Abstract
Despite evidence that deleterious variants in the same genes are implicated across multiple neurodevelopmental and neuropsychiatric disorders, there has been considerable interest in identifying genes that, when mutated, confer risk that is largely specific for autism spectrum disorder (ASD). Here, we review the findings and limitations of recent efforts to identify relatively “autism-specific” genes, efforts which focus on rare variants of large effect size that are thought to account for the observed phenotypes. We present a divergent interpretation of published evidence; discuss practical and theoretical issues related to studying the relationships between rare, large-effect deleterious variants and neurodevelopmental phenotypes; and describe potential future directions of this research. We argue that there is currently insufficient evidence to establish meaningful ASD specificity of any genes based on large-effect rare-variant data.
Keywords: ▪▪▪
Main Text
Autism spectrum disorder (ASD) is clinically and etiologically heterogeneous, and a unifying pathophysiology has not yet been identified for either the disorder as a whole or its core behavioral components. Heritability estimates are high (0.65–0.91) based on family and twin studies,1, 2, 3 and elucidation of the complex genetic architecture of ASD is revealing contributions from both rare and common variants. Chromosomal microarray and next-generation sequencing studies have identified many de novo and inherited rare variants of large effect size that contribute substantially to the etiology of ASD. It has also become clear that pathogenic variants in the same genes are identified in individuals with a variety of different clinically defined brain disorders, including ASD, intellectual disability (ID), epilepsy, schizophrenia, and other neurodevelopmental and neuropsychiatric conditions.4, 5, 6, 7, 8, 9 The known collective contribution of rare, large-effect pathogenic variants is greatest for neurodevelopmental disorders (NDDs) such as ID, ASD, and epilepsy, but they are also important etiologic factors in other conditions with onset in childhood (e.g., attention-deficit/hyperactivity disorder [ADHD]) or adolescence (e.g., schizophrenia) and, to a lesser degree, to later-onset neuropsychiatric conditions such as mood disorders.
Despite the evidence that deleterious variants in the same genes are implicated across multiple disorders, the recent literature reflects considerable interest in identifying genes that, when mutated, confer risk that is largely specific for ASD. Here, we review and comment on recent efforts to identify “autism genes,” efforts which focus on rare variants of large effect size that are thought to account for the observed phenotype in participants. We discuss practical and theoretical limitations to studying the relationships between rare, large-effect deleterious variants and ASD and other NDD phenotypes, along with the current lack of sufficient evidence to establish meaningful ASD specificity, as well as the possibility that other sources of genetic variation, such as common variant-related polygenic risk, may confer ASD-specific risk.
Categorical Diagnosis-Based Cohort Studies
To date, no genes have been identified that, when mutated, confer only ASD risk and not risk for ID or other NDDs. However, several recent studies have attempted to identify genes that are relatively ASD-specific (ASD-predominant or ASD-biased) by comparing the distribution of likely gene-disruptive de novo mutations between cohorts ascertained based on ASD or ID and/or developmental delay (ID/DD).10, 11, 12 For example, Satterstrom et al.11 asserted that among 102 genes implicated in ASD risk, some genes are relatively ASD-predominant, and others are associated with more global developmental impairment, including both ASD and ID and/or severe neurodevelopmental delay (ASD with ID/DD), based on comparison of the frequency of disruptive de novo variants among individuals ascertained for ASD (n = 11,986) to the frequency of such variants among those ascertained for severe ID/DD (n = 5,264). In this study, the authors defined “ASD-predominant genes” as those for which the ratio of the frequency of de novo disruptive variants identified in cohorts of individuals ascertained for ASD compared to the frequency of de novo disruptive variants in cohorts of individuals ascertained for ID/DD was greater than 1.0. Conversely, genes were classified as “ASD with neurodevelopmental delay genes” (referred to here as “ASD with ID/DD”) when the ratio of the frequency of disruptive de novo variants in ASD-ascertained participants compared to that in ID/DD-ascertained participants was less than 1.0. In this manner, 50 of the 102 genes were classified as ASD-predominant and 49 as ASD with ID/DD. Three additional genes were assigned to the ASD-predominant group on the basis of case-control data, bringing the total to 53 genes.11
Coe et al.12 also compared the distribution of likely gene-disruptive mutations between ASD- and ID/DD-ascertained cohorts but did not find evidence of ASD specificity for any of the 253 genes they identified as candidate NDD genes based on evidence of excess of de novo mutations through the use of two statistical models. In fact, 72% of genes predicted to be significant by the two statistical models showed evidence of excess de novo variants in both ASD and ID/DD cohorts. This study included fewer individuals ascertained for ASD (n = 5,624) than did the study by Satterstrom and colleagues,11 but a similar number ascertained for ID/DD (n = 5,303). In fact, the ID/DD cohorts evaluated in these two studies overlapped almost completely; each study used samples from five previously published studies, and four of the five were the same studies, accounting for 99% of the samples.11, 12, 13, 14, 15, 16
Although the issue of whether loss-of-function variants in certain genes confer risk that is relatively ASD-specific has been explored mainly in relation to ID risk, the question applies to other NDDs as well. Recently, the burden of rare protein-truncating variants in evolutionarily constrained genes was shown to be similar among individuals with ASD and those with ADHD.17 One analysis was limited to individuals with only one diagnosis each (ASD or ADHD, but not both; with no ID or other comorbid diagnoses) and was confined to a set of 212 constrained genes with a published rare protein-truncating variant in ASD. Even in this ASD-derived gene set, the rates of constrained rare protein-truncating variants among those with ASD and those with ADHD were not significantly different.17
Limitations of Categorical Diagnosis-Based Cohort Comparisons
Although pragmatic in terms of data availability, a significant problem with the cohort-ascertainment-based approach used in the Satterstrom et al.11 and Coe et al.12 studies is the potential bias introduced by the unequal opportunity for each participant to receive each diagnosis (i.e., ASD and ID) due to the lack of uniform phenotyping across studies. Because of this bias, the phenotypic overlap between the groups is unclear; the prevalence of ASD in several of the ID/DD-ascertained cohorts is not quantified,13, 14, 15,18,19 and the prevalence of ID in the ASD-ascertained cohorts is known only for a minority subset of participants.11,12 For example, standardized ASD diagnostic measures were frequently utilized in the ASD cohorts but not in the ID/DD cohorts.11,12 The majority of individuals in the group ascertained for ID/DD phenotype came from the Deciphering Developmental Disorders Study, for which the recruitment criteria included phenotypes such as multiple congenital anomalies, dysmorphic features, and abnormal growth in addition to neurodevelopmental diagnoses. Differences in age distribution between the cohorts may also impact the opportunity for an individual to receive each diagnosis. Another issue with this type of analysis is the scientifically arbitrary threshold used to define ASD-predominance (relative frequency of disruptive de novo variants of a gene in ASD-ascertained exomes versus ID/DD-ascertained exomes); a cutoff of >1.0 was used by Satterstrom and colleagues,11 meaning that a simple majority could establish ASD-predominance. This is a narrow distinction of dubious clinical significance. For example, if exome sequencing of two equally sized cohorts yielded 13 disruptive de novo variants in a particular gene among ASD-ascertained cases and 12 disruptive de novo variants in that gene among ID/DD-ascertained cases, the relative frequency would be 1.08 (13/12), and the criteria for ASD-predominance would be met. Even if the cohorts were non-overlapping in terms of diagnoses (i.e., none of the ASD-ascertained subjects had ID and none of the ID/DD-ascertained individuals had ASD), the rate of ASD among those with a pathogenic variant in this gene would be 52% (13/25), and the rate of ID/DD would be 48% (12/25), which is certainly not a clear indication of clinically significant ASD specificity. In fact, it is possible that in the same scenario, all 13 individuals ascertained for ASD could also have ID. In this case, the gene would still be classified as ASD-predominant even though all 25 individuals (100%) would have ID and only 52% would have ASD. This poses a challenge to the validity of the ascertainment-based approach.
It is also important to be able to assess the impact of intelligence quotient (IQ) on any differences between cohorts ascertained for autism and those ascertained for ID/DD. Individuals who participate in studies in which ascertainment is based on ID/DD may not have the same opportunity for diagnosis of ASD, not only because of differences in phenotyping methods (e.g., tests administered), but also because of the impact of very low IQ. For example, someone with an IQ of 40 may have the same ASD characteristics as someone with an IQ of 70, yet they may be discrepant for ASD diagnosis becausee one of the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-5) criteria for ASD specifies that “to make comorbid diagnoses of autism spectrum disorder and intellectual disability, social communication should be below that expected for general developmental level.”20 The same degree of social communication and interaction impairment may be below what is expected for someone with an IQ of 70, but not below the expectation for someone with an IQ of 40, resulting in the former individual meeting DSM-5 ASD criteria and the latter not meeting criteria for the diagnosis.
Satterstrom and colleagues11 demonstrated that disruptive de novo variants, including those in the 102 ASD genes, occur more commonly than expected even among individuals with ASD and higher IQ (defined as IQ > 70 and IQ > 82 in separate analyses); this suggests that de novo variants do not solely impair cognition. However, this finding does not eliminate the possibility that a large difference in mean IQ between the ASD-ascertained cohorts and the ID/DD-ascertained cohorts could be the primary factor responsible for the classification of genes as ASD-predominant or not. Among the subset of ASD probands with a detected de novo variant and available full-scale IQ (which represented 46.8% of the ASD probands from familyb ased samples and 25.1% of the total number of individuals with ASD), the rate of ID (defined as IQ < 70) was 30.6%, and there were significant mean IQ differences among the three groups: ASD + ID/DD genes (mean IQ 62) < ASD-predominant genes (mean IQ 74) < idiopathic ASD (mean IQ 82).11 All three group IQ means were significantly below the general population mean of 100, and it is clear that even the genes classified as ASD-predominant have a deleterious impact on cognition when mutated (mean IQ 74), though not as great as that of the ASD + ID/DD genes (mean IQ 62).11
Similarly, among individuals with ASD from the Simons Simplex Collection (SSC), the group with de novo likely gene disruptive (LGD) variants in any of 173 high-confidence ASD-associated genes (n = 74) had a significantly lower mean IQ (69.1) and higher ASD severity than did those with no such variants (n = 2,216; mean IQ 81.9).21 When the subgroup of individuals with ASD and IQ > 100 (n = 337) was compared to those with ASD and IQ < 70 (n = 562), the high IQ group had a significantly lower rate of de novo LGD variants overall and in the 173 ASD-associated genes than did the group with ASD and ID.21 Among this ASD cohort, the risk of coexisting ID was substantially increased by the presence of a deleterious variant in an ASD-associated gene.
Genotype-Based Cohorts
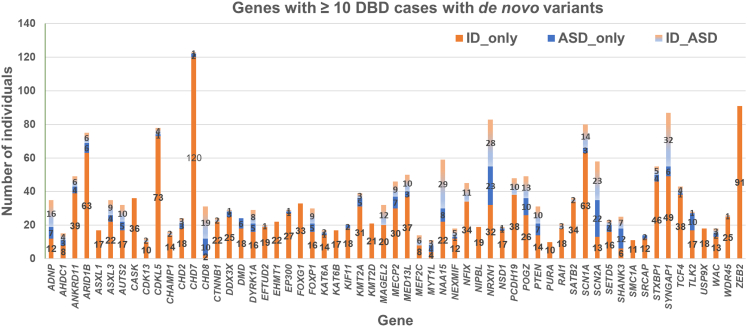
The high rate of ID among individuals with pathogenic variants in ASD-associated genes is also evident in data from other sources. For example, the Geisinger Developmental Brain Disorder Genes Database (DBD Genes Database) integrates data from exome and genome sequencing, copy number array, and targeted gene studies with phenotype data for six clinically defined brain disorders (including ASD and ID) to examine the phenotypes associated with de novo pathogenic loss-of-function (pLoF) variants (see Web Resources).22 At the time it was accessed, the database included 5,031 cases with loss-of-function variants in 553 genes based on information from 923 unique articles published between March 2003 and March 2019 that met curation criteria.22 Among 59 genes for each of which there are at least 10 total probands with de novo pLOF variants in the database, none are associated exclusively with ASD (Figure 1). For three genes, CHD8 [MIM: 610528], SHANK3 [MIM: 606230], and SCN2A [MIM: 182390], there are more individuals with ASD only than with ID only, but many have both ASD and ID, and for each of these genes, there are more individuals with ID than without ID (ID rates 68%, 52%, and 62% for CHD8, SHANK3, and SCN2A, respectively). However, this database is also limited by the variability in phenotypic information available from published studies, and therefore subject to potential ascertainment bias.
Figure 1.
Genes with ≥10 cases with de novo pLOF variants in the Geisinger Developmental Brain Disorder Genes Database
The number of individuals with intellectual disability (ID) without autism spectrum disorder (ASD) (ID_only), ASD without ID (ASD_only), and both ID and ASD (ID_ASD) is plotted for each of the 59 genes for which there are at least 10 total probands with de novo pLOF variants in the Geisinger Developmental Brain Disorder (DBD) Genes Database (see Web Resources). The DBD Genes Database is a curated resource providing genotype and phenotype data from six neurodevelopmental disorders (ID, ASD, attention-deficit/hyperactivity disorder, schizophrenia, bipolar disorder, and epilepsy) obtained from published literature.
Detailed phenotyping studies of individuals with pathogenic variants in specific genes are another source of data for evaluating ASD specificity. For example, CHD8 is frequently discussed as a model “ASD gene” because mutations in this gene are associated with a high rate of ASD.23 However, among 89 individuals with pathogenic variants in CHD8 for whom diagnostic information is available, the frequency of ID (62/89, 70%) is similar to that suggested by the DBD Genes Database query (Bernier, unpublished data). Specific IQ scores are available for a subset of these individuals: the mean nonverbal IQ is 67.8 (SD = 28.6) (n = 40) and the mean verbal IQ is 65.5 (SD = 28.2) (n = 35). Similarly, Douzgou and colleagues reported that among 25 individuals with protein-truncating CHD8 variants, 17 (68%) had ID (81% of the 21 with available clinical information about intellectual functioning).24 Twenty-one of the 25 (84%) had a diagnosis of ASD.24 However, ascertainment bias is also a concern with clinical cohorts such as these because individuals with mild phenotypes are less likely to be identified. For example, Guo et al. 25 assessed three families in which likely gene-disrupting CHD8 mutations were transmitted from parents with full-scale IQ scores between 80 and 87 (verbal IQ 88–-95, nonverbal IQ 75–79), establishing the possibility of relative sparing of cognition. Still, it is difficult to determine how meaningful it is to describe any gene as being ASD-specific, or even ASD-predominant, when pathogenic variants in that gene are also associated with cognitive impairment, including ID, in such a high proportion of individuals.
Theoretical Underpinnings and Unanswered Questions
A key question is: What would be necessary to demonstrate meaningful ASD specificity (or predominance) of large-effect rare variants? If loss of function of a particular gene conferred risk that was purely specific to ASD, the mean IQ associated with de novo pathogenic variants in that gene would not be significantly different from the population mean (100), or at least from the familial background mean, and the expected rate of ID would be no different from the ID rate in the general population (∼1.0%–1.3%).26,27 This is clearly not the case for the genes and variants identified to date and is very unlikely given the high rate of comorbidity. Rare, large-effect variants that increase risk for neurodevelopmental and neuropsychiatric disorders are also associated with deleterious effects on cognition,28, 29, 30 and ASD is no exception. Perhaps relative ASD specificity could be inferred if it were demonstrated that the ASD rate among individuals with pathogenic variants involving a given gene were significantly greater than the ASD rate among individuals matched for IQ and other appropriate demographics. However, any valid assessment would still require enough uniformity in phenotyping to allow equal opportunity for each participant to receive each diagnosis (i.e., ASD and ID), and even a statistically significant difference may not be a meaningful definition of ASD specificity if there is also a deleterious impact of the variant on IQ.
Another important pragmatic question is whether genotype-phenotype relationships should be evaluated and curated for individual neurodevelopmental or neuropsychiatric diagnoses, or whether these clinically defined disorders should be thought of not as causally and pathophysiologically distinct, but rather as manifestations of underlying developmental brain dysfunction and “lumped” accordingly for the purpose of assessing pathogenicity of variants and exploring variation in phenotypic expression.4,22,31 The latter approach, unlike the categorical, phenomenological nosology of the Diagnostic and Statistical Manual of Mental Disorders (DSM) and International Classification of Diseases (ICD), is consistent with existing data regarding within-disorder etiologic heterogeneity (“one disorder, multiple causes”), overlapping symptoms and high rates of comorbidity among disorders, high frequency of intermediate or subthreshold cases, and shared risk factors and etiologies across disorders (including variable expressivity of pathogenic gene and copy number variants—“one gene, multiple disorders”).4,32, 33, 34, 35 Efforts to elucidate the contributions of genomic, environmental, and stochastic developmental variation to phenotypic variability in neurodevelopmental and neuropsychiatric disorders have been hampered by reliance on categorical diagnoses, which are effective heuristics that may enhance interrater reliability, but do not align well with genomic, neuroimaging, and other neurobiological findings.33,34,36, 37, 38
Beyond the issues of design and methodology in genetic studies, the diagnosis of ASD in the presence of ID is fundamentally complicated. Many of the core social communication deficits that characterize ASD and are necessary for diagnosis represent a failure to acquire developmentally expected skills and, therefore, are expected to be present to some extent in individuals with ID.39 DSM-5 classification of ID severity is based on adaptive functioning across conceptual, social, and practical domains, and the social ability expectations overlap with the social deficits that define ASD.20,39 For example, the DSM-5 description of social domain impairments in moderate ID includes the examples “individuals may not perceive or interpret social cues accurately” and “social judgement and decision-making abilities are limited.”20 This makes determination of whether the social communication and interaction deficits are beyond what can be attributed to the level of general intellectual functioning (a requirement for DSM-5 ASD diagnosis) very difficult. In addition, many instruments developed for the assessment of social communication and interaction were developed primarily in populations without ID, and this complicates assessment of individuals with ID.39,40 Even the most comprehensive and well-researched tools used in the diagnosis of ASD, such as the Autism Diagnostic Interview-Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS-2) are far less specific when used for individuals with very low mental ages, and over-diagnosis of autism in individuals with low IQ is common with the Diagnostic Interview for Social Communication Disorders (DISCO).39,41 The complexities of clinical diagnosis, limitations of available diagnostic tools, and somewhat arbitrary nature of categorical cutoff points suggest caution with reliance upon dichotomous categorical ASD classification (i.e., ASD/no ASD) in studies of populations which include individuals with ID.
Implications for Clinical Genetic Testing
Despite the lack of strong evidence of ASD specificity of large-effect rare variants, many clinical laboratories market next-generation sequencing test panels for ASD, and it has been asserted that there is a need to develop a validated list of genes appropriate for inclusion in ASD-specific clinical laboratory test panels.42 This approach may provide marketing value, but it does not provide scientific value. In some respects, the situation with neurodevelopmental and neuropsychiatric disorders is analogous to that of structural congenital heart defects (CHDs). Each major anatomical type of CHD can be caused by multiple different genetic variants (etiologic heterogeneity) and each genetic variant identified as a cause of CHDs is associated with multiple different specific defects (variable expressivity).43 For example, tetralogy of Fallot (ToF) is associated with several dozen causal genetic abnormalities, including single gene variants, copy-number variants (CNVs), and chromosomal aneuploidies, and each of these pathogenic variants is also associated with multiple other CHD phenotypes. A 1:1 correspondence between genotype and specific CHD phenotype does not exist. It is not advantageous, for example, to consider each of the many heart defects associated with the recurrent 22q11.2 deletion (or TBX1 within this CNV region) separately to determine a causal relationship. Similarly, there would be no justification for marketing separate gene sequencing panels for most individual cardiac phenotypes (e.g., ToF, transposition of the great arteries, hypoplastic left heart syndrome, interrupted aortic arch, etc.).
Although gene panel testing may offer some advantages, such as sensitivity for detection of mosaicism and single exon-level deletions and/or duplications (when targeted deletion/duplication analysis is included), the set of genes included requires continual revision as new associations are discovered, and reanalysis is limited to the genes originally included in the panel.44 Recently, exome sequencing has been recommended as a first-tier clinical test for individuals with unexplained NDDs by a multidisciplinary expert group following completion of scoping review and meta-analysis of diagnostic yield.44 A comprehensive genomic analysis provides greater flexibility for reanalyzing the data as our knowledge and understanding of NDDs expands.
Moving Forward
Ascertainment bias and lack of uniform phenotyping severely limit the conclusions that can be drawn about the specificity of large-effect rare genetic variants for the ASD phenotype from the currently available cohorts of convenience. It is premature to declare that large-effect rare variants in any genes confer risk that is ASD-specific or meaningfully ASD-predominant, and it is possible that no such variants will ever be found. Among individuals whose developmental brain dysfunction is attributable to a rare variant of large effect size, factors such as background polygenic risk (conferred by a large number of common variants of small individual effect size) and stochastic developmental variation may be more important determinants of the specific neurodevelopmental phenotypes expressed than the gene(s) involved in the primary genetic etiology.
Effect sizes of deleterious variants on traits relevant to ASD, ID, and other neurodevelopmental and neuropsychiatric phenotypes can be quantified. The neurodevelopmental phenotype, whether pathological or not, depends on the profile of quantitative deleterious effects associated with the rare variant, other sources of genetic variation such as polygenic and oligogenic background risk, and environmental and stochastic variation. The possibility that for a given effect size of deleterious impact on IQ, rare variants in some genes may have a substantially greater or lesser effect size on core ASD features is worth exploring, because it may facilitate elucidation of the genomics and neurobiology of social communication and interaction. However, the evidence so far indicates that rare, large-effect mutations that cause ASD also cause cognitive impairment, including ID, in a high proportion of individuals. Answering the question of whether there are genes that, when mutated, confer risk that is meaningfully ASD-specific will require large-scale studies that cross diagnostic boundaries and include adequate phenotyping of all affected individuals, providing equal opportunity for diagnosis of each condition. Phenotyping should include quantitative measurement of continuously distributed traits (e.g., cognitive, behavioral, and neuroimaging traits), not just dichotomous categorical diagnoses.4,45, 46, 47, 48 Study designs that include analyses of the impact of quantified parental phenotypes and the individual’s own polygenic scores for various traits will facilitate accurate interpretation of rare genomic variants. Such studies should be conducted in a variety of samples including disease cohorts and unselected birth and population-based cohorts. Accumulation of larger cohorts of individuals with the same rare, large-effect pathogenic variants will facilitate evaluation of within-group genotype-phenotype correlations and allow comparisons across genetic diagnoses.48 It is also possible that grouping of patients with variants that are expected to impact the same molecular pathways may identify some degree of ASD specificity at the pathway level, rather than the individual gene level, although no molecular pathways are currently known to be uniquely associated with ASD when disrupted.49
Mutation severity also remains an important factor to consider. Variants that might cause ASD alone could be so mild (partial loss of function) that we might not even recognize them as consequential. In some people, they might manifest as ASD, possibly due to an additional hypomorphic allele in another gene (polygenic model) or simply because they impact gene function, and thus neural circuits, albeit mildly. These same variants might lead to a neurotypical phenotype in some people due to the genetic background. Such alleles might explain some of the isolated ASD cases, but information about functionality is limited. There is evidence that ASD risk of smaller effect size can be conferred by rare, inherited CNVs and protein-truncating mutations that disrupt genes intolerant to functional variation.50,51 It is important to note that a particular class of variant may confer different effects, even within the same gene. For example, missense variants may have severe consequences within certain protein domains, or even just at certain amino acid positions of a gene, and mild consequences elsewhere. In the case of SHANK3, functional and phenotypic modularity has been demonstrated; an ASD-associated missense mutation has been shown to interfere with one aspect of protein function and cause a subset of the phenotypes found with loss-of-function mutations.52 Elucidation of the role of rare variants of smaller effect size in ASD will require “top down,” large, carefully designed studies of people with ASD and “bottom up” functional studies using animal models (e.g., for characterizing missense variants).
In contrast to the situation with large-effect rare variants, genetic model fitting in twins53 and the consistent positive correlation of ASD with polygenic scores for IQ and educational attainment54, 55, 56, 57 suggest that common variant-associated risk may load on cognitive and behavioral dimensions that are distinct from those affected by rare variants and may be more specific to ASD. By definition, de novo variants do not account for the considerable heritability of autism, and in considering inherited genetic factors that mediate familial transmission of autism, Xie et al. recently showed in a population-based cohort that the aggregation of ID among first-degree relatives of non-intellectually disabled individuals with ASD was relatively low (odd ratio 2.3) compared to that of first-degree relatives of individuals with ASD and ID (OR 7.6).58 Moreover, Grove et al. showed that the positive correlation between polygenic scores for IQ and ASD is principally driven by the subgroup diagnosed with autism without ID.57 A recent observation that ASD is genetically correlated with empathy (negative correlation) and systemizing (positive correlation) suggests that social and non-social core ASD symptoms are partially genetically dissociable,59 and it is possible that such distinct genetic backgrounds might influence the diagnostic classification and clinical trajectory of individuals. Common variant-related polygenic risk and rare inherited variants in the genetic background also impact the phenotypic expression of rare, large-effect pathogenic variants in NDDs, and their potential role in conferring ASD-specific risk remains to be explored.47,55,60, 61, 62, 63
Declaration of Interests
S.M.M., T.D.C., R.B., T.B., J.N.C., S.J., D.T.M., K.J.M., H.Y.Z., and C.L.M. declare no competing interests. W.K.C. is on the scientific advisory board of the Regeneron Genetics Center. E.E.E. is on the scientific advisory board of DNAnexus. D.H.L. is on the scientific advisory boards of Clear Genetics (past), Natera, and X-Therma.
Acknowledgments
We thank Hermela Shimelis for assistance with the Geisinger DBD Genes Database data and the creation of Figure 1. This work was supported, in part, by the National Institute of Mental Health (NIMH) and Eunice Kennedy Shriver National Institute of Child Health and Human Development of the US National Institutes of Health (NIH), under award numbers R01MH074090, R01MH107431, and U01MH11970510 (D.H.L., C.L.M., S.M.M.); Institut Pasteur, Université de Paris, Fondation Bettencourt-Schueller (T.B.); the Simons Foundation Autism Research Initiative (W.K.C.); the Eunice Kennedy Shriver National Institute of Child Health and Human Development award number U54 HD087011 (J.N.C.); NIH grant MH101221 (E.E.E.); and the Canadian Institute of Health Research Canada Research Chair, Canadian Institute of Health Research award number 400528, and NIMH award number U01 MH119690-01 (S.J.). E.E.E. is an investigator of the Howard Hughes Medical Institute. D.T.M. receives an honorarium to serve on the Medical Genetics Committee of the Simons Foundation Powering Autism Research (SPARK) project and receives salary support from NIH grant U41 HG006834 (a Unified Clinical Genomics Database). H.Y.Z. is an investigator of the Howard Hughes Medical Institute.
Contributor Information
Scott M. Myers, Email: smyers1@geisinger.edu.
David H. Ledbetter, Email: dhledbetter@geisinger.edu.
Web Resources
Geisinger Developmental Brain Disorder Genes Database, 4/29/19 update, accessed 10/29/19, https://dbd.geisingeradmi.org/
Online Mendelian Inheritance in Man, https://omim.org/
References
- 1.Sandin S., Lichtenstein P., Kuja-Halkola R., Larsson H., Hultman C.M., Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tick B., Bolton P., Happé F., Rutter M., Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatry. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai D., Yip B.H.K., Windham G.C., Sourander A., Francis R., Yoffe R., Glasson E., Mahjani B., Suominen A., Leonard H. Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry. 2019 doi: 10.1001/jamapsychiatry.2019.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-De-Luca A., Myers S.M., Challman T.D., Moreno-De-Luca D., Evans D.W., Ledbetter D.H. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stessman H.A., Turner T.N., Eichler E.E. Molecular subtyping and improved treatment of neurodevelopmental disease. Genome Med. 2016;8:22. doi: 10.1186/s13073-016-0278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donovan M.C., Owen M.J. The implications of the shared genetics of psychiatric disorders. Nat. Med. 2016;22:1214–1219. doi: 10.1038/nm.4196. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan P.F., Geschwind D.H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177:162–183. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W., Corominas R., Lin G.N. De novo mutations from whole exome sequencing in neurodevelopmental and psychiatric disorders: From discovery to application. Front. Genet. 2019;10:258. doi: 10.3389/fgene.2019.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarrei M., Burton C.L., Engchuan W., Young E.J., Higginbotham E.J., MacDonald J.R., Trost B., Chan A.J.S., Walker S., Lamoureux S. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom. Med. 2019;4:26. doi: 10.1038/s41525-019-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stessman H.A., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.-Y., Peng M., Collins R., Grove J., Klei L. ). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020 doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coe B.P., Stessman H.A.F., Sulovari A., Geisheker M.R., Bakken T.E., Lake A.M., Dougherty J.D., Lein E.S., Hormozdiari F., Bernier R.A., Eichler E.E. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat. Genet. 2019;51:106–116. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 14.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 15.Lelieveld S.H., Reijnders M.R., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 16.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satterstrom F.K., Walters R.K., Singh T., Wigdor E.M., Lescai F., Demontis D., Kosmicki J.A., Grove J., Stevens C., Bybjerg-Grauholm J., iPSYCH-Broad Consortium Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 2019;22:1961–1965. doi: 10.1038/s41593-019-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 19.Halvardson J., Zhao J.J., Zaghlool A., Wentzel C., Georgii-Hemming P., Månsson E., Ederth Sävmarker H., Brandberg G., Soussi Zander C., Thuresson A.C., Feuk L. Mutations in HECW2 are associated with intellectual disability and epilepsy. J. Med. Genet. 2016;53:697–704. doi: 10.1136/jmedgenet-2016-103814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Psychiatric Association . Fifth Edition. American Psychiatric Association; 2012. Diagnostic and Statistical Manual of Mental Disorders, DSM-5. [Google Scholar]
- 21.Jensen M., Smolen C., Girirajan S. Gene discoveries in autism are biased towards comorbidity with intellectual disability. bioRxiv. 2019 doi: 10.1101/715755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Mantilla A.J., Moreno-De-Luca A., Ledbetter D.H., Martin C.L. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry. 2016;73:275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douzgou S., Liang H.W., Metcalfe K., Somarathi S., Tischkowitz M., Mohamed W., Kini U., McKee S., Yates L., Bertoli M., Deciphering Developmental Disorders Study The clinical presentation caused by truncating CHD8 variants. Clin. Genet. 2019;96:72–84. doi: 10.1111/cge.13554. [DOI] [PubMed] [Google Scholar]
- 25.Guo H., Wang T., Wu H., Long M., Coe B.P., Li H., Xun G., Ou J., Chen B., Duan G. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol. Autism. 2018;9:64. doi: 10.1186/s13229-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Van Naarden Braun K., Christensen D., Doernberg N., Schieve L., Rice C., Wiggins L., Schendel D., Yeargin-Allsopp M. Trends in the prevalence of autism spectrum disorder, cerebral palsy, hearing loss, intellectual disability, and vision impairment, metropolitan atlanta, 1991-2010. PLoS ONE. 2015;10:e0124120. doi: 10.1371/journal.pone.0124120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefansson H., Meyer-Lindenberg A., Steinberg S., Magnusdottir B., Morgen K., Arnarsdottir S., Bjornsdottir G., Walters G.B., Jonsdottir G.A., Doyle O.M. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 29.Huguet G., Schramm C., Douard E., Jiang L., Labbe A., Tihy F., Mathonnet G., Nizard S., Lemyre E., Mathieu A., IMAGEN Consortium Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA Psychiatry. 2018;75:447–457. doi: 10.1001/jamapsychiatry.2018.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganna A., Satterstrom F.K., Zekavat S.M., Das I., Kurki M.I., Churchhouse C., Alfoldi J., Martin A.R., Havulinna A.S., Byrnes A., GoT2D/T2D-GENES Consortium. SIGMA Consortium Helmsley IBD Exome Sequencing Project. FinMetSeq Consortium. iPSYCH-Broad Consortium Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am. J. Hum. Genet. 2018;102:1204–1211. doi: 10.1016/j.ajhg.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Lancet Neurology A shifting view of neurodevelopmental disability. Lancet Neurol. 2013;12:323. doi: 10.1016/S1474-4422(13)70063-2. [DOI] [PubMed] [Google Scholar]
- 32.Sebat J., Levy D.L., McCarthy S.E. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyman S.E. The diagnosis of mental disorders: the problem of reification. Annu. Rev. Clin. Psychol. 2010;6:155–179. doi: 10.1146/annurev.clinpsy.3.022806.091532. [DOI] [PubMed] [Google Scholar]
- 34.Lilienfeld S.O., Treadway M.T. Clashing diagnostic approaches: DSM-ICD versus RDoC. Annu. Rev. Clin. Psychol. 2016;12:435–463. doi: 10.1146/annurev-clinpsy-021815-093122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark L.A., Cuthbert B., Lewis-Fernández R., Narrow W.E., Reed G.M. Three Approaches to Understanding and Classifying Mental Disorder: ICD-11, DSM-5, and the National Institute of Mental Health’s Research Domain Criteria (RDoC) Psychol. Sci. Public Interest. 2017;18:72–145. doi: 10.1177/1529100617727266. [DOI] [PubMed] [Google Scholar]
- 36.Vogt G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J. Biosci. 2015;40:159–204. doi: 10.1007/s12038-015-9506-8. [DOI] [PubMed] [Google Scholar]
- 37.Tikhodeyev O.N., Shcherbakova О.V. The problem of non-shared environment in behavioral genetics. Behav. Genet. 2019;49:259–269. doi: 10.1007/s10519-019-09950-1. [DOI] [PubMed] [Google Scholar]
- 38.Insel T.R., Cuthbert B.N. Medicine. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 39.Thurm A., Farmer C., Salzman E., Lord C., Bishop S. State of the field: Differentiating intellectual disability from autism spectrum disorder. Front. Psychiatry. 2019;10:526. doi: 10.3389/fpsyt.2019.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constantino J.N. 2nd Edition Manual. Western Psychological Services; 2012. Social Responsiveness Scale. [Google Scholar]
- 41.Maljaars J., Noens I., Scholte E., van Berckelaer-Onnes I. Evaluation of the criterion and convergent validity of the Diagnostic Interview for Social and Communication Disorders in young and low-functioning children. Autism. 2012;16:487–497. doi: 10.1177/1362361311402857. [DOI] [PubMed] [Google Scholar]
- 42.Hoang N., Buchanan J.A., Scherer S.W. Heterogeneity in clinical sequencing tests marketed for autism spectrum disorders. NPJ Genom. Med. 2018;3:27. doi: 10.1038/s41525-018-006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierpont M.E., Brueckner M., Chung W.K., Garg V., Lacro R.V., McGuire A.L., Mital S., Priest J.R., Pu W.T., Roberts A., American Heart Association Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; and Council on Genomic and Precision Medicine Genetic basis for congenital heart disease: Revisited: A scientific statement from the American Heart Association. Circulation. 2018;138:e653–e711. doi: 10.1161/CIR.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K., Firth H.V., Frazier T., Hansen R.L., Prock L., NDD Exome Scoping Review Work Group Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019;21:2413–2421. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markon K.E., Chmielewski M., Miller C.J. The reliability and validity of discrete and continuous measures of psychopathology: a quantitative review. Psychol. Bull. 2011;137:856–879. doi: 10.1037/a0023678. [DOI] [PubMed] [Google Scholar]
- 46.Simmons J.M., Quinn K.J. The NIMH Research Domain Criteria (RDoC) Project: implications for genetics research. Mamm. Genome. 2014;25:23–31. doi: 10.1007/s00335-013-9476-9. [DOI] [PubMed] [Google Scholar]
- 47.Moreno-De-Luca A., Evans D.W., Boomer K.B., Hanson E., Bernier R., Goin-Kochel R.P., Myers S.M., Challman T.D., Moreno-De-Luca D., Slane M.M. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72:119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 48.Sanders S.J., Sahin M., Hostyk J., Thurm A., Jacquemont S., Avillach P., Douard E., Martin C.L., Modi M.E., Moreno-De-Luca A. A framework for the investigation of rare genetic disorders in neuropsychiatry. Nat. Med. 2019;25:1477–1487. doi: 10.1038/s41591-019-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iakoucheva L.M., Muotri A.R., Sebat J. Getting to the cores of autism. Cell. 2019;178:1287–1298. doi: 10.1016/j.cell.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Pang K., Han K., Adamski C.J., Wang W., He L., Lai J.K., Bondar V.V., Duman J.G., Richman R. An autism-linked missense mutation in SHANK3 reveals the modularity of Shank3 function. Mol. Psychiatry. 2019 doi: 10.1038/s41380-018-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douard E., Zeribi A., Schramm C., Tamer P., Loum M.A., Nowak S., Saci Z., Lord M.-P., Rodríguez-Herreros B., Jean-Louis M. Effects-sizes of deletions and duplications on autism risk across the genome. bioRxiv. 2020 doi: 10.1101/2020.03.09.979815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoekstra R.A., Happé F., Baron-Cohen S., Ronald A. Limited genetic covariance between autistic traits and intelligence: findings from a longitudinal twin study. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:994–1007. doi: 10.1002/ajmg.b.31066. [DOI] [PubMed] [Google Scholar]
- 54.Clarke T.K., Lupton M.K., Fernandez-Pujals A.M., Starr J., Davies G., Cox S., Pattie A., Liewald D.C., Hall L.S., MacIntyre D.J. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol. Psychiatry. 2016;21:419–425. doi: 10.1038/mp.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiner D.J., Wigdor E.M., Ripke S., Walters R.K., Kosmicki J.A., Grove J., Samocha K.E., Goldstein J.I., Okbay A., Bybjerg-Grauholm J., iPSYCH-Broad Autism Group. Psychiatric Genomics Consortium Autism Group Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niemi M.E.K., Martin H.C., Rice D.L., Gallone G., Gordon S., Kelemen M., McAloney K., McRae J., Radford E.J., Yu S. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., Pallesen J., Agerbo E., Andreassen O.A., Anney R., Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium. BUPGEN. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. 23andMe Research Team Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie S., Karlsson H., Dalman C., Widman L., Rai D., Gardner R.M., Magnusson C., Schendel D.E., Newschaffer C.J., Lee B.K. Family history of mental and neurological disorders and risk of autism. JAMA Netw. Open. 2019;2:e190154. doi: 10.1001/jamanetworkopen.2019.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warrier V., Toro R., Won H., Leblond C.S., Cliquet F., Delorme R., De Witte W., Bralten J., Chakrabarti B., Børglum A.D. Social and non-social autism symptoms and trait domains are genetically dissociable. Commun Biol. 2019;2:328. doi: 10.1038/s42003-019-0558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olszewski A.K., Radoeva P.D., Fremont W., Kates W.R., Antshel K.M. Is child intelligence associated with parent and sibling intelligence in individuals with developmental disorders? An investigation in youth with 22q11.2 deletion (velo-cardio-facial) syndrome. Res. Dev. Disabil. 2014;35:3582–3590. doi: 10.1016/j.ridd.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tansey K.E., Rees E., Linden D.E., Ripke S., Chambert K.D., Moran J.L., McCarroll S.A., Holmans P., Kirov G., Walters J. Common alleles contribute to schizophrenia in CNV carriers. Mol. Psychiatry. 2016;21:1085–1089. doi: 10.1038/mp.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pizzo L., Jensen M., Polyak A., Rosenfeld J.A., Mannik K., Krishnan A., McCready E., Pichon O., Le Caignec C., Van Dijck A. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet. Med. 2019;21:816–825. doi: 10.1038/s41436-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergen S.E., Ploner A., Howrigan D., O’Donovan M.C., Smoller J.W., Sullivan P.F., Sebat J., Neale B., Kendler K.S., CNV Analysis Group and the Schizophrenia Working Group of the Psychiatric Genomics Consortium Joint contributions of rare copy number variants and common SNPs to risk for schizophrenia. Am. J. Psychiatry. 2019;176:29–35. doi: 10.1176/appi.ajp.2018.17040467. [DOI] [PMC free article] [PubMed] [Google Scholar]