Abstract
Nucleoporins (NUPs) are an essential component of the nuclear-pore complex, which regulates nucleocytoplasmic transport of macromolecules. Pathogenic variants in NUP genes have been linked to several inherited human diseases, including a number with progressive neurological degeneration. We present six affected individuals with bi-allelic truncating variants in NUP188 and strikingly similar phenotypes and clinical courses, representing a recognizable genetic syndrome; the individuals are from four unrelated families. Key clinical features include congenital cataracts, hypotonia, prenatal-onset ventriculomegaly, white-matter abnormalities, hypoplastic corpus callosum, congenital heart defects, and central hypoventilation. Characteristic dysmorphic features include small palpebral fissures, a wide nasal bridge and nose, micrognathia, and digital anomalies. All affected individuals died as a result of respiratory failure, and five of them died within the first year of life. Nuclear import of proteins was decreased in affected individuals’ fibroblasts, supporting a possible disease mechanism. CRISPR-mediated knockout of NUP188 in Drosophila revealed motor deficits and seizure susceptibility, partially recapitulating the neurological phenotype seen in affected individuals. Removal of NUP188 also resulted in aberrant dendrite tiling, suggesting a potential role of NUP188 in dendritic development. Two of the NUP188 pathogenic variants are enriched in the Ashkenazi Jewish population in gnomAD, a finding we confirmed with a separate targeted population screen of an international sampling of 3,225 healthy Ashkenazi Jewish individuals. Taken together, our results implicate bi-allelic loss-of-function NUP188 variants in a recessive syndrome characterized by a distinct neurologic, ophthalmologic, and facial phenotype.
Keywords: nucleoporin, NUP188, dendritic arborization, negative geotaxis, nuclear pore complex, Ashkenazi Jewish, congenital cataracts, microcephaly, congenital heart defects, autosomal recessive
Introduction
The nuclear pore complex (NPC) is a massive channel that spans the nuclear envelope and controls the flow of mRNA and proteins between the cytoplasm and the nucleus. NPCs are macromolecular assemblies composed of ∼30 proteins called nucleoporins (NUPs).1 Although complete loss of nuclear pore function is hypothesized to be incompatible with cell viability, pathogenic variants in a number of nucleoporins resulting in partial loss of pore function have been implicated in inherited human disorders including nephrotic syndromes,2, 3, 4 triple A syndrome (adrenal insufficiency, achalasia, and alacrima with a variable neurologic phenotype),5 fetal akinesia deformation sequence,6 and progressive neurologic disease.7, 8, 9, 10
NUP188 (MIM: 615587) is a scaffold nucleoporin and a NUP93 (MIM: 614351) subcomplex member that forms the inner ring of the NPC.1 Disruption of NUP188 in yeast has been shown to inhibit both mRNA export and protein trafficking through the NPC.11 NUP188 has also been implicated in a diverse range of cellular functions outside of its role in nuclear transport; such functions include ciliogenesis,12 chromatin organization,13 transcriptional regulation,13 and chromosome segregation.14 A rare NUP188 duplication was reported in an individual with heterotaxy, and knockdown of Nup188 disrupted left-right patterning in Xenopus.15
We present a series of six individuals who come from four unrelated families and who show multisystem involvement characterized by cardiorespiratory, neurologic, and ophthalmologic abnormalities along with dysmorphic features due to bi-allelic pathogenic variants in NUP188.
Subjects and Methods
Cohort
Affected individuals were identified in the clinical setting, and pathogenic variants were identified through exome sequencing on a research or clinical basis. GeneMatcher16 facilitated identification of unrelated cases at four institutions. Parents or legal guardians of each individual gave informed consent for participation in this study. The study was performed according to the standards of the ethics committees and the institutional review boards at each center.
Carrier Screening of the Ashkenazi Jewish Population
To determine the carrier frequency of the p.Ile302Valfs∗7 and p.Tyr1048∗ variants in NUP188 within the Jewish population, we screened a total of 4,815 Jewish individuals as follows: 3,225 Ashkenazi individuals, 815 Sephardi individuals, and 775 individuals of mixed Ashkenazi and Sephardic descent; classification was by self-identification. Anonymous blood samples were obtained from worldwide Dor Yeshorim screening program17 locations, including the United States of America (New York, New Jersey, Maryland, California, Illinois, Florida, Ohio, and Michigan), Canada, Mexico, Argentina, Brazil, the United Kingdom, Belgium, France, Switzerland, Austria, Australia, South Africa, and Israel. All participants provided written consent for their samples to be used for research purposes. The consent form noted that patient material would be used for clinical testing and that excess material would be de-identified and used for research purposes to characterize single-gene disorders in the Ashkenazi Jewish population. Genomic DNA was extracted, and a multiplex PCR assay was used for separately targeting each of the two aforementioned NUP188 variants in barcode-indexed samples via high-throughput sequencing on a MiSeq instrument (Illumina). Resultant sequencing reads were aligned to the reference genome (hg38), and relevant NUP188 variants were genotyped via the “emit_all_sites” flag within the GATK UnifiedGenotyper package (Broad Institute). The assay was validated with positive-control blood samples from the heterozygous mother and heterozygous father of affected individual 3 from our cohort.
Cell Culture
Primary fibroblast cultures were established from skin biopsies for individuals 1 and 2, as well as from skin biopsies for two unrelated, unaffected control subjects. Fibroblasts were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. All experiments were performed on cells whose passage number did not exceed 8.
Immunoblotting of Cell Nuclei
To prepare nuclear lysates for immunoblotting, we grew fibroblasts to 80%–90% confluence and performed nuclei isolation as previously described.18 SDS-PAGE and immunoblotting were performed with polyvinylidene fluoride (PVDF) membranes (Millipore) and standard techniques. Blots were probed with anti-NUP188 (1:150, Abcam) and anti-laminin (1:1000, Abcam). All immunoblots were performed in triplicate.
Immunofluorescence Microscopy
Fibroblasts were plated on glass coverslips and grown to 80%–90% confluence before fixation with 2% paraformaldehyde. Immunostaining was performed according to standard protocols. Primary and secondary antibodies were incubated for 1 h at room temperature. The primary antibodies used were anti-NUP188 (1:150, Abcam), anti-actTUB (1:2000, Sigma-Aldrich), and anti-eGFP (1:500, ThermoFisher Scientific). The secondary antibodies used were Alexa 488-conjugated goat anti-rabbit IgG (1:1000, ThermoFisher Scientific) and Alexa 568-conjugated goat anti-mouse IgG (1:1000, ThermoFisher Scientific). DAPI (4’,6-diamidino-2-phenylindole) was used to visualize the nuclei. Fluorescent images were captured using a Zeiss LSM 880 laser scanning microscope equipped with Airyscan technology. All immunostaining was performed in triplicate.
Real-Time PCR
RNA was extracted from fibroblast cell lines with Trizol (Ambion) and the RNA Clean and Concentrator kit (Zymo Research). 500 ng of RNA was reverse transcribed with oligo(dT) nucleotides and SuperScript II (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed with primers for NUP188 (5′-GCACAGCACAGACACCTAGT-3′, forward; and 5′-ACTGCGTTGAGGCACTGTAA-3′, reverse) and GAPDH (5′-CTTTGTCAAGCTCATTTCCTGG-3′, forward; and 5′-TCTTCCTCTTGTGCTCTTGC-3′, reverse).
Nuclear-Import Assay
Nuclear import was assayed with a protocol adapted from yeast.11 A lentiviral vector containing three GFP molecules attached to the SV40 nuclear localization signal (NLS) (pHAGE-EFS-PCP-3XGFPnls) was obtained as gift from Thoru Pederson (Addgene plasmid #75385).19 To generate GFP constructs with no NLSs and to generate the hnRNPA1 NLS, double-stranded DNA oligos (5′-5Phos/GCCGCCGCCTAA-3′ and 5′-5Phos/TCAAATTTTGGACCCATGAAGGGAGGAAATTTTGGAGGCAGAAGCTCTGGCCCCTATTAA-3′, respectively) were cloned into pHAGE-EFS-PCP-3XGFPnls between the XhoI and XbaI sites. Fibroblasts from individual 1, individual 2, and control individuals were infected with GFP-SV40-NLS, GFP-hnRNPA1-NLS, or GFP-no-NLS virus. It was determined via live-cell imaging that on the sixth day after infection, GFP signal was visible in the cytoplasm and nuclei of infected cells. At this point, cells were incubated with Alexa 555-conjugated wheat germ agglutinin to stain cell membranes (ThermoFisher Scientific) and fixed in 2% paraformaldehyde for immunofluorescence microscopy. 2D images of individual cells were manually captured using a Zeiss LSM 880 laser scanning microscope, and images were quantified using ImageJ software. DAPI was used for defining the nuclei, and wheat germ agglutinin was used for defining the cell boundaries. After background subtraction, total GFP fluorescent intensity was measured in the nuclei and cytoplasm of infected cells and normalized to the respective cell compartment area before calculation of the ratio of nuclear to cytoplasmic GFP intensity.
Generation of Nup188KO Flies
Genomic DNA of vas-Cas9 flies was used as a template for PCR of the Nup188 genomic fragments. The fragments upstream of the Nup188 encoding region were amplified with the primers “Nup188 gDNA 5′ forward” (5′-TTGATCTCACGAGCATTTCG-3′), and “Nup188 gDNA 5′ reverse” (5′-GTCCAATCTGCCTTTTCTGC-3′) via PCR. The resulting template sequence was used to select target “Nup188 5′gRNA” (5′−GCACCGATTGGAAGCGCCTGTGG-3′). For gRNA synthesis, “Nup188 5′ sense oligo” (5′-5Phos/CTTCGCACCGATTGGAAGCGCCTG-3′) and “Nup188 5′ antisense oligo” (5′/5Phos/AAACCAGGCGCTTCCAATCGGTGC-3′) were annealed and ligated into a BbsI-digested pU6-BbsI chiRNA vector. The overhang sequences 5′-CTTC-3′ (sense oligo) and 5′-AAAC-3′ (anti-sense oligo) are complementary to the overhangs generated by BbsI digestion. The “Nup188 5′ gRNA” was injected into vas-Cas9 flies, and the resultant potential mutants were screened and verified by PCR.20
Live Imaging of Drosophila Larvae
Live imaging was performed as described.21,22 Embryos were collected for 2–24 h on yeasted grape juice agar plates and were aged at 25°C. At the appropriate time, a single larva was mounted in 90% glycerol under coverslips sealed with grease, imaged with a Zeiss LSM 880 microscope, and returned to grape juice agar plates between imaging sessions.
Behavioral Analyses in Adult Flies
Wild-type (WT) adults and Nup18810B homozygous adult survivors were collected daily from crosses, with males and females stored in separate vials. To prevent confounding effects on locomotor performance, we gave flies 72 h to recover from the CO2 used during handling. Negative geotaxis was tested 3 days after eclosion.23 Males and females from each group were placed in separate 20 cm vials, which were then secured in the geotaxis apparatus. The apparatus was tapped against a table three times so that flies would be knocked to the bottom of their vials. The response of the flies was recorded by video. The average height climbed by the flies in each vial 3 s after tapping was scored via ImageJ. Each vial was assayed 8–10 times and averaged. Each vial contained fewer than 30 flies.
The number of flies undergoing a seizure in each vial in the 10 s after tapping was manually counted. Seizure percentage was calculated as the number of flies experiencing a seizure divided by the total of flies in each vial. One-way ANOVA was used for comparing the average height climbed and percent of flies undergoing a seizure between groups.
Statistical Analysis
All data are expressed as means ± SEM. Student’s t tests were used for comparing differences between two experimental conditions unless otherwise noted; one-way ANOVA was used for comparison of three or more groups. All statistical tests were two sided. Statistical significance was set at p < 0.05. For all animal experiments, n ≥ 3 animals were analyzed for each group. For experiments using cell lines, all experiments were performed in triplicate.
Results
Identification of Bi-allelic NUP188 Variants in Affected Individuals
We identified six individuals with bi-allelic truncating variants in NUP188 (GenBank: NM_015354.3) by exome sequencing; the individuals are from four unrelated families (Table 1, Figure 1, Table S1). Three individuals from two different families are of Ashkenazi Jewish descent, and all three individuals share the same two compound heterozygous variants: c.904_907delATTT (p.Ile302Valfs∗7) and c.3144C>G (p.Tyr1048∗). Two siblings from family 3 have a homozygous variant predicted to cause a frameshift and premature truncation: c.5032dupC (p.Arg1678Profs∗13). Individual 6 has compound heterozygous nonsense variants: c.1890G>A (p.Trp630∗) and c.4078C>T (p.Gln1360∗).
Table 1.
Summary of Clinical Features of Individuals with Bi-allelic Truncating NUP188 Variants
| Family | 1 | 2 | 3 | 4 | ||
|---|---|---|---|---|---|---|
| Individual | 1 | 2 | 3 | 4 | 5 | 6 |
| Variants (NM_015354) | p.Ile302Valfs∗7 | p.Ile302Valfs∗7 | p.Arg1678Profs∗13 (hmz) | p.Trp630∗ | ||
| p.Trp1048∗ | p.Trp1048∗ | p.Gln1360∗ | ||||
| Gender | F | F | F | F | F | F |
| Age at death | 5 months | 5 weeks | 7 months | 2 years 7 months | 2.5 months | 1 month |
| Cause of death | respiratory failure | respiratory failure | respiratory failure | respiratory failure | respiratory failure | respiratory failure |
| Ophthalmologic abnormalities | + | + | + | + | − | + |
| Congenital fetal nuclear cataracts | + | + | + | − | − | + |
| Congenital heart defects | + | + | + | not tested | + | + |
| Respiratory abnormalities | + | + | + | + | + | + |
| Central hypoventilation | + | + | + | − | − | + |
| Neurological abnormalities | + | + | + | + | − | + |
| Hypotonia | + | + | + | + | − | + |
| Seizures | − | − | + | + | − | + |
| Poor oral co-ordination | + | + | + | − | − | − |
| MRI abnormalities | + | + | + | + | + | + |
| Thin corpus callosum | + | + | + | + | + | − |
| Delayed myelination | + | + | − | + | + | − |
| Loss of white matter | − | − | + | − | + | − |
| Ventriculomegaly | + | + | + | + | − | + |
Abbreviations: F, female; hmz, homozygous; +, present; and −, absent.
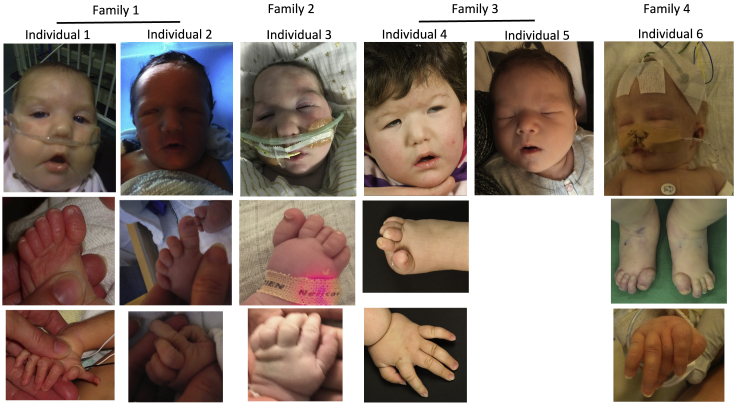
Figure 1.
Facial and Digital Features of Affected Individuals
Note the high anterior hair line, broad forehead, small and downslanting palpebral fissures, wide nasal bridge with bulbous tip, and down-turned corners of the mouth in the facial photographs. The feet are notable for broad halluces, overlapping toes, and camptodactyly. The hand photographs show long gracile fingers and camptodactyly, as well as preaxial polydactyly in individual 1.
The variants identified in families 1 and 2 are reported only in the Ashkenazi Jewish population in the heterozygous state in the gnomAD database: c.904_907delATTT (p.Ile302Valfs∗7) (referred to in gnomAD as c.902_905delTTAT) is present at an allele frequency of 0.075% (n = 5 alleles) and c.3144C>G (p.Tyr1048∗) at 0.040% (n = 4 alleles). The c.5032dupC (p.Arg1678Profs∗13) variant in individuals 4 and 5 is present in a single non-Finnish European, and the two variants in individual 6 are not found in gnomAD. There are no homozygous truncating variants in NUP188 in gnomAD. We screened an additional cohort of 4,815 healthy Jewish individuals for the two pathogenic variants that were restricted to the Ashkenazi Jewish population in gnomAD. Of the 3,225 Ashkenazi-only individuals, four were heterozygous carriers for c.904_907delATTT and five were heterozygous carriers for c.3144C>G, resulting in the following allele frequencies: 0.124% and 0.155%, respectively, in these additional Ashkenazi Jewish individuals. No heterozygous carriers were found among the Sephardi individuals (n = 815) or the individuals of mixed Ashkenazi and Sephardic descent (n = 775).
All six affected individuals have strikingly similar clinical features (Figure 1, Table 1, Supplemental Information). Prenatally, mild ventriculomegaly or suspected brain malformation was noted in 4/6 cases. All individuals were small at birth; their growth parameters measured at <10th percentile for gestational age. Dysmorphic features included a broad nasal bridge with a tubular shaped nose, small palpebral fissures, epicanthal folds, micrognathia, and digital anomalies, including long, gracile fingers, camptodactyly, and broad halluces. Congenital cataracts were present in 4/6 individuals, and an additional individual had vision impairment, poor eye contact, and myopia (−4.25/−1.25 diopters). Congenital heart anomalies, which included bicuspid aortic valve, partial anomalous pulmonary venous return, and patent ductus arteriosus, were observed in 5/6 individuals. All six individuals presented with congenital hypotonia and abnormal brain imaging, showing delayed myelination and white-matter abnormalities. A thin or absent corpus callosum was noted in most cases, and progressive microcephaly with suspicion for a neurodegenerative process was noted in at least three cases. All six individuals died of respiratory failure or respiratory-related illness prior to the age of 3 years. Five of the six affected individuals died within the first seven months of life. The sixth individual lived to 2 years and 7 months; she had severe intellectual disability and was non-ambulatory. Two individuals underwent autopsy. Autopsy findings for individual 2 included diffuse gliosis and patchy depletion of Purkinje cells in the brain, polysplenia, streak ovaries, and lung hyperinflation and congestion. Autopsy for individual 6 revealed streak gonads as well as pulmonary capillary hemangiomatosis.
Full-Length NUP188 Is Undetectable in Fibroblasts from Affected Individuals
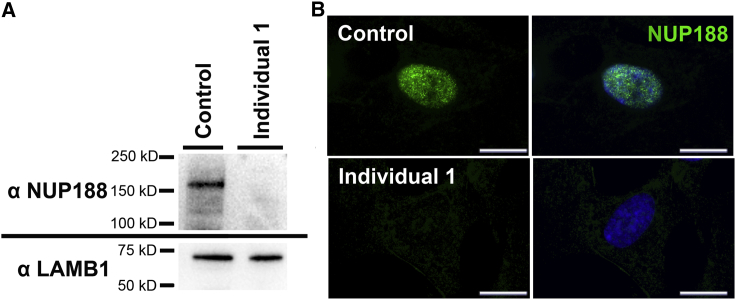
Individuals 1 and 2 had compound heterozygous NUP188 variants, p.Ile302Valfs∗7 and p.Tyr1048∗, that were shared by one other affected individual in our cohort; this individual was also of Ashkenazi Jewish descent. Immunoblot analysis of nuclei isolated from fibroblasts from individuals 1 and 2 and a control individual showed that full-length NUP188 is not present in affected individuals (Figures 2A and S1A). A smaller faint band present in nuclear lysates from individual 2 is most likely non-specific because it is not present in individual 1 or the control individual. Immunofluorescence studies in fibroblasts show that NUP188 localizes to the nucleus of control cells but is absent from cells from affected individuals (Figures 2B and S1B), suggesting that even if truncated protein product is present in affected individuals, it does not localize properly and is unlikely to be functional. Real-time PCR analysis shows that the amount of NUP188 transcript is reduced by 98% in affected individuals compared to controls, suggesting the truncated alleles are targeted by nonsense-mediated decay (Figure S1C).
Figure 2.
NUP188 p.Ile302Valfs∗7 and p.Tyr1048∗ Result in Loss of Full-Length NUP188
(A) Western blot of nuclei isolated from individual 1 and control fibroblasts stained with an antibody against NUP188 reveals full-length NUP188 (~188 kDa) is undetectable in individual 1. LAMB1 is used as a loading control. Individual 1 has compound heterozygous truncating variants NUP188 p.Ile302Valfs∗7 and p.Tyr1048∗. Experiment was performed in triplicate, and representative images are shown.
(B) Immunofluorescent staining of NUP188 in fibroblasts from individual 1 and a control individual. NUP188 (green) localizes to the nucleus in control fibroblasts but is undetectable in fibroblasts from individual 1. The experiment was performed in triplicate (n = 20 cells each), and representative images are shown. The scale bar represents 20 μm. Immunoblot and immunofluorescence data for individual 2 corroborate staining patterns seen in individual 1 and can be found in in Figure S1.
Affected Individuals Have a Disruption in Importinα-Importinβ1- and Transportin-1-Mediated Nuclear Import
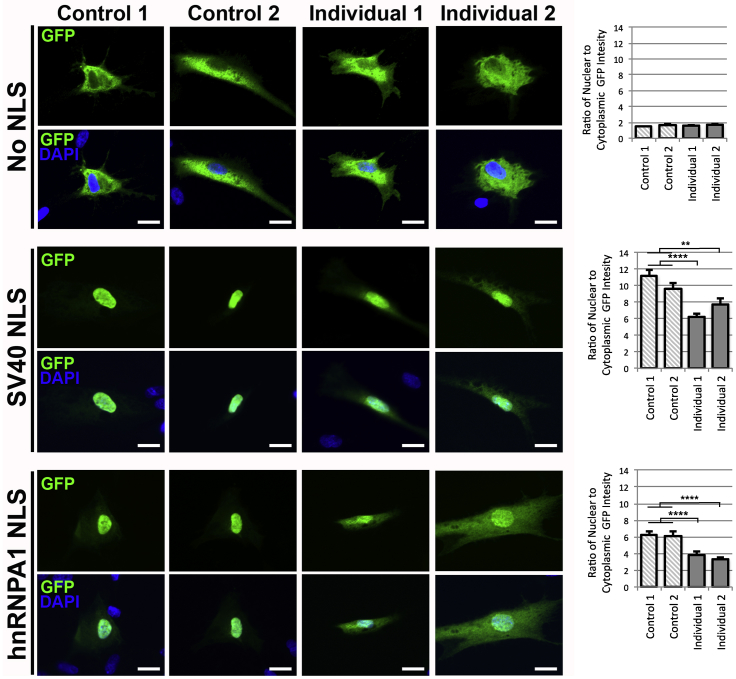
Protein import into the nucleus occurs through the NPC and is mediated by carrier proteins that recognize protein NLS. Loss of NUP188 results in a reduction of nuclear import in yeast.11 To determine whether nuclear import is also disrupted in affected individuals, we infected fibroblasts from individuals 1 and 2 and unrelated control individuals with a construct that encodes three sequential GFP molecules attached to an NLS and measured GFP fluorescence intensity in the cytoplasm and nucleus of infected cells. To test both the “classic” importinα-importinβ1 nuclear import pathway and the transportin-1 pathway, we created constructs containing the SV40 NLS and hnRNPA1 NLS, respectively. We observed a 33% lower ratio of nuclear GFP fluorescence intensity to cytoplasmic GFP fluorescence intensity in affected individuals compared to control individuals when the “classic” importinα-importinβ1 nuclear import pathway was used and a 43% lower ratio under the transportin-1 pathway (Figure 3). To account for differences in the rate of protein diffusion across the nuclear membrane, we infected cells with a GFP construct containing no NLS. Although a small amount of GFP diffused into nuclei, the rate of diffusion was similar between affected individuals and control individuals. Together, these data indicate that disruption of NUP188 in affected individuals is associated with less active protein transport into the nucleus.
Figure 3.
Impaired SV40 NLS-GFP and hnRNPA1 NLS-GFP Import into the Nucleus in Affected Individuals
Representative immunofluorescent images showing less GFP (green) transport into the nuclei of affected individuals’ fibroblasts compared to the GFP transport into the nuclei of control fibroblasts. Disruption of NUP188 in affected individuals inhibited the “classic” importinα-importinβ1 nuclear import pathway (SV40 NLS) and the transportin-1 pathway (hnRNPA1 NLS) but not the rate of diffusion (no NLS). DAPI (blue) was used to define the boundaries of the nucleus, and wheat-germ agglutinin (not shown) was used to define the cell boundaries for quantification of the ratio of nuclear GFP fluorescent intensity to cytoplasmic GFP fluorescent intensity. Error bars represent SEM. The scale bar represents 20 μm. The experiment was performed in triplicate with a combined n = 47–70 per condition, representative images shown. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by two-tailed unpaired Student’s t test.
Drosophila Studies
Generation of the Nup188-Knockout (Nup188KO) Alleles
To determine the physiological function of Nup188 in vivo, we generated two knockout alleles in Drosophila by using the CRISPR-Cas9 system.20 A guide RNA (gRNA) targeting the Nup188 translation start site (Figure S2) was injected into Cas9-expressing flies, resulting in a one-nucleotide insertion (Nup1886A) or a seven-nucleotide deletion (Nup18810B). Each allele is predicted to cause a frameshift and produce a nonfunctional protein with early truncation (Figure S2). When flies were raised at 25°C, homozygosity for either the Nup1886A or the Nup18810B allele was lethal with homozygous flies dying at the pupal stage. However, when raised at 22°C, approximately 55% of Nup1886A and Nup18810B homozygous flies survived. The lethality phenotype was further confirmed by use of the deficiency allele −Df(2R)BSC485, which covers the Nup188 locus. Transheterozygotes of Nup1886A-Df(2R)BSC485 and Nup18810B-Df(2R)BSC485 showed similar penetrance to the homozygotes. We then used Nup18810B in the following analyses to determine the neural phenotypes.
Nup188 Is Required for Proper Dendrite Tiling
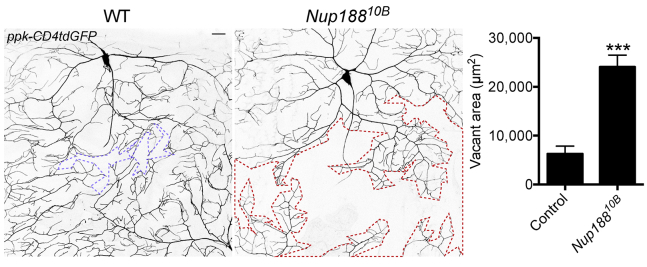
Given the neurological symptoms in affected individuals, we investigated whether Nup188KO flies have neurodevelopmental deficits. Drosophila dendritic arborization (da) sensory neurons cover the entire body wall of fly larvae, show stereotyped dendrite patterning, and have been extensively used to study neuronal development.24,25 In particular, class IV da (C4da) neuron dendrites branch out to completely cover each segment by forming close interfaces with the arbors of nearby neurons to tile the body wall (Figure 4). However, in vivo imaging of Nup18810B larvae revealed large vacant areas at the interface between two subtypes of C4da neurons—ddaC and v’ada (Figure 4). Quantification revealed a 2-fold larger vacant area in Nup18810B larvae than in the WT control (Figure 4). This result suggests an important role for Nup188 in dendrite tiling and, potentially, neural circuit formation.
Figure 4.
Nup188 Deficiency Results in Impaired Sensory Dendrite Tiling
Representative images of the interface between C4da neurons ddaC and v’ada. C4da neurons are genetically labeled with ppk-CD4tdGFP. The WT image on the left shows extensive dendrite tiling; a minimal vacant area between the dendrite arbors is outlined in purple. On the right is the Nup18810B mutant, and a larger vacant area is outlined in red. The scale bar represents 20 μm. The vacant area at the interface not covered by dendrites (the area inside the outline) is quantified. Error bars, SEM. n = 10 neurons from four larvae. ∗∗∗p < 0.001 by two-tailed unpaired Student’s t test.
Nup188 Deficiency Leads to Behavioral Deficits
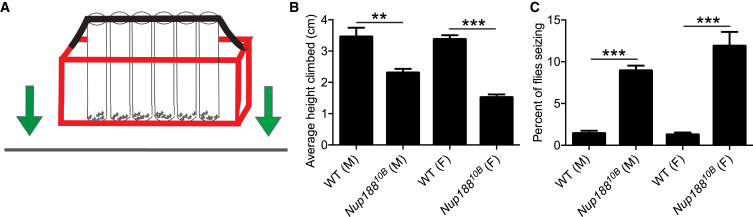
We next inquired as to whether Nup188KO flies show behavioral phenotypes. Although the majority of the Nup18810B flies failed to eclose and died at the pupal stage, a small fraction survived to adulthood as escapees. Given the hypotonia present in affected individuals, we investigated whether these Nup18810B escapees displayed weaker motor function. Showing a negative geotaxis response, WT adult flies crawl up the walls of a vial in response to being tapped to the bottom. The height climbed in a given time has been used as an assay of locomotor activity.23,26 We found that Nup18810B flies climbed a shorter distance than WT flies (Figures 5A and 5B and Video S1), suggestive of motor deficits. Furthermore, mechanical stimuli such as tapping are known to trigger paralysis and seizures in certain mutant flies.27,28 We observed seizures, during which flies twitched and spun on their backs (Video S1), in Nup18810B flies after tapping (Figure 5C).
Figure 5.
Nup188 Deficiency Leads to Decreased Negative Geotaxis and Seizures
(A) Schematic depiction of the negative geotaxis apparatus. Vials containing flies are secured in the red container, which is then tapped downward so that flies are knocked to the bottom. The response of flies is video recorded.
(B) The average height climbed up from the bottom of the vial by the flies was measured three seconds after tapping. Males (M) and females (F) in each group were tested separately. WT flies climbed significantly farther up the vial than Nup18810B flies.
(C) The average percent of flies showing seizure activity in a 10 s period after tapping. WT males = 9 vials, 210 flies total; Nup18810B males = 9 vials, 205 flies total; WT females = 6 vials, 149 flies total; Nup18810B females = 6 vials, 119 flies total. Error bars, SEM. ∗∗p < 0.01, ∗∗∗p < 0.001 by one-way ANOVA followed by Tukey’s test.
Discussion
Here, we describe a disorder caused by bi-allelic truncating variants in NUP188 and characterized by hypotonia, congenital cataracts, microcephaly, abnormal brain imaging, and early death due to respiratory failure, usually within the first year after birth. All five pathogenic NUP188 variants identified in this study were truncating variants. NUP188 is intolerant to loss of function variants (pLI = 0.95, probability of being loss-of-function intolerant),29 and there are no homozygous truncating variants in NUP188 in gnomAD. Immunoblot and immunofluorescence analysis of fibroblasts carrying NUP188 p.Ile302Valfs∗7 and p.Tyr1048∗ indicates that these variants result in either complete loss of NUP188 or a nonfunctional truncated product. The other bi-allelic truncating variants identified in the study most likely represent complete loss of NUP188 function in the affected individuals as well. Further studies of additional families with affected individuals will be needed if we are to determine whether bi-allelic missense variants or variants causing only a partial reduction in NUP188 function result in a similar phenotype.
Of the four unrelated families we identified in this study, two were of Ashkenazi Jewish descent. The three affected individuals from these two unrelated Ashkenazi Jewish families presented with the same two pathogenic NUP188 variants, p.Ile302Valfs∗7 and p.Tyr1048∗, each of which was restricted to the Ashkenazi Jewish population in gnomAD. These variants are present in the Ashkenazi Jewish population at allele frequencies in gnomAD of 0.075% and 0.040%, respectively—allele frequencies that are higher than those seen for any other NUP188 truncating variant in other populations. The higher prevalence of these two variants in the Ashkenazi Jewish population was confirmed by additional targeted screening of a large internationally diverse cohort of Ashkenazi (and Sephardi) healthy Jewish individuals (n = 4,815). This genetic disorder, therefore, will most likely be more prevalent in individuals of Ashkenazi Jewish descent than in the other populations and might have implications for future updates to Ashkenazi Jewish carrier screening.
Our results also show that active transport of proteins into the nucleus via both the “classic” importinα-importinβ1 nuclear import pathway and the transportin-1 pathway are lower in affected individuals’ cells. This result is congruent with the finding that loss of NUP188 results in a reduction of nuclear import and mRNA export in yeast.11 In human cells and in yeast, loss of NUP188 did not result in total absence of nuclear import. However, because nuclear transport is integral to the central cell processes of DNA replication, gene expression, and protein production, even a modest reduction in transport capacity could have a major impact on cell homeostasis. Interestingly, NUP214 disruption, which causes susceptibility to acute febrile encephalopathy, also results in a modest reduction in nuclear transport in affected individuals’ cells,10 suggesting that this may be a common mechanism through which disruption of nucleoporins results in human disease.
Knockdown of Nup188 has been shown to disrupt left-right patterning in Xenopus.15 NUP188 has also been associated with other cellular functions, including ciliogenesis,12 chromatin organization,13 transcriptional regulation,13 and chromosome segregation.14 Notably, some of the patients had clinical features that can be seen in heterotaxy or ciliopathies such as polysplenia, pre-axial polydactyly, possible rib anomalies, and cerebellar malformations. Although we hypothesize that reduction in nuclear transport is at least partially responsible for the phenotype of the affected individuals in this study, we cannot rule out the possibility that disruption of one or more of these other functions also contributes to disease progression.
Although pathogenic NUP188 variants cause a multisystem disorder, affected individuals had significant nervous system involvement. Complications included generalized hypotonia, progressive microcephaly, seizures, a thin corpus callosum, and other abnormal MRI findings. The early death due to respiratory failure was most likely secondary to nervous system complications because it occurred without signs of pulmonary or tracheal disease in 4/6 affected individuals. Furthermore, loss of Nup188 in fruit flies resulted in pronounced nervous system defects, including a reduced dendritic tiling, motor deficits, and seizures.
Several other neurological diseases in humans are due to pathogenic variants in genes encoding nucleoporin proteins. Triple A syndrome (MIM: 231550), caused by bi-allelic variants in AAAS (MIM: 605378), is characterized by adrenocorticotropic hormone (ACTH)-resistant adrenal insufficiency, achalasia, and alacrima with a variable neurologic phenotype including developmental delay, autonomic dysfunction, and muscle weakness.5 Recessive infantile striatonigral degeneration (MIM: 271930), caused by variants in NUP62 (MIM: 605815), is a progressive neurodegenerative disorder characterized by seizures, developmental delay, and brain atrophy.8 Bi-allelic variants in NUP214 (MIM: 114350) and heterozygous variants in RANBP2 (MIM: 601181) can cause susceptibility to acute infection-induced encephalopathy (MIM: 618426 and MIM: 608033, respectively) characterized by febrile-induced encephalopathy resulting in neurodevelopmental regression, seizures, and other neurological abnormalities.7,9,10 Because most nucleoporins are ubiquitously expressed,30 this suggests that the cells in the nervous system might be particularly susceptible to loss of nuclear-pore function.
All individuals with NUP188-associated disease presented with a strikingly similar phenotype and poor prognosis. Identification and characterization of additional affected individuals and additional pathogenic alleles will be necessary if we are to determine whether the phenotype is consistently as severe as in the individuals described here, all of whom experienced early respiratory failure and death in infancy or early childhood. Recently, homozygous truncating variants in NUP188 were reported in two additional individuals who had a similar severe phenotype and who also died of respiratory failure within the first year of life.31 Evaluation of NUP188 should be considered in infants with neurologic deficits, congenital cataracts, congenital heart defects, and unexplained respiratory failure, and particularly heightened attention should be given to infants of Ashkenazi Jewish descent.
Declaration of Interests
H.C.M. is a member of scientific advisory boards for Lennox Gastaut Syndrome Foundation, Dravet Syndrome Foundation, and SPARK. All other authors declare no competing interests.
Acknowledgments
We thank the families for their participation in this study. Y.S. and E.J.B. are funded by a Rapid Translation Award from the Roberts Collaborative, CHOP. H.C.M. received support from the National Institutes of Health (NIH R01 NS069605). N.W. received support from the Mary Gates Endowment for Students, University of Washington. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number TL1TR001880 (S.E.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Published: April 9, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.009.
Contributor Information
Yuanquan Song, Email: songy2@email.chop.edu.
Heather C. Mefford, Email: hmefford@uw.edu.
Web Resources
GeneMatcher, https://www.genematcher.org/
gnomAD v2.1.1, https://gnomad.broadinstitute.org/
ImageJ, https://imagej.nih.gov/ij/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Beck M., Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 2.Miyake N., Tsukaguchi H., Koshimizu E., Shono A., Matsunaga S., Shiina M., Mimura Y., Imamura S., Hirose T., Okudela K. Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am. J. Hum. Genet. 2015;97:555–566. doi: 10.1016/j.ajhg.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun D.A., Lovric S., Schapiro D., Schneider R., Marquez J., Asif M., Hussain M.S., Daga A., Widmeier E., Rao J. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J. Clin. Invest. 2018;128:4313–4328. doi: 10.1172/JCI98688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun D.A., Sadowski C.E., Kohl S., Lovric S., Astrinidis S.A., Pabst W.L., Gee H.Y., Ashraf S., Lawson J.A., Shril S. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 2016;48:457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronshaw J.M., Matunis M.J. The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc. Natl. Acad. Sci. USA. 2003;100:5823–5827. doi: 10.1073/pnas.1031047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnin E., Cabochette P., Filosa A., Jühlen R., Komatsuzaki S., Hezwani M., Dickmanns A., Martinelli V., Vermeersch M., Supply L. Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet. 2018;14:e1007845. doi: 10.1371/journal.pgen.1007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neilson D.E., Adams M.D., Orr C.M., Schelling D.K., Eiben R.M., Kerr D.S., Anderson J., Bassuk A.G., Bye A.M., Childs A.M. Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am. J. Hum. Genet. 2009;84:44–51. doi: 10.1016/j.ajhg.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basel-Vanagaite L., Muncher L., Straussberg R., Pasmanik-Chor M., Yahav M., Rainshtein L., Walsh C.A., Magal N., Taub E., Drasinover V. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann. Neurol. 2006;60:214–222. doi: 10.1002/ana.20902. [DOI] [PubMed] [Google Scholar]
- 9.Shamseldin H.E., Makhseed N., Ibrahim N., Al-Sheddi T., Alobeid E., Abdulwahab F., Alkuraya F.S. NUP214 deficiency causes severe encephalopathy and microcephaly in humans. Hum. Genet. 2019;138:221–229. doi: 10.1007/s00439-019-01979-w. [DOI] [PubMed] [Google Scholar]
- 10.Fichtman B., Harel T., Biran N., Zagairy F., Applegate C.D., Salzberg Y., Gilboa T., Salah S., Shaag A., Simanovsky N. Pathogenic Variants in NUP214 Cause “Plugged” Nuclear Pore Channels and Acute Febrile Encephalopathy. Am. J. Hum. Genet. 2019;105:48–64. doi: 10.1016/j.ajhg.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruyn Kops A., Guthrie C. Identification of the Novel Nup188-brr7 Allele in a Screen for Cold-Sensitive mRNA Export Mutants in Saccharomyces cerevisiae. G3. 2018;8:2991–3003. doi: 10.1534/g3.118.200447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Viso F., Huang F., Myers J., Chalfant M., Zhang Y., Reza N., Bewersdorf J., Lusk C.P., Khokha M.K. Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia. Dev. Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labade A.S., Karmodiya K., Sengupta K. HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin. 2016;9:54. doi: 10.1186/s13072-016-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh G., Sugino S., Ikeda M., Mizuguchi M., Kanno S., Amin M.A., Iemura K., Yasui A., Hirota T., Tanaka K. Nucleoporin Nup188 is required for chromosome alignment in mitosis. Cancer Sci. 2013;104:871–879. doi: 10.1111/cas.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fakhro K.A., Choi M., Ware S.M., Belmont J.W., Towbin J.A., Lifton R.P., Khokha M.K., Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc. Natl. Acad. Sci. USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekstein J., Katzenstein H. The Dor Yeshorim story: community-based carrier screening for Tay-Sachs disease. Adv. Genet. 2001;44:297–310. doi: 10.1016/s0065-2660(01)44087-9. [DOI] [PubMed] [Google Scholar]
- 18.Nabbi A., Riabowol K. Rapid Isolation of Nuclei from Cells In Vitro. Cold Spring Harb. Protoc. 2015;2015:769–772. doi: 10.1101/pdb.prot083733. [DOI] [PubMed] [Google Scholar]
- 19.Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016;34:528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gratz S.J., Rubinstein C.D., Harrison M.M., Wildonger J., O’Connor-Giles K.M. CRISPR-Cas9 Genome Editing in Drosophila. Curr. Protoc. Mol. Biol. 2015;111:1–20. doi: 10.1002/0471142727.mb3102s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emoto K., Parrish J.Z., Jan L.Y., Jan Y.N. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- 22.Parrish J.Z., Emoto K., Kim M.D., Jan Y.N. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- 23.Nichols C.D., Becnel J., Pandey U.B. Methods to assay Drosophila behavior. J. Vis. Exp. 2012;(61):3795. doi: 10.3791/3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grueber W.B., Jan L.Y., Jan Y.N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 25.Jan Y.N., Jan L.Y. Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali Y.O., Escala W., Ruan K., Zhai R.G. Assaying locomotor, learning, and memory deficits in Drosophila models of neurodegeneration. J. Vis. Exp. 2011;(49):2504. doi: 10.3791/2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burg M.G., Wu C.F. Mechanical and temperature stressor-induced seizure-and-paralysis behaviors in Drosophila bang-sensitive mutants. J. Neurogenet. 2012;26:189–197. doi: 10.3109/01677063.2012.690011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J., Wu C.F. Electroconvulsive seizure behavior in Drosophila: analysis of the physiological repertoire underlying a stereotyped action pattern in bang-sensitive mutants. J. Neurosci. 2002;22:11065–11079. doi: 10.1523/JNEUROSCI.22-24-11065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandestig A., Engström K., Pepler A., Danielsson I., Odelberg-Johnsson P., Biskup S., Holz A., Stefanova M. NUP188 Biallelic Loss of Function May Underlie a New Syndrome: Nucleoporin 188 Insufficiency Syndrome? Mol. Syndromol. 2020;10:313–319. doi: 10.1159/000504818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.