Abstract
How mutations in the non-coding U8 snoRNA cause the neurological disorder leukoencephalopathy with calcifications and cysts (LCC) is poorly understood. Here, we report the generation of a mutant U8 animal model for interrogating LCC-associated pathology. Mutant U8 zebrafish exhibit defective central nervous system development, a disturbance of ribosomal RNA (rRNA) biogenesis and tp53 activation, which monitors ribosome biogenesis. Further, we demonstrate that fibroblasts from individuals with LCC are defective in rRNA processing. Human precursor-U8 (pre-U8) containing a 3′ extension rescued mutant U8 zebrafish, and this result indicates conserved biological function. Analysis of LCC-associated U8 mutations in zebrafish revealed that one null and one functional allele contribute to LCC. We show that mutations in three nucleotides at the 5′ end of pre-U8 alter the processing of the 3′ extension, and we identify a previously unknown base-pairing interaction between the 5′ end and the 3′ extension of human pre-U8. Indeed, LCC-associated mutations in any one of seven nucleotides in the 5′ end and 3′ extension alter the processing of pre-U8, and these mutations are present on a single allele in almost all individuals with LCC identified to date. Given genetic data indicating that bi-allelic null U8 alleles are likely incompatible with human development, and that LCC is not caused by haploinsufficiency, the identification of hypomorphic misprocessing mutations that mediate viable embryogenesis furthers our understanding of LCC molecular pathology and cerebral vascular homeostasis.
Keywords: U8 snoRNA, zebrafish, SNORD118, snoRNA, leukoencephalopathy with calcifications and cysts, Labrune syndrome, ribosomopathy
Introduction
Leukoencephalopathy with calcifications and cysts (LCC [MIM: 614561]), also known as Labrune syndrome, is a Mendelian neurological disorder of the cerebral small blood vessels, and it is associated with increased morbidity and early mortality, presenting at any age from early infancy to late adulthood. Characterized by the radiological triad of cerebral white matter disease, intracranial calcifications, and cysts, LCC was recently shown to be an autosomal recessive genetic disorder caused by bi-allelic mutations in the gene SNORD118 (MIM: 616663), which encodes the box C/D U8 small nucleolar RNA (snoRNA).1
Ribosomes, the apparati of protein synthesis, consist of 28S, 18S, 5.8S, and 5S ribosomal RNA (rRNA) and 80 core ribosomal proteins distributed into 40S and 60S subunits.2 SnoRNAs are an evolutionarily conserved group of non-protein-coding RNAs required for the modification and processing of rRNA. The U8 snoRNA, a vertebrate-specific factor, is essential for maturation of the 28S and 5.8S rRNAs, components of the 60S large subunit.3 U8 snoRNA is required for removal of the 3′ external transcribed spacer (3′-ETS) sequence, one of a series of cleavage steps required to liberate the 28S, 5.8S, and 18S rRNA sequences from the polycistronic precursor-rRNA (pre-rRNA).4 Precursor-U8 (pre-U8) snoRNA contains an m7G cap and a short 3′ extension. Hypermethylation of the m7G cap to m3G and removal of the 3′ extension, through a series of steps that appear to involve nucleo-cytoplasmic shuttling and concurrent ordered association and dissociation of multiple protein components (including the LSm proteins, see below), results in the production of mature U8 snoRNA.5, 6, 7, 8 The box C/D motif of U8 is bound by four core proteins: 15.5K, NOP56, NOP58, and fibrillarin, contributing to the formation of the U8 small nucleolar ribonucleoprotein (U8 snoRNP) complex and its localization to the nucleolus, the site of pre-rRNA processing.9 The role of U8 in rRNA maturation implicates LCC as a recently discovered ribosomopathy. Ribosomopathies are a group of disorders caused by ribosome biogenesis dysfunction that manifest as a diverse set of highly stereotyped clinical syndromes.10
Of 33 mutation-positive families identified by Jenkinson et al.,1 31 probands with LCC were compound heterozygotes for two different SNORD118 variants; this implies the existence of one severe (null) and one milder (hypomorphic) mutation, with bi-allelic null mutations likely incompatible with life. In total, Jenkinson et al., recorded seven putatively causal mutations in the invariant box C/D motif, three within the stem of a conserved hairpin loop which would be predicted to decrease the stability of this structure, three within the highly conserved GAUU motif of the LSm-binding site, and four mutations in the short 3′ extension of the precursor. Promoter mutations were also found that reduced expression levels of U8. Presumably, LCC-associated variant combinations reduce U8 function below a critical level while allowing for viable embryogenesis, thus maintaining sufficient levels of production of functional ribosomes. However, because U8 is a non-coding RNA, in silico algorithms cannot be used to predict the functional consequences of U8 variants and, therefore, the precise molecular pathology of LCC remains unknown.
Here, we report a vertebrate mutant model system for studying U8 snoRNA function. Zebrafish U8 mutants were found to exhibit defective rRNA biogenesis and activation of the tumor suppressor p53 (tp53), which monitors ribosome biogenesis dysfunction in a regulatory loop known as “nucleolar stress surveillance.”11, 12, 13 Functional assessment of LCC disease-associated U8 alleles confirmed the importance of combinatorial null and functional mutations. We show that the 3′ extension of U8 is critical for U8 biological activity, an observation reflected in the fact that mutations within the 3′ extension, or in nucleotides predicted to base-pair with the 3′ extension, were recorded in 29 of 33 individuals. Assays using HeLa nuclear cell extracts demonstrated that these mutations alter the processing of pre-U8; this result supports the proposed secondary structure of the human pre-U8 snoRNA. Importantly, fibroblasts from individuals with LCC also exhibit rRNA processing defects, and human pre-U8 snoRNA was found to rescue the zebrafish U8 mutant; this finding indicates conserved biological function. Taken together, these data support the characterization of LCC as a ribosomopathy whose effects are restricted to the cerebral vessels, and of the utility of zebrafish to provide insight into the pathology of human disease and U8 biology.
Material and Methods
Zebrafish Strains and Husbandry
Establishment and characterization of the tp53M214K/M214K and Tg(kdrl:GFP)s843 strains have been described elsewhere.14,15 Embryos and adults were maintained under standard laboratory conditions as described previously,16 and experiments were approved by the University of Manchester Ethical Review Board and performed according to UK Home Office regulations. No statistical method was used to predetermine sample size for experimental groups.
Real-Time Quantitative PCR
RNA was isolated from homogenized zebrafish embryos through the use of TRIzol (Thermo Fisher Scientific), and genomic DNA was removed using the TURBO DNA-free Kit (Thermo Fisher Scientific). Reverse transcriptase was performed using the ProtoScript II First Strand cDNA synthesis kit (New England BioLabs) using 1μg of total RNA with random hexamer primers. qRT–PCR analysis was performed with the primers described in Table S2 and using a 60°C annealing temperature, primers with efficiencies from 95%–105%, and the SensiFAST SYBR No-ROX kit (Bioline) and the Mx3000P system (Stratagene).
Genome Editing
Capped nls-zCas9-nls, mKate2, or H2B-mCerulean3 mRNA was synthesized using a mMESSAGE mMACHINE SP6 kit (Life Technologies) from a linearized pCS2 construct and purified using a RNeasy mini kit (QIAGEN). Zebrafish guides were designed using the CHOPCHOP program. Guide RNA (gRNA) incorporating this target sequence was generated from a polymerase chain reaction (PCR) amplification product (see Table S2 for primer sequences) including the remaining sequence of S. pyogenes chimeric single gRNA through in vitro transcription using a HiScribe T7 Quick kit (New England Biolabs). The gRNA was then precipitated in a 1/10 volume of 3M sodium acetate and two volumes of 100% ethanol by chilling the reaction at −20°C for 15 min, then spinning in a microcentrifuge (Sigma) at 13K for 15 min, and finally the RNA pellet was resuspended in 15 μL of RNase-free water. Cas9 mRNA (250 pg) and gRNA (30 pg) and mKate2 (100 pg) or H2B-Cerulean3 (100 pg) were injected into the yolks of one-cell-stage embryos, and fluorescence was used to identify successfully injected embryos. We identified working guides by PCR amplifying the target region and running the PCR product on a 3% agarose gel to identify INDEL events that produced visible shifts or smearing of the amplification product.
Genotyping
Embryos or fin-clips were placed in PCR tubes with 50 μL of 50 mM NaOH and denatured for 20 min at 95°C. A volume of 20 μL of Tris-HCl pH 8 was added to each tube and 1 μL of the genomic DNA used for PCR amplification.
PCR Conditions
PCR was performed in a 25–50 μL reaction mix containing DNA template (0.1–100 ng DNA), sense and antisense primer 0.8 μm each, 0.25 mM dNTPs (Bioline), 1X HF buffer (New England Biolabs), 1U Phusion Taq polymerase (New England Biolabs), 0.5 mM MgCl2 (New England Biolabs), and 1.5 μL DMSO (New England Biolabs) per 50 μL reaction. PCR was performed in a Techne TC-PLUS or Alpha Thermal Cycler PCRMAX machine with an initial denaturing step at 98°C for 3 min followed by 35 cycles of denaturing at 98°C for 10 s, annealing at 60°C for 30 s, and amplification at 72°C for 45 s/1 kb. A final 5–10 min cycle at 72°C was routinely performed to allow the complete extension phase to occur.
Imaging and Embryo Measurement
Zebrafish embryos were anesthetized using MS-222 (Sigma-Aldrich) and imaged on an M165FC fluorescent stereomicroscope (Leica) with a DFC310 FX camera (Leica). For measuring the length of embryos, images were taken at 2.5× magnification, and embryo length was quantified in the CorelDRAW graphic suite. For confocal microscopy, images were taken from anesthetized embryos through the use of a Leica TCS SP8 AOBS upright confocal using a 20× 0.50 Plan Fluortar objective and processed using LAS X (Leica version 3.5.2.18963).
Quantitation of 28S:18S Ratios
Total RNA was run on a TapeStation 4200 (Agilent) according to manufacturers’ instructions. Total RNA with integrity values in excess of 9 were selected for 28S:18S quantitation, and quantitation was performed using the TapeStation Analysis Software A.01.05 (SR1).
ASO-Knockdown of U8 and rRNA Processing Assays in Fibroblasts
Antisense oligonucleotide (ASO)-mediated depletion of U8 in control cells (HCT116, colon carcinoma) was performed as described previously.17 RNA extraction and pre-rRNA processing analysis of U8 depleted cells and fibroblasts derived from individuals with LCC was performed as described previously.17,18 The ATCC fibroblast control PCS-201-012 (primary dermal fibroblasts from a normal human adult) was used. The sequence of anti-U8 ASO used in depletion was mGmGmAmUmUATCCCACCTGmAmCmGmAmU. N and mN are deoxynucleotide and 2′-O-methoxyethylribonucleotide, respectively. Phosphodiester backbones are phosphorothioates.
In Vitro Transcription of U8 RNA Variants
U8 DNA templates containing a T7 consensus sequence were PCR amplified from human or zebrafish genomic DNA (see Table S2 for primer sequences) and, after agarose gel electrophoresis, were purified using a QIAEX II kit (QIAGEN). Human and zebrafish U8 snoRNAs were generated using 400–1,000 ng of template DNA and a mMESSAGE mMACHINE T7 kit (Life Technologies) followed by lithium chloride precipitation and quantitation using a NanoDrop (Thermo Fisher Scientific).
Microinjection and Transgenesis
The yolks of fertilized one-cell-stage embryos were microinjected with 2 nl of synthetic mRNA/snoRNA, or for transgenesis, with 40 pg of both DNA and tol2 transposase mRNA,19 through the use of a PLI-90 pico-injector (Harvard Apparatus) and a Leica MZ6 stereomicroscope.
Electrophoretic Mobility Shift Assays
For electrophoretic mobility shift assays (EMSAs), recombinant His-15.5K was incubated with 50,000 dpm 32P-end-labeled U8 snoRNA in EMSA buffer (20 mM HEPES-KOH, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 0.1% Triton X-100) for 30–45 min on ice. Resulting RNA-protein complexes were resolved on a native 7% acrylamide gel for 8.5 h at 4°C. Gels were dried and exposed to X-ray film for approximately 8 h at −80°C in the presence of an intensifying screen.
Bioinformatic Analyses of U8 Secondary Structures
Determination of the minimum free energy secondary structures of human and zebrafish mature and precursor U8 sequences was performed by uploading the sequences to the RNAfold webserver using the default settings.
3′-Processing Assays
For 3′-processing assays, 32P-end-labeled U8 snoRNA was incubated with HeLa nuclear extract (CIL Biotech) at 30°C in buffer containing 0.25 mM ATP, 10 mM phosphocreatine, 3.2 mM MgCl2, 20 mM HEPES KOH, pH 7.9, 2.6% polyvinyl alcohol, and 240 U RNasin (Promega). At 0, 30, and 60 min, 10 μL of the reaction was removed and added to a tube containing 4 μL of stop solution (1 mg/mL proteinase K, 50 mM EDTA, and 1% sodium dodecyl sulfate). Reactions were then incubated at 37°C for 15 min, phenol extracted, precipitated, and resolved on a 5% acrylamide/7 M urea gel. Gels were dried and exposed to X-ray film at −80°C in the presence of an intensifying screen.
Statistics and Reproducibility
All statistical analyses were performed using GraphPad Prism 8 or Microsoft Excel software. Results are presented as mean ± SD. For all analyses, p < 0.05 was considered statistically significant. Statistical methods were not used to predetermine sample size, which varies between experiments. Experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. For Figure 1B, significance was determined using one-way ANOVA and post hoc Tukey’s multiple comparisons test. For Figure S3, significance was determined using a Mantel-Cox test. For all other statistical analyses, significance was determined using an unpaired t test. The number of biological replicates upon which significance was determined is specified in the figure legend. For oligonucleotides used in this study, refer to Table S2.
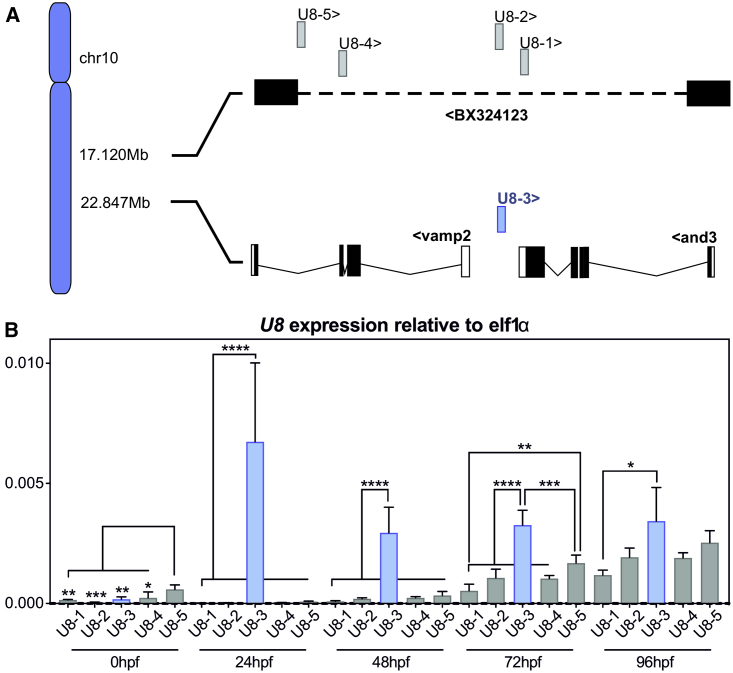
Figure 1.
U8-3 Is the Predominantly Expressed U8 snoRNA during Zebrafish Embryogenesis
(A) Schematic depicting the five U8 zebrafish small nucleolar RNA (snoRNA) gene copies located on chromosome 10. U8-3 (blue) is located between the vamp2 and and3 genes, whereas the other four copies (gray) are clustered within the intron (dashed line) of the non-coding transcript BX324123. < or > indicates direction of transcription.
(B) Quantitative RT-PCR to U8-1, U8-2, U8-3, U8-4, and U8-5 snoRNA transcripts at the indicated developmental time points. n = 4 biological replicates per time point. hpf—h post-fertilization. Error bars indicate SD from the mean.
Results
U8-3 Is the Predominantly Expressed Zebrafish U8 during Embryogenesis
Zebrafish contain five copies of U8 located on chromosome 10, four copies clustered within the intron of the transcript BX324123, and the remaining copy located between the genes vamp2 and and3 (Figure 1A). Quantitative RT-PCR analysis, exploiting the single-nucleotide polymorphisms present between the five copies of U8 for specificity, identified minimal maternal deposition of U8 transcripts in zebrafish, and U8-5 was the most highly deposited (Figure 1B; see Figure S1 for alignment of zebrafish U8 copies). At 24 h post-fertilization (hpf), U8-3 was the sole U8 species identified in the zebrafish embryo, with only weak expression of the clustered U8-1, U8-2, U8-4, and U8-5 induced at 48 hpf (albeit increasing thereafter) (Figure 1B). These data provided a rationale for targeted disruption of the U8-3 gene locus to interrogate U8 function during early embryogenesis.
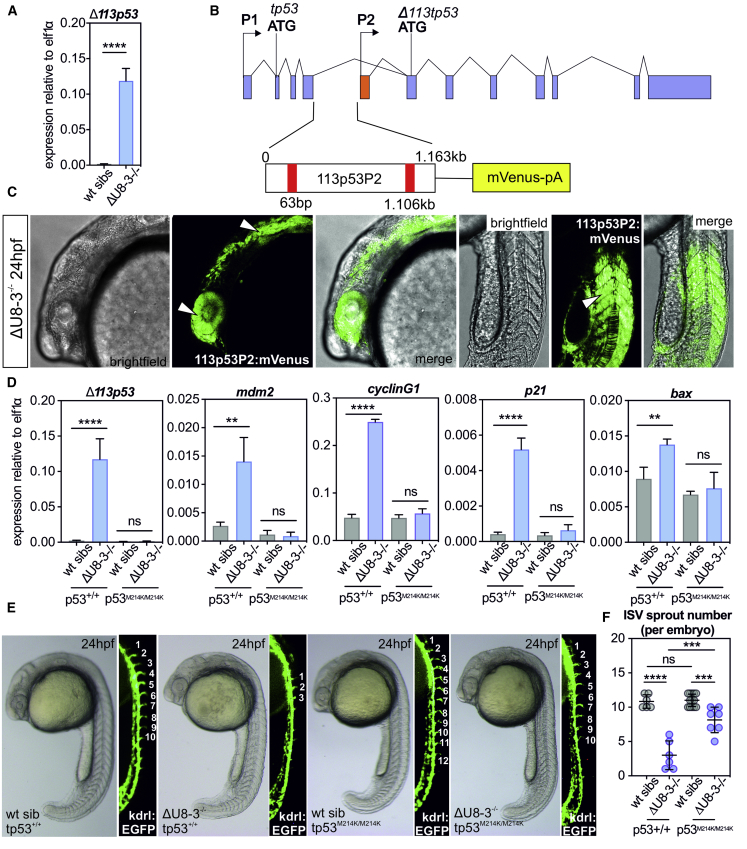
CRISPR/Cas9 was employed to disrupt the U8-3 locus. Creating insertion and/or deletion events in a non-coding RNA could lead to unpredictable consequences for U8-3 function. Consequently, two guides were used to excise U8-3 from the genome, producing a null allele, herein referred to as ΔU8-3 (Figure S2). By 24 hpf, ΔU8-3 mutants exhibited a less defined midbrain-hindbrain boundary and reduced angiogenic sprouting from the dorsal aorta (Figure 2A). By 48 hpf, ΔU8-3 mutants demonstrated swelling of the fourth ventricle, consistent with abnormal development of the central nervous system (CNS), reduced melanocyte development, smaller eye size, impaired yolk resorption, disturbed branching of the trunk vasculature, and a reduction in embryo length consistent with developmental delay (Figure 2A and 2B). A time course analysis indicated that the ΔU8-3 mutant comes to a developmental standstill which is reflected in a failure to resorb yolk and expand the swim bladder (Figure S3). Death was observed from 6 days post fertilization (dpf), and 100% mortality was recorded by 9 dpf (Figure S4). Quantitative RT-PCR analysis confirmed that U8-3 expression is lost in ΔU8-3 mutants (Figure 2C).
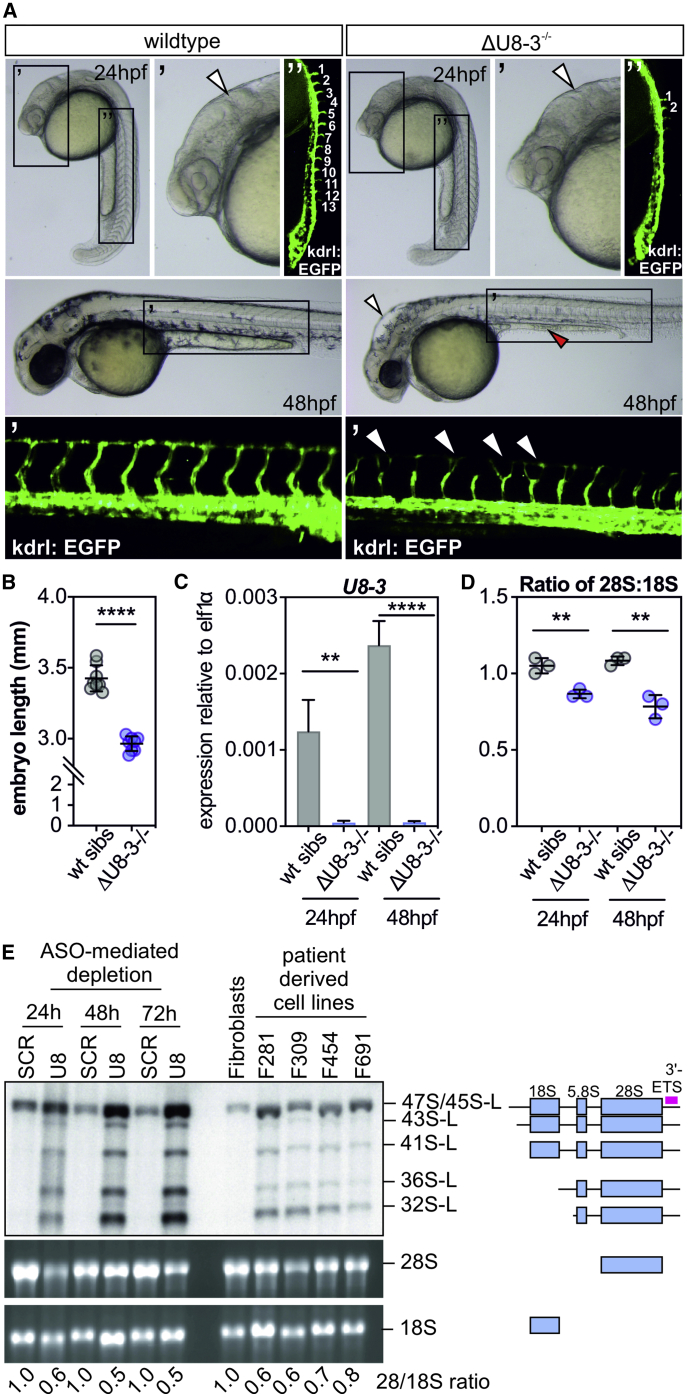
Figure 2.
U8-3 Is Required for Correct Development of the Central Nervous System and Vasculature in the Zebrafish and 28S Biogenesis in Fish and LCC Individual-Derived Cells
(A) ΔU8-3 mutants exhibit a less-defined midbrain-hindbrain boundary (arrowheads) and reduced angiogenic sprouting (numbered in white, visualized with the kdrl:EGFP transgene) from the dorsal aorta compared to wild-type siblings at 24 hpf. By 48 hpf, ΔU8-3 mutant embryos exhibit hindbrain swelling (arrowhead), an underdeveloped yolk extension (red arrowhead), reduced eye size, impaired melanocyte development, and disorganized trunk vasculature compared to wild-type siblings. hpf—h post-fertilization.
(B) Quantitation of embryo length of the indicated genotype 48 hpf. n = 8 embryos per genotype. Error bars indicate SD from the mean.
(C) qRT-PCR of U8-3 small nucleolar RNA (snoRNA) in the indicated genotype and developmental time point. n = 4 biological replicates per genotype. Error bars indicate SD from the mean.
(D) Tapestation analysis demonstrates ΔU8-3 mutant embryos display a preferential reduction in 28S biogenesis consistent with defective 3′ external transcribed spacer (3′-ETS) processing. n = 3 biological replicates per genotype. Error bars indicate SD from the mean.
(E) Antisense oligonucleotide (ASO)-mediated depletion of human U8 snoRNA results in accumulation of 3′ extended forms of pre-ribosomal RNA (rRNA) intermediates (identified by a 3′-ETS specific probe, pink bar) which are required for biogenesis of 28S rRNA. Likewise, these same 3′ extended intermediates accumulate in leukoencephalopathy with calcifications and cysts (LCC) fibroblast cell lines F281, F309, F454, and F691 when compared to wild-type fibroblasts (ATCC). SCR—non-targeting ASO control. The mature 28S:18S ratio, established by densitometry, is indicated.
U8 is required for removal of the 3′-ETS sequence, and in particular for the biogenesis of 28S and 5.8S rRNAs.17,20,21 Quantification of 28S:18S ratios via TapeStation assay found that ΔU8-3 mutants exhibit a preferential reduction in 28S biogenesis compared to 18S (Figure 2D). Significantly, Northern blotting with a probe specific to the 3′-ETS region of the pre-rRNA demonstrated that fibroblasts derived from individuals with LCC, and control cells in which U8 had been knocked down by antisense oligonucleotides, accumulated aberrant unprocessed 3′ extended rRNA precursors required for 28S biogenesis, precursors which are also impaired for removal of ITS2 (see Figure 2E and 2L for longform). Further, these pre-rRNA processing defects alter the ratios of 28S to 18S similarly to the ΔU8-3 mutant, preferentially impairing biogenesis of 28S (Figure 2D and 2E). Northern blotting analysis with a probe specific to U8 confirmed antisense oligonucleotide-mediated knockdown of U8, and that fibroblasts from individuals with LCC have reduced levels of total U8; this is most likely due to reduced stability of one or both mutant alleles (Figure S5).1 Taken together, these data support the possibility of a conserved biological function for U8 in rRNA processing and ribosome biogenesis, and these data show that cells derived from individuals with LCC expressing U8 mutations are indeed defective for ITS2 and 3′-ETS maturation.
A portion of the U8-3 promoter was deleted in the ΔU8-3 allele, which potentially contains regulatory elements required for the function of other genes. To confirm the specificity of the ΔU8-3 mutant phenotype, a complementation test with an independent U8-3 mutant allele was performed. A guide specific to U8-3, the design of which was facilitated by the absence of a relevant protospacer adjacent motif (PAM) sequence in the other zebrafish U8 gene copies, was used to delete 54bp from the U8-3 gene locus, herein referred to as Δ54U8-3 (see Figure S6A and S6B). Δ54U8-3 mutants demonstrated morphology indistinguishable from that of ΔU8-3 mutants, with both alleles displaying Mendelian autosomal recessive inheritance (Figure S6C). A failure of complementation (i.e., the production of 100% wild-type progeny) was observed when a ΔU8-3 heterozygote zebrafish was crossed to a Δ54U8-3 heterozygote zebrafish, demonstrating that the two mutants are associated with loss of function (LoF) of the same gene (Figure S6C).
Precursor Zebrafish U8-3 and Human U8 snoRNAs Are Functionally Equivalent
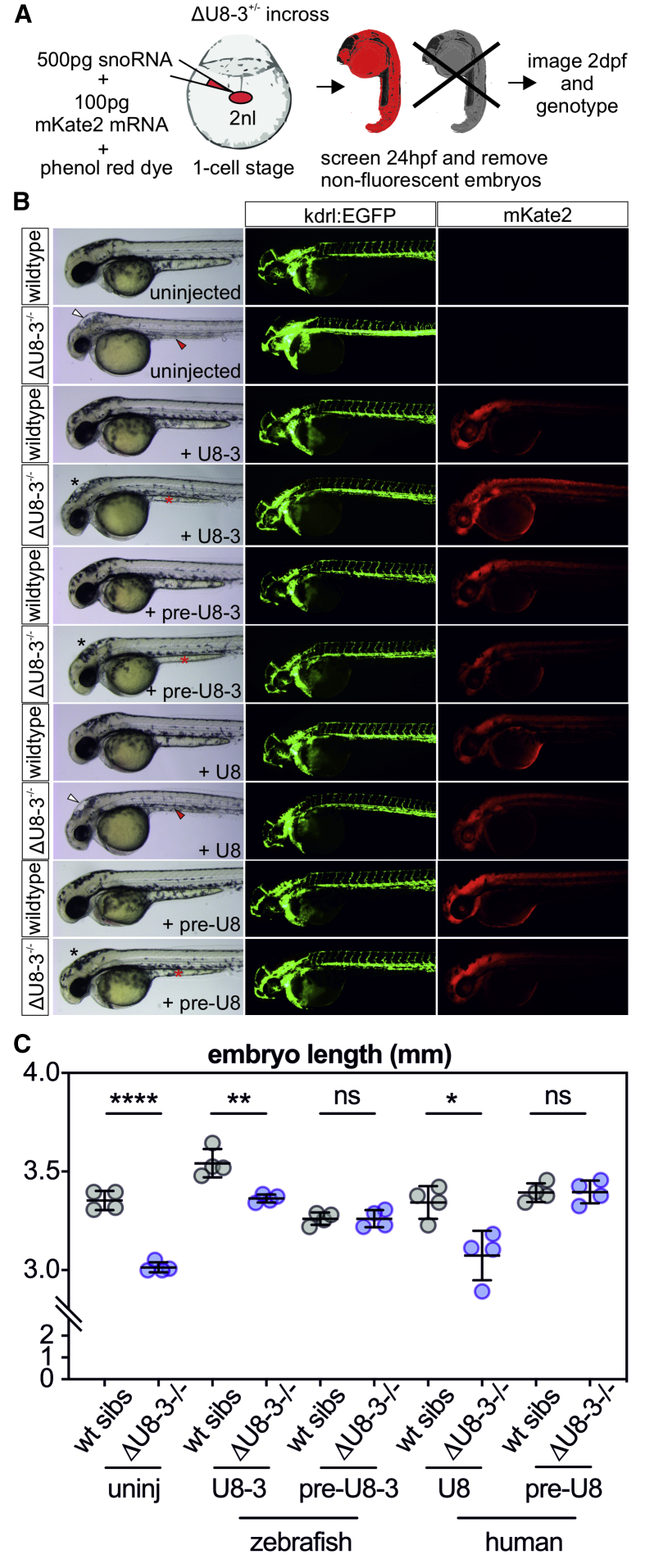
Mutations that lie within the short 3′ extension of human U8 in individuals with LCC imply that this region is of functional significance.1 However, it has previously been reported that exogenous mature U8 snoRNA, which lacks the 3′ extension sequence, localizes to the nucleolus and rescues endogenous U8 depletion in Xenopus oocytes.22 We first performed an EMSA with in vitro transcribed mature zebrafish U8-3 and the highly conserved human 15.5K to confirm the ability of in vitro synthesized U8-3 to interact with a key U8 snoRNP factor. Addition of 15.5K was found to shift U8-3 migration, and also migration of zebrafish U8-1, 2, 4, and 5, demonstrating that these zebrafish mature U8 species bind 15.5K (Figure S7). Alignment of human SNORD118 and zebrafish U8-3 gene loci identified a putative 3′ extension and 3′ box in zebrafish U8 (Figure S8). A transient rescue assay was performed, comparing the capacity of exogenous in vitro transcribed mature U8-3 and the putative pre-U8-3 snoRNA to rescue the gross morphological abnormalities observed in the ΔU8-3 mutant. Mature or pre-U8-3 snoRNAs were co-injected into the yolk of one-cell-stage zebrafish ΔU8-3 mutants or wild-type siblings with an mRNA encoding a fluorescent protein acting as a tracer, enabling ubiquitous expression of the transcripts throughout the embryo over the first two days of development (Figure 3A). Mature U8-3 snoRNA rescued the yolk extension and hindbrain swelling of the ΔU8-3 mutant, but not the effect on embryo length (Figure 3B and 3C). The addition of 14 nucleotides 3′ to the mature sequence of U8-3 (constituting the pre-U8-3) resulted in a rescue of the hindbrain swelling, yolk extension, and embryo length of ΔU83 mutants, thereby demonstrating the importance of this 3′ extension sequence of U8 in generating functional ribosomes (Figure 3).
Figure 3.
The 3′ Extension of Precursor U8 Is Required for Optimal U8 Biological Activity
(A) Schematic depicting the experimental design for transient ubiquitous expression of U8 small nucleolar RNA (snoRNA) variants and mKate2 mRNA. Bolus size is monitored through the use of a phenol red dye, and successful uptake of the microinjected solution into the animal pole is traced using red fluorescence.
(B) Representative brightfield and fluorescent images of the genotype, exogenous snoRNA, and transgenic or fluorescent protein is indicated, taken at 48 hpf. U8 denotes human sequence whereas U8-3 denotes zebrafish. White arrowheads denote hindbrain swelling, red arrowheads denote aberrant yolk extension, black asterisks denote rescued hindbrain, and red asterisks denote rescued yolk extension.
(C) Quantitation of embryo length for the genotypes and introduced snoRNAs indicated. uninj—uninjected. n = 4 embryos per genotype. Error bars indicate SD from the mean.
The above rescue assay provided the means to investigate whether human U8 snoRNA is functional in the zebrafish. As for zebrafish pre-U8-3, pre-U8 human snoRNA rescued ΔU8-3-associated gross morphological abnormalities, whereas mature human U8 snoRNA failed to do so (Figure 3B and 3C), indicating that zebrafish and human U8 snoRNA are functionally equivalent. However, although a number of the ΔU8-3 mutant features were rescued by exogenous pre-U8, the eye and head size did not recover to wild-type sibling levels. This lack of complete rescue likely reflects the technical challenge of introducing enough non-coding RNA at the one-cell-stage to sustain a rapidly growing zebrafish embryo over two days of development.
Rescue Experiments in Zebrafish Identifies One Null and One Functional U8 Allele in Individuals with LCC
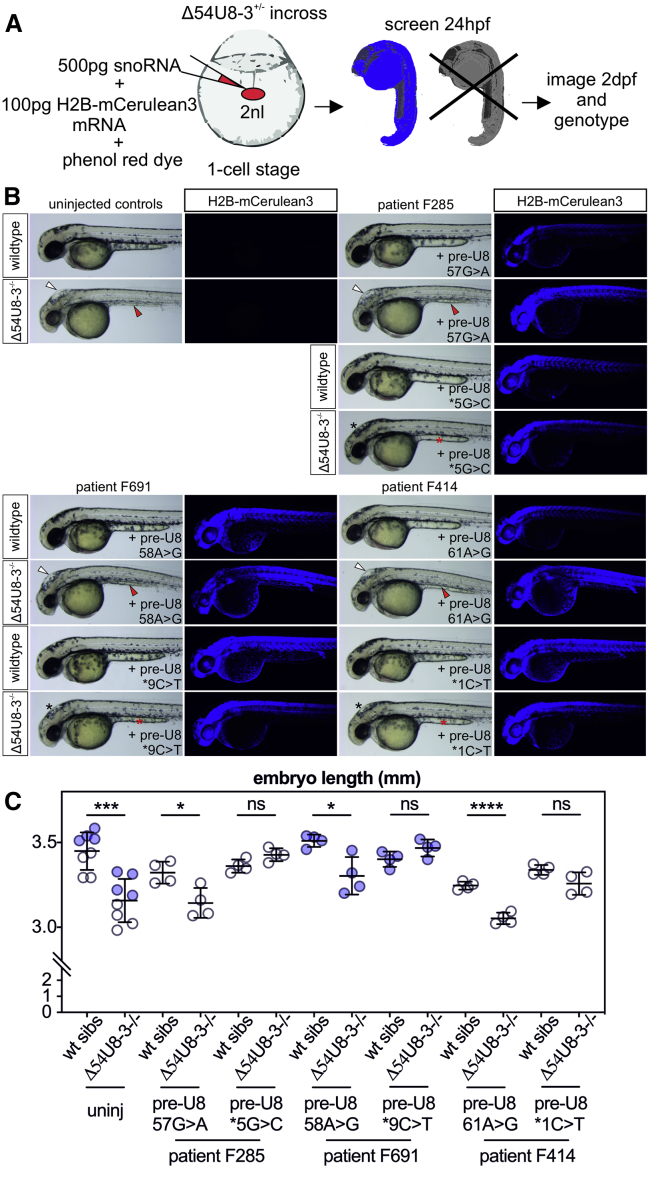
Having demonstrated that human pre-U8 snoRNA is functional in the zebrafish, we next wanted to test the effect of LCC-disease-associated mutations in U8-3 null embryos. Six alleles were chosen in which a molecular defect in functionality had previously been shown in vitro: specifically, either a complete loss of (n57G>A and n58A>G) or reduced (n61A>G) ability to interact with 15.5K compared to wild type, or disrupted 3′ end processing in HeLa nuclear extracts (n∗1C>T, n∗5C>G, and n∗9C>T; the asterisk denotes that the nucleotide in question is located in the 3′ extension of human pre-U8).1 Despite molecular evidence suggesting that the n61A>G variant might be hypomorphic, in each case, variants affecting nucleotides required for binding to 15.5K were found to act as functional null alleles in that they failed to alter the Δ54U8-3 mutant phenotype; this result demonstrates the essential nature of this domain for U8 function (Figure 4). In contrast, all of the LCC mutations in the 3′ extension rescued the morphological abnormalities observed in the Δ54U8-3 mutant, including the embryo length defect (Figure 4 B and C); this finding indicates a preserved function in ribosome biogenesis. These data likely reflect the null and functional, though hypomorphic, status of distinct alleles.
Figure 4.
Functional Testing of LCC Mutant U8 snoRNAs Identifies One Null and One Functional Allele
(A) Schematic depicting the experimental design for transient ubiquitous expression of U8 small nucleolar RNA (snoRNA) mutants and H2B-mCerulean3 mRNA. Bolus size is monitored through the use of phenol red dye and successful uptake of the microinjected solution into the animal pole is traced using nuclear cerulean fluorescence.
(B) Leukoencephalopathy with calcifications and cysts (LCC) mutant snoRNAs specific to the region required for 15.5K binding (n57, n58, and n61) fail to rescue the hindbrain swelling (white arrowheads) and yolk extension (red arrowheads) of Δ54U8-3 mutants, whereas LCC mutants specific to the 3′ extension (n∗1, n∗5, and n∗9) rescue hindbrain swelling (black asterisks), yolk abnormalities (red asterisks), and pigmentation defects.
(C) LCC mutant snoRNAs specific to the region required for 15.5K binding (n57, n58, and n61) do not rescue the reduced embryo length of Δ54U8-3 mutants, whereas LCC mutants specific to the 3′ extension restore Δ54U8-3 mutant embryo length to that of wild-type siblings. n = 4–8 embryos per genotype. Blue and black data points represent embryos collected from two different pairs of heterozygote Δ54U8-3 adults. Error bars indicate SD from the mean.
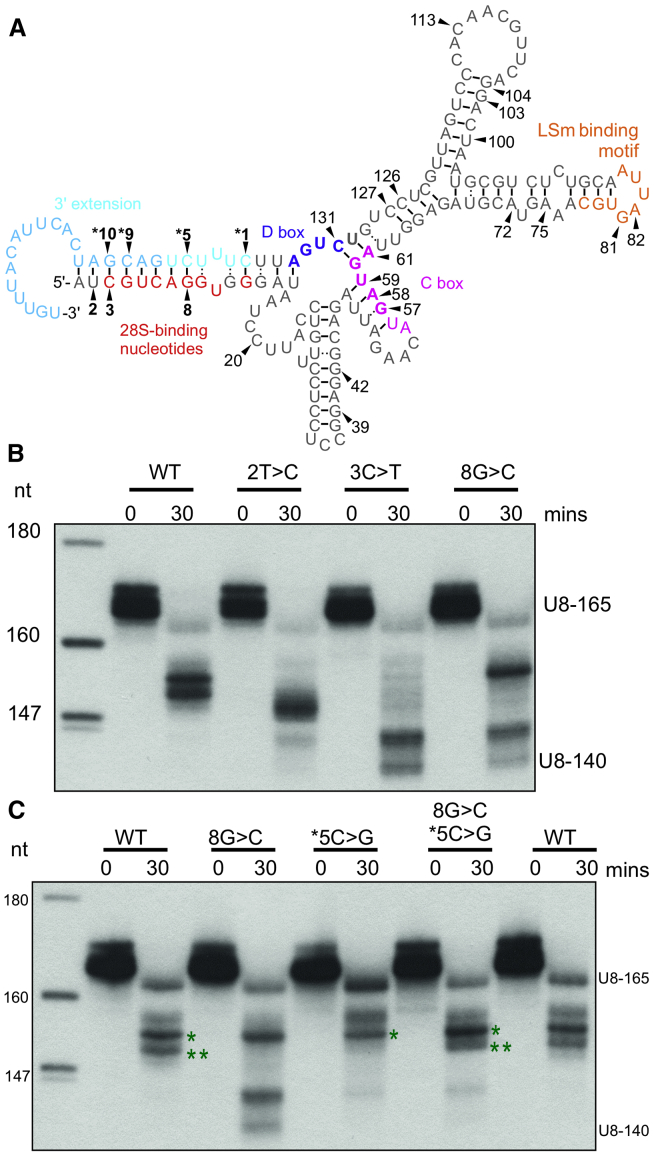
Mutation of Seven Distinct U8 snoRNA Nucleotides Alter Processing of pre-U8 and Are Present in Most Individuals with LCC
To our knowledge, the secondary structure of the human pre-U8 snoRNA has yet to be reported in the literature. The minimum free energy secondary structure for the human pre-U8, determined by RNAfold,23 suggested a high probability for duplex formation between the 3′ extension and the 5′ end of the human pre-U8 snoRNA (Figure 5A and Figure S9A). Intra-molecular base-pairing of the 5′ end of mature human U8 with its 3′ end is only suggested to occur with low probability in the minimum free energy state, in keeping with the widely accepted and reported role for the 5′ end of mature U8 to base-pair with rRNA sequence (Figure S9B).24 Mapping of all the LCC individual variants onto the human pre-U8 secondary structure revealed that another three disease-associated-mutations, located at the 5′ end of U8, lie precisely within the proposed base-paired region (Figure 5A). Strikingly, one of the seven mutated nucleotides found within this hitherto unappreciated duplex was observed in 29 of 33 individuals with LCC overall (Table S1). We have previously reported that the n∗1C>T, n∗5C>G, n∗9C>T, and n∗10G>T U8 mutants demonstrate defective 3′ end processing in HeLa nuclear extracts.1 The predicted secondary structure of human pre-U8, which indicates base-pairing between the 5′ end and 3′ extension, combined with the knowledge that n∗1C>T, n∗5C>G, n∗9C>T, and n∗10G>T mutants cause disrupted processing, suggests that the n2T>C, n3C>T, and n8G>C mutations might also affect U8 precursor processing. The human pre-U8 snoRNA is processed to the mature U8 snoRNA in HeLa nuclear extracts after 60 min (Figure S10). When we examined U8 processing intermediates in HeLa nuclear extracts at 30 min, a time point before mature U8 snoRNA is produced, we observed that each of the n2T>C, n3C>T, and n8G>C mutants conferred an aberrant, increased, rate of 3′ end processing when compared to wild type (Figure 5B).
Figure 5.
Aberrantly Processed Mutant U8 snoRNA Alleles Are Present in 29 of 33 Individuals with LCC
(A) Schematic depicting the secondary structure of human precursor U8 predicted by RNAfold software (18). Black arrowheads and bold numbering identify seven mutated nucleotides that lie within a duplex encompassing the 3′ extension (denoted in blue) in individuals with leukoencephalopathy with calcifications and cysts (LCC). The remaining mutated nucleotides recorded in individuals with LCC are numbered (refer to Table S1 for a complete list of LCC-associated mutant alleles). The 5′ end/3′ extension base-pairing may preclude formation of the kink-turn that is mediated by the D box (purple nucleotides) and C box (pink nucleotides) until U8 has matured sufficiently. The kink-turn interaction encompasses nucleotides 57–61 and 130–134 (bold). Important functional regions of U8 are also annotated: the nucleotides that interact with the 28S ribosomal RNA (rRNA; in red) and LSm-binding nucleotides (in orange). A black bar identifies a Watson-Crick base pairing interaction and : represents a non-Watson-Crick base pairing interaction.
(B) Processing of 5′ end-radiolabeled in vitro transcribed precursor U8 wild-type/mutant small nucleolar RNA (snoRNA; U8-165) was assessed in HeLa nuclear extracts. At 30 min, 2T>C, 3C>T, and 8G>C mutant U8 snoRNAs exhibit an enhanced rate of processing when compared to wild type as demonstrated by the presence of mature U8 (U8-140).
(C) Processing of 5′ end-radiolabeled in vitro transcribed precursor U8 wild-type/mutant snoRNA (U8-165) was assessed in HeLa nuclear extracts. At 30 min, the n∗5C>G mutant U8 is blocked in processing when compared to wild type (∗∗ band is absent), in contrast to 8G>C mutant U8 snoRNA that exhibits production of mature U8 (U8-140). When base-pairing complementarity is restored by combining the two mutations, the pattern of processing is restored to wild type.
If the proposed interaction of the 5′ and 3′ ends of human pre-U8 is correct, then restoring base-pairing complementarity for the n8G>C mutation would be predicted to return processing to wild type. Thus, the n8G>C and n∗5C>G mutations were combined to test this hypothesis. n∗5C>G alone appeared to exhibit slowed processing when compared to wild type at the early 30 min time point, whereas n8G>C was again associated with an increased rate of processing and production of mature U8 (U8-140) after 30 min (Figure 5C). As predicted by the model, the n8G>C/n∗5C>G double mutant pre-U8 was found to confer almost identical processing when compared to wild type, providing functional evidence for the proposed base-pairing between the 5′ end and 3′ extension of the human pre-U8 snoRNA (Figure 5C).
The very survival of individuals with LCC beyond embryogenesis suggests that, like the 3′ extension mutations tested in Figure 4, the n2T>C, n3C>T, n8G>C, and n∗10G>T mutant pre-U8 snoRNAs also retain some degree of functional competence necessary for ribosome biogenesis. To test this, rescue experiments using the n2T>C, n3C>T, n8G>C, and n∗10G>T U8 mutants were performed in the zebrafish Δ54U8-3 mutant. Consistent with our hypothesis, n2T>C, n8G>C, and n∗10G>T pre-U8 all rescued Δ54U8-3 mutant morphology, including embryo length (Figure S11A and S11B). In contrast, the n3C>T pre-U8 did not salvage the yolk extension defect or embryo length (Figure S11A and S11B). However, it should be noted that almost half of the nucleotides required for 28S binding in human differ from zebrafish (Figure S11C), and such a lack of conservation might adversely affect the capacity of human U8 to completely substitute for U8-3 function in the zebrafish when additional single base pair changes are introduced within the 5′ end of human U8. As a consequence, caution must be exercised when interpreting a failure to rescue in these circumstances.
tp53 Is Activated in Response to Loss of U8-3 in Zebrafish
Perturbation of ribosome biogenesis activates the transcription factor TP53, a tumor suppressor with a role in a wide range of biological processes, including DNA damage, mitochondrial stress, autophagy, and oncogenesis.25 Trans-activated tp53 increases expression of different effectors depending on the biological context, including Δ113tp53, transcribed from intron 4 of the tp53 gene in zebrafish. Δ113tp53 was found to be upregulated 50-fold in ΔU8-3 mutants compared to wild-type siblings at 24 hpf, as were the tp53 target genes mdm2, cyclinG1, p21, and bax (Figure 6A, Figure S12A). To determine which tissues were affected by loss of U8-3 in zebrafish, a spatio-temporal reporter of tp53 activity was generated that utilized the Δ113tp53 promoter containing two tp53-binding sites (Figure 6B).26 At 17 hpf, before any morphological abnormalities are observable, tp53 trans-activation activity can be observed already in ΔU8-3 mutants (Figure S12B). By 24 hpf, tp53 trans-activation activity was detected in the eye, CNS, and somites of ΔU8-3 mutants (Figure 6C, Figure S12C), and by 48 hpf, the CNS and somites were the most highly fluorescent tissues (Figure S12D). Importantly, somite-derived vegf expression is critical for correct patterning of the dorsal aorta and for arterial development.27 As such, the delayed sprouting and abnormal branching of the trunk vasculature is unlikely to be a cell autonomous effect, but rather is likely to be secondary to the impairment of somitogenesis. The tissues in which tp53 is activated all display obviously impaired development in the ΔU8-3 mutant as evidenced by the reduced eye size, shortened body length, and fourth ventricular swelling (Figure S12D).
Figure 6.
tp53 Is Trans-activated in Response to Loss of U8-3 in Zebrafish
(A) Quantitative RT-PCR of Δ113tp53 transcripts at 24 hpf for indicated phenotype. Error bars indicate SD from the mean.
(B) Schematic depicting 1.163kb of intron 4 of the tp53 gene that contains two tp53-binding sites (red bars) used to drive expression of mVenus when tp53 is trans-activated.
(C) tp53 activity is observed in numerous tissues that develop abnormally in ΔU8-3 mutants, including the eye, hindbrain, and somites (arrowheads) at 24 hpf.
(D) Quantitative RT-PCR demonstrates significant upregulation of the tp53 target genes Δ113tp53, mdm2, cyclinG1, p21, and bax in ΔU8-3 mutants when compared to wild-type siblings at 48 hpf; this upregulation is abrogated by genetic inactivation of tp53. hpf—h post-fertilization. n = 4 biological replicates per genotype. Error bars indicate SD from the mean.
(E) Representative brightfield and corresponding fluorescent images of the indicated genotype and transgenic reporter. Angiogenic sprout number is indicated by white numbering.
(F) Quantitation of angiogenic sprouts in the indicated genotype. n = 6-13 embryos per genotype. Error bars indicate SD from the mean.
Inactivation of tp53 Signaling Partially Rescues the ΔU8-3 Mutant
To characterize the consequences of tp53 signaling in the ΔU8-3 mutant, ΔU8-3 mutant embryos were crossed onto a tp53 mutant background.15 Notably, genetic inactivation of tp53 prevented induction of Δ113tp53, mdm2, cyclinG1, p21, and bax expression in ΔU8-3 mutants, induction that is typically observed at 48 hpf (Figure 6D). This result confirms that the increased expression of these mRNAs is tp53-dependent. Genetic inactivation of tp53 was found to partially restore angiogenic sprouting in ΔU8-3 mutants (Figure 6E, F), and to rescue ventricular swelling 48 hpf (Figure S13A). Although tp53 clearly contributes to the gross morphological abnormalities of ΔU8-3 mutants, inactivation of tp53 signaling would not be predicted to restore the defect in rRNA processing. Indeed, quantification of 28S:18S ratios by TapeStation assay demonstrated that the preferential inhibition of 28S biogenesis is not rescued in tp53 mutant ΔU8-3 mutant embryos (Figure S13B), and these embryos remain significantly shorter than their U8-3 wild-type sibling counterparts (Figure S13C). Taken together, these data indicate that the reduced embryo length of ΔU8-3 mutants results from impaired ribosome function.
Discussion
Informed by the genetic architecture of the Mendelian disease LCC, here we describe the use of a vertebrate mutant animal model of U8 dysfunction to shed light on the processing of the snoRNA U8, its function in ribosome biogenesis, and the molecular pathology of LCC.
Our data indicate that both zebrafish U8 mutant animals and fibroblasts from individuals with LCC exhibit rRNA processing defects. The majority of human syndromes linked to defective ribosome biogenesis are thought to result from haploinsufficiency for, or partial LoF of, ribosomal proteins or ribosome assembly factors.28, 29, 30 In our previously published series, 31 of 33 probands with LCC were compound heterozygous for two different SNORD118 mutant alleles.1 Given that most rare autosomal recessive traits demonstrate enrichment for consanguinity and allelic homozygosity, these molecular data indicate that bi-allelic null mutations in SNORD118 are likely incompatible with development. In keeping with this, we show that the U8-3 zebrafish mutant is embryonic lethal. Our genetic data further imply that disease results from the combination of one severe (null) and one milder (hypomorphic) mutation. We suggest that the latter acts to mediate viable embryogenesis, being characterized by a rescue of the gross morphology of zebrafish U8 mutants, which is apparently indistinguishable from wild-type human U8, while conferring a defect in U8 processing in HeLa nuclear cell extracts.
The biogenesis and processing of essential, independently transcribed snoRNAs is dependent on highly organized secondary structures and their sequential association with core snoRNP proteins that halt advancing exonucleases, and nucleolytic trimming of the precursor RNA is necessary to achieve functionality and to provide metabolic stability and nucleolar localization.7,8,31 Using our zebrafish model, we were able to shed further light on the processing of human U8, identifying a base-pairing interaction between the 5′ end and 3′ extension of human pre-U8. Twenty-one of 33 individuals with LCC identified by Jenkinson et al. harbored at least one mutation within the 3′ extension, and a further eight individuals were positive for a mutation in a 5′ situated nucleotide predicted to base-pair with this 3′ extension.1 Human pre-U8 snoRNA containing a 3′ extension rescued the zebrafish U8 mutant, indicating a conserved biological function. To our knowledge, the secondary structure of human pre-U8 and the requirement for such nucleotide base-pairing within this structure has not been described previously. Close examination of the five zebrafish pre-U8 snoRNAs does not reveal an extensive potential for base-pairing between the 5′ end and the 3′ extension as proposed for the human pre-U8 snoRNA. Any base-pairing potential of the zebrafish pre-U8 snoRNAs would require exact mapping of the 5′ and 3′ ends of the zebrafish pre-U8 snoRNAs and experimental validation. We hypothesize that in human cells, the base-pairing between the 5′ and 3′ ends of the human pre-U8 snoRNA may be important to regulate the timing of final maturation of U8 by either masking its functionally relevant 5′ end, and/or by preventing premature formation of a canonical kink-turn, and associated protein binding, until the U8 snoRNP has sufficiently matured. Indeed, the vast majority of the precursor forms of human U8 are detected within cytoplasmic and nucleoplasmic fractions, whereas the bulk of the mature U8 is found in the nucleolus, the site of pre-rRNA processing.7,8 Ultimately, such regulation may be important for multiple aspects of U8 biology, including the correct processing of U8, its metabolic stability, its nucleo-cytoplasmic trafficking,7,32 its final targeting to the nucleolus, or even possibly facilitation of extra-ribosomal functions of U8 independent of its interaction with the pre-rRNAs. Previous work from the Lührmann laboratory identified four U8 snoRNA precursor processing intermediates, leading them to hypothesize either that the U8 pre-snoRNA is processed by more than one exonuclease activity, or that the sequence of the 3′ extension may determine the kinetics of 3′ end processing.7 Our data, as presented in Figure 5, are in agreement with the latter hypothesis, as LCC-associated single nucleotides changes located within the 3′ extension, or that pair with this region, appear to alter the kinetics of processing of the U8 pre-snoRNA. The assembly, processing, and export of small nuclear RNPs (snRNPs) also follows a well-defined pathway like that described for snoRNPs.33, 34, 35 Precursor forms of the spliceosomal U2 small nuclear RNA (snRNA) pair the 3′ extension with an internal sequence, this base-pairing being essential for correct processing into a mature snRNP.36 It is possible that base-pairing of 3′ extension sequences may be a more general feature of snRNA and/or snoRNA maturation.
In cell culture models, tp53 is required for activation of cell cycle arrest within 24 h from the onset of ribosomal stress before, over time, impaired ribosome function becomes rate limiting for cellular growth and division.11,37, 38, 39 Depletion of U8 has recently been shown to result in potent induction of TP53 in human cells,13,17 and tp53 signaling in the U8-3 mutant would be predicted to induce an earlier and more complete inhibition of the cell cycle. In keeping with this, the angiogenic sprouting defect observed in the U8-3 mutant at 24 hpf was largely, but not completely, rescued by genetic inactivation of tp53; the partial rescue is probably explained by a preferential reduction of 28S in U8-3 mutants, indicating that ribosomal dysfunction is already manifested by this time. By 48 hpf, when ribosome dysfunction is likely more pronounced, the morphology of the U8-3 mutant was only minimally salvaged. TP53 is normally constitutively degraded by the proteasome following Hdm2-mediated ubiquitination. When ribosome biogenesis is compromised, unassembled ribosomal components accumulate; this is notably the case for a trimeric complex consisting of the two ribosomal proteins uL5 and uL18 and the 5S rRNA which captures Hdm2, titrating it away from TP53, resulting in a net stabilization of TP53 and the activation of a cell death program.40,41 Although knockdown of U8 in human cells would, therefore, indirectly activate TP53, it remains to be determined whether such activation occurs in the affected neurological tissue of individuals with LCC, and whether this plays a mechanistic role in disease pathology.
Alterations in ribosomal components underlie a heterogeneous class of diseases referred to as the ribosomopathies, with a diversity in associated clinical phenotype providing an indication of the multiple specialized roles of the ribosome in normal physiology. Despite ubiquitous expression of the U8 snoRNA, germline mutations in SNORD118 cause a progressive microangiopathy apparently limited to the cerebral vasculature. Although the phenotype of the exclusively neurological disease LCC is highly distinctive, it is not pathognomonic, as a remarkably similar radiological association is seen in the context of the multisystem disorder Coats plus. Coats plus is caused by mutations in CTC142 and STN1,43 both components of the conserved heterotrimeric telomeric capping complex, and in the telomeric protein POT1.44 Interestingly then, an unbiased enChip-RNaseq approach identified U8 as a telomere-associated RNA.45 As such, the precise link between U8 and cerebral vascular homeostasis awaits elucidation, and may conceivably involve both ribosomal defects (that activate TP53 and/or induce selective impairment of translation of distinct mRNAs in a cell lineage specific context) and currently undefined non-ribosomal functions of U8.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the following people for reagents: pDest-γcrystallin:mCherry, pcs2+-mKate2 and pcs2+-H2B-mCerulean3 plasmids were gifts from Dr. Emily Don, pcs2+-nCas9n (Addgene plasmid #47929) was a gift from Dr. Adam Hurlstone. We thank Dr. Martin Reijns (Edinburgh) for helpful discussions. This study was supported by a grant to Y.J.C and R.T.O. from the Great Ormond Street Hospital Charity (V4017). Y.J.C. also acknowledges a state subsidy managed by the National Research Agency (France) under the "Investments for the Future" program bearing the reference ANR-10-IAHU-01 and the MSDAvenir fund (DEVO-DECODE Project). P.R.K was supported by the Stroke Association (TSA LECT 2017/02). Research in the Lab of D.L.J.L. is supported by the Belgian Fonds de la Recherche Scientifique (F.R.S./FNRS), the Université Libre de Bruxelles (ULB), the Région Wallonne (DGO6) [grant RIBOcancer n°1810070], the Fonds Jean Brachet, and the International Brachet Stiftung.
Published: April 30, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.003.
Contributor Information
Andrew P. Badrock, Email: andrew.badrock@igmm.ed.ac.uk.
Raymond T. O’Keefe, Email: rokeefe@manchester.ac.uk.
Web Resources
CHOPCHOP, http://chopchop.cbu.uib.no/
Online Mendelian Inheritance in Man, https://www.omim.org
RNAfold, http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi
Supplemental Information
References
- 1.Jenkinson E.M., Rodero M.P., Kasher P.R., Uggenti C., Oojageer A., Goosey L.C., Rose Y., Kershaw C.J., Urquhart J.E., Williams S.G. Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat. Genet. 2016;48:1185–1192. doi: 10.1038/ng.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Cruz J., Karbstein K., Woolford J.L., Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peculis B.A., Steitz J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 4.Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015;6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstein L.B., Steitz J.A. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 6.Peculis B.A., Steitz J.A. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 7.Watkins N.J., Lemm I., Lührmann R. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol. Cell. Biol. 2007;27:7018–7027. doi: 10.1128/MCB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., Watkins N.J. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terns M.P., Terns R.M. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 10.Yelick P.C., Trainor P.A. Ribosomopathies: Global process, tissue specific defects. Rare Dis. 2015;3:e1025185. doi: 10.1080/21675511.2015.1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestov D.G., Strezoska Z., Lau L.F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golomb L., Volarevic S., Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014;588:2571–2579. doi: 10.1016/j.febslet.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas E., Parisot P., Pinto-Monteiro C., de Walque R., De Vleeschouwer C., Lafontaine D.L.J. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016;7:11390. doi: 10.1038/ncomms11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin S.-W., Beis D., Mitchell T., Chen J.N., Stainier D.Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- 15.Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D.M., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert S.P., Huisken J., Kim T.N., Feldman M.E., Houseman B.T., Wang R.A., Shokat K.M., Stainier D.Y.R. Arterial-venous segregation by selective cell sprouting: an alternative mode of blood vessel formation. Science. 2009;326:294–298. doi: 10.1126/science.1178577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langhendries J.-L., Nicolas E., Doumont G., Goldman S., Lafontaine D.L.J. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget. 2016;7:59519–59534. doi: 10.18632/oncotarget.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.J. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 20.Peculis B.A., DeGregorio S., McDowell K. The U8 snoRNA gene family: identification and characterization of distinct, functional U8 genes in Xenopus. Gene. 2001;274:83–92. doi: 10.1016/s0378-1119(01)00596-0. [DOI] [PubMed] [Google Scholar]
- 21.Peculis B.A. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 1997;17:3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange T.S., Borovjagin A.V., Gerbi S.A. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA. 1998;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn188. W70-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 25.Kastenhuber E.R., Lowe S.W. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Ng S.M., Chang C., Zhang Z., Bourdon J.C., Lane D.P., Peng J. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23:278–290. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson N.D., Weinstein B.M. Arteries and veins: making a difference with zebrafish. Nat. Rev. Genet. 2002;3:674–682. doi: 10.1038/nrg888. [DOI] [PubMed] [Google Scholar]
- 28.Boocock G.R.B., Morrison J.A., Popovic M., Richards N., Ellis L., Durie P.R., Rommens J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 29.Valdez B.C., Henning D., So R.B., Dixon J., Dixon M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert B.L., Pretz J., Bosco J., Chang C.Y., Tamayo P., Galili N., Raza A., Root D.E., Attar E., Ellis S.R., Golub T.R. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Lührmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Massenet S., Bertrand E., Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017;14:680–692. doi: 10.1080/15476286.2016.1243646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 35.Becker D., Hirsch A.G., Bender L., Lingner T., Salinas G., Krebber H. Nuclear Pre-snRNA Export Is an Essential Quality Assurance Mechanism for Functional Spliceosomes. Cell Rep. 2019;27:3199–3214.e3. doi: 10.1016/j.celrep.2019.05.031. [DOI] [PubMed] [Google Scholar]
- 36.Huang Q., Jacobson M.R., Pederson T. 3′ processing of human pre-U2 small nuclear RNA: a base-pairing interaction between the 3′ extension of the precursor and an internal region. Mol. Cell. Biol. 1997;17:7178–7185. doi: 10.1128/mcb.17.12.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindström M.S., Nistér M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS ONE. 2010;5:e9578. doi: 10.1371/journal.pone.0009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G.F., Bernardi R., Pandolfi P.P., Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilkes D.M., Chen L., Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sloan K.E., Bohnsack M.T., Watkins N.J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donati G., Peddigari S., Mercer C.A., Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O’Sullivan J. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 2012;44:338–342. doi: 10.1038/ng.1084. [DOI] [PubMed] [Google Scholar]
- 43.Simon A.J., Lev A., Zhang Y., Weiss B., Rylova A., Eyal E., Kol N., Barel O., Cesarkas K., Soudack M. Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J. Exp. Med. 2016;213:1429–1440. doi: 10.1084/jem.20151618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai H., Jenkinson E., Kabir S., Babul-Hirji R., Najm-Tehrani N., Chitayat D.A., Crow Y.J., de Lange T. A POT1 mutation implicates defective telomere end fill-in and telomere truncations in Coats plus. Genes Dev. 2016;30:812–826. doi: 10.1101/gad.276873.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita T., Yuno M., Okuzaki D., Ohki R., Fujii H. Identification of non-coding RNAs associated with telomeres using a combination of enChIP and RNA sequencing. PLoS ONE. 2015;10:e0123387. doi: 10.1371/journal.pone.0123387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.