Abstract
The new Corona-virus, recently called the severe acute respiratory syndrome Coronavirus (SARS-CoV-2) appears for the first time in China and more precisely in Wuhan (December 2019). This disease can be fatal. Seniors, and people with other medical conditions (diabetes, heart disease…), may be more vulnerable and become seriously ill. This is why research into drugs to treat this infection remains essential in several research laboratories. Natural herbal remedies have long been the main, if not the only, remedy in the oral tradition for treating illnesses. Modern medicine has known its success thanks to traditional medicine, the effectiveness of which derives from medicinal plants. The objective of this study is to determine if the components of natural origin have an anti-viral effect and which can prevent humans from infection by this coronavirus using the most reliable method is molecular docking, which used to find the interaction between studied molecules and the protein, in our case we based on the inhibitor of Coronavirus (nCoV-2019) main protease. The results of molecular docking showed that among 67 molecules of natural origin, three molecules (Crocin, Digitoxigenin, and β-Eudesmol) are proposed as inhibitors against the coronavirus based on the energy types of interaction between these molecules and studied protein. 
Communicated by Ramaswamy H. Sarma
Highlights
Determine natural compounds that can have an anti-viral effect and which can prevent humans from infection by this coronavirus;
Molecular docking to find interaction between the molecules studied and the receptor of COVID-19;
The synthesis of these molecules and the evaluation of their in vitro activity against SARS-Cov-2 could be interesting.
Keywords: Molecular docking, CoV-2019, natural herbal, crocin, digitoxigenin, β-eudesmol
Introduction
Coronaviruses are non-segmented positive-sense RNA viruses which are part of the family Coronaviridae distributed in humans (Richman et al., 2016), and the subfamily Orthocoronaviridae, there are four genera of coronaviruses; Betacoronavirus, Alpha coronavirus, Gamma coronavirus and Deltacorona virus (Schwartz & Graham, 2020).
In humans, especially common colds affecting children and adults, are called mild illnesses. However, there are two zoonotic coronaviruses which are cited: The Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) which can especially affect the respiratory system through serious infections. In addition, the latter have the same characteristics: production factors, nosocomial transmission, replication in the lower respiratory tract and viral immunopathology. MERS-CoV and SARS-CoV result from serious public health problems which ultimately lead to epidemics resulting in significant loss of life (Hui, 2017; Perlman, 2020; Hui & Zumla, 2019). When these two zoonotic coronaviruses infect pregnant women, they can lead to poor obstetric outcomes, including maternal morbidity and death. There is currently no specific vaccine or treatment approved for coronavirus infection (Hui & Zumla, 2019; Song et al., 2019).
In late December 2019, an outbreak of a new disease, the coronavirus (COVID-19; previously known as 2019-nCoV) (Wu et al., 2020; Huang et al., 2020), was reported in Wuhan, the capital from Hubei province and a large city of around 11 million people in the central region of the People’s Republic of China (Zhu, 2020), which subsequently affected 26 countries around the world. China immediately declared the epidemic to the World Health Organization (WHO) and also shared the sequence information with the international community after the discovery of the causative agent. WHO has done its part by coordinating the development of the diagnosis; issue guidelines for patient monitoring, sample collection and treatment; and provide up-to-date information on the epidemic (Munster et al., 2020). To date, the primary source of infection has been pneumonia patients infected with COVID-19. Transmission of respiratory droplets is the main route of transmission, which can also be transmitted by contact (G. O. of N. H. Committee & Office of State Administration of Traditional Chinese Medicine, 2020). The source of the virus and its ability to spread between people remains unknown, with an increasing number of cases showing signs of human-to-human transmission (Zhu, 2020; Chan et al., 2020).
Currently, there are still no effective treatments that target the coronavirus and the development of these treatments requires months and years, so we must be oriented towards treatments of natural origin based on aromatic and medicinal plants to have the compounds having the ability to inhibit COVID-19 (Chen & Nakamura, 2004). The appearance of SARS, pushes a category of researchers to find anti-coronavirus agents, including certain natural compounds that exist in Morocco herbal medicines.
In our study we made a selection of plants based on two principles: the first one is oral efficacy, this means that the majority of Moroccan plants should be absorbable by the oral route, the second one is the compatibility of traditional use, then we have used molecular docking study to selected potential compounds that could have an anti-coronavirus effect by fighting their energy and type of interactions in studied enzyme.
2. Materials and methods
2.1. Data set
For this study we have selected 67 compounds extracted from different aromatic and medicinal plants, Table 1 shows the origin for each studied compound and the percentage present in each plant. These molecules were considered to molecular docking study.
Table 1.
Chemical Composition and Percentages of several plants.
| N° Compound | Name | Plant | % |
|---|---|---|---|
| 1 | Estragol | Foeniculum Vulgare | 5.29 |
| 2 | β-Phellandren | Nerium Oleander | 4.84 |
| 3 | β-Eudesmol | Lauris Nobilis L | 2.39 |
| 4 | Amorphan | Nerium Oleander | 8.11 |
| 5 | α-Terpinyl acetate | Lauris Nobilis L | 10.49 |
| 6 | α-Terpeneol | Myrtus Communis L Lauris Nobilis L |
3.84 9.68 |
| 7 | Myrtenyl Acetat | Myrtus Communis L Lavandula Stoechas |
25.05 11.54 |
| 8 | β-Thujone | Sauge Officinal | 3-8.5 |
| 9 | Camphre | Sauge Officinal | 4.5-24.5 |
| 10 | α-Thujone | Sauge Officinal | 18-43 |
| 11 | Isoamyl-2-methyl butyrate | Ammi Visnaga L | 27.68 |
| 12 | Cis- linalool oxide | Ammi Visnaga L | 2.14 |
| 13 | α-Terpinene | Ammi Visnaga L | 3.97 |
| 14 | Amyl valerate | Ammi Visnaga L | 9.98 |
| 15 | Amyl isobutyrate | Ammi Visnaga L | 16.04 |
| 16 | Allyl-methyl disulfide | Allium Sativum | 1.71 |
|
17 |
Limonene |
Thymelea Tartonraira Artemisia Vulgaris Foeniculum Vulgare Nerium Oleander |
2.96 2.58 8.14 5.01 |
| 18 | Diallyl sulfide | Allium Sativum | 0.66 |
| 19 | Diallyl disulfide | Allium Sativum | 14.30 |
| 20 | Diallyl trisulfide | Allium Sativum | 46.52 |
| 21 | Tricyclene | Thymelea Tartonraira | 7.11 |
|
22 |
β-Carophyllene |
Thymelea Tartonraira Eugenia Caryophyllus |
3.1 5-14 |
|
23 |
D-Camphor |
Thymelea Tartonraira Lavandula Stoechas Artemisia Vulgaris |
37.92 3.47 16.72 |
| 24 | Camphene | Thymelea Tartonraira Artemisia Vulgaris |
14.66 8.35 |
| 25 | Borneol | Thymelea Tartonraira Artemisia Vulgaris |
2.45 19.65 |
|
26 |
α-Pinene |
Thymelea Tartonraira Myrtus Communis L Nerium Oleander |
10.98 10 5.54 |
|
27 |
1-8 Cineol |
Thymelea Tartonraira Myrtus Communis L Nerium Oleander Artemisia Vulgaris |
8.39 43.03 6.58 3.23 |
|
28 |
Eugenol |
Eugenia Caryophyllus Lauris Nobilis L |
80 2.15 |
| 29 | Acetyleugenol | Eugenia Caryophyllus | 6.6 |
|
30 |
Terpine-4-ol | Nerium Oleander Lauris Nobilis L |
3.98 2.39 |
|
31 |
Sabinene | Nerium Oleander Lauris Nobilis L |
3.22 3.15 |
| 32 | Pulegone | Mentha Rotundifolia | 85.47 |
| 33 | Methyl-Eugenol | Lauris Nobilis L | 4.10 |
| 34 | Linalool | Artemisia Vulgaris Lauris Nobilis L Ammi Visnaga L |
6.77 8.78 22.71 |
| 35 | Fenchone | Foeniculum Vulgare Lavandula Stoechas |
9.55 6.94 |
| 36 | Eucalyptol | Lauris Nobilis L | 30.52 |
| 37 | Trans-Cinnamylacetat | Cinnamomum Cassia | 4.69 |
| 38 | Trans-Cinnamaldehyde | Cinnamomum Cassia | 90.08 |
| 39 | Undecan | Allium Cepa L | 8.09 |
| 40 | Undecane-2-6-dimethyl | Allium Cepa L | 6.86 |
| 41 | 2-carboxylic acid,3- methyl thiophene | Allium Cepa L | 8.96 |
| 42 | Safranal | Crocus Sativus L | n.d |
| 43 | pi-Cymene | Thymus Broussoneti | 11 |
| 44 | Picrocrocin | Crocus Sativus L | n.d |
| 45 | Neriine | Nerium Oleander | 22.56 |
| 46 | ɣ -Terpinene | Thymus Broussoneti | 8.99 |
| 47 | Dodecan | Allium Cepa L | 28.69 |
| 48 | Digitoxigenine | Nerium Oleander | 11.25 |
| 49 | Crocin | Crocus Sativus L | N.D |
| 50 | Crocetin | Crocus Sativus L | N.D |
| 51 | Calarene | Nerium Oleander | 5.12 |
| 52 | 1,2,4-Trithiolane,3,5-dimethyl | Allium Cepa L | 5.82 |
| 53 | Trans-Anethole | Foeniculum Vulgare | 53.2 |
| 54 | Undecan,2methyl | Allium Cepa L | 3.59 |
| 55 | 6-Isopropenyl-4,8a-Dimethyl-1,2,3,5,6,7,8, 8a-Octahydronaphthalene-2,3-diol | Lavandula Stoechas | 4.56 |
| 56 | α-Terpinene | Myrtus CommunisL | 2.90 |
| 57 | β-Ocimene | Artemisia Vulgaris | 2.21 |
| 58 | Bicyclogermacrene | Artemisia Vulgaris | 3.18 |
| 59 | Acetat Bornyl | Lavandula Stoechas | 8.86 |
| 60 | Cubenol | Lavandula Stoechas | 2.55 |
| 61 | D-Fenchol | Lavandula Stoechas | 6.62 |
| 62 | Geranyl Acetat | Myrtus CommunisL | 2.85 |
| 63 | Germacrene D | Artemisia Vulgaris | 3.82 |
| 64 | 2-Methyl-9-(prop-1-en-3-ol-2-yl)Bicyclo[4.4.0] dec-2-en-4-ol | Lavandula Stoechas | 4.50 |
| 65 | Mycrene | Artemisia Vulgaris | 3.8 |
| 66 | Thymol | Thymus Broussoneti | 63.09 |
| 67 | Viridifloral | Lavandula Stoechas | 6.10 |
2.2. Molecular docking
Molecular docking analysis was used to study the binding affinity and the type of interactions between all compounds (67 molecules) and the target (Coronavirus (2019-nCoV) main protease).
The steps for preparing ligands and proteins for docking protocol were done in the Autodock 1.5.4 tools from MGL Tools package employing default settings (Morris et al., 1998), a grid box (x = −26,283, y = 12,599, z = 58,965 at 1 Angstrom spacing), the bioactive conformations were simulated employing Autodock vina (Trott & Olson, 2010). For autodock vina study, an extended PDB format, termed PDBQT, is used for cordonnante files, which includes atomic partial charges and atom types. Torsion angles were calculated to assign the flexible and non-bonded rotation of molecules. The results were subsequently analyzed using Discovery studio 2016 (Pilot, 2016) and PyMol (DeLano, 2002).
The crystal structure of Coronavirus(2019-nCoV) main protease (PDB entry code: 6lu7) was downloaded from the protein databank (http://www.rcsb.org), and its original ligand and water were eliminated, then all compounds from our data set were docked in the active site of the studied protein. The preparation of the PDB file was done using Discovery Studio 2016 (Pilot, 2016).
3. Results and discussion
Totally, we have docked 67 components, Table 2 shows the binding affinity of natural compounds toward main protease, and Table 3 mentioned the 11 top flavor agent docking result based on binding energy.
Table 2.
Flavor agents docking results.
| N° compound | Binding Energy(Kcal/mol) |
|---|---|
| 1 | −4.7 |
| 2 | −4.7 |
| 3 | −7.1 |
| 4 | −5.8 |
| 5 | −5.7 |
| 6 | −6.1 |
| 7 | −5.9 |
| 8 | −5.6 |
| 9 | −5.8 |
| 10 | −5.6 |
| 11 | −4.8 |
| 12 | −4.4 |
| 13 | −4.9 |
| 14 | −4.3 |
| 15 | −4.3 |
| 16 | −3.2 |
| 17 | −4.6 |
| 18 | −2.9 |
| 19 | −2.9 |
| 20 | −3.3 |
| 21 | −4.6 |
| 22 | −6.1 |
| 23 | −4.7 |
| 24 | −5 |
| 25 | −4.8 |
| 26 | −4.8 |
| 27 | −5.1 |
| 28 | −5.5 |
| 29 | −5.5 |
| 30 | −5 |
| 31 | −5.1 |
| 32 | −5.2 |
| 33 | −5.1 |
| 34 | −4.4 |
| 35 | −5.3 |
| 36 | −5.3 |
| 37 | −5.4 |
| 38 | −5.1 |
| 39 | −3.9 |
| 40 | −4.5 |
| 41 | −4.1 |
| 42 | −5.3 |
| 43 | −5 |
| 44 | −6.8 |
| 45 | −6.9 |
| 46 | −4.9 |
| 47 | −3.9 |
| 48 | −7.2 |
| 49 | −8.2 |
| 50 | −6.2 |
| 51 | −6.1 |
| 52 | −3.1 |
| 53 | −4.5 |
| 54 | −4 |
| 55 | −6.4 |
| 56 | −4.9 |
| 57 | −4.3 |
| 58 | −6.1 |
| 59 | −5.5 |
| 60 | −5.9 |
| 61 | −5 |
| 62 | −5.1 |
| 63 | −5.9 |
| 64 | −6.1 |
| 65 | −4.1 |
| 66 | −4.9 |
| 67 | −5.8 |
Table 3.
11Top compounds docking results.
| N° | Name | Structure | Binding Energy |
|---|---|---|---|
| 3 | β-Eudesmol | 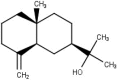 |
−7.1 |
| 6 | α-Terpeneol | 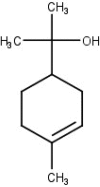 |
−6.1 |
| 22 | β-Carophyllene | 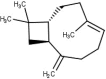 |
−6.1 |
| 44 | Picrocrocin |  |
−6.8 |
| 48 | Digitoxigenine |  |
−7.2 |
| 49 | Crocin |  |
−8.2 |
| 50 | Crocetin |  |
−6.2 |
| 51 | Calarene |  |
−6.1 |
| 55 | Bicyclogermacrene | 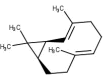 |
−6.1 |
| 58 | 6-Isopropenyl-4,8a-Dimethyl-1,2,3,5,6,7,8,8a-Octahydronaphthalene-2,3-diol | 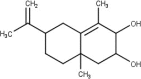 |
−6.4 |
| 64 | 2-Methyl-9-(prop-1-en-3-ol-2-yl)Bicyclo[4.4.0] dec-2-en-4-ol | 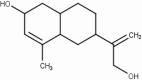 |
−6.1 |
Table 2 shows the obtained results from the molecular docking study carried out on all the molecules present in different medicinal plants (67 molecules), by giving the interaction energy for each compound, there is a difference in energy between each ligand and nCOV-19 main protease.
By comparing all studied molecules with Chloroquine on the basis of the interaction energy criterion. Knowing that the energy value of interaction of the molecule referred (Chloroquine) is (-6 kcal/mol), 11 molecules which have a good interaction with the studied enzyme are mentioned in Table 2. For example, the Crocin at interaction energy equal to (-8.2 kcal/mol), Digitoxigenin at a value of (-7.2 kcal/mol), and β-Eudesmol at a value of (-7.1 kcal/mol). The 3 D binding mode of these compounds is shown in Figures 1 and 2.
Figure 1.
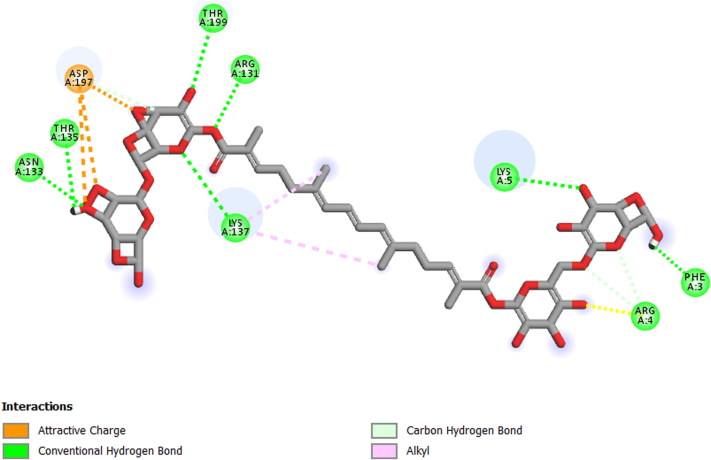
2 D View of the binding conformation of the Crocin inhibitor at the active site of Coronavirus (2019-nCoV) main protease.
Figure 2.
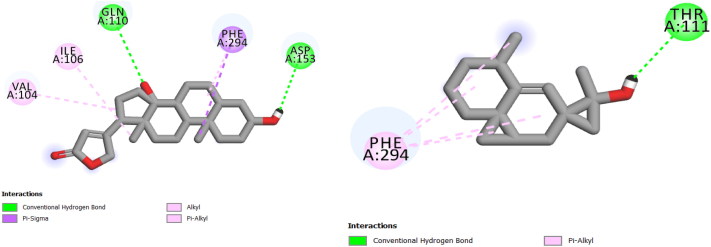
2 D View of the binding conformation of the Digitoxigenin and β–Eudesmol inhibitors at the active site of Coronavirus (2019-nCoV) main protease.
From a biological or pharmacological point of view, these first three molecules which are proposed as inhibitors of Coronavirus main protease are molecules having a significant antiviral power and according to bibliographical research and experiments which have already done, the results found for each molecule of natural origin is as following:
Crocin is an important compound in Crocus Sativus L, it has the capacity to inhibit the replication of HSV before and after the entry of virions in Vero cells. Crocin could be a promising anti-HSV and anti-HIV agent for herbal medicine against viral infections (Soleymani et al., 2018).
Digitoxigenin: represents 11.25% of the quantity present in Nerium Oleander, the derivatives of these molecules are used as antiviral and anti-cancer inhibitors (Boff et al., 2019).
β–Eudesmol despite its low amount of Lauris Nobilis L which contains only 2.39%, but this compound has a good interaction with the target, and it has significant antibacterial and antiviral power (Astani et al., 2011).
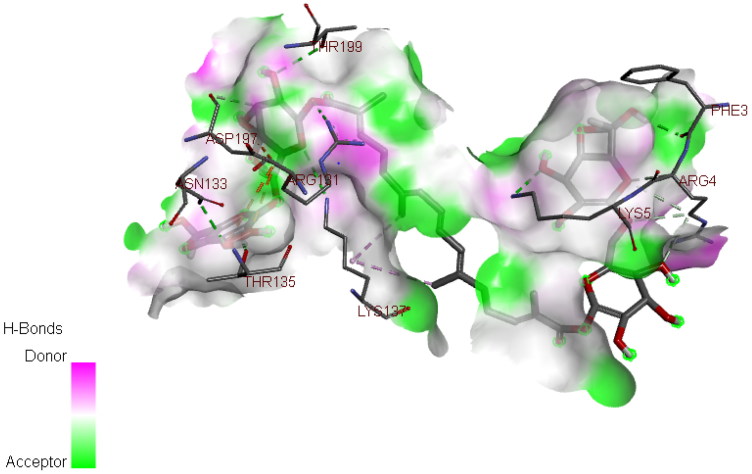
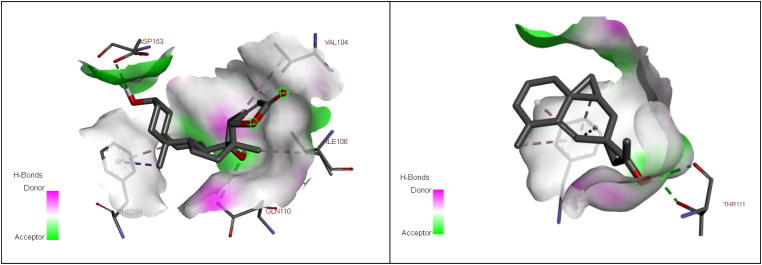
In our study, we took into account the interaction energies, so the interactions of the 1st level concern the hydrogen bonds, those of the 2nd level concerning the interactions between π systems and cation -π interactions, while the interactions of the 3rd level are hydrophobic contacts and non-specific Van der Waals interactions between aliphatic or aromatic carbon atoms. These interactions are generally spherical with a radius of 4 Å and cover most of the ligand. So, for the displacement of ligand in the binding site of our enzyme, the first level of interactions was considerate as the most important, the presence of Hydrogen bond interaction in selected complex explains that we have a good interaction between the three molecules and the studied protein (Table 4, Figure 3 and 4) (Adnan, 2019).
Table 4.
Hydrogen bond interaction between the three natural compounds and Coronavirus (2019-nCoV) main protease.
| Molecules | Residue formed Hydrogen bond interaction with studied compounds |
|---|---|
| β–Eudesmol | THR 111 |
| Digitoxigenin | GLN 110, ASP 135 |
| Crocin | THR 135, ASN 133, THR 199, LYS 137, LYS 5, PHE 3, ARG 4, ARG 131 |
Figure 3.
3 D View of the binding conformation of the Crocin inhibitor at the active site of Coronavirus (2019-nCoV) main protease (Hydrogen Bond interaction).
Figure 4.
3 D View of the binding conformation of the Digitoxigenin and β–Eudesmol inhibitor at the active site of Coronavirus (2019-nCoV) main protease (Hydrogen Bond interaction).
4. Conclusion
Today, the search for new molecules with a preservative power of natural origin is based on ethnobotanical studies which make it possible to carry out inventories of plants in a zone or a country, then on phytochemical and pharmacological studies and well other scientific aspects, so, the importance of the use of these medicinal plants which pushed us to seek and find the molecules which can prevent SARS-CoV-2 infection. Based on molecular docking, the results is very satisfactory, we have found three molecules among 67 which are very interesting either on the chemical side or on the biological side and therefore we propose these three molecules as inhibitor of SARS-CoV-2 main protease. The synthesis of the these molecules and the evaluation of their in vitro and in vivo activity against SARS-Cov-2 main protease could be interesting, before clinical essay.
Acknowledgements
We are grateful to the “Association Marocaine des Chimistes Théoriciens” (AMCT) for its pertinent help concerning the programs.
Disclosure statement
The authors declare that they have no competing interests.
References
- Adnan A. (2019). Nouvelles molécules organiques bioactives liées à l’inhibition des kinases. Utilisation Des Méthodes QSAR-3D, Docking Moléculaire et ADMET. Chimie Physique et Modélisation, Meknès, Université Moulay Ismail. [Google Scholar]
- Astani A., Reichling J., & Schnitzler P. (2011). Screening for antiviral activities of isolated compounds from essential oils. Evidence-Based Complementary and Alternative Medicine, 2011, 1–8. . 10.1093/ecam/nep187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boff L., Munkert J., Ottoni F. M., Zanchett Schneider N. F., Ramos G. S., Kreis W., Fernandes de Andrade S., Dias de Souza Filho J., Braga F. C., Alves R. J., Maia de Pádua R., & Oliveira Simões C. M. (2019). Potential anti-herpes and cytotoxic action of novel semisynthetic digitoxigenin-derivatives. European Journal of Medicinal Chemistry., 167, 546–561. 10.1016/j.ejmech.2019.01.076 [DOI] [PubMed] [Google Scholar]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., Xing F., Liu J., Yip C. C.-Y., Poon R. W.-S., Tsoi H.-W., Lo S. K.-F., Chan K.-H., Poon V. K.-M., Chan W.-M., Ip J. D., Cai J.-P., Cheng V. C.-C., Chen H., Hui C. K.-M., & Yuen K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., & Nakamura T. (2004). Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytotherapy Research, 18(7), 592–594. 10.1002/ptr.1485 [DOI] [PubMed] [Google Scholar]
- DeLano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr, 40(1), 82–92. [Google Scholar]
- G. O. of N. H. Committee, Office of State Administration of Traditional Chinese Medicine (2020). Notice on the issuance of a programme for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (Trial Version 4). 2020.
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., … Cao B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D. S. (2017). Epidemic and Emerging coronaviruses (severe acute respiratory syndrome and middle east respiratory syndrome. Clinics in Chest Medicine, 38(1), 71–86. 10.1016/j.ccm.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D. S., & Zumla A. (2019). Severe Acute Respiratory Syndrome: Historical, Epidemiologic, and Clinical Features. Infectious Disease Clinics of North America, 33(4), 869–889. 10.1016/j.idc.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., & Olson A. J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry, 19(14), 1639–1662. [DOI] [Google Scholar]
- Munster V. J., Koopmans M., van Doremalen N., van Riel D., & de Wit E. (2020). A novel coronavirus emerging in China—key questions for impact assessment. New England Journal of Medicine, 382(8), 692–694. 10.1056/NEJMp2000929 [DOI] [PubMed] [Google Scholar]
- Perlman S. (2020). Another decade, another coronavirus. Massachusetts Medical Society, 382, 760–762. 10.1056/NEJMe2001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot P. (2016). Dassault Systèmes BIOVIA, discovery studio modelling environment. Release.
- Richman D. D., Whitley R. J., & Hayden F. G. (2016). Clinical Virology. John Wiley & Sons. [Google Scholar]
- Schwartz D. A., Graham A. L. (2020). Potential maternal and infant outcomes from coronavirus 2019-nCoV (SARS-CoV-2) infecting pregnant women: Lessons from SARS, MERS, and other human coronavirus infections. Viruses, 12(2), 194 https://www.mdpi.com/1999-4915/12/2/194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani S., Zabihollahi R., Shahbazi S., & Bolhassani A. (2018). Antiviral effects of saffron and its major ingredients. Current Drug Delivery, 15(5), 698–704. 10.2174/1567201814666171129210654 [DOI] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., & Qin C. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses, 11(1), 59 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., & Zhang Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382, 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]