Abstract
Because polygenic risk scores (PRSs) for coronary heart disease (CHD) are derived from mainly European ancestry (EA) cohorts, their validity in African ancestry (AA) and Hispanic ethnicity (HE) individuals is unclear. We investigated associations of “restricted” and genome-wide PRSs with CHD in three major racial and ethnic groups in the U.S. The eMERGE cohort (mean age 48 ± 14 years, 58% female) included 45,645 EA, 7,597 AA, and 2,493 HE individuals. We assessed two restricted PRSs (PRSTikkanen and PRSTada; 28 and 50 variants, respectively) and two genome-wide PRSs (PRSmetaGRS and PRSLDPred; 1.7 M and 6.6 M variants, respectively) derived from EA cohorts. Over a median follow-up of 11.1 years, 2,652 incident CHD events occurred. Hazard and odds ratios for the association of PRSs with CHD were similar in EA and HE cohorts but lower in AA cohorts. Genome-wide PRSs were more strongly associated with CHD than restricted PRSs were. PRSmetaGRS, the best performing PRS, was associated with CHD in all three cohorts; hazard ratios (95% CI) per 1 SD increase were 1.53 (1.46–1.60), 1.53 (1.23–1.90), and 1.27 (1.13–1.43) for incident CHD in EA, HE, and AA individuals, respectively. The hazard ratios were comparable in the EA and HE cohorts (pinteraction = 0.77) but were significantly attenuated in AA individuals (pinteraction= 2.9 × 10−3). These results highlight the potential clinical utility of PRSs for CHD as well as the need to assemble diverse cohorts to generate ancestry- and ethnicity PRSs.
Keywords: risk prediction, polygenic risk scores, multiethnic, ischemic heart disease, coronary heart disease, coronary artery disease, African American, hispanic, genome-wide polygenic score
Introduction
Coronary heart disease (CHD) is a genetically complex disease with an estimated heritability of 40%–60%.1,2 Over the past decade, genome-wide association studies (GWAS) have revealed numerous genetic susceptibility loci for CHD,3, 4, 5, 6, 7, 8, 9, 10 generating interest in the use of polygenic risk scores (PRSs) to improve prediction of adverse CHD events.11,12 “Restricted” PRSs12, 13, 14, 15, 16 typically include variants reaching genome-wide significance (p < 5 × 10−8) and account for only a small proportion of heritability, suggesting there might be additional information in variants that are below the genome-wide significance threshold.1,17Genome-wide PRSs select from millions of variants across the genome by applying more lenient type I error thresholds and accounting for linkage disequilibrium (LD) between variants.18, 19, 20 Such PRSs for CHD in European ancestry (EA) individuals outperform restricted polygenic scores, identifying individuals who have a 2–3 times higher risk of developing CHD than does the general population in the UK Biobank dataset.18,19
These reports have generated enthusiasm about the use of genome-wide PRSs to improve prediction of adverse CHD events, but their validity in minority populations is unclear. Differences in allele frequencies and LD patterns at risk loci limit the generalizability of variants identified in EA cohorts to other racial and ethnic groups.7,9,21, 22, 23, 24, 25 Lack of diversity in many of the available datasets, and consequently limited sample sizes and power for minority-specific GWASs, have hindered development of racial and ethnicity specific PRSs for CHD.4,7, 8, 9,23,25, 26, 27, 28 To prevent exacerbation of health disparities in the context of genomic medicine, researchers will need to assess the predictive utility of different types of PRSs across ancestral and ethnic groups.
We therefore investigated the strengths of associations of available PRSs with CHD in EA, African ancestry (AA), and Hispanic ethnicity (HE) adults by using a high-density genotype dataset linked to electronic health record (EHR) data from the electronic medical records and genomics (eMERGE) network.29,30 We hypothesized that PRSs derived from EA cohorts would be less strongly associated with CHD in non-EA cohorts because of reduced specificity and lower effect sizes of risk alleles.
Methods
Study Cohort
The eMERGE network is a US-based consortium of cohorts in which DNA samples are linked to EHR data to enable large-scale, high-throughput genomic studies.29,30 At the time of our analysis, the network, in its third phase, included 99,185 genotyped participants from 12 geographically distinct healthcare institutions located across the US, each with its own biorepository and site-specific eligibility criteria for enrollment. The majority of the eMERGE sites recruited primarily from the outpatient setting (Table S1). The Mayo Clinic cohort partially included individuals referred for non-invasive vascular evaluation or cardiac stress testing (n = 3,640, 35% of the Mayo Clinic genotyped cohort). Details of the enrollment process of each biorepository contributing to the eMERGE network have been previously published.29,30 Each member site obtained approval from its respective institutional review board.
Genotype Data
High-density genotype data were available for 99,185 participants from the eMERGE network. To harmonize the genotype data from 12 member sites, we imputed each of the 80 Illumina and Affymetrix genotype batches31 via the Michigan Imputation Server by using the minimac3 algorithm and the genotype reference panel from the Haplotype Reference Consortium.32,33 A detailed description of quality-control procedures has been previously published.31
Ascertaining CHD Events
We limited our study cohort to adult participants (≥18 years) with at least 1 year of EHR history. To ascertain CHD, defined as occurrence of either myocardial infarction (MI) or coronary revascularization events (such as percutaneous coronary intervention or coronary artery bypass grafting), we used an electronic phenotyping algorithm based on International Classification of Diseases, 9th and 10th revisions, Clinical Modification (ICD-9-CM and ICD-10-CM) codes and Current Procedural Terminology (CPT) codes. Individuals with MI were defined as those whose EHR included at least two related diagnostic codes on separate occasions within a 5-day window, and individuals with coronary revascularization were defined as those who had at least one relevant procedural code in the EHR. The CHD-related diagnostic and procedural codes were obtained from two previously validated eMERGE electronic phenotyping algorithms34,35 and adapted for this study. A list of diagnostic and procedural codes is provided in the Supplemental Data.
DNA sample collection dates were not always available for participants; therefore, follow-up was started from the time of their first EHR. For each participant, we identified the first CHD event and classified it as “incident” if the event occurred at least 6 months after the participant’s first record in the EHR and if there were no previous ICD-9-CM or ICD-10-CM codes associated with CHD. All other CHD events were classified as “prevalent.” Individuals without an index event based on the above definition but who had other CHD-related codes in their record (i.e., participants with uncertain CHD event data) were excluded. After applying these criteria, we further excluded eMERGE sites that had very few participants (<15) or that had no CHD events (Table S1).
To validate the CHD phenotyping algorithm, we conducted a manual EHR review, at one eMERGE site (Mayo Clinic), of 25 individuals with incident CHD, 25 individuals with prevalent CHD, and 25 non-CHD control individuals. The algorithm had a positive-predictive value of 88% and 96% for individuals with incident and prevalent CHD, respectively, and a negative-predictive value of 100% for non-CHD control individuals.
Ascertaining Conventional Risk Factors
To ascertain hypertension and diabetes, we used ICD-9-CM and ICD-10-CM codes (obtained from previously validated eMERGE algorithms34,36,37 on the Phenotype KnowledgeBase (PheKB);38 see Supplemental Information) and required the presence of at least two codes on separate days in an individual’s EHR. Hypercholesterolemia was defined as the presence of related diagnostic codes (at least two codes on separate days) or having a low-density lipoprotein cholesterol (LDL-C) >160 mg/dl or non-high-density lipoprotein cholesterol (non-HDL-C) >190 mg/dl. Statin therapy was defined as the presence of at least two statin medication records on two different days. For incident cases, the presence of conventional CHD risk factors during the period preceding the index event was ascertained. For the remaining participants without an index event, these risk factors were ascertained in the window between the earliest record and the median time until index CHD events in the incident cases of the corresponding racial-ethnic group.
Calculating PRS and Quality Control
We identified three mutually exclusive racial and ethnic groups, namely, non-Hispanic EA, non-Hispanic AA, and HE. We identified EA and AA individuals by matching genetic ancestry (with principal component analysis (PCA)-based k-means) and self-reported ancestry in which individuals self-identified to be “non-Hispanic,” whereas HE individuals were identified by self-report (Figure S1).31 We excluded any ambiguous AT/GC single-nucleotide variants (SNVs) as well as any SNVs demonstrating allele mismatch between the eMERGE dataset and published PRSs and applied the following quality-control metrics to the genotype data of each racial and ethnic group: SNV and per-individual call rate > 95%, Hardy-Weinberg equilibrium p value > 1 × 10−5, and imputation quality r2 > 0.3 via PLINK.39,40 To estimate ancestry-specific genetic principal components robust to admixture and cryptic relatedness, we used PC-AiR41 and KING-robust42 separately in each group.
We assessed two restricted PRSs (i.e., Tikkanen et al. and Tada et al., 28 and 50 variants, respectively)13,14and two genome-wide PRSs (i.e., metaGRS and LDPred, 1.7 M and 6.6 M variants, respectively)18,19 for CHD in EA populations, hereafter denoted as PRSTikkanen, PRSTada, PRSmetaGRS, and PRSLDPred, respectively (Table S2). In brief, PRSTikkanen and PRSTada included variants that, at the time these PRSs were developed, were associated with CHD at genome-wide significance in the literature. PRSLDPred is based on a Bayesian approach that assumes that a fraction of variants in the genome are causal for CHD and then infers the posterior mean effect size of each GWAS marker, while taking into account this assumption and LD between neighboring variants.18,20 PRSmetaGRS is derived with a meta-analytic approach that combines three different previously developed PRSs: GRS46K19,43 (nSNV = 46,000 after LD-based pruning), a PRS based on a GWAS conducted by Nikpay et al.19,27 (nSNV = 1.7 M after LD-based pruning), and FDR20219,27 (nSNV = 202), which was derived by application of a false-discovery-rate threshold of <0.05 to the previously published CHD GWAS summary statistics. PRSmetaGRS was constructed as an average of all three PRSs within a training set in UK Biobank (n = 3000) and was weighted by the logarithm of hazard ratios (HRs) for CHD of each PRS and inter-PRS correlation coefficients.
The effect-size estimates for the SNVs used in these scores were obtained from the relevant studies. We used the PRSice pipeline44 to calculate PRSs as a weighted sum of all the effect alleles on the basis of allele dosages and the provided SNV effect sizes. All scores were standardized to zero-mean and unit-variance within each racial and ethnic group.
Statistical Analyses
In our primary analyses, we excluded prevalent CHD (as defined above) and evaluated PRSs separately in each racial and ethnic group by using Cox proportional hazards regression. Participants were right censored at age 75 years (because of an insufficient number of CHD events in non-EA cohorts beyond that age) or at the age of last observation in the EHR (whichever was first). In Cox models, we included PRS as a continuous variable; adjusted for sex, eMERGE site, and first five ancestry-specific principal components; and used age as the timescale. Associations of PRSs with incident CHD independent of conventional risk factors were also assessed after adjustment for these risk factors. Schoenfeld residuals and interaction terms for PRS and time were assessed in all Cox regression models for any significant departure from the proportional-hazards assumption.
We additionally evaluated the association of a PRS with CHD separately in each group by including all CHD-affected individuals (i.e., those with either incident or prevalent CHD) and using multivariable logistic regression with disease as the outcome and PRS as the predictor; we adjusted for age at first EHR record, sex, eMERGE site, duration of EHR, and first five ancestry-specific principal components. For these models, we computed the c-index (i.e., area under the receiver-operator characteristic curve) by using the pROC package.
To investigate whether the strength of association of a PRS with CHD was modified by an individual’s race or ethnicity, we performed analyses including all three racial and ethnic groups. In these regression models, we included a racial and ethnic group indicator as an adjustment variable as well as an interaction term for this variable and PRS. We further adjusted for population structure within each group by using a design matrix of the first five ancestry-specific principal components estimated separately for each racial and ethnic cohort.
To explore the potential clinical utility of EA-based PRSs that were significantly associated with CHD in non-EA groups, we estimated the 10-year absolute risk of MI as a function of PRS for EA and AA individuals. We used the iCARE statistical package45,46 to compute absolute risk estimates by combining the following: (1) HRs of PRSs (per 1 SD increase) for incident CHD, (2) PRS distributions in the eMERGE cohort, and (3) age-, sex-, race-, and ethnicity-specific rates of incident MI in the US and respective non-CHD mortality rates as competing risks. We used the 2019 heart disease and stroke statistics reported by the American Heart Association (AHA)47 to obtain rates of incident MI and the Centers for Disease Control and Prevention’s WONDER online database to obtain mortality rates in the U.S. (see Web Resources; Tables S3 and S4). Because of the lack of data on age- and sex-specific rates of incident MI in the HE cohort, these analyses were limited to EA and AA individuals aged 35–74 years.
Simulation-based power calculations for both race- and ethnic-group-stratified and joint cohort analyses were performed via logistic regression models with 1,000 iterations. All statistical tests were two sided, and a p value of <0.05 was considered statistically significant. All analyses were performed with R statistical computing software (version 3.5.2).48 A detailed list of QC pipelines and statistical packages used in this study is provided in Table S5.
Results
Characteristics of the eMERGE Phase III Genotyped Cohort
After implementation of the CHD phenotyping algorithm and quality-control measures, our study cohort consisted of 55,735 participants (mean age 48 ± 14 years, 58% female), including 45,645 non-Hispanic EA, 7,597 non-Hispanic AA, and 2,493 HE individuals from 10 eMERGE sites. In comparison to the EA cohort, the AA and HE cohorts were younger and included a higher proportion of women. Additional characteristics of the study cohorts are shown in Table 1. During a median follow up of 11.1 years (interquartile range [IQR] 6.0–17.5 years), 2,652 (4.8%) incident CHD events were noted. Distributions of PRSs stratified by race, ethnicity, and CHD status are shown in Figures S2–S5.
Table 1.
Participant Characteristics
| Characteristic | EA (n = 45,645) | AA (n = 7,597) | HE (n = 2,493) |
|---|---|---|---|
| Age, years (mean ± SD) | 49.0 ± 14.1 | 43.6 ± 12.5 | 41.1 ± 13.2 |
| Female, n (%) | 25,301 (55.4) | 5245 (69.0) | 1590 (63.8) |
| Diabetes, n (%) | 3388 (7.4) | 875 (11.5) | 292 (11.7) |
| Hypertension, n (%) | 5561 (12.2) | 1086 (14.3) | 246 (9.9) |
| Hypercholesterolemia, n (%) | 9702 (21.3) | 1200 (15.8) | 552 (22.1) |
| Statin use, n (%) | 12,521 (27.4) | 1045 (13.8) | 201 (8.1) |
| Incident CHD events, n (%) | 2221 (4.9) | 311 (4.1) | 120 (4.8) |
| Age at incident CHD event, years (mean ± SD) | 59.5 ± 9.5 | 56.0 ± 10.4 | 57.9 ± 10.1 |
| Prevalent CHD cases, n (%) | 5887 (12.9) | 527 (6.9) | 299 (12.0) |
| Follow-up in years, median (IQR) | 11.7 (6.0–18.5) | 9.2 (5.5–13.0) | 10.4 (5.7–14.7) |
| Person years | 595,896 | 75,370 | 27,191 |
Association Results for CHD
In EA individuals, restricted PRSs and genome-wide PRSs were associated with up to 1.20-fold and 1.53-fold increased risk of incident CHD per 1 SD increase, respectively (Table 2). Models that included genome-wide PRSs had higher c-indices than did models with restricted PRSs (0.719 versus 0.697–0.698). Estimated HRs were similar to the original reports, suggesting good generalizability among different EA cohorts (Table S6).
Table 2.
Hazard Ratios for Incident CHD per 1 SD Increase in PRS
| HR (95% CI)a | p Valuea | ModelBasec-indexb | ModelPRSc-indexc | pinteractiond | |
|---|---|---|---|---|---|
| EA | |||||
| PRSTikkanen | 1.18 (1.13–1.23) | 6.54 × 10−14 | 0.690 | 0.697 | ref |
| PRSTada | 1.20 (1.15–1.25) | <2 × 10−16 | 0.690 | 0.698 | ref |
| PRSLDPred | 1.50 (1.43–1.56) | <2 × 10−16 | 0.690 | 0.719 | ref |
| PRSmetaGRS | 1.53 (1.46–1.60) | <2 × 10−16 | 0.690 | 0.719 | ref |
| AA | |||||
| PRSTikkanen | 1.11 (0.99–1.24) | 0.07 | 0.649 | 0.652 | 0.39 |
| PRSTada | 1.05 (0.94–1.17) | 0.41 | 0.649 | 0.649 | 0.02 |
| PRSLDPred | 1.19 (1.07–1.33) | 2.2 × 10−3 | 0.649 | 0.656 | 1.6 × 10−4 |
| PRSmetaGRS | 1.27 (1.13–1.43) | 4.1 × 10−5 | 0.649 | 0.663 | 2.9 × 10−3 |
| HE | |||||
| PRSTikkanen | 1.14 (0.94–1.37) | 0.19 | 0.654 | 0.655 | 0.77 |
| PRSTada | 1.13 (0.93–1.36) | 0.22 | 0.654 | 0.654 | 0.53 |
| PRSLDPred | 1.16 (0.96–1.41) | 0.13 | 0.654 | 0.659 | 0.02 |
| PRSmetaGRS | 1.53 (1.23–1.90) | 1.1 × 10−4 | 0.654 | 0.683 | 0.77 |
Age-as-time-scale Cox regression models separate for each racial and ethnic group adjusted for sex, eMERGE site, and first five ancestry-specific principal components.
c-indices for base models without PRSs across racial and ethnic groups: age-as-time-scale Cox model with sex, eMERGE site, and first five ancestry-specific principal components.
c-indices for base model + PRSs for each racial and ethnic group.
Age-as-time-scale Cox regression models on the joint cohort adjusted for sex, eMERGE site, design matrix of first five ancestry-specific principal components, racial and ethnic group, and an interaction term with PRSs. ref, referent.
In AA individuals, HRs for all PRSs were lower than in EA individuals (Table 2). All PRSs except PRSTikkanen (pinteraction = 0.39) showed statistically significant heterogeneity of effect in AA individuals in reference to the EA cohort (pinteraction; PRSTada, 0.02; PRSLDPred, 1.6 × 10−4; PRSmetaGRS, 2.9 × 10−3). Genome-wide PRSs were more strongly associated with incident CHD (HR per 1 SD increase; 1.19–1.27, p ≤ 2.2 × 10−3) than were restricted PRSs (HR per 1 SD increase; 1.05–1.11, p ≥ 0.07) and resulted in a greater model discrimination (c-index; genome-wide PRSs 0.656–0.663 versus restricted PRSs 0.649–0.652).
In HE individuals, as in the EA and AA cohorts, genome-wide PRSs, especially PRSmetaGRS, were more strongly associated with incident CHD. PRSLDPred was the only PRS that demonstrated significant heterogeneity of effect in the HE individuals compared to the EA individuals, and there was a large difference in estimated HRs between the two cohorts (pinteraction = 0.02). In contrast, PRSmetaGRS had the strongest association with incident CHD (HR 1.53 per 1 SD increase) and the highest c-index in comparison to other PRSs in the HE cohort (Table 2).
After adjustment for cardiovascular risk factors (diabetes, hypertension, and hyperlipidemia) and statin use in Cox models, we observed minimal attenuation in risk estimates for statistically significant PRSs across all racial and ethnic groups (Table S7).
When we evaluated PRSs in analyses that included all CHD cases, genome-wide PRSs were associated with higher odds of CHD per 1 SD increase and higher model c-indices than were the restricted PRSs across the three groups (Table 3). Odds ratios (ORs) in the EA and HE cohorts were comparable, and there was no evidence of heterogeneity between these cohorts except for PRSLDPred (pinteraction = 0.03). In AA individuals, relative to the EA cohort, estimated ORs for CHD were significantly lower (pinteraction ≤ 3.7 × 10−4) (Table 3).
Table 3.
Odds Ratios for (All) CHD per 1 Standard Deviation Increase in PRS
| OR (95% CI)a | p valuea | ModelBasec-indexb | ModelPRSc-indexc | pinteractiond | |
|---|---|---|---|---|---|
| EA | |||||
| PRSTikkanen | 1.24 (1.21–1.28) | <2 × 10−16 | 0.742 | 0.748 | ref |
| PRSTada | 1.28 (1.25–1.32) | <2 × 10−16 | 0.742 | 0.750 | ref |
| PRSLDPred | 1.66 (1.62–1.71) | <2 × 10−16 | 0.742 | 0.770 | ref |
| PRSmetaGRS | 1.73 (1.68–1.78) | <2 × 10−16 | 0.742 | 0.772 | ref |
| AA | |||||
| PRSTikkanen | 1.07 (0.99–1.16) | 0.08 | 0.762 | 0.763 | 3.7 × 10−4 |
| PRSTada | 1.05 (0.98–1.14) | 0.19 | 0.762 | 0.763 | 1.7 × 10−6 |
| PRSLDPred | 1.30 (1.21–1.41) | 1.6 × 10−11 | 0.762 | 0.771 | 5.4 × 10−9 |
| PRSmetaGRS | 1.40 (1.30–1.52) | <2 × 10−16 | 0.762 | 0.775 | 3.6 × 10−6 |
| HE | |||||
| PRSTikkanen | 1.27 (1.12–1.42) | 1.0 × 10−4 | 0.765 | 0.771 | 0.69 |
| PRSTada | 1.20 (1.06–1.35) | 3.4 × 10−3 | 0.765 | 0.769 | 0.28 |
| PRSLDPred | 1.42 (1.25–1.61) | 4.9 × 10−8 | 0.765 | 0.776 | 0.03 |
| PRSmetaGRS | 1.93 (1.67–2.22) | <2 × 10−16 | 0.765 | 0.794 | 0.09 |
Multivariable logistic-regression models, separate for each racial and ethnic group, adjusted for age at first EHR record, duration of EHR, sex, eMERGE site, and first five ancestry-specific principal components.
c-indices for base models without PRSs across racial and ethnic groups: multivariable logistic-regression model with age at first EHR record, duration of EHR, sex, eMERGE site, and first five ancestry-specific principal components.
c-indices for base model + PRSs for each racial and ethnic group.
Multivariable logistic regression models on the joint cohort adjusted for age at first EHR record, duration of EHR, sex, eMERGE site, design matrix of first five ancestry-specific principal components, racial and ethnic group, and an interaction term with PRSs. ref, referent.
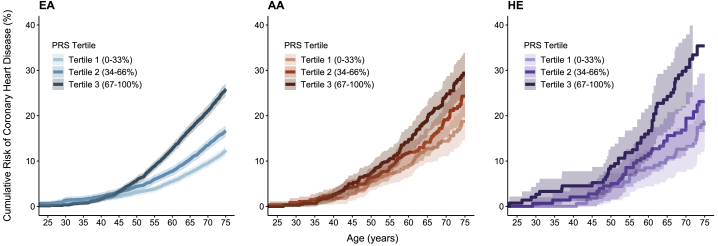
PRSmetaGRS demonstrated the strongest association with CHD in all three groups, resulting in different cumulative risk trajectories when individuals were grouped into tertiles of PRS distribution. Those in the bottom tertile of PRSmetaGRS distribution reached a cumulative CHD risk of 3.9%, 7.3%, and 6.9%, whereas those in the top tertile reached 8.9%, 10.5%, and 11.8% risk by age 55 years in the EA, AA, and HE cohorts, respectively (Figure 1). The cumulative CHD risks estimated with other PRSs are shown in Figures S6–S8.
Figure 1.
Cumulative Risk of CHD by PRSmetaGRS in Three Racial and Ethnic Groups
Cumulative risk of CHD by tertiles of PRSmetaGRS in European ancestry (EA), African ancestry (AA), and Hispanic ethnicity (HE) cohorts, represented by the colors blue (left), orange (middle), and purple (right), respectively. Shaded regions denote 95% confidence intervals.
Estimation of Absolute Risk of MI
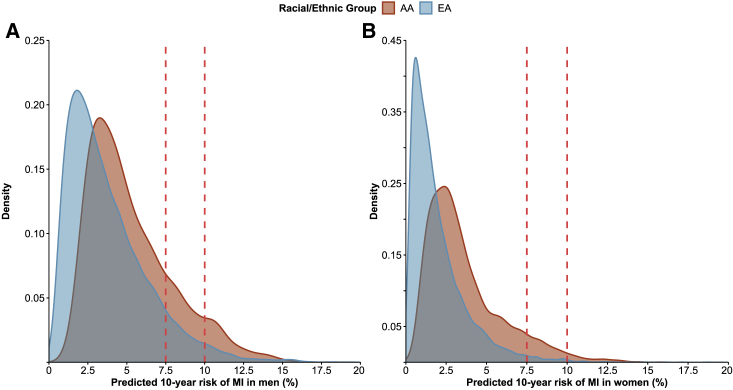
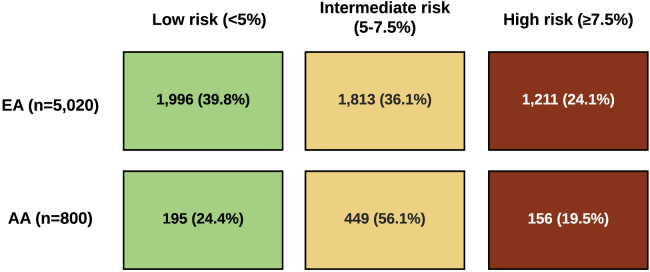
Because genome-wide PRSs were more strongly associated with CHD in both EA and AA groups, we used HRs per 1 SD increase in PRSmetaGRS to estimate 10-year absolute risk of MI on the basis of age- and sex-specific rates of incident MI and respective non-CHD mortality rates in the U.S (Tables S3 and S4). Figure 2 depicts the distribution of the estimated 10-year absolute risk of MI for EA and AA participants aged 35–74 years as well as commonly recommended thresholds for initiation of statin therapy (≥7.5% or ≥10% 10-year risk).49,50 Even though PRSmetaGRS associated less strongly with CHD in the AA cohort than in the EA cohort (HR 1.27 in AA versus 1.53 in EA; per 1 SD increase), higher incidence of MI in the AA cohort across age and sex groups in the US (2.4–15.9 and 1.1–12.0 for AA men and women, respectively, versus 0.8–9.4 and 0.3–8.5 for EA men and women, respectively; per 1,000 person years) resulted in a higher distribution of predicted 10-year absolute risk of MI in AA individuals than in EA individuals as determined from PRSmetaGRS. Adding PRSmetaGRS to the 10-year absolute risk estimates for MI resulted in reclassification of risk categories in both EA and AA individuals at “intermediate” risk (5%–7.5% 10-year risk; Figure 3); 39.8% of the EA participants were reclassified to lower (<5%) and 24.1% were reclassified to higher (≥7.5%) 10-year risk groups, and 24.4% of the AA participants were reclassified to a lower risk group, and 19.5% were reclassified to a higher risk group.
Figure 2.
Distributions of the Absolute CHD Risk Predicted by PRSmetaGRS in EA and AA Individuals
Distributions of predicted 10-year absolute risk of MI based on PRSmetaGRS in EA and AA participants aged 35–74 years. (A) shows distribution of risk in men, whereas (B) shows the distribution of risk in women. Dotted vertical lines represent commonly accepted risk thresholds for statin therapy (≥7.5% and ≥10% 10-year absolute CHD risk).
Figure 3.
Reclassification of CHD Risk Category by PRSmetaGRS in EA and AA Individuals
Reclassification rates after incorporating PRSmetaGRS in individuals aged 35–74 and at intermediate 10-year risk of MI (5%–7.5%) calculated on the basis of age, sex, race- and ethnicity-specific incidence of MI, and corresponding non-CHD mortality rates as competing risks. Note: Conventional CHD risk factors (other than age and sex) were not included in the risk estimates.
Power Calculations for PRS Associations
We assessed the statistical power for testing associations of PRSs with CHD given our sample size and the number of incident CHD events for each racial and ethnic group in the eMERGE cohort. For an OR of 1.25 per 1 SD increase in a restricted PRS for incident CHD in the EA cohort, we assumed an OR of 1.20 in the HE cohort and 1.10 in the AA cohort and expected that effect sizes would attenuate as a result of differences in the proportion of European ancestry51 in these groups. We had 38% and 56% power to detect an association with CHD and 52% and 8% power to detect any heterogeneity of PRS effect in the AA and HE cohorts compared to the EA cohort, respectively. When testing a genome-wide PRS, and assuming a greater effect size in each group (OR 1.50 in EA, 1.20 in AA, and 1.40 in HE), we had greater power to detect associations with CHD (power 85% in AA and 97% in HE). To detect heterogeneity of a genome-wide PRS effect in the AA and HE cohorts with reference to the EA cohort, we had 95% power in the AA cohort and 11% in the HE cohort.
Discussion
In this study, we quantified the strengths of associations of four published PRSs (derived largely from EA cohorts) with CHD in the three major racial and ethnic groups in the US. We replicated previously reported associations of PRSs with CHD in EA individuals and demonstrated that PRSs were similarly associated with CHD in HE individuals but that the associations were significantly weaker in AA individuals.
The generalizability of a PRS across ancestral and ethnic groups depends on LD between the causal and the tagging variant, frequencies of variants, and the genetic architecture of the trait of interest in such groups.21,24,53, 54, 55 HE individuals in the US have a greater proportion of European ancestry than do AA individuals51 and allele frequencies similar to those of EA individuals, which might be why PRS associations were less attenuated in this group. In AA individuals, despite attenuation, genome-wide PRSs remained significantly associated with CHD and had up to ∼1.3-fold increased risk per 1 SD increase in PRS. Inclusion of a large number of variants in the genome-wide PRSs (albeit with effect size estimates derived from EA-based GWAS) might have led to inclusion of variants associated with CHD in both EA and non-EA groups, partially capturing the risk of CHD through shared risk alleles. Restricted PRSs include a much smaller number of variants and therefore might not replicate well across ancestries.23,55,56
A few studies investigated generalizability of genome-wide PRSs in EA and non-EA cohorts. Wünneman et al.57 confirmed the association of PRSmetaGRS and PRSLDPred with prevalent CHD in French-Canadian individuals from three different cohorts. Khera et al.58 evaluated PRSLDPred in a multiethnic cohort of 2,081 early-onset MI cases and 3,761 population-based controls and found attenuated but significant associations of the PRS with prevalent early-onset MI in minority groups—HE individuals, AA individuals, and Asians. The ORs reported58 were higher than our estimates for the association of PRSLDPred with CHD, possibly due to a stronger genetic contribution to early-onset CHD.
We previously demonstrated that disclosure of a PRS for CHD led to lower LDL-C levels11 in a randomized clinical trial and that such disclosure was associated with higher likelihood of information seeking and sharing on cardiovascular disease.59 Other studies have demonstrated that statins and healthy lifestyle factors reduce adverse CHD events in those with a high PRS for CHD.60,61 Although PRSs are beginning to be used in clinical practice,11,62,63 their application to non-EA individuals is unclear. Until race- and ancestry-specific PRSs become available, our results suggest that EA-derived genome-wide PRSs for CHD could be adapted for use in AA individuals while taking into account race-specific CHD risk.
When using PRSs in different racial and ethnic groups, one must keep in mind that epidemiological differences in CHD risk across these groups will influence estimates of absolute risk. In our study, relative to the EA cohort, a lower yet substantial proportion of AA participants were reclassified from the intermediate to high 10-year risk category (24.1% versus 19.5%) on the basis of PRSmetaGRS. Despite a narrower relative risk gradient in the AA cohort, genome-wide PRSs could facilitate decision-making regarding prevention and treatment based on estimates of absolute risk of CHD. Individuals likely to be impacted the most would be those at intermediate risk of CHD, where initiation of lipid-lowering therapy is subject to uncertainty, and those at the extremes of PRS distribution because their risk category is more likely to change. Large and diverse cohorts are needed for the construction of both relative and absolute risk models as well as for evaluation of model validity and calibration46 prior to clinical implementation.
The development of minority-specific PRSs based on empirical data has been challenging because these groups are underrepresented in genomic studies;26 in 2009 ∼96% of GWAS participants were of European ancestry, although this had decreased to ∼80% by 2016.64 Efforts are underway to increase racial and ethnic diversity in research cohorts such as the National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine (TOPMed), the National Institutes of Health’s All of Us program, and the Million Veteran Program65 from the Department of Veterans Affairs healthcare system. Future large multiethnic cohorts will enable construction of racial- and ethnicity-specific PRSs, potentially leading to performances comparable to what is currently available for EA cohorts.
Study Limitations
Several limitations of our study should be noted. The AA and HE cohorts were from a limited number of geographic regions and institutions, were smaller in sample size, and the number of CHD events was lower than in the EA cohort, potentially reducing the precision of risk estimates. We did not have sufficient numbers of individuals of Asian or Native American ancestry to perform the relevant comparative analyses. For time-to-event analyses, follow-up was initiated from the first EHR record because of incomplete data on the DNA sample collection date. CHD events occurring between first EHR record and the DNA sample collection date can bias the hazard ratios toward the null because individuals who were healthy enough to survive until blood collection for genotyping are included. Smoking status and family history of CHD were not available as structured data in EHR systems, possibly resulting in incomplete adjustment for these risk factors in our analyses.
Conclusion
The results of our study represent one of the largest comparative analyses of PRSs for CHD in three major racial and ethnic groups in the US. Genome-wide PRSs were more strongly associated with CHD than were restricted PRSs, and PRSmetaGRS, had the strongest association with CHD in all three groups. The strengths of associations of PRSs with CHD were similar in EA and HE individuals but lower for AA individuals. On the basis of a genome-wide PRS, a substantial proportion of individuals at intermediate risk for CHD were reclassified to either a lower or higher absolute risk category. Our results highlight the potential utility of PRSs for CHD in the clinical setting and suggest that until ancestry- and ethnicity-specific PRSs become available, a genome-wide PRS could be adapted for use in AA individuals.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are indebted to the investigators and participants of the electronic Medical Records and Genomics (eMERGE) Network. The eMERGE Network was initiated and funded by National Human Genome Research Institute (NHGRI) through the following grants: U01HG006828 (Cincinnati Children's Hospital Medical Center and Boston Children's Hospital); U01HG006830 (Children's Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation, and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative and the University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center). This phase of the eMERGE network was initiated and funded by the NHGRI through the following grants: U01HG8657 (Group Health Cooperative/University of Washington); U01HG8685 (Brigham and Women's Hospital); U01HG8672 (Vanderbilt University Medical Center); U01HG8666 (Cincinnati Children's Hospital Medical Center); U01HG6379 (Mayo Clinic); U01HG8679 (Geisinger Clinic); U01HG8680 (Columbia University Health Sciences); U01HG8684 (Children's Hospital of Philadelphia); U01HG8673 (Northwestern University); U01HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG8676 (Partners Healthcare and the Broad Institute); and U01HG8664 (Baylor College of Medicine). This work was also supported by the CTSA grant UL1 TR002377 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). I.J.K. was additionally supported by NIH grant K24 HL137010. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Published: May 7, 2020
Footnotes
Twitter: @iftikhar_kullo
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.002.
Accession Numbers
The dbGaP accession number for the imputed genotype data reported in this paper is phs001584.v1.p1.
Web Resources
CDC Wonder, Underlying Cause of Death 1999–2017, http://wonder.cdc.gov/ucd-icd10.html
Michigan Imputation Server, https://imputationserver.sph.umich.edu/
R for Statistical Computing, https://www.r-project.org/
Supplemental Information
List of ICD and CPT Codes Used in the Electronic Phenotyping Algorithms
References
- 1.McPherson R., Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ. Res. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 2.Kullo I.J., Ding K. Mechanisms of disease: The genetic basis of coronary heart disease. Nat. Clin. Pract. Cardiovasc. Med. 2007;4:558–569. doi: 10.1038/ncpcardio0982. [DOI] [PubMed] [Google Scholar]
- 3.Verweij N., Eppinga R.N., Hagemeijer Y., van der Harst P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci. Rep. 2017;7:2761. doi: 10.1038/s41598-017-03062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Harst P., Verweij N. The identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 6.Howson J.M.M., Zhao W., Barnes D.R., Ho W.-K., Young R., Paul D.S., Waite L.L., Freitag D.F., Fauman E.B., Salfati E.L., CARDIoGRAMplusC4D. EPIC-CVD Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehghan A., Bis J.C., White C.C., Smith A.V., Morrison A.C., Cupples L.A., Trompet S., Chasman D.I., Lumley T., Völker U. Genome-wide association study for incident myocardial infarction and coronary heart disease in prospective cohort studies: The CHARGE Consortium. PLoS ONE. 2016;11:e0144997. doi: 10.1371/journal.pone.0144997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lettre G., Palmer C.D., Young T., Ejebe K.G., Allayee H., Benjamin E.J., Bennett F., Bowden D.W., Chakravarti A., Dreisbach A. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franceschini N., Carty C., Bůzková P., Reiner A.P., Garrett T., Lin Y., Vöckler J.-S., Hindorff L.A., Cole S.A., Boerwinkle E. Association of genetic variants and incident coronary heart disease in multiethnic cohorts: the PAGE study. Circ Cardiovasc Genet. 2011;4:661–672. doi: 10.1161/CIRCGENETICS.111.960096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kullo I.J., Jouni H., Austin E.E., Brown S.-A., Kruisselbrink T.M., Isseh I.N., Haddad R.A., Marroush T.S., Shameer K., Olson J.E. Incorporating a genetic risk score into coronary heart disease risk estimates: Effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding K., Bailey K.R., Kullo I.J. Genotype-informed estimation of risk of coronary heart disease based on genome-wide association data linked to the electronic medical record. BMC Cardiovasc. Disord. 2011;11:66. doi: 10.1186/1471-2261-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tada H., Melander O., Louie J.Z., Catanese J.J., Rowland C.M., Devlin J.J., Kathiresan S., Shiffman D. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur. Heart J. 2016;37:561–567. doi: 10.1093/eurheartj/ehv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikkanen E., Havulinna A.S., Palotie A., Salomaa V., Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2013;33:2261–2266. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thanassoulis G., Peloso G.M., Pencina M.J., Hoffmann U., Fox C.S., Cupples L.A., Levy D., D’Agostino R.B., Hwang S.-J., O’Donnell C.J. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripatti S., Tikkanen E., Orho-Melander M., Havulinna A.S., Silander K., Sharma A., Guiducci C., Perola M., Jula A., Sinisalo J. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., UK Biobank CardioMetabolic Consortium CHD Working Group Genomic risk prediction of coronary artery disease in 480,000 adults: Implications for primary prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.-R., Bhatia G., Do R., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurdasani D., Barroso I., Zeggini E., Sandhu M.S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 2019;20:520–535. doi: 10.1038/s41576-019-0144-0. [DOI] [PubMed] [Google Scholar]
- 23.Ke W., Rand K.A., Conti D.V., Setiawan V.W., Stram D.O., Wilkens L., Le Marchand L., Assimes T.L., Haiman C.A. Evaluation of 71 coronary artery disease risk variants in a multiethnic cohort. Front. Cardiovasc. Med. 2018;5:19. doi: 10.3389/fcvm.2018.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi L., Ma J., Qi Q., Hartiala J., Allayee H., Campos H. Genetic risk score and risk of myocardial infarction in Hispanics. Circulation. 2011;123:374–380. doi: 10.1161/CIRCULATIONAHA.110.976613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manolio T.A. Using the data we have: improving diversity in genomic research. Am. J. Hum. Genet. 2019;105:233–236. doi: 10.1016/j.ajhg.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikpay M., Goel A., Won H.-H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbalic M., Reiner A.P., Wu C., Hixson J.E., Franceschini N., Eaton C.B., Heiss G., Couper D., Mosley T., Boerwinkle E. Genome-wide association analysis of incident coronary heart disease (CHD) in African Americans: a short report. PLoS Genet. 2011;7:e1002199. doi: 10.1371/journal.pgen.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesman O., Kuivaniemi H., Tromp G., Faucett W.A., Li R., Manolio T.A., Sanderson S.C., Kannry J., Zinberg R., Basford M.A., eMERGE Network The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M., eMERGE Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanaway I.B., Hall T.O., Rosenthal E.A., Palmer M., Naranbhai V., Knevel R., Namjou-Khales B., Carroll R.J., Kiryluk K., Gordon A.S., eMERGE Network The eMERGE genotype set of 83,717 subjects imputed to ∼40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet. Epidemiol. 2019;43:63–81. doi: 10.1002/gepi.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safarova M.S., Liu H., Kullo I.J. Rapid identification of familial hypercholesterolemia from electronic health records: The SEARCH study. J. Clin. Lipidol. 2016;10:1230–1239. doi: 10.1016/j.jacl.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei W.-Q., Feng Q., Weeke P., Bush W., Waitara M.S., Iwuchukwu O.F., Roden D.M., Wilke R.A., Stein C.M., Denny J.C. Creation and validation of an EMR-based algorithm for identifying major adverse cardiac events while on statins. AMIA Jt. Summits Transl. Sci. Proc. 2014;2014:112–119. [PMC free article] [PubMed] [Google Scholar]
- 36.Dumitrescu L., Ritchie M.D., Denny J.C., El Rouby N.M., McDonough C.W., Bradford Y., Ramirez A.H., Bielinski S.J., Basford M.A., Chai H.S. Genome-wide study of resistant hypertension identified from electronic health records. PLoS ONE. 2017;12:e0171745. doi: 10.1371/journal.pone.0171745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kho A.N., Hayes M.G., Rasmussen-Torvik L., Pacheco J.A., Thompson W.K., Armstrong L.L., Denny J.C., Peissig P.L., Miller A.W., Wei W.-Q. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J. Am. Med. Inform. Assoc. 2012;19:212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirby J.C., Speltz P., Rasmussen L.V., Basford M., Gottesman O., Peissig P.L., Pacheco J.A., Tromp G., Pathak J., Carrell D.S. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J. Am. Med. Inform. Assoc. 2016;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham G., Havulinna A.S., Bhalala O.G., Byars S.G., De Livera A.M., Yetukuri L., Tikkanen E., Perola M., Schunkert H., Sijbrands E.J. Genomic prediction of coronary heart disease. Eur. Heart J. 2016;37:3267–3278. doi: 10.1093/eurheartj/ehw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Euesden J., Lewis C.M., O’Reilly P.F. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal Choudhury P., Maas P., Wilcox A., Wheeler W., Brook M., Check D., Garcia-Closas M., Chatterjee N. iCARE: An R package to build, validate and apply absolute risk models. PLoS ONE. 2020;15:e0228198. doi: 10.1371/journal.pone.0228198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee N., Shi J., García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics-2019 update: A report from the American Heart Association. Circulation. 2019;139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 48.R Core Team . R Foundation for Statistical Computing; 2018. R: A language and environment for statistical computing.https://www.R-project.org [Google Scholar]
- 49.Arnett D.K., Blumenthal R.S., Albert M.A., Buroker A.B., Goldberger Z.D., Hahn E.J., Himmelfarb C.D., Khera A., Lloyd-Jones D., McEvoy J.W. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bibbins-Domingo Kirsten, US Preventive Services Task Force Statin Use for the Primary Prevention of Cardiovascular Disease in Adults: US Preventive Services Task Force Recommendation Statement. JAMA. 2016;316:1997–2007. doi: 10.1001/jama.2016.15450. [DOI] [PubMed] [Google Scholar]
- 51.Bryc K., Durand E.Y., Macpherson J.M., Reich D., Mountain J.L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grinde K.E., Qi Q., Thornton T.A., Liu S., Shadyab A.H., Chan K.H.K., Reiner A.P., Sofer T. Generalizing polygenic risk scores from Europeans to Hispanics/Latinos. Genet. Epidemiol. 2019;43:50–62. doi: 10.1002/gepi.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iribarren C., Lu M., Jorgenson E., Martínez M., Lluis-Ganella C., Subirana I., Salas E., Elosua R. Weighted multi-marker genetic risk scores for incident coronary heart disease among individuals of African, Latino and East-Asian ancestry. Sci. Rep. 2018;8:6853. doi: 10.1038/s41598-018-25128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Márquez-Luna C., Loh P.-R., Price A.L., South Asian Type 2 Diabetes (SAT2D) Consortium. SIGMA Type 2 Diabetes Consortium Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 2017;41:811–823. doi: 10.1002/gepi.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bressler J., Folsom A.R., Couper D.J., Volcik K.A., Boerwinkle E. Genetic variants identified in a European genome-wide association study that were found to predict incident coronary heart disease in the atherosclerosis risk in communities study. Am. J. Epidemiol. 2010;171:14–23. doi: 10.1093/aje/kwp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franceschini N., Hu Y., Reiner A.P., Buyske S., Nalls M., Yanek L.R., Li Y., Hindorff L.A., Cole S.A., Howard B.V. Prospective associations of coronary heart disease loci in African Americans using the MetaboChip: the PAGE study. PLoS ONE. 2014;9:e113203. doi: 10.1371/journal.pone.0113203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wünnemann F., Sin Lo K., Langford-Avelar A., Busseuil D., Dubé M.-P., Tardif J.-C., Lettre G. Validation of genome-wide polygenic risk scores for coronary artery disease in French Canadians. Circ Genom Precis Med. 2019;12:e002481. doi: 10.1161/CIRCGEN.119.002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khera A.V., Chaffin M., Zekavat S.M., Collins R.L., Roselli C., Natarajan P., Lichtman J.H., D’Onofrio G., Mattera J., Dreyer R. Whole genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. 2019;139:1593–1602. doi: 10.1161/CIRCULATIONAHA.118.035658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown S.N., Jouni H., Marroush T.S., Kullo I.J. Effect of disclosing genetic risk for coronary heart disease on information seeking and sharing: The MI-GENES Study (Myocardial Infarction Genes) Circ Cardiovasc Genet. 2017;10:e001613. doi: 10.1161/CIRCGENETICS.116.001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khera A.V., Emdin C.A., Drake I., Natarajan P., Bick A.G., Cook N.R., Chasman D.I., Baber U., Mehran R., Rader D.J. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N. Engl. J. Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natarajan P., Young R., Stitziel N.O., Padmanabhan S., Baber U., Mehran R., Sartori S., Fuster V., Reilly D.F., Butterworth A. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knowles J.W., Ashley E.A. Cardiovascular disease: The rise of the genetic risk score. PLoS Med. 2018;15:e1002546. doi: 10.1371/journal.pmed.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 64.Mensah G.A., Jaquish C., Srinivas P., Papanicolaou G.J., Wei G.S., Redmond N., Roberts M.C., Nelson C., Aviles-Santa L., Puggal M. Emerging concepts in precision medicine and cardiovascular diseases in racial and ethnic minority populations. Circ. Res. 2019;125:7–13. doi: 10.1161/CIRCRESAHA.119.314970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of ICD and CPT Codes Used in the Electronic Phenotyping Algorithms