Mesenchymal stromal cells (MSCs) are multipotent progenitor cells that are studied as a treatment for inflammatory bowel disease. Local injection of MSCs stimulates closure of perianal fistulas in Crohn’s disease.1,2 Previously, we found that local injections of bone marrow–derived MSCs alleviated experimental colitis in mice.3 MSCs are thought to work via modulating immune responses and stimulating tissue regeneration via secreted proteins and cell–cell contacts. In addition, recent studies have indicated that MSCs also exert effects via exosomes, which are small membrane-enclosed vesicles containing proteins, DNA, and (micro)RNAs.4 The objective of this study was to evaluate if MSC-derived exosomes contribute to the therapeutic effects of local MSC therapy. We investigated whether MSC exosomes stimulate epithelial regeneration and if local application of MSC exosomes, as a cell-free alternative for MSC therapy, can alleviate colitis in epithelial damage–driven models.
MSC exosomes were isolated from murine, bone marrow–derived MSCs (Supplementary Figure 1A and B), using ultracentrifugation of MSC-conditioned medium (CM), containing 1.2 μg exosomes per milliliter. The presence of MSC exosomes was confirmed by the markers flotillin-1 and alix (Supplementary Figure 1C), and visualization of 50- to 150-nm vesicles using transmission electron microscopy (Supplementary Figure 1D). The uptake of fluorescently labeled MSC exosomes by CT26 mouse colonic epithelial cells was confirmed by a red fluorescent signal upon addition of MSC exosomes to CT26 cells (Figure 1A, Supplementary Figure 2A). To determine the effects of MSC exosomes on epithelial regeneration, CT26 cells were first damaged by exposure to dextran sulfate sodium (DSS) (Supplementary Figure 2B). A significantly higher cell number was detected when DSS-damaged CT26 cells were cultured with 20 μg/mL MSC exosomes (Figure 1B). The high-dose MSC exosomes reduced levels of the apoptotic marker cleaved caspase-3 in CT26 cells upon damage with 2% DSS (Figure 1C, Supplementary Figure 2C), and 3% DSS (Supplementary Figure 2D), indicating decreased apoptosis. Because epithelial repair is a combination of proliferation and migration, we also assessed the effects of MSC exosomes on cell migration using a scratch assay. CT26 cells treated with CM with exosomes showed the fastest wound closure, but CM without exosomes and 20 μg/mL exosomes also significantly increased wound healing compared with non-CM (Figure 1D, Supplementary Figure 2E). In addition, also cytokine-stimulated human MSC exosomes showed increased wound closure in human epithelial cells compared with non-CM (Supplementary Figure 2F). Non-damaged murine epithelial cells stimulated with CM with exosomes showed a slight but significant increase in proliferation in CT26 cultures (Supplementary Figure 2G). Cell-cycle analysis showed that MSC exosomes increased the percentage of epithelial cells in both the S- and G2-phases (Figure 1E, Supplementary Figure 2H). Next, we evaluated the effects in 3-dimensional mouse colonic organoids. We confirmed that PKH26-labeled exosomes were taken-up by the epithelial organoids (Figure 1F, Supplementary Figure 3A) and induced organoid proliferation without changing the number of Ki67-positive cells (Figure 1G, Supplementary Figure 3B and C). Mucin 2 and cytokeratin 20 (Supplementary Figure 3D) were down-regulated in colonic organoids cocultured with MSC exosomes, suggesting that the increase in organoid proliferation by MSC exosomes was not leading directly to more differentiation. No differences in expression of the stem cell marker, Leucine-rich repeat-containing G-protein coupled receptor 5, and enteroendocrine marker, chromogranin A, were found (Supplementary Figure 3C). Finally, we showed that cyclo-oxygenase 2, an enzyme described to be up-regulated in colonic epithelial cells from inflammatory bowel disease patients,5 was down-regulated significantly in colonic organoids 72 hours after exosome treatment (Supplementary Figure 3C).
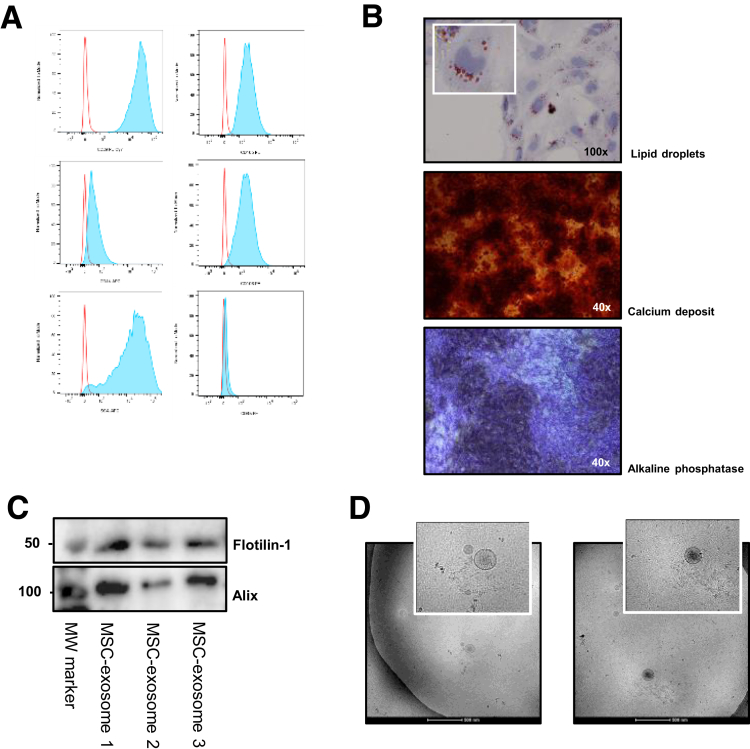
Supplementary Figure 1.
Characterization of murine MSCs and MSC exosomes. (A) MSCs were characterized by flow cytometry analysis for surface markers (anti-CD105–PE, clone MJ7/18 [562759], anti-CD106–PE, clone 429 [561613], anti-CD44–APC, clone IM-7 [561862]; all BD Biosciences; anti-CD45–PE, clone 30-F11 [12-0451-82], anti-CD29–PECy7, clone HMb1-1 [25-0291-80], anti-stem cells antigen-1–APC, clone D7 [17-5981-81]; all eBioscience, Vienna, Austria) using the LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star, Inc, Ashland, OR). (B) MSC differentiation staining for adipocytes (lipid droplets) and osteoblasts (calcium deposit and alkaline phosphatase activity).5 (C) Western blot for exosomal markers flottilin-1 and alix. Western blot analysis was performed using 25 μg protein loaded on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis8 and transferred to polyvinylidene difluoride membranes (0.45-μm pore size, 10401196; Whatman, Maidstone, UK). Ponceau staining (3504; Sigma-Aldrich) was used to confirm equal loading. Blots were incubated with primary antibodies to rabbit anti-alix, clone 3A9 (MCA2493; Bio-Rad Laboratories, Hercules, CA) and mouse anti–flotillin-1, clone EPR6041 (1333497; Abcam, Cambridge, UK). (D) Transmission electron microscopy images of exosomes isolated from MSCs. From a purified exosome suspension, 3 μL was put on a copper electron microscopy grid that was glow-discharged in air. By using an electron microscopy grid plunger freezer (Leica), excess suspension was removed by blotting for 1 second using filter paper (Whatman) at 95% humidity at room temperature and the grid was plunged into liquid ethane at -183°C. Samples were transferred to a Tecnai F20 Transmission Electron Microscope (ThermoFisher Scientific) using a Gatan cryo holder and images were recorded at 15,000× magnification on a Gatan 2kx2k CCD camera (Gatan, Pleasanton, CA) behind a 2001 energy filter operating at 20 electron-volts slit width. MW, molecular weight.
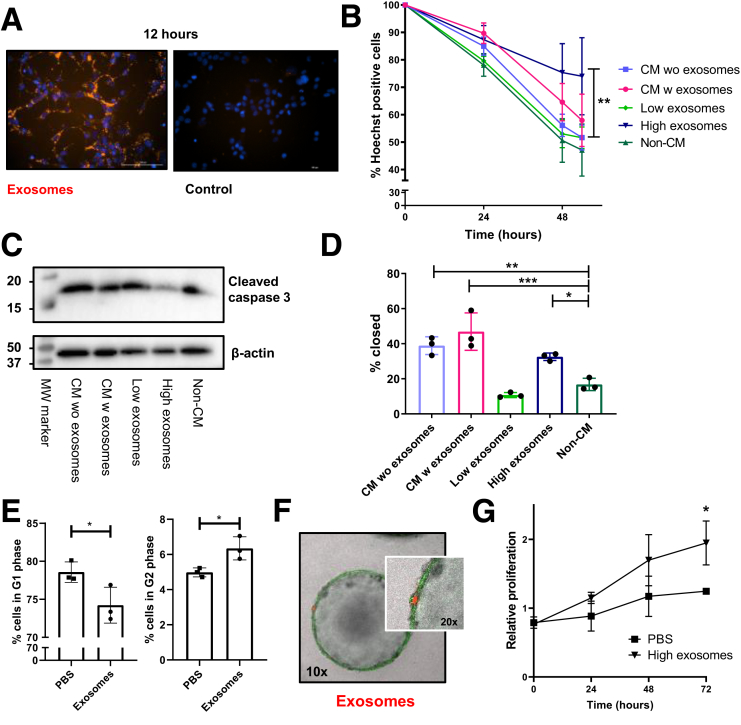
Figure 1.
MSC exosomes stimulate epithelial regeneration in vitro. (A) Images of CT26 cells treated for 12 hours with PKH26-labeled exosomes. (B) Percentage of Hoechst-positive DSS-damaged CT26 cells after treatment with the indicated conditions. Data represent the average of 3 independent experiments in triplicate. One-way analysis of variance, Dunnett multiple comparison with non-CM. (C) Western blot analysis for cleaved caspase-3 in 2% DSS-treated CT26 cells under the indicated conditions. Representative blot from 3 independent experiments. (D) Relative wound closure (after 27 h) of CT26 cells stimulated with the indicated conditions. Data represent the mean of 3 independent experiments in triplicate. One-way analysis of variance, the Dunnett multiple comparison with non-CM. (E) Percentage of CT26 cells in G1- and G2-phases after co-culture with exosomes. Representative data are shown from 2 independent experiments in triplicate. Student t test was used. (F) Images of green fluorescent protein-positive colon organoids cultured with PKH26-labeled exosomes for 1 week. Images are representative of 2 independent experiments. (G) MTS assay of colon organoids cultured with exosomes. Data represent 2 independent experiments performed in triplicate. Student t test. ∗P ≤ .05, ∗∗P < .01, and ∗∗∗P < .001. MW, molecular weight; PBS, phosphate-buffered saline; w, with; wo, without.
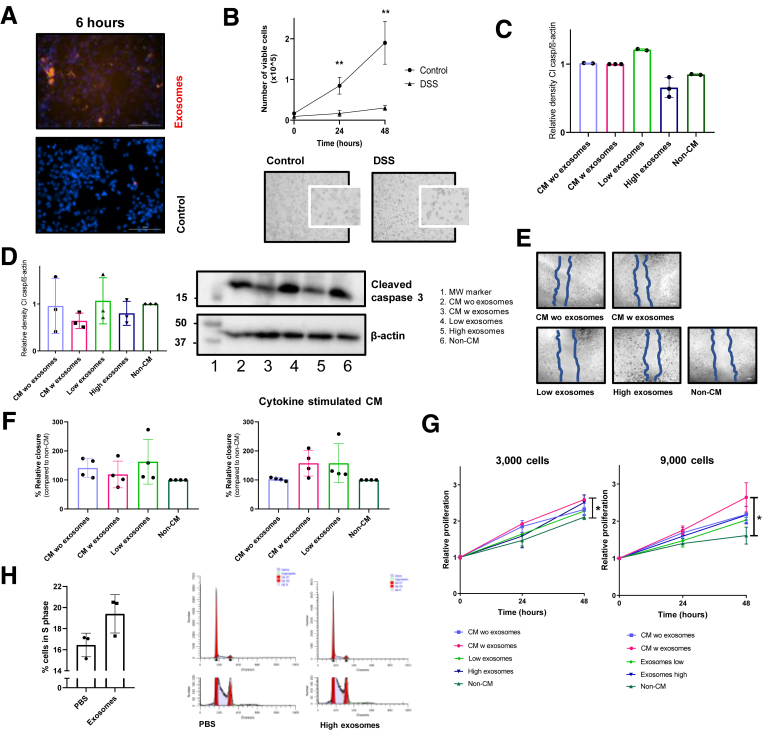
Supplementary Figure 2.
In vitro models for inflammatory bowel disease and the effects of MSC-derived exosomes in these models. (A) Fluorescent images of CT26 cells treated with PKH26-labeled exosomes after 6 hours. Murine MSC exosomes were labeled using PKH26 (PKH26GL; Sigma-Aldrich) according to the manufacturer’s protocol. In short, exosomes were incubated for 4 minutes with PKH26. After this period, the staining was stopped with vesicle-free fetal calf serum and exosomes were harvested with centrifugation (4 h at 100,000 × g) and washed with phosphate-buffered saline. To exclude nonspecific fluorescent staining, a mock control containing vesicle-free fetal calf serum was included in the entire staining procedure. To verify if MSC exosomes could be taken up by epithelial cells, CT26 cells were co-cultured with PKH26-labeled MSC exosomes and fluorescent images were obtained after 6 and 12 hours using the Cytation5 live cell imaging system (Biotek). (B) Number of viable cells in DSS exposed CT26 cells over time. Pictures of 4% DSS-treated CT26 cells for 3 hours. Data represent 3 independent experiments in triplicate. Student t test. (C) Densitometric analysis of the cleaved caspase-3 bands corrected for the loading control. (D) Western blot analysis for cleaved caspase-3 in CT26 cells incubated with 3% DSS for 6 hours and treated with CM with or without exosomes, a low or high dose of exosomes or non-CM for 24 hours. Representative blot from 3 independent experiments. Graph of the relative density of the cleaved caspase-3 bands corrected for the loading control β-actin. (E) Representative images to Figure 1D of wound healing assay in CT26 cells after 27 hours. (F) Relative wound closure after 40 hours of DLD1 cells stimulated with the indicated conditions. To mimic the IBD environment, human MSCs also were stimulated in fetal calf serum–free medium supplemented with proinflammatory cytokines (1 μg/mL interleukin 17 (IL17, 167200-17-B), 1 μg/mL oncostatin M (OSM, 167300-10-B), 1 μg/mL interferon-γ (IFN-γ, 167300-02-B), and 1 μg/mL tumor necrosis factor-α (TNF-α, 167300-01A-B) (all Peprotech, London, UK) for 3 days. This cytokine-stimulated CM was used to obtain the exosomes used in the second graph. Representative data from 4 independent experiments performed in triplicate. (G) MTS assay of 3000 and 9000 CT26 cells cultured with CM with or without exosomes, low or high exosomes, or non-CM. Data represent means of 2 independent experiments in triplicate for every time point. One-way analysis of variance and the Dunnett multiple comparison with non-CM were used. (H) Percentage of CT26 cells in S-phase after culture with and without exosomes. Two representative images from ModFit cell-cycle analysis after co-culture of CT26 cells with PBS or 20 μg exosomes. Representative data from 2 independent experiments. ∗P < .05, ∗∗P < .01. MW, molecular weight; w, with; wo, without.
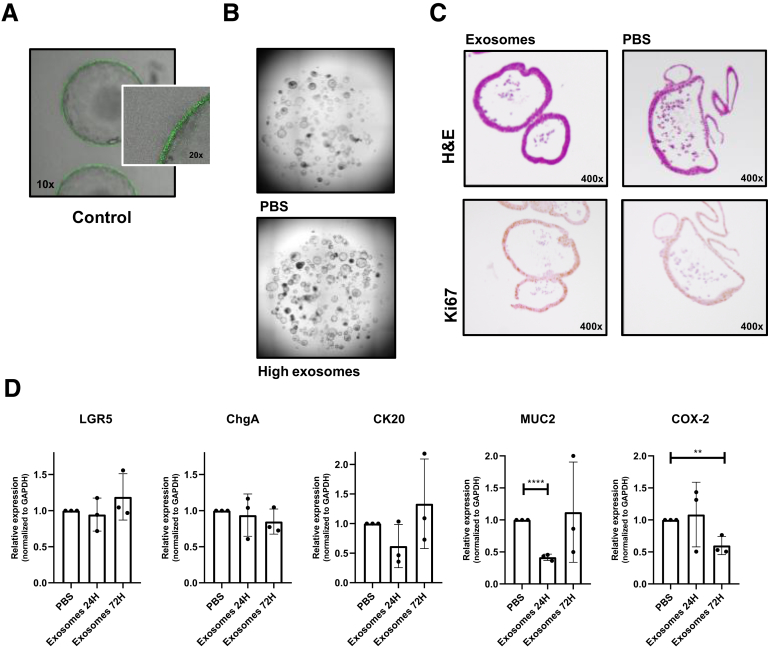
Supplementary Figure 3.
Gene expression and proliferation in MSC exosome–treated organoids. (A) Images of green fluorescent protein-positive colon organoids cultured with a mock control containing vesicle-free fetal calf serum for 1 week. Images are representative of 2 independent experiments. (B) Representative images of colon organoids cultured without and with MSC exosomes for 72 hours. To obtain colon organoids, after killing the mice, the colonic tissue was transferred to 20 mmol/L EDTA (324503; Merck Millipore) in Hanks’ balanced salt solution (14175-053; Gibco) and incubated for 30 minutes at 37°C with repeated vortexing to release the colonic crypts. The crypts were washed in Ad-DF+++, consisting of Advanced Dulbecco's modified Eagle medium/Ham’s F-12 (12634-010; Gibco) supplemented with 1% Glutamax (35050-038; Gibco), 1% penicillin/streptomycin (15140-122; Gibco) and 1% HEPES (15630-056; Gibco). After centrifugation, the colonic crypts were plated in 20 μL Matrigel (growth factor–reduced Matrigel, 356231; Corning, Corning, NY) in 48-well culture plates. After polymerization of the Matrigel, 250 μL of complete growth medium was added, consisting of Ad-DF+++, supplemented with B27 (11530536; Invitrogen, Carlsbad, CA), N-acetylcysteine (A9165-5; Sigma-Aldrich), nicotinamide (n0636; Sigma-Aldrich), A83-01 (2939; Tocris, Bristol, UK), p38 inhibitor (s7067; Sigma-Aldrich), epidermal growth factor (PMG8041; Invitrogen), 20% Noggin CM, 10% R-spondin CM (both kindly provided by the Tytgat Institute, Amsterdam, The Netherlands), and 50% Wnt3a CM (kindly provided by the Hubrecht Institute, Utrecht University, The Netherlands). During the first 4 days, medium was supplemented with Rho-K inhibitor (Y0503; Sigma-Aldrich). Colonic organoids were maintained in a humidified incubator at 37°C containing 5% CO2. (C) H&E and Ki67 staining of colon organoids cultured with exosomes or phosphate-buffered saline (PBS) for 72 hours. Colonic organoids treated with or without exosomes were stained immunohistochemically for the proliferation marker rabbit anti-Ki67 (clone SP6, 16667; Abcam) as described before.1 (D) Relative messenger RNA expression of LGR5, chromogranin A (ChgA), cytokeratin 20 (CK20), cyclo-oxygenase-2 (COX-2), and mucin (MUC2) in dissociated colon organoids co-cultured with PBS or exosomes for 24 and 72 hours. Data represent the means of 3 independent experiments performed in triplicate. The Student t test was used. Messenger RNA was isolated using the nucleospin RNA kit (740955250; Macherey-Nagel, Düren, Germany) after 24 and 72 hours. Complementary DNA was generated using the RevertAid First Strand Complementary DNA Synthesis Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. Quantitative polymerase chain reactions were performed using SYBR green (1708886; Bio-Rad, Hercules, CA) and primers (all Invitrogen) for a stem cell marker (leucine-rich repeat-containing G-protein–coupled receptor 5: forward: TGGTGGCTTTGACCGTGTT; reverse: CGATTACCCCAATTAGCAGCTTT), differentiation markers (mucin 2: forward: CCCAGAGAGTTTGGAGAGCA; reverse: CTCCTCACATGTGGTCTGGT; chromogranin A: forward: GGCCCAGCAGCCGCTGAAGCAGCA; reverse: CTCTGCGGTTGGCGCTGCCCTCCTC; cytokeratin 20: forward: CGCATCTCTGTCTCCAAAGC; reverse: TTCTGCATTGCCAGTTTCCC), and the prostaglandin pathway (cyclo-oxygenase 2: forward: CCGTGCTGCTCTGTCTTAAC; reverse: TTGGGAACCCTTCTTTGTTC). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene: forward: AACTTTGGCATTGTGGAAGG; reverse ACACATTGGGGGTAGGAACA. **P < .01, ****P < .0001.
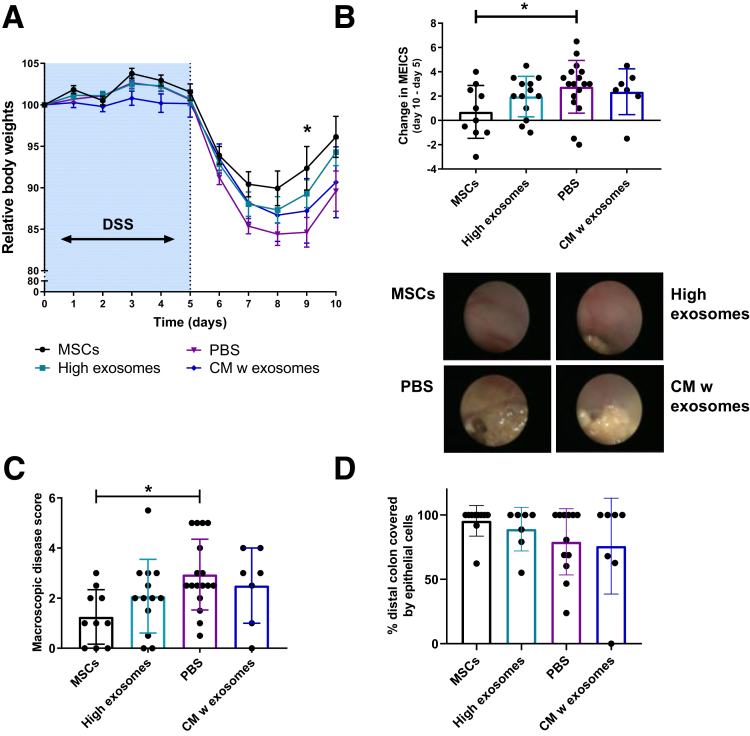
Next, we used the DSS mouse colitis model to investigate if MSC exosomes are responsible for the beneficial effects of local MSC therapy. DSS-treated mice were injected endoscopically with MSCs (2 × 106), 20 μg MSC exosomes, CM (containing ∼0.24 μg exosomes), or solvent control at day 5. In vitro, 2 × 106 MSCs will produce approximately 9.6 μg of exosomes every 3 days. Local MSC therapy and, to some extent, MSC exosome therapy alleviated DSS-induced colitis, as shown by a higher relative body weight, lower murine endoscopic index of colon severity, lower macroscopic disease score, increased colon length, and decreased epithelial damage, compared with control or CM-treated mice. However, local MSC exosome therapy was less effective compared with MSC therapy (Figure 2A–D, Supplementary Figure 4). This suggests that MSCs also exert their efficacy through other mechanisms or that continuous production of exosomes is needed for profound therapeutic effects. Because locally injected MSCs are thought to be licensed in vivo by the proinflammatory milieu, it might be that cytokine-stimulated MSCs produce more efficient vesicles,6 which also is supported by our human MSC data (Supplementary Figure 2F). The effects of MSC exosomes might be mediated by microRNAs because it was shown that microRNAs involved in cell death and growth were enriched in exosomes.7 In conclusion, our results show that MSC-derived exosomes may contribute to the amelioration of colitis by stimulation of epithelial repair and decreasing epithelial apoptosis.
Figure 2.
Locally applied MSC exosomes partially alleviate experimental colitis. (A) Relative body weights of mice with DSS-induced colitis, endoscopically treated with the indicated conditions. Means ± SEM. One-way analysis of variance, Dunnett multiple comparison with PBS. (B) Difference in murine endoscopic index of colitis severity (MEICS) between day 10 and day 5 for the treatment groups. One-way analysis of variance, Dunnett multiple comparison with PBS. (C) Macroscopic colonic disease score at day 10. One-way analysis of variance, the Dunnett multiple comparison with PBS. (D) Percentage of distal colon covered by cytokeratin-positive epithelial cells. Data represent 2 independent mouse experiments, n = 7–19 mice/group. ∗P < .05. PBS, phosphate-buffered saline; w, with.
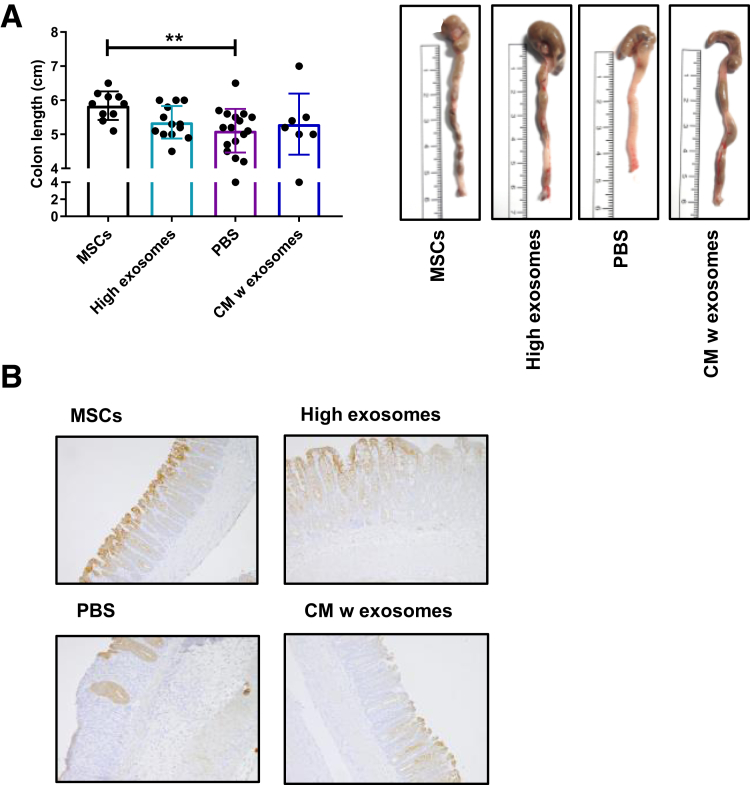
Supplementary Figure 4.
(A) Colon length. Representative pictures of colons from the different treatment groups. (B) Representative images showing pan-cytokeratin–positive epithelial cells to identify epithelial cells. Data represent 2 independent mouse experiments, n = 7–19 mice/group. PBS, phosphate-buffered saline; w, with. **P < .01.
Acknowledgment
The authors thank the staff of the Central Animal Facility of the Leiden University Medical Center for animal care and the group of Professor Clevers, and especially Dr van Es, from the Hubrecht Institute, and Dr Muncan from the Tytgat Institute for providing WNT3a, Noggin, and R-spondin cell lines.
Footnotes
Author contributions M. C. Barnoorn designed the study, performed data acquisition, analysis, and interpretation, and drafted the manuscript; L. Plug performed data acquisition, analysis, and interpretation; E. S. M. Muller-de Jonge, D. Molenkamp, E. Bos, and W. E. Corver acquired and analyzed the data; M. J. A. Schoonderwoerd interpreted the data and critically revised the manuscript for intellectual content; A. E. van der Meulen-de Jong and H. W. Verspaget designed and advised in the execution of the study and critically revised the manuscript for intellectual content; and L. J. A. C. Hawinkels interpreted the data, designed and supervised the study, and critically revised the manuscript for intellectual content.
Conflicts of interest The authors disclose no conflicts.
Supplementary Methods
MSC Isolation
Animal experiments were approved by the Central Authority for Scientific Procedures on Animals and the Animal Welfare Body of the Leiden University Medical Center (AVD116002017860). MSCs were isolated from the bone marrow of Tg(s100a4-cre)1Egn mice (Jackson Laboratory, Bar Harbor, ME) as described previously.1 Bone marrow–derived MSCs were cultured in α-MEM (32561-029; Gibco, Gaithersburg, MD) with 1% penicillin/streptomycin (15140-122; Gibco) and 10% fetal calf serum (10270-106; Gibco). Human MSCs were obtained from the bone marrow of healthy volunteers, with informed consent for clinical application and research, and cultured and analyzed as described previously.2
MSC CM and Exosome Isolation
CM was obtained by culturing confluent MSCs in fetal calf serum–free medium for 3 days. CM was centrifuged at 300 and 2000 × g for 10 minutes to remove cell debris and the supernatant was used for experiments (CM with exosomes). For isolation of exosomes the CM was concentrated by ultrafiltration over a 100-kilodalton molecular weight cut-off filter (Amicon Ultra-15 tubes, UFC910024; Merck Millipore, Burlington, MA) at 5000 × g for 40 minutes (Heraeus multifugeX1R; ThermoFisher, Waltham, MA). The flow-through contained the CM without exosomes. The pellet was resuspended in phosphate-buffered saline and consequently centrifugated at 100,000 × g for 8 hours (Optima XE-90 ultracentrifuge; Beckman Coulter, Pasadena, CA), after which pelleted exosomes were visible. The concentration of MSC exosomes was determined by the Pierce BCA Protein Assay Kit (ThermoFisher). MSC exosomes were characterized for exosome markers by Western blot and electron microscopy.
In Vitro Colitis Models
To induce epithelial damage, 2% to 4% DSS (molecular weight, 36,000–50,000 kilodaltons, 160110; MP Biomedicals, Brussels, Belgium) in fetal calf serum–free RPMI1640 (21875-034; Gibco) was used in CT26 cells for 3, 6, 12, or 24 hours. MSC CM with exosomes (∼1.2 μg/mL exosomes), MSC CM without exosomes, non-CM, 2 μg/mL exosomes (low) or 20 μg/mL exosomes (high) in non-CM was added to the damaged epithelial cells. The cell number over time was measured by Hoechst staining (33342; Cell Signaling, Danvers, MA) using the Cytation5 and Gen5 software (Biotek, Winooski, VT) for up to 54 hours. The percentage of Hoechst-positive cells was given relative to 0 hours. Proteins from CT26 cells treated with different exosome conditions were extracted after 24 hours. A total of 25 μg protein was loaded on a 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and, after transfer, Western blot was performed for rabbit anti-cleaved caspase-3 (clone5A1E, 9661S; Cell Signaling) and rabbit anti–β-actin (clone I-19, 1616; Santa Cruz Biotechnology, Dallas, TX) as a loading control. For densitometric analysis, cleaved caspase-3 bands were corrected for β-actin.
To assess the effect of MSC exosomes on the migration of epithelial cells, a wound healing assay was performed. CT26 (mouse) or DLD1 cells (human) were seeded in 48-well plates (25,000 cells/well) and after overnight incubation a wound was created using a 200-μL pipet tip. MSC CM with exosomes, MSC CM without exosomes, non-CM, 2 μg/mL exosomes (low) or 20 μg/mL exosomes (high) in non-CM were added to the damaged epithelial cells. Images were obtained after 15, 27, 65, and 73 hours for CT26 and 40 hours for DLD1, using Cytation5. Wound closure was determined by an average of 5 measurements per image and made relative to the start of the experiment. Proliferation of nondamaged CT26 cells was determined by a MTS assay. In short, 3000 or 9000 CT26-cells were seeded and stimulated with the previous mentioned conditions. MTS substrate (CellTiter, G3580; Promega, Madison, WI) was added to the wells and the absorbance was measured at 490 nm using Cytation5.
Cell-Cycle Analysis
CT26 cells (250,000 or 500,000 cells/well) were stimulated with 20 μg/mL exosomes in non-CM. After 24 hours, cells were harvested, fixated with methanol,3 and stained with 10 μmol/L 4′,6-diamidino-2-phenylindole (D9542; Sigma-Aldrich, St Louis, MO) to analyze the percentage of cells in each phase of the cell cycle. A LSRII flow cytometer (BD Biosciences, San Diego, CA) was used for data acquisition. The 488-nm laser was used to generate forward scatter and side scatter signals. The 405-nm violet laser was used to generate 4′,6-diamidino-2-phenylindole fluorescence using a 450-/50-nm band pass filter. A 450-/50-pulse width vs a 450-/50-pulse area was used to select for single cells. Data were analyzed using WinList 8.0 (Verity Software House, Topsham, ME) to select for single cells and to generate a DNA histogram remotely linked to ModFit LT 4.1 (Verity Software House, Topsham, ME). A trapezoid S-phase model was used, providing a best fit with the data.
Organoid Models
Colonic organoids were generated form colonic crypts of wild-type C57BL/6J and C57BL/6-Tg(UBC-GFP)30Scha/J mice (both Jackson Laboratory) to study the effect of exosomes on epithelial cells.4 To confirm that MSC exosomes are taken up by colonic organoids, mechanically disrupted organoids were cultured with 60 μg PKH26-labeled exosomes for 1 week. The Leica (Wetzlar, Germany) SP8 microscope was used to capture both the GFP-positive organoids (488–509 nm) and PKH26-labeled exosomes (551–567 nm). To determine the effects of MSC exosomes on colonic organoids, organoids (5 wells) were cultured with either 60 μg exosomes in phosphate-buffered saline or in phosphate-buffered saline only, after induction of epithelial damage by mechanical disruption. Organoids were processed for paraffin embedding or messenger RNA isolation. For proliferation assays, MTS substrate was added to the wells with organoids, with or without exosomes after.
In Vivo Colitis Model
Experimental colitis was induced in female C57BL/6Jico mice by adding 2% DSS to the drinking water for 7 days. Mice were treated endoscopically at day 5, using a colonoscope system (Karl Storz, Tuttlingen, Germany), as described previously,5 with MSCs (2 × 106 cells), MSC exosomes (20 μg), or 200 μL MSC CM containing approximately 1.2 μg/mL exosomes (n = 7–19 mice/group). The control mice received local injections with 200 μL phosphate-buffered saline. On the day of treatment, the murine endoscopic index of colitis severity6 was scored. Five days after treatment, endoscopy and the murine endoscopic index of colitis severity scoring were repeated and mice were euthanized. The colon length and macroscopic disease score7 were determined. The experiment was performed twice and, except for the murine endoscopic index of colitis severity during treatment, all parameters were scored blinded to treatment groups. To evaluate colonic epithelial damage, the percentage of distal colon covered by pan-cytokeratin–positive cells (mouse anti–pan-cytokeratin, clone PCK-26, C5992; Sigma-Aldrich1) was scored blinded to treatment groups.
Statistical Analysis
Data are presented as means ± SD, except for data in Figure 2A, which are presented as means ± SEM. Unpaired Student t tests were used to compare the 2 groups. Differences between more than 2 groups were measured using 1-way analysis of variance or Kruskal–Wallis tests followed by multiple comparison tests. All analyses were performed using GraphPad Prism software (San Diego, CA). P values of .05 or less were considered statistically significant. All authors had access to the study data and have reviewed and approved the final manuscript.
References
- 1.Panes J. Lancet. 2016;388:1281–1290. doi: 10.1016/S0140-6736(16)31203-X. [DOI] [PubMed] [Google Scholar]
- 2.Molendijk I. Gastroenterology. 2015;149:918–927 e6. doi: 10.1053/j.gastro.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Barnhoorn M. Inflamm Bowel Dis. 2018;24:1755–1767. doi: 10.1093/ibd/izy130. [DOI] [PubMed] [Google Scholar]
- 4.Phinney D.G. Gastroenterology. 1998;35:297–306. [Google Scholar]
- 5.Singer I.I. Gastroenterology. 1998;115:297–306. doi: 10.1016/s0016-5085(98)70196-9. [DOI] [PubMed] [Google Scholar]
- 6.Harting M.T. Stem Cell. 2018;36:79–90. doi: 10.1002/stem.2730. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson S.W. Sci Rep. 2018;8:1419. doi: 10.1038/s41598-018-19581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Molendijk I. J Crohns Colitis. 2016;10:953–964. doi: 10.1093/ecco-jcc/jjw047. [DOI] [PubMed] [Google Scholar]
- 2.Molendijk I. Gastroenterology. 2015;149:918–927 e6. doi: 10.1053/j.gastro.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 3.van Haaften C. J Exp Clin Cancer Res. 2015;34:38. doi: 10.1186/s13046-015-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 5.Barnhoorn M. Inflamm Bowel Dis. 2018;24:1755–1767. doi: 10.1093/ibd/izy130. [DOI] [PubMed] [Google Scholar]
- 6.Becker C. Gut. 2005;54:950–954. doi: 10.1136/gut.2004.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper H.S. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 8.Hawinkels L.J. Oncogene. 2014;33:97–107. doi: 10.1038/onc.2012.536. [DOI] [PubMed] [Google Scholar]