Abstract
Topologically Associating Domains (TADs) have been proposed to both guide and constrain enhancer activity. Shh is located within a TAD known to contain all its enhancers. To investigate the importance of chromatin conformation and TAD integrity on developmental gene regulation, we have manipulated the Shh TAD – creating internal deletions, deleting CTCF sites, and deleting and inverting sequences at TAD boundaries. Chromosome conformation capture and fluorescence in situ hybridisation assays were used to investigate the changes in chromatin conformation that result from these manipulations. Our data suggest that these substantial alterations in TAD structure have no readily detecx effect on Shh expression patterns or levels of Shh expression during development – except where enhancers are deleted - and result in no detectable phenotypes. Only in the case of a larger deletion at one TAD boundary could ectopic influence of the Shh limb enhancer be detected on a gene (Mnx1) in the neighbouring TAD. Our data suggests that, contrary to expectations, the developmental regulation of Shh expression is remarkably robust to TAD perturbations.
Keywords: chromosome conformation capture, fluorescence in situ hybridisation, Shh, enhancers, genome organisation, development, topologically associated domains
Introduction
At the megabase-scale, the mammalian genome is partitioned into self-interacting topologically associated domains (TADs) (Dixon et al., 2012; Nora et al., 2012). Mammalian TAD boundaries are enriched in CTCF sites with their relative orientation appearing crucial to function (Narendra et al., 2015; Rao et al., 2014; Sanborn et al., 2015). TADs are formed by dynamic cohesin-driven loop extrusion (Fudenberg et al., 2016; Nora et al., 2017; Rao et al., 2017; Schwarzer et al., 2017; Vian et al., 2018) and convergent CTCF sites act to impede loop extrusion by enabling WAPL-mediated release of cohesin from the chromosome (Haarhuis et al., 2017).
The regulatory landscapes of developmental genes are frequently found to be contained within the same TAD (Dixon et al., 2012; Rao et al., 2014). TADs have, therefore, been proposed to act as functional regulatory units within which contacts between enhancers and their target gene are favoured while limiting aberrant interactions of enhancers across TAD boundaries (Fudenberg et al., 2016; Sun et al., 2019). In support of this hypothesis, some studies have found that deletion or inversion of CTCF sites at TAD boundaries can promote TAD boundary crosstalk and re-wire enhancer-promoter contacts (de Wit et al., 2015; Guo et al., 2015; Narendra et al., 2015; Rodríguez-Carballo et al., 2017). Moreover, a number of recent studies have suggested that changes to TAD structure can disrupt gene regulation through enhancer-rewiring in human disease (Flavahan et al., 2016; Franke et al., 2016; Lupiáñez et al., 2015). However, other studies report that, although depletion of CTCF erases the insulation between TADs, it has limited effects on gene expression (Nora et al., 2017; Soshnikova et al., 2010).
To further study the CTCF mediated function of TADs in developmental gene regulation, we have exploited the sonic hedgehog (Shh) regulatory domain – a paradigm locus for long-range regulation. The SHH morphogen controls the growth and patterning of many tissues during embryonic development, including the brain, neural tube and limbs. Spatial and temporal Shh expression is regulated by tissue-specific enhancers located within the gene, and upstream in a large gene desert and within neighbouring genes (Jeong et al., 2006; Anderson and Hill, 2014). Shh and its cis-acting elements are all contained within a well-characterised ~960kb TAD (Anderson et al., 2014; Williamson et al., 2016). In the developing mouse limb bud, Shh expression is solely determined by the ZRS enhancer (Lettice et al., 2003; Sagai et al, 2005) located 850kb upstream of Shh within an intron of the widely expressed Lmbr1 (Fig. 1A). Fluorescence in situ hybridisation (FISH) showed that Shh and the ZRS are consistently located in relatively close proximity to each other in all cell types and tissues examined, which we infer to be a consequence of the underlying invariant TAD structure. In contrast, we observed increased ZRS-Shh co-localisation in the Shh-expressing posterior portion of developing limb buds (Williamson et al., 2016), consistent with a specific gene-enhancer contact.
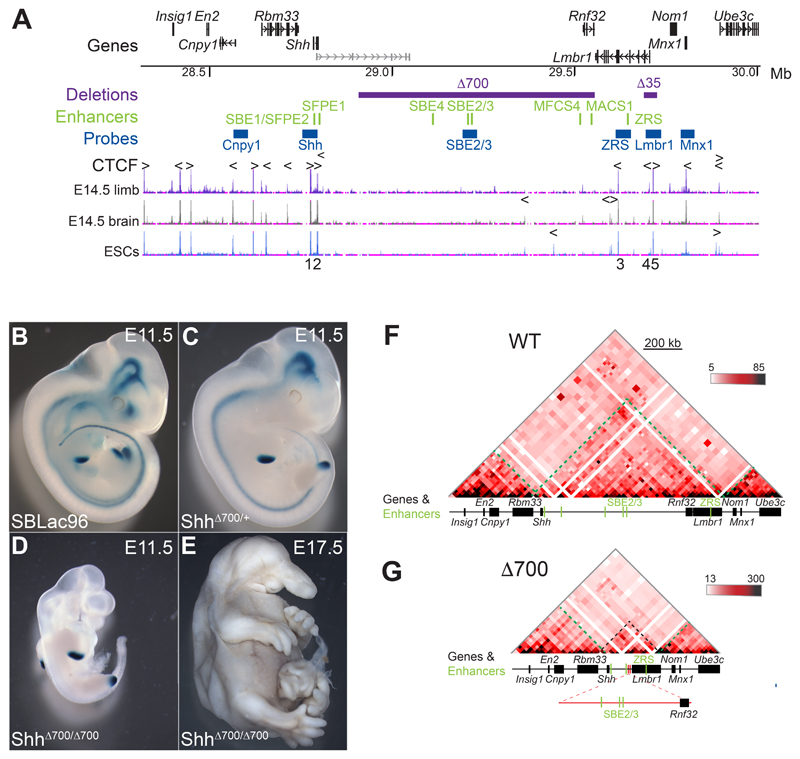
Figure 1. A 700-kb intra-TAD deletion has no adverse effects on limb-specific expression of Shh.
(A) (Top) Location of genes in the 1.7 Mb murine genomic locus (chr5: 28317087-30005000; mm9) containing Shh analysed by 5C and FISH, with the position of tissue-specific Shh enhancers shown below in green. Locations to which the fosmid FISH probes hybridize are shown in blue, and the purple bars indicate deleted genomic regions (Δ700 and Δ35). The bottom three tracks show UCSC ENCODE CTCF ChIP-seq profiles in E14.5 limb buds, brain and in ESCs. Arrowheads above the tracks indicate the orientation of CTCF-binding motifs and the deleted CTCF binding sites (1-5) are numbered below. (B-D) Staining for the LacZ reporter gene carried by the sleeping beauty transposon in E11.5 embryos, (B) carries the intact SBLac96 insertion (Anderson et al, 2014) while (C) shows the remaining sites of expression after Cre mediated deletion of 700Kb (purple bar in A) and (D) shows the phenotype of an embryo homozygous for the 700kb deletion, with LacZ staining only evident in the ZPA of the limb buds. A homozygous deletion embryo (E) at E17.5 showing craniofacial and brain defects but normally formed limbs. (F and G) 5C heat-maps from cells of the bodies of E11.5 wild-type embryos (F) and embryos homozygous for the 700kb deletion (G). Heat map intensities represent the average of interaction frequency for each window, binned over 21kb windows, with read counts summed from two biological replicates and colour-coded according to the scale shown. Green dashed lines highlight TAD boundaries, black dashed lines indicate the Shh TAD boundaries and the reduced size of the TAD. Data for individual biological replicates are in Figure S1.
Here, we genetically manipulate the Shh TAD and its TAD boundaries to investigate the importance of chromatin architecture on TAD structure and on the regulation of gene expression. We use a chromosome conformation assay (5C) and FISH to investigate how these manipulations affect structures within the Shh TAD and its interactions with adjacent TADs. We determine how these alterations affect the expression pattern of Shh and nearby developmentally regulated genes in vivo. We also examine the phenotypic consequences of these manipulations. Our results question the importance of TADs for correct spatial and temporal gene regulation.
Results
A large deletion within the Shh TAD does not disrupt local genome organisation or limb-specific activation of Shh
Prominent features of the Shh TAD include five CTCF binding sites preserved across multiple cell types (Fig. 1A), and two sub-TADs with overlapping boundaries located within the gene desert between the forebrain enhancers and Rnf32 (Fig. 1F and 2A). This region of the gene desert includes less well defined CTCF peaks that differ across cell types but due to their location may have some role in defining these sub-TADs (Fig. 1A) (Rosenbloom et al., 2013).
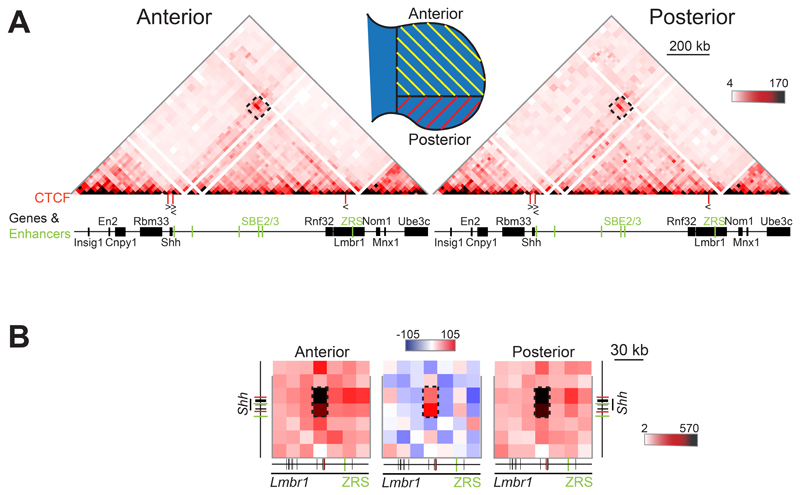
Figure 2. 5C analysis in E11.5 distal anterior and posterior limb tissue.
(A) 5C heat maps showing data from distal anterior and posterior limb bud cells of E11.5 embryos, across the Shh region shown in Fig. 1A. The schematic indicates the limb bud portions dissected for anterior and posterior cell populations. Interactions highlighted by the black dashed boxes locate the region of the heat maps shown in (B) at higher resolution. (B) Higher resolution (15kb binning) 5C heat maps from data displayed in (A) showing interactions encompassing Shh and ZRS. The comparison heatmap (centre), with compared data sets normalised by read count, shows interactions enriched in posterior cells (red) or anterior cells (blue). Enriched interactions between loci containing CTCF binding sites are indicated by the black dashed boxes. Data for individual biological replicates are in Fig. S2.
To determine the contribution of TAD internal sequence to 3D chromatin organisation and gene expression, we exploited our previous work that used the local hopping activity of the sleeping beauty (SB) transposon to probe the Shh regulatory domain (Anderson et al., 2014). Transposition of the SB leaves one LoxP site at the initial integration site and inserts a second site where it re-integrates, enabling Cre recombinase to create deletions of the intervening DNA. The orientation of the re-integration means the LacZ gene carried by the SB is retained in the deleted chromosome allowing remaining enhancer activity to be monitored. Using this approach, we deleted approximately 700kb (~70%) of the internal Shh TAD sequence, including the sub-TAD boundaries, but leaving the five CTCF binding sites at the TAD extremities intact (Fig. 1A). The Δ700 deletion removes many of the known Shh enhancers and relocates the ZRS to within 96kb of the Shh promoter (Fig. 1A). Removal of the Shh forebrain and epithelial enhancers in the Δ700 deletion is shown by changes in the LacZ staining of ShhΔ700/+ embryos. Staining is observed only within the floor plate and hind brain, presumably driven by the proximal enhancers SFPE1/2 and SBE1, and within the limbs driven by the ZRS (Compare the wildtype in Fig. 1B with the ShhΔ700/+ embryo in Fig. 1C). Homozygous ShhΔ700/Δ700 embryos show phenotypes very similar to those of Shh-/- embryos but with normal limb and digit patterning (Chiang et al., 1996). These data indicate that, despite its incorrect position now only 96kb from the Shh promoter, ZRS is able to function normally to drive Shh expression in limb development (Fig. 1D & 1E).
5C on whole E11.5 Shh Δ700/Δ700 and wild type embryos shows that the Shh TAD boundaries and the adjacent TADs are unaffected by the Δ700 deletion (Fig. 1F, 1G & S1). Therefore, neither sequence elements nor chromatin interactions within the deleted region are needed for maintaining the location of the TAD boundaries. Additionally, the large genomic distance between Shh and its limb enhancer ZRS is not required for correct function.
Interactions within the Shh TAD are delineated by CTCF sites either side of Shh and within Lmbr1
Our previous 5C analyses on cells dissected from whole limbs, bodies and heads of E11.5 embryos showed enriched interactions between the genomic region containing Shh, located at one TAD boundary, and a genomic region within Lmbr1 close to ZRS, located ~70kb from the other TAD boundary (Williamson et al., 2016). That this enrichment can be identified throughout the E11.5 embryo, a stage when we have shown that high levels of Shh-ZRS co-localisation occur only in the posterior distal limb, excludes active Shh-ZRS co-localisation as the sole driver of this apparent chromatin loop (Williamson et al., 2016). To gain further insight into the nature of these interactions, we dissected E11.5 limb buds to compare cell populations with no ZRS activity (anterior 2/3 of bud) to those in which ZRS is active (posterior 1/3) (Fig. 2A and S2A & C).
The 5C heatmaps of the Shh TAD are similar in both anterior and posterior limb bud cell populations, and comparable to dissected E11.5 bodies (compare Fig. 1F and Fig. 2A). At high (15kb) resolution, the strongest enrichment involved interactions between both Shh and the genomic region immediately 3ʹ of Shh, and a locus ~20kb from ZRS in intron 5 of Lmbr1 (Fig. 2B and S2B & D, left- and right-hand heatmaps). ENCODE data (Rosenbloom et al., 2013) indicates these three loci are all bound by CTCF across a range of cell and tissue types (Fig. 1A), with the underlying DNA containing CTCF-binding motifs in a convergent orientation consistent with a role blocking loop extrusion (Fig. 2A).
CTCF site deletions reduce Shh intra-TAD interactions and disrupts Shh/ZRS proximity
To examine the role of CTCF sites on the architecture of the Shh TAD we used CRISPR-Cas9 to make small (~1kb) deletions of sequences containing the 5 major CTCF binding sites in mouse ESCs (Table S1). We first generated ESC lines homozygous for deletions of the CTCF binding regions 3ʹ and 5ʹ of Shh (sites 1 and 2 respectively in Fig. 1A. and assayed chromatin conformation by 5C and FISH.
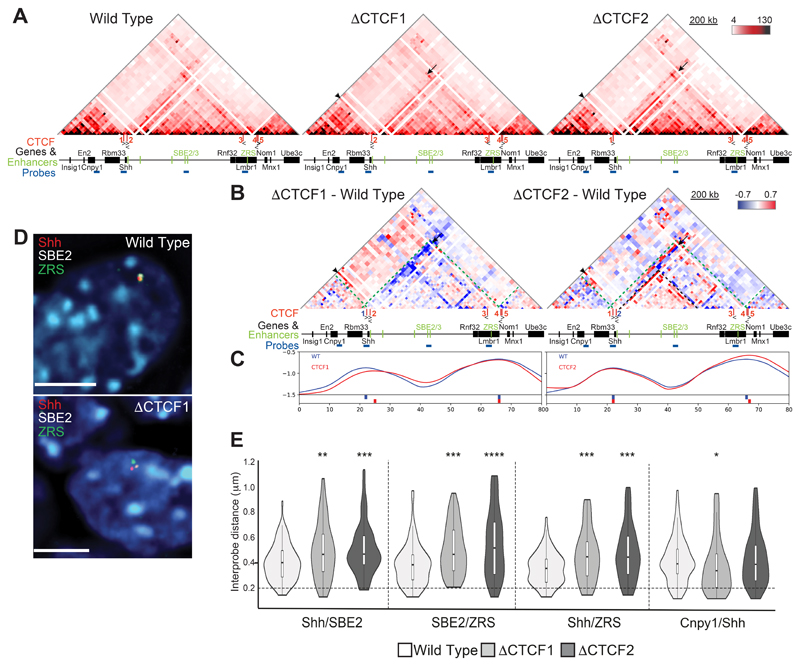
The Shh TAD structure in wild-type ESCs is similar to that in E11.5 embryos (Fig. 3A and S3A & C). Deletion of CTCF site 1 (ΔCTCF1), which delineates the TAD boundary 3ʹ of Shh, results in Shh losing interactions (arrows) with the rest of its own TAD and gaining interactions (arrowheads) with regions just 5ʹ of En2 and Rbm33 (Fig. 3A and S3A & C). The TAD boundary re-locates by ~60kb to 5ʹ of Shh beyond CTCF2 (Fig. 3B and C and S3B & D). There is also loss of interactions with a locus upstream of the forebrain enhancers near the sub-TAD boundary within the larger Shh sub-TAD (strong blue diagonals located between SBE3 and Rnf32). These data are consistent with CTCF1 forming the Shh TAD boundary by blocking loop extrusion emanating from within the En2 TAD.
Figure 3. 5C and 3D FISH identifies perturbations to chromatin conformation in ΔCTCF1 and ΔCTCF2 ESCs.
(A) 5C heat maps showing data from wild type, ΔCTCF1 and ΔCTCF2 ESCs. (B) Heat maps comparing ΔCTCF1 or ΔCTCF2 enrichment (red) with wild type (blue). Green dashed lines indicate TAD boundaries in wild-type, black arrows highlight loss of interactions in ΔCTCF1 or ΔCTCF2 cells and black arrowheads indicate enriched interactions in ΔCTCF1 or ΔCTCF2 cells. (C) Insulation score graphs that identify the location of TAD boundaries using raw summed 5C matrices. Peaks of insulation are shown below the plots, ΔCTCF1 or ΔCTCF2 (red) and wild type (blue). Data for biological replicates are in Supplemental Fig. S3. (D) Images of representative nuclei from wild type and ΔCTCF1 ESCs showing FISH signals for Shh/SBE2/ZRS probes. Scale bars = 5 μm. (E) Violin plots show the distribution of interprobe distances (μm) between Shh/SBE2, SBE2/ZRS, <Shh/ZRS and Cnpy1/Shh probes in wild type, ΔCTCF1 and ΔCTCF2 ESCs. Horizontal dashed line shows the proportion of alleles that are co-localised (< 200 nm). The statistical significance between data sets was examined by Mann-Whitney U Tests, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001.
The left hand Shh TAD boundary is not affected by deletion of CTCF site 2, however, the 5ʹ Shh region does gain contacts (arrowheads) with the En2 TAD in a similar manner to the loss of CTCF1 (Fig. 3A-C and S3A - D), suggesting that both CTCF1 and 2 are necessary to optimally block loop extrusion emanating from the En2 TAD. While loss of CTCF2 results in decreased interactions between the Shh locus and the rest of its own TAD (arrows), this is compensated for by increased interactions with CTCF1, thereby maintaining the boundary position (Fig. 3A & B and S3, right-hand heatmaps). There are also enriched interactions within the Shh sub-TAD in ΔCTCF2 cells (Fig. 3B and S3B & D).
We also analysed possible alterations of chromosome conformation due to the CTCF site deletions with 3D-FISH using probes for Shh, ZRS and SBE2, (Shh Brain Enhancer 2) an enhancer which is located 460 kb upstream of the Shh coding sequence in the middle of the TAD (Jeong et al., 2006) (Fig 1A and Fig. 3D). Interprobe distances between all three probe pairs were significantly increased in CTCF deletion cells compared to wild type ESCs (Fig. 3E and Table S4), consistent with the reduced interactions between Shh and the rest of its TAD identified by 5C. Conversely, distances between Shh and Cnpy1 (in the neighbouring En2 TAD) were significantly decreased in ΔCTCF1 cells compared to wild type (Fig. 3E and Table S4), consistent with relocation of the TAD boundary.
Deleting either CTCF1 or CTCF2 disrupts Shh-ZRS spatial proximity in ESCs and, more generally, results in reduced 5C interactions between Shh and the rest of the regulatory TAD that may be due to the re-location of the TAD boundary (ΔCTCF1) or greater sub-division of the TAD (ΔCTCF2). The TAD boundary adjacent to Shh is sharply defined by CTCF1 whereas the boundary location of the neighbouring En2 TAD cumulatively results from both CTCF 1 and 2, possibly by blocking loop extrusion emanating from this TAD. However, neither of these deletions on their own is sufficient to cause merging of the two neighbouring TADs.
Shh-ZRS proximity is disrupted by the deletion of ZRS/Lmbr1 CTCF sites
Both CTCF1 and CTCF2 have enriched interactions with the CTCF site ~20 kb from ZRS in intron 5 of Lmbr1 (CTCF3) (Fig. 1A). Therefore, we deleted both copies of CTCF3 (ΔCTCF3), described as i5 in (Paliou et al., 2019).
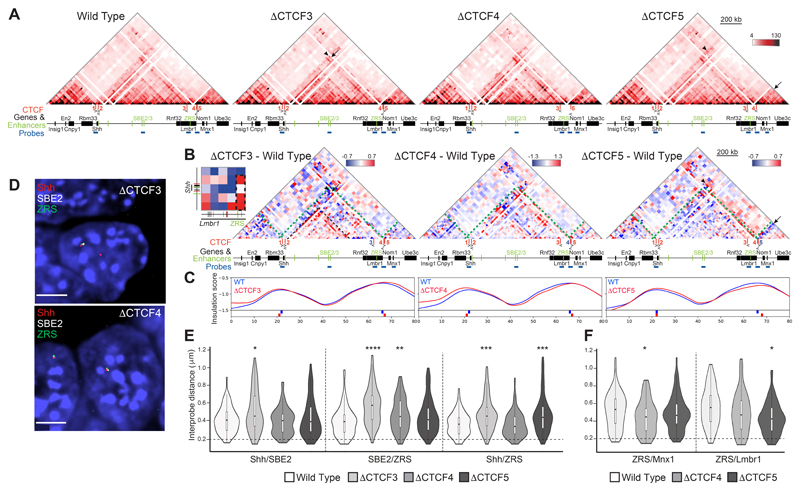
Whilst whole TAD integrity was unaffected by ΔCTCF3 (Fig. 4A & C and S4A & C), intra-TAD reorganisation occurred in a similar manner to the loss of CTCF2, with enriched interactions within the sub-TADs (Fig. 4B and S4B & D). Loss of interactions between CTCF3 and Shh/CTCF2 in ΔCTCF3 cells appears to be somewhat compensated for by enriched contacts (arrowheads) between the ZRS locus and the Shh region of the TAD, particularly CTCF1 (See inset heatmap adjacent to ΔCTCF3). Ectopic CTCF binding at ZRS has been recently identified following the loss of neighbouring CTCF sites including CTCF3 (Paliou et al., 2019). Despite this compensation identified by 5C, FISH showed significantly increased inter-probe distances between Shh, SBE2 and ZRS in ΔCTCF3 cells compared to wild type (Fig. 4D & E and Table S4). These data suggest that loss of any one of the three CTCF binding sites (1, 2 or 3) can disrupt the spatial proximity of Shh, SBE2 and ZRS (Fig. 3E & 4E).
Figure 4. 5C and 3D FISH identifies perturbations to local chromatin conformation in ΔCTCF3,ΔCTCF4 andΔCTCF5 ESCs.
(A) 5C heat maps from wild type, ΔCTCF3, ΔCTCF4 and ΔCTCF5 ESCs. (B) Heat maps comparing ΔCTCF3, ΔCTCF4 or ΔCTCF5 enrichment (red) with wild type (blue). Green dashed lines indicate the TAD boundaries in wild-type cells, black arrows highlight loss of interactions in ΔCTCF3 or ΔCTCF4 or ΔCTCF5 cells and black arrowheads indicate enriched interactions in ΔCTCF3 or ΔCTCF4 or ΔCTCF5 cells. Inset heatmap highlights enriched contacts between Shh and ZRS loci in ΔCTCF3. Black dashed rectangle indicate interactions between ZRS bin and bins covering the Shh locus. (C) Insulation score graphs that identify the location of TAD boundaries using raw summed 5C matrices. Peaks of insulation are shown below the plots, ΔCTCF3 or ΔCTCF4 or ΔCTCF5 (red) and wild type (blue). Data for biological replicates are in Supplemental Fig. S4. (D) Images of representative nuclei from ΔCTCF3 and ΔCTCF4 ESCs showing FISH signals for Shh/SBE2/ZRS probes. Scale bars = 5 μm. (E) Violin plots show the distribution of interprobe distances (μm) between Shh/SBE2, SBE2/ZRS and Shh/ZRS probes in wild type, ΔCTCF3, ΔCTCF4 and ΔCTCF5 ESCs. Horizontal dashed line shows the proportion of alleles that are colocalised (< 200 nm). The statistical significance between data sets was examined by Mann-Whitney U Tests, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. (F) As in (E) but for ZRS/Mnx1 and ZRS/Lmbr1 probes in wild type, ΔCTCF4 and ΔCTCF5 ESCs.
Finally, we generated ESC lines with deletions of CTCF binding sites at the Lmbr1 promoter (ΔCTCF4) and 5ʹ Lmbr1 (ΔCTCF5), both of which are located at the boundary between the Shh TAD and the adjacent TAD containing Mnx1 (Fig. 1A). The CTCF motif within CTCF5 is oriented towards the Mnx1-containing TAD and deletion of CTCF5 caused a loss of interactions between this boundary region and the Mnx1 TAD (Fig. 4A & B and S4B & D) with the TAD boundary shifted toward Nom1 (Fig. 4C). 5C also reveals increased interactions in the ZRS-Lmbr1 region in ΔCTCF5 (Fig. 4B) and FISH also shows increased spatial proximity between ZRS and Lmbr1 (Fig. 4E and Table S4).
FISH revealed significantly increased inter-probe distances between Shh and ZRS in ΔCTCF5 cells and between SBE2 and ZRS in ΔCTCF4 cells, which is not detected by 5C (Fig. 4E). There are also decreased distances seen between ZRS and Mnx1 in the adjacent TAD in the absence of Shh-CTCF4, also not apparent in the 5C data (Fig. 4F).
We conclude that deletion of CTCF binding sites at either of the Shh TAD and sub-TAD boundaries, especially CTCF1, 2 and 3, affects local chromatin organisation in ESCs and disrupts Shh/ZRS spatial proximity.
Reduced Shh-ZRS colocalisation in the limb upon the loss of CTCF1, 2 and 3
To test how disrupted TAD organisation impacts on chromosome conformation and Shh gene expression during embryonic development, we generated mouse lines carrying each of the homozygous CTCF deletions. We previously reported enhanced Shh-ZRS colocalisation in the limb bud at the time and place of Shh expression that depends on a fully functional ZRS (Lettice et al., 2014; Williamson et al., 2016). Therefore, we assayed the spatial proximity of Shh, SBE2 and ZRS by FISH in E11.5 embryo sections that include posterior (ZPA) and anterior distal limb tissue from wild-type and homozygous ΔCTCF mutant embryos (Fig. 5A).
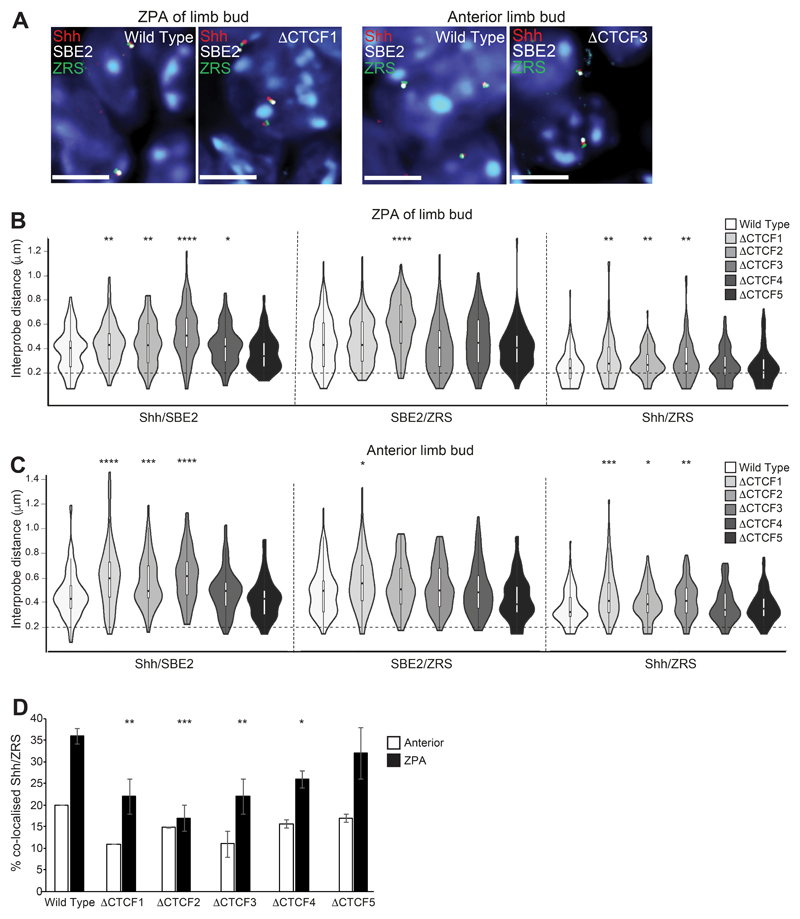
Figure 5. Perturbation of chromatin conformation within theShhTAD in the distal limb bud of ΔCTCF mutant embryos.
(A) Images of representative nuclei from E11.5 ZPA and distal anterior limb bud in wild type, ΔCTCF1 and ΔCTCF3 embryos showing FISH signals for Shh/SBE2/ZRS probes. Scale bars = 5 μm. (B) & (C) Violin plots show the distribution of interprobe distances (μm) between Shh/SBE2, SBE2/ZRS and Shh/ZRS probes in E11.5 wild type and ΔCTCF mutant embryos in (B) ZPA/distal posterior and (C) distal anterior limb bud. Horizontal dashed lines show the proportion of alleles that are colocalised (< 200 nm). The statistical significance between data sets was examined by Mann-Whitney U Tests, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. (D) Histograms show the percentage of colocalised Shh/ZRS probe pairs (<200nm) in wild type and each of the ΔCTCF E11.5 embryos for distal anterior and ZPA limb bud tissue. Error bars represent SEM obtained from two or three different tissue sections. The statistical significance between data sets was examined by Fisher’s Exact Tests, * < 0.05, ** < 0.01, *** < 0.001.
In both regions of the wild type limb bud analysed (ZPA and anterior), Shh-ZRS distances were shorter, than between either Shh-SBE2 or SBE2-ZRS, consistent with Shh and ZRS being maintained in spatial proximity across the limb bud (Fig. 5B & C). Similar to our observation in ESCs (Fig. 3 and 4), distances between Shh and both SBE2 and ZRS were significantly increased in ΔCTCF1, 2 and 3, but not ΔCTCF4 and 5 embryos, (Fig. 5B & C and Table S5 & S6). The frequency of Shh-ZRS colocalisation (<200nm) in the ZPA of ΔCTCF1, 2, and 3 mutant embryos was reduced to levels seen in non-expressing parts of the wild-type limb bud (Fig. 5D and Table S7).
Shh expression patterns and development are unaffected in CTCF site deletion mice
Our data indicate that deletion of individual CTCF sites can affect TAD boundaries, intra- and inter-TAD interactions and enhancer-promoter co-localisation frequencies. These alterations in 3D chromosome conformation might be predicted to affect gene expression. However, we found that mice homozygous for any of the ΔCTCF deletions are viable, fertile and have no overt deleterious phenotype. In situ hybridisation in homozygous mutant embryos showed a normal pattern of Shh expression in the brain (Fig. 6A), and body (Fig. 6B) at similar levels to wild type. At E11.5 expression is detected only within the developing midline of the brain, the Zli and the medial ganglionic eminence in the head and staining is visible in the floor plate and notochord, the ZPA of the limb buds and umbilicus in the body. No ectopic expression is detected at the midbrain / hindbrain junction driven by neighbouring En2 or Cnpy1 enhancers (Fig. 6B). Conversely, in embryos homozygous for either ΔCTCF1 or ΔCTCF2 there is no evidence for ectopic En2 and Cnpy1 expression in any of the normal sites of Shh expression in the brain (Fig. 6C and D).
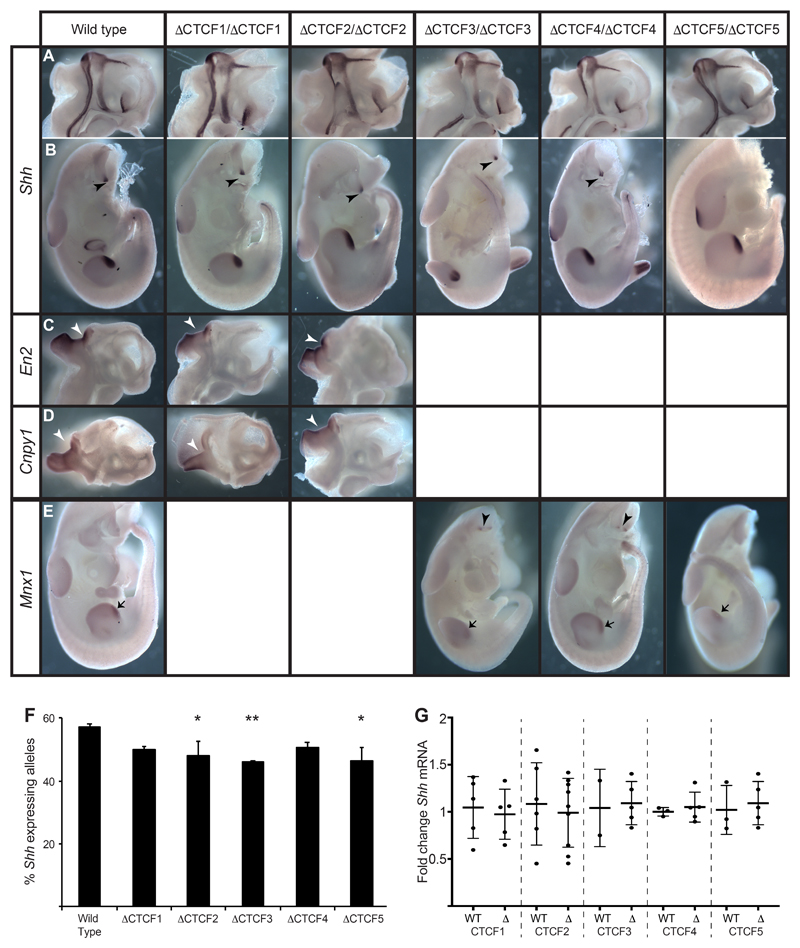
Figure 6. Expression patterns of Shh and genes in neighbouring TADs are unaffected by CTCF site deletions.
In situ analysis of gene expression at E11.5 in wild type and embryos homozygous for each of the ΔCTCF lines. (A & B) Normal expression of Shh in the midline of bisected heads (A) and in the bodies (B) where expression is detected in the ZPA of the limb bud and in the floorplate and notochord (arrowheads). (C & D) Expression of En2 (C) or Cnpy1 (D) in bisected heads. Expression is detected only at the mid brain hindbrain junction (arrowheads). (E) Expression of Mnx1 in the ZPAs of the limb buds (arrows) and in the motor neurons (arrowheads). (F) Percent of expressing Shh alleles detected by RNA FISH within the region of the brain where expression is controlled by SBE2. The statistical significance between data sets was examined by Fisher’s Exact Tests, * < 0.05, ** < 0.01. (G) Fold change of Shh expression determined by qRT-PCR of cDNA made from E11.5 heads. Mutants from each of ΔCTCF lines are compared to wildtype litter mates. Each dot represents a single embryo and data is graphed as mean +/- SD. The statistical significance between data sets was examined using unpaired Student t-tests.
Similarly, no ectopic Shh expression is detected in motor neurons driven by Mnx1 enhancers in the TAD beyond ZRS/Lmbr1 (Lee et al., 2004) (Fig. 6A), and Mnx1 was not expressed ectopically in any of the normal sites of Shh expression in embryos carrying homozygous deletions of CTCF3, 4 or 5 (Fig. 6E). These findings indicate that despite the alterations to Shh TAD architecture and chromosome conformation, enhancer/promoter specificity is maintained in the ΔCTCF embryos and that, in the absence of these CTCF sites, there is no cross-talk across TAD boundaries resulting in ectopic expression driven by Shh enhancers.
In situ hybridisation is good for determining spatial expression patterns but is at best a semi-quantitative technique. Therefore, we used RNA FISH to detect nascent Shh transcripts in regions of the developing brain in order to quantify the number of expressing alleles in individual cells. (Representative images are shown in Fig. S5A.) In a region where Shh expression is driven by SBE2 (Z Crane-Smith, pers. com.) we detect a small but significant reduction in the percentage of Shh expressing alleles in embryos carry homozygous deletions of CTCF2, 3 and 5 (Fig. 6F). However, when we examine RNA by qRT-PCR from entire heads where expression is be controlled by multiple enhancers, no significant changes in Shh mRNA levels are observed (Fig. 6G).
Shh expression in the limb bud driven by ZRS lasts only 48 hours from initiation to down regulation. qRT-PCR performed from limb buds at E10.5 can detect changes in expression in response to deletions within the Shh TAD (Paliou et al., 2019). However, we detect no differences in the percentage of expressing Shh alleles by RNA FISH on E11.5 developing limb buds (Fig. S5B) and qRT-PCR shows no significant differences in Shh mRNA levels expression levels, with the exception of ΔCTCF5 mutants (Fig. S5C).
Mice heterozygous for a Shh null allele express only 50-60% wild type levels of Shh in the limb bud but develop normally and in fact in the limb Shh levels must fall to about 20% of wildtype before development is perturbed and digits are lost (Lettice et al., 2017). As the TAD boundary moves beyond the 5’ end of Shh in ΔCTCF1 cells, arguably separating the coding region from its enhancers, we also made compound heterozygotes carrying both the ΔCTCF1 and Shh null alleles to uncover subtle effects on Shh expression. These ShhΔCTCF1/- mice develop normally and are viable and fertile, further suggesting that deletion of CTCF1 results in no deleterious changes in Shh expression.
A 35kb deletion that removes the Lmbr1 promoter and TAD boundary disrupts chromatin conformation with no deleterious phenotype
Deletion of CTCF1 3ʹ of Shh showed that this position was important for the TAD boundary location and for Shh physical proximity with its regulatory domain (Fig. 3 & S3), but the loss of this site had no apparent phenotypic consequence (Fig. 6). In a similar manner, deleting CTCF5 at the Lmbr1 TAD boundary affected the Shh TAD boundary location but resulted in a minimal loss of proximity between ZRS and Shh, at least in E11.5 limb tissue (Fig. 4 & S4 and 5). Loss of TAD boundary regions can result in the merging of adjacent TADs and the ectopic activation of genes in one TAD by enhancers in the other merged TAD, with phenotypic consequences (Fabre et al., 2017; Lupiáñez et al., 2015). However, this involved the deletion of sizeable stretches of DNA across the boundaries in question, tens of kilobases rather than individual CTCF sites. In addition to CTCF binding sites, a number of features are found enriched at TAD boundaries including those associated with active promoters (Dixon et al., 2012). To determine if a more extensive deletion across the Lmbr1 boundary results in the merging of adjacent TADs, a homozygous 35kb deletion (Δ35) was generated in mice which removed CTCF4 and CTCF5 and covering a region containing the first two exons of Lmbr1 and 13kb upstream (Fig. 1A). RT-PCR in mouse embryos confirmed that this deletion eliminates transcription throughout the 5ʹ end of Lmbr1 in both isolated limb buds and the rest of the body (Fig. S6A).
5C from homozygous Δ35 ES cells showed that this deletion caused relocation of the TAD boundary a further ~60kb 5ʹ of the Lmbr1 promoter towards the promoter of Nom1 rather than a merging of the adjacent TADs (Fig. 7A and S6B & C). Interactions within the region extending from CTCF3 to Nom1 are enriched in Δ35 cells compared to wild type and the CTCF3/ZRS genomic region gains interactions into the adjacent TAD up to Mnx1 (arrowheads). Nom1 loses interactions within its own TAD (arrows) (Fig. 7A and S6B & C). Consistent with this, 3D-FISH (Fig. 7B) showed that distances between ZRS and Shh, SBE2 and Mnx1 were all significantly decreased in Δ35 (Fig. 7C and Table S8). The reduced spatial distance between ZRS and Mnx1 was not due to reduction of the linear genomic distance caused by the 35kb deletion, as similar effects were seen in cells carrying an inversion of this 35kb of DNA (Fig. 7C).
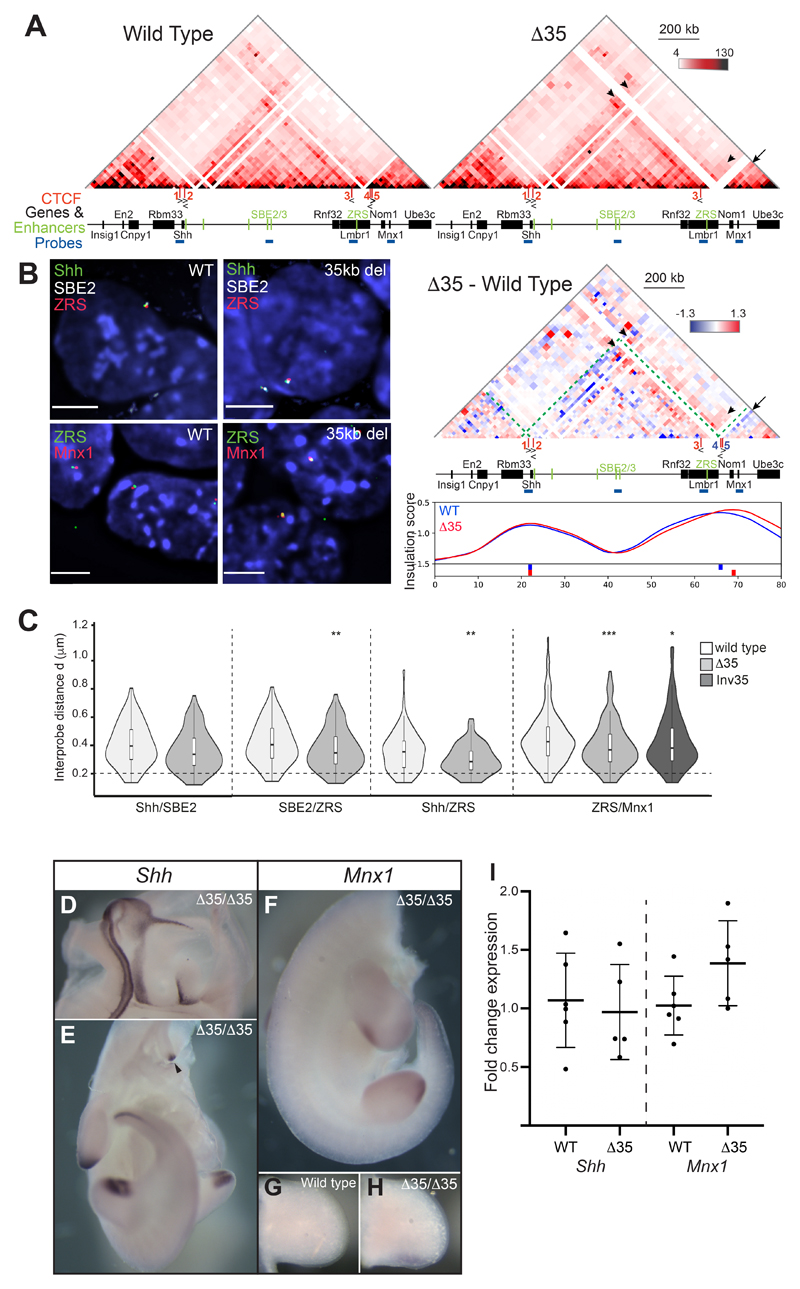
Figure 7. Chromosome conformation and gene expression as a consequence of a 35-kb deletion (Δ35) at the Lmbr1 TAD boundary.
(A) 5C heat maps from wild type and Δ35 ESCs. Below is a heat map comparing Δ35 5C enrichment (red) with wild type (blueGreen dashed lines indicate the Shh TAD boundaries, black arrows highlight loss of interactions in Δ35 cells and black arrowheads indicate enriched interactions in Δ35 cells. At the bottom are insulation score graphs that identify the location of TAD boundaries using raw summed 5C matrices. Peaks of insulation are shown below the plots, Δ35 (red) and wild type (blue). Data for biological replicates are in Supplemental Fig. S6. (B) Images of representative nuclei from wild type and Δ35 ESCs showing FISH signals for Shh/SBE2/ZRS and ZRS/Mnx1 probes. Scale bars = 5 μm. (C) Violin plots show the distribution of interprobe distances (μm) between Shh/SBE2, SBE2/ZRS, Shh/ZRS and ZRS/Mnx1 probes in wild type and Δ35 ESCs, and ZRS/Mnx1 distances in 35kb inversion ESCs. Horizontal dashed line shows the proportion of alleles that are colocalised (< 200 nm). The statistical significance between data sets was examined by Mann-Whitney U Tests, * < 0.05, ** < 0.01, ** < 0.001. (D & E) In situ hybridisations showing normal Shh expression in a bisected head and body, respectively, of a Δ35/Δ35 E11.5 embryo. (F-H) In situ hybridisations for Mnx1 in a Δ35/Δ35 homozygote (F) and limb bud (H) and for comparison lower levels of staining in a wildtype limb bud is shown in (G). The staining in (G) and (H) was stopped before wildtype signal was apparent to highlight Mnx1 up-regulation. (I) Fold change of Shh and Mnx1 expression determined by qRT-PCR of cDNA made from E11.5 limb buds. Mutants Δ35/Δ35 embryos are compared to wildtype litter mates. Each dot represents a single embryo and data is graphed as mean +/- SD. The statistical significance between data sets was examined using unpaired Student t-tests.
Deletions of the Shh TAD boundary at the Lmbr1 promoter relocates the boundary to the promoter of Nom1, and the ZRS has enhanced ability to contact sequences both within its own TAD and the Mnx1 TAD. Despite these differences, Δ35 homozygous mice were viable, fertile and had no apparent phenotype. The Shh expression pattern is also indistinguishable from wild type (Fig. 7D & E) - in particular midline expression is detected in the floor plate and notochord as one stripe down the body (Fig. 7E, arrow head), with no evidence for expression as two more lateral stripes driven by Mnx1 motor neuron enhancers (Lee et al., 2004) (as seen in Fig. 6E). Because of the changes in interactions across the TAD boundary observed in Δ35, we made compound heterozygotes with the Shh null chromosome to highlight subtle changes in expression. Even in this sensitised Shh background, compound ShhΔ35/- mice are phenotypically normal.
Interestingly, given the decreased distances measured by FISH between ZRS and Mnx1, in situ hybridisations indicate that the limb expression of Mnx1 is increased in ShhΔ35/Δ35 embryos in comparison to wild-type embryos (Fig. 7F-H). This is supported by qRT-PCR which detects a modest, but not significant, up regulation of Mnx1 expression in limb buds while Shh expression is unchanged. These data suggest the deletion of 35kb encompassing the TAD boundary, enhances the ability of the Mnx1 promoter to respond to the ZRS. However, no upregulation of Mnx1 expression is seen in the pharyngeal endoderm and developing lungs which would be driven by the enhancers neighbouring ZRS, MACS1 and MFCS4 (Fig. 1A) (Sagai et al., 2009).
Discussion
A systematic genetic approach to delete individual CTCF sites, and to delete or invert large regions, including those encompassing a TAD boundary, has enabled us to use chromosome conformation capture and imaging to assay the resulting perturbations to chromosome organisation within the Shh regulatory TAD, and between this and neighbouring TADs. Analysing CTCF deletions, we detected little or no disruption to gene regulation during embryonic development and no detectable phenotype in animals that can be attributed to this altered chromosome conformation. While deletions of the majority of DNA responsible for the interactions detected by 5C (Δ700) had no effect on boundary formation or activity of a distant enhancer.
ZRS activity is not distance dependent
5C analysis confirmed that TAD boundaries were unaffected by removal of most of the internal region of the Shh TAD (Δ700) (Fig. 1 & S1), with Shh and its remaining enhancers still located within the same, but smaller, TAD. This large deletion did cause extensive disruption to the developing embryo, mainly, it can be assumed, due to the loss of several known forebrain and epithelial enhancers within the deleted region. However, even in embryos homozygous for the 700kb deletion, which relocates ZRS to less than 100kb distant from Shh, ZRS function is maintained with no detrimental effects on limb bud-specific Shh activation and normal development of the limbs occurs. Therefore, the large genomic distance from Shh is not intrinsic to the function of the ZRS. This is in contrast to the loss of interactions following similar perturbations between a limb-specific enhancer and Hoxd13 that resulted in loss of Hoxd13 activity (Fabre et al., 2017).
Loss of CTCF sites at the Shh TAD boundaries disrupts chromatin architecture, and impacts Shh/ZRS spatial proximity
We have previously shown that Shh and ZRS are in spatial proximity (~300nm) in the early embryo in both expressing limb tissue and the non-expressing adjacent flank (Williamson et al., 2016). Here, using 5C on cells dissected from E11.5 anterior and posterior limb buds we show that this is driven by an interaction between the sites 3ʹ and 5ʹ of Shh (containing CTCF1 and CTCF2 sites) and a region within intron 5 of Lmbr1 about 20kb from ZRS (CTCF3) (Fig. 2 & S2). This loop is also present in ESCs, and spatial proximity of Shh and ZRS is lost upon the deletion of any one of the three CTCF sites in both ESCs and E11.5 limb bud tissue (Fig. 3, 4, 5, S3 & S4). Deleting CTCF sites at the Lmbr1 promoter TAD boundary (ΔCTCF4 and ΔCTCF5) had a lesser effect on Shh/ZRS spatial proximity. Increased inter-probe distances between either Shh or ZRS and the forebrain enhancer SBE2 located at the centre of the TAD suggest that the loss of spatial proximity may be due to a general decompaction throughout the TAD, rather than a loss of interactions which could be detected by 5C.
Shh responds to its developmental enhancers regardless of TAD disruption
5C analysis in ESCs suggests that the disruption caused by CTCF site deletions can remove Shh from its regulatory TAD (ΔCTCF1) or re-enforces contacts within sub-TAD domains such that the Shh forebrain enhancers are sequestered in one and ZRS and the long-range epithelial enhancers in the other, with a loss of interactions between both sub-TADs and either sub-TAD with Shh (ΔCTCF2 and ΔCTCF3). Nevertheless, in all of these configurations, the expression pattern of Shh during embryonic development appears to be normal and mRNA levels are largely unchanged with the resulting mice having no detectable phenotype. This indicates that communication between Shh and its extensive set of developmental enhancers is remarkably robust to TAD perturbation.
Ectopic expression across disrupted TAD boundaries is not common
Loss of CTCF1 not only moves the TAD boundary ~60kb to beyond the 5ʹ end of Shh but also enables greater interactions between Shh and the adjacent TAD which contains other genes and their enhancers active during brain development, but in a pattern distinct from Shh. En2 is expressed at the mid-hindbrain boundary, a pattern at least partly dependent on an enhancer binding Pax2/5/8 (Li Song and Joyner, 2000). Similarly, Cnpy1 expression at the mid-hindbrain boundary is thought to be important for FGF signalling (Hirate and Okamoto, 2006). Despite increased chromatin interactions over the Shh TAD boundary in ΔCTCF1, there is no ectopic expression of Shh in the mid-hindbrain driven by the En2/Cnpy1 enhancers and, vice versa, there is no ectopic expression of En2/Cnpy1 at sites driven by Shh enhancers (Fig. 6).
The Lmbr1 TAD boundary has been suggested to be less precise than that at the Shh end of the TAD from both a structural and regulatory point of view (Anderson et al., 2014 Symmons et al., 2016). Deletion of CTCF5 weakened the boundary of the neighbouring Mnx1 TAD and increased proximity between ZRS and Mnx1 was detected in ΔCTCF4. However, in neither case was there evidence for enhanced expression of Mnx1 – e.g. in limb buds driven by ZRS – beyond that detected in wild-type embryos. Interestingly, even in wildtype situations, Mnx1 has a weak expression domain concomitant with the limb bud ZPA, suggesting that this gene may be influenced by ZRS activity emanating from the adjacent TAD. Nor was there evidence of the Mnx1 motor-neuron enhancer (Zelenchuk and Brusés, 2011) driving expression of Shh in motor neurons of the developing neural tube in any of the mutant embryos.
A larger (35kb) deletion of this boundary removing CTCF4, CTCF5 and the promoter/first two exons of Lmbr1, enhanced ZRS contacts across both the Shh TAD and into the neighbouring Mnx1 TAD (Fig. 7 & S6). Increased Mnx1 expression in the ZPA of embryos homozygous for the 35kb deletion suggests that the potentially increased contacts between Mnx1 and ZRS identified in ESCs could be enabling greater activation of this gene by the Shh limb enhancer.
Perturbations of the Shh TAD boundaries can negatively impact on gene-enhancer co-localisation but are insufficient to cause a deleterious phenotype
It is commonly held that enhancer driven gene-activation requires ‘contact’ or very close apposition of the enhancer and promoter. Inversions encompassing the Shh TAD boundaries that disrupted TAD integrity and significantly increased the genomic distance between Shh and ZRS result in severe limb malformations, suggesting that these rearrangements prevent ZRS from contacting/regulating the Shh promoter (Symmons et al., 2016). These data and our 5C and FISH analyses which show that the Shh TAD forms a compact, discrete regulatory hub (Williamson et al., 2016) suggest that 3D organisation of the Shh TAD could allow distal enhancers to come into close proximity to selectively regulate Shh expression. However, in the functionally relevant cells of the limb bud ZPA, ZRS co-localisation (<200 nm) with Shh was reduced to levels of the non-expressing distal anterior levels in ΔCTCF1, ΔCTCF2 and ΔCTCF3 homozygous embryos without adversely affecting Shh expression (Fig. 6) and with no subsequent phenotypic effects. This is consistent with evidence showing reduced Shh neural enhancer-promoter co-localisation in expressing cells and tissues (Benabdallah et al., 2019).
All embryos homozygous for one of the five CTCF binding domain deletions or the 35kb deletion of the Lmbr1 boundary, developed normally and were able to reproduce. Moreover, sufficient Shh expression was maintained for compound heterozygote embryos carrying either ΔCTCF1 or the 35kb deletion opposite a Shh null allele to have no abnormal phenotype. A contemporaneous study on the same genomic territory has largely re-capitulated these results – deletions of Lmbr1 CTCF sites and the gene promoter caused perturbations to local chromatin conformation but Shh expression, although reported to be reduced, was enough to drive normal limb development (Paliou et al., 2019). The Shh regulatory landscape is set up to ensure optimal activation of the gene and here we have shown that this is robust to perturbations of TAD integrity and structure. Similarly, recent work on the the Sox9-Kcnj2 locus suggests that even manipulations that result the fusion of neighbouring TADs have no major effects on gene expression (Depang et al., 2019). However, large-scale disruptions incorporating boundaries which cause TADs to merge do result in developmental defects (Lupiáñez et al., 2015).
Our data suggest that CTCF binding has a role in TAD structure and loss of sites perturbs internal interactions and the position of boundaries. However, at the Shh locus these major disruptions have no effect on gene expression patterns and little effect on expression levels. We speculate that the largely unvarying organisation of TADs could have provided the necessary stable genomic environment for the accumulation of regulatory elements over evolutionary time rather than being essential for target gene activation.
Materials and Methods
Cell Culture and CRISPR/cas9 mediated deletions
E14TG2A mouse embryonic stem cells (ESCs) (a kind gift from Austin Smith)were cultured under standard conditions (Anderson et al., 2014). CRISPR guides were made by cloning annealed oligos (Table S1) into pSpCas9(BB)-2A-GFP (PX458), a gift from Feng Zhang (Addgene plasmid # 48138 ; http://n2t.net/addgene:48138; RRID:Addgene_48138). 2μg of vector DNA were transfected into 8x105 ESCs using Lipofectamine 2000 (ThermoFisher) following the manufacturer’s instructions. After 48 hours, GFP positive cells were sorted by FACS and plated at low density. Ten days later, individual clones were picked and screened for correct deletion by PCR and Sanger sequencing (primers are listed in Table S1).
Mouse lines and embryo analysis
The ShhΔ700 deletion was created by crossing the line SBLac96 (Anderson et al., 2014) to a line carrying a pCAGGS-Cre recombinase gene (Araki et al., 2006). With the exception of the Δ35kb and Inv35kb mouse lines, which were made by injection of the ESCs in to blastocysts, all of the other mouse lines were created as in Lettice et al., (2017) by direct microinjection into C57Bl6/CBA F2 zygotes of the same guides as were used in ESCs. Resultant G0 mice are screened by PCR using flanking primers (Supp Table 1) and the deletions confirmed by Sanger sequencing. Lines were then established by crossing founder mice to C57Bl6 wildtypes. LacZ expression analysis, in situ hybridisations and RT-PCR reactions were conducted as in Anderson et al. (2014).
All mouse work has been ethical approved by the University of Edinburgh Animal Welfare and Ethics Review board and is conducted under the authority of Home Office Licences.
qRT-PCR
Heads and limb buds were dissected from individual E11.5 embryos, snap frozen in separate tubes and stored at -80°C. The rest of the embryo was used to make DNA and the genotype established by PCR using the primers listed in Table S1. RNA was extracted from the tissues of mutant and wildtype embryos using TRIzol (Invitrogen) and first strand cDNA synthesised with a Transcriptor First strand cDNA synthesis kit (Roche) following the manufacturers’ instructions. qRT-PCR was run on an LC480 lightcycler (Roche) and made use of the Universal ProbeLibrary (probe 32 for Shh, probe 60 for Mnx1 and the Universal ProbeLibrary Mouse GAPD Gene Assay). PCR primers are listed in Table S2. Each gene was assayed in separate wells and each sample run in triplicate. Gene expression data was analysed by the del del Ct method with mutants from each line compared to their wildtype littermates. (n equals between 3 and 9 embryos.) Students’ unpaired t-test was used for statistical validations.
DNA FISH
E11.5 embryos were collected, fixed, embedded, sectioned, antibody stained for SHH expression and processed for FISH as previously described (Morey et al., 2007, Lettice et al., 2014), except that sections were cut at 8 μm. Regions expressing Shh were identified by antibody staining with an anti-Shh antibody (Ab86462, Batch GR182460-5, Abcam. This antibody has been shown to give the correct expression pattern in immunostaining- see https://www.abcam.com/sonic-hedgehog-antibody-rm0128-4a37-ab86462.html). Fosmid clones (Fig. 1A, Table S3) were prepared and labelled as previously described (Morey et al., 2007). Between 160-240 ng of biotin- and digoxigenin-labelled fosmid probes were used per slide, with 16-24 μg of mouse Cot1 DNA (Invitrogen) and 10 μg salmon sperm DNA. For 4-colour FISH, similar quantities of the additional fosmid was labelled with either Green496-dUTP (Enzo Life Sciences) or red-dUTP (Alexa Fluor™ 594-5-dUTP, Invitrogen).
For 3D FISH on ESCs, 1x106 cells were seeded on slides for overnight. Cells were fixed in 4% paraformaldehyde (pFA) for 10 mins at room temperature and then permeabilized using 0.5% TritonX for 10 mins (Eskeland et al., 2010).
RNA FISH
Custom Stellaris® RNA FISH Probes were designed against Shh nascent mRNA (pool of 48 unique 22-mer probes) by utilizing the Stellaris® RNA FISH Probe Designer (Biosearch Technologies, Inc., Petaluma, CA) available online at www.biosearchtech.com/stellarisdesigner (version 4.2). The slides were hybridized with the Shh Stellaris FISH Probe set labelled with Quasar 570 (Biosearch Technologies, Inc.), following the manufacturer’s instructions available online at www.biosearchtech.com/stellarisprotocols. Briefly, FFPE tissue sections from E11.5 embryos were deparaffinised in xylene, hydrated in ethanol and permeabilised in 70% ethanol overnight at 4°C. Slides were incubated in 10 μg/mL proteinase K in 1X PBS for 20 minutes at 37°C followed by washes in 1X PBS and wash buffer (2X SSC, 10% deionised formamide). Shh RNA FISH probes were diluted in Stellaris RNA FISH hybridisation buffer (#SMF-HB1-10) to 125 nM and hybridised to slides overnight in humidified chamber at 37°C. Slides were washed 2 x 30 minutes in wash buffer (2X SSC, 10% deionised formamide) at 37°C, counterstained with 5 ng/mL DAPI, washed in 1X PBS and mounted in Vectashield.
Image analysis
Slides were imaged using a Photometrics Coolsnap HQ2 CCD camera and a Zeiss AxioImager A1 fluorescence microscope with a Plan Apochromat 100x 1.4NA objective, a Nikon Intensilight Mercury based light source (Nikon UK Ltd, Kingston-on-Thames, UK) and either Chroma #89014ET (3 colour) or #89000ET (4 colour) single excitation and emission filters (Chroma Technology Corp., Rockingham, VT) with the excitation and emission filters installed in Prior motorised filter wheels. A piezoelectrically driven objective mount (PIFOC model P-721, Physik Instrumente GmbH & Co, Karlsruhe) was used to control movement in the z dimension. Step size for z stacks was set at 0.2 μm. Hardware control, image capture and analysis were performed using Nikon Nis-Elements software (Nikon UK Ltd, Kingston-on-Thames, UK). Images were deconvolved using a calculated point spread function with the constrained iterative algorithm of Volocity (Perkinelmer Inc, Waltham, MA). Image analysis was carried out using the Quantitation module of Volocity (Perkinelmer Inc, Waltham, MA). For DNA FISH, only alleles with single probe signals were analysed to eliminate the possibility of measuring sister chromatids.
3C library preparation
Limbs buds and bodies (with the limbs and heads removed) from wild type embryos, and entire ShhΔ700/Δ700 embryos were dissected at E11.5 and the tissue dissociated by pipetting in just enough PBS to cover them. The cells were fixed with 1% formaldehyde for 10 min at room temperature. For ESCs, 5 x106 – 1 x107 cells were fixed. Crosslinking was stopped with 125 mM glycine, for 5 min at room temperature followed by 15 min on ice. Cells were centrifuged at 400 g for 10 min at 4°C, supernatants removed, and cell pellets flash frozen on dry ice before storage at -80°C.
Cell pellets were treated as previously described (Dostie and Dekker, 2007; Ferraiuolo et al., 2010; Williamson et al., 2014). HindIII-HF (NEB) was the restriction enzyme used to digest the crosslinked DNA.
5C primer and library design
5C primers covering the Usp22 (mm9, chr11: 60,917,307-61,003,268) and Shh regions (mm9, chr5: 28,317,087-30,005,000) were designed using 'my5C.primer' (Lajoie et al., 2009) with the following parameters: optimal primer length of 30 nt, optimal TM of 65°C, default primer quality parameters (mer:800, U-blast:3, S-blasr:50). Primers were not designed for large (>20 kb) and small (<100 bp) restriction fragments, for low complexity and repetitive sequences, or where there were sequence matches to >1 genomic target. The Usp22 region was used to assess the success of each 5C experiment but was not used for further data normalization or quantification.
The universal A-key (CCATCTCATCCCTGCGTGTCTCCGACTCAG-(5C-specific)) and the P1-key tails ((5C-specific)-ATCACCGACTGCCCATAGAGAGG) were added to the Forward and Reverse 5C primers, respectively. Reverse 5C primers were phosphorylated at their 5ʹ ends. An alternating design consisting of 365 primers in the Shh region (182 Forward and 183 Reverse primers) was used. Primer sequences are listed in Table S9.
5C library preparation
5C libraries were prepared and amplified with the A-key and P1-key primers as described in (Fraser et al., 2012). Briefly, 3C libraries were first titrated by PCR for quality control (single band, absence of primer dimers, etc.), and to verify that contacts were amplified at frequencies similar to that usually obtained from comparable libraries (same DNA amount from the same species and karyotype) (Dostie and Dekker, 2007; Dostie et al., 2007; Fraser et al., 2010). We used 1 - 10 μg of 3C library per 5C ligation reaction.
5C primer stocks (20 μM) were diluted individually in water on ice and mixed to a final concentration of 2 nM. Mixed diluted primers (1.7 μl) were combined with 1 μl of annealing buffer (10X NEBuffer 4, New England Biolabs Inc.) on ice in reaction tubes. 1.5 μg salmon testis DNA was added to each tube, followed by the 3C libraries and water to a final volume of 10 μl. Samples were denatured at 95°C for 5 min and annealed at 55°C (48°C ESCs) for 16 hours. Ligation with Taq DNA ligase (10 U) was performed at 55°C (48°C ESCs) for one hour. One tenth (3 μl) of each ligation was then PCR-amplified individually with primers against the A-key and P1-key primer tails. We used 26 cycles based on dilution series showing linear PCR amplification within that cycle range. The products from 3 to 5 PCR reactions were pooled before purifying the DNA on MinElute columns (Qiagen).
5C libraries were quantified by bioanalyser (Agilent) and diluted to 26 pmol (for Ion PGM™ Sequencing 200 Kit v2.0). One microlitre of diluted 5C library was used for sequencing with an Ion PGM™ Sequencer. Samples were sequenced onto Ion 316™ Chips following the Ion PGM™ Sequencing 200 Kit v2.0 protocols as recommended by the manufacturer (Life TechnologiesTM).
5C data analysis
Analysis of the 5C sequencing data was performed as described in (Berlivet et al., 2013). The sequencing data was processed through a Torrent 5C data transformation pipeline on Galaxy (https://main.g2.bx.psu.edu/). Before normalizing, interactions between adjacent fragments were removed due to the high noise: signal ratio likely to occur here. Average read count values over 21kb bins were calculated from the raw sequencing data and 5C data were further processed for visualization. First, the matrices were normalized to sum up to 50,000 reads (excluding the first two diagonals of the matrix). Then adaptive coarsegraining of the matrices was performed to reduce noise using cooltools.numutils.adaptive_coarsegrain, with the three lowest coverage bins masked and cutoff of 10 reads. Level of coarsegraining for all ES cell matrices was determining using the merged wild type data to ensure identical bin sizes across conditions. For comparison of 5C matrices across conditions, we additionally performed observed/expected normalization by dividing each diagonal of the matrix by its mean. For high resolution, zoomed in, (15kb) heatmaps in Fig. 2 & S2 raw data was used, with comparison zoomed in heatmaps normalised to total read count of compared limb anterior and posterior tissue samples. All 5C heatmaps in the figures contain the summed read counts of at least two biological replicates apart from E11.5 embryos in Fig. 1, each individual replicates are shown in supplemental figures associated with the main figures The number of total reads and of used reads is provided for each experiment in Table S10. 5C datasets have been uploaded to NCBI GEO under accession number GSE135840.
For insulation score analysis, we used raw 5C data without coarsening and applied cooltools.numutils._insul_diamond_dense to it with window=25 and without normalization by median. The curves were further smoothed using LOWESS implementation from statsmodels.nonparametric.smoothers_lowess.lowess with frac=0.2, and plotted after inversion, since in raw insulation score valleys correspond to peaks of insulation, and peaks are easier to interpret visually. cooltools.lib.peaks.peakdet was used to determine location of peaks in inverted smoothed data with prominence of at least 0.2, and they were shown below the plots.
Supplementary Material
Summary statement.
Genetic manipulation of the Shh TAD results in chromatin organisation changes without effects on expression patterns or phenotypes. We suggest that Shh developmental regulation is robust to TAD perturbations.
Acknowledgements
We thank the staff of the IGMM advanced imaging resource and technical services for their assistance with imaging and sequencing. We would also like to thank Lorraine Rose, Kyle Davies and the staff at the BRF/Evans Building for expert technical assistance. We thank Maxim Imakaev for help with coarsegraining.
L.K and E.A were funded by PhD studentship from the UK Medical Research Council (MRC). IMF is funded by a PhD studentship from the Darwin Trust. Work in the WAB lab is funded by an MRC University Unit grant MC_UU_00007/2 and in the REH lab by an MRC University Unit grant MM_UU_00007/8.
References
- Anderson E, Hill RE. Long range regulation of the sonic hedgehog gene. Curr Opin Genet Dev. 2014;27:54–59. doi: 10.1016/j.gde.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Anderson E, Devenney PS, Hill RE, Lettice LA. Mapping the Shh long-range regulatory domain. Development. 2014;707 doi: 10.1242/dev.108480. 108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Araki M, Miyazaki J, Vassalli P. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci. 2006;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. Decreased enhancer-promoter proximity accompanying enhancer activation. Mol Cell. 2019 doi: 10.1016/j.molcel.2019.07.038. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of HoxA Genes in Developing Limbs. PLoS Genet. 2013;9:e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M, Mundlos S, Ibrahim DM. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet. 2019;51:1263–1271. doi: 10.1038/s41588-019-0466-z. [DOI] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- Dostie J, Zhan Y, Dekker J. Current Protocols in Molecular Biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2007. Chromosome Conformation Capture Carbon Copy Technology; pp. 21.14.1–21.14.13. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi I, Wutz A, et al. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PJ, Leleu M, Mormann BH, Lopez-Delisle L, Noordermeer D, Beccari L, Duboule D. Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol. 2017;18:149. doi: 10.1186/s13059-017-1278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, Wang XQD, Nadler M, Blanchette M, Dostie J. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38:7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan W-L, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Fraser J, Rousseau M, Blanchette M, Dostie J. Computing Chromosome Conformation. Humana Press; Totowa, NJ: 2010. pp. 251–268. [DOI] [PubMed] [Google Scholar]
- Fraser J, Ethier SD, Miura H, Dostie J. A torrent of data: Mapping chromatin organization using 5C and high-throughput sequencing. Methods Enzymol. 2012;513:113–141. doi: 10.1016/B978-0-12-391938-0.00005-7. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten S, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Okamoto H. Canopy1, a novel regulator of FGF signaling around the midbrain-hindbrain boundary in zebrafish. Curr Biol. 2006;16:421–427. doi: 10.1016/j.cub.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Funahashi J, Ruiz EC, Pfaff SL. Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Williamson I, Devenney PS, Kilanowski F, Dorin J, Hill RE. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141:1715–25. doi: 10.1242/dev.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Devenney P, De Angelis C, Hill RE. The Conserved Sonic Hedgehog Limb Enhancer Consists of Discrete Functional Elements that Regulate Precise Spatial Expression. Cell Rep. 2017;20:1396–1408. doi: 10.1016/j.celrep.2017.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Song D, Joyner AL. Two Pax2/5/8-binding sites in Engrailed2 are required for proper initiation of endogenous mid-hindbrain expression. Mech Dev. 2000;90:155–165. doi: 10.1016/s0925-4773(99)00242-7. [DOI] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–19. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science (80-) 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum L, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-L, Dekker J, Mirny LA, Bruneau Correspondence BG, Gibcus JH, Uebersohn A, Abdennur N, Bruneau BG. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–933.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliou C, Guckelberger P, Schöpflin R, Heinrich V, Esposito A, Chiariello AM, Bianco S, Annunziatella C, Helmuth J, Haas S, et al. Preformed Chromatin Topology Assists Transcriptional Robustness of Shh during Limb Development. Proc Natl Acad Sci U S. 2019;116:12390–12399. doi: 10.1073/pnas.1900672116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, Nguyen Huynh TH, Ozadam H, Dekker J, Duboule D. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 2017;31:2264–2281. doi: 10.1101/gad.307769.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE Data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sagai T, Amano T, Tamura M, Mizushina Y, Sumiyama K, Shiroishi T. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136:1665–1674. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, H Haering C, Mirny L, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional Analysis of CTCF During Mammalian Limb Development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Sun F, Chronis C, Kronenberg M, Chen XF, Su T, Lay FD, Plath K, Kurdistani SK, Carey MF. Promoter-Enhancer Communication Occurs Primarily within Insulated Neighborhoods. Mol Cell. 2019;73:250–263.e5. doi: 10.1016/j.molcel.2018.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, Spitz F. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev Cell. 2016;39:529–543. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pekowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. Spatial genome organization: Contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Lettice LA, Hill RE, Bickmore WA. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development. 2016;143:2994–3001. doi: 10.1242/dev.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenchuk TA, Brusés JL. In Vivo labeling of zebrafish motor neurons using an mnx1 enhancer and Gal4/UAS. Genesis. 2011;49:546–554. doi: 10.1002/dvg.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.