV constructs each harbouring one DMD-specific gRNA (gRNA DMD-5’ and gRNA DMD-3’, respectively) under the control of a U6 promoter and either the N-terminal or the C-terminal half of the Cas9 nuclease (N-Cas9(2-573) or C-Cas9(574-1368), respectively), fused to N- or C-terminal split-intein domains (N-intein and C-intein, respectively), under the control of a CBh promoter. NLS, nuclear localization sequence. FLAG, flag-tag. HA, HA-tag. e, Immunocytochemistry for both the N- and the C-terminal Cas9 peptides after AAV co-transduction of primary porcine myoblasts using a pair of AAV constructs (AAV9_5’_1 and AAV9_3’_1) harbouring gRNAs 5’-1 and 3’-1, respectively (representative for n=2). Scale bar, 15 μm. f, PCR analysis of genomic editing as in panel c in primary porcine kidney cells co-transduced with the above-mentioned pair of AAV constructs, with another pair (AAV9_5’_3 and AAV9_3’_3, harbouring gRNAs 5’-3 and 3’-3) or with control AAV, as indicated (representative for n=2 transductions).
Extended Data Figure 2. System analyses of DMD exon 51 deletion in pig skeletal muscles and heart after injection of AAV9-Cas9-gE51.
a,b, Top, genomic PCR analysis of DMD gene editing in samples from indicated skeletal muscles of DMD pigs treated by intramuscular (i.m) (a) or high dose intravenous (b) injection with G2-AAV9-Cas9-gE51, representative of 2 (a) and 3 (b) animals. Percentages of edited (ΔEx51+52) relative to total (ΔEx52 + ΔEx51+52) amplicon are shown. Quantifications by RT-PCR of the ratio of edited to total DMD mRNA expression (Δ51DMD / total DMD, middle) and mass spectrometry-based quantification of dystrophin protein (Dys) expression (bottom) are shown. C.l. = contralateral. c, Top, genomic PCR assessing cardiac DMD exon 51 editing in DMD pigs (five specimens of left ventricle (LV), left atrium (LA), right atrium (RA), right ventricle (RV), representative of 3 animals, are shown) treated with high dose intravenous injection of G2-AAV9-Cas9-gE51. Expected band sizes corresponding to unedited and edited DNA are indicated. Bottom, quantification of the ratio of edited to total DMD transcript (Δ51DMD/total DMD) by quantitative RT-PCR. d, Immunofluorescence for dystrophin (Dys) with wheat germ agglutinin (WGA) membrane staining in heart tissue of wildtype and untreated or high dose i.v. treated DMD pigs, representative of 8 images collected from 2 animals per group. Scale bars, 20 μm. e, left, immunoblotting for Cas9 in M. quadriceps muscle from 4 AAV9-Cas9-gE51 treated pigs as indicated, using an antibody against the Cas9 N-terminus. The expected band sizes corresponding to N-Cas9-N-intein (N-Cas9) and full-length Cas9 protein (Full Cas9) are indicated. Right, immunofluorescence staining of M. quadriceps muscle cells with antibodies detecting N-Cas9 (green) and the HA-tag (HA-C-Cas9) (yellow) with WGA membrane staining and DAPI nuclear labelling (DNA) (representative for n=2 pigs). Arrows indicate nuclei with overlapping fluorescence. Scale bar, 10 μm.
Extended Data Figure 3. Analysis of off-target effects in porcine tissue samples by targeted deep sequencing.
For each off-target region, the reference sequence shows the gRNAs and PAM sequence marked by a black rectangle and additional 5 nucleotides up- and downstream. The tables show in the first line the number of sequence reads matching the reference sequence and in the following lines the number of INDELS found in each sample. Description of samples: Qc = quadriceps muscle; LV = left ventricle; Liv = liver; WT = wildtype, non-injected; i.m. = intramuscularly-injected; i.v. high = high dose intravenously-injected.
Extended Data Figure 4. Colocalisation of dystrophin-associated glycoprotein complex (DGC) and restored dystrophin in DMD pig skeletal muscle after i.m. and i.v. injection of AAV9-Cas9-gE51.
a,b, Immunofluorescence co-staining for dystrophin and γ-sarcoglycan (a) or dystrophin and β-dystroglycan (b) in biceps femoris of wildtype, untreated DMD (DMD untr.) and intramuscularly (DMD i.m.) or intravenously (DMD i.v.) AAV9-Cas9-gE51-treated DMD pigs. Top and bottom rows for i.m. treated DMD in (a) are of areas close and distant to the injection site, respectively. Scale bars, 200 μm (left 20x merge column), 20 μm (right merge column), and 10 μm (detail column in b). c, Quantification of colocalization of dystrophin with either γ-sarcoglycan or β-dystroglycan. A threshold overlap score (TOS) was calculated giving a dimensionless number reflecting the degree of co-occurrence of signals between dystrophin and γ-sarcoglycan (TOS Dys-γSG, n=4 images except DMD i.v. n=6, collected from 2 pigs) or dystrophin and β-dystroglycan (TOS Dys-βDG, n=3 images from 2 pigs), with values ranging from 0 (no colocalisation) to 1 (perfect colocalisation). Data (Source Data Extended Data Fig. 4) are mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (TOS Dys-γSG F=34.92, df=14; TOS Dys-βDG F=11.33, df=8).
Extended Data Figure 5. In-vivo electro-mapping and ex-vivo single-cell Ca2+ analyses of DMD hearts.
a, Schematic drawing of the I8.5F IntellaMap-Orion catheter used for high-resolution 3D-mapping, containing 64 flat microelectrodes (0.8 mm diameter) in a basket configuration with 8 splines that is steerable in 2 directions and can be opened and closed to provide appropriate wall contact for detection of electrophysiological signals. b, Mean voltage measured by in-vivo electro-mapping of the heart of wildtype (WT, n=3 animals), untreated DMD (n=2) and high dose intravenously (i.v.) G2-AAV9-Cas9-gE51 treated DMD (n=3) pigs (Source Data Extended Data Fig. 5), indicated as mean±SEM with p values from a one-way ANOVA with Tukey’s multiple comparison test (F=11.59, df=5). c, Size of endocardial low voltage area, expressed as percentage of the whole region, in indicated regions of the heart of WT (n=3 animals), untreated DMD (n=2) and high dose i.v. treated DMD (n=3) pigs (Source Data Extended Data Fig. 5), indicated as mean±SEM with p values from a two-way ANOVA with Tukey’s multiple comparison test (F=38.31, df=15). d, Schematic diagram of the experimental procedure for ex-vivo single-cell Ca2+ measurements, achieved by processing left-ventricular transmural sections to 1.0 x 0.5 cm myocardial tissue slices of 300 μm thickness, which were then submitted to physiological preload and continuous electrical field stimulation in biomimetic culture chambers. The right panel shows a pseudocolor image of Fluo-4 fluorescence recorded from a slice loaded with this calcium sensor and a region of interest (ROI) over which the average fluorescence signal was calculated to investigate intracellular calcium dynamics.
Extended Data Figure 6. Generation of patient-specific DMD iPSC isogenic lines.
a, Left, schematic representation of the DMD exon 52 deletion in the patient-specific hDMDΔ52 hiPSCs and position of the primers (DMD exon 52 fwd and DMD exon 52 rev) used for PCR verification of the mutation. Right, a gel showing the 370 bp amplicon specific for the exon 52 deletion. Bottom, results from Sanger sequencing of the hDMDΔ52 hiPSCs. b, Bright field image of alkaline phosphatase staining in hDMDΔ52 hiPSC colonies at passage 6. Scale bar, 100 μm. c, Normal karyotype in hDMDΔ52 hiPSCs at passage 23. d, RT-PCR analysis of the Sendai vector (SeV) and transgenes OCT4, SOX2, KLF4 and c-MYC in untransduced peripheral blood mononuclear cells (PBMCs, negative control), Sendai-transduced PBMCs (positive control) and hDMDΔ52 hiPSCs at passage 13, using GAPDH as an endogenous control. e, Immunofluorescence analysis of the pluripotency markers NANOG and TRA-1-81 in hDMDΔ52 hiPSCs at passage 24. Scale bar, 50 μm. f, RT-qPCR analysis of the pluripotency markers OCT4, SOX2, NANOG, REX1 and TDGF-1 in hDMDΔ52 hiPSCs. The relative mean fold change expression normalized to GAPDH is indicated, n=2 (passages 13 and 20). g, RT-qPCR analysis of markers of endoderm (SOX7, AFP), mesoderm (CD31, DES, ACTA2, SCL, CDH5) and ectoderm (KRT14, NCAM1, TH, GABRR2) after 21 days of spontaneous embryoid body differentiation of hDMDΔ52 hiPSCs. The relative mean fold change expression normalized to GAPDH is indicated, n=2 independent differentiations. h, Left, schematic diagram of the deletion of DMD exon 51 in hDMDΔ52 hiPSCs and primers used for PCR verification of the deletion (right). i, Normal karyotype after CRISPR/Cas9 editing confirmed in hDMDΔ51-52 hiPSCs at passage 14. Uncropped gels for (a), (d), and (h) and statistics for (f) and (g) are shown in Source Data Extended Data Fig. 6.
Extended Data Figure 7. Generation of control iPSCs from a healthy, young male donor.
a, Bright field image of alkaline phosphatase staining performed on control hiPSC colonies at passage 12. Scale bar, 100 μm. b, Normal male karyotype confirmed in control hiPSCs at passage 21. c, RT-PCR analysis of the Sendai vector (SeV) and transgenes OCT4, SOX2, KLF4 and c-MYC in untransduced peripheral blood mononuclear cells (PBMCs, negative control), Sendai-transduced PBMCs (positive control) and control hiPSCs at passage 24, using GAPDH as an endogenous control. d, Immunofluorescence analysis of the pluripotency markers NANOG and TRA-1-81 in hiPSCs at passage 21. Scale bar, 50 μm. e, RT-qPCR analysis of the pluripotency markers OCT4, SOX2, NANOG, REX1 and TDGF-1 in hiPSCs. The mean fold change expression relative to parental patient PBMCs and normalized to GAPDH is indicated, n=2 (passages 15 and 21). f, RT-qPCR analysis of markers of endoderm (SOX7, AFP), mesoderm (CD31, DES, ACTA2, SCL, CDH5) and ectoderm (KRT14, NCAM1, TH, GABRR2) in control hiPSCs after 21 days of spontaneous embryoid body differentiation. The mean fold change expression relative to hiPSCs and normalized to GAPDH is indicated, n=2 independent differentiations. Uncropped gels for (c) and statistics for (e) and (f) are shown in Source Data Extended Data Fig. 7.
Extended Data Figure 8. Infection of hDMDΔ52 hiPSC-derived skeletal myoblasts and cardiomyocytes with AAV6-Cas9-gE51 restores expression of a re-framed dystrophin.
a, Bright field images of skeletal myoblasts from control, hDMDΔ52 or hDMDΔ51-52 hiPSCs, representative of >10 images (3 independent differentiations). Scale bars, 100 μm. b, RT-qPCR analysis of MYOD1, MYOG and DES in control (n=5 independent differentiations except DES n=4), untreated hDMDΔ52 (n=4 except MYOD1 n=3), hDMDΔ52 6 days after AAV6-Cas9/gE51 transduction (hDMDΔ52+AAV, n=3) or hDMDΔ51-52 (n=2) myoblasts. Relative fold change expression normalized to GAPDH is shown as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (MYOD1 F=12.86, df=10; MYOG F=7.159, df=9; DES F=103, df=9). c, Images of hDMDΔ52 skeletal myoblasts (top) or cardiomyocytes (bottom) 10 days after transduction with AAV6-Cas9/gE51 vectors encoding eGFP or mCherry (AAV6-N-Cas9/gRNA5’-eGFP and AAV6-C-Cas9/gRNA3’-mCherry), representative of >20 images (2 independent differentiations). Scale bars, 100 μm. d, Percentages of double-positive, single-positive and double-negative skeletal myoblasts (top) or cardiomyocytes (bottom) 4 and 10 days after transduction. e, Genomic PCR analysis of DMD exon 51 excision after AAV6-Cas9/gE51 transduction of hiPSC-derived skeletal myoblasts or cardiomyocytes (3 independent differentiations). f, Percentage of exon 51 excision based on relative PCR band intensity (edited versus total), indicated as mean±SEM. g, Dystrophin detection by capillary-based immunoassay after myotube induction of control, untreated or AAV6-Cas9/gE51-transduced hDMDΔ52 and hDMDΔ51-52 skeletal myoblasts (top) and in control, untreated or AAV6-Cas9/gE51-transduced hDMDΔ52 and hDMDΔ51-52 cardiomyocytes (bottom) from 3 independent differentiations. Bands represent the main (Dp427) and a shorter dystrophin isoform (Dp71). β-actin, loading control. h, Dystrophin (Dp427) levels normalized to β-actin expressed as percentage of mean level in control cells are depicted for skeletal muscle cells (top) and cardiomyocytes (bottom) as mean±SEM (p values from one-way ANOVA with Bonferroni’s multiple comparison test; Skeletal cells F=63.46, df=8, Cardiomyocytes F=21.59, df=8).
Extended Data Figure 9. AAV6-Cas9/scrambled-gRNA transduction of hDMDΔ52-iPSC-derived myoblast fails to restore dystrophin expression and capability of the cells to differentiate into myotubes.
a, Immunofluorescence staining for myosin heavy chain β (MyHC-β), α-actinin and dystrophin 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts, hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or hDMDΔ52 myoblasts transduced with AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr) (representative for n=3 independent differentiations). Scale bars, 100 μm. b, Genomic PCR analysis of DMD exon 51 excision 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts, hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or hDMDΔ52 myoblasts transduced with AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr), representative of 2 independent differentiations. The expected band sizes corresponding to edited and unedited genomic DNA are indicated. Uncropped gel is shown in Source Data Extended Data Fig. 9. c, Capillary-based immunoassay of dystrophin 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts and hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr), using β-actin as a loading control, representative of 2 independent differentiations. The antibody detected both the main dystrophin isoform (Dp427) and a shorter isoform (Dp71). Uncropped blots are shown in Source Data Extended Data Fig. 9.
Extended Data Figure 10. Levenshtein analysis of distance of guide RNAs around variants identified by whole-genome sequencing of isogenic hDMDΔ51-52 iPSCs compared to the parental hDMDΔ52 iPSCs.
Histogram of all minimal Levenshtein distances obtained by aligning the two DMD-E51 guide RNAs to all variants identified by whole-genome sequencing in isogenic hDMDΔ51-52 iPSCs compared to the parental hDMDΔ52 iPSC line, applying a sliding window starting 25 bp upstream and ending 25 bp downstream of each variant. For any candidate region around a variant at least 7 operations (base exchanges, deletions, insertions) were required to match one of the gRNAs, indicating that the variants were not off-target effects of the CRISPR-Cas treatment.
Supplementary Material
Acknowledgements
We thank the DMD patient and the healthy volunteer who provided us with blood for iPSC reprogramming. We would like to acknowledge Christina Scherb for technical assistance in molecular cloning, and Gabi Lederer (Cytogenetic Department, TUM) for karyotyping, as well as Manfred Ogris (MMCT Laboratory for Macromolecular Cancer Therapeutics, Department of Pharma-ceutical Chemistry, University of Vienna, Austria) for advice with G2 coating and Andreas Blutke (Helmholtz Zentrum München, Research Unit Analytical Pathology) for help with pathological workup. Annette Frank established the capillary Western blot (Department of Apoptosis Research, Helmholtz Centre Munich), Miwako Kösters (Gene Center, LMU) provided technical assistance in mass spectrometry. The Muscular Dystrophy Association (USA) supports monoclonal antibody development in the laboratory of Glenn E. Morris, whom we would like to thank for providing the MANDAG antibody against β-dystroglycan.
This work was supported by grants from: the Else-Kröner-Fresenius foundation (2015/180 and 2018/T20 to C.K., E.W. and W.W.); the European Research Council, ERC 788381 (to A.M.), ERC 681524 (to I.J.); the German Research Foundation (DFG), Transregio Research Units 152 (to A.M., K-L.L.), 127 (to E.W., A.S., A.B., N.K. and C.K.), 267 (to A.M., A.S. and C.K.); and the German Center for Cardiovascular Research, Munich Heart Alliance (to A.M., K.-L.L., and C.K.); the generous support of A. Klesius and coworkers (Boston Scientific) for the Rhythmia system is gratefully acknowledged.
Footnotes
Author contributions
C.K., E.W. and W.W. designed the pig study. M.K., V.Z., N.K., B.K. and E.W. generated the DMD pigs and raised the cohort. L.F., A.B., K.K., R.H. and C.K. conducted the pig transduction, structural and functional analyses. P.H., C. J. and E.M. performed high-resolution electrophysiological mapping and analysed data. T.B., K.K., R.H., I.J., K.V., V.J., F.A.R., S.R. and S.K. performed and analyzed expression assays and histology of pig tissues. ,. F.G., W.W. generated intein-split Cas9 and gRNAs, H.B, A.G., S.K., G.S. and F.G. sequenced and analyzed DNA samples for genome editing and off-target studies. T.B., T.Z. and A.W. generated and raised AAV9-vectors. S.L., T.Z., and M.O. introduced G2-optimization in vitro and in vivo. A.S. generated and analysed dTomato pigs for AAV-Cre transduction. A.M. and K.-L.L. conceived and supervised the iPSC study and provided financial support. A.B.M., D.S., T.H. and S.S. performed all experiments with iPSCs and their muscle derivatives. B.C. generated, characterized, and differentiated the iPSC lines. A.B.M. generated isogenic hDMDΔ51-52 hiPSCs. D.S., R.D., and T.D. analysed data. T.F. and F.F. performed mass spectrometry. C.M.S., A.D. and D.S. performed the ex-vivo experiments on heart slices and analysed data. S.K. and M.W. provided human patient blood for reprogramming and conceptual advice. C.K. and A.M. wrote the paper. All authors commented on and edited the manuscript.
References
- 1.Klymiuk N, et al. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Human Molecular Genetics. 2013;22:4368–4382. doi: 10.1093/hmg/ddt287. [DOI] [PubMed] [Google Scholar]
- 2.Truong DJ, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle & nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 4.White S, et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. American journal of human genetics. 2002;71:365–374. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser H. Duchenne muscular dystrophy: pathogenetic aspects and genetic prevention. Human genetics. 1984;66:17–40. doi: 10.1007/BF00275183. [DOI] [PubMed] [Google Scholar]
- 6.Sharp PS, Bye-a-Jee H, Wells DJ. Physiological characterization of muscle strength with variable levels of dystrophin restoration in mdx mice following local antisense therapy. Mol Ther. 2011;19:165–171. doi: 10.1038/mt.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Deutekom JC, et al. Local Dystrophin Restoration with Antisense Oligonucleotide PRO051. New England Journal of Medicine. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 8.Goemans NM, et al. Systemic Administration of PRO051 in Duchenne's Muscular Dystrophy. New England Journal of Medicine. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 9.Verhaart IE, et al. The Dynamics of Compound, Transcript, and Protein Effects After Treatment With 2OMePS Antisense Oligonucleotides in mdx Mice. Molecular therapy Nucleic acids. 2014;3:e148. doi: 10.1038/mtna.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengtsson NE, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8 doi: 10.1038/ncomms14454. 14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.EL Refaey M, et al. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circulation Research. 2017 doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amoasii L, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018 doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punnoose AR, et al. Cardiac Disease Burden and Risk of Mortality in Hospitalized Muscular Dystrophy Patients. Pediatric Cardiology. 2016;37:1290–1296. doi: 10.1007/s00246-016-1432-5. [DOI] [PubMed] [Google Scholar]
- 17.Feingold B, et al. Management of Cardiac Involvement Associated With Neuromuscular Diseases: A Scientific Statement From the American Heart Association. Circulation. 2017;136:e200–e231. doi: 10.1161/CIR.0000000000000526. [DOI] [PubMed] [Google Scholar]
- 18.Vetter A, et al. Adenoviral vectors coated with PAMAM dendrimer conjugates allow CAR independent virus uptake and targeting to the EGF receptor. Mol Pharm. 2013;10:606–618. doi: 10.1021/mp300366f. [DOI] [PubMed] [Google Scholar]
- 19.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV Serotypes 1-9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 20.Pleger ST, et al. Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci Transl Med. 2011;3:92ra64. doi: 10.1126/scitranslmed.3002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson CE, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nature medicine. 2019 doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner DL, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nature Medicine. 2018 doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 23.Chew WL, et al. A multifunctional AAV–CRISPR–Cas9 and its host response. Nature Methods. 2016;13:868. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyva-Leyva M, Sandoval A, Felix R, González-Ramírez R. Biochemical and Functional Interplay Between Ion Channels and the Components of the Dystrophin-Associated Glycoprotein Complex. The Journal of Membrane Biology. 2018;251:535–550. doi: 10.1007/s00232-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 25.Thajudeen A, et al. Correlation of scar in cardiac MRI and high-resolution contact mapping of left ventricle in a chronic infarct model. Pacing and clinical electrophysiology : PACE. 2015;38:663–674. doi: 10.1111/pace.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer CMH, Fein E, Reiser E, Lu K, Seidel T, Schinner C, Schwarzmayr T, Schramm R, Tomasi R, Husse B, Cao-Ehlker X, et al. Long-term functional and structural preservation of precision-cut human myocardium under continuous electromechanical stimulation in vitro. Nat Commun. 2019 doi: 10.1038/s41467-018-08003-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 28.Jiwlawat N, Lynch E, Jeffrey J, Van Dyke JM, Suzuki M. Current Progress and Challenges for Skeletal Muscle Differentiation from Human Pluripotent Stem Cells Using Transgene-Free Approaches. Stem Cells Int. 2018;2018 doi: 10.1155/2018/6241681. 6241681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi In Y, et al. Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts Derived using a Human iPSC-Based Model. Cell Reports. 2016;15:2301–2312. doi: 10.1016/j.celrep.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Young Courtney S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell. 2016;18:8. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar S, et al. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem J. 1990;272:557–560. doi: 10.1042/bj2720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi T, et al. Detection of Dystrophin Dp71 in Human Skeletal Muscle Using an Automated Capillary Western Assay System. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19061546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinderer C, et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an AAV vector expressing human SMN. Human gene therapy. 2018 doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter MC, Reilich P. Recent developments in Duchenne muscular dystrophy: facts and numbers. 2017;8:681–685. doi: 10.1002/jcsm.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aartsma-Rus A, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Human mutation. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 36.Richter A, et al. Potential of primary kidney cells for somatic cell nuclear transfer mediated transgenesis in pig. BMC biotechnology. 2012;12:84. doi: 10.1186/1472-6750-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurome M, Kessler B, Wuensch A, Nagashima H, Wolf E. Nuclear transfer and transgenesis in the pig. Methods in molecular biology (Clifton, N.J.) 2015;1222:37–59. doi: 10.1007/978-1-4939-1594-1_4. [DOI] [PubMed] [Google Scholar]
- 38.Concordet JP, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kupatt C, et al. Cotransfection of Vascular Endothelial Growth Factor-A and Platelet-Derived Growth Factor-B Via Recombinant Adeno-Associated Virus Resolves Chronic Ischemic Malperfusion: Role of Vessel Maturation. Journal of the American College of Cardiology. 2010;56:414–422. doi: 10.1016/j.jacc.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 40.Martin P, Bateson P. Measuring Behaviour: An introductory guide. Cambridge University Press; 2007. [Google Scholar]
- 41.Kupatt C, et al. Endothelial nitric oxide synthase overexpression provides a functionally relevant angiogenic switch in hibernating pig myocardium. Journal of the American College of Cardiology. 2007;49:1575–1584. doi: 10.1016/j.jacc.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 42.Hinkel R, et al. Diabetes Mellitus-Induced Microvascular Destabilization in the Myocardium. J Am Coll Cardiol. 2017;69:131–143. doi: 10.1016/j.jacc.2016.10.058. [DOI] [PubMed] [Google Scholar]
- 43.Pathik B, et al. New Insights Into an Old Arrhythmia: High-Resolution Mapping Demonstrates Conduction and Substrate Variability in Right Atrial Macro–Re-Entrant Tachycardia. JACC: Clinical Electrophysiology. 2017;3:971–986. doi: 10.1016/j.jacep.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Childers MK, Grange RW, Kornegay JN. In vivo canine muscle function assay. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauffer W, Sheng H, Lim HN. EzColocalization: An ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Scientific Reports. 2018;8 doi: 10.1038/s41598-018-33592-8. 15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–d450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gramlich M, et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO molecular medicine. 2015 doi: 10.15252/emmm.201505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorn T, et al. Interplay of cell-cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 2018;37 doi: 10.15252/embj.201798133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer B, et al. A complete workflow for the differentiation and the dissociation of hiPSC-derived cardiospheres. Stem cell research. 2018;32:65–72. doi: 10.1016/j.scr.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 52.Labun K, et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2018;47:W171–w174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11.10.11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE database with the dataset identifier PXD014893. Unprocessed full scans of agarose gels and Western blots for Figs. 1 and 3, Extended Data Figs. 1, 2, 6, 7, 8 and 9 and Supplementary Figs. 3 and 5 are available online. Source data for Figs. 1-4, Extended Data Figs. 4-8 and Supplementary Figs. 2 and 3 are available online. A “Life Sciences Reporting Summary” with detailed information on experimental design and reagents is accompanying this publication.