Abstract
Mucolipin-1 is a membrane protein encoded by the gene MCOLN1, mutations in which result in the lysosomal storage disorder Mucolipidosis type IV (MLIV). Efficient lysosomal targeting of mucolipin-1 requires di-leucine motifs in both the N-terminal and C-terminal cytosolic tails. We have shown that aberrant lactosylceramide trafficking in MLIV cells may be rescued by wild type mucolipin-1 expression but not by mucolipin-1 mistargeted to the plasma membrane, nor by lysosome-localised mucolipin-1 mutated in its predicted ion pore-selectivity region. Our data demonstrate that the correct localisation of mucolipin-1 and the integrity of its ion pore are essential for its physiological function in the late endocytic pathway.
Keywords: endocytosis, lysosomes, organelle fusion, transient receptor potential
Introduction
Mucolipidosis type IV (MLIV) is an autosomal recessive neurodegenerative lysosomal storage disorder caused by mutations in the gene MCOLN1 which encodes the 580 amino acid, multi-spanning, membrane protein mucolipin-1 (1–4). Mucolipin-1 is a member of the large family of TRP (transient receptor potential) cation channels. Studies of mucolipin-1 overexpressed in Xenopus oocytes, mammalian cells, or incorporated into liposomes, have suggested that mucolipin-1 is a non-specific cation channel modulated by changes in Ca2+ concentration and pH (5–9). Experiments on mammalian cells have also implied that its H+ -permeability may be important in regulating lysosomal pH (9).
The lysosomal storage defect in MLIV is consistent with abnormalities of membrane traffic and organelle dynamics in the late endocytic pathway (10–12). Fibroblasts from MLIV patients are characterised by swollen lysosomes containing multi-concentric lamellae and the accumulation of both lipids and water soluble substances. Lactosylceramide traffic is severely impaired in MLIV cells and in other sphingolipid storage diseases (13). Thus, endocytosed LacCer (N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl) sphingosyl 1-β-D-lactoside; BODIPY FL C5-lactosylceramide) and its fluorescent metabolites accumulate in endocytic organelles/lysosomes in MLIV cells rather than concentrating in the Golgi complex as in normal cells (9,10). Lysosomal accumulation of LacCer is not due to a block in its hydrolysis to fluorescent ceramide (10).
The swollen lysosomes in MLIV cells may be a consequence of defective lysosome re-formation after fusion events. Endocytic delivery to lysosomes involves kissing between late endosomes and lysosomes as well as direct fusion to produce endosome-lysosome hybrid organelles from which lysosomes are re-formed (14). The mechanisms of fusion and lysosome re-formation are partially understood (15,16), and it has been suggested that the CUP-5 protein, which is the C. elegans orthologue of mucolipin-1, is required for lysosome biogenesis from hybrid organelles (17,18).
Although early experiments localising tagged mucolipin-1 protein in transiently transfected cells suggested a predominantly plasma membrane localisation (3,11) it has more recently been shown that green fluorescent protein-tagged mucolipin-1 localises to lysosomes (19,20). Given the potential significance of mucolipin-1 in traffic through the late endocytic pathway we decided to investigate the importance of its intracellular localisation and the role of the predicted ion pore in its function.
Results
Localisation and targeting of mucolipin-1
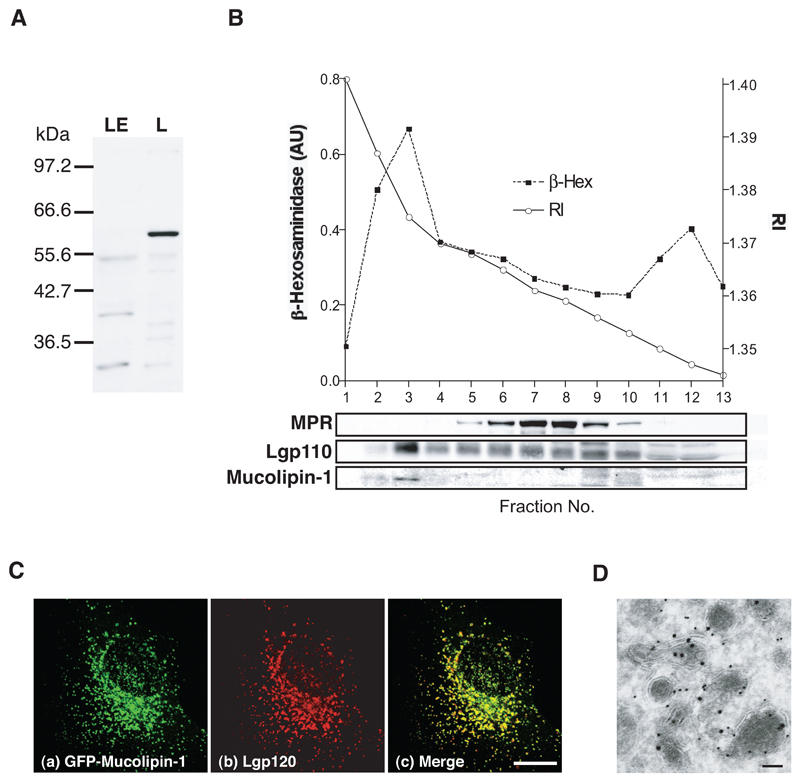
Recent reports have shown that mucolipin-1 is proteolytically cleaved after amino acid 200 in the lumenal loop between the first two predicted trans-membrane domains (21,22). However, the cleaved protein fragments remain associated with each other. To address the localisation of endogenous full length mucolipin-1, we generated an antiserum to a bacterially expressed mucolipin-1 fusion protein that was useful for immunoblotting (but not immunofluorescence). We immunoblotted rat liver subcellular fractions enriched in lysosomes or late endosomes. Endogenous full length mucolipin-1, migrating as a 60kDa band (identical to a band observed after in vitro translation of human mucolipin-1 cRNA, data not shown), was present only in the lysosome-enriched fraction (Fig 1A). Furthermore, we found that when NRK cell homogenates were fractionated by ultracentrifugation on a Ficoll gradient, then full length mucolipin-1 showed a peak in the same fraction as the lysosomal enzyme ß-hexosaminidase and the lysosomal membrane protein lgp110, but not the cation-independent mannose 6-phosphate receptor (MPR), which in these cells is mostly in late endosomes (Fig 1B).
Figure 1. Mucolipin-1 is a lysosomal membrane protein.
(A) Immunoblot of rat mucolipin-1 in rat liver fractions enriched in late endosomes (LE) and lysosomes (L). Proteins (10 μg total per track) were separated by electrophoresis on a 10%SDS polyacrylamide gel using a discontinuous buffer and, after immunoblotting were visualised by ECL. (B) NRK cells were fractionated and the homogenate separated on a 1-22% Ficoll gradient with a 45% Nycodenz cushion. 3 ml fractions were collected from the bottom of the gradient. The refractive index (RI) of the gradient fractions is shown. Each fraction was assayed for β-hexosaminidase activity and immunoblotted for MPR, lgp110 and mucolipin-1. (C) Confocal images (shown as maximum intensity Z projections) using indirect immunofluorescence of (a)GFP-mucolipin-1, (b)lgp120 and (c)extent of co-localisation in NRK cells stably expressing GFP-mucolipin-1. Bar, 10 μm. (D) Immunoelectron micrograph showing GFP-mucolipin-1 (15 nm gold) and lgp110 (10 nm gold) in NRK cells stably expressing GFP-mucolipin-1. Bar, 100 nm.
When expressed in NRK cells, mucolipin-1, tagged with enhanced green fluorescent protein (GFP) at its N-terminus, showed no significant co-localisation with MPR and almost completely co-localised with a lysosomal membrane protein, lgp120 (Fig 1C), in agreement with the lysosomal localisation reported by others (19,20). The lysosomal localisation in NRK cells was confirmed by immunoelectron microscopy where GFP-mucolipin-1 was observed in electron dense organelles labelled with antibodies to lgp110 (Fig 1D). In contrast to one report of alteration of late endocytic organelle distribution as a result of over-expression of mucolipin-1 (19), we saw no gross morphological changes to lysosomes after moderate expression of GFP-mucolipin-1. This agrees with the data of Miedel et al. (22).
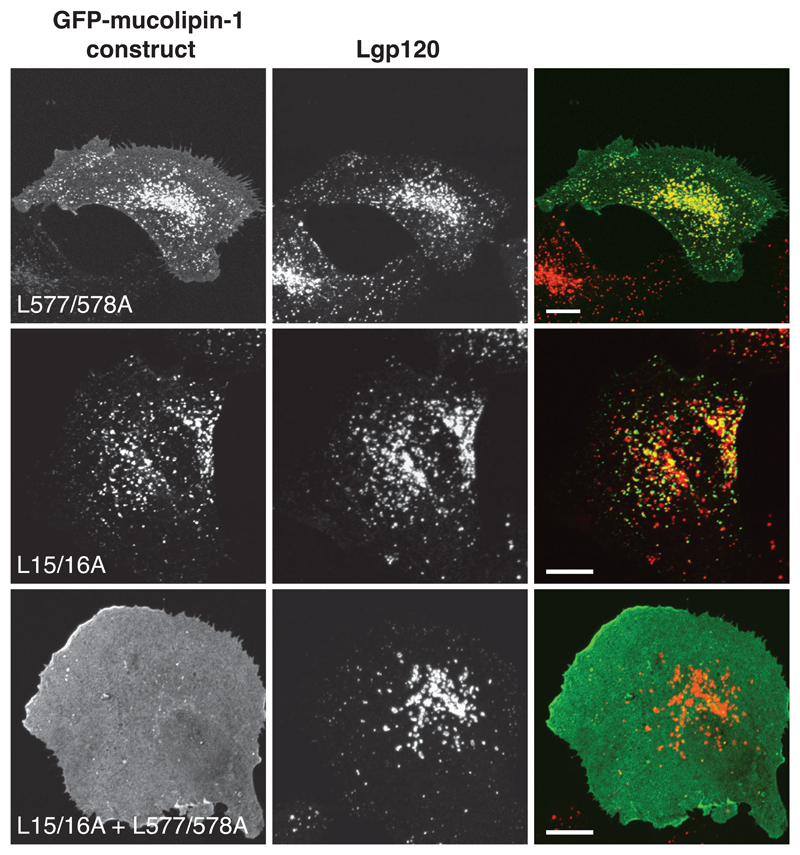
The predicted aminoterminal and carboxyterminal cytoplasmic tails of mucolipin-1 contain di-leucine motifs (leucines 15&16 and leucines 577&578 respectively) preceded by acid patches, that conform to adaptor binding motifs (23). We transiently expressed GFP-mucolipin-1 in which the aminoterminal di-leucine motif was replaced with alanines (L577/578A). This mutated protein partially localised to lysosomes but targeting was not efficient since a significant proportion of the GFP-tagged protein was observed at the cell surface (Fig 2). In contrast, when GFP-mucolipin-1 in which the aminoterminal di-leucine motif was replaced with alanines (L15/16A) and transiently expressed it still localised to lysosomes (Fig 2) suggesting that this di-leucine motif is not a strong lysosomal targeting motif. Nevertheless, when the double di-leucine mutant GFP-mucolipin-1 (L15/16A + L577/578A) was expressed it was found to be exclusively at the plasma membrane (Fig 2), implying that in the context of the full length protein the two di-leucine motifs function together to achieve efficient lysosomal localisation of wild type mucolipin-1. Our data are consistent with the recent report of Vergarajauregui and Puertollano (20). In contrast, a role for the carboxyterminal di-leucine motif in lysosomal targeting was not detected by Miedel et al. (22) who examined the localisation of a mutant mucolipin-1 in which the four carboxyterminal amino acids were deleted (ΔL577LVN). In separate experiments we confirmed that the carboxyterminal LL577/588 motif was not sufficient on its own for lysosomal targeting by studying the localisation of chimeras consisting of the ecto- (lumenal) and transmembrane-domains of the plasma membrane protein CD8α with either the wild type mucolipin-1 tail (amino acids 522-580) or a mutant tail in which the di-leucine motif was replaced with alanines (L577/578A) (Supplementary Information Fig S1). The LL577/588 motif was responsible for intracellular localisation of the chimera but did not target the chimera efficiently to lysosomes.
Figure 2. Two di-leucine signals are required for efficient targeting of mucolipin-1 to lysosomes.
cDNAs encoding human mucolipin-1, single di-leucine mutants L577/578A and L15/16A, and a double di-leucine mutant L15/16A + L577/578A, were sub-cloned into pEGFP-C3 and were transiently expressed for 24h in NRK cells. Confocal images show the extent of co-localisation between the various GFP-mucolipin-1 constructs and lgp120 as indicated by yellow in the panels in the right hand column. Bars, 10 μm.
Lactosylceramide trafficking in MLIV cells is rescued by correctly localised mucolipin-1
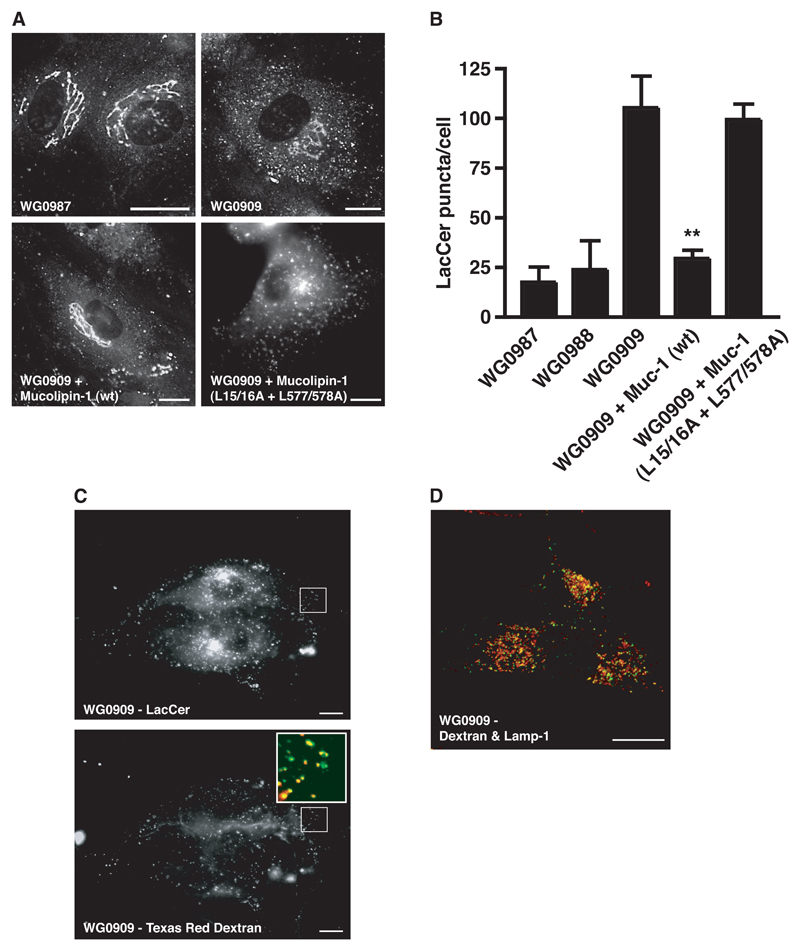
We wanted to test whether expression of mucolipin-1 would rescue trafficking defects in MLIV cells (previously characterised by transmission electron microscopy and measuring lysosomal pH; see Supplementary Information) and whether such rescue depended on correct localisation of the mucolipin-1. We observed a clear difference in LacCer trafficking between the MLIV (WG0909) cells and the heterozygous parental cells (WG0987 and WG0988), consistent with previous reports (10,24). After endocytosis of LacCer, the parental WG0987 and WG0988 cells showed primarily a tubular peri-nuclear pattern of fluorescence, typical of the Golgi compartment (Fig 3A, equivalent image of WG0988 cells not shown). In contrast, in the WG0909 fibroblasts the LacCer accumulated mostly in numerous puncta with only a small amount of fluorescent lipid accumulation in the Golgi (Fig 3A). When the MLIV fibroblasts were transfected with HcRed-tagged wild type mucolipin-1 the defect of Lac Cer trafficking was rescued with fluorescent lipid showing a Golgi distribution and very few puncta (Fig 3A). The rescue of LacCer trafficking in MLIV cells was dependent on mucolipin-1 being targeted to lysosomes since it did not occur in MLIV cells transfected with HcRed-mucolipin-1 mutated in both di-leucine motifs which is targeted to the plasma membrane (L15/16A + L577/578A) (Fig 3A). Quantitation of these experiments was achieved by counting the LacCer puncta (Fig 3B). Although LacCer trafficking was rescued by expression of wild type mucolipin-1 in MLIV cells the morphology of the swollen lysosomes was not altered during the time course of these experiments (data not shown).
Figure 3. Aberrant LacCer trafficking in MLIV cells is rescued by expression of lysosome-localised mucolipin-1.
(A) Fluorescence images of LacCer in cells heterozygous for a mutation in the mucolipin-1 gene (parental cells, WG0987), in untransfected MLIV cells (WG0909) and MLIV cells (WG0909) transiently transfected for 24h with either HcRed-tagged mucolipin-1 (wt) or a double di-leucine mutant (L15/16A + L577/578A). Endocytosed LacCer was observed after 3 h uptake in untransfected or transfected cells, the latter identified by the presence of HcRed. (B) Quantitation of LacCer puncta (excluding Golgi fluorescence) in the parents' cells heterozygous for mutations in the mucolipin-1 gene (WG0987 & WG0988), the MLIV patient's cells (WG0909) and MLIV (WG0909) cells transfected with HcRed-tagged wildtype (wt) or double di-leucine mutant (L15/16A + L577/578A) mucolipin-1. Quantitation is from analysis of ≥10 cells for each cell type from ≥ 3 separate experiments. **, P< 0.01 (WG0909 vs. WG0909 + mucolipin-1 (wt)). (C) MLIV, WG0909 cells were pulsed for 1 h with BODIPY-LacCer and chased for 2 h. Both the pulse and chase were in the presence of 1 mg/ml Texas Red dextran. The enlarged region shows co-localisation (yellow) between the LacCer (green) and the dextran (red). (D) WG0909 cells were pulsed with Oregon green 488–dextran (green) for 4 h then chased for 20 h. Cells were fixed and stained for lamp-1 (red). Co-localisation is shown by yellow. Bar = 10 μm.
To confirm that the puncta in which LacCer accumulated in MLIV cells were endocytic organelles we pulse-chased the cells with LacCer (1h pulse, 2h chase) in the continuous presence of Texas Red dextran and found that the LacCer-positive puncta co-localised with endocytosed dextran (Fig 3C). The entire endocytic pathway was accessible to endocytosed dextran because when the MLIV cells were incubated with dextran for 4h followed by a 20h chase, good co-localisation was observed with the lysosomal membrane protein Lamp-1 which marks late endocytic compartments and lysosomes (Fig 3D). Under the conditions of our experiments the majority of the LacCer was not lysosomal (data not shown).
Cholesterol distribution and the kinetics of lactosylceramide traffic in MLIV cells
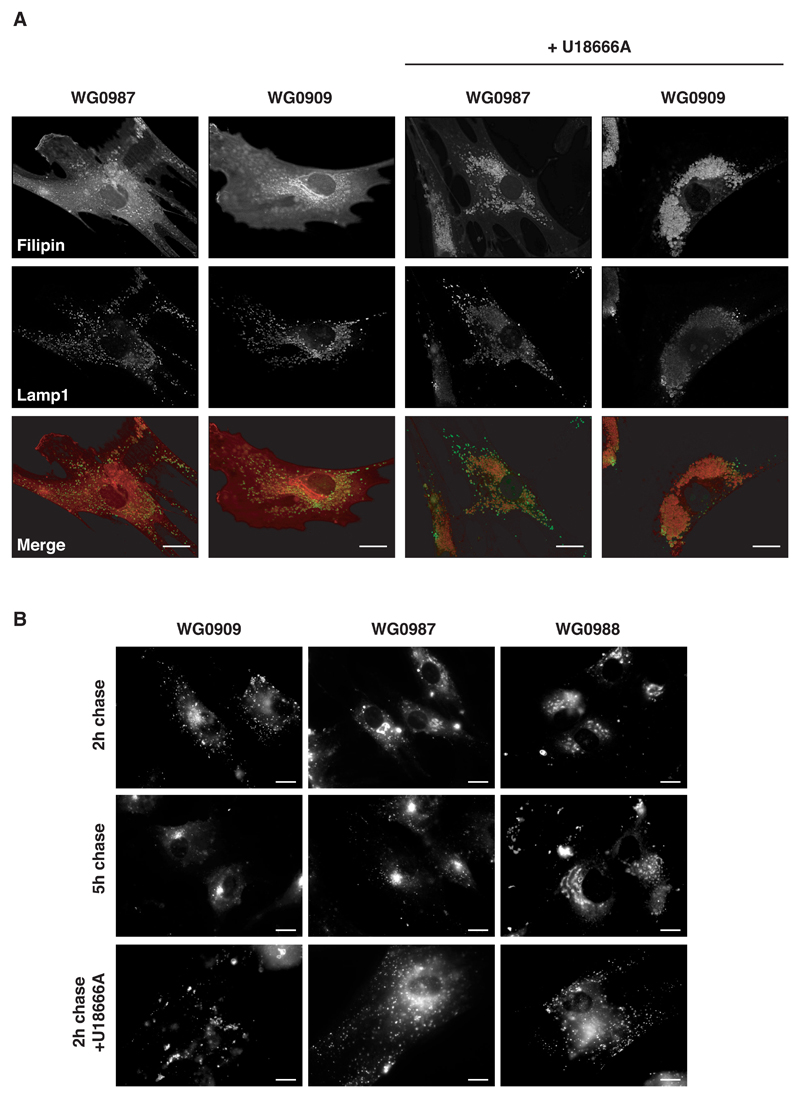
In addition to LacCer uptake we also investigated cholesterol distribution in MLIV cells by staining with filipin, which forms a fluorescent complex with unesterified cholesterol. We found that the distribution of unesterified cholesterol in MLIV cells (WG0909) did not differ markedly from the parental cells and was primarily at the plasma membrane (Fig 4A). We observed no significant visible accumulation of unesterified cholesterol in the lysosomes of MLIV cells (Fig 4A), although Soyombo et al. (9) have reported a three-fold increase in cellular accumulation of filipin in MLIV cells. However, when the parental and MLIV cells were pre-treated with the amphipathic tertiary amine U18666A, unesterified cholesterol accumulated in the lysosomes of all cells examined (Fig 4A). U18666A interferes with the removal of unesterified cholesterol from lysosomes and causes accumulation of cholesterol in lysosomes, thus mimicking Niemann-Pick type C lipid storage disease, a genetic condition characterised by the deficiency of the integral membrane protein NPC-1 (25).
Figure 4. Cholesterol distribution and LacCer traffic kinetics.
(A) MLIV, WG0909 and parental, WG0987 cells treated for 24 h with or without U18666A were stained for unesterified cholesterol (filipin) and lamp-1. Merged pictures show lamp-1 (green) and filipin (red). Bars = 10 μm. (B) Parental and MLIV fibroblasts were pulsed for 1 h with LacCer and then chased for either 2 or 5 h (top and middle panels respectively). Cells treated for 24 h with U18666A were also pulsed for 1 h with LacCer and then chased for 2 h (bottom panels). Bars = 10 μm.
Since the steady state distribution of cholesterol was not grossly perturbed in the MLIV cells we speculated that the accumulation of LacCer in mucolipidosis IV, under our experimental conditions, was the result of slower traffic through the late endocytic pathway and not a complete block of traffic per se. Indeed, when the LacCer was chased for 5 h as opposed to 2 h as in previous experiments, the fluorescent lipid in the MLIV cells (WG0909) gave a staining pattern consistent with labelling of the Golgi as seen with the parental cells (WG0987 & WG0988; Fig 4B). U18666A inhibited LacCer traffic (1h pulse, 2 h chase) to the Golgi in all cells (Fig 4B). Whilst the mode of action of U18666A is unclear, it has been reported that it may disrupt membranes by affecting lipid order (26), which may explain the latter result.
The mucolipin-1 ion pore and rescue of lactosylceramide trafficking
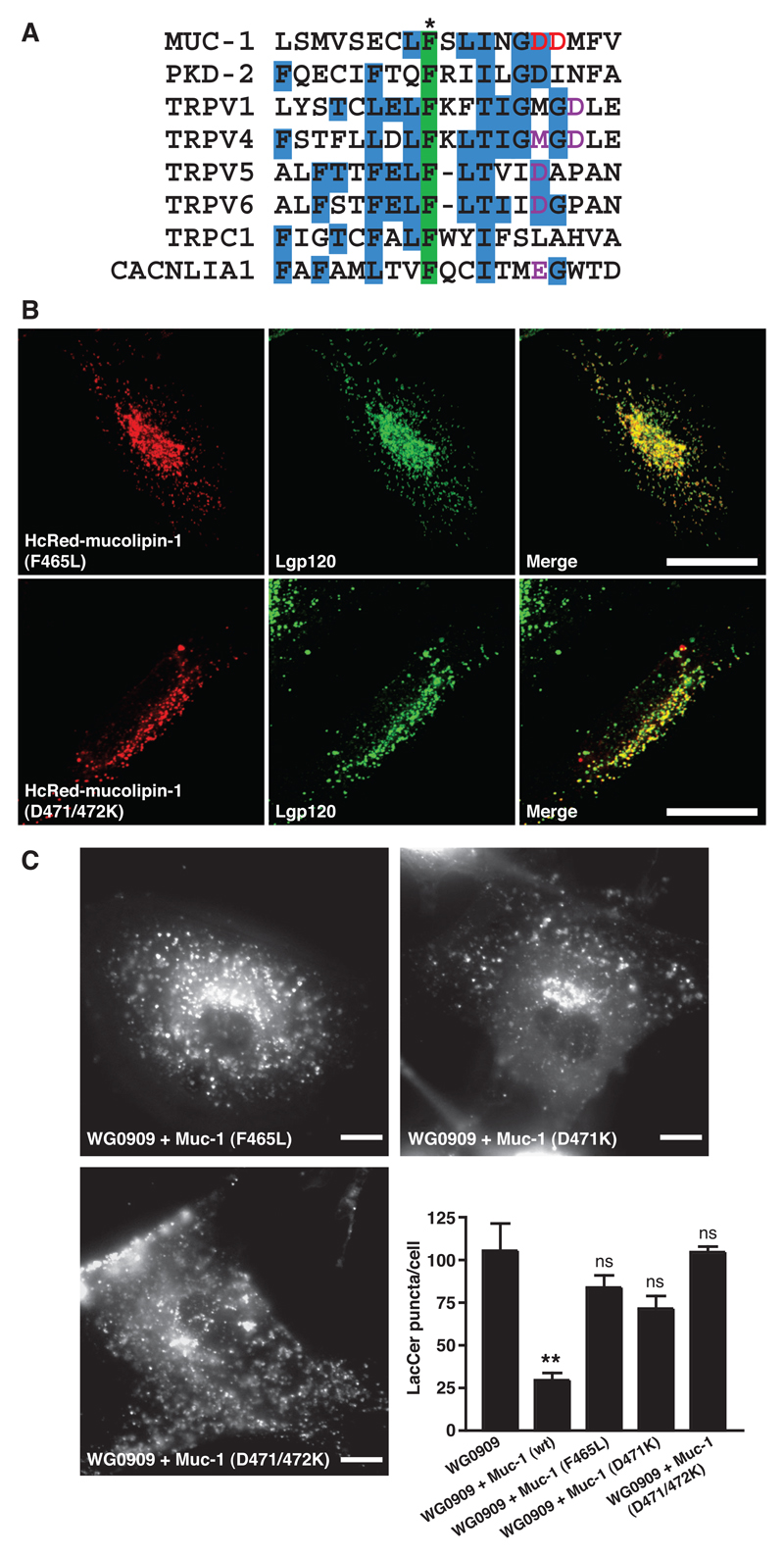
Having established that correct targeting of mucolipin-1 to lysosomes is important for recovery of LacCer trafficking in MLIV cells, we wanted to assess the effects of mutations in the predicted ‘ion pore region’ on the ability to recover LacCer trafficking. Mucolipin-1 contains six putative transmembrane domains with the predicted ion pore between the fifth and sixth of these (12). We aligned part of the pore region of mucolipin-1 with other members of the TRP super-family and a voltage-gated Ca2+ channel α-subunit (CACNLIA1), where mutations in the putative pore region have been shown to affect ion channel characteristics (Fig 5A). Based on this protein alignment we mutated one or both of two aspartate residues to lysine (D471K and D471/472K), to interfere with pore selectivity. In addition to the resultant mucolipin-1 mutants, we also investigated the human MLIV mutation F465L, since this is in the putative pore region and the phenylalanine is highly conserved in the sequences we aligned (Fig 5A). When transiently expressed in NRK cells as HcRed fusion proteins all of these mutants extensively co-localised with lgp120 (Fig 5B, data not shown for D471K ). Despite their ability to be targeted to lysosomes, none of these mutants were able to rescue significantly LacCer trafficking in the MLIV WG0909 cells (Fig 5C cf Fig 3).
Figure 5. Transfection of MLIV cells with mucolipin-1 constructs containing mutations in the pore region fail to rescue LacCer trafficking.
(A) Alignment of part of the pore region of human mucolipin-1 (Muc-1) with members of the TRP super-family and a voltage-gated Ca2+ channel, CACNLIA1 (for references see Supplementary Information). Three or more identical residues are shaded blue. A conserved phenylalanine (shaded green) includes residue F465 of mucolipin-1 (*) which is mutated in at least one case of MLIV (2). Residues which have been previously shown to affect pore channel activity are coloured purple. Residues which we predicted to affect pore channel activity and which we subsequently mutated (aspartates 471 and 472) are coloured red. Accession numbers for sequences (in order) Q9GZU1, Q13563, Q8NER1, Q9HBC0, Q9NQA5, Q9H1D0, P48995, Q13936. (B) NRK cells were transiently transfected for 24h with cDNA encoding different HcRed-tagged mutated mucolipin-1 constructs, F465L, single aspartate mutant D471K or double aspartate mutant D471/472K. Confocal images show the extent of co-localisation between the HcRed-tagged mucolipin mutants and lgp120 as indicated by yellow in the panels in the right hand column. Bars, 10 μm. (C) Fluorescence images of LacCer and quantitation of LacCer puncta in MLIV WG0909 cells transfected with the HcRed-tagged mutant mucolipin-1 constructs F465L, D471K and D471/472K (data from WG0909 and WG0909+mucolipin-1 (wt) cells from Fig 3B is included for comparison). Quantitation is from analysis of ≥10 cells for each cell type from ≥ 3 separate experiments. ns (not significant, P>0.05; data compared to WG0909). **, P< 0.01 (WG0909 vs. WG0909 + mucolipin-1 (wt)).
Discussion
Our data show that mucolipin-1 is a lysosomal membrane protein with two dileucine targeting motifs that are required for efficient lysosomal targeting. We also provide the first evidence that correct targeting of mucolipin-1 to lysosomes and the integrity of its predicted ion pore are essential to its physiological function. Our localisation and targeting data are in agreement with Vergarajauregui and Puertollano (20), who further showed that the carboxyterminal di-leucine motif functions as an AP-2-dependent internalisation motif. They also suggested that the relative lack of accumulation of an L577/578A mutant at the cell surface implies that a direct intracellular route of traffic is the major route of delivery of newly synthesised mucolipin-1 from the Golgi complex to lysosomes. Miedel et al. (22) have also recently shown that mucolipin-1 is mostly delivered to lysosomes by a direct route that does not require AP-3 but is dependent on AP-1.
We have demonstrated that the rate of LacCer traffic through the late endocytic pathway is slower in an MLIV patient’s cells than in parental cells. We rescued this defect by expression of wild type mucolipin-1. However, we only saw rescue if the mucolipin-1 protein was correctly targeted to lysosomes. Mutation of the two di-leucine motifs to generate a mutant mucolipin-1 that was localised to the cell surface resulted in a mutant that could not rescue the MLIV cell defect. The reduced rate of LacCer traffic through the endocytic pathway in MLIV cells was consistent with there being a perturbation in the membrane fusion/fission events of late endocytic organelles. This has been suggested as the underlying defect in cup-5 mutants of C. elegans (17,18), which show a reduction in the rate of degradation of cell surface receptors (27). Under steady-state conditions we did not observe any significant lysosomal accumulation of unesterified cholesterol in the lysosomes of MLIV fibroblasts which implies that its removal, presumably with the aid of NPC1, was proceeding despite other abnormalities of late endocytic traffic. The description of the defect in LacCer trafficking as being kinetic rather than an absolute block may explain why under some of our experimental conditions (LacCer 1h pulse, 2h chase) the majority of LacCer in MLIV cells was in endocytic organelles other than lysosomes whereas others have reported greater lysosomal localisation (9,10,24).
Correctly localising mucolipin-1 to lysosomes is essential for normal LacCer traffic and simply having mucolipin-1 protein present in the cell is not sufficient for normal cell function. Thus, the mucolipin-1 mutant V446L, which causes retention of mucolipin-1 in the endoplasmic reticulum (12,19;our unpublished data) results in MLIV disease despite being predicted to have a functional ion pore. Retention of the V446L mutant in the endoplasmic reticulum is likely to be because it is incorrectly folded, which is also a probable explanation of the retention in the endoplasmic reticulum of many other mutant mucolipin-1s encoded by MLIV genes with mutations, e.g. the MLIV mutant T232P (19). The rate of lactosylceramide traffic through the late endocytic pathway is dependent on an intact mucolipin-1 ion pore since mutations altering charge within the putative pore or a nearby disease mutation (F465L) results in proteins that cannot rescue the trafficking defect despite them being correctly localised to lysosomes. It should be noted that the lack of rescue by predicted ion pore mutants, correctly targeted to lysosomes, argues strongly that it is the ion pore, but only when localised to lysosomes, that is essential for mucolipin-1 function. Despite a recent proposal that MLIV is primarily due to a metabolic effect caused by inactivation of lipases (9) possibly affecting phospholipid synthesis, our data are more consistent with it being a disease of membrane traffic as previously suggested (10–12). It should also be noted that it has previously been shown that phosphatidylcholine metabolism in MLIV cells is normal (28).
The preferred cation passing through the mucolipin-1 channel and the mechanism(s) by which the mucolipin-1 ion channel may be regulated remain unclear. Although others have determined some characteristics of the channel by electrophysiology experiments after expression from cRNA in Xenopus oocytes (5), we were unable to achieve reproducible data showing channels different to endogenous oocyte channels by patch clamping such cells. This situation did not improve when we increased the concentration of oocyte cell surface mucolipin by expressing the double di-leucine mutant GFP-mucolipin-1 (L15/16A + L577/578A) (Supplementary Information Fig S2 A-G). We were also unable to obtain electrophysiological data from HEK-293 cells expressing GFP-mucolipin-1 (L15/16A + L577/578A) that were different from cells expressing GFP alone (Supplementary Fig S2 H-J). The difficulty of obtaining electrophysiological data on the mucolipin-1 channel in transfected cells may be due to several factors. Firstly, it could be due to proteolytic cleavage inactivating the channel. Recent work has shown that mucolipin-1 is cleaved in the lumenal loop between transmembrane domains 1 and 2 (21,22). When the protease cathepsin B was added to mammalian cells overexpressing mucolipin-1, ion channel activity was reduced (21). This suggests that mucolipin-1 cleavage inactivates the ion channel. However, there are conflicting reports on whether cathepsin B is responsible for mucolipin-1 cleavage. Kiselyov et al. inhibited mucolipin-1 cleavage with CA-074-Me, an inhibitor of cathepsins B and L (21), whereas Miedel et al. observed no block of mucolipin-1 cleavage with the same inhibitor (22). Miedel et al. have further suggested that cleavage of mucolipin-1 occurs in a prelysosomal compartment, possibly the trans-Golgi network (22). Whether proteolytic cleavage plays any role in physiological regulation of the cation channel is not known.
A second possible reason for the difficulty in obtaining electrophysiological data from transfected cells is that there are missing subunits of the channel. In this context it should be noted firstly that there is some evidence that mucolipin-1 oligomerises and/or forms complexes with other proteins (19,22) and secondly, that other TRP channels are heteromeric in order to form cation permeable pores (29). A third reason is that unknown proteins or other molecules may be required to gate the channel.
The preferred cation for the mucolipin-1 channel remains to be firmly established although most reports in the literature favour Ca2+ or H+ (5–9). It might therefore be assumed that the concentration of Ca2+ and/or H+ ions in the lysosomes of MLIV cells would be altered. We have seen no reports of a direct measurement of the Ca2+ concentration in the lysosomes of MLIV cells and our own attempts at measurement, using the endocytosed fluorescent probe Fura-2-dextran (30), failed because of interfering autofluorescence (31) and because the concentration of calcium being measured was below the Kd of the probe at acidic pH. Measurements of lysosomal H+ concentration in MLIV fibroblasts have produced confusing data with conflicting reports that it is either elevated by ~1pH unit (32), normal (33 and this study, see Supplementary Information), or reduced (9). Published data supporting the hypothesis that mucolipin-1 is a H+-selective channel are not completely compelling since for high H+-selectivity one would expect that for one unit change in pH the reversal potential would alter by 40-50 mV (34), however mucolipin-1 only shows a change in the reversal potential of 15mV/pH unit (9).
If mucolipin-1 is a Ca2+ channel, it should be noted that it is unlikely to be the only transporter/channel playing a role in regulating the lumenal concentration of Ca2+ in lysosomes. For instance, Galione and colleagues (35) have described a lysosomal NAADP regulated Ca2+ channel. However this is unlikely to be mucolipin-1 because membranes from NRK cells over-expressing mucolipin-1 showed no increased binding of radiolabelled NAADP compared to membranes from untransfected cells (data not shown) and addition of NAADP did not cause an increase in current in electrophysiology experiments on oocytes expressing mucolipin-1 (Supplementary Fig S2 G). A recent proteomics study of lysosome membranes has identified the presence of a ligand gated P2X4 cation channel (36), which could also play a role in regulating the lumenal concentration of Ca2+.
Methods
Reagents
N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)sphingosyl 1-β-D-lactoside (BODIPY FL C5-lactosylceramide; LacCer), nigericin, fluorescent dextran-conjugates and fluorescent-conjugated antibodies were purchased from Invitrogen (Paisley, UK). Fatty acid- free bovine serum albumin (BSA) was purchased from Roche Diagnostics (Lewes, UK). Filipin was from Sigma-Aldrich (Poole, UK) and U18666A was from Biomol (Exeter, UK)
cDNA constructs
cDNA encoding human mucolipin-1 (IMAGE clone 4179987) was from the HGMP Resource Centre, Hinxton, UK. All mutations were generated using a QuikChange site-directed in vitro mutagenesis kit according to the manufacturer’s protocol (Stratagene, Amsterdam, Netherlands). GFP- and HcRed-mucolipin-1 constructs were made by inserting cDNA encoding mucolipin-1 into the XhoI and KpnI restriction enzyme sites of pEGFP-C3 and pHcRed-C1 (BD Biosciences, Oxford, UK) respectively. GFP-mucolipin-1 constructs were prepared by inserting the GFP-mucolipin-1 cDNA into the NdeI sites of the plasmid ΔpMEP4 (37). The cDNA encoding the rat mucolipin-1 (accession number XP_213684) fragment (amino acids 99-282) used for antibody production was cloned from a rat liver 5'-STRETCH PLUS λgt11 cDNA library (RL 5001b; BD Biosciences, Oxford, UK) using primers 5’-ATTGCCTTCCGACATCTCTTCCTG-3’ and 5’-GTGTTTGCACTACTGGATGTG-3’.
Antibodies
Monoclonal anti-rat lgp120 (GM10), polyclonal anti-rat mannose 6-phosphate receptor (MPR) and polyclonal anti-rat lgp110 (580) have been previously described (38). Rabbit anti-rat mucolipin-1 was generated by immunisation of Murex Lop rabbits (Murex Biotech Limited, Dartford, Kent) with a fragment of rat mucolipin-1 (amino acids 99-282) fused to glutathione-S-transferase (GST). The rabbit polyclonal antibody to GFP was from Abcam Ltd (Cambridge, UK).
Cell culture and transfection
Normal Rat Kidney (NRK) cells were cultured in DMEM containing 10% (v/v) foetal calf serum (FCS) and 2 mM L-glutamine. Skin fibroblasts from an MLIV patient (WG0909) and the heterozygous parental cells (WG0987 and WG0988) were obtained from ‘The Repository for Mutant Human Cell Strains’, Canada (www.cellbank.mcgill.ca). The MCOLN1 gene in these cells has been characterized previously (3). Human fibroblasts were cultured in modified Eagle’s medium (MEM) with Earle’s salts supplemented with 10% FCS and 2 mM L-glutamine. All cells were grown in a humidified 5% CO2 atmosphere at 37ºC. NRK and HEK-293 cells were transfected using Fugene 6 transfection reagent at a ratio of 2μg DNA: 6 μl Fugene. Stable cell lines were generated by incubating cells with 0.2 mg/ml hygromycin (ΔpMEP4 transfected cells) or 0.5 mg/ml G418 (pEGFP transfected cells). Expression of the ΔpMEP4 constructs was induced by adding 10 μM CdCl2 for 16 h to the tissue culture medium before experiments. Human fibroblast transfection was undertaken by passaging cells one week before transfection so that confluency was obtained on the day of transfection. For each transfection the cells from a confluent 75 cm2 flask were transfected with 5 μg of Qiagen midiprep DNA using an Amaxa Biosystems (Cologne, Germany) human dermal fibroblast Nucleofector kit (Nucleofector program U-23). Following transfection, cells were cultured in fresh MEM medium (see tissue culture) and used in LacCer uptake experiments 24 h post-transfection. For any given experiment all cells were of the same passage number and no cells beyond passage number 10 were used for transfection.
Cells treated with U18666A were incubated with 3μg/ml U18666A for 24 h before analysis.
Fractionation and immunoblotting
Fractions enriched in late endosomes (LE) and lysosomes (L) from rat liver were prepared as previously described (15). Briefly, rat livers were quickly flushed with ice cold STM buffer (250 mM sucrose, 10 mM TES, pH 7.4, 1 mM MgCl2) and then homogensised in 3 ml of STM/g of liver in a Potter-Elvehjem homogeniser. The liver homogenate was centrifuged at 500g for 10 min and then the post-mitochondrial supernatant was loaded on top of a Ficoll gradient (late endosomes) or a Nycodenz step gradient (lysosomes). For late endosomes, 5 ml of the post mitochondrial supernatant was loaded on top of 30ml of a 1-22% continuous Ficoll gradient on a 4 ml cushion of 45% Nycodenz cushion. For lysosomes 14 ml of the post mitochondrial supernatant was overlayed on top of a step gradient consisting of 21 ml 20 % Nycodenz and 4 ml 45% Nycodenz. Gradients were centrifuged at 206,000g for 1h in a Beckman vertical rotor (VTi50). Fractions (1ml) were collected from the bottom and those fractions enriched in either late endosomes or lysosomes were isolated. Typically lysosomes were found in fractions 4-10, and late endosomes were found in the part of the gradient corresponding to refractive indexes 1.366-1.373.
Confluent NRK cells (1 m2) were scraped into ice cold PBS and pelleted by centrifugation. The cells were resuspended in 5 ml of STM containing protease inhibitors and passed 5 times through a Balch homogeniser (specific clearance of 14 μm). The homogenate was centrifuged at 1,500g for 10 min and the post nuclear supernatant overlayed onto a 1-22% Ficoll gradient and centrifuged as above. 3 ml fractions were collected from the bottom of the gradient. β-hexosaminidase was assayed by incubating equal volumes of substrate (5 mM p-nitrophenyl N-acetyl-β-D-glucosaminidase, 50 mM sodium citrate, pH 4.8, 0.2% Triton X-100) with gradient fractions at 37°C for 1 h. The reaction was stopped by adding an equal volume of a carbonate buffer (83 mM Na2CO3, pH 10.7, 133 mM glycine, 67 mM NaCl) and then the absorbance was measured at 410 nm.
Proteins were separated by electrophoresis on a 10%SDS polyacrylamide gel using a discontinuous buffer, immunoblotted using antibodies to either rat mucolipin-1, lgp110 or the mannose 6-phosphate receptor and bands visualised by enhanced chemiluminescence (ECL; GE Healthcare, Little Chalfont, UK).
Microscopy
For immunofluorescent labelling, cells grown on coverslips were washed with PBS then fixed with 4% paraformaldehyde in PBS for 20 minutes. Cells were subsequently permeabilised with 0.1% Triton X-100 in PBS for 10 min. All cells were incubated in 0.2% (w/v) BSA in PBS (BSA-PBS) for 10 min at room temperature before incubating with primary antibodies in BSA-PBS for 1 h at room temperature. Cells were washed 3 x 5 min with BSA-PBS then incubated with fluorescently-conjugated (Alexa Fluor 488 or Alexa Fluor 555) secondary antibodies as appropriate in BSA-PBS for 30 min at room temperature. Cells were washed as previously described and coverslips mounted on slides with Mowiol containing 2.5% (w/v) 1,4-diazobicyclo-[2,2,2]-octane (DABCO).
Cells stained with filipin were fixed with paraformaldehyde (as above) but not permeabilised with Triton X-100. Cells were then stained as above but with the PBS-BSA solution containing 0.5mg/ml filipin at all stages.
Fluorescent dextran images were obtained by incubating cells with a 1mg/ml lysine-fixable fluorescent dextran-conjugate (Invitrogen, Paisley, UK) in the tissue culture medium for 4 h and then chased for 20h before fixation (figure 3D). Cells shown in figure 3C were continuously pulsed with the dextran conjugate for 3 h before live-cell imaging.
All fixed-cell immunofluorescence pictures (apart from figure 4A) were acquired with a Zeiss LSM510 confocal microscope using a 63x Plan-Apochromat objective (numerical aperture 1.4). Images were collected at a resolution of 1024 x 1024 pixels and are shown as maximum intensity Z-projections. Adobe Photoshop software was used for image processing. The images for figure 4A were obtained on a microscope as detailed in the LacCer section below. Immunoelectron microscopy was as described previously (37).
LacCer trafficking
Cells for LacCer uptake experiments were grown on 35 mm glass-bottomed tissue culture dishes. Cells were washed twice with PBS then incubated with 5μM LacCer (complexed to BSA as per the manufacturer’s instructions) in serum free MEM for 1 h at 37ºC. Cells were then washed twice with PBS and then incubated with normal MEM (MEM + 10% FCS) for 2 h, or 5 h where stated, at 37ºC. Cells were washed with PBS, then plasma membrane LacCer was back-extracted by washing cells (3 x 15 min) with normal MEM containing 2 % (w/v) fatty acid free BSA at 4ºC. Cells were then washed with phenol red- and sodium bicarbonate-free MEM medium containing 2 mM L-glutamine, 1% (v/v) FCS and 10 mM HEPES, pH 7.4 at 20°C. Endocytosed lipid was immediately visualised (at room temperature) using a 63x Plan-Apochromat objective (numerical aperture 1.4) on a Zeiss Axiovert 200M microscope fitted with a mercury vapour lamp and filter set XF26 (Omega Optical, Vermont, USA). Filter set XF26 consists of a single excitation filter (485DF22) and dichroic (505DRLP) and two emission filters (OG590 and 530DF30). The two excitation filters take into account that the BODIPY FL fluorophore conjugated to the lactosylceramide moiety gives both green fluorescence and an aggregation-dependent shift to red fluorescence due to intermolecular excimer formation. Under the experimental conditions used, little to no LacCer red fluorescence was observed, hence all LacCer images (figures 3, 4 & 5) were acquired using the 530nm discriminating filter. Significant green auto-fluorescence was observed in some cell types which was eliminated by using a Zeiss 1.0 ND filter in front of the mercury vapour light path but still allowed LacCer to be detected. Fluorescent images were captured with a Hamamatsu Orca ER cooled CCD camera. Images were analysed using Openlab software by Improvision (Coventry, UK). For each image captured, a binary mask was generated using thresholds that selected brightness values that corresponded to the observed pattern of fluorescence (eliminating background signals and out of focal plane fluorescence). Using this procedure, pixels with an intensity over the threshold value had a value of 1, all other pixels had a value of 0. Pixels clearly corresponding to Golgi fluorescence were selected and deleted from the binary images. The Openlab software was then used to count the number of individual regions (corresponding to positive pixels) in the binary layer. Comparison of masks generated with various threshold values did not significantly alter the outcome. Cells transfected with HcRed-mucolipin-1 constructs were first identified using filter set 00 (Carl Zeiss, Welwyn Garden City, UK) before acquiring the LacCer image. Adobe Photoshop software was used for all subsequent image processing.
Supplementary Material
Acknowledgements
This work was funded by MRC Grant G9310915. CIMR is supported by a Strategic Initiative Grant (066438) and FMG by a Senior Clinical Research Fellowship from the Wellcome Trust. FR was supported by a Diabetes UK R.D. Lawrence Fellowship. We thank Abigail Brown for the immunoelectron micrograph shown in Fig 1B, Simon McCallum for assistance with flow cytometry and Katherine Bowers, Folma Buss, Antony Galione, Ruth Murrell-Lagnado and Rob Piper for helpful discussions.
References
- 1.Slaugenhaupt SA, Acierno JS, Helbling LA, Bove C, Goldin E, Bach G, Schiffmann R, Gusella JF. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am J Hum Genet. 1999;65:773–778. doi: 10.1086/302549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 3.Bassi MT, Manzoni M, Monti E, Pizzo MT, Ballabio A, Borsani G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Bove C, Kaneski CR, Nagle J, Bromley MC, Colman M, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 5.LaPlante JM, Falardeau J, Sun M, Kanazirska M, Brown EM, Slaugenhaupt SA, Vassilev PM. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 2002;532:183–187. doi: 10.1016/s0014-5793(02)03670-0. [DOI] [PubMed] [Google Scholar]
- 6.Raychowdhury MK, Gonzalez-Perrett S, Montalbetti N, Timpanaro GA, Chasan B, Goldmann WH, Stahl S, Cooney A, Goldin E, Cantiello HF. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum Mol Genet. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 7.LaPlante JM, Ye CP, Quinn SJ, Goldin E, Brown EM, Slaugenhaupt SA, Vassilev PM. Functional links between mucolipin-1 and Ca2+-dependent membrane trafficking in mucolipidosis IV. Biochem Biophys Res Comm. 2004;322:1384–1391. doi: 10.1016/j.bbrc.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 8.Cantiello HF, Montalbetti N, Goldmann WH, Raychowdhury MK, Gonzalez-Perrett S, Timpanaro GA, Chasan B. Cation channel activity of mucolipin-1: the effect of calcium. Pflügers Archiv – Eur J Physiol. 2005;451:304–312. doi: 10.1007/s00424-005-1448-9. [DOI] [PubMed] [Google Scholar]
- 9.Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, Kiselyov K. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J Biol Chem. 2006;281:7294–7301. doi: 10.1074/jbc.M508211200. [DOI] [PubMed] [Google Scholar]
- 10.Chen CS, Bach G, Pagano RE. Abnormal transport along the lysosomal pathway in mucolipidosis, type IV disease. Proc Natl Acad Sci USA. 1998;95:6373–6378. doi: 10.1073/pnas.95.11.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bach G. Mucolipidosis type IV. Mol Genet Metab. 2001;73:197–203. doi: 10.1006/mgme.2001.3195. [DOI] [PubMed] [Google Scholar]
- 12.Bach G. Mucolipin 1: endocytosis and cation channel-a review. Pflügers Archiv. 2004 doi: 10.1007/s00424-004-1361-7. on line 27/11/04. [DOI] [PubMed] [Google Scholar]
- 13.Chen CS, Patterson MC, Wheatley CL, O’Brien JF, Pagano RE. Broad screening test for sphingolipid-storage diseases. Lancet. 1999;354:901–905. doi: 10.1016/S0140-6736(98)10034-X. [DOI] [PubMed] [Google Scholar]
- 14.Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 15.Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP. The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol. 2000;149:1053–1062. doi: 10.1083/jcb.149.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, Stewart A, James DE, Piper RC, Luzio JP. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piper RC, Luzio JP. CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 2004;14:471–473. doi: 10.1016/j.tcb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Manzoni M, Monti E, Bresciani R, Bozzato A, Barlati S, Bassi MT, Borsani G. Overexpression of wild-type and mutant mucolipin proteins in mammalian cells: effects on the late endocytic compartment organization. FEBS Lett. 2004;567:219–224. doi: 10.1016/j.febslet.2004.04.080. [DOI] [PubMed] [Google Scholar]
- 20.Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–353. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiselyov K, Chen J, Rbaibi Y, Oberdick D, Tjon-Kon-Sang S, Shcheynikov N, Muallem S, Soyombo A. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J Biol Chem. 2005;280:43218–43223. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- 22.Miedel MT, Weixel KM, Bruns JR, Traub LM, Weisz OA. Posttranslational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J Biol Chem. 2006 doi: 10.1074/jbc.M511104200. [DOI] [PubMed] [Google Scholar]
- 23.Bonifacino JS, Traub LM. Signals for Sorting of Transmembrane Proteins to Endosomes and Lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 24.Puri V, Watanabe R, Dominguez M, Sun X, Wheatley CL, Marks DL, Pagano RE. Cholesterol modulates membrane traffic along the endocytic pathway in sphingolipid-storage diseases. Nat Cell Biol. 1999;1:386–388. doi: 10.1038/14084. [DOI] [PubMed] [Google Scholar]
- 25.Liscum L. Niemann-Pick type C mutations cause lipid traffic jam. Traffic. 2000;1:218–225. doi: 10.1034/j.1600-0854.2000.010304.x. [DOI] [PubMed] [Google Scholar]
- 26.Cenedella RJ, Sexton PS, Krishnan K, Covey DF. Comparison of effects of U18666A and enantiomeric U18666A on sterol synthesis and induction of apoptosis. Lipids. 2005;40:635–640. doi: 10.1007/s11745-005-1426-9. [DOI] [PubMed] [Google Scholar]
- 27.Schaheen L, Dang H, Fares H. Basis of lethality in C. elegans lacking CUP-5, the mucolipidosis type IV orthologue. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Bargal R, Bach G. Mucolipidosis type IV: Abnormal transport of lipids to lysosomes. J Inher Metab Dis. 1997;20:625–632. doi: 10.1023/a:1005362123443. [DOI] [PubMed] [Google Scholar]
- 29.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 30.Christensen KA, Myers JT, Swanson JA. pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci. 2002;115:599–607. doi: 10.1242/jcs.115.3.599. [DOI] [PubMed] [Google Scholar]
- 31.Goldin E, Blanchette-Mackie EJ, Dwyer NK, Pentchev PG, Brady RO. Cultured skin fibroblasts derived from patients with mucolipidosis 4 are auto-fluorescent. Pediatr Res. 1995;37:687–692. doi: 10.1203/00006450-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Bach G, Chen CS, Pagano RE. Elevated lysosomal pH in Mucolipidosis type IV cells. Clin Chim Acta. 1999;280:173–179. doi: 10.1016/s0009-8981(98)00183-1. [DOI] [PubMed] [Google Scholar]
- 33.Kopitz J, Gerhard C, Hofler P, Cantz M. [14C]Methylamine accumulation in cultured human skin fibroblasts--a biochemical test for lysosomal storage and lysosomal diseases. Clin Chim Acta. 1994;227:121–133. doi: 10.1016/0009-8981(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 34.DeCoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 35.Kinnear NP, Boittin FX, Thomas JM, Galione A, Evans MA. Lysosome-sarcoplasmic reticulum junctions. J Biol Chem. 2004;279:54319–54326. doi: 10.1074/jbc.M406132200. [DOI] [PubMed] [Google Scholar]
- 36.Bagshaw RD, Mahuran DJ, Callahan JW. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol Cell Proteomics. 2005;4:133–143. doi: 10.1074/mcp.M400128-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Girotti M, Banting G. TGN38-green fluorescent protein hybrid proteins expressed in stably transfected eukaryotic cells provide a tool for the real-time, in vivo study of membrane traffic pathways and suggest a possible role for ratTGN38. J Cell Sci. 1996;109:2915–2926. doi: 10.1242/jcs.109.12.2915. [DOI] [PubMed] [Google Scholar]
- 38.Reaves BJ, Bright NA, Mullock BM, Luzio JP. The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J Cell Sci. 1996;109:749–762. doi: 10.1242/jcs.109.4.749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.