Abstract
The heterodimeric cytokine interleukin-23 (IL-23 or IL23A/IL12B) is produced by dendritic cells and macrophages and promotes the proinflammatory and regenerative activities of T helper 17 (Th17) and innate lymphoid cells. A recent study has reported that IL-23 is also secreted by lung adenoma cells and generates an inflammatory and immune-suppressed stroma. Here, we observed that proinflammatory tumor necrosis factor (TNF)/NF-κB and mitogen-activated protein kinase (MAPK) signaling strongly induce IL23A expression in intestinal epithelial cells. Moreover, we identified a strong crosstalk between the NF-κB and MAPK/ERK kinase (MEK) pathways, involving the formation of a transcriptional enhancer complex consisting of proto-oncogene c-Jun (c-Jun), RELA proto-oncogene NF-κB subunit (RelA), RUNX family transcription factor 1 (RUNX1), and RUNX3. Collectively, these proteins induced IL23A secretion, confirmed by immunoprecipitation of endogenous IL23A from activated human colorectal cancer (CRC) cell culture supernatants. Interestingly, IL23A was likely secreted in a noncanonical form, as it was not detected by an ELISA specific for heterodimeric IL-23 likely because IL12B expression is absent in CRC cells. Given recent evidence that IL23A promotes tumor formation, we evaluated the efficacy of MAPK/NF-κB inhibitors in attenuating IL23A expression and found that the MEK inhibitor trametinib and BAY 11–7082 (an IKKα/IκB inhibitor) effectively inhibited IL23A in a subset of human CRC lines with mutant KRAS or BRAFV600E mutations. Together, these results indicate that proinflammatory and mitogenic signals dynamically regulate IL23A in epithelial cells. They further reveal its secretion in a noncanonical form independent of IL12B and that small-molecule inhibitors can attenuate IL23A secretion.
Keywords: cytokine induction, inflammation, colorectal cancer, intestinal epithelium, mitogen-activated protein kinase (MAPK), IL23A, MAPK, NF-κB, RUNX, IL23A secretion, intestinal epithelial cells, mitogenic signal, carcinogenesis, non-canonical IL-23, innate immunity, cytokine signaling.
Introduction
In the intestinal epithelium, the maintenance of homeostatic balance between pathogens' surveillance and commensal tolerance is reliant on an intricate network of cytokines. Of these, dendritic cell- and macrophage-derived IL-23 provides a crucial microenvironmental cue for the effector functions of interleukin-17 secreting helper T cells (Th17)4 and innate lymphoid cells that coordinate immune response and tissue repair (1, 2).
IL-23 is a heterodimeric cytokine consisting of IL12B and IL23A (3). It is a member of the IL-12 family of cytokines that also includes IL-12 (IL12A/IL12B), IL-35 (IL12A/EBI3), IL-27 (EBI3/IL27A), and the recently reported member IL-39 (IL23A/EBI3) (4). IL-23 is a primary driver of Th17-mediated immune response during infection and inflammation (5). In humans, IL-23 is critical for Th17 differentiation and activities (6, 7), and a polymorphism in its receptor, IL23R, protects against a range of autoimmune and autoinflammatory conditions (8–11). Importantly, overproduction of IL-23 during chronic inflammation is a potent promoter of cancer incidence and growth (12, 13). However, it is unclear what the cellular sources of IL-23 are, especially in light of recent reports (14–16). Although significant evidence has been presented in the past that dendritic cell- and macrophage-derived IL-23 is indispensable for disease progression and carcinogenesis in mouse models (12, 13, 17), other sources of IL23A/IL-23 have been identified. In particular, IL23A expression has been reported in a number of nonhematopoietic sources, including keratinocytes (18, 19) and gastric (14, 20), intestinal (15), lung (16), and ovarian epithelial cells (12). Indeed, lung adenoma–derived IL-23 was found responsible for the rapid remodeling of the tumor niche into a proinflammatory and immune suppressive microenvironment (16). Similarly, epithelial-derived IL-23 has been attributed to mediating regeneration in DSS-damaged intestinal epithelium (15). In contrast, IL23A is strongly induced in gastric epithelial cells by TNFα and Helicobacter pylori in the absence of IL12B (14).
The potent activities of IL-23 in normal biology and disease have attracted interests in its regulation, especially that of the IL23R-interacting IL23A subunit. Upstream signals directing IL23A expression have been investigated in vitro by several groups. These efforts revealed a central role for NF-κB in the regulation of IL23A initially in leukocytes (21–23), but also in keratinocytes (24) and epithelial cells (14, 15). This is supported by other transcription factors in a context-specific manner (14, 23, 25). Of note, strong synergy was observed between the canonical TNFα/NF-κB pathway and the tumor suppressor RUNX3 in gastric epithelial cells, in concert with the activation of SHP2 by H. pylori (14). In contrast, the transient induction of IL23A in intestinal epithelial cells that is necessary for intestinal regeneration is supported by the noncanonical LTβR/NF-κB pathway (15). These observations highlight the complex role played by NF-κB, in partnership with specific stimuli, in regulating IL23A under different physiological conditions and cell types.
Although the link between IL-23 and carcinogenesis is well-established, its tumorigenic activity is generally considered an adverse consequence of its perpetuation of chronic inflammation (12, 13). Furthermore, these studies have shown that this is sourced from bone marrow–derived dendritic cells and macrophages (12, 13). However, a recent study reports that epithelial-derived IL-23 during Kras/c-Myc–driven lung carcinogenesis potently fashioned an immune-suppressed and proinflammatory niche (16). In addition to describing a new source of IL-23, an important question that arises from that study is how cell-intrinsic driver mutations like KRASG12V may impact tumor immunity through the direct regulation of IL23A. As driver mutations were inherited by each tumor cell, their impact on IL23A expression would increase during tumor progression to alter the immune microenvironment. Therefore, there is a need to identify cell-intrinsic mitogenic signals that regulate IL23A expression in partnership with extracellular cues, as it could uncover novel application for existing therapeutics to interrupt a proinflammatory and immunosuppressive tumor microenvironment.
In the current study, we investigate the upstream signaling that regulates IL23A expression in colorectal carcinoma cells with respect to driver mutations. We observed that the MAPK and the canonical NF-κB pathways, prominent in intestinal homeostasis and carcinogenesis, play key roles in driving IL23A expression in this cell type. This involves a strong crosstalk between these two pathways acting on a proximal promoter enhancer complex consisting of NF-κB, c-Jun, and RUNX1/3. Surprisingly, the expression and secretion of IL23A in this cell type is independent of the IL12B partner subunit. These observations indicate that intestinal epithelial cells secrete IL23A under mitogenic and inflammatory conditions, but not as a heterodimer with IL12B. Lastly, we showed that the secretion of IL23A could be intervened by inhibitors against the MAPK and NF-κB pathways in MAPK mutant colorectal carcinoma cells, which could be of therapeutic potential.
Results
IL23A is widely expressed in human colorectal carcinoma lines
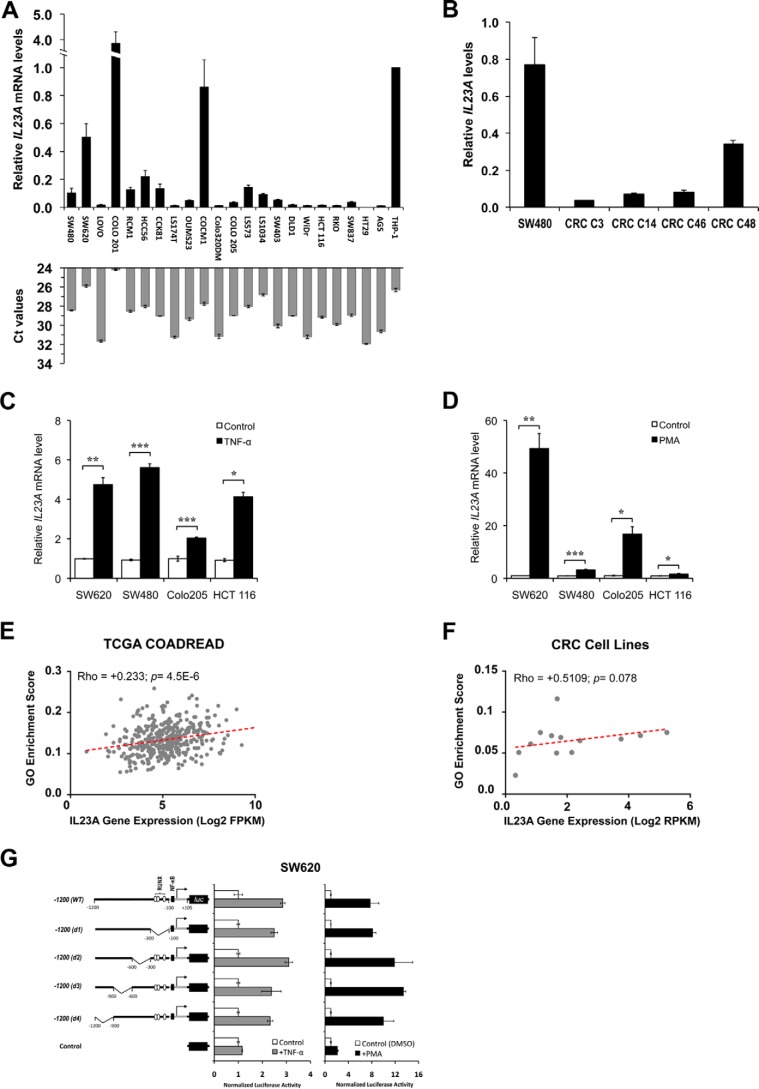
It was previously reported that IL23A is transiently induced in mouse intestinal epithelial cells following dextran sulfate sodium (DSS)–induced injury through the LTβR–NF-κB pathway (15). To investigate if IL23A is expressed in human intestinal epithelial cells, we measured the basal expression of IL23A mRNA in 21 human colorectal carcinoma (CRC) lines. This revealed that IL23A is widely expressed in intestinal epithelial cells of diverse mutation profiles (Fig. 1A). The expression of IL23A is further confirmed in four separate human patient–derived colorectal cancer organoids (Fig. 1B).
Figure 1.
Basal and inducible expression of IL23A expression in CRC lines. A and B, IL23A is widely expressed in diverse human colorectal carcinoma lines and organoids. A, IL23A mRNA levels in 21 CRC cell lines were evaluated by qRT-PCR using an IL23A-specific Taqman probe. IL23A expression is normalized with that of GAPDH and compared against that of AGS human gastric epithelial cells and THP-1 human monocytes, known producers of IL23A (14). The corresponding Ct values of each sample (with a cut-off limit of 35 cycles) are presented in gray bars. Normalised IL23A mRNA levels are expressed relative to that of THP-1. Data are presented as mean ± SEM of four replicates. B, the expression levels of four human CRC-derived organoids relative to that of SW480. Normalised IL23A mRNA levels are expressed relative to that of SW480. Data are presented as mean ± SEM of four replicates. C, TNFα-induced IL23A expression in colorectal cancer cell lines. SW620, SW480, COLO 205, and HCT 116 cells were treated with TNFα (50 ng/ml) for 10 h and harvested for qRT-PCR measurement of IL23A mRNA levels. Normalized IL23A mRNA levels are expressed relative to that of untreated controls. Data are presented as mean ± S.E. of four replicates. D, PMA strongly induced IL23A expression. Four CRC cell lines were treated with PMA (1 μm) for 10 h and harvested for qRT-PCR measurement of IL23A mRNA. Normalized IL23A mRNA levels are expressed relative to those of DMSO-treated control samples (n = 4; mean ± S.E.). E and F, correlation plot of Gene Ontology (GO) MAPK pathway enrichment score (y axis) and IL23A Log2 gene expression value (x axis) in TCGA COADREAD cohort (E) and CRC cell lines (F). Correlation Rho and p value are computed using Spearman's correlation coefficient rank test. G, activating effects of TNFα and PMA are mediated via the proximal promoter. A firefly luciferase reporter construct containing the −1200 to +105 region of the IL23A promoter and its deletion variants were transiently transfected into SW620 cells, which were treated with TNFα (50 ng/ml) or PMA (1 μm) for 8 h and harvested for luciferase assay. Normalized luciferase activities are expressed relative to the values of samples transfected with an empty control vector. Data presented are as mean ± S.E. from four replicates. The p values are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
It has been reported that IL23A is under the regulation of canonical NF-κB pathway in gastric epithelial cells (14). To test if this pathway promotes IL23A expression in intestinal epithelial cells, four CRC lines, SW620, SW480, COLO 205, and HCT 116, were treated with TNFα for 24 h. This resulted in the significant induction of IL23A in all four lines (Fig. 1C), confirming that the canonical NF-κB pathway regulates IL23A in this cell type, in addition to the noncanonical LTβR–NF-κB pathway previously reported (15).
The mitogenic KRAS-BRAF-MAPK pathway is often aberrantly activated by somatic mutations during intestinal carcinogenesis (26, 27). To determine whether this central oncogenic pathway regulates IL23A, CRC cells were treated with phorbol 12-myristate 13-acetate (PMA), a pharmacological activator of protein kinase C (PKC) and the MAPK pathway (28). Treatment with PMA strongly induced IL23A expression in the tested CRC lines to levels much higher than those following TNFα treatment (Fig. 1D). Concordant with this, IL23A expression level is positively correlated to the activation of MAPK in the TCGA COADREAD gene expression dataset (Fig. 1E) and a published panel of CRC cell lines (Fig. 1F) (29).
To investigate the molecular mechanisms underlying the inducible expression of IL23A, we generated a firefly reporter gene construct containing the −1200 to +105 region of the IL23A promoter and transfected it into CRC lines. In SW620 and other CRC lines, this reporter construct effectively recapitulated the induction of IL23A promoter (Fig. 1G and Fig. S1). Sequential deletion of four upstream promoter regions did not affect PMA and TNFα responsiveness of this reporter construct, indicating that the necessary cis-acting elements reside within the proximal promoter (nucleotides −100 to +105) (Fig. 1G).
AP-1, NF-κB, and RUNX3 contribute to basal and induced IL23A transcription
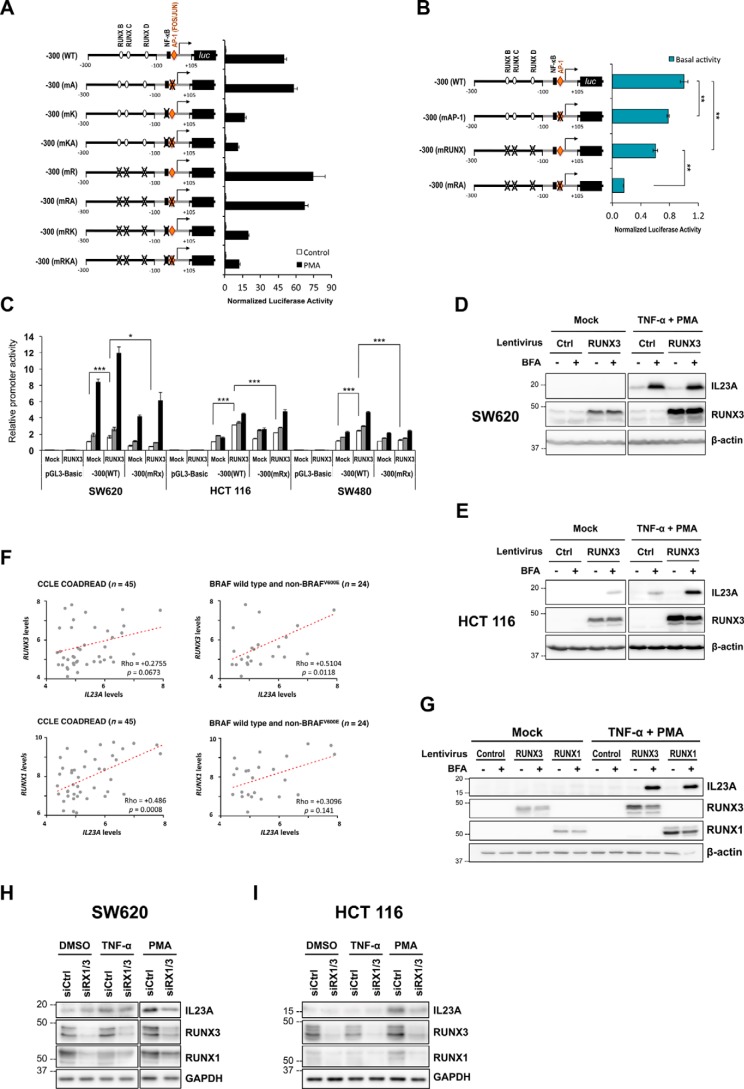
Detailed examination of the minimal IL23A promoter region (30–32) identified a putative AP-1 (FOS/JUN)-binding site (Fig. S2), a known effector of the MAPK pathway (33). To study its functional contribution, a series of mutant reporter constructs was generated (Fig. 2A), in which the AP-1 site was mutated in combination with previously validated NF-κB site and RUNX sites (14). In SW620 cells, the mutation of the consensus AP-1 site alone did not significantly affect the PMA-responsiveness of the minimal IL23A promoter (Fig. 2A). However, the simultaneous disruption of the NF-κB (mKA) synergistically attenuated PMA induction (Fig. 2A). In addition, the AP-1 site cooperated strongly with the RUNX sites to maintain basal IL23A promoter activity (Fig. 2B). These results indicate that the AP-1 and RUNX sites contribute to the strong baseline IL23A expression in SW620 cells (Fig. 1A). Concurrently, the AP-1 site primes the promoter for cooperative induction by PMA through the NF-κB site.
Figure 2.
cis- and trans-elements required for the basal and induced IL23A expression. A, SW620 cells were transiently transfected with a series of reporter constructs containing the WT minimal IL23A promoter (−300 to +105) or mutant variants, in which AP-1 (mA), NF-κB (mK) and three functional RUNX sites (mR) are mutated in combinations as indicated. Transfected cells were rested for 18 h post transfection and treated with DMSO (Control) or PMA (50 ng/ml). After 8 h of treatments, the cells were harvested for luciferase assays. Normalized luciferase values are expressed relative to DMSO-treated values for each reporter construct and presented as mean ± S.E. (n = 4). B, the effect of AP-1 and RUNX site mutations on basal promoter activity in the same experiment as (A). C, ectopic RUNX3 increased the basal activity of the WT −300 (WT) reporter construct and augmented its TNFα and PMA responsiveness in SW620, HCT 116, and SW480 cells. Transfected cells were rested for 24 h and treated with TNFα (50 ng/ml, with DMSO) or PMA (1 μm) or DMSO (Mock). After 8 h of treatment, they were harvested for luciferase reporter assay. Firefly (SW620) or normalized (HCT 116 and SW480) luciferase activities are expressed relative to basal −300 (WT) reporter values and charted as mean ± S.E.; n = 4. D and E, ectopic RUNX3 has differential effects on IL23A protein expression. SW620 cells (D) and HCT 116 cells (E) were transduced with the indicated lentiviruses for 48 h and treated with TNFα (5 ng/ml) or PMA (20 ng/ml) for another 24 h. Cell lysates were subjected to Western blotting for the detection of IL23A, RUNX3, and β-actin (loading control). F, correlation between the expression levels of IL23A and RUNX3 (top) and RUNX1 (bottom) in all CRC lines profiled by the CCLE (left) and those with WT or non-BRAFV600E mutant (i.e. MAPKlow). Spearman's correlation coefficient test was used to assess correlation of gene expressions. G, RUNX1 synergizes with PMA/TNFα to induce IL23A. HCT 116 cells were transduced with Control, RUNX1, or RUNX3 lentiviruses for 48 h and treated with TNFα (5 ng/ml) or PMA (20 ng/ml) for another 24 h. Cell lysates were subjected to Western blotting for the detection of IL23A, RUNX3, RUNX1, and β-actin (loading control). The p values are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. H and I, effects of RNAi of RUNX1/3 on basal, TNFα-, or PMA-induced IL23A expression. SW620 (H) and HCT 116 (I) cells were transfected with either a nontargeting Control or RUNX1 and RUNX3 siRNAs. Cells were rested for 48 h and treated with PMA for 24 h. Cells were treated with brefeldin A for 18 h prior before they harvested for Western blot analyses for the detection of IL23A, RUNX3, RUNX1, and GAPDH (loading control). The two panels in (H) represent two exposures of a single membrane to account for the strong induction of IL23A by PMA in SW620 cells.
As RUNX3 is a crucial regulator of IL23A expression in gastric epithelial cells during inflammation and H. pylori infection (14, 34), a RUNX3 expression construct was co-transfected with the reporter constructs in SW620, HCT 116, and SW480 cells. This significantly increased basal IL23A promoter activities (Fig. 2C), which was abolished when three previously validated RUNX-binding sites (14) were mutated (Fig. 2C). Moreover, the presence of ectopic RUNX3 augmented the induction of IL23A promoter by TNFα and PMA (Fig. 2C). However, this modest effect is unlike the strong cooperativity previously observed between RUNX3 and TNFα/NF-κB in gastric epithelial cells, indicating tissue-specific differences (14).
To determine the overall contribution of RUNX3 to IL23A expression, CRC lines were transduced with RUNX3-encoding lentiviruses and treated with TNFα and PMA. In SW620 cells that express high levels of endogenous RUNX3 (34), ectopic expression of RUNX3 had little impact on basal or PMA/TNFα-induced IL23A protein expression (Fig. 2D). In contrast, in HCT 116 where RUNX3 expression is low, exogenous RUNX3 enhanced basal and PMA/TNFα-induced IL23A protein expression (Fig. 2E). Moreover, a strong increase in intracellular IL23A protein was observed following the blockade of protein export by brefeldin A, indicating that IL23A is actively secreted (Fig. 2, D and E).
To gain a clearer picture of the context specificity of RUNX3's involvement in epithelial IL23A expression cells, we correlated the basal expression levels of RUNX3 and IL23A in 45 human CRC cell lines characterized by the Cancer Cell Line Encyclopedia (CCLE) (35). This revealed that RUNX3 expression is more closely correlated to IL23A expression in CRC lines with WT BRAF or non-BRAFV600E mutations (n = 24) and therefore low intrinsic MAPK activities, whereas overall correlation is not statistically significant (Fig. 2F). In contrast, RUNX1 expression is significantly correlated to IL23A levels overall but surprisingly not in CRC lines with low MAPK activities. These data implicate RUNX3 to be engaged for the maintenance of basal IL23A transcription when intrinsic MAPK activity is low, whereas RUNX1 is utilized when background MAPK activity is high (Fig. 2F). To demonstrate an involvement of RUNX1, we compared the effects of exogenous RUNX1 and RUNX3 in inducing IL23A in HCT 116 line. This showed that RUNX1 is functionally comparable with RUNX3 in mediating PMA/TNFα induction of IL23A protein (Fig. 2G). Moreover, although the difference is subtle, RUNX1 appears to increase basal IL23A more than RUNX3 (Fig. S3). Similarly, the overexpression of RUNX1 had a stronger effect than RUNX3 in SW480 and SW620 cells (Fig. S4)
Lastly, to confirm the involvement of endogenous RUNX1 and RUNX3 in IL23A regulation, we conducted RNAi knockdown experiments in SW620 and HCT 116 cells. These show that the knockdown of RUNX1/3 in both cell lines significantly attenuated the induction of IL23A by PMA while not having clear effects on basal or TNFα-induced IL23A expression (Fig. 2, H and I).
NF-κB and MAPK pathways cooperate to transcriptionally regulate IL23A
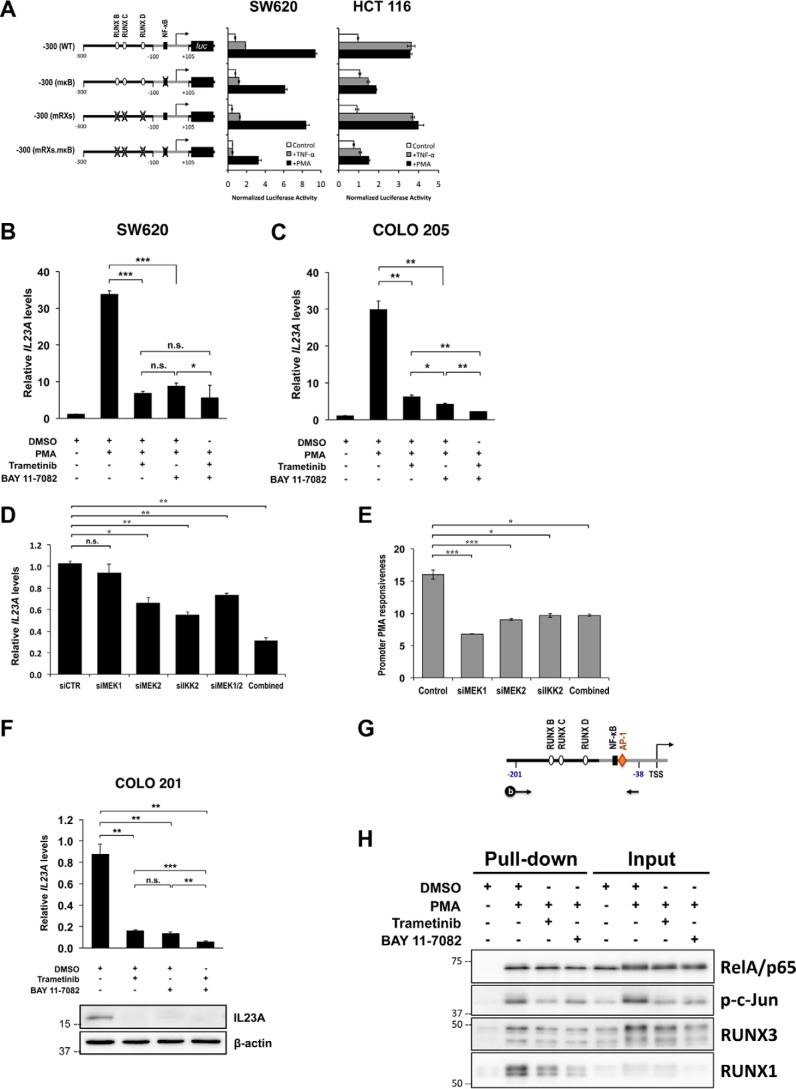
The mutation of the NF-κB site appeared to reduce the PMA responsiveness of the IL23A promoter in Fig. 2A. This was confirmed in SW620 and HCT 116 cells, where the mutation of NF-κB site attenuated PMA and TNFα responsiveness (Fig. 3A). Crosstalk between the MAPK and NF-κB pathways is known to occur downstream of protein kinase C (36, 37). To assess their functional interplay in the regulation of IL23A in CRC cells, we employed inhibitors against MEK1/2 (trametinib) and IKKα/IκB (BAY 11-7082) at doses that displayed little cytotoxicity (Fig. S5). Notably, trametinib and BAY 11-7082 were equally effective in blocking PMA induction of IL23A mRNA in SW620 cells (Fig. 3B). Moreover, combined treatment of both inhibitors resulted in no further blockade, indicating strong cooperativity between the two pathways (Fig. 3B). A similar pattern was observed in COLO 205 cells, indicating that the cooperation between these two pathways is a recurrent mechanism in the regulation of IL23A in intestinal epithelial cells (Fig. 3C).
Figure 3.
Strong cooperation between the MAPK/MEK and NF-κB pathways in the transcriptional regulation of IL23A in intestinal epithelial cells. A, SW620 and HCT 116 cells were transiently transfected with WT reporter construct or variants with mutated RUNX and NF-κB sites (as indicated) and rested for 24 h. Subsequently, cells were treated with TNFα (50 ng/ml) or PMA (1 μm) for 8 h and harvested for luciferase assays. Normalized luciferase activities are expressed relative to those of untreated control samples (mean ± S.E.; n = 4). B and C, SW620 (B) and COLO 205 (C) were pretreated with either DMSO (Mock), trametinib (10 nm), BAY 11-7082 (10 μm), or in combination for 2 h before treatment with PMA (1 μm) or DMSO (Mock) for 24 h. Changes in IL23A mRNA levels were measured by qRT-PCR, normalized against corresponding GAPDH values, and presented relative to those of untreated controls (mean ± S.E.; n = 4). D and E, effects of RNAi of MEK1/MEK2 and the IKKα/β complex. D, SW620 cells were transfected with the indicated siRNAs. Cells were rested 36 h post transfection for effective RNAi knockdown and then treated with PMA for 10 h and harvested for qRT-PCR analyses. IL23A mRNA levels were normalized against 18S rRNA and expressed relative to the DMSO-treated control (siCTR) (mean ± S.E.; n = 3). E, WT −300 (WT) reporter construct was co-transfected with the indicated siRNAs into SW620 cells. Cells were rested 36 h post transfection for effective RNAi knockdown and then treated with PMA for 12 h and harvested for luciferase assay. The data presented are -fold increases in relative promoter activity following PMA treatment, relative to the DMSO-treated samples (mean ± S.E.; n = 3). F, COLO 201 were treated with DMSO (mock), trametinib (10 nm), BAY 11-7082 (10 μm), or in combination for 24 h and harvested for qRT-PCR measurements and Western blotting. Expression of IL23A mRNA was normalized against GAPDH values and charted relative to mock values (mean ± S.E.; n = 4). The p values are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. G and H, promoter pulldown assay reveals a PMA-inducible transcription enhancer complex at the proximal IL23A promoter. G, schematics illustrating the coupling of a 5′ biotinylated amplicon spanning the −201 to −38 region of the IL23A proximal promoter to paramagnetic beads and used to enrich RelA/p65, p-c-Jun, RUNX3, and RUNX1 from nuclear extracts prepared from SW620 cells treated as indicated. H, PMA-induced nuclear p-c-Jun, RUNX3, and RUNX1 and their binding to the IL23A promoter fragment strongly enhanced RelA/p65 recruitment. Co-treatment with trametinib and BAY 11-7082 attenuated to varying degrees the induction of these transcription factors, thereby disrupting the formation of the transcription enhancer complex.
To confirm the involvement of MEK1/2 and the IKK complex, RNAi knockdown experiments were performed in SW620 cells. The knockdown of MEK2 and IKK2, but not MEK1, significantly reduced IL23A mRNA expression in resting cells (Fig. 3D). In comparison, the knockdown of the three kinases individually or in combination effectively reduced PMA responsiveness of the IL23A promoter in reporter assays (Fig. 3E). To clarify the involvement of MEK1, we treated SW620 cells with two additional MEK1/2 inhibitors, PD98059 and PD0325901. Our data show that both inhibitors were efficacious, attenuating PMA induction to a similar degree to trametinib (Fig. S6). Of note, PD98059 has a greater specificity against MEK1 (IC50 4 μm) than MEK2 (IC50 50 μm). We observed that PD98059 effectively blocked PMA at 10 μm, providing additional evidence for MEK1 involvement (Fig. S6). Overall, our data indicate that both MEK1 and MEK2 are utilized downstream of PMA to transactivate the IL23A promoter.
The effects of intrinsic mutations that activate MAPK pathway on basal IL23A transcription were investigated next. For this, resting CRC cells were treated with inhibitors. In COLO 201 cells, which carry the BRAFV600E mutation, trametinib and BAY 11-7082 strongly suppressed basal IL23A mRNA and protein expression (Fig. 3F), mimicking their effects in blocking PMA induction (Fig. 3, B and C). Collectively, these data clearly show that MAPK/MEK and IKK/NF-κB cooperate to maintain basal and induced IL23A expression.
PMA induces a transcription enhancer complex for the recruitment of NF-κB
To understand the molecular mechanism underlying the MAPK–NF-κB cooperation, we conducted affinity pulldown assays with a 164-bp biotinylated IL23A promoter fragment that spans the RUNX, NF-κB, and AP-1 sites (Fig. 3G). By conjugation with paramagnetic beads, this promoter fragment readily enriched RelA/p65 (NF-κB), phosphorylated c-Jun (p-c-Jun; AP-1), RUNX3, and RUNX1 from SW620 nuclear extracts (Fig. 3H). Treatment with PMA markedly increased the binding of RelA/p65, p-c-Jun, RUNX3, and RUNX1 to the promoter fragment (Fig. 3H). Interestingly, increased RelA/p65 binding occurred without a significant change in nuclear RelA/p65 level. In contrast, PMA strongly induced nuclear p-c-Jun, RUNX3, and, to a lesser extent, RUNX1, which explains their increased binding. These data suggest that PMA increases nuclear p-c-Jun, RUNX3, and RUNX1, and their binding to the promoter fragment in turn recruits the relatively abundant RelA/p65 (Fig. 3H).
Concordant with the literature, treatment with trametinib attenuated the induction of p-c-Jun by PMA (38–40). Likewise, BAY 11-7082 reduced both the nuclear accumulation and the binding of NF-κB, in line with its known activity. In addition, both trametinib and BAY 11-7082, significantly blocked PMA induction of RUNX3 and RUNX1, resulting reduced promoter binding. These observations suggest that BAY 11-7082 inhibits PMA-induced IL23A transcription by acting on multiple transcription factors.
Collectively, these observations support a molecular mechanism whereby PMA induces p-c-Jun, RUNX3, and RUNX1 to form a transcription enhancer complex at the proximal IL23A promoter that efficiently recruits NF-κB, hence providing a molecular basis for the cooperativity between the MAPK and NF-κB pathways. Moreover, components of this enhancer complex were effectively targeted by trametinib and BAY 11-7082, which provides an important link between the efficacies of these inhibitors in blocking PMA induction of IL23A (Fig. 3B) and the functional contributions of the proximal transcription factor binding sites (Figs. 2A and 3A and Fig. S7).
IL23A is secreted by intestinal epithelial cells independent of IL12B
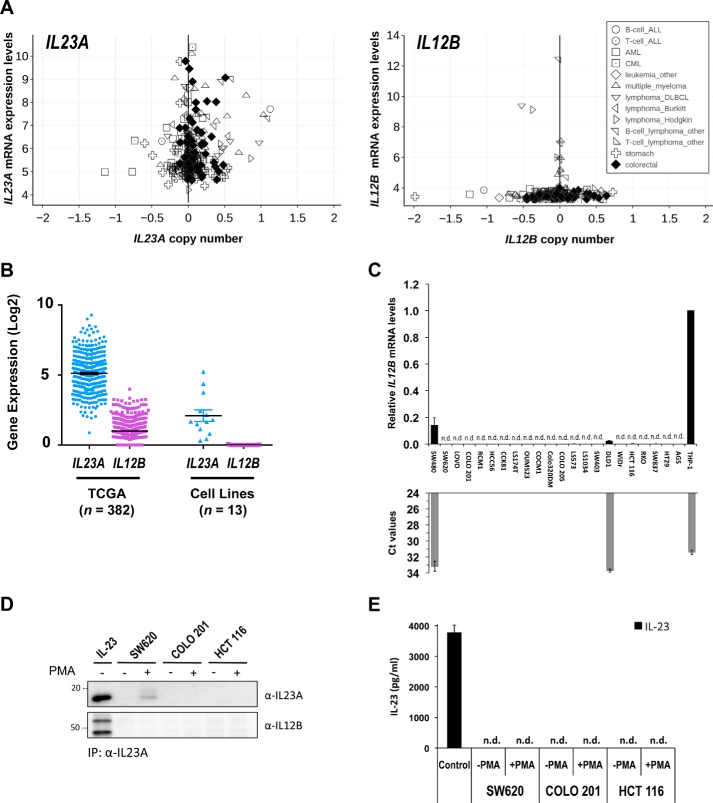
Having observed strong accumulation of intracellular IL23A protein in induced SW620 and HCT 116 cells, we sought to clarify the nature of the secreted IL23A. Canonically, IL23A is secreted as a heterodimer with IL12B by macrophages and dendritic cells. However, IL23A expression is also widely observed in epithelial cells of different tissues profiled by the CCLE, including intestinal epithelial cells (Fig. 4A). Of note, the relative expression levels of IL23A in epithelial cell lines are comparable with those of hematopoietic lines (Fig. 4A). In stark contrast, the expression of IL12B and EBI3, another potential binding partner, is far more restricted (Fig. 4A and Fig. S8). Similarly, the expression of IL23A is substantially higher than that of IL12B in mixed tissue samples from the TCGA database or in a published panel of CRC cell lines (Fig. 4B). To provide a more definite assessment of IL12B expression levels, we conducted quantitative RT-PCR assays with IL12B-specific Taqman hydrolysis probe in the same CRC lines used in Fig. 1A. This confirms that the IL12B transcript is either completely absent in intestinal cells or detectable at far lower levels than in THP-1 monocytes (Fig. 4C). Similarly, EBI3 expression was very low or undetectable by Taqman qRT-PCR in resting or PMA-induced CRC cells (Fig. S9 and Fig. S10). These observations suggest that IL23A is secreted independent of IL12B and EBI3. To check this, we performed immunoprecipitation of endogenous IL23A from the supernatant of uninduced or PMA-induced SW620, COLO 201, and HCT 116 cells. Using a human IL23A-specific antibody, endogenous IL23A was readily precipitated from supernatants of PMA-induced SW620 cells, but not in COLO 201 and HCT 116 cells, likely because of the high concentration necessary for effective immunoprecipitation. As a positive control, we prepared supernatant from transfected HEK293T cells expressing exogenous IL23A and IL12B (Fig. 4D). Consistent with the lack of IL12B mRNA expression in the CRC lines, IL12B was not co-immunoprecipitated with IL23A (Fig. 4D). To further demonstrate that IL23A is secreted independent of IL12B, these supernatants were analyzed by “sandwich” ELISA specific for IL23A/IL12B, which readily detected the secretion of exogenous and endogenous IL-23 (Fig. 4E and Fig. S11). Strikingly, canonical IL23A/IL12B was completely absent in the supernatants of activated SW620, COLO 201, or HCT 116 cells. Collectively, these data provide evidence that intestinal epithelial cells secrete a novel form of IL23A that is activated by mitogenic and inflammatory signals, independent of IL12B and in the absence of EBI3.
Figure 4.
CRC cells secrete IL23A in the absence of IL12B. A, relative expression levels of IL23A and IL12B in selected cancer cell lines. Normalized RNA-Seq data derived from cancer cell lines of hematopoietic, gastric, and colorectal (large intestine) lineages were extracted from the CCLE database (35) and charted against copy number. B, dot plot of IL23A and IL12B gene expression levels (y axis; mean ± S.E.) in TCGA COADREAD cohort (left panel; n = 382) and CRC cell line (right panel; n = 13). C, IL12B mRNA levels were evaluated by qRT-PCR using a specific Taqman hydrolysis probe. IL12B expression is normalized against that of GAPDH and compared with that of AGS human gastric epithelial cells and THP-1 human monocytes, the latter is known for producing IL23A/IL12B. The corresponding Ct values of every sample (with a cut-off limit of 35 cycles) are presented in gray bars. D, immunoprecipitation of endogenous IL23A from the culture supernatants of activated SW620, COLO 201, and HCT 116 cells. CRC cells were treated with DMSO (mock) or PMA (100 ng/ml) for 36 h before collection for immunoprecipitation (IP) by a monoclonal IL23A-specific antibody. The immunoprecipitation of IL23A and co-immunoprecipitation of IL12B were measured by reducing (IL23A) and nonreducing (IL12B) Western blotting. Culture supernatant of HEK293T cells expressing exogenous IL23A and IL12B was used as a positive control for canonical IL-23. E, the same supernatants used in immunoprecipitation experiment (C) were analyzed by ELISA for the presence of IL-23 (IL23A/IL12B).
Discussion
Immunity in the periphery is reliant on a finely tuned network of cytokines and chemokines contributed by cells of different lineages. Recent advances in cancer genome sequencing and gene expression profiling has deepened our appreciation of tumor heterogeneity in terms of the multi-clonality within a tumor and its complex cellular composition. Nevertheless, the contribution of epithelial cell–derived cytokines to inflammation, infection, and tumor immunity remains underappreciated. Recent studies have pointed to the expression and secretion of IL23A by the epithelial cells of several tissues, including gastric (14, 20), intestine (15, 41), and lung epithelial cells (16).
In the intestine, a previous study reported that epithelial expression of IL23A was triggered by DSS-induced damage in a manner dependent on lymphotoxin β receptor (LTβR) and the noncanonical NF-κB pathway. This in turn instructs the secretion of IL-22 by type 3 innate lymphoid cells, as part of a tissue regeneration program (15). Hence, IL23A serves a protective function against intestinal injury. However, this beneficial role of IL-23 in intestinal repair was recently reported to have potent tumor-promoting activities during Kras/c-Myc–mediated lung carcinogenesis (16). Epithelial-derived IL-23 was identified as an effector that enables lung adenoma cells to fashion a tumor stroma that greatly suppressed tumor immunity (16). Given these contrasting activities, it is important to understand how IL23A is regulated by inflammatory and mitogenic signals in epithelial cells.
In the current study, we identified the canonical NF-κB and MAPK pathways as two important drivers of IL23A expression in intestinal epithelial cells. The TNFα/NF-κB pathway is a central mediator of intestinal inflammation and its direct involvement further implicates IL23A to be part of an epithelial response toward intestinal inflammation and infection. On the other hand, the activation of MAPK is a key event downstream of growth factor signaling and hence its involvement couples the expression IL23A with tissue growth and regeneration. Importantly, the KRAS-MAPK pathway is frequently targeted by activating mutations during intestinal carcinogenesis (26, 27). Therefore, it is possible that epithelial cell–intrinsic driver mutations may lead to aberrant production of IL23A during intestinal carcinogenesis.
We further observed a significant crosstalk between the MAPK and the NF-κB pathways. There have been numerous reports of crosstalk between PKC-MAPK and the NF-κB pathways, involving interplay between several MAPK signaling axes, including PKCθ (42–44) and JNK (45), as reviewed elsewhere (46, 47). Here, we observed that the activation of IL23A promoter by PMA is dependent in part on an intact NF-κB site in the proximal region of the IL23A promoter. Furthermore, PMA induction of IL23A mRNA is strongly attenuated by the IKKα/IκB inhibitor BAY 11-7082 in some CRC lines. Confirming the involvement of MEK1/2, PMA induction was similarly blocked by several MEK1/2 inhibitors, including trametinib. Tellingly, the combined treatment of trametinib and BAY 11-7082 achieved little or no additional blockade, indicating a common mechanism downstream of MEK1/2 and IKK. Our investigation into the underlying molecular mechanism reveals functional partnership between closely situated AP-1, NF-κB, and RUNX binding sites on the IL23A promoter. Promoter pulldown analyses further revealed that PMA induces p-c-Jun, RUNX3, and RUNX1 to facilitate the formation of a proximal transcription enhancer complex that strongly recruits NF-κB. These observations provide a molecular basis to the observed responsiveness of IL23A to mitogenic and inflammatory signals and the inhibitory effects of trametinib and BAY 11-7082. In addition, we observed that PMA induced nuclear RUNX3 and RUNX1. Although ERK phosphorylation of RUNX1 has been reported in the past (48–50), our data suggest that it could promote RUNX nuclear localization, as total cellular RUNX3 levels have remained largely unchanged over a 12-h treatment (Fig. S12).
Trametinib and BAY 11-7082 were also effective in attenuating basal IL23A expression in certain CRC lines with activating MAPK mutation, such as COLO 201 (BRAFV600E). This indicates that heightened MAPK signaling in these cells maintains the constitutive expression of IL23A. It is possible that cell-intrinsic mutations within intestinal carcinoma cells could fuel aberrant IL23A secretion into the tumor microenvironment and alter tumor immunity. As such, these inhibitors have the potential to intervene excessive epithelial IL23A production to restore tumor immunity or augment immune checkpoint therapy, in addition to their demonstrated effects on T cells (51). Of note, trametinib is in Phase III clinical trials with BRAF inhibitors (e.g. dabrafenib) to treat colorectal cancer patients with activating BRAFV600E mutation (52, 53). Although it remains unclear if epithelial-derived IL23A suppresses tumor immunity as observed in mouse models of lung cancer (16), the effectiveness of trametinib and BAY 11-7082 in blocking IL23A production offers a timely example on how classical small chemical inhibitors could be deployed in combination with immune checkpoint therapeutics.
There remains ambiguity in the nature of the IL23A secreted by epithelial cells. Recent studies in the intestine and lung report that epithelial cells secrete the canonical IL23A/IL12B heterodimer. However, only the expression of IL23A was clearly demonstrated in the epithelial cells (15, 16). In both studies, the interpretation of the data is complicated by the presence of other sources of IL23A/IL12B within the tissue microenvironment, such as macrophages. The IL-23 measured in whole tissue homogenates by ELISA or immunoblotting cannot be conclusively attributed to epithelial origins because they contained leukocytes. Moreover, as these assays are designed specifically for the IL23A/IL12B heterodimer, they cannot detect IL23A secreted in any other form. Most importantly, the production of IL23A/IL12B would be inconsistent with the absence of IL12B mRNA expression in human intestinal epithelial cells. In this study, we observed a striking difference between the expression pattern of IL23A and those of IL12B and EBI3. Whereas IL23A expression was widely expressed in CRC cell lines, at levels comparable with human monocytes, IL12B, in particular, was conspicuously absent. This was further supported by our analysis of CCLE and TCGA expression datasets.
To provide direct evidence of IL23A secretion, we successfully immunoprecipitated endogenous IL23A from culture supernatants of activated CRC cells. Consistent with the absence of IL12B expression, IL12B was not co-immunoprecipitated with IL23A. Moreover, despite being present at high levels, the secreted IL23A could not be detected by IL-23–specific ELISA. Contrary to the saturating values from exogenous IL23A/IL12B produced by transfected HEK293T cells and the clear detection of endogenous IL23A/IL12B from THP-1 monocytes, the values from CRC supernatants are indistinguishable from background noise. Collectively, these observations suggest that the intestinal epithelial cells secrete a form of IL23A that is independent of IL12B. Although yet to be fully validated in vivo, such a notion would make sense as the tissue microenvironment already has prolific producers of canonical IL-23, as highlighted above. Therefore, the production of a distinct form of IL-23, under the regulation of common inflammatory and mitogenic cues, would ensure timely feedback from epithelial cells in the maintenance of homeostatic balance during inflammation and repair. Such a possibility should be taken into account when we contemplate the enigmatic association of epithelial-derived IL23A with tissue regeneration and cancer.
Experimental procedures
Cell culture and treatment
The human CRC cell lines SW620, SW480, COLO 201, COLO 205, and HCT 116 were acquired from the ATCC (Manassas, VA) maintained in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin-streptomycin, and 2 mm l-glutamine under standard cell culture conditions. During induction studies, these lines were treated with either TNFα (50 ng/ml) (PeproTech, Rocky Hill, NJ) or PMA (at indicated concentrations) (Tocris Bioscience, Bristol, UK) for 10 h prior to qRT-PCR analysis. For reporter gene assays, transiently transfected cells were treated at 24 h post transfection with either TNFα or PMA for 8–12 h and harvested for firefly and Renilla luciferase assays. For inhibitor experiment, cells were pretreated for 2 h with trametinib (10 nm) (Selleck Chemicals, Houston, TX) or BAY 11-7082 (10 μm) (Tocris), PD98059 (10 μm) (Tocris), PD0325901 (1 μm) (Sigma-Aldrich) or in combination. Subsequently, cells were treated with either TNFα or PMA for 12 or 24 h and harvested for qRT-PCR or Western blotting analyses, respectively. The pBOBI-RUNX3 and pBOBI-RUNX1 lentiviral transfer vectors encoding the p44 isoform of RUNX3 and isoform 1B of RUNX1 were generated as described in the supporting information. Lentiviruses were generated in HEK293T cells by co-transfecting pBOBI-RUNX3, pBOBI-RUNX1, or nonencoding pBOBI-control transfer vectors with ViraPower Lentiviral Packaging Mix (Thermo Fisher Scientific), following a protocol described previously (54).
Human colorectal tumor organoids
Human CRC organoids were cultured in conditions detailed in Ref. 55. Briefly, mechanically disrupted organoids were embedded in 30 μl of Matrigel (Corning, New York) and seeded in 48-well plates. Following polymerization, the mixture was overlaid with 300 μl of Advanced DMEM/F-12 medium (Thermo Fisher Scientific) supplemented with 1% penicillin-streptomycin, GlutaMAX (Thermo Fisher Scientific), B27 supplement (Thermo Fisher Scientific), 10 mmol/liter HEPES (Sigma-Aldrich), 10 nmol/liter gastrin (Sigma-Aldrich), 1 mmol/liter N-acetylcysteine (Sigma-Aldrich), 50 ng/ml mouse EGF (Thermo Fisher Scientific), 100 ng/ml murine Noggin (PeproTech), 500 nmol/liter A83–01 (Sigma-Aldrich), 10 μmol/liter SB202190 (Sigma-Aldrich), and 1:1 conditioned medium prepared from L-fibroblasts stably expressing R-spondin1/Wnt3a/noggin (CRL-3276, ATCC; a kind gift from Dr. Kazuhiro Murakami). Human CRC tissues were obtained from the Ishikawa Prefecture Hospital where informed and written consents were obtained prior to specimen collection. The protocol used in this study was approved by the ethics committees at Ishikawa Prefecture Hospital (Approval Number: 1166) and Kanazawa University (Approval Number: 2016-086(433)). This study abides by the Declaration of Helsinki principles.
Promoter pulldown assay
Promoter pulldown assay is performed using a protocol described in Ref. 56 and adapted for the use of a longer promoter fragment, as described in Ref. 37. A 164-nucleotide 5′ biotinylated PCR amplicon containing the nucleotide −201 to −38 region of the proximal IL23A promoter was generated using oligonucleotide primers (Table S1) and the −300 (WT) or −300 (mRKA) minimal reporter constructs as template DNA. The purified amplicons were coupled to streptavidin-coated paramagnetic beads (Dynabeads M-280, Thermo Fisher Scientific) as per the accompanying instructions. Nuclear extracts were prepared from SW620 cells treated with DMSO or PMA (50 ng/ml), together with trametinib (10 nm) or BAY 11-7082 (10 μm) for 5 h, using the method described by Li et al. (57). Nuclear extracts (30 μg) were pre-incubated for 10 min with 0.2 μg/ml of poly(dI-dC) (Santa Cruz Biotechnology) and 0.8 μg/ml of sonicated salmon sperm DNA (Thermo Fisher Scientific) in binding buffer (20 mm Hepes, pH 7.9, 2 mm MgCl2, 50 mm NaCl, 1 mm DTT, 20% glycerol). DNA-bound paramagnetic beads were added and incubated with the binding reaction for 30 min. The beads were then washed three times in binding buffer with 0.05% Tween 20 (Sigma-Aldrich) and analyzed by Western blotting.
Reporter gene assay
The firefly luciferase reporter vectors (pGL3-Basic, Promega, Madison, WI) containing the long and short IL23A promoters, and their deletion or mutation variants were generated as described in the supporting information. These reporter constructs were transfected into SW620, SW480, COLO 201, COLO 205, and HCT 116 cells together with a modified Renilla luciferase encoding vector (phRL-SV40, Promega), and control or RUNX3 expression constructs using FuGENE HD (Promega) or Lipofectamine 2000 (Thermo Fisher Scientific) for 24 h. Firefly luciferase activities were determined using the Dual Luciferase Reporter Assay system (Promega), following the accompanying instructions. Firefly luciferase data were normalized against the corresponding Renilla luciferase values and presented as graphs. In RNAi experiments, cells were co-transfected with 40–60 nmol/ml siRNA against MEK1 (MAP2K1, SI00300699; Qiagen, Hilden, Germany), MEK2 (MAP2K2, SI02225090; Qiagen), IKK2 (IKBKB, 280309; Dharmacon, Lafayette, CO) and a nontargeting control siRNA (SIC001, Sigma-Aldrich). Cells were rested for 36 h and treated with PMA or DMSO for a further 24 h.
Quantitative RT-PCR
Total RNAs were isolated using the RNeasy Mini Kit (Qiagen) or ISOGEN (Nippon Gene; Toyama, Japan). Complementary DNA was synthesized with the iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad Laboratories; Hercules, CA). Quantitative PCR was conducted with the Precision Fast 2× qPCR MasterMix or SYBR Green MasterMix (Primerdesign; Southhampton, UK) using gene-specific Taqman hydrolysis probes (Thermo Fisher Scientific) or oligonucleotide primers, respectively (Table S1). All procedures were performed in accordance with standard manufacturer's instructions. Samples were analyzed on an ABI QuantStudio 3 Real Time PCR machine (Thermo Fisher Scientific) or AriaMx Real-Time PCR System (Agilent Technologies; Santa Clara, CA). The expression levels of specific mRNA are normalized against those of GAPDH and presented as graphs.
Western blotting and immunoprecipitation
Whole-cell lysates were resolved in SDS-polyacrylamide gel and immunoblotted with the following antibodies at their respective dilutions: anti-RUNX3 (1:2500; R3–5G4, MBL; Nagoya, Japan), anti-IL23A (1:2000; eBio 473P19, Thermo Fisher Scientific), anti-IL23A (1:500; clone C-3, Santa Cruz Biotechnology; Dallas, TX), anti-IL12B (1:1000; clone C8.6, Thermo Fisher Scientific), anti-β-actin (1:10,000; clone AC-74, Sigma-Aldrich), anti-phospho-c-Jun (1:1000; 3270, Cell Signaling Technology, Danvers, MA); anti–NF-κB p65 (8242, Cell Signaling Technology); anti-RUNX1 (clone 1H9); anti-mouse IgG-HRP (1:5000; GE Healthcare Life Sciences), and anti-rabbit IgG-HRP (1:20,000; GE Healthcare Life Sciences). Cells were transduced with lentiviruses encoding RUNX3 or RUNX1 for 48 h prior to treatment with TNFα or PMA for 24 h. To inhibit protein secretion, cells were treated with brefeldin A (Thermo Fisher Scientific) 12–18 h prior to harvesting. Immunoprecipitation of IL23A from CRC culture supernatants was performed using anti-IL23A antibodies coupled with Dynabeads Protein G (Invitrogen), according to the manufacturer's protocol. Supernatant was prepared from CRC cells induced with PMA (100 ng/ml) for 36 h prior to harvesting.
ELISA of IL-23
To measure IL-23 in the supernatants harvested from cultured cells, ELISA was performed using the Human IL-23 Uncoated ELISA Kit (Invitrogen) following the manufacturer's instructions. Supernatant was prepared from CRC cells stimulated with PMA (100 ng/ml) for 36 h prior to harvesting.
Gene expression and pathway analyses
The IL23A and IL12B gene expression data were extracted from TCGA COADREAD (Broad GDAC version 2016_01_28) (58) and those published by Mouradov et al. (29). Gene expressions were compared and analyzed in terms of FPKM and RPKM. Prior to pathway analysis, low expressed genes were removed (mean expression <1). We applied R version 3.3.1, GSVA version 1.20, and Msigdb v6.1 C5 geneset to estimate the pathway enrichment for each sample in the TCGA and Mouradov et al. cohorts. Pre-processed microarray gene expression (data version 2010–09-29) and hybrid capture mutation data (data version 2012–02-20) of the Cancer Cell Line Encyclopedia (CCLE) were downloaded from CCLE portal (RRID: SCR_01383635). mRNA expression levels and mutation status of selected genes were extracted. Spearman's correlation coefficient test was used to assess correlation of gene expressions.
Statistical analysis
All charted data are presented as mean ± S.D. or S.E. Comparative analyses of two datasets were conducted either by the Student's t test (for parametric data sets) or Mann-Whitney U test (for nonparametric data sets). The p values were indicated on the figures as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
Data availability
The gene expression data of colorectal cancer samples from TCGA COADREAD cohort are publicly available in Broad Institute GDAC (58). The gene expression data of colorectal cell lines are publicly available (29). The mutation data of colorectal cell lines from CCLE are publicly available in CCLE portal (35). All experimental data generated in this manuscript are contained within the manuscript.
Author contributions
K. S. L., Z. W. E. Y., T. Z. T., and D. C.-C. V. data curation; K. S. L., Z. W. E. Y., and D. C.-C. V. formal analysis; K. S. L., D. Y., and D. C.-C. V. validation; K. S. L., Z. W. E. Y., H. W., T. Z. T., M. H., M. O., and D. C.-C. V. methodology; Z. W. E. Y., H. W., T. Z. T., M. O., Y. I., and D. C.-C. V. investigation; T. Z. T., R. Y.-J. H., D. Y., N. I., M. H., R. W. W., H. O., M. O., and D. C.-C. V. resources; T. Z. T., H. O., M. O., and D. C.-C. V. software; R. Y.-J. H., N. I., R. W. W., H. O., M. O., Y. I., and D. C.-C. V. supervision; Y. I. and D. C.-C. V. conceptualization; Y. I. and D. C.-C. V. funding acquisition; Y. I. and D. C.-C. V. writing-review and editing; D. C.-C. V. writing-original draft; D. C.-C. V. project administration.
Supplementary Material
Acknowledgments
We thank Li Ren Kong, Boon Cher Goh, Sha Shi, Atsushi Hirao, Eun Myoung Shin, Vinay Tergaonkar, Hiromichi Ebi, Ryu Imamura, and Kunio Matsumoto for the generous provision of reagents and technical help.
This work was supported by Ministry of Education Academic Research Fund (AcRF) Tier 2 Grant MOE2014-T2-2-143 (to Y. I.); a Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Scientific Research JP18K07228 (to D. C.-C. V.); and an Institute for Frontier Science Initiative, Kanazawa University, inter-disciplinary research grant. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12 and Table S1 and supporting Experimental procedures.
- Th17
- T helper 17 cell
- DSS
- dextran sulfate sodium
- TNF
- tumor necrosis factor
- MAPK
- mitogen-activated protein kinase
- CRC
- colorectal carcinoma
- PMA
- phorbol 12-myristate 13-acetate
- nt
- nucleotide
- CCLE
- Cancer Cell Line Encyclopedia
- p-c-Jun
- phosphorylated c-Jun
- TCGA
- The Cancer Genome Atlas
- qRT
- quantitative RT.
References
- 1. Croxford A. L., Kulig P., and Becher B. (2014) IL-12-and IL-23 in health and disease. Cytokine Growth Factor Rev. 25, 415–421 10.1016/j.cytogfr.2014.07.017 [DOI] [PubMed] [Google Scholar]
- 2. Morrison P. J., Ballantyne S. J., and Kullberg M. C. (2011) Interleukin-23 and T helper 17-type responses in intestinal inflammation: From cytokines to T-cell plasticity. Immunology 133, 397–408 10.1111/j.1365-2567.2011.03454.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Floss D. M., Schroder J., Franke M., and Scheller J. (2015) Insights into IL-23 biology: From structure to function. Cytokine Growth Factor Rev. 26, 569–578 10.1016/j.cytogfr.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa H., Mizoguchi I., Chiba Y., Ohashi M., Xu M., and Yoshimoto T. (2016) Expanding diversity in molecular structures and functions of the IL-6/IL-12 heterodimeric cytokine family. Front. Immunol. 7, 479 10.3389/fimmu.2016.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGeachy M. J., and Cua D. J. (2007) The link between IL-23 and Th17 cell-mediated immune pathologies. Semin. Immunol. 19, 372–376 10.1016/j.smim.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 6. Ivanov I. I., Zhou L., and Littman D. R. (2007) Transcriptional regulation of Th17 cell differentiation. Semin. Immunol. 19, 409–417 10.1016/j.smim.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Korn T., Oukka M., Kuchroo V., and Bettelli E. (2007) Th17 cells: Effector T cells with inflammatory properties. Semin. Immunol. 19, 362–371 10.1016/j.smim.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duerr R. H., Taylor K. D., Brant S. R., Rioux J. D., Silverberg M. S., Daly M. J., Steinhart A. H., Abraham C., Regueiro M., Griffiths A., Dassopoulos T., Bitton A., Yang H., Targan S., Datta L. W., et al. (2006) A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 10.1126/science.1135245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tremelling M., Cummings F., Fisher S. A., Mansfield J., Gwilliam R., Keniry A., Nimmo E. R., Drummond H., Onnie C. M., Prescott N. J., Sanderson J., Bredin F., Berzuini C., Forbes A., Lewis C. M., et al. (2007) IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology 132, 1657–1664 10.1053/j.gastro.2007.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Momozawa Y., Mni M., Nakamura K., Coppieters W., Almer S., Amininejad L., Cleynen I., Colombel J. F., de Rijk P., Dewit O., Finkel Y., Gassull M. A., Goossens D., Laukens D., Lémann M., et al. (2011) Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 43, 43–47 10.1038/ng.733 [DOI] [PubMed] [Google Scholar]
- 11. Beaudoin M., Goyette P., Boucher G., Lo K. S., Rivas M. A., Stevens C., Alikashani A., Ladouceur M., Ellinghaus D., Törkvist L., Goel G., Lagacé C., Annese V., Bitton A., Begun J., et al. (2013) Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 10.1371/journal.pgen.1003723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langowski J. L., Zhang X., Wu L., Mattson J. D., Chen T., Smith K., Basham B., McClanahan T., Kastelein R. A., and Oft M. (2006) IL-23 promotes tumour incidence and growth. Nature 442, 461–465 10.1038/nature04808 [DOI] [PubMed] [Google Scholar]
- 13. Grivennikov S. I., Wang K., Mucida D., Stewart C. A., Schnabl B., Jauch D., Taniguchi K., Yu G. Y., Osterreicher C. H., Hung K. E., Datz C., Feng Y., Fearon E. R., Oukka M., Tessarollo L., et al. (2012) Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hor Y. T., Voon D. C., Koo J. K., Wang H., Lau W. M., Ashktorab H., Chan S. L., and Ito Y. (2014) A role for RUNX3 in inflammation-induced expression of IL23A in gastric epithelial cells. Cell Rep. 8, 50–58 10.1016/j.celrep.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Macho-Fernandez E., Koroleva E. P., Spencer C. M., Tighe M., Torrado E., Cooper A. M., Fu Y. X., and Tumanov A. V. (2015) Lymphotoxin beta receptor signaling limits mucosal damage through driving IL-23 production by epithelial cells. Mucosal Immunol. 8, 403–413 10.1038/mi.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kortlever R. M., Sodir N. M., Wilson C. H., Burkhart D. L., Pellegrinet L., Brown Swigart L., Littlewood T. D., and Evan G. I. (2017) Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171, 1301–1315.e1314 10.1016/j.cell.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langrish C. L., Chen Y., Blumenschein W. M., Mattson J., Basham B., Sedgwick J. D., McClanahan T., Kastelein R. A., and Cua D. J. (2005) IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201, 233–240 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopp T., Lenz P., Bello-Fernandez C., Kastelein R. A., Kupper T. S., and Stingl G. (2003) IL-23 production by cosecretion of endogenous p19 and transgenic p40 in keratin 14/p40 transgenic mice: Evidence for enhanced cutaneous immunity. J. Immunol. 170, 5438–5444 10.4049/jimmunol.170.11.5438 [DOI] [PubMed] [Google Scholar]
- 19. Piskin G., Sylva-Steenland R. M., Bos J. D., and Teunissen M. B. (2006) In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: Enhanced expression in psoriatic skin. J. Immunol. 176, 1908–1915 10.4049/jimmunol.176.3.1908 [DOI] [PubMed] [Google Scholar]
- 20. Al-Sammak F., Kalinski T., Weinert S., Link A., Wex T., and Malfertheiner P. (2013) Gastric epithelial expression of IL-12 cytokine family in Helicobacter pylori infection in human: Is it head or tail of the coin? PLoS One 8, e75192 10.1371/journal.pone.0075192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carmody R. J., Ruan Q., Liou H. C., and Chen Y. H. (2007) Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J. Immunol. 178, 186–191 10.4049/jimmunol.178.1.186 [DOI] [PubMed] [Google Scholar]
- 22. Mise-Omata S., Kuroda E., Niikura J., Yamashita U., Obata Y., and Doi T. S. (2007) A proximal κB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J. Immunol. 179, 6596–6603 10.4049/jimmunol.179.10.6596 [DOI] [PubMed] [Google Scholar]
- 23. Utsugi M., Dobashi K., Ishizuka T., Kawata T., Hisada T., Shimizu Y., Ono A., and Mori M. (2006) Rac1 negatively regulates lipopolysaccharide-induced IL-23 p19 expression in human macrophages and dendritic cells and NF-κB p65 trans activation plays a novel role. J. Immunol. 177, 4550–4557 10.4049/jimmunol.177.7.4550 [DOI] [PubMed] [Google Scholar]
- 24. Ramnath D., Tunny K., Hohenhaus D. M., Pitts C. M., Bergot A. S., Hogarth P. M., Hamilton J. A., Kapetanovic R., Sturm R. A., Scholz G. M., and Sweet M. J. (2015) TLR3 drives IRF6-dependent IL-23p19 expression and p19/EBI3 heterodimer formation in keratinocytes. Immunol. Cell Biol. 93, 771–779 10.1038/icb.2015.77 [DOI] [PubMed] [Google Scholar]
- 25. Al-Salleeh F., and Petro T. M. (2008) Promoter analysis reveals critical roles for SMAD-3 and ATF-2 in expression of IL-23 p19 in macrophages. J. Immunol. 181, 4523–4533 10.4049/jimmunol.181.7.4523 [DOI] [PubMed] [Google Scholar]
- 26. Vogelstein B., Fearon E. R., Kern S. E., Hamilton S. R., Preisinger A. C., Nakamura Y., and White R. (1989) Allelotype of colorectal carcinomas. Science 244, 207–211 10.1126/science.2565047 [DOI] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Network. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voon D. C., Subrata L. S., and Abraham L. J. (2001) Regulation of lymphotoxin-β by tumor necrosis factor, phorbol myristate acetate, and ionomycin in Jurkat T cells. J. Interferon Cytokine Res. 21, 921–930 10.1089/107999001753289532 [DOI] [PubMed] [Google Scholar]
- 29. Mouradov D., Sloggett C., Jorissen R. N., Love C. G., Li S., Burgess A. W., Arango D., Strausberg R. L., Buchanan D., Wormald S., O'Connor L., Wilding J. L., Bicknell D., Tomlinson I. P., Bodmer W. F., Mariadason J. M., and Sieber O. M. (2014) Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 74, 3238–3247 10.1158/0008-5472.CAN-14-0013 [DOI] [PubMed] [Google Scholar]
- 30. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Zhuang J., Iyer S., Lin X., Whitfield T. W., Greven M. C., Pierce B. G., Dong X., Kundaje A., Cheng Y., Rando O. J., Birney E., Myers R. M., Noble W. S., Snyder M., and Weng Z. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerstein M. B., Kundaje A., Hariharan M., Landt S. G., Yan K. K., Cheng C., Mu X. J., Khurana E., Rozowsky J., Alexander R., Min R., Alves P., Abyzov A., Addleman N., Bhardwaj N., et al. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eferl R., and Wagner E. F. (2003) AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- 34. Ito K., Lim A. C., Salto-Tellez M., Motoda L., Osato M., Chuang L. S., Lee C. W., Voon D. C., Koo J. K., Wang H., Fukamachi H., and Ito Y. (2008) RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell 14, 226–237 10.1016/j.ccr.2008.08.004 [DOI] [PubMed] [Google Scholar]
- 35. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subrata L. S., Voon D. C., Yeoh G. C., Ulgiati D., Quail E. A., and Abraham L. J. (2012) TNF-inducible expression of lymphotoxin-β in hepatic cells: An essential role for NF-κB and Ets1 transcription factors. Cytokine 60, 498–504 10.1016/j.cyto.2012.05.029 [DOI] [PubMed] [Google Scholar]
- 37. Voon D. C., Subrata L. S., Karimi M., Ulgiati D., and Abraham L. J. (2004) TNF and phorbol esters induce lymphotoxin-β expression through distinct pathways involving Ets and NF-κB family members. J. Immunol. 172, 4332–4341 10.4049/jimmunol.172.7.4332 [DOI] [PubMed] [Google Scholar]
- 38. Yao W., Oh Y. T., Deng J., Yue P., Deng L., Huang H., Zhou W., and Sun S. Y. (2016) Expression of death receptor 4 is positively regulated by MEK/ERK/AP-1 signaling and Suppressed upon MEK inhibition. J. Biol. Chem. 291, 21694–21702 10.1074/jbc.M116.738302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leppä S., Saffrich R., Ansorge W., and Bohmann D. (1998) Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 17, 4404–4413 10.1093/emboj/17.15.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng Z., Sui G., Rosa P. M., and Zhao W. (2012) Radiation-induced c-Jun activation depends on MEK1-ERK1/2 signaling pathway in microglial cells. PLoS One 7, e36739 10.1371/journal.pone.0036739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ciccia F., Bombardieri M., Principato A., Giardina A., Tripodo C., Porcasi R., Peralta S., Franco V., Giardina E., Craxi A., Pitzalis C., and Triolo G. (2009) Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis Rheum. 60, 955–965 10.1002/art.24389 [DOI] [PubMed] [Google Scholar]
- 42. Lin X., O'Mahony A., Mu Y., Geleziunas R., and Greene W. C. (2000) Protein kinase C-θ participates in NF-κB activation induced by CD3-CD28 costimulation through selective activation of IκB kinase β. Mol. Cell. Biol. 20, 2933–2940 10.1128/MCB.20.8.2933-2940.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moscat J., Diaz-Meco M. T., and Rennert P. (2003) NF-κB activation by protein kinase C isoforms and B-cell function. EMBO Rep. 4, 31–36 10.1038/sj.embor.embor704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun S. C. (2011) Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 10.1038/cr.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Smaele E., Zazzeroni F., Papa S., Nguyen D. U., Jin R., Jones J., Cong R., and Franzoso G. (2001) Induction of gadd45β by NF-κB downregulates pro-apoptotic JNK signalling. Nature 414, 308–313 10.1038/35104560 [DOI] [PubMed] [Google Scholar]
- 46. Oeckinghaus A., Hayden M. S., and Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 10.1038/ni.2065 [DOI] [PubMed] [Google Scholar]
- 47. Hoesel B., and Schmid J. A. (2013) The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12, 86 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., and Hirai H. (1996) The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 16, 3967–3979 10.1128/MCB.16.7.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hamelin V., Letourneux C., Romeo P. H., Porteu F., and Gaudry M. (2006) Thrombopoietin regulates IEX-1 gene expression through ERK-induced AML1 phosphorylation. Blood 107, 3106–3113 10.1182/blood-2005-07-2953 [DOI] [PubMed] [Google Scholar]
- 50. Imai Y., Kurokawa M., Yamaguchi Y., Izutsu K., Nitta E., Mitani K., Satake M., Noda T., Ito Y., and Hirai H. (2004) The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol. Cell. Biol. 24, 1033–1043 10.1128/MCB.24.3.1033-1043.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu L., Mayes P. A., Eastman S., Shi H., Yadavilli S., Zhang T., Yang J., Seestaller-Wehr L., Zhang S. Y., Hopson C., Tsvetkov L., Jing J., Zhang S., Smothers J., and Hoos A. (2015) The BRAF and MEK inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–1651 10.1158/1078-0432.CCR-14-2339 [DOI] [PubMed] [Google Scholar]
- 52. Corcoran R. B., André T., Atreya C. E., Schellens J. H. M., Yoshino T., Bendell J. C., Hollebecque A., McRee A. J., Siena S., Middleton G., Muro K., Gordon M. S., Tabernero J., Yaeger R., O'Dwyer P. J., et al. (2018) Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Dis. 8, 428–443 10.1158/2159-8290.CD-17-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Corcoran R. B., Atreya C. E., Falchook G. S., Kwak E. L., Ryan D. P., Bendell J. C., Hamid O., Messersmith W. A., Daud A., Kurzrock R., Pierobon M., Sun P., Cunningham E., Little S., Orford K., et al. (2015) Combined BRAF and MEK Inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 33, 4023–4031 10.1200/JCO.2015.63.2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Voon D. C., Wang H., Koo J. K., Nguyen T. A., Hor Y. T., Chu Y. S., Ito K., Fukamachi H., Chan S. L., Thiery J. P., and Ito Y. (2012) Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells 30, 2088–2099 10.1002/stem.1183 [DOI] [PubMed] [Google Scholar]
- 55. Sato T., Stange D. E., Ferrante M., Vries R. G., Van Es J. H., Van den Brink S., Van Houdt W. J., Pronk A., Van Gorp J., Siersema P. D., and Clevers H. (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 56. Oh Y. S., Gao P., Lee K. W., Ceglia I., Seo J. S., Zhang X., Ahn J. H., Chait B. T., Patel D. J., Kim Y., and Greengard P. (2013) SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell 152, 831–843 10.1016/j.cell.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li Y. C., Ross J., Scheppler J. A., and Franza B. R. Jr. (1991) An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol. Cell. Biol. 11, 1883–1893 10.1128/MCB.11.4.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Broad Institute TCGA Genome Data Analysis Center. (2016) Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Havard. Dataset [Google Scholar]
- 59. Michaud J., Wu F., Osato M., Cottles G. M., Yanagida M., Asou N., Shigesada K., Ito Y., Benson K. F., Raskind W. H., Rossier C., Antonarakis S. E., Israels S., McNicol A., Weiss H., Horwitz M., and Scott H. S. (2002) In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: Implications for mechanisms of pathogenesis. Blood 99, 1364–1372 10.1182/blood.V99.4.1364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data of colorectal cancer samples from TCGA COADREAD cohort are publicly available in Broad Institute GDAC (58). The gene expression data of colorectal cell lines are publicly available (29). The mutation data of colorectal cell lines from CCLE are publicly available in CCLE portal (35). All experimental data generated in this manuscript are contained within the manuscript.