Abstract
Background/Aims:
Regulation of mRNA translation is central to protein homeostasis and is optimized for speed and accuracy. Spontaneous recoding events occur virtually at any codon but at very low frequency and are commonly assumed to increase as the cell ages.
Methods:
Here, we leveraged the polyglutamine(polyQ)-frameshifting model of huntingtin exon 1 with CAG repeat length in the pathological range (Htt51Q), which undergoes enhanced non-programmed translational −1 frameshifting.
Results:
In body muscle cells of Caenorhabditis elegans, −1 frameshifting occured at the onset of expression of the zero-frame product, correlated with mRNA level of the non-frameshifted expression and formed aggregates correlated with reduced motility in C. elegans. Spontaneous frameshifting was modulated by IFG-1, the homologue of the nutrient-responsive eukaryotic initiation factor 4G (eIF4G), under normal growth conditions and NSUN-5, a conserved ribosomal RNA methyltransferase, under osmotic stress.
Conclusion:
Our results suggest that frameshifting and aggregation occur at even early stages of development and, because of their intrinsic stability, may persist and accelerate the onset of age-related proteinopathies.
Keywords: Translation, Frameshifting, Aging, CAG repeat, Polyglutamine, C. elegans, Aggregation
Introduction
Translation is central to genetic decoding and is orchestrated by the ribosome which translates, codon by codon, the information stored in the mRNA. Translation is highly dynamic and optimized for speed and accuracy [1]. However, the cost of maintaining high translation levels is reduced accuracy resulting in some aberrancies in translation, like recoding or misincorporation of amino acids. Translational recoding, or frameshifting, is due to local “out-of-register” decoding in which the ribosomes shift from the normal reading frame to either the −1 frame or the +1 frame. Both −1 and +1 frameshifting result in different trans-frame protein products. Spontaneous recoding events may occur virtually at any codon with relatively low frequency, resulting in expression of an aberrant protein product [2, 3]. In contrast, programmed recoding occurs more frequently at specific signals in the open reading frame (e.g. slippery site, secondary structure) [2–5]. Programmed frameshifting is frequently employed in viral genomes and is seen as an evolutionary tool to expand protein repertoire and maintain stoichiometry of protein complexes [4, 6, 7].
Incidental or non-programmed frameshifting is a stochastic process that uses atypical mechanisms of recoding. For example, mutation-based extension of a homopolymeric polyCAG stretch of huntingtin (Htt) exon 1 or ataxin-3, both implicated in neurodegenerative repeat pathologies, undergo −1 and +1 frameshifting with different underlying mechanisms [8–14]. +1 frameshifting is induced at an atypical slippery sequence 5’-upstream of the CAG repeat and this ribosomal slippage is facilitated by the increased secondary structure propensity of the expanded CAG-repeat. In contrast, −1 frameshifting is more frequent and is triggered by depletion of charged tRNAGlnCUG that reads the CAG codon [10, 11]. Despite mechanistic differences in their origin, the trans-frame products (e.g. polyS from +1 frameshifting or polyQ/A from −1 frameshifting) are refractory to degradation and aggregate in the cell [8, 10, 15, 16].
Decades ago, the hypothesis was put forward that translation efficiency decreases with age and errors in translation accumulate [17]. Yet, direct experimental evidence of this longstanding assumption is missing. In general, aging is defined as a time-dependent progressive loss of physiological integrity and molecular activities which increases the vulnerability to death [18, 19]. One of the key players in physiological integrity, protein homeostasis or proteostasis, deteriorates with aging [18, 20]. Translation is intimately linked to proteostasis, hence compromised proteostasis integrity with age has been also extrapolated to alterations in translation fidelity. Furthermore, the level of many translation-associated factors, e.g. some ribosomal proteins, initiation factors, and modifying enzymes, declines with age, leading to general downregulation of protein synthesis (reviewed in [21]). But does the decreased translation that occurs with age correlate with errors in translation?
Using lysates prepared from young and old human fibroblasts, Luce and Bunn observed a sevenfold increase in cysteine misincorporation in aged human fibroblast cell line IMR-90 [22]. By contrast, other studies do not find any correlation between translation errors and aging [23, 24]. Furthermore, a recent study demonstrated that longer-lived species had lower basal rates of translation errors [25], suggesting that translation fidelity coevolved with the longevity of the organism. Measurements of the global protein synthesis in germ cells of Drosophila melanogaster show that differentiating cells have higher protein output compared to stem cells [26]. Does this increased translation activity in differentiated cells lead to higher translational errors?
To address whether and how translation fidelity changes with age in differentiated cells, we used Caenorhabditis elegans, a widely-used model organism to study aging. Under a muscle-specific promoter, we integrated a reporter with an expanded polyCAG stretch which is sensitive to translation recoding errors and reports on −1 frameshifting (Htt51Q(−1)). By analyzing the frameshifting frequency as a function of age, from larvae to late adult state, we found that translational recoding errors accompany the zero-frame product from the onset of its expression in a manner that correlates with the overall level of gene expression. To test the influence of translational regulation that increases lifespan and resistance to age-related disease, we used knockdown of the nutrient-responsive eukaryotic translation initiation factor 4G/ifg-1 [27] or ribosomal RNA methyltransferase gene nsun-5 [28]. We find that the toxicity associated with the Htt51Q(−1) aggregates is modulated by IFG-1 under normal growth conditions and by NSUN-5 under osmotic stress.
Materials and Methods
Design of a frameshifting reporter and generation of C. elegans transgenic strain
The reporter of frameshifting was adopted from [10], containing eCFP in zero frame to Htt exon 1 with 51 CAG repeats and mCherry in −1 (eCFP-Htt51Q(−1)-mCherry) generating the strain ANR105 (rogEx103[unc54p::CFP::Q51::mCherry]). It was cloned into pPD30.38 containing the promoter and enhancer elements from unc-54 for body wall muscle expression. To bypass an independent translational start for mCherry, its N-terminal Met was mutated to Val. Integration of extrachromosomal arrays was carried out into the Bristol N2 nematodes using UV irradiation. Integrated lines were backcrossed three times into N2 wild-type to guarantee a wild-type genetic background generating the strain ANR144 (rogEx103[unc54p::CFP::Q51(2 151_2152insC)::mCherry]). Three independent lines were isolated and they all showed same phenotype.
C. elegans strains and maintenance
Besides ANR105, ANR144 and N2, the strain AM141 (rmIs133[unc-54p::Q40::YFP]) expressing a polyCAG-stretch alone and C-terminally fused to YFP was used. Furthermore, we included the strain AM881 (rmEx336[unc54p::htt171Q75::YFP] as a control (a kind gift from R.I. Morimoto, Northwestern University). AM881 expressed Htt exon 1 with 75 CAG repeats fused to YFP. All Htt variants were under the control of the body wall muscle specific promoter unc-54. ANR127 (rogIs113[sca-1p::GFP + myo-2::mCherry]) expressing mCherry and GFP only was used as a positive control. Strains were maintained at 20°C on nematode growth media (NGM) plates seeded with Escherichia coli OP50. For age synchronization, young adult animals were placed on NGM plates and were allowed to lay eggs. After about eight hours adult nematodes were removed and hatchlings were used for experiments. All experiments were carried out at 20°C. During reproductive stage, C. elegans individuals were transferred to fresh NGM plates every second day to avoid contamination by progeny.
Imaging
For light and fluorescence microscopy, synchronized nematodes were immobilized using either 5 mM levamisole or 10 mM NaN3. The animals were imaged with a Leica DMi8 or Leica DFC310 FX (Leica Microsystems) and BZ-9000 (Keyence), respectively. It should be noted, that in general the mCherry fluorescence was hardly detectable under the same settings used to detect the zero-frame CFP-labeled Htt51Q, hence for the animals were usually longer exposed when recording the mCherry fluorescence. Intensity of fluorescence was quantified by using ImageJ 1.44p software.
RNAi experiments
RNAi bacteria strains expressing dsRNA ifg-1(M110.4), daf-2 (Y55D5A_391.b), daf-16 (R13H8.1), nsun-5 (Y53F4B.4), hsf-1 (Y53C10A.12), qars-1 (Y41E3.4), yars-1 (Y105E8A.19) (from the Ahringer library and from the Vidal library), including empty vector L4440 (Addgene, Cambridge, MA, USA.), were cultured as previously described [29]. 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was used for induction of dsRNA. C. elegans strains were synchronized via timed-egg lay, transferred to RNAi plates 48 hours after hatching and exposed to RNAi for 48 hours. Compared to N2 wild-type, the animals expressing Htt51Q exhibited delayed developing time. Therefore, they were monitored over a longer period until they reached adulthood phase comparable to that of the N2 animals. The knockdown of the gene of interest was verified by qRT-PCR or in case of ifg-1 by immunoblot and nsun-5 by COBRA.
Life span assay
Synchronized populations of 100 individual C. elegans nematodes were used for life-span assays starting from first day of adulthood. Life-span assays were carried out at 20°C on NGM plates spotted with E. coli OP50. Nematodes were scored every second day for survival and animals that failed to respond to a touch-provoked stimulus were referred as dead. Worms were censored if they died because of vulva burst or internal hatching of progeny. For life-span assays of AM141, the survival was scored starting from the first day of adulthood. 100 or more animals were used per condition (RNAi ifg-1 or control RNAi L4440) and put on seeded RNAi plates containing 200 μM fluorodeoxyuridine (FUDR). Survival was plotted by means of Kaplan-Meier survival curves and statistically analyzed by log-rank test, both integrated in GraphPad Prism 5.
Motility assay
Thrashing rate of individually synchronized nematodes was determined as described previously [30]. Briefly, one individual worm was transferred into 15 μl of M9 or S-basal medium. After a recovery of 1 min the body bends were counted for 30 s. A body bend was defined as a change in direction of bending at midbody of the animals as described in [31]. Statistical analysis was performed by unpaired Student’s t-test integrated in GraphPad Prism 5. Differences with p < 0.05 were considered as significant.
To determine motility traces, single one-day old adult worms of wild-type N2 and ANR144 were placed onto NGM plates with E. coli OP50 and allowed to move freely for 60 min at 20°C. Afterwards, their traces were imaged using Leica DMi8 (Leica Microsystems) with 10x objective. Assembling of one mosaic image from multiple smaller images was done to obtain a defined field size (here 6×6, software module “Tile Scan”).
RNA isolation and qRT-PCR
From synchronized C. elegans populations grown on NGM or RNAi plates the total RNA was isolated with TRIzol reagent (Ambion, ThermoFisher Scientific) following manufacturer’s protocol for chloroform extraction. The integrity of the RNA was analyzed with RNA Nano Chip 2100 (Bioanalyzer, Agilent Technologies). 0.5 μg total RNA was treated with RNase-free DNase I (Thermo Scientific) and reverse transcribed using oligo(dT)-primers and Revert Aid Reverse Transcriptase (Thermo Scientific). qRT-PCR was performed in a SYBR green-based approach (Qiagen) on a real-time PCR system (TOptical, Analytic Jena). mRNA expression of target genes was normalized to the housekeeping gene act-2.
Immunoblotting analysis and filter-retardation assay
A large population of animals (i.e. several thousands) were synchronized via timed egg lay and collected at different time points, starting from the first day of adulthood. C. elegans individuals were placed in 100 μl standard lysis buffer (8 M urea, 2% SDS, 50 mM DTT, 50 mM Tris pH 7.4, 1x Halt Protease Inhibitor Cocktail (Thermo Scientific)) on ice for several hours and then frozen. The protein concentrations of the lysates were determined using DC Protein Assay (Bio-Rad) and analyzed by immunoblotting using anti-GFP monoclonal antibody (1:1000, Thermo Scientific) or anti-mCherry (1:1000, St. John’s laboratory). For loading control, membranes were stripped with buffer containing 0.4 M NaOH, washed twice (5 minutes each), and immunostained with antibodies against beta-tubulin (1:500, Developmental Studies Hybridoma Bank). As a positive control, protein lysate of mChery-expressing ANR127 strain were loaded.
For filter-retardation assays [32] a defined number of individual synchronized C. elegans were picked and lysed by boiling for 15 min in 15 μl of lysis buffer (50 mM DTT, 2% SDS). The lysates were loaded onto a cellulose acetate membrane with a defined pore size of 0.2 μm and polyQ variants were detected by immunoblotting.
Osmotic stress test
Nematodes were synchronized via timed egg lay on nsun-5 RNAi plates or control RNAi, respectively. At one day of adulthood, C. elegans were moved to plates containing 50 mM, 100 mM or 200 mM NaCl. After 24 hours, animals were harvested for filter-retardation assay.
Detection of NSUN-5-specific methylation at C2381 of 26S rRNA by COBRA
C2381 methylation was detected by COBRA as previously described [28, 33] with minor modifications. Worms exposed to nsun-5 or control RNAi were subjected to total RNA extraction and on-column DNase I digestion using the Direct-zol™ RNA Miniprep Kit (Zymo Research). 0.5 μg total RNA were bisulfite-converted using the EZ RNA Methylation™ Kit (Zymo Research) and subsequently cDNA was generated using Super Script IV (Life Technologies). PCR using OneTaq® DNA Polymerase was performed to generate a 101 bp PCR product surrounding C2381 with forward and reverse primers specific for the cDNA after bisulfite conversion (i.e., 5’-GGGAGTAATTATGATTTTTCTAAGGTAG-3’ and 5’-ATAATAAATAAAAACAATAAAAATCTCACTAATCCATTCATACAC-3’). The PCR product was digested with MseI (New England Biolabs) and loaded on a Novex™ TBE Gel, 20%, 10 well (Life Technologies). Two products of 45 and 56 bp were formed, if C2381 was methylated and three products of 15, 30 and 56 bp, if C2381 was not methylated.
Results
−1 frameshifted species appears early in the development of C. elegans
To determine the age-dependent effect on frameshifting in C. elegans, we generated a reporter construct with the fluorescent mCherry protein fused C-terminally in −1 reading frame to human Htt exon 1 with a repeat in the pathological range encoding 51 glutamines (Htt51Q) (Fig 1A). N-terminally of Htt, the reporter carries eCFP in the zero-reading frame, enabling detection of the overall expression level (Fig 1A). For simplicity, we named the full-length constructs eCFP::Htt_exon1(Q51)::mCherry(−1) as Htt51Q(−1). The reporter was cloned under the control of the unc-54 promoter, which normally drives expression of the myosin heavy chain in body muscle (Fig 1A). Expression in muscle cells was chosen as expression of polyQ proteins produces a toxic effect that reduces locomotion in C. elegans [34]. Using the mCherry fluorescence as a read-out of frameshifting, we detected abundant −1 frameshifting (Fig 1B). In parallel, we produced a similar construct that reports on +1 frameshifting (Htt51Q(+1)) to compare both recoding events. However, +1 frameshifting was extremely rare (i.e. far less than 1%), which precluded further analysis and so, we focused only on −1 frameshifting. This observation agrees with results from mammalian cell cultures in which +1 frameshifting was also much more rarely detected than −1 [10, 11].
Fig. 1.
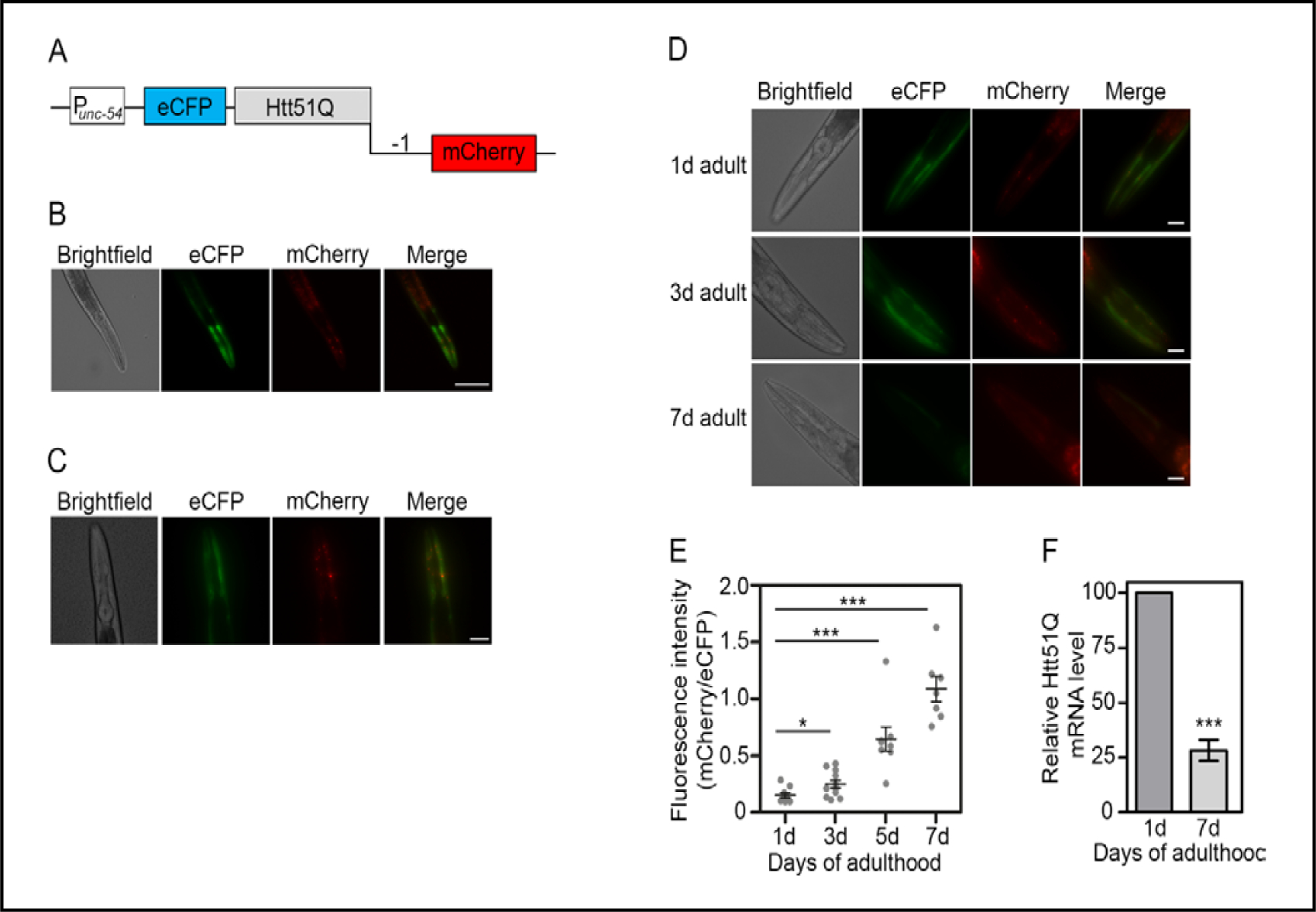
The frequency of −1 frameshifting with age. (A) Schematic of the frameshift reporter Htt51Q(−1) for expression in body wall muscle cells under control of unc-54 promoter. N-terminal eCFP is expressed in zero frame and mCherry reports on −1 frame. (B) Representative fluorescence images of one day-old nematodes with genome-integrated expression of Htt51Q(−1) in the body wall muscle cells. For better visualization, eCFP fluorescence is depicted in green. Scale bar, 25 μm. (C) Representative fluorescence images of synchronized nematodes at larval stage L4. Scale bar, 25 μm. (D) Representative fluorescence images of synchronized nematodes imaged at different ages. Starting at day 5, autofluorescence in the intestine becomes visible. Scale bar, 25 μm. (E) Quantification of the ratio between mCherry and CFP fluorescence of the animals expressing Htt51Q(−1) at different age (n = 7–11 animals). Fluorescence was integrated over the head area since the expression was more prominent in the head and only this region was considered in the quantifications. Note that the mCherry channel was disproportionally enhanced in panels B-D compared to the blue CFP channel for better visualization. The quantification, however, was performed with non-enhanced images. *, p<0.05; ***, p<0.001, Student’s t-test. See also Table S1. (F) qPCR analysis of adult nematodes with genome-integrated Htt51Q(−1) expression. Htt51Q(−1) mRNA was normalized to act-2. Expression in day 1 adults was set to 100%. Data are means ± SEM (n = 3). ***, p<0.001, Student’s t-test.
We used two different C. elegans strains, ANR144 with the integrated −1 frameshift reporter and ANR105 strain expressing Htt extrachromosomally. Overall, the expression of the integrated frameshift reporter was similar to that already observed in nematodes expressing Htt extrachromosomally, but with fewer mCherry-positive –1-frameshifted species (Fig. 1B and Fig. S1A - for all supplemental material see www.cellphysiolbiochem.com). On average, we detected 1.9 vs 4.6 red fluorescent spots per animal with integrated Htt51Q(−1) gene vs extrachromosomal array, respectively (Fig S1B). The mCherry fluorescence reporting on −1 frameshifting was detectable in every animal tested and appeared during larval development as hyper-fluorescent puncta (Fig. 1C and S1C). Expression of both Htt51Q(−1) and Htt51Q was more prominent in the head of the transgenic animals and rarely detectable in the rest of the body wall muscle cells (Fig. 1B, C and Fig. S1A, C). Notably, the zero-frame Htt51Q, detected through eCFP, exhibited rather diffuse fluorescence throughout the body wall muscle cells, whereas the foci were exclusively red (Fig. 1B). mCherry-foci that were clearly visible in young adults and characterized by sharp and distinct boundaries which disappeared with age (Fig. 1D and Fig. S1D). Instead, the red fluorescence was dominated by mostly intestinal age–associated autofluorescence from day 5 on (Fig. 1D and Fig. S1D). Limited expression to the head region was also observed for the AM881 strain (Fig. S1G), another C. elegans model expressing Htt exon 1 with a polyQ stretch within the pathological range (Htt75Q).
To determine the relative accumulation of the −1 trans-frame product with age, we measured the ratio of mCherry to eCFP fluorescence of the main zero-frame Htt51Q product by integrating the fluorescence over the head area of the animals (Fig. 1E and Fig. S1E). The ratio of mCherry to CFP fluorescence increased with age (Fig. 1E and Fig. S1E). But when analyzing the zero- and trans-frame products separately, we realized that changes in the ratio were mostly due to a decrease of CFP fluorescence, while the mCherry fluorescence changed only marginally (Table S1 and S2), suggesting higher stability or higher aggregation propensity of polyA stretches compared to polyQ. Supportive of this, in the polyA pathologies, much shorter expansions of polyA confer cellular dysfunction [35].
The amount of the zero-frame protein Htt51Q decreased with age in both animals with genome-integrated or extrachromosomal expression (Fig. 1D, E, Fig. S1D, E and Table S1, S2). Compared to the C. elegans strain AM141 expressing solely the polyQ stretch under the control of the same muscle-specific promoter the average number of the zero-frame Htt51Q(−1) aggregates per young animal was much lower (Fig S1B) [34]. The decrease in fluorescence of the zero-frame Htt51Q strongly correlated with the decline in mRNA levels over time for both animal groups with genome-integrated and extrachromosomal expression of Htt51Q(−1) (Fig 1F and Fig S1F) which corroborates previous observations for age-dependent decrease in mRNA levels in muscle tissues [36].
−1 frameshifted protein alone forms aggregates that persist with age
The mCherry fluorescence reporting on −1 frameshifting remained relatively unchanged with age on the background of decreased zero-frame Htt51Q product (Table S1 and S2), which raised the question as to whether zero- and trans-frame proteins exhibit different stability, and/or the CFP fluorescence might be quenched by interactions with cellular proteins or through the Htt51Q aggregation itself. Hence, we performed immunoblotting to monitor the expression of SDS-soluble proteins (Fig. 2A, C) and filter-retardation assays to detect detergent-insoluble aggregates (Fig. 2B). The zero-frame Htt51Q protein was detectable at early adulthood and declined with age (Fig 2C). By contrast, we were not able to detect mCherry biochemically in the first days of adulthood (Fig. 2A, C) despite visible detection of mCherry fluorescence during this time (Fig. 1B, D); Htt51Q(−1) protein expression might be under the detection limit of the immunoblots (Fig. 2A). At a later age, between day 5 and 11 of adulthood, we detected frameshifted species (Fig. 2A, C), which in the mCherry-immunostained blot exhibited higher molecular weights, most likely oligomers and persisted over time (Fig. 2A). Furthermore, the SDS-insoluble aggregates were mCherry-positive (Fig. 2B). Together, these results demonstrate that frameshifting in Htt51Q occurs early in life, presumably due to the high level of expression driven by the unc-54 promoter at this time, and that the frameshifted product formed aggregates that were present throughout the life cycle of the animal. Our data suggests that the increase in aggregates with age might be due at least in part, to decreased turnover of frameshifted products.
Fig. 2.
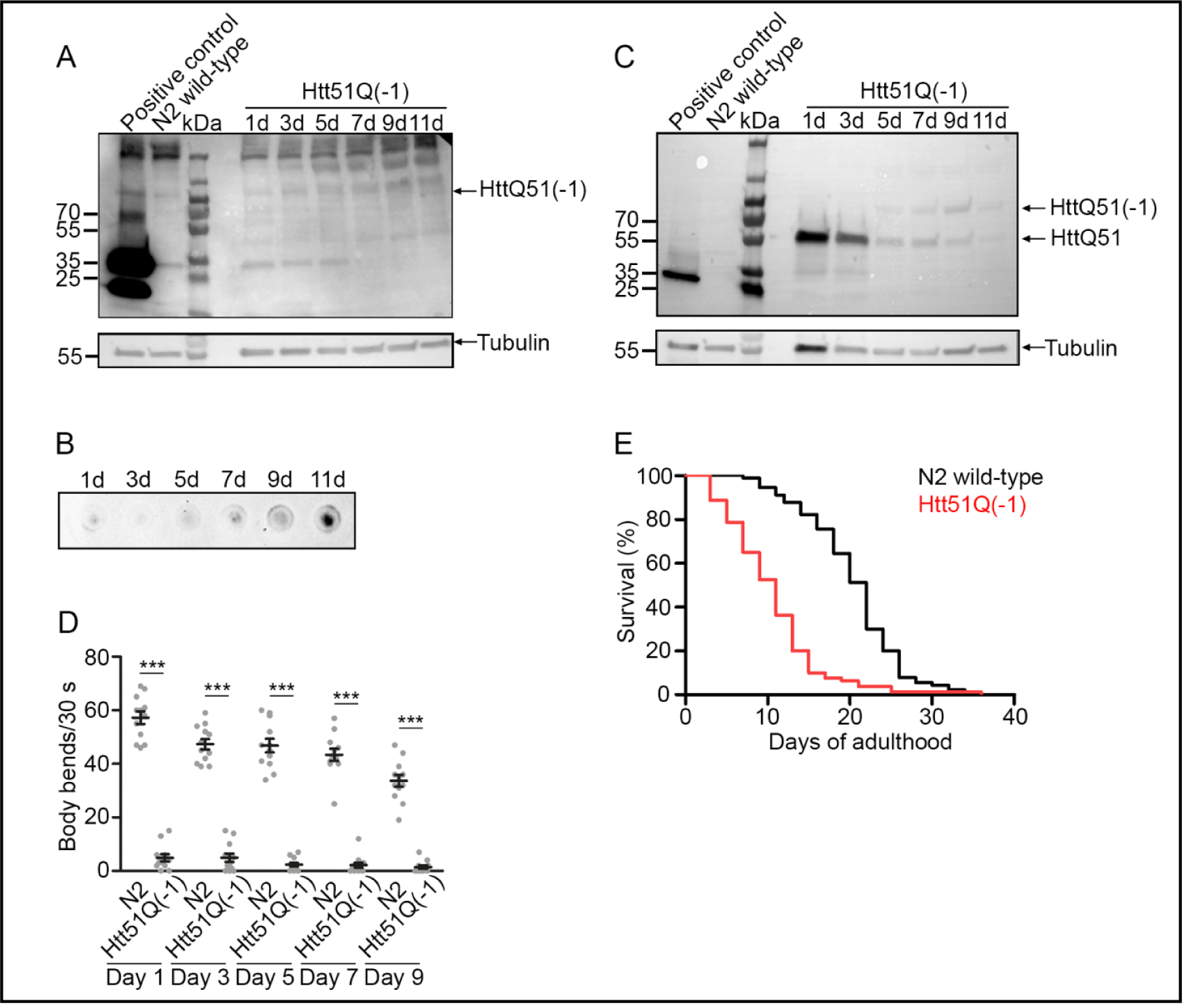
Aggregates containing −1 frameshifted Htt51Q persist with age in C. elegans with genome-integrated frameshift reporter. (A,C) Immunoblot of synchronized nematodes at different ages to detect soluble trans-frame and zero-frame proteins. 50 μg of protein were loaded per lane and trans-frame proteins were detected with anti-mCherry antibodies (A) and zero-frame proteins with anti-GFP antibody (C). Arrows indicate the position of the Htt51Q(−1) or Htt51Q monomers, respectively. Note the extended exposure in mCherry channel (A) compared to CFP channel (C) to enable detection of frameshift products, which were produced in very low amounts. Immunoblot membranes were stripped and reprobed with anti-tubulin antibody (A,C). As a positive control animals expressing both, GFP and mCherry, were used (ANR127). The signal at 250 kDa results from unspecific binding of mCherry antibodies. As a negative control, N2 animals were loaded. Note, the tubulin signal overlaps with CFP-polyQ in size; hence, the enhanced tubulin amount at 1d might be due to inefficient stripping in this part of the blot (C). (B) Filter-retardation assay of synchronized animals at different ages. 30 animals were loaded per sample and frameshifted Htt51Q(−1) was detected with anti-mCherry antibodies. (D) Motility assay on different days of adulthood. Each dot represents one single nematode, the horizontal line indicates the mean thrashing rate (± SEM, n = 10 animals for each strain and time point). Data among one time-point are normalized to the body bends of N2 wild-type. ***p < 0.001 (Student’s t-test). (E) Life span assay of wild-type N2 and nematodes expressing Htt51Q(−1). Kaplan-Meier survival curves were compared using log-rank test. Median lifespan of 12.5 days for animals with integrated expression of Htt51Q(−1) vs. 22 days for N2. p<0.001 for animals with integrated Htt51Q(−1) vs. N2, (log-rank test). See also Fig. S2B.
In parallel, some phenotypic observations on the effect of Htt51Q(−1) expression on nematode behavior were made. When placed in liquid, C. elegans thrash about as they try to get traction. This is frequently used to measure motility, which decreases with age and which is malleable under certain genetic or environmental conditions. The impact on morphological development of C. elegans with integrated Htt51Q(−1) was severe and locomotion was severely limited (Fig 2D). When placing animals on NGM plates spotted with E. coli OP50 to track their traces within the bacteria lawn, after 60 min transgenic animals with genome-integrated Htt51Q(−1) expression remained in close proximity to the start point, while the wild-type N2 had traversed almost the entire E. coli lawn (Fig S2A). This defect in exploratory locomotion was observed beginning from young larvae right after hatching; they remained near their hatching spot. Although wild-type nematodes moved slower with age, they retained coordinated thrashing behavior. In contrast, movements of transgenic animals with the integrated reporter were characterized by uncoordinated thrashing even at young age with some animals exhibiting paralysis. Impaired locomotion was also detectable by the egg laying pattern with eggs laid in clusters or straight lines. Very often, egg-laying adults stayed within a close proximity to the eggs. We isolated three independent transgenic lines with genome-integrated Htt51Q(−1) and these severe phenotypic changes were observed in all three which argues that the phenotype was not caused by the point of chromosomal insertion.
The mean survival was significantly reduced by ~40% compared to wild-type N2 (p < 0.001, log-rank test, Fig 2E). Many animals were censored during life-span assays because of internal hatching of larvae (Fig S2B). In contrast, the overall life span of the Htt51Q(−1) animals with the extrachromosomal array was not altered (Fig. S2C), although the thrashing rate of Htt51Q(−1) animals with the extrachromosomal array decreased compared to N2 wild-type animals (Fig S2D), despite exhibiting typical sinusoidal movements. With increasing age, the cuticles of transgenic animals became crumpled and the locomotion decreased. The Htt51Q(−1) animals with extrachromosomal array showed comparable motility with different polyglutamine-expressing strains (e.g. Htt75Q-YFP and poly40Q-YFP; Fig S2E).
IFG-1 and NSUN-5 modulate toxicity associated with aggregation
Next, we tested which cellular proteins influence translational frameshifting. We silenced several genes involved in regulation of translation and lifespan using interfering RNA (RNAi), e.g. daf-16 and daf-2 (abnormal dauer formation), hsf-1 (heat shock factor 1), ifg-1 (homologue of the eukaryotic initiation factor 4G (eIF4G)), rsks-1 (worm homologue of S6 kinase), qars-1 (glutaminyl-tRNA synthetase, QARS) yars-1 (tyrosyl-tRNA synthetase, YARS) and nsun-5 (NOP2/Sun RNA methyltransferase family member 5). daf-2 and daf-16 were chosen for their role in regulating life span and longevity: daf-16 encodes a FOXO transcription factor and daf-2 an insulin-like receptor tyrosine kinase. Attenuation of DAF-2 activates DAF-16, which translocates to the nucleus and activates expression of stress response and longevity assurance genes [37]. HSF-1 is the heat shock transcription factor regulating transcription of heat shock genes, e. g. chaperones, assisting in refolding misfolded proteins [38]. IFG-1 was selected as its knock-down has been shown to decrease global translation rates [39, 40] and increases life span in C. elegans [27, 37, 39–41]. RSKS-1 activates ribosomal protein S6 through phosphorylation and also decreases global translation rates. Similar to ifg-1, knockdown of rsks-1 results in life span extension of C. elegans [40]. Our previous studies in mammalian cell culture models have shown that frameshifting frequency depends on the availability of charged glutaminyl-tRNAGlnCUG that reads the CAG codon [10]. Hence, to decrease the amount of charged glutaminyl-tRNAGlnCUG, we down regulated qars-1 encoding the QARS which aminoacylates tRNAGlnCUG with glutamine. As a control, yars-1 encoding tyrosyl-tRNA synthetase (YARS) was selected, which charges tRNAs unrelated to the translation of CAG repeats. The down-regulation of the tested genes was verified by qPCR (Fig S3A), Western blot (Fig. S3B), or Combined Bisulfite Restriction Analysis (COBRA) methylation activity assay (Fig S3C, D). NSUN-5 introduces an m5C modification at C2381 of 26S ribosomal RNA and its depletion extends the mean lifespan and increases paraquat resistance, without affecting growth, pharyngeal pumping or locomotion [28].
The low intensity of the frameshifted Htt51Q(−1) species in the immunoblots precluded quantifiable comparison of the frameshifting yields following the knockdown of various genes. Instead, we compared the thrashing rates of individual nematodes subjected to different selected RNAi as readout of changes in proteotoxicity and associated changes in frameshifting products. In both nematodes strains with extrachromosomal or integrated expression of Htt51Q(−1) we observed the highest accumulation of frameshifted species at one day of adulthood, which coincides with the highest amount of Htt51Q(−1) mRNA (Fig 1F, S1F). Thus, we compared the thrashing rates following exposure to selected RNAi only at that age. In general, the motility was very low and the animals exhibited uncoordinated movements as also seen for nematodes grown on bacteria without any RNAi background (Fig 2D, 3A). For the majority of the genes with downregulated expression, we did not observe any effect on thrashing (Fig 3A). Day-one adult animals subjected to ifg-1 knockdown in the previous 48 hours exhibited the highest variation in their body bends and the motility increased significantly compared to empty RNAi vector controls (Fig 3A). This may be due to enhanced activation of heat shock responsive genes when translation is attenuated in this manner [27]. Furthermore, many individuals presented the same sinusoidal movements on solid media and the same coordinated thrashing behavior in liquid characteristic for the N2 wild-type strain albeit with a slower frequency, showing the protective effect of the ifg-1 knockdown. Furthermore, the ifg-1 knockdown significantly reduced the fluorescence ratio of mCherry to eCFP over the head area of the animals (Fig S3E). Moreover, a positive and protective effect of ifg-1 knockdown was also observed on the life span of nematodes expressing poly40QYFP (Fig S3F). A downregulation of aminoacyl-tRNA-synthetases (here QARS and YARS) may influence translation globally and consequently it was difficult to disentangle the effect of QARS on the overall lowered translation from that on the frameshifting frequency.
Fig. 3.
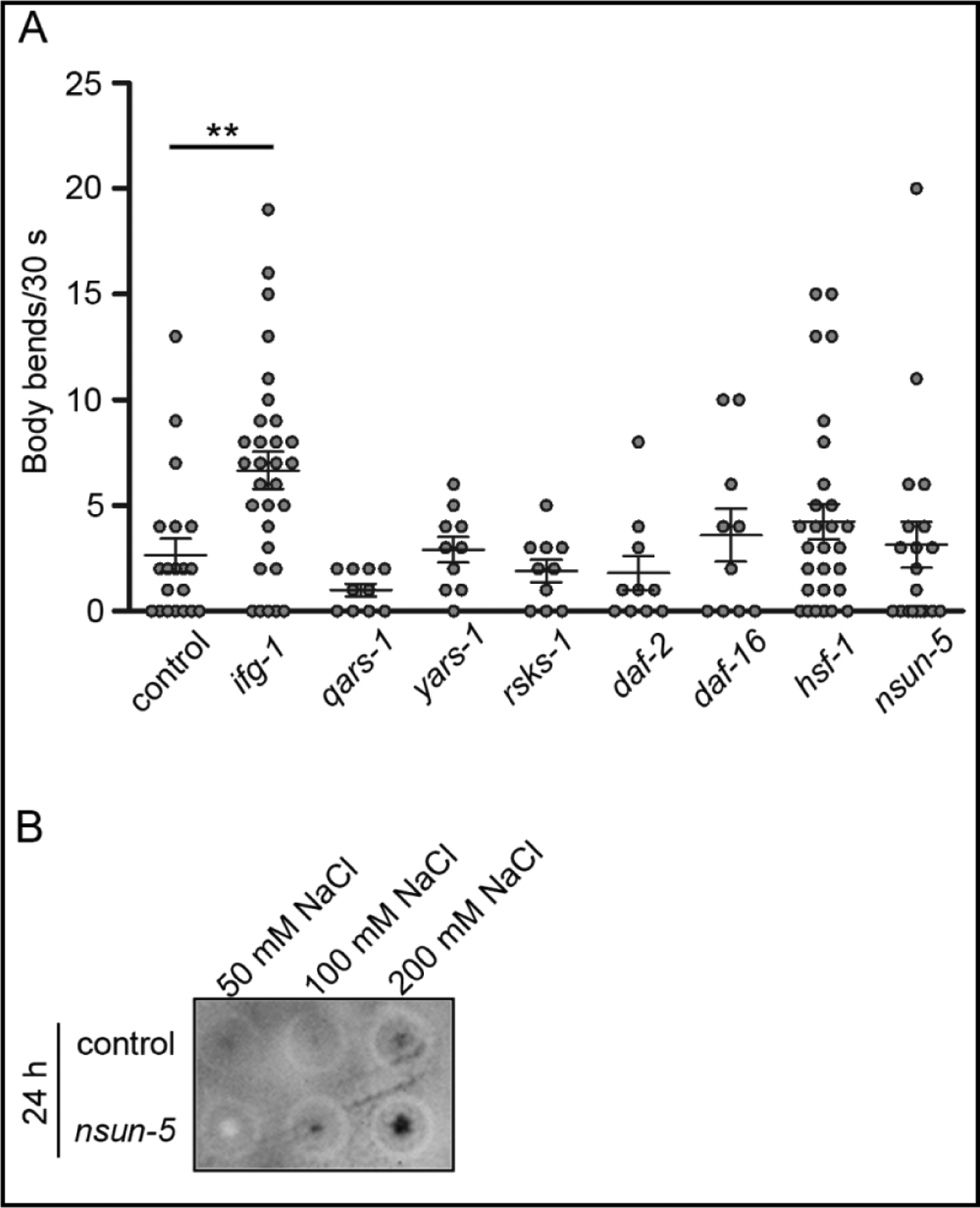
Thrashing rate of 1d-old Htt51Q(−1) C. elegans after RNAi knockdown. (A) Motility was analyzed by counting the body bends for each nematode within 30 s. Each dot represents one individual, the horizontal bar represents mean ± SEM (n = 10 for qars-1, yars-1, rsks-1, daf-2, daf-16; n = 20 for control RNAi/empty L4440 vector, n = 30 for hsf-1, ifg-1). Data were normalized to the thrashing rate of control animals. ** p<0.01 for ifg-1 compared to the control, differences between all other knockdowns were not significant. (B) Filter-retardation assay of synchronized animals expressing Htt51Q(−1) grown on control RNAi/L4440 vector or nsun-5 RNAi and subjected at 1d of adulthood to various doses of osmotic stress. 30 animals were loaded onto a cellulose-acetate membrane and frameshifted Htt51Q(−1) detergent-insoluble aggregates were detected using anti-mCherry antibodies.
Following knockdown of nsun-5, we observed only a marginal impact on the thrashing rate in day 1 adult nematodes causing more heterogeneity without changing the mean (Fig 3A). Since it is known that loss of RCM1, the yeast orthologue of NSUN-5, decreases ribosome stability at high salt concentrations or oxidative stress and alters translation fidelity [28, 33], we next tested the effect of nsun-5 depletion on aggregation of frameshifted products under osmotic stress. After one day, animals with reduced nsun-5 levels formed more mCherry-positive aggregates when subjected to osmotic stress suggesting higher frameshifting frequency when rRNA methylation is altered (Fig 3B). This increase was visible under high salt conditions (200 mM), whereas at lower salt concentration (100 mM) the amount of the aggregates containing frameshifted species was comparable to the control (Fig 3B). These results demonstrate that the presence of m5C2381 of 26S rRNA as mediated by NSUN-5 is required for translation fidelity and maintenance of protein homeostasis under osmotic stress. Furthermore, relief of the toxic effect of aggregates on locomotion is specific to low IFG-1 and not shared with other RNAi treatments known to promote longevity and decrease translation, like rsks-1 and daf-2.
Discussion
Translation fidelity plays an important role in maintaining proteostasis which, in turn, is a key player in protecting cellular function. Here, we took advantage of the multicellular model organism C. elegans to explore the frequency of translation recoding as a function of age and mRNA expression level. Spontaneous recoding is a relatively rare event, which occurs at rates comparable to the background rate [42] of translational errors ~5 × 10−4 per codon [43]. Such low rates are difficult to detect, and hence we leveraged the polyQ-frameshifting model with an expanded glutamine stretch which undergoes an enhanced and detectable non-programmed +1 and –1 frameshifting [10, 11]. By expressing the Htt51Q(−1) frameshift reporter either extrachromosomally or through genome integration we determined the incidental frameshifting frequency in the body wall muscle cells as the nematode aged. Intriguingly, the frameshifting is readily detectable during larval development, but changes little during the remaining life cycle of the nematodes. Frameshifting correlates with the expression level of the zero-frame protein. The latter declines because of a decrease in the steady-state Htt51Q mRNA levels with age (Fig 1F). Earlier studies demonstrate that mRNA levels of transgenic polyCAG and endogenous genes expressed in muscle cells (under the control of the unc-54 promoter) decrease after the first day of adulthood in C. elegans [36, 44]. Moreover, decline in the expression of polyQ protein in muscle cells has been reported earlier: In nematodes expressing polyQ fragments only, aggregation did not increase over time [34] suggesting that the polyQ expression was most likely largely diminished with age. Nematodes with the integrated reporter show a shorter life span as well as reduced motility rates compared to the ones with the extrachromosomal array suggesting that the genetic background or expression level in general may also influence the turnover. In light of our observations, the spontaneous frameshifting occurs at the onset of Htt51Q expression during larval development, but the aggregation frequency remains constant with age likely due to a global decrease in Htt51Q expression.
The modulatory effect of IFG-1 on locomotion under normal growth was modest, albeit significant and easily detected. The IFG-1 effect on translation depends on the mRNA steady-state levels [39, 40], hence the decline in Htt51Q(−1) mRNA steady-state levels with aging likely weakens the IFG-1 effect. Similarly, the NSUN-5 effect is more pronounced at early time points of growth and weakens with aging because of the general decline in the Htt51Q mRNA steady-state levels.
The animals expressing the Htt51Q(−1) frameshift reporter either extrachromosomally or genome-integrated show significant locomotion alterations implying a toxic effect, which likely comes from both zero-frame and trans-frame proteins in light of the gain-of-toxicity effect described for polyQ and polyA containing aggregates [45–47], which result in reduced motility when expressed in muscle cells [34]. We do not see an increase of the amount of aggregates, but rather a persistent small fraction that remains unchanged with age implying that the effect may originate from the onset of Htt51Q(−1) expression. Moreover, the hyperfluorescent aggregate foci of the trans-frame Htt51Q(−1) disappear early in adulthood suggesting a different turnover as compared to aggregates formed by polyQ stretch only, which are refractory to degradation [30, 34, 48]. Lee and coworkers, who also used full-length exon 1 instead of the polyQ stretch by itself, observed very modest aggregation behavior [49]. The native sequences flanking polyQ within exon 1 largely influence its aggregation propensity [50, 51].
Conclusion
Together, our results suggest that frameshifting is exacerbated when polyQ-containing mRNA is expressed at higher levels. Furthermore, spontaneous translational recoding occurs at the onset of expression. Some trans-frame products might be efficiently cleared by proteasome- or autophagy-mediated pathways, while trans-frame products of some transcripts may produce more persistent or even toxic products, like the example here, whose repetitive expanded polyamino acid stretch is refractory to clearance by the proteasome [52] and the aggregates of it likely alter the proteostasis network [53]. Trans-frame products produced at early age may risk accumulation because of their intrinsic stability or age-related deregulation of proteostasis, which, in turn, might be of importance in age-related proteinopathies.
Supplementary Material
Acknowledgements
The work was funded by the Innovation Award from the MDI Biological Laboratory Maine to Z.I., the James L. Boyer Fellowship at the MDI Biological Laboratory and the Austrian Science Fund (FWF: P30623-B26) to M.S. We thank Kevin Strange and Dustin Updike from the MDI Biological Laboratory in Maine, USA, for providing RNAi libraries. We thank R. Morimoto from the Northwestern University for the kind gift of the C. elegans strain AM881. N2 wild-type C. elegans was provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- 1.Rodnina MV, Wintermeyer W: The ribosome as a molecular machine: The mechanism of trna-mrna movement in translocation. Biochem Soc Transact 2011;39:658–662. [DOI] [PubMed] [Google Scholar]
- 2.Advani VM, Dinman JD: Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. BioEssays: News Rev Mol Cell Develop Biol 2016;38:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinman JD: Control of gene expression by translational recoding. Adv Prot Chem Struct Biol 2012;86:129–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV: Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucl Acids Res 2016;44:7007–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinman JD: Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA 2012;3:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giedroc DP, Cornish PV: Frameshifting rna pseudoknots: Structure and mechanism. Virus Res 2009;139:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namy O, Rousset JP, Napthine S, Brierley I: Reprogrammed genetic decoding in cellular gene expression. Mol Cell 2004;13:157–168. [DOI] [PubMed] [Google Scholar]
- 8.Davies JE, Rubinsztein DC: Polyalanine and polyserine frameshift products in huntington’s disease. J Med Genet 2006;43:893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaspar C, Jannatipour M, Dion P, Laganiere J, Sequeiros J, Brais B, Rouleau GA: Cag tract of mjd-1 may be prone to frameshifts causing polyalanine accumulation. Human Mol Genet 2000;9:1957–1966. [DOI] [PubMed] [Google Scholar]
- 10.Girstmair H, Saffert P, Rode S, Czech A, Holland G, Bannert N, Ignatova Z: Depletion of cognate charged transfer rna causes translational frameshifting within the expanded cag stretch in huntingtin. Cell Rep 2013;3:148–159. [DOI] [PubMed] [Google Scholar]
- 11.Saffert P, Adamla F, Schieweck R, Atkins JF, Ignatova Z: An expanded cag repeat in huntingtin causes +1 frameshifting. J Biol Chem 2016;291:18505–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stochmanski SJ, Therrien M, Laganiere J, Rochefort D, Laurent S, Karemera L, Gaudet R, Vyboh K, Van Meyel DJ, Di Cristo G, Dion PA, Gaspar C, Rouleau GA: Expanded atxn3 frameshifting events are toxic in drosophila and mammalian neuron models. Human Mol Genet 2012;21:2211–2218. [DOI] [PubMed] [Google Scholar]
- 13.Toulouse A, Au-Yeung F, Gaspar C, Roussel J, Dion P, Rouleau GA: Ribosomal frameshifting on mjd-1 transcripts with long cag tracts. Human Mol Genet 2005;14:2649–2660. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Fedyunin I, Miekley O, Valleriani A, Moura A, Ignatova Z: Global and local depletion of ternary complex limits translational elongation. Nucl Acid Res 2010;38:4778–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R: Inhibition of the ubiquitin-proteasome system in alzheimer’s disease. Proc Natl Acad Sci USA 2000;97:9902–9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, Grosveld FG, Peter J, Burbach H, Hol EM: Frameshift mutants of beta amyloid precursor protein and ubiquitin-b in alzheimer’s and down patients. Science 1998;279:242–247. [DOI] [PubMed] [Google Scholar]
- 17.Orgel LE: The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci USA 1963;49:517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G: The hallmarks of aging. Cell 2013;153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard Smith J: Review lectures on senescence - i. The causes of ageing. Proc Royal Soc B 1962; DOI: 10.1098/rspb.1962.0065. [DOI] [Google Scholar]
- 20.Balch WE, Morimoto RI, Dillin A, Kelly JW: Adapting proteostasis for disease intervention. Science 2008;319:916–919. [DOI] [PubMed] [Google Scholar]
- 21.Gonskikh Y, Polacek N: Alterations of the translation apparatus during aging and stress response. Mech Ageing Develop 2017;168:30–36. [DOI] [PubMed] [Google Scholar]
- 22.Luce MC, Bunn CL: Decreased accuracy of protein synthesis in extracts from aging human diploid fibroblasts. Exp Gerontol 1989;24:113–125. [DOI] [PubMed] [Google Scholar]
- 23.Mori N, Hiruta K, Funatsu Y, Goto S: Codon recognition fidelity of ribosomes at the first and second positions does not decrease during aging. Mech Ageing Develop 1983;22:1–10. [DOI] [PubMed] [Google Scholar]
- 24.Filion AM, Laughrea M: Translation fidelity in the aging mammal: Studies with an accurate in vitro system on aged rats. Mech Ageing Develop 1985;29:125–142. [DOI] [PubMed] [Google Scholar]
- 25.Ke Z, Mallik P, Johnson AB, Luna F, Nevo E, Zhang ZD, Gladyshev VN, Seluanov A, Gorbunova V: Translation fidelity coevolves with longevity. Aging Cell 2017;16:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R: Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 2016;18:276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard A, Rogers AN: Role of translation initiation factor 4g in lifespan regulation and age-related health. Ageing Res Rev 2014;13:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, Brandl C, Sonntagbauer M, Kriegner A, Linder A, Weinhausel A, Mohr T, Steiger M, Mattanovich D, Rinnerthaler M, Karl T et al. : Methylation of ribosomal rna by nsun5 is a conserved mechanism modulating organismal lifespan. Nat Commun 2015;6:6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J: Effectiveness of specific rna-mediated interference through ingested double-stranded rna in caenorhabditis elegans. Genome Biol 2001;2:RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brignull HR, Moore FE, Tang SJ, Morimoto RI: Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal caenorhabditis elegans model. J Neurosci 2006;26:7597–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB: A genetic selection for caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA 1996;93:12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE: Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 1997;90:549–558. [DOI] [PubMed] [Google Scholar]
- 33.Gigova A, Duggimpudi S, Pollex T, Schaefer M, Kos M: A cluster of methylations in the domain iv of 25s rrna is required for ribosome stability. RNA 2014;20:1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morley JF, Brignull HR, Weyers JJ, Morimoto RI: The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in caenorhabditis elegans. Proc Natl Acad Sci USA 2002;99:10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S: Polyalanine expansions in human. Human Mol Genet 2004;13 Spec No 2:R235–243. [DOI] [PubMed] [Google Scholar]
- 36.Adamla F, Ignatova Z: Somatic expression of unc-54 and vha-6 mrnas declines but not pan-neuronal rgef-1 and unc-119 expression in aging caenorhabditis elegans. Sci Rep 2015;5:10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson ST, Johnson TE: Daf-16 integrates developmental and environmental inputs to mediate aging in the nematode caenorhabditis elegans. Curr Biol 2001;11:1975–1980. [DOI] [PubMed] [Google Scholar]
- 38.Hsu AL, Murphy CT, Kenyon C: Regulation of aging and age-related disease by daf-16 and heat-shock factor. Science 2003;300:1142–1145. [DOI] [PubMed] [Google Scholar]
- 39.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C: Lifespan extension by conditions that inhibit translation in caenorhabditis elegans. Aging Cell 2007;6:95–110. [DOI] [PubMed] [Google Scholar]
- 40.Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P: Inhibition of mrna translation extends lifespan in caenorhabditis elegans. Aging Cell 2007;6:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curran SP, Ruvkun G: Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 2007;3:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farabaugh PJ: Translational frameshifting: Implications for the mechanism of translational frame maintenance. Prog Nucl Acid Res Mol Biol 2000;64:131–170. [DOI] [PubMed] [Google Scholar]
- 43.Parker J: Errors and alternatives in reading the universal genetic code. Microbiol Rev 1989;53:273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B: Decline of nucleotide excision repair capacity in aging caenorhabditis elegans. Genome Biol 2007;8:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, Nance M, Ross CA, Scahill RI, Wetzel R, Wild EJ, Tabrizi SJ: Huntington disease. Nat Rev Dis Primers 2015;1:15005. [DOI] [PubMed] [Google Scholar]
- 46.Labbadia J, Morimoto RI: Huntington’s disease: Underlying molecular mechanisms and emerging concepts. Trends Biochem Sci 2013;38:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Eyk CL, McLeod CJ, O’Keefe LV, Richards RI: Comparative toxicity of polyglutamine, polyalanine and polyleucine tracts in drosophila models of expanded repeat disease. Human Mol Genet 2012;21:536–547. [DOI] [PubMed] [Google Scholar]
- 48.Mohri-Shiomi A, Garsin DA: Insulin signaling and the heat shock response modulate protein homeostasis in the caenorhabditis elegans intestine during infection. J Biol Sci 2008;283:194–201. [DOI] [PubMed] [Google Scholar]
- 49.Lee AL, Ung HM, Sands LP, Kikis EA: A new caenorhabditis elegans model of human huntingtin 513 aggregation and toxicity in body wall muscles. PLoS One 2017;12:e0173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R: Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol 2006;355:524–535. [DOI] [PubMed] [Google Scholar]
- 51.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R: Polyglutamine disruption of the huntingtin exon 1 n terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol 2009;16:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL: Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell 2004;14:95–104. [DOI] [PubMed] [Google Scholar]
- 53.Labbadia J, Morimoto RI: The biology of proteostasis in aging and disease. Ann Rev Biochem 2015;84:435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


