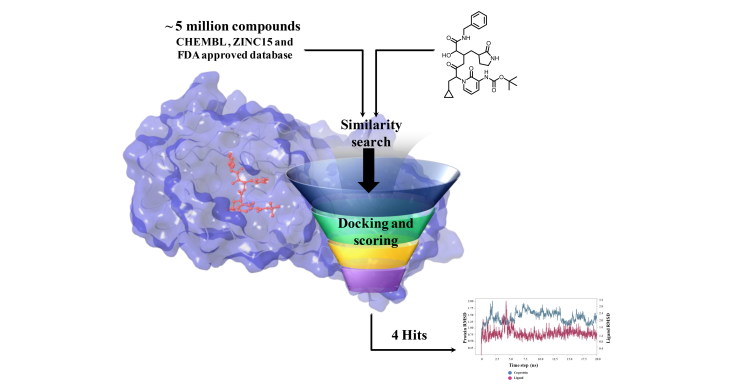
Abstract
Coronavirus disease strain (SARS-CoV-2) was discovered in 2019, and it is spreading very fast around the world causing the disease Covid-19. Currently, more than 1.6 million individuals are infected, and several thousand are dead across the globe because of Covid-19. Here, we utilized the in-silico approaches to identify possible protease inhibitors against SARS-CoV-2. Potential compounds were screened from the CHEMBL database, ZINC database, FDA approved drugs and molecules under clinical trials. Our study is based on 6Y2F and 6W63 co-crystallized structures available in the protein data bank (PDB). Seven hundred compounds from ZINC/CHEMBL databases and fourteen hundred compounds from drug-bank were selected based on positive interactions with the reported binding site. All the selected compounds were subjected to standard-precision (SP) and extra-precision (XP) mode of docking. Generated docked poses were carefully visualized for known interactions within the binding site. Molecular mechanics-generalized born surface area (MM-GBSA) calculations were performed to screen the best compounds based on docking scores and binding energy values. Molecular dynamics (MD) simulations were carried out on four selected compounds from the CHEMBL database to validate the stability and interactions. MD simulations were also performed on the PDB structure 6YF2F to understand the differences between screened molecules and co-crystallized ligand. We screened 300 potential compounds from various databases, and 66 potential compounds from FDA approved drugs. Cobicistat, ritonavir, lopinavir, and darunavir are in the top screened molecules from FDA approved drugs. The screened drugs and molecules may be helpful in fighting with SARS-CoV-2 after further studies.
Keywords: Covid-19, MD Simulations, Virtual Screening, Drug Repurposing
Graphical Abstract
Communicated by Ramaswamy H. Sarma
Introduction
The entire world in late December 2019 witnessed a sudden outbreak of an emerging disease named coronavirus (Covid-19) that started in Wuhan city of Hubei province of China and then got spread worldwide rapidly (Chen et al., 2020, Kupferschmidt & Cohen, 2020, Lai et al., 2020). The epidemic of Covid-19 was declared a pandemic on 12th March 2020 by the world health organization (WHO) (Zhang et al., 2020). Coronaviruses are a large family of viruses causing mild to severe cold such as severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) (Cascella et al., 2020). Many coronaviruses are zoonotic, i.e. they get transmitted from animals to humans. However, the SARS coronavirus is believed to be an animal-oriented virus (Cascella et al., 2020). In 2002, the first SARS infection in a human was reported in Guangdong province of South China. MERS coronavirus was transferred to humans from a camel in Saudi Arabia in 2012 (Omrani et al., 2015).
The SARS-CoV-2 is a novel virus that is spherical and has mushroom-shaped proteins termed as spikes that give this virus the shape of a crown (Vankadari & Wilce, 2020). The SARS-CoV-2 virus spreads primarily through droplets, saliva, or discharges from the nose of an infected person after sneezing or coughing. Worldwide more than 1.6 million cases have been registered, and several thousand people lost their lives due to Covid-19 (Worldometer 10/04/2020). Important to note that approximately 370,000 people recovered from this disease to date (Worldometer 10/04/2020). At this time, there are no specific vaccines or treatments available for this disease. Many clinical trials and efforts are being made to provide potential therapies (ECT Register, 2020). Any traditional drug discovery program is a costly and time-consuming process that sometimes takes decades to complete (Romano & Tatonetti, 2019). Thus, we cannot afford to wait for the discovery of medicines for Covid-19 from traditional drug discovery. In recent times computational drug discovery program has achieved popularity and success due to its ability to predict potent molecules before their synthesis (Gahtori et al., 2019). Data mining, machine learning, high-level quantum mechanical (QM), quantum-mechanical/molecular-mechanical (QM/MM), and quantitative structure-activity relationship (QSAR) techniques are useful to accelerate the drug discovery program (Lin et al., 2008, Murty et al., 2008, Srivastava & Sastry, 2012).
One of the best-characterized targets for SARS-CoV-2 is its main protease, and various groups have reported the crystal structure with or without the inhibitor (Zhang et al., 2020). Many efforts were made to provide molecular insight into SARS-CoV-2 protein and ligand interactions, which will help to design newer drugs (Ziebuhr et al., 2000). The protease was widely exploited as a therapeutic option for viral diseases like human immunodeficiency virus (HIV) (Xu et al., 2020). Currently, darunavir, lopinavir, and ritonavir are being used as a supportive medication against SARS-CoV-2 (Wang et al., 2020). Sahu et al. performed molecular docking calculations to identify various protease inhibitors from a pool of 2827 molecules under clinical trials (Sahu et al., 2020).
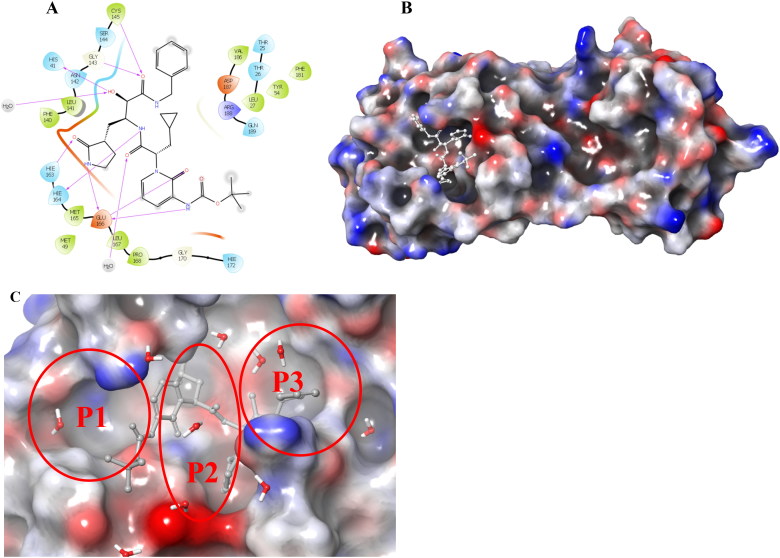
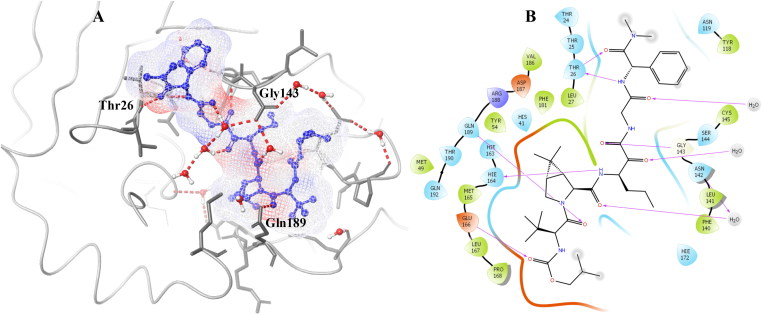
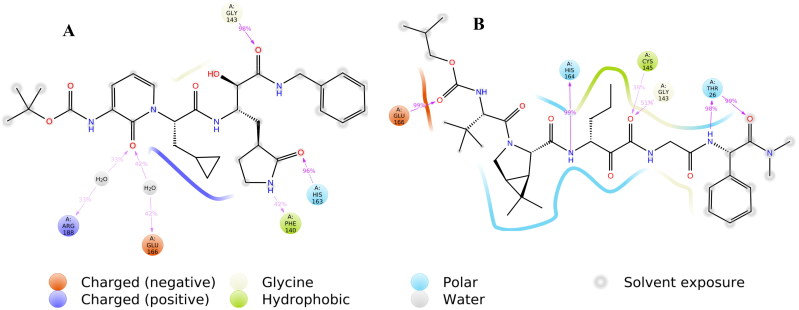
Junmei Wang utilized a computational approach to screen FDA approved drugs for SARS-CoV-2 main protease inhibitors (Wang, 2020). She also proposed a few compounds that could possibly be repurposed for the treatment of Covid-19. Fischer et al. utilized the computational methods to screen 687 million molecules to find inhibitors against SARS-CoV-2 main protease (Fischer et al., 2020). Zhang et al. published a crystal structure for SARS-CoV-2 main protease co-crystallized with a peptide-like inhibitor (Zhang et al., 2020). Peptides are known for their structural diversity with ease of synthesis and unique mode of action with limited off-target activity. Peptide-like inhibitors are widely used to treat several diseases like cancer, diabetes, autoimmune diseases, etc. and have high success rates in commercial development (Beeley 1994). Structural analysis of the co-crystallized molecule suggests that compound binding to SARS-CoV-2 main protease hampers the function of the protease and could be a possible target for drug development. The binding site of SARS-CoV-2 main protease is a combination of hydrophobic, hydrophilic and charged residues holding with hydrogen bonds in excess, Figure 1. The main protease of SARS-CoV-2 is important for the maturation of viral particles, which makes it a potential target for antiviral drugs.
Figure 1.
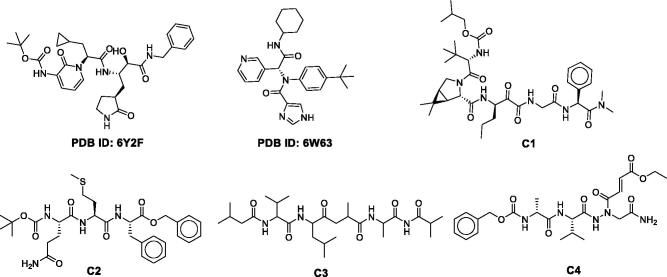
The chemical structure of the four best compounds and two co-crystallized inhibitors. C-1: CHEMBL206650; C-2: CHEMBL303543; C-3: CHEMBL127888; C-4: CHEMBL573507.
We utilized the reported co-crystallized structures (PDB IDs: 6Y2F and 6W63) and performed the similarity search on CHEMBL and ZINC databases to find potential inhibitors. We also screened the database of FDA approved drugs to repurpose a few available drugs as inhibitors against SARS-CoV-2 main protease (Lionta et al., 2014). The resulting compounds were used in the virtual screening workflow, followed by free energy calculations (Srivastava & Sastry, 2013). All the top selected compounds were rescored using MM-GBSA free energy calculations. Interactions and binding strength of four best molecules were validated by molecular dynamics (MD) simulations, Figure 1. Finally, we screened 300 potential molecules from databases, and 66 molecules from FDA approved drugs. These molecules may be tested for their applicability against SARS-CoV-2.
Materials and method
All the calculations were carried out using the Schrodinger 2018-4 package (SCHRODINGER 2018). SARS-CoV-2 main protease protein structure, along with peptide-like inhibitor (PDB ID: 6Y2F) (Zhang et al., 2020) and small-molecule inhibitor (PDB ID: 6W63) (Mesecar, 2020), was retrieved from the PDB (Berman et al., 2000). Until date, 83 structures are available in PDB related to SARS-Cov-2 main protease. The majority of these structures are bound with a small fragment and are suitable for the fragment-based drug discovery approach. Based on available information on bound inhibitors, missing residues, and resolution of the structure, we have selected 6Y2F and 6W63 for our studies.
Protein and ligand preparation
Protein was prepared in protein preparation wizard, hydrogen atoms were added, water molecules beyond 5 Å of the binding site were removed. Sidechains and loops were built using the prime module. All the collected ligand from CHEMBL (Gaulton et al., 2012) Zinc (Sterling & Irwin, 2015) and drug-bank (Wishart et al., 2008)were prepared using the pH value of 7.4.
Docking and free energy calculations
The grid of 20 Å was generated over the co-crystallized peptide-like inhibitor (PDB ID: 6Y2F) and small-molecule inhibitor (PDB ID: 6W63). Re-docking of the co-crystallized compounds was performed to validate the docking protocols. The docked complexes were superimposed to the original crystal structure to calculate the root mean square deviation (RMSD). The re-docking of peptide-like structure and small-molecule inhibitor reproduces the original pose with 1.23 Å and 0.75 Å RMSD, respectively. Lower RMSD indicates that our docking methodology is adequate and can be utilized to search small molecule inhibitors.
Docking calculations were carried out in three different modes, virtual screening followed by standard-precision (SP) and extra-precision (XP) docking using the Glide program. Only top survived compounds obtained from virtual screening and SP docking were subjected to XP docking. After XP docking, compounds were re-scored using prime MM-GBSA free energy calculations (Schrödinger, 2020).
Molecular dynamics simulations
MD simulations for protein-ligand complexes were performed using the Desmond package (Desmond Research, 2020). The OPLS3e force field was used to model the protein interactions, and the SPC mode was used for water molecules. Long-range electrostatic interactions were calculated using the Particle-mesh Ewald (PME) method with a grid spacing of 0.8 Å. Nose-Hoover thermostatic was used for maintaining the constant temperature and the Martina-Tobias-Klein method was used for the constant pressure. The equations of motion were integrated using the multistep RESPA integrator with an inner time step of 2.0 fs for bonded and non-bonded interactions within the short-range cutoff. An outer time step of 6.0 fs was used for non-bonded interactions beyond the cutoff. Periodic boundary conditions (PBC) were applied. After minimization, all the complexes were subjected to the production run for 20 ns in the NPT ensemble.
Results and discussion
Docking
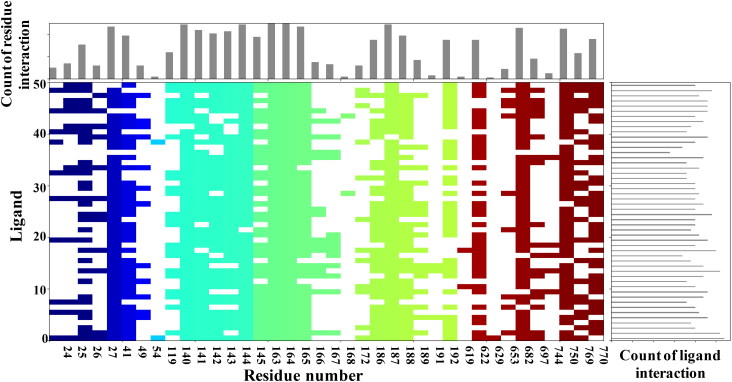
SARS-CoV-2 main protease ligand-binding pocket could be divided into three sub pockets (P1, P2, and P3), Figure 2. The interaction map and surface diagram of SARS-Cov-2 are depicted in Figure 2. Four thousand compounds were screened based on the similarity search on CHEMBL and ZINC databases, which were docked inside the binding site. Table 1 contains the docking scores of the top fifty selected compounds. Names of the top 300 compounds, along with their scores, are given in Table S1. The binding site for SARS-CoV-2 is surrounded with hydrophilic as well as hydrophobic residues with two negatively charged (Glu-166 and Asp-187) and one positively charged (Arg-188) residues, Figure 2. Gln-192, Glu-166, Hie-163 and Hie-41 residues with water surrounding molecules form a bridge between ligand and protein. This observation correlates with the interactions available in the original PDB (PDB ID: 6Y2F) structure. Besides those residues, Gln-192, and Thr-26 residues also interact with the ligands. An interaction map of the top 50 selected compounds is depicted in Figure 3.
Figure 2.
A. 2 D interaction map of the co-crystallized ligand of SARS-CoV-2 main protease. B. Surface diagram of SARS-CoV-2 main protease (ligand is shown in white stick). C. Three pockets of SARS-CoV-2 main protease (P1, P2, and P3).
Table 1.
Docking score, glide emodel score and MM-GBSA ΔG-bind values of top fifty compounds.
| IDs | Score | emodel | ΔG-bind | IDs | Score | emodel | ΔG-bind |
|---|---|---|---|---|---|---|---|
| 6Y2F# | −12.20 | −152.88 | −84.48 | ZINC42929983 | −7.97 | −82.35 | −76.11 |
| C-1 | −11.02 | −120.87 | −104.89 | ZINC03243503 | −7.91 | −76.36 | −66.77 |
| C-2 | −9.58 | −111.13 | −89.08 | ZINC09006550 | −7.90 | −83.78 | −50.77 |
| ZINC51316638 | −9.55 | −73.25 | −53.57 | CHEMBL2396925 | −7.89 | −95.00 | −64.06 |
| C-3 | −9.28 | −98.78 | −77.63 | ZINC09934205 | −7.82 | −93.45 | −67.24 |
| ZINC51316553 | −9.24 | −79.28 | −61.63 | ZINC11166391 | −7.82 | −93.09 | −69.80 |
| ZINC05225437 | −9.16 | −93.83 | −83.46 | ZINC23566409 | −7.82 | −84.34 | 82.08 |
| ZINC09601385 | −9.10 | −93.22 | −74.55 | ZINC09343712 | −7.80 | −78.32 | −70.81 |
| CHEMBL152104 | −9.09 | −73.20 | −78.03 | ZINC11909002 | −7.75 | −94.64 | −73.56 |
| ZINC09797971 | −8.89 | −78.47 | −64.31 | ZINC15532605 | −7.74 | −79.87 | −73.83 |
| ZINC01031116 | −8.84 | −70.63 | −57.98 | ZINC71759840 | −7.74 | −76.87 | −81.02 |
| CHEMBL2316587 | −8.72 | −93.67 | −93.55 | ZINC13127908 | −7.72 | −97.78 | −62.45 |
| CHEMBL2371798 | −8.72 | −88.83 | −85.65 | ZINC10912500 | −7.69 | −91.05 | −84.75 |
| C-4 | −8.72 | −110.18 | −84.79 | ZINC26538087 | −7.69 | −89.68 | −66.28 |
| ZINC46569546 | −8.71 | −84.44 | −67.01 | ZINC09934141 | −7.68 | −76.72 | −77.00 |
| ZINC00702508 | −8.66 | −87.40 | −78.12 | ZINC03192567 | −7.67 | −89.71 | −70.62 |
| CHEMBL207579 | −8.56 | −111.34 | −76.61 | ZINC09934209 | −7.66 | −105.46 | −111.29 |
| CHEMBL200490 | −8.51 | −108.67 | −75.82 | ZINC97058430 | −7.66 | −81.69 | −65.30 |
| CHEMBL422440 | −8.40 | −103.04 | −77.02 | ZINC09934209 | −7.66 | −105.46 | −111.29 |
| ZINC08926270 | −8.21 | −70.64 | −85.44 | ZINC46087259 | −7.60 | −86.97 | −76.60 |
| ZINC01108942 | −8.20 | −83.66 | −61.22 | ZINC46087259 | −7.60 | −86.97 | −76.60 |
| ZINC07238596 | −8.16 | −90.99 | −71.00 | ZINC46087170 | −7.58 | −85.30 | −70.51 |
| CHEMBL40589 | −8.09 | −94.85 | −73.47 | ZINC46087170 | −7.58 | −85.30 | −70.51 |
| ZINC12053378 | −8.05 | −88.25 | −81.30 | ZINC14732819 | −7.57 | −75.20 | −65.42 |
| ZINC29975718 | −8.02 | −78.69 | −56.88 | ZINC14732819 | −7.57 | −75.20 | −65.42 |
Co-crystallized ligand of PDB ID 6Y2F.
CHEMBL shows compounds were selected from CHEMBL.
ZINC shows compounds were chosen from ZINC15 database.
IDs are compound IDs from databases.
The score is the glide docking score.
emodel is the glide emodel score.
ΔG-bind is MM-GBSA based binding free energy.
Figure 3.
Interaction fingerprint for the top fifty selected compounds. In the interaction histogram, each contact of a particular residue with the ligand is indicated by a color. Residue number is given below the color.
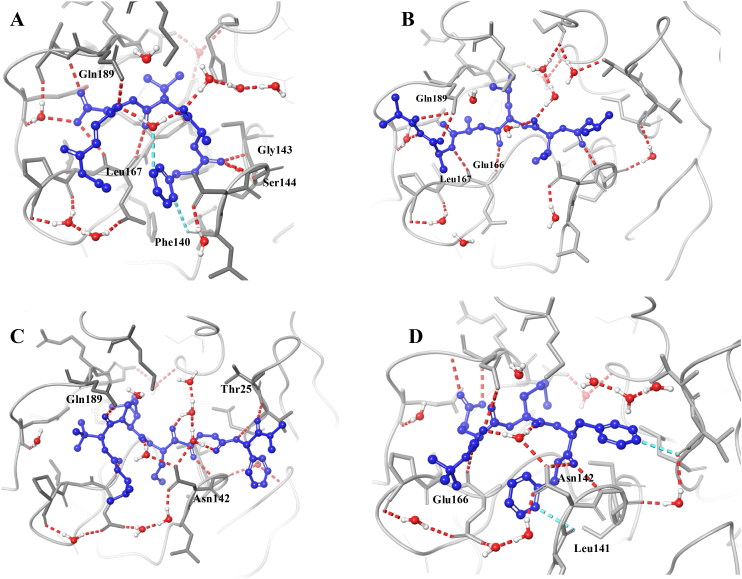
C-1 shows the comparable docking score of -11.02 and able to make hydrogen bond interactions with Hie-164, Glu-166, Gln-189, Gly-143, Thr-26 and water bridges, Figure 4. C-2, C-3, and C-4 also have a good docking score of -9.58, −9.28, −8.72, respectively Figure 5. As the docking scores were not able to distinguish between the molecules, we utilized glide emodel, and MM-GBSA based binding free energy (ΔG-bind) values for selecting the best complexes for MD simulations.
Figure 4.
2 D and 3 D interaction map of C-1 with interacting residue names.
Figure 5.
Binding modes of four selected complexes. C-1 (A), C-2 (B), C-3 (C) and C-4 (D).
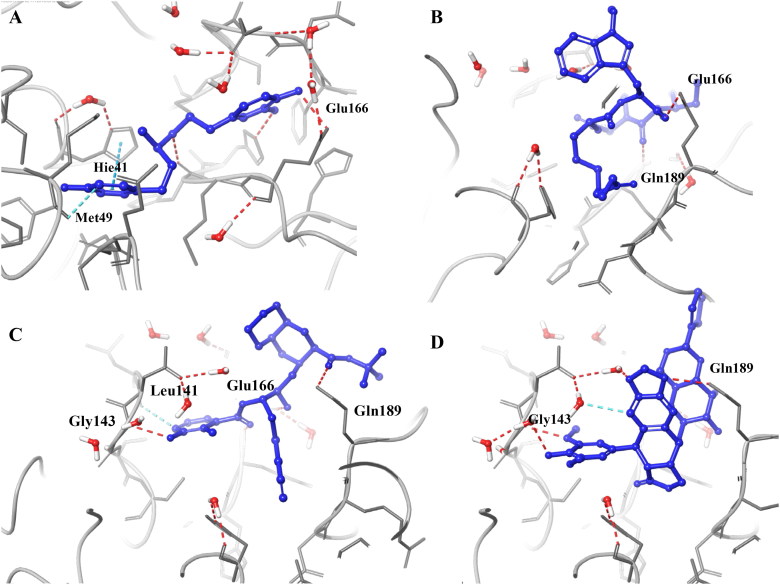
FDA approved drugs were docked inside the SARS-CoV-2 main protease (PDB ID: 6W63). Interestingly, HIV protease inhibitors (ritonavir, lopinavir, and darunavir) were among the top 20 compounds with a docking score of -8.878, −8.358 and -7.208, respectively. These three inhibitors are being tested against SARS-CoV-2 (Harrison, 2020). Apart from these three, a few antivirals (nelfinavir, saquinavir) and some others namely dobutamine, carfilzomib, teniposide (Gordaliza et al., 1994), and apicidine (Gordon et al., 2020) were also scored well and made positive interactions with important residues, Figure 6. Docking scores and MM-GBSA ΔG-bind values for selected FDA approved drugs are given in Table 2 and Table S2. Nevertheless, some compounds like apicidine were reported to inhibit SARS-CoV-2 main protease, and further research is required to use this against Covid-19 disease.
Figure 6.
Docking poses of a few selected FDA approved drugs. Dobutamine (A); Apicidin (B); Nelfinavir (C); Teniposide (D).
Table 2.
Docking score, glide emodel score and MM-GBSA ΔG-bind values for the top 50 FDA approved and under clinical trialdrugs.
| Name | Score | emodel | ΔG-bind | Name | Score | emodel | ΔG-bind |
|---|---|---|---|---|---|---|---|
| 6W63# | −5.987 | −58.637 | −52.57 | Scopolamine | −7.42 | −49.98 | −41.05 |
| Lactulose | −10.10 | −57.79 | −67.92 | Gluconolactone | −7.41 | −36.27 | −27.69 |
| Oxytocin | −9.79 | −86.17 | −69.80 | Bortezomib | −7.38 | −71.28 | −60.70 |
| Boceprevir | −9.12 | −74.76 | −86.03 | Indinavir* | −7.35 | −78.14 | −83.27 |
| Saquinavir | −8.64 | −85.66 | −51.98 | Latanoprost | −7.35 | −72.68 | −64.93 |
| Adenosine | −8.47 | −54.09 | −59.15 | Teniposide | −7.30 | −50.85 | −79.63 |
| Masoprocol | −8.45 | −58.19 | −58.79 | Bicalutamide | −7.30 | −70.90 | −48.75 |
| Doxorubicin | −8.40 | −63.70 | −93.50 | Apremilast | −7.28 | −68.76 | −65.67 |
| Cromolyn sodium | −8.37 | −83.16 | −31.02 | Dobutamine | −7.28 | −67.44 | −79.11 |
| Lopinavir## | −8.36 | −86.41 | −73.33 | Benazepril | −7.25 | −63.55 | −53.44 |
| Dibucaine | −8.32 | −69.53 | −62.22 | Fluorometholone | −7.25 | −35.56 | −38.25 |
| Ritonavir## | −8.30 | −94.60 | −87.24 | Pravastatin sodium | −7.25 | −55.43 | −53.75 |
| Regadenoson | −8.23 | −77.58 | −69.04 | Darunavir## | −7.21 | −69.20 | −64.49 |
| Cladribine | −8.22 | −55.55 | −76.16 | Carfilzomib* | −7.20 | −85.26 | −79.74 |
| Daunorubicin* | −8.06 | −59.07 | −85.03 | Tropicamide | −7.16 | −58.45 | −74.11 |
| Albuterolsulfate | −7.97 | −55.62 | −65.78 | Elvitegravir* | −7.12 | −62.38 | −42.73 |
| Dapagliflozin | −7.94 | −58.45 | −67.69 | Azacitidine | −7.09 | −48.97 | −40.10 |
| Pravastatin sodium | −7.85 | −54.98 | −41.37 | Cangrelor | −7.00 | −80.23 | −16.84 |
| Pemetrexed | −7.72 | −78.87 | −46.25 | Telaprevir | −6.99 | −86.37 | −92.74 |
| Protirelin | −7.71 | −77.10 | −67.27 | Deferoxamine | −6.94 | −61.40 | −85.34 |
| Mupirocin | −7.65 | −57.45 | −62.64 | Ganciclovir | −6.89 | −51.13 | −46.66 |
| Entagastrin | −7.64 | −82.78 | −59.15 | Cangrelor | −6.89 | −86.11 | −34.33 |
| Nadolol | −7.64 | −51.19 | −67.71 | Pentagastrin | −6.88 | −100.37 | −79.88 |
| Dinoprost | −7.59 | −49.54 | −46.72 | Estradiol | −6.85 | −48.53 | −60.13 |
| Nelfinavir* | −7.44 | −73.97 | −68.63 | Apicidine** | −6.39 | −68.28 | −55.56 |
Co-crystallized ligand of PDB ID 6W63
Compound currently in supportive therapy of SARS-CoV-2 infection.
Compound reported for antiviral activity.
Compound reported inhibiting SARS-CoV-2 main protease in-vitro.
The score is glide-docking score.
emodel is glide emodel score.
ΔG-bind is MM-GBSA binding free energy.
The prime MM-GBSA is widely accepted for re-scoring the docked complexes. All the selected complexes, after XP docking, were subjected to prime MM-GBSA calculations. MM-GBSA ΔG-bind scores for all the selected compounds are given in Tables 1 and 2. The negative values of ΔG-bind indicate that the selected compounds favorably interact with the receptor. The ligand binding energies for all the screened compounds are in the range of -50.77 kcal/mol to -111.29 kcal/mol. The binding energy for the co-crystallized inhibitor with SARS-CoV-2 main protease was -84.48 kcal/mol.
Interestingly, the binding energies for four selected compounds are -104.89, −89.08, −77.63 and -84.79 kcal/mol respectively for C-1, C-2, C-3, and C-4. Among the top hits from similarity search and molecular docking calculations, C-1 shows the lowest binding energy (-104.89 kcal/mol), which is considerably higher than the co-crystallized inhibitor -89.48 kcal/mol. Moreover, glide emodel scores correlate well with the MM-GBSA ΔG-bind values. These findings strongly suggest that the selected compounds may inhibit the SARS-CoV-2 main protease. Docking scores of all the compounds were incorporated in Tables S1 and S2.
Molecular dynamics simulations
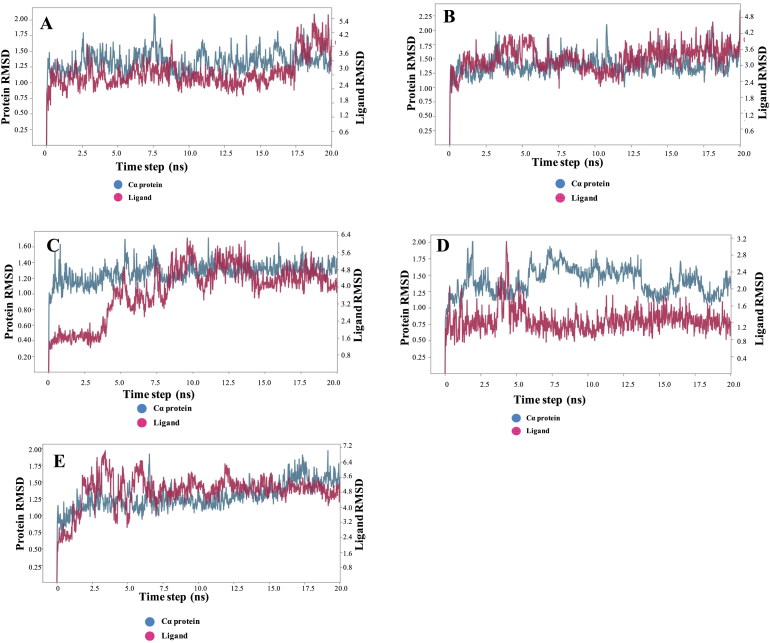
Based on the docking results, re-scoring by MM-GBSA and visual inspection, four compounds, and one co-crystallized ligand was selected for MD simulations. The backbone RMSD of the protein-ligand complex (PDB ID: 6Y2F) increased gradually until the 2.09 Å then gets stable till 20 ns, Figure 7. Low RMSD during the simulation indicates the stable complex formation. C-1 shows excellent stability as this complex is equilibrated at 1.5 ns and remains stable throughout the simulation with the least conformational changes, Figure 7. All the selected compounds remain stable throughout the simulation with the change in backbone RMSD within the acceptable range. As suggested via protein backbone RMSD, ligand RMSD was also found stable throughout the simulation with minimal fluctuation.
Figure 7.
RMSD of protein and ligand backbones during the simulation [A. SARS-CoV-2 main protease (PDB ID: 6Y2F); B. C-1; C. C-2; D. C-3; E. C-4].
The stable conformation obtained from MD simulations could be further utilized for screening large chemical libraries. C-1, C-2 and C-4 complex structures showed a slight deviation, which indicates that these ligands are much active inside the binding pocket and hydrogen bond and other interactions are durable and hold inside the SARS-CoV-2 main protease. The ligand RMSD for co-crystallized ligand fluctuates from 1.48 Å to 4.49 Å till 17 ns and then comes to 5.03 Å and stabilized till 20 ns. C-1 fluctuates at 4.5 ns and then is stable throughout the simulation. Figure 7 provides a detailed insight of the RMSD of all the selected complexes.
Interaction H-bond and interaction stability analysis
To understand the stability of predicted protein-ligand complexes, we analyzed the hydrogen bond formation during the 20 ns simulation. 6Y2F structure shows three direct hydrogen bonds and two hydrogen bond bridges between protein water ligand. Two of them were maintained throughout the simulation. C-1 shows the maximum interactions throughout the simulation as four out of six hydrogen bonds were intact throughout the simulation, Figure 8. The other three complexes also maintain the hydrogen bond contacts during the simulation, Figure S1. This suggests that hydrogen bonding plays a significant role in accommodating the ligand inside the binding site.
Figure 8.
Hydrogen bond interaction maintained throughout the MD simulation A. Peptide like inhibitor (6Y2F) B. C-1.
Further studies are necessary on the application of data mining techniques like the expert system to classify the peptide-like inhibitors, cluster analysis by self-organizing map (SOM) to make a group of inhibitors, and to understand similarity and dissimilarity should (Yan et al., 2013, Rein et al., 2019) (Zhavoronkov et al., 2020). Molecules screened from FDA approved drugs may undergo directly for clinical trials against Covid-19 disease. Molecules obtained from databases, need to go for pharmacological, toxicological, and preclinical studies before clinical trials.
Conclusions
We utilized the recently published co-crystallized structures of SARS-CoV-2 main protease (6Y2F and 6W63). Peptide-like molecules provide a basic pharmacophore for the design of SARS-CoV-2 main protease inhibitors. The amide linkage backbone gives them the flexibility to fit comfortably inside the binding site. Peptides may be an excellent alternative for small molecules, as they are easy to synthesize and less toxic when compared to small molecules. This study provides a detailed analysis of essential residues and ligand-receptor interactions for the development of peptide-like structures as SARS-CoV-2 main protease inhibitors. We screened 300 peptide-like structures from various databases. Interactions of four most potent peptide-like structures were further validated utilizing MD simulations. The chosen compounds showed strong binding affinities with residues inside the binding site and formed the strong H-bonding and salt bridge with Gln-192, Glu-166, Hie-166, and Hie-41 residues. Docking analysis suggests that most of the compounds which are in the top 50 are hydrophilic and tend to form hydrogen bonds. Drug re-purposing techniques are widely being explored to overcome the current outbreak of SARS-Cov-2. A few compounds were identified, and some of them are currently under clinical trials. We have identified 66 FDA approved drugs utilizing drug re-purposing approaches. Interestingly, a few of the selected drugs from this work match with the drugs under clinical trials (lopinavir, ritonavir, darunavir) against Covid-19 disease. Further experimental studies are necessary for validating our findings.
Supplementary Material
Acknowledgements
H.K.S is grateful to the Science and Engineering Research Board, Department of Science and Technology, New Delhi, for financial assistance through the Ramanujan (SB/S2/RJN/004-2015) grant. We thank NIPER Guwahati and NIPER Kolkata for necessary facilities. We thank Department of Chemistry, IIT Guwahati and Param-ishan for the support. We also acknowledge the useful discussion with Professor C. V. Sastri (IIT Guwahati).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., & Bourne P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S. C., & Napoli R. D. (2020). Features, evaluation and treatment Coronavirus (COVID-19). StatPearls. [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., & Zhang L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond Molecular Dynamics System, D. E. Shaw Research. (2020). Maestro-Desmond interoperability tools. New York, NY: Schrödinger. [Google Scholar]
- ECT Register (2020, April 10). https://www.clinicaltrialsregister.eu/ctr-search/search?query=covid-19.
- Fischer A., Sellner M., Neranjan S., Lill M. A., & Smieško M. (2020). Inhibitors for novel coronavirus protease identified by virtual screening of 687 million compounds. ChemRxiv. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahtori J., Pant, S., & Srivastava H. K. (2019). Modeling antimalarial and antihuman African trypanosomiasis compounds: A ligand- and structure-based approaches. Molecular Diversity. In Press. 10.1007/s11030-019-10015-y [DOI] [PubMed] [Google Scholar]
- Gaulton A., Bellis L. J., Bento A. P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., & Overington J. P. (2012). ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Research, 40(D1), D1100–1107. (Database issue): 10.1093/nar/gkr777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordaliza M., Castro M. A., Garcia-Gravalos M. D., Ruiz P., Miguel del Corral J. M., & San Feliciano A. (1994). Antineoplastic and antiviral activities of podophyllotoxin related lignans. Archiv Der Pharmazie, (Weinheim), 327(3), 175–179. 10.1002/ardp.19943270309 [DOI] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., O’Meara M. J., Guo J. Z., Swaney D. L., Tummino T. A., Hüttenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., … Krogan N. J. (2020). A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug repurposing. bioRxiv 2020.03.22.002386. [Google Scholar]
- Harrison C. (2020). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology, 38(4), 379–381. 10.1038/d41587-020-00003-1 [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K., & Cohen J. (2020). Will novel virus go pandemic or be contained? Science, 367(6478), 610–611. 10.1126/science.367.6478.610 [DOI] [PubMed] [Google Scholar]
- Lai C. C., Shih T. P., Ko W. C., Tang H. J., & Hsueh P. R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z. H., Long H. X., Bo Z., Wang Y. Q., & Wu Y. Z. (2008). New descriptors of amino acids and their application to peptide QSAR study. Peptides, 29(10), 1798–1805. 10.1016/j.peptides.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Lionta E., Spyrou G., Vassilatis D. K., & Cournia Z. (2014). Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Current Topics in Medicinal Chemistry, 14(16), 1923–1938. 10.2174/1568026614666140929124445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L. V., Watkins S. P., Carter L. J., Smoot J., Gregg A. C., Daniels A. D., Jervey S., & Albaiu D. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Science, 6(3), 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecar A. D. (2020). A taxonomically-driven approach to development of potent, broad-spectrum inhibitors of coronavirus main protease including SARS-CoV-2 (COVID-19). 10.2210/pdb6W63/pdb [DOI]
- Murty U. S., Srinivasa Rao M., & Misra S. (2008). Prioritization of malaria endemic zones using self-organizing maps in the Manipur state of India. Informatics for Health and Social Care, 33(3), 170–178. 10.1080/17538150802457687 [DOI] [PubMed] [Google Scholar]
- Omrani A. S., Al-Tawfiq J. A., & Memish Z. A. (2015). Middle East respiratory syndrome coronavirus (MERS-CoV): Animal to human interaction. Pathogens and Global Health, 109(8), 354–362. 10.1080/20477724.2015.1122852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein D., Ternes P., Demin R., Gierke J., Helgason T., & Schon C. (2019). Artificial intelligence identified peptides modulate inflammation in healthy adults. Food & Function, 10(9), 6030–6060. 10.1039/C9FO01398A [DOI] [PubMed] [Google Scholar]
- Romano J. D., & Tatonetti N. P. (2019). Informatics and computational methods in natural product drug discovery: A review and perspectives. Frontiers in Genetics, 10, 368 10.3389/fgene.2019.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu K., Noskov S., Tuszynski J., Houghton M., & Tyrrell D. L. (2020). Computational screening of molecules approved in phase-i clinical trials to identify 3CL protease inhibitors to treat COVID-19. Preprint 2020. 10.20944/preprints202004.0015.v2 [DOI]
- Schrodinger (2018). Maestro small-molecular drug discovery suite 2018-4. https://www.schrodinger.com/
- Schrödinger (2020). Schrödinger Release 2020-1: BioLuminate. New York, NY: https://www.schrodinger.com/ [Google Scholar]
- Srivastava H. K., & Sastry G. N. (2012). Molecular dynamics investigation on a series of HIV protease inhibitors: Assessing the performance of MM-PBSA and MM-GBSA approaches. Journal of Chemical Information and Modeling, 52(11), 3088–3098. 10.1021/ci300385h [DOI] [PubMed] [Google Scholar]
- Srivastava H. K., & Sastry G. N. (2013). Efficient estimation of MMGBSA-based BEs for DNA and aromatic furan amidino derivatives. Journal of Biomolecular Structure and Dynamics, 31(5), 522–537. 10.1080/07391102.2012.703071 [DOI] [PubMed] [Google Scholar]
- Sterling T., & Irwin J. J. (2015). ZINC 15–Ligand Discovery for Everyone. Journal of Chemical Information and Modeling, 55(11), 2324–2337. 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N., & Wilce J. A. (2020). Emerging WuHan (COVID-19) coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerging Microbes & Infections, 9(1), 601–604. 10.1080/22221751.2020.1739565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. (2020). Fast identification of possible drug treatment of Coronavirus disease—19 (COVID-19) through computational drug repurposing study. ChemRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., & Zhu W. (2020). Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv 2020.01.27.921627. [Google Scholar]
- Wang L., Wang Y., Ye D., & Liu D. (inpress, 2020). Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. International Journal of Antimicrobial Agents.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S., Knox C., Guo A. C., Cheng D., Shrivastava S., Tzur D., Gautam B., & Hassanali M. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Research, 36(suppl_1), D901–906. (Database issue): 10.1093/nar/gkm958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometer. (2020, April 10). https://www.worldometers.info/coronavirus/.
- Yan A., Nie X., Wang K., & Wang M. (2013). Classification of Aurora kinase inhibitors by self-organizing map (SOM) and support vector machine (SVM). European Journal of Medicinal Chemistry, 61, 73–83. 10.1016/j.ejmech.2012.06.037 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science, 368(6489), 409-412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A., Aladinskiy V., Zhebrak A., Zagribelnyy B., Terentiev V., Bezrukov D.S., Polykovskiy D., Shayakhmetov R., Filimonov A., Orekhov P., … Yan Y. (2020). Potential COVID-2019 3C-like protease inhibitors designed using generative deep learning approaches. ChemRxiv. [Google Scholar]
- Ziebuhr J., Snijder E. J., & Gorbalenya A. E. (2000). Virus-encoded proteinases and proteolytic processing in the Nidovirales. Journal of General Virology, 81(4), 853–879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.