Abstract
Motor protein-based active transport is essential for mRNA localization and local translation in animal cells, yet how mRNA granules interact with motor proteins remains poorly understood. Using an unbiased yeast two–hybrid screen for interactions between murine RNA-binding proteins (RBPs) and motor proteins, here we identified protein interaction with APP tail-1 (PAT1) as a potential direct adapter between zipcode-binding protein 1 (ZBP1, a β-actin RBP) and the kinesin-I motor complex. The amino acid sequence of mouse PAT1 is similar to that of the kinesin light chain (KLC), and we found that PAT1 binds to KLC directly. Studying PAT1 in mouse primary hippocampal neuronal cultures from both sexes and using structured illumination microscopic imaging of these neurons, we observed that brain-derived neurotrophic factor (BDNF) enhances co-localization of dendritic ZBP1 and PAT1 within granules that also contain kinesin-I. PAT1 is essential for BDNF-stimulated neuronal growth cone development and dendritic protrusion formation, and we noted that ZBP1 and PAT1 co-locate along with β-actin mRNA in actively transported granules in living neurons. Acute disruption of the PAT1–ZBP1 interaction in neurons with PAT1 siRNA or a dominant-negative ZBP1 construct diminished localization of β-actin mRNA but not of Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) mRNA in dendrites. The aberrant β-actin mRNA localization resulted in abnormal dendritic protrusions and growth cone dynamics. These results suggest a critical role for PAT1 in BDNF-induced β-actin mRNA transport during postnatal development and reveal a new molecular mechanism for mRNA localization in vertebrates.
Keywords: neuron, kinesin, brain-derived neurotrophic factor (BDNF), synapse, RNA interference (RNAi), BDNF, kinesin, mRNA localization, PAT1, ZBP1
Introduction
mRNA localization is essential for localized protein synthesis within cells (1, 2), especially in neurons (3, 4). More mRNAs have been observed in hippocampal neuropil than expected (5), which supports localized demands for proteome plasticity at synapses (6–8). In neurons, the molecular motor protein and microtubule-dependent active transport are the most well-studied models that can facilitate mRNA transport (9), but the detailed mechanism remains elusive.
Previous live-cell imaging data indicated mRNAs (10–17) or RBPs3 (18–22) move along microtubules in dendrites, indicating a strong association between the conventional kinesin (kinesin-I) (23), RBPs, and mRNAs (11, 14, 24, 25). The effect of kinesin activity inhibition (11, 14, 26) revealed a role of kinesin-I in the anterograde-directed movement of dendritic mRNAs/RBPs complex. KLC (Fig. S3) is involved in a mechanism for the fragile X mental retardation protein (FMRP)-dependent mRNA transport in neurons (11), which is significant because KLC is a bona fide cargo-binding subunit of kinesin-I (27, 28). Kinesin heavy chain (KHC) mediates the transport of the large messenger ribonucleoprotein (mRNP) complex, which contains at least a dozen RBPs and other proteins, yet the KLC subunit was not identified within this complex (14). Multiple, distinct types of motor proteins have been shown to associate with mRNA cargoes, increasing the complexity of the mRNA-localizing mechanism (29, 30).
The most well-characterized mechanisms for mRNA transport are from yeast and flies (31). In yeast, the RBP She2p binds to the Ash1 mRNA and an She3p protein, which is an adapter protein that binds to the type-V myosin (32). This model was recently complicated by the fact that She3p binds directly to the Ash1 mRNA together with She2p (33, 34). In Drosophila, the egalitarian (Egl) protein was shown to bind directly to the bicaudal D (BicD) protein and several mRNAs that do not share consensus localization motifs (35). Previous work implicated BicD in binding to and stimulating dynein/dynactin motility; it was proposed that the interaction between the Egl/BicD complex and dynein activates mRNA localization. However, Egl is known to bind the dynein light chain (DLC) directly (36), and the details of the interaction between BicD and dynein/dynactin are not well-understood (37).
In this study, the PAT1 protein (Fig. S3) was identified to bind directly to ZBP1 (Fig. S3), a β-actin mRNA RBP. PAT1 also binds to KLC simultaneously. PAT1 and ZBP1 fluorescence fusion proteins and fluorescent β-actin mRNAs (38) were shown to transport together in living hippocampal neurons. PAT1 co-localized with KLC. According to this study, PAT1 is involved in the BDNF-induced transport of β-actin mRNA into dendrites. Suppression of PAT1 by PAT1 siRNA led to altered morphologies of dendritic protrusions and neuronal growth cones. These data pointed to an essential function for PAT1 in dendritic β-actin mRNA transport and highlighted a new molecular mechanism for the β-actin mRNA transport in vertebrates.
Results
ZBP1 associates with kinesin-I through PAT1
To probe mRNA transport in neurons, we looked at conventional kinesin and its subunits and cytoskeletal associations. Conventional kinesin is involved in the dendritic mRNA transport and associates with numerous (>42) RBPs, including Pur-α, Staufen, FMRP, and translocated in liposarcoma within a large (>1000 S) complex containing CaMKIIα and Arc mRNAs (14). No RBP has been shown to bind directly to any molecular motor subunit in vertebrates; therefore, the molecular mechanisms of how mRNA complex associates with motor proteins remain unknown. Abundant cellular RBPs are expressed in most cell types, among which ZBP1 traffics from the cell body into dendrites in response to N-methyl-d-aspartic acid receptor activation and neurotrophins (21, 39). ZBP1 binds directly to the conserved 54-nucleotide zipcode in the 3′ UTR of β-actin mRNA, making it a potential candidate to interact with motor proteins directly or indirectly.
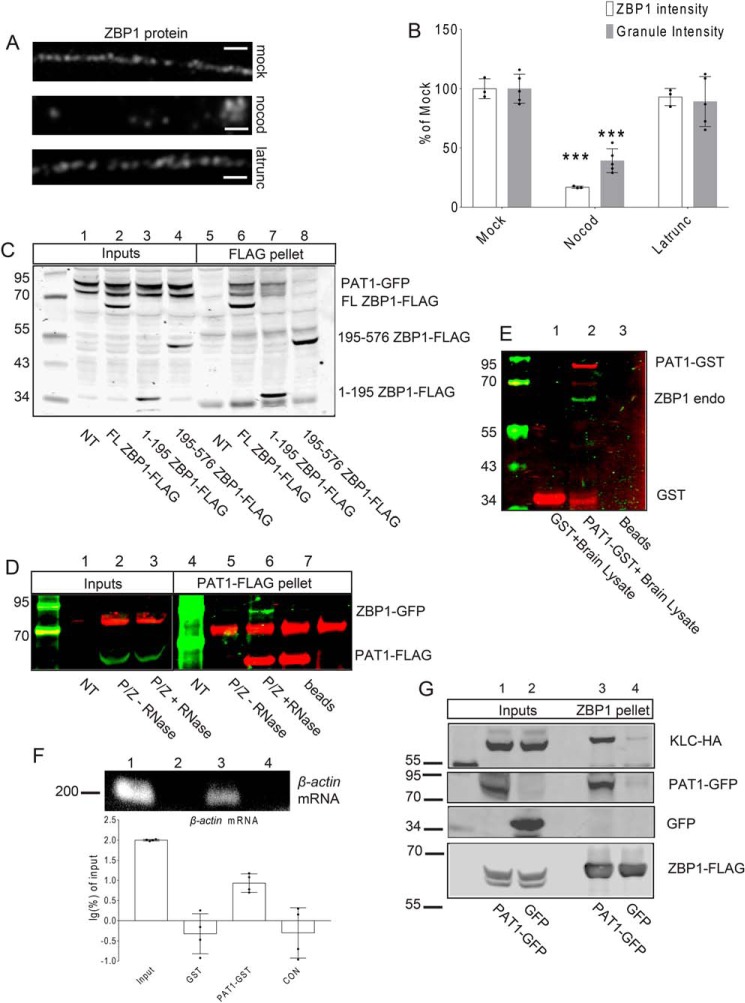
Brief depolymerization of microtubules in primary mouse hippocampal neurons at 7 DIV significantly reduced ZBP1 along dendrites, although interference with actin polymerization by latrunculin treatment had no apparent effect on the dendritic distribution of ZBP1 (Fig. 1, A and B; ***, p < 0.001, n = 12 cells, 2 dendrites per cell).
Figure 1.
A, mouse primary hippocampal neurons (7 DIV) were treated with the carrier (mock, upper panel), nocodazole (nocod, middle panel), or latrunculin (latrunc, lower panel) for 30 min; neurons were fixed and immunostained for ZBP1. Latrunculin has no effect on the distribution of dendritic ZBP1, whereas nocodazole treatment causes the reduced distribution of ZBP1 along dendrites. Scale bar, 5 μm. Three repeats were done independently with similar results for this experiment. B, histogram shows the quantification of dendritic ZBP1 intensities and granule intensities from A. Mock-, nocodazole (nocod)-, and latrunculin (latrunc)-treated dendrites are shown; nocodazole treatment decreases dendritic ZBP1 intensity, whereas latrunculin treatment has no effect on the distribution of ZBP1 along dendrites. ***, p < 0.001, n = 12 cells, two dendrites per cell, error bar, ±S.D. C, both the N-terminal and full-length (FL) ZBP1 interact with PAT1. Co-IPs were performed between PAT1–GFP and the full-length ZBP1–FLAG (FL ZBP1–FLAG, lanes 2 and 6) or the N terminus of ZBP1 (1–195 ZBP1–FLAG, lanes 3 and 7) or the C terminus of ZBP1 (195–576 ZBP1–FLAG, lanes 4 and 8) with anti-FLAG antibody-conjugated beads. NT denotes only PAT1–GFP-transfected cells (lanes 1 and 5). Both the full-length ZBP1 (lane 6) and the N terminus of ZBP1 (lane 7) are positive for interactions with PAT1–GFP. Six independent repeats were done for this experiment. D, interaction between PAT1 and ZBP1 requires RNA. ZBP1–GFP was co-transfected with PAT1–FLAG into HEK-293 cells (P/Z) and immunoprecipitated with anti-FLAG antibody-conjugated beads. ZBP1–GFP is precipitated by PAT1–FLAG in normal conditions (lane 5, −RNase), but not after RNase treatment (lane 6, + RNase). NT denotes nontransfected lysate; beads denote beads only; P/Z denotes transfected cell lysate. Six independent repeats were done for this experiment. E and F, recombinant PAT1–GST interacts with the endogenous ZBP1 protein and β-actin mRNA from mouse brain lysate. GSH–Sepharose-immobilized PAT1–GST interacts with the endogenous ZBP1 protein (E, lane 2) and β-actin mRNA (F, lane 3, 200 bp) from P0 mouse brain lysate. Histogram shows the log percent of β-actin mRNA associated with the recombinant PAT1–GST pellets, determined by RT-PCR. The amount of β-actin mRNA recovered from each group was normalized by the input and transformed into log percent of input. The result demonstrated that recombinant PAT1–GST interacts with endogenous ZBP1 and β-actin mRNA from the P0 brain lysate (F, lane1, lysate input; lane 2, GSH–Sepharose-immobilized GST only; lane 3, GSH–Sepharose-immobilized PAT1–GST; lane 4, CON denotes GSH–Sepharose only). Three independent experiments were performed. G, ZBP1 associates with KLC through PAT1. HEK-293 cells were co-transfected with KLC–HA, ZBP1–FLAG, and either GFP (lanes 2 and 4) or PAT1–GFP (lanes 1 and 3). Both lysates were immunoprecipitated with anti-FLAG antibody-conjugated beads. Blots indicate that the interaction between ZBP1–FLAG and KLC–HA requires the presence of PAT1–GFP (lane 3). Six independent repeats were done for this experiment.
To determine whether the molecular connection between ZBP1 and motor kinesin is direct or indirect, an unbiased screening of the mouse fetal brain library by yeast two-hybrid was performed with ZBP1 as the bait. Several positive clones revealed an interaction between ZBP1 and a protein (Table 1). This protein was identified as the β-amyloid precursor protein-binding protein 2 (APPBP2), also known as Drosophila protein interacting with APP tail-1 (PAT1) (40).
Table 1.
PAT1 interacts with ZBP1 N terminus and KLC, but not KHC or PTB
Results are from a directed yeast two-hybrid screen. After mating strains with the indicated prey or bait plasmids, the successful growth was scored as indicated. For each pair, + is given for robust colony growth; − is given for no colony growth; and ND indicates not determined. ZBP1 showed strong association with PAT1, but not KHC or KLC; PAT1 was positive for association with the KLC and ZBP1 N termini and was negative for KHC and ZBP1 C termini. PTB as a control showed no association with all baits.
| Bait/prey | PAT1 | ZBP1 N terminus | ZBP1 C terminus | KLC | KHC |
|---|---|---|---|---|---|
| ZBP1 | + | ND | ND | − | − |
| PAT1 | ND | + | − | + | − |
| KLC | + | − | − | ND | + |
| KHC | − | ND | ND | + | ND |
| PTB | − | − | − | − | − |
| No plasmid | − | − | − | − | − |
The N terminus of ZBP1 contains two conserved RNA recognition motif (RRM) domains in tandem and have not yet been implicated in a specific molecular interaction. The N terminus of ZBP1 showed a positive association with PAT1; however, the C terminus of ZBP1, which contains the β-actin mRNA-binding K homology (KH) domains, did not show an interaction with PAT1. ZBP1 was negative for direct interaction with both KLC and KHC and another KH domain–containing RBP, polypyrimidine tract-binding protein (PTB) (Table 1). An interesting result was a direct interaction between PAT1 and KLC, the cargo-binding domain of kinesin, indicating the possibility that PAT1 might be the adapter between ZBP1 and kinesin.
The interaction between full-length ZBP1 (FL ZBP1–FLAG) and full-length PAT1 (PAT1–GFP) was confirmed with co-immunoprecipitation (co-IP). The PAT1–GFP was co-transfected to HEK-293 cells with the full-length ZBP1 (FL ZBP1–FLAG). Anti-FLAG antibody-conjugated beads were able to precipitate PAT1 (PAT1–GFP, Fig. 1C, lane 6) along with ZBP1 (FL ZBP1–FLAG). To map the specific interaction between PAT1 and ZBP1 domains, co-IPs were performed between the full-length PAT1 (PAT1–GFP) and different domains of ZBP1 (FLAG constructs) after co-transfection. The full-length PAT1 (PAT1–GFP) was able to interact with both the full-length ZBP1 (FL ZBP1–FLAG) and the N-terminal 1–195 amino acids of ZBP1, which contain two RRM domains (1–195 ZBP1–FLAG, Fig. 1C, lanes 6 and 7). The N terminus of ZBP1 (1–195 ZBP1–FLAG) exhibited reduced binding with PAT1 (PAT1–GFP), likely due to the absence of RNA-binding domains. In contrast, the C terminus of ZBP1, amino acids 195–576 (195–576 ZBP1–FLAG), had no significant binding with PAT1 (PAT1–GFP, Fig. 1C, lane 8). These results confirmed an interaction exists between ZBP1 and PAT1, which requires the presence of RNAs (Fig. 1D). With the treatment of RNase during the co-IP, the interaction between ZBP1 (ZBP1–GFP) and PAT1 (PAT1–FLAG) disappeared. Furthermore, the purified recombinant PAT1–GST protein expressed from Escherichia coli was immobilized to GSH–Sepharose and applied to P0 mice brain lysate. The PAT1-GST was able to precipitate the endogenous ZBP1 (Fig. 1E) and β-actin mRNA (Fig. 1F) from P0 mice brain lysate, indicating the interaction between PAT1, ZBP1, and β-actin mRNA in a natural condition. Co-expressed PAT1 and ZBP1 showed a small but reproducible shift on the sucrose gradient. According to the standard curve, ZBP1 and PAT1 formed a complex together of about 4.5 S, which estimates a molecular size of 120 kDa, roughly equal to one PAT1 and one ZBP1 molecule per particle. Because the individual sedimentation coefficient values of both proteins are higher than expected at ∼3.7 S, it is likely that they are more unfolded when not bound together (Fig. S1, A and B). The purified recombinant PAT1–GST and ZBP1–MBP showed an association, but with low affinity, perhaps due to uncorrected folding and a lack of phosphorylation in the prokaryotic environment. For unknown reasons, the inclusion of in vitro transcribed β-actin mRNA 3′ UTR zipcode 1209–1441 caused a loss of the soluble PAT/ZBP1 complex in vitro (Fig. S1C, lane 3).
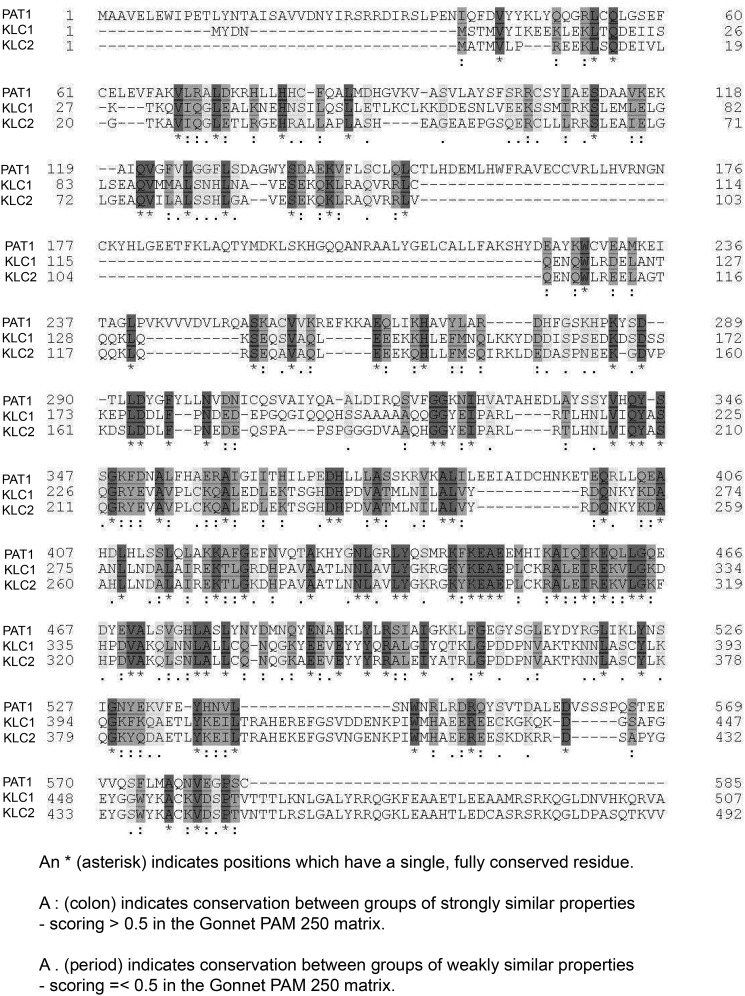
The amino acid sequence of mouse PAT1 was analyzed. PAT1 is highly similar to mouse KLC (Fig. 2). An interesting fact is that PAT1 stimulates kinesin motility through an unknown mechanism in Drosophila (40). Mouse KLC has two distinct protein domains, an HR domain and a TPR domain. The HR domain is required for the interaction between KLC and the tail of KHC, whereas the TPR domain of KLC binds to cargoes such as JNK-interacting protein (JIP) and APP vesicle-associated proteins (27, 28). Alignment and conserved amino acid comparisons showed that mouse KLC1/2 and PAT1 have rather highly-conserved domains. The PAT1–HR domain is 54–60% similar to the KLC1/2–HR domains and 26–28% identical, whereas the TPR domains from PAT1 and KLC1/2 are 59–62% similar and 30–32% identical. Because PAT1 is similar to KLC, it might act like KLC to facilitate mRNA granule transport by binding directly to KHC. However, the results from the yeast two-hybrid experiment showed that PAT1 has no direct interaction with KHC (Table 1). It is possible that instead of acting like KLC, PAT1 mediates the interaction between mRNA granule cargoes and kinesin through KLC, because PAT1 binds to both ZBP1 and KLC. Some primary co-IP data do support this assumption, and as both PAT1 and KLC have TPR domains, the co-IP experiment suggested that PAT1 interacts with KLC through the TPR domain of KLC (Fig. S1D). Co-IPs between PAT1–GFP, KLC–HA, and ZBP1–FLAG were performed, and the interaction between KLC (KLC–HA) and ZBP1 (ZBP1–FLAG) is more noticeable on the co-expression of PAT1 (PAT1–GFP) in HEK-293 cells (Fig. 1G). Without the co-expression of the PAT–GFP, the ZBP1–FLAG showed low binding to the KLC–HA, probably through the endogenous PAT1. These data showed that PAT1 associates with both the ZBP1 N terminus and KLC, and therefore, it may bridge between the kinesin and ZBP1/β-actin mRNA complex.
Figure 2.
PAT1 is a kinesin light-chain–like protein. Schematic shows the alignment between mouse PAT1 (NP_080101.1 amyloid protein-binding protein 2 (Mus musculus)), KLC1 (NP_032476.2 kinesin light chain 1 isoform 1A (M. musculus)), and KLC2 (NP_032477.2 kinesin light-chain 2 (M. musculus)). The result was generated from uniprot.com. Asterisk indicates positions that have a single, fully-conserved residue. Colon indicates conservation between groups of strongly-similar properties, scoring >0.5 in the Gonnet PAM 250 matrix. Period indicates conservation between groups of weakly similar properties scoring = <0.5 in the Gonnet PAM 250 matrix.
PAT1 co-transports with ZBP1 in dendritic shafts
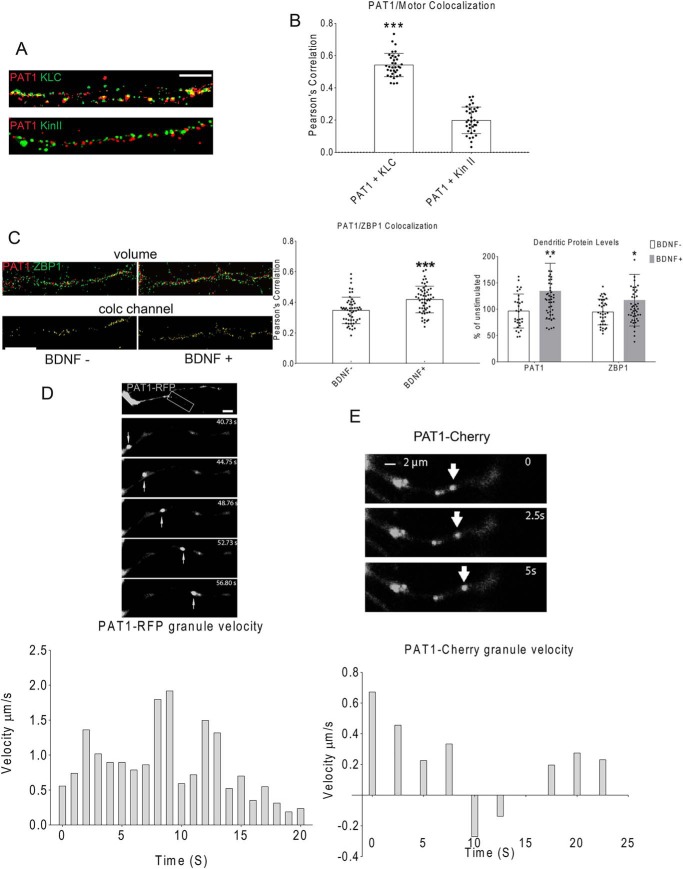
The spatial and temporal expression patterns of PAT1 in mouse brain were analyzed by semi-quantitative PCR. Total RNAs were isolated from P7 and P14 mouse hippocampi, cortices, and whole brain. 1 μg of total RNA from each sample was used to perform reverse transcription to yield cDNA, followed by PCR to detect the expression levels of PAT1. Based on the results, PAT1 is widely expressed in the brain and is expressed relatively lower within the hippocampus and cortex at P7 but is enriched at P14 in both regions (Fig. 3A). Next, the expression patterns of PAT1 in the cultured hippocampal neurons (5 and 14 DIV) were investigated. Punctate PAT1 staining in the cell body and dendritic shafts was observed, as evinced by the co-immunostaining of MAP2, both in developing (Fig. 3B, left, 5 DIV) and mature (Fig. 3B, right, 14 DIV) hippocampal neurons. PAT1 co-immunostaining with Tau showed accumulation of PAT1 in both axonal and dendritic growth cones of developing neurons but is not abundant in developing axonal shafts (Fig. 3C). Fluorescence images showed that ZBP1 and PAT1 co-localize along secondary and tertiary dendrites in mature, cultured neurons (14 DIV) (Fig. 3D and Fig. S4A). To confirm the overlap is co-localization, not a coincidence, co-localization controls were applied as described under “Materials and methods.” To obtain a molecular view of the ZBP1 and PAT1, super-resolution structured illumination microscopy (SIM) was used, and the results revealed that PAT1 and ZBP1 co-localize along dendrites (Fig. 3E, Pearson's correlation value is significant at 0.45, p < 0.001, n = 12 cells, 3–4 dendrites per cell, Fig. S4B). In contrast, amyloid precursor protein (APP), a proposed cargo for PAT1 in non-neuronal cells, and FMRP co-localize less significantly with PAT1 (Fig. 3, E–G, Pearson's correlation value is not significant at 0.25 for APP or at 0.16 for FMRP; ***, p < 0.001, n = 10 cells, 3–4 dendrites per cell). PAT1 co-localizes with KLC (Fig. 4, A and B, ***, p < 0.001, n = 20 cells, 2–3 dendrites per cell, Fig. S4C) in hippocampal dendrites. However, kinesin-II, a distinct kinesin isoform from the KIF3 kinesin family, showed less significant overlap with PAT1 (Fig. 4, A and B, p > 0.4, n = 12 cells, 3–4 dendrites per cell). Next, whether BDNF stimulation affects the co-localization between PAT1 and ZBP1 was tested because ZBP1 is known to transport with β-actin mRNA into axons in response to the BDNF treatment (41). Upon stimulation, both ZBP1 and PAT1 protein levels significantly increased in dendrites (Fig. 4C, *, p < 0.05 for ZBP1; **, p < 0.01 for PAT1, n = 43 cells, 2–3 dendrites per cell). There also was a significant increase in the overlap between ZBP1 and PAT1 after BDNF stimulation (Fig. 4C, ***, p < 0.001, n = 33 cells, 2–3 dendrites per cell).
Figure 3.
A, PAT1 increases its expression during development and is highly expressed in mouse hippocampus and cortex. Total RNA from P7 and P14 mouse hippocampus (HC), cortex (CO), or the whole brain (WB) were isolated, and 1 μg of total RNA from each sample was reverse-transcribed, followed by PCR with PAT1 primers (upper panel, 200 bp). PCR for β-actin was also performed in parallel as controls (lower panel, 200 bp). The histogram shows the ratio of PAT1 to β-actin mRNA within different brain regions and developmental stages. Two independent experiments were done for this analysis showing almost identical ratios. B, PAT1 is localized to dendritic shafts and growth cones in cultured neurons. Left panel, hippocampal neurons (5 DIV) were immunostained for PAT1 (red) and MAP2 (green), and fluorescence images were overlaid onto DIC images to visualize the cell outline. PAT1 localizes to the cell body, dendritic shafts (arrowheads), and developing growth cones and associated protrusions (arrows). Right panel, mature hippocampal neurons (14 DIV) were immunostained for PAT1 (green) and MAP2 (blue) to visualize PAT1 in dendrites. Scale bars, 10 μm (left panel) and 20 μm (right panels). Individual channels are shown in grayscale. Five independent repeats were done for this experiment with consistent findings. C, PAT1 is localized to dendritic and axonal growth cones. Hippocampal neurons (7 DIV) were immunostained for PAT1 (red) and tau (green), and fluorescence images were overlaid onto DIC images to visualize the cell outline, each individual channel is shown in grayscale. PAT1 is localized to the cell body, dendritic shafts, and growth cones in axons and dendrites (arrows). Scale bar, 10 μm (upper left panel and bottom panels) and 5 μm (upper right panel). This experiment was repeated five times independently with consistent findings. D, PAT1 co-localizes with ZBP1 in the secondary and tertiary dendrites in primary hippocampal cultures. Mouse primary hippocampal cultures (14 DIV) were immunostained for PAT1 and ZBP1. PAT1 (red) is highly punctate and co-localizes with ZBP1 (green) in dendrites (MAP2, blue), especially in secondary and tertiary segments (see arrows, lower image). Inset (lower image) shows a magnified view of the white boxed area of the dendrite. Scale bar, 5 μm (upper left panel and right panels) and 1 μm (lower panel); each individual channel is shown in grayscale. This experiment was repeated five times independently with consistent findings. E, 3D SIM images indicate that PAT1 and ZBP1 are significantly co-localized along dendrites. Hippocampal neurons (10 DIV) were stimulated with BDNF for 1 h, and then fixed and immunostained for either ZBP1 (green) or APP (green) and PAT1 (red). Images were taken by 3D SIM. Segments of dendrites were further analyzed for co-localization in 3D and displayed as maximum projection images (volume view). In the spots views, spots were generated to show only co-localized voxels that red (PAT1) and green (ZBP1 or APP) signals are overlapped within 100 nm. Scale bar, 10 μm (upper panels) and 1 μm (lower four panels). Three independent repeats were performed for these experiments with consistent findings. F, hippocampal neurons (10 DIV) were stimulated with BDNF for 1 h and immunostained for PAT1 (red) and FMRP (green). Images were taken in 3D with a SIM microscope. PAT1 and FMRP are not significantly co-localized with each other. Scale bar, 5 μm. Three independent repeats were performed for these experiments with consistent findings. G, histogram shows the Pearson's coefficient of correlation analysis for PAT1/ZBP1, PAT1/APP, and PAT1/FMRP pairs. PAT1 significantly co-localizes with ZBP1. (n = 6–7 cells, 2–3 dendrites per cell, ***, p < 0.001, **, p < 0.01; error bar, ±S.D.)
Figure 4.
A and B, PAT1 co-localizes with KLC, but not Kin-II. Primary hippocampal neurons (8 DIV) were immunostained for PAT1 (red) and KLC (green) or kinesin-II (Kin-II, green). The histogram shows the Pearson's correlation coefficient for PAT1/KLC (***, p < 0.001, n = 20 cells, 2–3 dendrites per cell, error bar, ±S.D.) and PAT1/Kin-II (p > 0.4, n = 10 cells, 2–3 dendrites per cell, error bar, ±S.D.) Scale bar, 5 μm. Three repeats were done independently for this experiment with consistent findings. C, BDNF stimulation significantly increases dendritic ZBP1 and PAT1 and their co-localization. Primary hippocampal cultures (10 DIV) were stimulated with BDNF for 1 h and immunostained for PAT1 (red), and ZBP1 (green); the co-localization (colc) channel (yellow) was generated. The histogram on the left shows dendritic ZBP1 and PAT1 increased after 1 h of BDNF stimulation (**, p < 0.01 for PAT1; *, p < 0.05 for ZBP1, n = 33 cells, 2–3 dendrites per cell, error bar, ±S.D.) The right histogram shows Pearson's correlation coefficient between PAT1 and ZBP1 increased after 1 h of BDNF stimulation (***, p < 0.001 for Pearson's correlation) compare with untreated. Scale bar, 5 μm. This experiment was repeated independently four times with consistent findings. D, PAT1 forms granules that transport in dendrites of primary hippocampal neurons. Primary hippocampal neurons (7 DIV) were transfected with PAT1–RFP and imaged with time-lapse video microscopy. Top, successive images (every 4 s) show a PAT1 granule is moving in the anterograde direction in dendrites. Bottom, granule was tracked for successive frames (every 1 s), and the velocities were recorded and plotted as positive displacements (anterograde). Scale bar, 2 μm. Six repeats were done independently for this experiment with consistent findings. E, primary hippocampal neurons (7 DIV) were transfected with PAT1–cherry and imaged with time-lapse video microscopy. Top, successive images (every 2.5 s) show several PAT1–Cherry granules. Bottom, a PAT1–Cherry granule (red, highlighted with arrows) moves in both directions. One granule was tracked for successive frames (every 1 s), and its velocities were recorded and plotted as positive displacements (anterograde) and negative displacements (retrograde). Scale bar, 2 μm. Three repeats were done independently for this experiment with consistent findings.
To visualize PAT1 in living neurons, fluorescent fusion proteins were introduced into the neurons by transfection. It is possible that the fluorescent tag proteins may alter the natural transport of PAT1 in neurons, so two different fluorescent tag proteins were utilized here. The PAT1–RFP fluorescent fusion protein was expressed in hippocampal cultures (7 DIV). The live-imaging data showed that PAT1–RFP moves rapidly in both the anterograde and retrograde directions in the dendrites, at an average rate of 0.5–1 μm/s (Fig. 4D), similar to the speeds reported for kinesin-I and the associated cargoes, as well as that of ZBP1 in dendrites (21). This observation was confirmed with a different PAT1 construct that fused to Cherry fluorescent protein (Fig. 4E). Both PAT1–RFP and PAT1–Cherry formed granules inside primary neurons and showed similar movement patterns. Other PAT1 fusion proteins, the PAT1–GFP and PAT1–FLAG were used in the co-IP experiments and also showed similar results in association with the ZBP1–FLAG, ZBP1–GFP, and KLC–HA. Strikingly, dual expression of the ZBP1–GFP and the PAT1–RFP/PAT1–Cherry showed some dendritic granules containing both the ZBP1 and PAT1 fluorescent fusion proteins moved in anterograde and retrograde directions together, whereas some oscillate without significant displacement (Fig. 5, A and B, Movies S1 and S2). Note that some stationary ZBP1–GFP granules contain PAT1–RFP, and some do not; also, some PAT1–RFP granules do not contain ZBP1–GFP yet they moved in dendrites, possibly with other unlabeled ZBP1/RNA cargoes. Taken together, both PAT1 and ZBP1 traffic together in dendrites, and their co-localization is increased in response to BDNF stimulation.
Figure 5.
A, PAT1 and ZBP1 co-transport in neurons. Primary hippocampal neurons (7 DIV) were co-transfected with PAT1–RFP (red) and ZBP1–GFP (green), and the granule movements were recorded 18–24 h later at 2 min per frame. Left, successive frames (every 2 min) show a PAT1–RFP (red, highlighted with arrows) granule is co-trafficking with a ZBP1–GFP (green) granule. Granule velocities were recorded for 15 frames and plotted over time as positive displacements (anterograde) and negative displacements (retrograde). Scale bar, 1 μm. Three repeats were done independently for this experiment with consistent findings. B, primary hippocampal neurons (7DIV) were co-transfected with PAT1–Cherry (red) and ZBP1–GFP (green), and the granule movements were recorded 18–24 h later at 30 s per frame. Left, successive frames (every 30 s) show a PAT1 and ZBP1 co-localized granule (arrow) moves in the anterograde direction, then in the retrograde direction, and continues in a saltatory manner. Right, granule was tracked, and the granule velocities for each component (ZBP1, black; PAT1, gray) were recorded and plotted as positive displacements (anterograde) and negative displacements (retrograde). Scale bar, 2 μm. Six repeats were done independently for this experiment with consistent findings. C, β-actin mRNA associates with PAT1 and ZBP1. Full-length ZBP1 (ZBP1–FLAG and ZBP1–GFP), the KH domain of ZBP1 (ZBP1 KH–FLAG), the RRM domain of ZBP1 (ZBP1–RRM–FLAG), PAT1 (PAT1–GFP), and FMRP (FMRP–GFP) were transfected to HEK-293 cells. mRNA extracts from the immunoprecipitates were analyzed. PCR was performed with primers on both β-actin (upper gels) and vinculin (lower gels, as negative controls). Blank PCR indicates no mRNA template was added to the reaction. Constructs used are indicated above lanes. Immunoprecipitation antibodies: lanes 1–4, anti-GFP antibody; lanes 5–7, anti-FLAG antibody; lane 8, anti-FLAG and anti-GFP antibodies. Dashed lines indicate the 500-bp marker. PAT1–GFP, ZBP1–GFP, ZBP1–FLAG, and ZBP1-KH–FLAG are positive for β-actin mRNA detection. Three repeats were done independently for this experiment with consistent findings. D, ZBP1 and kinesin associate with β-actin mRNA in a 3′-UTR zipcode-dependent manner. Semi-quantitative RT-PCR of mRNA from immunoprecipitates of ZBP1 and different kinesin subunits and isoforms were analyzed. Endogenous (Endog) or transfected proteins were precipitated, and the mRNAs were extracted and used as templates for RT-PCRs with primers to either β-actin (upper gels) or vinculin (lower gels, as a negative control). LacZ(+) indicates co-transfection with a competitive 3′ UTR zipcode construct that binds to ZBP1 and can displace endogenous β-actin mRNA. Blank PCR indicates no mRNA template was added to the reaction. Constructs used are indicated above lanes. Immunoprecipitation antibodies: lane 1, anti-FLAG and anti-mouse antibodies (nonimmune); lanes 2 and 3, anti-FLAG antibody; lanes 4 and 5, anti-KHC antibody; lanes 6 and 7, anti-KLC antibody; lanes 8 and 9, anti-kinesin II antibody. Dashed lines indicate 500-bp marker. β-Actin mRNA was approved to be associated with ZBP1–FLAG/LacZ− (lane 3), endogenous KHC/LacZ− (lane 5), and endogenous KLC/LacZ− (lane 7). Three repeats were done independently for this experiment with consistent findings. E, PAT1 co-localizes with β-actin mRNA in distal dendrites in neurons. Mouse primary hippocampal cultures (9 DIV) were immunostained for PAT1 (green) and MAP2 (blue), followed by FISH to visualize β-actin mRNAs. Endogenous PAT1 (green) co-localizes with β-actin mRNA (red) in the secondary and tertiary dendrites. Right panels show separate low magnification images of MAP2 (blue), PAT1 (green), and β-actin mRNA (red). The lower panel shows close up of the white-boxed region in the main image, with arrows indicating areas of PAT1 (green) and β-actin mRNA (red) are co-localized (yellow), each individual channel is shown in grayscale. Scale bar, 10 μm (upper right panel), 20 μm (left panels), and 5 μm (bottom panel). Six independent repeats were done for this experiment with consistent findings. F, PAT1 and β-actin mRNA co-transport in dendrites. Hippocampal neurons (7 DIV) were co-transfected with PAT1–RFP and β-actin–MS2–MCP–GFP, and granule movements were recorded 18–24 h later at 3 s per frame. The image panels show three consecutive frames of duel channels (merge, left) as well as the individual channel for PAT1 (red) and β-actin mRNA (green). Blue arrows highlight two individual moving particles relative to one that did not move during the Movie S3 (red arrowhead), each individual channel is shown in grayscale. Scale bar, 2 μm. The granule velocities were recorded and plotted as positive displacements (anterograde) and negative displacements (retrograde). Three repeats were done independently for these experiments with consistent findings.
BDNF-induced transport of β-actin mRNA requires the interaction between zipcode, PAT1, and the N terminus of ZBP1
To explore the function of PAT1 in β-actin mRNA localization, the requirement for the 3′ UTR zipcode of β-actin mRNA was tested. The C terminus of ZBP1 (aa 195–576) mediates the base pair–specific binding between the β-actin mRNA zipcode and ZBP1 (42). Accordingly, HEK-293 cells were transfected with different constructs, including PAT1 (PAT1–GFP), FMRP (FMRP–GFP), and ZBP1 domains (ZBP1–GFP, ZBP1–FLAG, ZBP1–KH–FLAG, and ZBP1–RRM–FLAG). Immunoprecipitations were performed, and the precipitates were analyzed for β-actin mRNA association (Fig. 5C, upper panel). RT-PCR showed that β-actin mRNA was present as expected with both the full-length (Fig. 5C, lanes 4 and 5) and C terminus (lane 6) but not the N terminus of ZBP1 (lane 7). PAT1 was also able to precipitate the β-actin mRNA to a lesser extent than ZBP1 (Fig. 5C, lane 3). The FMRP precipitate was negative for β-actin mRNA (Fig. 5C, lane 2), as was the GFP alone (lane 1) and the mock-transfected control (lane 8). The specificity for β-actin mRNA was evident because no proteins were shown to interact with the negative control vinculin mRNA (Fig. 5C, lower gel).
To determine the role of β-actin mRNA 3′ UTR zipcode in kinesin association, HEK-293 cells were transfected with constructs overexpressing the β-actin mRNA zipcode fused to a lacZ reporter, and the kinesin complexes were isolated. Hypothetically, if the zipcode is required for the formation of ZBP1/motor complexes, excess zipcode would compete with the endogenous β-actin mRNA. The ZBP1 immunoprecipitates from the HEK-293 cells overexpressing a zipcode-lacking construct (LacZ−) contained β-actin mRNA (Fig. 5D lane 3). In contrast, the ZBP1 immunoprecipitates from the cells expressing the zipcode (lacZ+) constructs showed little detectable β-actin mRNA (Fig. 5D lane 2). Interestingly, both the endogenous KHC and KLC immunoprecipitates showed β-actin mRNA (Fig. 5D lanes 5 and 7), yet in the presence of excess zipcode, these associations were significantly diminished (Fig. 5D lanes 4 and 6). These were in contrast to the kinesin-II immunoprecipitates, which did not show β-actin mRNA (Fig. 5D lanes 8 and 9) by PCR (Fig. 5D, upper panel). The negative controls confirmed that neither the ZBP1 nor the kinesin immunoprecipitate showed vinculin mRNA by PCR (Fig. 5D, lower panel).
Fluorescence in situ hybridization (FISH) images showed that PAT1 co-localizes significantly with β-actin mRNA in dendrites (Fig. 5E, Pearson's coefficient = 0.82, n = 12 cells, 3–4 dendrites per cell, 14 DIV) compared with the flipped control (Fig. S4D). Live-cell imaging data showed that β-actin mRNA and PAT1 co-transport within granules in living dendrites of hippocampal neurons at rates consistent with those observed for PAT1 and ZBP1 (Fig. 5F, Movie S3, 7 DIV, and Video S1). These data suggested that the associations between β-actin mRNA, PAT1, and kinesin-I are dependent on the interactions involving the β-actin mRNA 3′ UTR zipcode element.
Using small-interfering RNAs (siRNAs) to repress PAT1 expression in primary hippocampal neurons, the roles of PAT1 in the localizations of ZBP1 and β-actin mRNA to dendrites were investigated. Two distinct siRNA duplexes directed against PAT1 were tested in HEK-293 cells and mouse primary hippocampal neurons and were found to knock down the expression of PAT1 at both the mRNA and protein levels. The semi-quantitative PCR results showed siRNAs designed to target PAT1 caused a profound 90% repression of PAT1 mRNA, as determined by normalization to the total β-actin mRNA levels, which were not affected (Fig. 6A). The repression of PAT1 in dendrites was further confirmed by fluorescence quantification (Fig. 6B) and immunoblotting (Fig. S1D); both endogenous and overexpressed PAT1 protein decreased to 50% of control levels. Both the PAT1 siRNA duplexes and control duplexes appeared to enter >93% of neurons as determined by co-transfection of the fluorescent duplex RNA tracers.
Figure 6.
A, PAT1 siRNA inhibits PAT1 expression at the mRNA level. Mouse primary hippocampal neurons were transfected with PAT1/Control siRNAs. PAT1 mRNA (products 200 bp) was analyzed with RT-PCR, and β-actin mRNA (products 200 bp) was also analyzed here as an internal control. Three repeats were done independently for these experiments with consistent findings. B, PAT1 siRNA inhibits PAT1 expression at the protein level. Mouse primary hippocampal neurons were immunostained for PAT1 (green) after PAT1/Control siRNA treatment. The dendritic PAT1 was quantified. Scale bar, 10 μm. Three repeats were done independently for these experiments with consistent findings. C, histograms show the experimental data from A and B, with the PAT1 mRNA and protein levels from the Control siRNA–treated neurons as 100%. ***, p < 0.001, n = 28 cells, 2–3 dendrites per cell; error bar, ±S.D. D, BDNF induces β-actin mRNA localization to the dendrites. Mouse hippocampal neurons (10 DIV) were stimulated with BDNF or mock-stimulated for 1 h. Neurons were fixed and subjected to immunostaining for MAP2 (blue) and FISH for β-actin mRNA (red). The histogram shows dendritic β-actin mRNA increased after 1 h of BDNF stimulation. (***, p < 0.001, n = 28 cells, 2–3 dendrites per cell, error bar, ±S.D.) Scale bar, 10 μm. Six independent repeats were done for this experiment with consistent findings. E, histogram shows the ratio between the dendritic β-actin mRNA intensity and the somatic β-actin mRNA intensity from PAT1 siRNA and control (CON) siRNA-treated neurons. The result indicates that PAT1 repression causes a decrease in dendritic β-actin mRNA localization. F, PAT1 repression blocks the dendritic localization of β-actin mRNA and ZBP1. Mouse hippocampal neurons (7 DIV) were treated with PAT1/Control siRNA for 72 h and stimulated with BDNF for 1 h before fixation and FISH for β-actin mRNA (green). Immunostaining was also applied to visualize the dendritic ZBP1 simultaneously (white). Top, representative images of control and PAT1 siRNA-treated neurons immunostained for ZBP1 are shown. Bottom, representative images of control and PAT1 siRNA-treated neurons stained for β-actin mRNA are shown. Right, histograms show that the dendritic ZBP1 intensity, β-actin mRNA intensity, and β-actin mRNA granule density all decreased compared with the controls after PAT1 siRNA treatment. Scale bar, 10 μm. (**, p < 0.01; ***, p < 0.001, n = 40 cells, 2–3 dendrites per cell; error bar, ±S.D.) Six repeats were done independently for these experiments with consistent findings. G, PAT1 repression does not impair the dendritic localization of GABA-A-R-δ and CaMKIIα mRNA. Mouse hippocampal neurons (7 DIV) were treated with PAT1/Control siRNA for 72 h and stimulated with BDNF for 1 h before fixation and FISH. Top, representative images of control and PAT1 siRNA-treated neurons stained for GABA-A-R-δ mRNA are shown (red). Bottom, representative images of control and PAT1 siRNA-treated neurons stained for CaMKIIα mRNA are shown (red). Right, histograms show the quantification of dendritic GABA-A-R-δ and CaMKIIα mRNA intensities. After PAT1 siRNA treatment, the CaMKIIα mRNA intensity was not significantly changed compared with the control siRNA-treated neurons; the GABA-A-R-δ mRNA intensity was increased. Scale bar, 10 μm. (*, p < 0.05, n = 30 cells, 2–3 dendrites per cell; error bar, ±S.D.) Six repeats were done independently for these experiments with consistent findings.
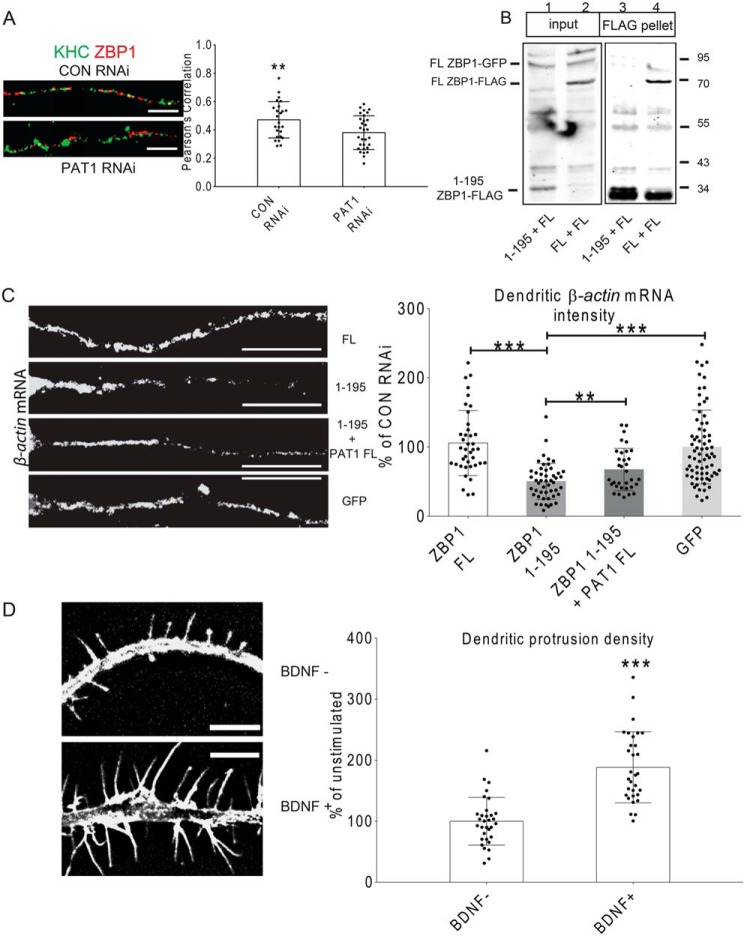
BDNF has been shown to signal the Src and ZBP1-dependent localized translation of β-actin protein in growth cones and to play a role in the ZBP1-mediated dendritic filopodial growth (43, 44). To determine the roles of PAT1 in the activity-dependent β-actin mRNA localization, quantitative FISH was performed after neurons were treated with siRNAs and stimulated with BDNF. In this study, for the control siRNA-treated neurons, 1 h of BDNF stimulation significantly increased dendritic β-actin mRNA intensities by ∼38% (Fig. 6D, ***, p < 0.001, n = 42 cells, 2–3 dendrites per cell). In PAT1 siRNA-treated neurons (in the presence of BDNF), dendritic β-actin mRNA intensity was diminished to ∼55% of the controls (Fig. 6F, ***, p < 0.001, n = 38 cells, 2–3 dendrites per cell). This was similar to the localization of dendritic ZBP1 after PAT1 repression, which decreased by ∼70% of control neurons (Fig. 6F, **, p < 0.01, n = 38 cells, 2–3 dendrites per cell). A notable fact is that PAT1 siRNA treatment did not affect the total ZBP1 level in neurons, as they were similar to that of the control siRNA-treated neurons. The ratio of dendritic to somatic β-actin mRNA intensities was measured instead of the absolute intensities. The ratio showed a similar ∼42% reduction upon PAT1 repression (Fig. 6E, *, p < 0.05, n = 22 cells, 2–3 dendrites per cell), indicating a significant decrease of the dendritic β-actin mRNA. More significantly, the total number of dendritic β-actin mRNA granules decreased by ∼63% upon PAT1 repression compared with the controls (Fig. 6F, ***, p < 0.001 all n = 33, 2–3 dendrites per cell). These effects of PAT1 repression were unique to the β-actin mRNAs as PAT1 repression did not diminish the dendritic localization of two other localized mRNAs, CaMKIIα and GABA-A-R-δ mRNAs (Fig. 6G, p > 0.3 for CaMKIIα mRNA; *, p < 0.05 for GABA-A-R-δ mRNA increase, n = 30 cells, 2–3 dendrites per cell). The PAT1 siRNA-mediated decrease in β-actin mRNA localization presumably affects the association between ZBP1 and kinesin. Therefore, the overlap between ZBP1 and kinesin in dendrites was tested. The fluorescence images showed that the co-localization between ZBP1 and KHC in dendrites was diminished upon reduction of PAT1 (Fig. 7A, **, p < 0.01, n = 18 cells, 3–4 dendrites per cell).
Figure 7.
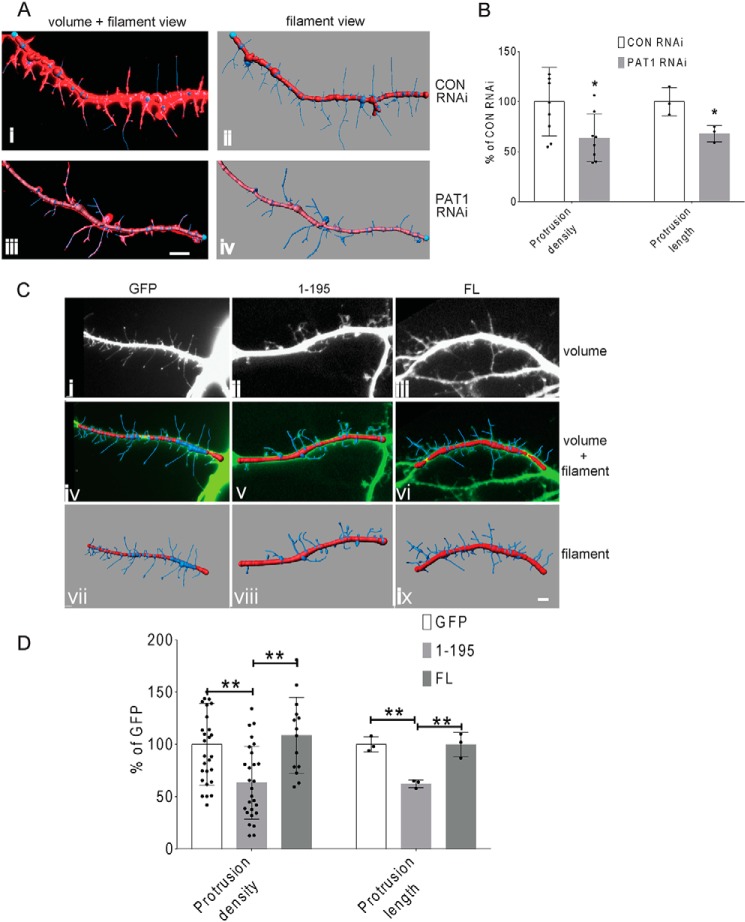
A, repression of PAT1 led to decreased co-localization between KHC and ZBP1. Mouse hippocampal neurons (7 DIV) were immunostained for KHC (green) and ZBP1 (red) after 72 h of control (CON) or PAT1 siRNA treatment. Pearson's correlation between KHC and ZBP1 was analyzed. The histogram represents the Pearson's correlation between ZBP1 and KHC after two treatments; results indicate deceased KHC/ZBP1 co-localization after PAT1 siRNA treatment (**, p < 0.01, n = 18 cells, 3–4 dendrites per cell, error bar, ±S.D.) Scale bar, 10 μm. Three repeats were done independently for these experiments with consistent findings. B, 1–195 ZBP1 acts as a dominant-negative protein because it is bound to PAT1 and has compromised interaction with the endogenous full-length ZBP1. HEK-293 cells were transfected with either full-length ZBP1–FLAG or 1–195 ZBP1–FLAG, and full-length ZBP1–GFP was co-transfected. Immunoprecipitation between the full-length ZBP1–GFP (FL ZBP1–GFP), full-length ZBP1–FLAG (FL ZBP1–FLAG), and 1–195 ZBP1–FLAG (1–195 ZBP1–FLAG) was carried out with anti-FLAG beads. Lanes 1 and 3, 1–195 ZBP1–FLAG + FL ZBP1–GFP; lanes 2 and 4, FL ZBP1–FLAG + FL ZBP1–GFP. Blots indicate that 1–195 ZBP1–FLAG cannot interact with full-length ZBP1–GFP and thus acts as in a dominant-negative manner. Three repeats were done independently for these experiments with consistent findings. C, dominant-negative 1–195 ZBP1 interferes with the dendritic localization of β-actin mRNA. Mouse hippocampal neurons (7 DIV) were transfected with different ZBP1 constructs for 24 h, stimulated with BDNF for 1 h, and then fixed. FISH was performed to visualize β-actin mRNA (red). Either GFP alone (GFP), GFP + full-length ZBP1–FLAG (FL), GFP+ 1–195 ZBP1–FLAG (1–195), or GFP + 1–195 ZBP1–FLAG+ full-length PAT1–FLAG (1–195 + PAT1 FL) were transfected into neurons, and GFP-positive neurons were quantified for dendritic β-actin mRNA intensity. Right, histogram shows dendritic β-actin mRNA intensities from different transfections. Results indicate that 1–195 ZBP1 overexpression causes decreased β-actin mRNA intensity in dendrites, which can be overridden by co-transfecting 1–195 ZBP1–FLAG with full-length PAT1. Scale bar, 10 μm. (**, p < 0.01; ***, p < 0.001, n = 40 cells, 3–4 dendrites per cell, error bar, ±S.D.) Three repeats were done independently for these experiments with consistent findings. D, BDNF stimulation significantly increases the density of dendritic protrusions in cultured hippocampal neurons. Mouse hippocampal neurons (7 DIV) were stimulated with BDNF for 1 h, then fixed, and immunostained for F-actin (phalloidin, red). Left, representative dendrites with protrusions from either treatment. Right, histogram shows the change in dendritic protrusion density after BDNF treatment, indicating an increase of the protrusion density in response to BDNF stimulation. Protrusion density was measured as the number of dendritic protrusions per 10-μm dendrite. Scale bar, 5 μm. (***, p < 0.001, n = 32 cells, 2–3 dendrites per cell, error bar, ±S.D.) Six repeats were done independently for this experiment with consistent findings.
To confirm the data gathered from the PAT1 repression experiments, a dominant-negative approach based on the ZBP1 1–195 was developed. Because the interaction between ZBP1 and PAT1 depends on the N terminus of ZBP1 (ZBP1 1–195), overexpression of the N terminus of ZBP1 might act in a dominant-negative manner to compete with the endogenous ZBP1/β-actin mRNA complexes for the PAT1 association. HEK-293 cells were transfected with either the full-length or the N terminus of ZBP1–FLAG, and full-length ZBP1–GFP was co-transfected. The co-IP result showed that ZBP1 1–195 (1–195 ZBP1–FLAG) does not multimerize with the full-length ZBP1 (FL ZBP1–GFP) (Fig. 7B), confirming the dominant-negative manner of the ZBP1 N terminus. Mouse hippocampal neurons were transfected with the dominant-negative 1–195 ZBP1–FLAG or full-length ZBP1–FLAG; GFP was co-transfected to identify positive transfections, and the GFP transfection alone served as the negative control. Based on the previous co-IP experiment, we estimated the expression level of the dominant-negative 1–195 ZBP1–FLAG is about 70–80% of the full-length ZBP1–FLAG. After 24 h expression of the dominant-negative 1–195 ZBP1–FLAG, an ∼46% reduction in the dendritic β-actin mRNA intensity was observed in response to the BDNF stimulation, compared with the control group that only expressed GFP alone (Fig. 7C, ***, p < 0.001, n = 30 cells, 2–3 dendrites per cell) and an ∼62% reduction compared with the group expressing the full-length ZBP1 (FL ZBP1–FLAG) (Fig. 7C, ***, p < 0.001, n = 32 cells, 2–3 dendrites per cell). There was a small increase in the dendritic β-actin mRNA intensity in the full-length ZBP1–FLAG–expressing neurons, which was not significant (Fig. 7C, p > 0.3, n = 28 cells, 2–3 dendrites per cell). If indeed the dominant-negative 1–195 ZBP1–FLAG competes with the endogenous ZBP1, then the overexpression of PAT1 should have the ability to override this effect. Upon co-expression of the dominant-negative 1–195 ZBP1–FLAG and PAT1–FLAG in neurons, the dominant-negative effect of 1–195 ZBP1–FLAG on dendritic β-actin mRNA localization was partially rescued by ∼32% (Fig. 7C, **, p < 0.01, n = 15 cells, 3–4 dendrites per cell). Another notable phenomenon is the uneven distribution of β-actin mRNA along the dendrites after the dominant-negative 1–195 ZBP1 transfection. Being proximal near the soma, the dendrite shows gradually decreased β-actin mRNA granules. The β-actin mRNA also accumulates at the proximal dendrite, similar to the pattern of ZBP1 accumulation after the depolymerization of microtubules. After the rescue effect of PAT1 overexpression, there is less proximal accumulation and more dendritic distribution of β-actin mRNA. Taken together, these data demonstrated that PAT1 is required for the BDNF-induced transport of β-actin mRNA/ZBP1 complex into neuronal dendrites.
Interference with PAT1 reduces BDNF-induced dendritic protrusion formation and growth cone development
ZBP1 is involved in dendritic protrusion growth in response to BDNF (46). Therefore, the changes in dendritic protrusions after BDNF treatment was examined to determine the role of PAT1 in this process. Hippocampal neurons were fixed and immunostained for phalloidin and MAP2, and cells were imaged under blind conditions to capture secondary and tertiary dendrites. A significant increase in the density of protrusions after BDNF stimulation was observed compared with unstimulated neurons (Fig. 7D, ***, p < 0.001, n = 32 cells, 2–3 dendrites per cell).
To determine the roles of PAT1-mediated β-actin mRNA transport in dendritic protrusion development, both the PAT1 siRNA and the dominant-negative ZBP1 1–195 approaches were applied to mouse primary hippocampal neurons. Imaris (Bitplane, Inc) was used to create a mask of the protrusions to facilitate the quantification of over 2000 protrusions. Upon PAT1 siRNA treatment, the increase in protrusion density caused by BDNF stimulation was inhibited by ∼40%. This was accompanied by a concomitant inhibition in the BDNF-induced lengthening of protrusions to a similar extent (Fig. 8, A and B, ***, p < 0.001, n = 34 cells, 2–3 dendrites per cell). Expression of the dominant-negative 1–195 ZBP1–FLAG in mouse primary hippocampal neurons resulted in an ∼42 and ∼49% reduction in protrusion density compared with GFP and full-length ZBP1–FLAG-transfected neurons, respectively (Fig. 8, C and D, ***, p < 0.001, n = 31 cells, 2–3 dendrites per cell). There was no significant difference in protrusion density between GFP alone and full-length ZBP1–FLAG-transfected neurons (p > 0.3, n = 29 cells, 2–3 dendrites per cell), although the average protrusion density was increased in the full-length ZBP1–FLAG-transfected neurons. The expression of dominant-negative 1–195 ZBP1–FLAG also caused a similar reduction in the length of protrusions (Fig. 8, C and D, ***, p < 0.001, n = 31 cells, 2–3 dendrites per cell), whereas the expression of GFP alone or full-length ZBP1–FLAG did not appreciably alter the lengths of protrusions (Fig. 8, C and D, p > 0.4, n = 28 cells, 2–3 dendrites per cell). The decrease in the protrusion density and length after perturbation of the PAT1–ZBP1 interaction suggested that the PAT1-mediated delivery of β-actin mRNA is important for BDNF-induced rapid changes in dendritic protrusions.
Figure 8.
A, PAT1 siRNA treatment decreases the dendritic protrusion density in cultured hippocampal neurons. Mouse hippocampal neurons (7 DIV) were treated with either control siRNA (panels i and ii) or PAT1 siRNA (panels iii and iv) for 72 h and stimulated with BDNF for 1 h. F-actin was stained with phalloidin to visualize dendritic protrusions (panels i and iii, red). Panels ii and iv, dendritic segments were captured in 3D, deconvolved, and reconstructed. The protrusions are traced, and the mask (filaments) was overlaid onto the volume view (panels i and iii) or shown alone (blue lines with red main dendrite branch) (panels ii and iv). Scale bar, 5 μm. B, histograms show the changes in dendritic protrusion density and protrusion length after control siRNA and PAT1 siRNA treatment. Left, protrusions densities were measured as per 10 μm dendrite and averaged. (n = 30 cells, 2–3 dendrites per cell, 1125 protrusions counted, *, p < 0.05.) Right, protrusion lengths were measured after each treatment (n = 30 cells, 2–3 dendrites per cell, 1125 protrusions counted, *, p < 0.05, error bars, ±S.D.). The data indicate that PAT1 repression significantly decreases the protrusion density and protrusion length. Three independent repeats were done for these experiments with consistent findings. C, expression of dominant-negative ZBP1 1–195 causes decreased density and length of dendritic protrusions in cultured hippocampal neurons. Mouse hippocampal neurons (7 DIV) were transfected with different ZBP1 constructs for 24 h and then stimulated with BDNF for 1 h. Either GFP alone (GFP) or GFP + full-length ZBP1–FLAG (FL) or GFP + 1–195 ZBP1–FLAG (1–195) were transfected into neurons, and the GFP-positive neurons were quantified for protrusion density and length. Dendritic segments were captured in 3D, deconvolved, and reconstructed (top panels, i–iii), and then the protrusions were traced. The protrusions mask filaments are overlaid (middle panels, blue lines with red main dendrite branch, panels iv–vi). The filament mask alone is shown in bottom panels (vii–ix) to highlight protrusions. Scale bar, 5 μm. D, histograms show protrusion density and length are decreased after the expression of dominant-negative ZBP1 1–195. Left, protrusion densities were measured per 10-μm dendrite for different transfections. (n = 32 cells, 2–3 dendrites per cell, 1255 protrusions; **, p < 0.01, error bar, ±S.D.) Right, protrusion lengths were measured (μm) for different transfections. (n = 32 cells, 2–3 dendrites per cell, 1255 protrusions; **, p < 0.01, error bar, ±S.D.) Quantification results indicate that the overexpression of the dominant-negative 1–195 ZBP1 causes significant decrease on protrusion density and length, and overexpression of full-length ZBP1 has no significant change on protrusion density and length. Six repeats were done independently for this experiment with consistent findings.
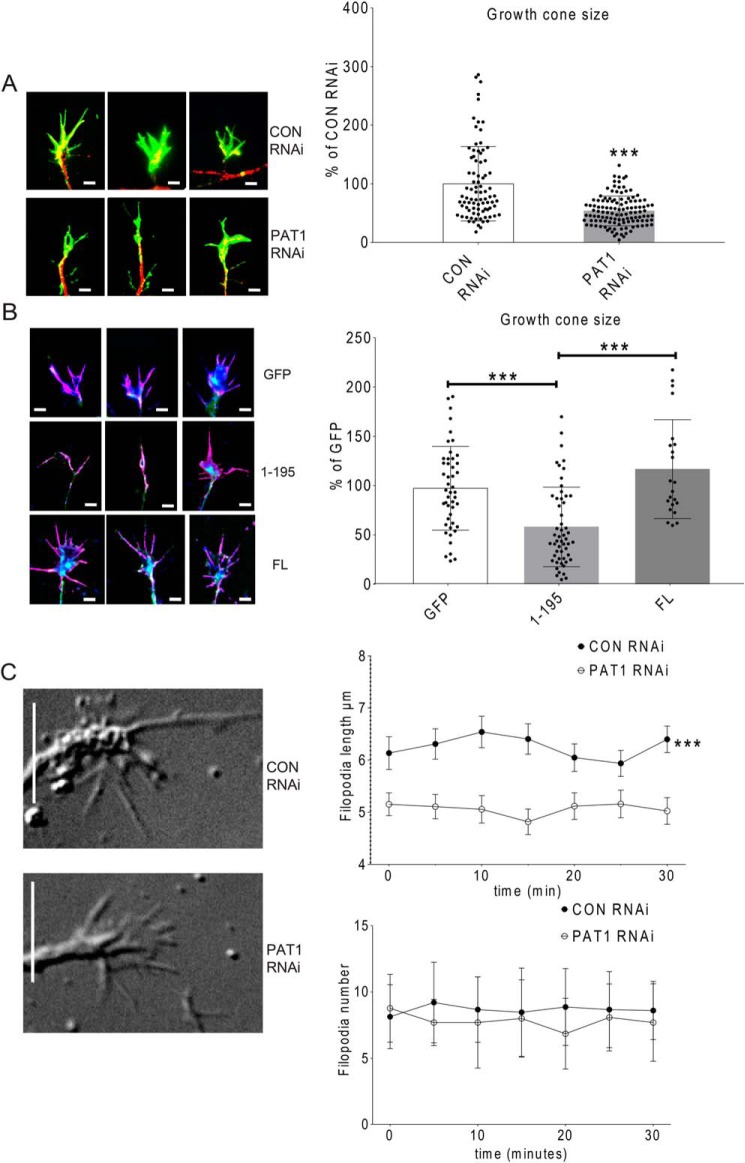
Because PAT1 repression affects dendritic protrusion dynamics, the filopodia on growth cones might be regulated by the PAT1-mediated β-actin mRNA transport as well. Very little is known about how dendritic growth cone morphology is regulated or how their dynamics are modified during development (45). Mouse hippocampal neurons (7 DIV) were transfected with PAT1 siRNA or the dominant-negative 1–195 ZBP1–FLAG construct. Neurons were stimulated with BDNF for 1 h, fixed, and processed with immunostaining for MAP2 and phalloidin. The size of growth cones was measured. On average, in the control siRNA-treated group, growth cones were ∼124 μm2 in size. Upon PAT1 siRNA treatment, the growth cone size on average was significantly reduced by ∼46% (from 124 to 67 μm2) (Fig. 9A, ***, p < 0.001, n = 30 cells, 2–3 growth cones per cell). The average growth cone size in the control siRNA-treated neurons was similar to that observed in the only GFP-transfected neurons (Fig. 9B), which on average was ∼135 μm2 in size. Upon the dominant-negative 1–195 ZBP1–FLAG transfection, the growth cone size decreased ∼43 and ∼66% compared with the GFP alone (Fig. 9B, ***, p < 0.001, n = 31 cells, 2–3 dendrites per cell) or the full-length ZBP1–FLAG-transfected neurons (Fig. 9B, ***, p < 0.001, n = 30 cells, 2–3 growth cones per cell). In contrast, the full-length ZBP1–FLAG-transfected neurons showed no significant change in growth cone size compared with the GFP-transfected neurons (Fig. 9B, p > 0.3, n = 28 cells, 2–3 growth cones per cell). Notable was the partial collapse of most growth cone lamellae in both PAT1 siRNA and dominant-negative 1–195 ZBP1–FLAG-treated neurons, accounting for the decrease in size.
Figure 9.
A, PAT1 siRNA treatment results in a decrease in the size of dendritic growth cones in cultured hippocampal neurons. Mouse hippocampal neurons (7 DIV) were treated with either control siRNA or PAT1 siRNA and stimulated with BDNF for 1 h. Neurons were fixed and immunostained for MAP2 (red) and phalloidin (green) to visualize dendrites and growth cones. Left, representative growth cones are shown, with MAP2 and phalloidin-stained, highlighting dendritic growth cones between the two treatments. Right, histogram shows the difference in dendritic growth cone size between the two treatments, indicating a decrease in the growth cone size from the PAT1 siRNA-treated neurons. Scale bar, 2 μm. (n = 38 cells, 2–3 growth cones per cell; ***, p < 0.001, error bar, ±S.D.) Four independent experiments were done for this analysis with consistent findings. B, dominant-negative ZBP1 1–195 overexpression results in the decreased size of dendritic growth cones. Mouse hippocampal neurons (7 DIV) were transfected with different constructs for 24 h and then immunostained for MAP2 (blue) and phalloidin (purple) to identify dendritic growth cones. Either GFP alone (GFP) or GFP + full-length ZBP1–FLAG (FL) or GFP + 1–195 ZBP1–FLAG (1–195) were transfected into neurons, and GFP-positive neurons were quantified for the growth cone size. Left, three representative images show growth cone morphology from each treatment. Right, histogram shows the growth cone size is different between different transfections, indicating a decrease in the growth cone size in the dominant-negative ZBP1 1–195 group compared with the control groups. Scale bar, 2 μm. (n = 40 cells, 2–3 dendritic growth cones for each treatment, **, p < 0.001, error bar, ±S.D.) Four repeats were done independently for this experiment with consistent findings. C, PAT1 siRNA treatment results in a shorter length of neuronal growth cone filopodia in living hippocampal neurons. Mouse hippocampal neurons cultured in chambered coverglass (2 DIV) were treated with either control siRNA or PAT1 siRNA for 72 h, stimulated with BDNF for 1 h, and then imaged using DIC video microscopy to record growth cones for 30 min. Left, representative growth cones from each treatment are shown (right). The histograms show the differences in the growth cone filopodial length and number. The results indicate shorter filopodia after PAT1 siRNA treatment (n = 28 cells, n = 1–2 growth cones per cell; ***, p < 0.001, error bar, ±S.D.) However, the number of filopodia is not significantly affected (n = 25 cells, n = 1–2 growth cones per cell; p > 0.4, error bar, ±S.D.) Scale bar, 5 μm. Four repeats were done independently for this experiment with consistent findings.
The effect of PAT1 repression on growth cone development could arise from either a decrease in the nucleation of new filopodia or from a decrease in the length of filopodia. To determine the change in filopodial dynamics in PAT1 siRNA-treated neurons, the growth cones were analyzed by live-cell imaging technology. 72 h after transfection, neurons were stimulated with BDNF for 1 h and recorded over 30 min by DIC microscopy. Growth cone filopodia in the PAT1 siRNA-treated neurons were significantly shorter over the time course compared with the control siRNA-treated neurons (Fig. 9C, ***, p < 0.001, n = 28 growth cones, 3–8 filopodia per growth cone). However, no significant difference was observed in the number of filopodia emerging from the growth cones over this time course in the PAT1 siRNA-treated neurons (Fig. 9C, p > 0.4, n = 28 cells, 3–8 filopodia per growth cone), although this number was smaller on average (16%) compared with that of the controls. These data indicated that PAT1 actively maintains the size of the growth cone in response to BDNF through sufficient β-actin mRNA delivery. However, the filopodial nucleation was less affected by the inhibition of PAT1.
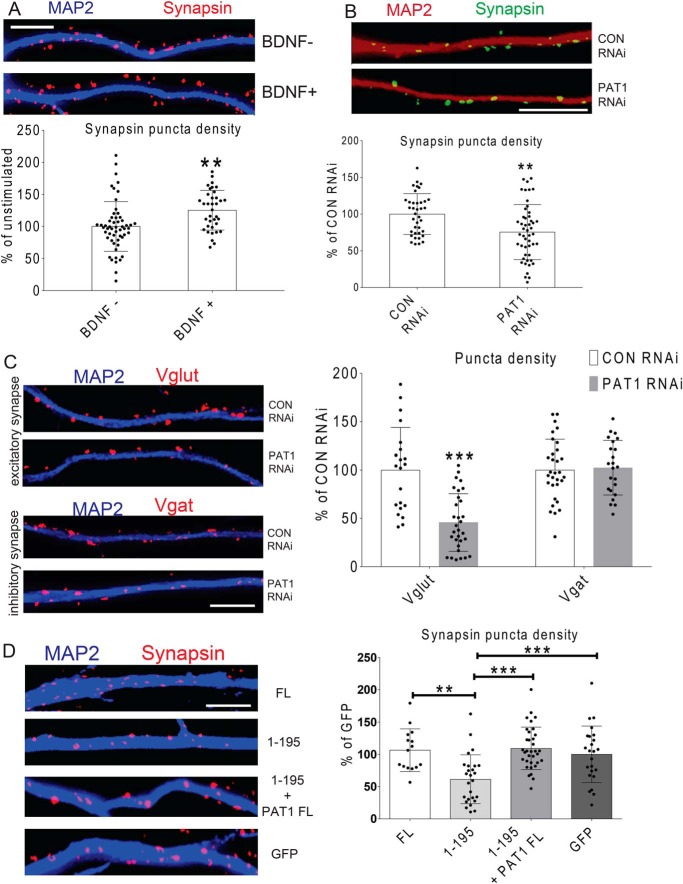
Given the role of PAT1 in regulating dendritic protrusion density, the role of PAT1-mediated β-actin mRNA delivery in the regulation of potential synapse development was explored. Because there is an inherent heterogeneity of synapse density that depends on cell culture density, low-density neuronal cultures that routinely showed single neurons in multiple fields that were comparable in numbers between treatments by blind analysis using DIC microscopy were used. Neurons were stained for MAP2 and synapsin, and the densities of dendritic synapsin puncta were automatically counted by thresholding of the synapsin staining and were equally applied to all cells. Somatic synapses were completely omitted in this analysis, as the dendritic synapsin puncta were counted starting 20 μm out from the cell body. Using these criteria, the densities of synapsin puncta after BDNF treatment were counted. The results indicated an ∼34% increase within just 1 h after BDNF application (Fig. 10A; **, p < 0.01, n = 36 cells, 2–3 dendrites per cell). In addition, upon PAT1 siRNA treatment, the density of synapsin puncta decreased significantly (∼33%) compared with control siRNA-treated neurons (Fig. 10B; **, p < 0.01, n = 31 cells, 2–3 dendrites per cell).
Figure 10.
A, BDNF stimulation results in a significant increase in the density of synapsin puncta in cultured hippocampal neurons. Mouse hippocampal neurons were treated with either control siRNA or PAT1 siRNA for 72 h at 7–9 DIV, stimulated with BDNF for 1 h, and then stained for MAP2 (blue) and synapsin (red) to visualize dendritic synapsin puncta. Top, representative dendrite shows MAP2 and synapsin staining highlighting potential dendritic synapses. Bottom, histogram shows the change in dendritic synapsin puncta density after BDNF treatment, showing an increase in response to the BDNF treatment. Synapsin puncta were measured per 10-μm dendrite. Scale bar, 10 μm. (n = 25 cells, 2–3 dendrites per cell; **, p < 0.01, error bar, ±S.D.) Three repeats were done independently for this experiment with consistent findings. B, PAT1 siRNA treatment results in a decrease in the dendritic synapsin puncta density in cultured hippocampal neurons. Mouse hippocampal neurons were cultured for 7–9 DIV, treated with either control siRNA or PAT1 siRNA, stimulated with BDNF for 1 h, and then stained for MAP2 (red) to visualize dendrites and synapsin (green) for synapsin puncta. Top, representative dendrite shows MAP2 and synapsin staining between the two treatments. Bottom, histogram quantified the change in dendritic synapsin puncta density after PAT1 siRNA treatment and showed a decrease in response to PAT1 RNAi. Synapsin puncta numbers were measured per 10-μm dendrite. Scale bar, 10 μm, (n = 32 cells, 2–3 dendrites per cell; **, p < 0.01, error bar, ±S.D.) Four independent repeats were done for this experiment with consistent findings. C, PAT1 siRNA treatment results in a decrease in the density of dendritic Vglut puncta in cultured hippocampal neurons. Mouse hippocampal neurons were cultured for 7–9 DIV, treated with either control siRNA or PAT1 siRNA, stimulated with BDNF for 1 h, and then stained for MAP2 (blue) to visualize dendrites and Vglut or Vgat (red) for potential excitatory or inhibitory synapses. Top panels, representative dendrites show MAP2 and Vglut staining between the two treatments. Bottom panels, representative dendrites show MAP2 and Vgat staining between the two treatments. Right, histograms show the change in Vglut/Vgat puncta density after PAT1 siRNA treatment, showing a decrease in Vglut puncta density but no significant decrease in the number of Vgat puncta. Puncta numbers are measured per 10-μm dendrite. Scale bar, 10 μm. (***, p < 0.001, n = 22 cells, 3–4 dendrites per cell, error bar, ±S.D.) Three independent repeats were done for this experiment with consistent findings. D, overexpression of dominant-negative ZBP1 decreases the number of synapsin puncta, which can be rescued by PAT1 overexpression. Mouse hippocampal neurons were cultured for 7–9 DIV and transfected with different constructs for 24 h, stimulated with BDNF for 1 h, and then fixed and stained for MAP2 (blue) and synapsin (red) to identify dendritic synapsin puncta. Left, representative dendrites show transfections from GFP alone (GFP), GFP + full-length ZBP1–FLAG (FL), GFP + 1–195 ZBP1–FLAG (1–195), or GFP + 1–195 ZBP1–FLAG + PAT1–FLAG (1–195 + PAT1). GFP-positive neurons were quantified for synapsin puncta density for each treatment. Right, histograms show synapsin puncta quantification from different transfections indicate a decrease of synapsin puncta density and the rescue by overexpression of PAT1. Synapsin puncta were measured per 10-μm dendrite. Scale bar, 10 μm (n = 30 cells, 2–3 dendrites per cell; **, p < 0.01; ***, p < 0.001, error bar, ±S.D.) Three independent repeats were performed for this experiment with consistent findings.
Because it is possible that knocking down PAT1 affects both the potential excitatory and inhibitory synapse development, the vesicular presynaptic markers Vglut (glutamatergic) and Vgat (GABAergic) were stained after PAT1 knockdown. Surprisingly, there was an ∼50% decrease in the density of Vglut puncta (Fig. 10C; ***, p < 0.001, n = 22 cells, 3–4 dendrites per cell) but no significant change in Vgat puncta onto hippocampal dendrites (Fig. 10C; p > 0.4, n = 20 cells, 3–4 dendrites). In a parallel approach with the dominant-negative 1–195 ZBP1–FLAG overexpression, the density of synapsin puncta was decreased by ∼45% compared with GFP-transfected control neurons (Fig. 10D; ***, p < 0.001, n = 34 cells, 2–3 dendrites per cell). In contrast, full-length ZBP1–FLAG overexpression had little effect on synapsin puncta density and about a 5% increase compared with GFP-transfected control neurons (Fig. 10D; p > 0.4, n = 30 cells, 2–3 dendrites per cell). If the dominant-negative 1–195 ZBP1 interferes with β-actin mRNA-mediated synapsin puncta formation through PAT1, then the overexpression of PAT1 should compensate for this effect. A rescue of synapsin puncta density in response to BDNF upon co-expression of PAT1–FLAG and 1–195 ZBP1–FLAG in neurons was observed (Fig. 10D; ***, p < 0.001, n = 28 cells, 3–4 dendrites per cell). Taken together, the data strongly supported the role of PAT1 in mediating dendritic transport of β-actin mRNA in the positive regulation of potential synapse formation and protrusion morphology in response to BDNF stimulation.
Discussion
This study aimed to reveal the role of a potential protein adapter between the motor proteins and the β-actin mRNA-transporting complex. This finding sheds light on kinesin-facilitated dendritic mRNAs transport during neuronal development and how BDNFs regulate this process. Here, we elucidate a direct link from kinesin to the β-actin mRNA/ZBP1 complex through a ZBP1–PAT1–KLC interaction that can be regulated by BDNF. This is the first example of a motor adapter that mediates mRNA localization and direct molecular linkage of a specific RBP to kinesin. This model suggests PAT1 as a new link between localized dendritic mRNAs and motors. In this model, ZBP1 binds to the 3′ UTR zipcode of the β-actin mRNA; the N terminus of ZBP1 binds to PAT1, which associates to KLC and activates kinesin for microtubule-dependent β-actin mRNA transport. We also studied PAT1's potential role in BDNF-activated dendritic protrusion formation and growth cone development. Many mRNAs are potentially actively transported to dendrites (5), so PAT1 may deliver other mRNAs to the dendrites, and this study opens the path to discover the mechanisms for diverse mRNAs in neurotrophin-dependent signaling to distinct molecular motor adapters during development.
PAT1 as a new adapter protein for mRNA granule transport through kinesin
Because the PAT1 sequence is similar to KLC, it may be that PAT1 is a novel subunit of kinesin-I in vertebrate neurons. However, the yeast two-hybrid and multiple co-IP experiments failed to show a direct interaction between KHC and PAT1. Our preliminary data showed that PAT1 interacts with the TPR domain of KLC (Fig. S1D), probably through the PAT1–HR domain (data not shown). It is suspected that the binding site on KLC to PAT1 is distinct from that of other KLC-associated vesicular cargoes such as the JIPs mentioned earlier. Exploring the details of this KLC/PAT1/ZBP1 complex will require further experiments. Nevertheless, the current data illustrate a model that may facilitate future experiments (Fig. S2).
The identification of PAT1 as a direct adapter between an RBP and KLC is the first observation of such a linkage between mRNA granule transport and a plus-end–directed motor. Kinesin is implicated in mRNA transport in various cell types (46, 47) and specifically in primary neurons for several dendritic transcripts (11, 19, 26, 48). KHC was shown to associate with over a dozen RBPs in neurons, including Pur-α, Staufen, FMRP, and hnRNP family members, and is required for the transport of CaMKIIα and Arc mRNAs (14). mRNA granules are heterogeneous, as Kanai et al. (14) found that kinesin family member 5B (KIF5b) associates with large RNA granules that contain 42 proteins and two well-studied dendritically-localized mRNAs, CaMKIIα and Arc (14), and another paper (49) that biochemically isolated RNA granules in developing rat brains that are enriched for β-actin mRNA but not for CaMKIIα mRNA. Elvira et al. (49) identified ZBP1 in their proteomic analysis of developing rat brain, whereas Kanai et al. (14) did not. The heterogeneity of RNA granules makes it hard to determine whether or not PAT1 exclusively transports β-actin mRNA. PAT1 repression diminishes β-actin mRNA transport, but not CaMKIIα and GABA-A-R-δ mRNA. These data pointed out the specificity of the PAT1–ZBP1 interaction, although it remains possible that both the CaMKIIα and GABA-A-R-δ mRNAs transport through PAT1, but BDNF stimulation works with a distinct signaling pathway to regulate their transport. The complexity of CaMKIIα mRNA transport likely derives from its interactions with numerous RBPs, such as translin (26, 50), CPEB (19), FMRP (11, 51, 52), hnRNPA2 (13, 53), and Staufen (25, 54). Given their responses to BDNF, it is curious that they belong to distinct mRNPs (55). Several Staufen isoforms influence the dendritic localization of β-actin mRNA (18, 56), suggesting an overlap because CaMKIIα mRNA is also localized to Staufen mRNPs (25). ZBP1 inhibition resulted in a reduced dendritic β-actin mRNA localization in cultured hippocampal neurons, whereas the localization of CaMKIIα mRNA was unaffected (43). Neurons also showed reduced growth of dendritic protrusions after ZBP1 inhibition, similar to our results. Here in this paper, we show the interaction between ZBP1 and PAT1, and interference with PAT1 through siRNA and dominant-negative 1–195 ZBP1 led to similar morphological changes as expected. Although these data provided insightful understanding of the different RNA granules and the overlapped components between them, the heterogeneity of the RNA granules isolated by Kanai et al. (14) and Elvira et al. (49) makes it hard to assemble a coherent view of these macromolecules. The RNA-transporting granules were isolated by the immobilized kinesin tail, whereas the RNA granules isolated by Elvira et al. (49) were based on differential centrifugation and sucrose gradient. Perhaps other approaches should be utilized, such as the MS2 system that was used to visualize the β-actin mRNA granule transport. Knocking in multiple MS2 motifs into the β-actin mRNA or any other mRNAs for that matter should allow isolation of homogeneous mRNA granules, along with its binding proteins and transport complexes, potentially enriching PAT1. How does BDNF signal through TrkB to regulate mRNA localization? BDNF increases the proportion of motile dendritic mRNA granules (49) and stimulates the Src-dependent phosphorylation of ZBP1, which is required for the β-actin protein synthesis in growth cones (44). The phosphorylation at Tyr-396 appears to be specific to translational regulation (44) rather than mRNA-binding regulation (57). Other ZBP1 potential phosphorylation sites may regulate the interaction between ZBP1 and PAT1 once mRNAs are loaded onto ZBP1, similar to that reported for fasciculation and elongation protein ζ1 (FEZ1)-mediated transport of syntaxin to presynaptic terminals (58).
Another observation from these live-imaging experiments is that the granules move bidirectionally or oscillate, which implies other motors are contained in the complex. Kinesin and dynein are often coupled together, and the interactions between dynactin and kinesin-2 and kinesin-5 have been reported, where dynactin increases the processivity of kinesin-2 through microtubule binding (59, 60), but the function of this interaction is unclear (61). One model to explain the presence of both motor proteins during transport implies the functions of both motors are controlled by their engagement (62, 63), by modulating the on/off rate of motors to the cytoskeleton or via adapters or post-translational modification of motor proteins to achieve such regulation of transport. Another “tug of war” model proposes that motor proteins fight over cargoes and the strongest force wins, supported by high-resolution analysis of vesicles. The stoichiometric ratio of kinesin-2 and dynein could determine vesicle transport direction (64). The directionality of mRNA transport may apply a similar “tug of war” model.
The identification of other RBPs that bind to PAT1 for dendritic mRNA transport is important. PAT1 was first identified as a basolateral sorting signal of APP. It is possible that PAT1 functions dual roles for both mRNA and vesicular transport because purification of RBPs can yield both of them (25). The question of whether distinct cargo types can co-transport is an important area to delineate. This question also applies to distinct mRNA cargoes because mRNA granules are quite large (∼0.3–0.6 μm diameter) (65, 66), and RBPs associate with multiple, diverse mRNAs. Still, it is not clear whether or not other RBPs and their mRNA targets ride along with ZBP1/β-actin mRNA complexes through heteromeric interactions, which have been proposed for several dendritic mRNAs, including CaMKIIα, Arc, and MAP1b through FMRP (52).
Models for mRNA transport
In yeast, the RBP She2p binds to the Ash1 mRNA and simultaneously to the She3p protein, which recruits the myosin Myo4p (67). Functionally, She3p is analogous to PAT1 as an adapter between RBPs and the motor proteins. In Drosophila, kinesin is essential for the posterior oskar mRNA localization in oocytes (46, 68). PAT1 was found to play a role in the oskar mRNA localization; KLC overexpression can rescue the defects in oskar mRNA localization in PAT1 mutants (40). However, the role of Drosophila PAT1 in mediating a direct link between mRNA complexes and KHC remains undetermined, as KLC appears to be dispensable for some aspects of the KHC-dependent oskar mRNA transport in Drosophila (69). The essential function of KLC in mice (70) suggests a more significant role for the PAT1-mediated kinesin function in vertebrates. Drosophila Egl binds directly to several mRNAs localized in the syncytial blastoderm embryo that requires dynein and BicD (35). As a dynein-interacting protein that can specify some minus-end transport in vertebrates (37), BicD serves as the adapter protein between dynein and the mRNAs/Egl complex, but the fact that Egl also binds directly to DLC is confounding (35). It is interesting that BicD negatively regulates dynein, as does KLC for kinesin function in vertebrates (71). Drosophila FMRP is implicated in neuronal mRNA transport (72), and BicD was shown to interact with FMRP. However, there is no evidence for a role of BicD in the FMRP-mediated mRNA transport (73). Future studies will determine how PAT1 and BicD mediate kinesin- and dynein-specific mRNA localization, and whether they act simultaneously on these motor complexes that show alternating plus-end- and minus-end–directed mRNAs movements (11, 12, 15, 17, 74).
PAT1's function in BDNF induced neuronal development
BDNF induces synaptic plasticity in neurons through a translation-dependent mechanism (75, 76) and structurally remodels synapses (77). BDNF plays a vital role in the regulation of the translation machinery itself (76, 78, 79). Several plasticity transcripts are implicated in this process, although the details are unknown. Arc mRNA is increased by BDNF and bound by hnRNPA2 (13) and FMRP (52), although its interaction with FMRP through the noncoding RNA BC1 is controversial (80).
The transport of both Arc and CaMKIIα mRNAs requires functional kinesin (11, 14). It is interesting that BDNF-induced delivery and translation of β-actin mRNA may regulate the F-actin–dependent Arc mRNA anchoring at active synapses (81), providing spatially controlled translation events in response to BDNF-induced long-term potentiation. Because translation is sequestered to F-actin (82, 83), the β-actin mRNA transport and translation may be critical for highly-accurate positioning of transcripts in response to signaling. BDNF regulates actin-dependent spine and synapse formation during development (43, 84–86). However, the molecular mechanism is not clear (75). BDNF signaling to β-actin mRNA may result in the convergence of localized signals to trigger the motor-dependent transport and translation of synaptic scaffold transcripts. ZBP1 has been suggested to play a role in the development of neuromuscular junction (87). Indeed, a role for localized forms of protein synthesis in synapse development and function is just emerging (89).
Our data showed that interfering with β-actin mRNA transport by repression of PAT1 resulted in decreased dendritic protrusion and synapsin puncta density. Furthermore, altered β-actin mRNA localization only seems diminished at excitatory synapses and not inhibitory synapses based on our data. It is reasonable to expect fewer spines and excitatory synapses were developing from PAT1-repressed neurons. This result suggested that PAT1-mediated β-actin localization has certain roles in filopodia/protrusion and synapse remolding, but future research is required to elucidate the details. We examined the role of actin dynamics in neuronal growth cones; upon PAT1 siRNA or dominant-negative 1–195 ZBP1 transfection, growth cones were impaired compared with the controls in the presence of BDNF; the length of growth cone filopodia was diminished after PAT1 siRNA treatment, but not the number of filopodia. These data are consistent with a study showing that axonal growth cone guidance is impaired in neurons lacking ZBP1 (41). It is interesting to determine the roles for PAT1 in β-actin–mediated turning of dendritic growth cones and how ZBP1 and PAT1 may regulate this transport- and translation-dependent process in asymmetrical turning assays in the future.
Materials and methods
Animals and dissociated hippocampal cultures
FVB/129 mice with either sex were kept in a controlled environment. Animal facility protocols were approved by the Institutional Animal Care and Use Committee. Mice were housed in the AALAC-approved animal facility at Hunter College. Low-density–dissociated neuronal cell cultures were prepared as described from P0 mice, with some modifications (11). Hippocampi from P0 mice were separated from the rest of the brain, collected, and digested with trypsin for 15 min, dissociated in plating medium (minimum Eagle's medium, 10% fetal bovine serum, 10 mm HEPES, 33 mm glucose) with glass pipettes, and plated at low density (25,000 cells/cm2) on poly-l-lysine–coated (0.1 mg/ml) and waxed coverslips. 2 h after plating, the coverslips were inverted from the plating media to the maintenance media (astrocytes-conditioned Neurobasal/B27/GlutaMAX). Cortices were digested with trypsin and DNase I for 30 min, dissociated in the plating media with glass pipettes, and plated at high density (150,000 cells/cm2) in poly-l-lysine–coated (0.1 mg/ml) 60-mm plates. The plating media were replaced with the maintenance medium 2 h after plating. 48–72 h after plating, 1-β-d-arabinofuranosylcytosine (AraC) was added to a final concentration of 2.5 μm to curb glial proliferation. Depending on the experimental application, cultures were transfected or fixed or harvested. Neurons were treated with BDNF (50 ng/ml) or heat-inactivated BDNF for 1 h if needed and fixed (4% paraformaldehyde in 1× PBS with 4% glucose) for 20 min at room temperature.
Yeast two-hybrid
The yeast two-hybrid screen was carried out with a MATCHMAKER GAL4 two-hybrid system 3 (Clontech) according to the manufacturer's protocol. In brief, PJ69-4A cells were transformed with pGBD-ZBP1 1–195 and selected for growth on media lacking uracil. The PJ69-4A ZBP1 1–195 yeast was transformed with a mouse brain cDNA library in pACT2 (Clontech). Positive clones were selected and cultured on plates lacking uracil, leucine, and histidine; all plates were supplemented with 10 mm 3-aminotriazole. Positive clones were tested on plates lacking uracil, leucine, and adenine. The β-gal activity was tested on positive clones. To confirm the specificity of this interaction, PJ69-4A yeast (expressing only the library plasmid) were mated to Y187 yeast expressing the ZBP1 1–195, PTB, or the bait plasmid pGBD alone. To further determine the interaction between ZBP1 and PAT1, directed two-hybrid–binding tests between PAT1 and ZBP1, KHC, KLC, and PTB were carried out by yeast mating. Yeast strain AH109 expressing pGBD clones (bait, PAT1, ZBP1 N terminus, aa 1–195, and ZBP1 C terminus, aa 196–576, KLC, and KHC aa 692–833) were mated to yeast strain Y189 expressing pACT2 clones (prey, ZBP1, PAT1, KLC, KHC, and PTB) in 96-well plates. Diploid yeasts were sequentially plated on double (−leu and −trp) and triple (−leu, −trp, −his + 10 mm 3-aminotriazole and −leu, −trp, and −ade) drop-out plates. Successful mating was evidenced by growth on double drop-out (−leu and −trp) plates. A positive association between the prey and bait was evidenced by growth on −leu, −trp, and −his and on −leu, −trp, and −ade drop-out plates.
Expression and purification of recombinant PAT1–GST and in vitro binding assay
Recombinant GSH–S-transferase (GST) fusion protein was constructed by inserting the mouse PAT1 cDNA into a pGEX-6P-1 vector (GE Healthcare). pGEX-6P-1-PAT1 was expressed in E. coli BL21-Gold strain cells (Stratagene, La Jolla, CA). The recombinant protein was extracted with 1× PBS-containing protease inhibitors (PI), 1 mm 1,4-dithiothreitol (DTT), 0.1% Triton X-100, 2% glycerol, 850 mm NaCl, and 10 mm EDTA. The extract was applied to a GSH–SepharoseTM 4 Fast Flow (GE Healthcare). The Sepharose beads were eluted with 20 mm GSH. GSH and extra salt in the elute were eliminated by further purification using a disposable PD-10 desalting column (GE Healthcare). To prepare PAT1–GST for the in vitro binding experiment from the brain lysate, three pairs of P0 fresh cortex and hippocampi were homogenized into a final volume of 4 ml in IP buffer (10 mm HEPES (pH 7.3), 200 mm NaCl, 50 mm KCl, 30 mm EDTA, 1% Triton X-100, PI, and RNase inhibitor (RI)), and centrifuged at 3000 × g for 10 min at 4 °C to remove nuclei and debris. Supernatants were further centrifuged at 70,000 × g at 4 °C; the samples were pooled together and pre-cleared with GSH beads. The recombinant GST–PAT1 was bound to the GSH–SepharoseTM 4 Fast Flow beads and rotated overnight, with PI and RI included. The GSH beads with GST–PAT1 were washed three times with the brain IP buffer and rotated with the P0 brain lysate for 2 h at 4 °C. Half of the GSH beads were used for Western blotting, and the other half was kept for RT-PCR for β-actin mRNA
Immunostaining and FISH
Immunostaining was performed as described (Antar et al. (91)). Antibodies against ZBP1 (Abcam, ab82968, RRID:AB_1860674; Santa Cruz Biotechnology, sc-166344, RRID:AB_2122660), PAT1 (Abcam, ab151305), MAP2 (Sigma, HM-2, RRID:AB_477193), KHC (Millipore MAB1614, RRID:AB_94284), and KLC (Millipore, MAB1617, RRID:AB_94287) were used. F-actin was labeled with Alexa-488, -546, or -647–conjugated phalloidin, and 4′,6-diamidino-2-phenylindole was from Invitrogen. Vybrant® CM DiI cell-labeling solution from Molecular Probes was used to visualize the cell membrane. Alexa Fluoro®-488, -546, and -647 goat anti-mouse and rabbit IgG (H+L) (Invitrogen) were used as secondary antibodies. 48 h after vector transfection, or 72 h after siRNA transfection, neurons were fixed (4% paraformaldehyde in 1× PBS with 4% glucose) for 20 min at room temperature. For endogenous staining, neurons were typically fixed at DIV 7–9. After fixation, the cell membranes were removed with PBST (1× PBS with 0.1% Triton X-100), and the cells were blocked with PBSAT (PBST with 3% BSA); primary and secondary antibodies were diluted in blocking buffer. After staining, coverslips were mounted on micro slides with ProLong® gold anti-fade reagent overnight before any images were taken.
The FISH protocol was adapted as described (11, 90) with modifications; DNAsis and Oligo 6 were used to design probes. Probes against mouse β-actin, GABA-A-R-δ, and CaMKII α mRNAs were used as described previously (11). In situ hybridizations were performed as described (91), and sense or scrambled probes were used as negative controls. The 1-h BDNF/heat-inactivated BDNF stimulation, fixation, and FISH were performed 48–72 h after transfection.
Co-IPs and Western blotting
HEK-293 (RRID:CVCL_0045) cells were used for co-IP experiments. Antibodies against FLAG (Sigma, M2, RRID:AB_439685; SIGI-25, RRID:AB_796202), HA (Sigma, HA-7, RRID:AB_262051; H6908, RRID:AB_260070), GFP (Abcam ab290 RRID:AB_303395; Clontech 8362-1), GST (Santa Cruz Biotechnology, sc-138, RRID:AB_627677), KHC (Millipore MAB1614, RRID:AB_94284), and KLC (Millipore, MAB1617, RRID:AB_94287) were used. For the β-actin mRNA-dependent experiment, PAT1–FLAG and ZBP1–GFP were co-transfected into HEK cells for 36–48 h. Cells were lysed in IP buffer (50 mm Tris-Cl (pH 7.3), 100 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 5% glycerol, 1% Triton X-100, and PI) (91) for 5 min on ice, and RNase I was added as needed. Supernatants were acquired after 10 min of centrifugation (16,000 × g) at 4 °C. Supernatants were immunoprecipitated by mouse anti-FLAG® M2 Magnetic Beads (Sigma, M-8823, RRID:AB_2637089) for 12 h at 4 °C. Precipitates were resolved in 2× SDS-PAGE loading buffer and run on 4–12% BisTris gels. Blots were probed with anti-FLAG and GFP antibodies. For the ZBP1 1–195 dominant-negative experiments, either full-length ZBP1–FLAG or 1–195 ZBP1–FLAG was co-transfected with ZBP1–GFP, and co-IPs and blottings were performed as described previously. For experiments between PAT1 and ZBP1 domains, PAT1–HA and different ZBP1 domain constructs (full-length ZBP1–FLAG, 1–195 ZBP1–FLAG, and 195–576 ZBP1–FLAG) were co-transfected into HEK-293 cells, and anti-FLAG and anti-HA antibodies were used for immunoprecipitations. Prior to gel loading, the total protein concentration of cell lysate was evaluated by UV-spec 280 nm (Nanodrop); 20 μg of protein/well was loaded, and the loading volume of each well was adjusted accordingly. If the loading volume exceeded the well capacity, the total protein/well was adjusted to 10 μg or less.
Live-cell imaging of PAT1/ZBP1 and growth cone dynamics
To observe the transport of PAT1 in dendrites, PAT1–Cherry or PAT1–RFP was transfected into primary mouse neurons. ZBP1–GFP was co-transfected into mouse primary hippocampal neurons to observe ZBP1/PAT1 co-transport. For the observation of PAT1 and β-actin mRNA co-transport, MS2-capsid protein (MCP)–GFP and β-actin 3′UTR-MS2 were used to track β-actin mRNA granules. Using the MS2–MCP system, where the mRNA contains eight repeats of the MS2–hairpin loop motif in the 3′ UTR that bound to the MCP–GFP, PAT1–RFP was co-transfected at the same time. Mouse primary hippocampal neurons were plated in chambered coverglass and imaged 12–16 h after the transfection. Chambers were kept at 37 °C during the experiment, and CO2 and humidity were maintained with the TOKAI HIT microscopy closed stage incubation system. Images were acquired with a Nikon Eclipse Ti inverted microscope and were converted into videos using NIS-elements software (Nikon). For growth cone dynamic experiments, dissociated hippocampal cultures in chambered coverglass (Thermo Fisher Scientific) were transfected with PAT1 siRNA at DIV 1 and imaged 72 h later. 2 h before fixation, the complete medium in the chambers was replaced with fresh neurobasal medium without supplement. Cultures were stimulated with BDNF/heat-inactivated BDNF for 1 h before the fixation. A ×60 PlanApo TIRF 1.49 NA lens was used with a ×1.5 built-in magnifier (total magnification of ×90); images were taken every 1 min for 30 min. DIC images were captured and analyzed using NIS-Element. The growth cone filopodial length was measured from the central domain out to the tip of each filopodium on the growth cone (only those >2 μm were counted), and the lengths of the filopodia on growth cones were averaged. Every 5 min, the number of filopodia on one growth cone was counted, and at each point, the filopodia numbers of each growth cone were averaged. Finally, five sets of data were obtained from this experiment.
Neuronal transfections
Using Lipofectamine 2000, following the provided protocols from Invitrogen, neurons were transfected between 5 and 7 DIV. For PAT1 siRNA experiments, neurons were transfected with PAT1 stealth siRNA (Invitrogen, Stealth RNAi siRNAs, catalogue no. 1320001, and three sets of siRNA targeted to exons 3, 6, and 13) at 5 DIV. 40 pmol of PAT1 stealth siRNA or control siRNA (Invitrogen, Stealth RNAiTM siRNA negative control, catalogue no. 12935300) was applied to each coverslip in 12-well plates containing 1 ml of media in each well. BLOCK-iTTM fluorescent oligonucleotides from Invitrogen were used to visualize the delivery of siRNA into hippocampal neurons. For the dominant-negative ZBP1 experiments, either full-length ZBP1–FLAG or dominant-negative 1–195 ZBP1–FLAG were transfected to primary hippocampal neurons at 6–8 DIV. GFP was co-transfected at a ratio of 1: 5 (GFP: ZBP1 constructs) at the same time to identify transfection-positive neurons. For the transfection of both the full-length ZBP1–FLAG and 1–195 ZBP-FLAG, the plasmid concentration was adjusted to a ratio of 4:5. For the co-transfection of PAT1–RFP, MCP–GFP, and β-actin--MS2 3′ UTR mRNA, a ratio of 1:1:8 (PAT1–RFP, MCP–GFP, and β-actin--MS2 3′UTR mRNA) was used.
Dendritic protrusion density analysis
Quantification of dendritic protrusions was performed as described (11). For PAT1/CON siRNA experiments, 72 h after the PAT1/CON siRNA transfection, neurons were fixed and stained with Vybrant® CM DiI cell-labeling solution. Secondary and tertiary dendrites were randomly selected during dendritic quantification; if primary dendrites had to be quantified, they must be at least 20 μm away from the cell body. Quantification of protrusion density was performed by dividing the total number of protrusions from one segment of the dendrite by the length of the segment and then averaged for the group. For the ZBP1 1–195 dominant-negative experiment on protrusion density, either full-length ZBP1–FLAG or 1–195 ZBP1–FLAG was co-transfected with GFP to neurons at DIV 7, and neurons were fixed 48 h later. MAP2 staining was used to select dendrites randomly. The protrusion density of each dendritic segment was acquired and averaged for the group as described earlier.
Semi-quantitative reverse transcript-PCR of PAT1 and β-actin mRNA
To detect β-actin mRNA in the PAT1 in vitro binding experiment, mRNA was extracted from the PAT1–GST precipitate with TRIzol. mRNA pellets were resuspended in RNase-free water and then reverse-transcribed, followed by PCR using β-actin primers (products 200 bp). For the PAT1 knockdown experiments, PAT1/CON siRNA-transfected neurons were treated with lysate buffer, followed by mRNA extraction with TRIzol and reverse transcription as described before; PCR was performed with PAT1 primers (products 200 bp). The same procedures were carried out for PAT1 expression experiments with P7 and P14 brain lysate. For the reverse transcript, 1 μg of RNA was used for each sample, and either oligo(dT) or random hexamer primers was used. The concentrations of RNA and cDNA were determined by UV-spec 280/260 nm (Nanodrop). For the mRNA-binding experiments between β-actin mRNA, ZBP1 domains, and the kinesin domains, similar procedures were applied, and β-actin mRNA (products 500 bp) and vinculin mRNA (products 500 bp) were detected with PCR.
Quantification analysis of fluorescence images
The images for dendritic quantification were captured using 0.1-μm Z-steps in three-dimensions on a Nikon ECLIPSE TE200-U–inverted fluorescence microscope with a ×60 1.40 Oil Plan ApoVC lens. All images were taken using the NIS-elements software (Nikon) and deconvolved using AutoQuant (Media Cybernetics). Images were restored and analyzed with the NIS-elements software in 3D as described previously (21). Fluorescence quantifications of dendrites were performed in an unbiased way by looking for all MAP2-positive dendrites (or DIC for other parameters) and auto-capturing them after verification of normal morphology. Images were acquired under the same settings using the NIS-Element software. By defining a region of interest, the total fluorescence was acquired and normalized by length. Typically, 1–3 dendrites were taken from each neuron, mostly from secondary or tertiary dendritic segments, and if not then at least 20 μm away from the cell body. The background of all images was subtracted from a region outside of the neurons. Histograms show average values with error bars reflecting standard deviation, and statistical significances were acquired from unpaired Student' t test.
Morphological assay, co-localization analysis, and Imaris software
For morphological assays, protrusion length was measured in Imaris software using the filament tracer module. Protrusion lengths were exported to excel for statistical analysis. For co-localization assays, fluorescence images were analyzed using the co-localization module of the Imaris software. Volume views were created for each antigen channel (ZBP1, PAT1, and APP); co-localization channels were created, and “spot” views were created for each antigen channel (ZBP1, PAT1, or APP), and spot distances from different channels were calculated by the Imaris software. Only spots with distance <0.2 μm were shown between different channels. Pearson's correlation coefficients were obtained in the NIS-Elements or Imaris from maximum projection images for each antigen (PAT1, ZBP1, and APP). To serve as co-localization controls, random dendrites without obvious curves were selected and the dual channel images were split to two separate images. One of the images was flipped 180° and merged back to the other image to construct a dual-channel image, and it served as the control for the original dual-channel image. Histograms for all figure legends show average values with error b ars reflecting standard deviation, and statistical significances were acquired from unpaired Student's t test.
SIM
3D SIM images were acquired on a Nikon SIM–total internal reflection fluorescence (TIRF) microscopy with a ×100 1.49 NA Apo TIRF oil lens (Nikon) using 488- and 561-nm laser lines. Dendritic images were acquired with multimode optical fibers and a 100 EX V-R diffraction grating block, and 15 images in each channel were captured with 0.1-μm steps in Z and a pixel size of 64 nm on an Andor 897 EMCCD. The raw images were reconstructed using a structured illumination contrast of 1, an apodization filter of 0.3, and a 3D SIM filter width of 0.03. Following point spread function (PSF) reconstruction, we obtained a full-width half-maximum (XY lateral resolution) of ∼100 nm.
Statistical analysis
The one-way analysis of variance was performed on experiments with multiple groups. If multiple comparisons were required, Tukey's multiple comparisons test was carried out. For p value significance, * denotes <0.05, ** denotes < 0.01, and *** denotes < 0.001. All values are provided with error bars ±S.D.; p values are given in the figure legends. Numbers denote repeat or sample size and are provided in the figure legends or under “Results.” All data processing and statistical analyses were carried out in GraphPad Prism 7.0.
Author contributions
H. W. and J. D. conceptualization; H. W., J. Z., T. Z., and I. C. data curation; H. W., J. Z., T. Z., and I. C. software; H. W., J. Z., and T. Z. formal analysis; H. W. and J. Z. validation; H. W., J. Z., T. Z., and I. C. investigation; H. W. and J. Z. visualization; H. W., J. Z., T. Z., and I. C. methodology; H. W. and J. D. writing-original draft; H. W., J. Z., and J. D. writing-review and editing; J. D. resources; J. D. supervision; J. D. funding acquisition.
Supplementary Material
Acknowledgments
We thank S. Huttelmaier for sharing yeast two-hybrid results for PAT1–ZBP1 interactions, and I. Palacios for sharing information on PAT1–KLC interactions in Drosophila. We thank Kristen Verhey (University of Michigan Medical School) for help with the two-hybrid studies for PAT1/kinesin binding, Robert Singer (Albert Einstein College of Medicine) for β-actin mRNA and ZBP1 constructs and suggestions for those experiments, and Gary Bassell (Emory University Medical School) for neuronal mRNA and ZBP1 tools and suggestions for imaging and neurotrophin stimulation studies.
This work was supported by National Institutes of Health Grants GM084805 (to J. D.). Also supported by National Science Foundation (NSF) Grants 0819022 and 0960367 (to J. D.), and by Hunter College, Lehman College, and the Graduate Center of the City University of New York. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4, Movies S1–S3, and supporting Video S1.
- RBP
- RNA-binding protein
- KLC
- kinesin light chain
- SIM
- structured illumination microscopy
- BDNF
- brain-derived neurotrophic factor
- DIV
- days in vitro
- DIC
- differential interference contrast
- FMRP
- fragile X mental retardation protein
- RFP
- red fluorescent protein
- FISH
- fluorescence in situ hybridization
- DLC
- the dynein light chain
- PTB
- polypyrimidine tract-binding protein
- JIP
- JNK-interacting protein
- mRNP
- messenger ribonucleoprotein
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- KHC
- kinesin heavy chain
- Egl
- egalitarian
- RRM
- RNA recognition motif
- KH
- K homology
- FL
- full-length
- co-IP
- co-immunoprecipitation
- APP
- amyloid precursor protein
- aa
- amino acid
- PI
- protease inhibitor
- RI
- RNase inhibitor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MCP
- MS2-capsid protein
- TIRF
- total internal reflection fluorescence
- HR
- Heptad repeat
- TPR
- Tetratricopeptide repeat
- CM
- chloromethyl
- DiI
- 1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindocarbocyanine Perchlorate.
References
- 1. Blower M. D. (2013) Molecular insights into intracellular RNA localization. Int. Rev. Cell Mol. Biol. 302, 1–39 10.1016/B978-0-12-407699-0.00001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. St Johnston D. (2005) Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363–375 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- 3. Bramham C. R., and Wells D. G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8, 776–789 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- 4. Kiebler M. A., and Bassell G. J. (2006) Neuronal RNA granules: movers and makers. Neuron 51, 685–690 10.1016/j.neuron.2006.08.021 [DOI] [PubMed] [Google Scholar]
- 5. Cajigas I. J., Tushev G., Will T. J., tom Dieck S., Fuerst N., and Schuman E. M. (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin K. C., and Ephrussi A. (2009) mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richter J. D., and Klann E. (2009) Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 23, 1–11 10.1101/gad.1735809 [DOI] [PubMed] [Google Scholar]
- 8. Sutton M. A., and Schuman E. M. (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58 10.1016/j.cell.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 9. Hirokawa N. (2006) mRNA transport in dendrites: RNA granules, motors, and tracks. J. Neurosci. 26, 7139–7142 10.1523/JNEUROSCI.1821-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brechbiel J. L., and Gavis E. R. (2008) Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr. Biol. 18, 745–750 10.1016/j.cub.2008.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., and Bassell G. J. (2008) A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939 10.1016/j.devcel.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dynes J. L., and Steward O. (2007) Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J. Comp. Neurol. 500, 433–447 10.1002/cne.21189 [DOI] [PubMed] [Google Scholar]
- 13. Gao Y., Tatavarty V., Korza G., Levin M. K., and Carson J. H. (2008) Multiplexed dendritic targeting of α calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell 19, 2311–2327 10.1091/mbc.e07-09-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanai Y., Dohmae N., and Hirokawa N. (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 15. Rook M. S., Lu M., and Kosik K. S. (2000) CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 20, 6385–6393 10.1523/JNEUROSCI.20-17-06385.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shan J., Moran-Jones K., Munro T. P., Kidd G. J., Winzor D. J., Hoek K. S., and Smith R. (2000) Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 275, 38286–38295 10.1074/jbc.M007642200 [DOI] [PubMed] [Google Scholar]
- 17. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J. L., and Moine H. (2011) G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 12, 697–704 10.1038/embor.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goetze B., Tuebing F., Xie Y., Dorostkar M. M., Thomas S., Pehl U., Boehm S., Macchi P., and Kiebler M. A. (2006) The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 172, 221–231 10.1083/jcb.200509035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y. S., Carson J. H., Barbarese E., and Richter J. D. (2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 17, 638–653 10.1101/gad.1053003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Köhrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C. G., and Kiebler M. A. (1999) Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10, 2945–2953 10.1091/mbc.10.9.2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tiruchinapalli D. M., Oleynikov Y., Kelic S., Shenoy S. M., Hartley A., Stanton P. K., Singer R. H., and Bassell G. J. (2003) Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 23, 3251–3261 10.1523/JNEUROSCI.23-08-03251.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang W., and Benson D. L. (2001) Stages of synapse development defined by dependence on F-actin. J. Neurosci. 21, 5169–5181 10.1523/JNEUROSCI.21-14-05169.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., Gindhart J. G., and Goldstein L. S. (2000) Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103, 583–594 10.1016/S0092-8674(00)00162-8 [DOI] [PubMed] [Google Scholar]
- 24. Brendel C., Rehbein M., Kreienkamp H. J., Buck F., Richter D., and Kindler S. (2004) Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 384, 239–246 10.1042/BJ20040812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mallardo M., Deitinghoff A., Müller J., Goetze B., Macchi P., Peters C., and Kiebler M. A. (2003) Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U.S.A. 100, 2100–2105 10.1073/pnas.0334355100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Severt W. L., Biber T. U., Wu X., Hecht N. B., DeLorenzo R. J., and Jakoi E. R. (1999) The suppression of testis-brain RNA-binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J. Cell Sci. 112, 3691–3702 [DOI] [PubMed] [Google Scholar]
- 27. Kamal A., Almenar-Queralt A., LeBlanc J. F., Roberts E. A., and Goldstein L. S. (2001) Kinesin-mediated axonal transport of a membrane compartment containing β-secretase and presenilin-1 requires APP. Nature 414, 643–648 10.1038/414643a [DOI] [PubMed] [Google Scholar]
- 28. Verhey K. J., Meyer D., Deehan R., Blenis J., Schnapp B. J., Rapoport T. A., and Margolis B. (2001) Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152, 959–970 10.1083/jcb.152.5.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bullock S. L. (2011) Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem. Soc. Trans. 39, 1161–1165 10.1042/BST0391161 [DOI] [PubMed] [Google Scholar]
- 30. Messitt T. J., Gagnon J. A., Kreiling J. A., Pratt C. A., Yoon Y. J., and Mowry K. L. (2008) Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell 15, 426–436 10.1016/j.devcel.2008.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gagnon J. A., and Mowry K. L. (2011) Molecular motors: directing traffic during RNA localization. Crit. Rev. Biochem. Mol. Biol. 46, 229–239 10.3109/10409238.2011.572861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paquin N., and Chartrand P. (2008) Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18, 105–111 10.1016/j.tcb.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 33. Landers S. M., Gallas M. R., Little J., and Long R. M. (2009) She3p possesses a novel activity required for ASH1 mRNA localization in Saccharomyces cerevisiae. Eukaryot. Cell 8, 1072–1083 10.1128/EC.00084-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Müller M., Heym R. G., Mayer A., Kramer K., Schmid M., Cramer P., Urlaub H., Jansen R. P., and Niessing D. (2011) A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 9, e1000611 10.1371/journal.pbio.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dienstbier M., Boehl F., Li X., and Bullock S. L. (2009) Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546–1558 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Navarro C., Puthalakath H., Adams J. M., Strasser A., and Lehmann R. (2004) Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6, 427–435 10.1038/ncb1122 [DOI] [PubMed] [Google Scholar]
- 37. Hoogenraad C. C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J. V., Grosveld F., de Zeeuw C. I., and Akhmanova A. (2003) Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22, 6004–6015 10.1093/emboj/cdg592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dictenberg J. (2012) Genetic encoding of fluorescent RNA ensures a bright future for visualizing nucleic acid dynamics. Trends Biotechnol. 30, 621–626 10.1016/j.tibtech.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 39. Zhang H. L., Eom T., Oleynikov Y., Shenoy S. M., Liebelt D. A., Dictenberg J. B., Singer R. H., and Bassell G. J. (2001) Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31, 261–275 10.1016/S0896-6273(01)00357-9 [DOI] [PubMed] [Google Scholar]
- 40. Loiseau P., Davies T., Williams L. S., Mishima M., and Palacios I. M. (2010) Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development 137, 2763–2772 10.1242/dev.048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Welshhans K., and Bassell G. J. (2011) Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 31, 9800–9813 10.1523/JNEUROSCI.0166-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farina K. L., Huttelmaier S., Musunuru K., Darnell R., and Singer R. H. (2003) Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160, 77–87 10.1083/jcb.200206003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eom T., Antar L. N., Singer R. H., and Bassell G. J. (2003) Localization of a β-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23, 10433–10444 10.1523/JNEUROSCI.23-32-10433.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sasaki Y., Welshhans K., Wen Z., Yao J., Xu M., Goshima Y., Zheng J. Q., and Bassell G. J. (2010) Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J. Neurosci. 30, 9349–9358 10.1523/JNEUROSCI.0499-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Libersat F., and Duch C. (2004) Mechanisms of dendritic maturation. Mol. Neurobiol. 29, 303–320 10.1385/MN:29:3:303 [DOI] [PubMed] [Google Scholar]
- 46. Brendza R. P., Serbus L. R., Duffy J. B., and Saxton W. M. (2000) A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289, 2120–2122 10.1126/science.289.5487.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carson J. H., Worboys K., Ainger K., and Barbarese E. (1997) Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton 38, 318–328 [DOI] [PubMed] [Google Scholar]
- 48. Falley K., Schütt J., Iglauer P., Menke K., Maas C., Kneussel M., Kindler S., Wouters F. S., Richter D., and Kreienkamp H. J. (2009) Shank1 mRNA: dendritic transport by kinesin and translational control by the 5′-untranslated region. Traffic 10, 844–857 10.1111/j.1600-0854.2009.00912.x [DOI] [PubMed] [Google Scholar]
- 49. Elvira G., Wasiak S., Blandford V., Tong X. K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A. W., Boismenu D., Lacaille J. C., McPherson P. S., DesGroseillers L., and Sossin W. S. (2006) Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics 5, 635–651 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- 50. Wu X. Q., and Hecht N. B. (2000) Mouse testis brain ribonucleic acid-binding protein/translin co-localizes with microtubules and is immunoprecipitated with messenger ribonucleic acids encoding myelin basic protein, α calmodulin kinase II, and protamines 1 and 2. Biol. Reprod. 62, 720–725 10.1095/biolreprod62.3.720 [DOI] [PubMed] [Google Scholar]
- 51. Muddashetty R. S., Kelić S., Gross C., Xu M., and Bassell G. J. (2007) Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 27, 5338–5348 10.1523/JNEUROSCI.0937-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S., Oostra B., and Bagni C. (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327 10.1016/S0092-8674(03)00079-5 [DOI] [PubMed] [Google Scholar]
- 53. Shan J., Munro T. P., Barbarese E., Carson J. H., and Smith R. (2003) A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 23, 8859–8866 10.1523/JNEUROSCI.23-26-08859.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeong J. H., Nam Y. J., Kim S. Y., Kim E. G., Jeong J., and Kim H. K. (2007) The transport of Staufen2-containing ribonucleoprotein complexes involves kinesin motor protein and is modulated by mitogen-activated protein kinase pathway. J. Neurochem. 102, 2073–2084 10.1111/j.1471-4159.2007.04697.x [DOI] [PubMed] [Google Scholar]
- 55. Tübing F., Vendra G., Mikl M., Macchi P., Thomas S., and Kiebler M. A. (2010) Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J. Neurosci. 30, 4160–4170 10.1523/JNEUROSCI.3537-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vessey J. P., Macchi P., Stein J. M., Mikl M., Hawker K. N., Vogelsang P., Wieczorek K., Vendra G., Riefler J., Tübing F., Aparicio S. A., Abel T., and Kiebler M. A. (2008) A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 16374–16379 10.1073/pnas.0804583105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., and Singer R. H. (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- 58. Chua J. J., Butkevich E., Worseck J. M., Kittelmann M., Grønborg M., Behrmann E., Stelzl U., Pavlos N. J., Lalowski M. M., Eimer S., Wanker E. E., Klopfenstein D. R., and Jahn R. (2012) Phosphorylation-regulated axonal dependent transport of syntaxin 1 is mediated by a Kinesin-1 adapter. Proc. Natl. Acad. Sci. U.S.A. 109, 5862–5867 10.1073/pnas.1113819109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berezuk M. A., and Schroer T. A. (2007) Dynactin enhances the processivity of kinesin-2. Traffic 8, 124–129 10.1111/j.1600-0854.2006.00517.x [DOI] [PubMed] [Google Scholar]
- 60. Deacon S. W., Serpinskaya A. S., Vaughan P. S., Lopez Fanarraga M., Vernos I., Vaughan K. T., and Gelfand V. I. (2003) Dynactin is required for bidirectional organelle transport. J. Cell Biol. 160, 297–301 10.1083/jcb.200210066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blangy A., Arnaud L., and Nigg E. A. (1997) Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 272, 19418–19424 10.1074/jbc.272.31.19418 [DOI] [PubMed] [Google Scholar]
- 62. Gross S. P., Vershinin M., and Shubeita G. T. (2007) Cargo transport: two motors are sometimes better than one. Curr. Biol. 17, R478–R486 10.1016/j.cub.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 63. Welte M. A. (2004) Bidirectional transport along microtubules. Curr. Biol. 14, R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- 64. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W 3rd, Tokito M., and Holzbaur E. L. (2010) Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20, 697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Knowles R. B., Sabry J. H., Martone M. E., Deerinck T. J., Ellisman M. H., Bassell G. J., and Kosik K. S. (1996) Translocation of RNA granules in living neurons. J. Neurosci. 16, 7812–7820 10.1523/JNEUROSCI.16-24-07812.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krichevsky A. M., and Kosik K. S. (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32, 683–696 10.1016/S0896-6273(01)00508-6 [DOI] [PubMed] [Google Scholar]
- 67. Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., and Jansen R. P. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- 68. Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., Davis I., and St Johnston D. (2008) In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134, 843–853 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Palacios I. M., and St Johnston D. (2002) Kinesin light chain-independent function of the kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development 129, 5473–5485 10.1242/dev.00119 [DOI] [PubMed] [Google Scholar]
- 70. Rahman A., Kamal A., Roberts E. A., and Goldstein L. S. (1999) Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J. Cell Biol. 146, 1277–1288 10.1083/jcb.146.6.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Verhey K. J., Lizotte D. L., Abramson T., Barenboim L., Schnapp B. J., and Rapoport T. A. (1998) Light chain-dependent regulation of kinesin's interaction with microtubules. J. Cell Biol. 143, 1053–1066 10.1083/jcb.143.4.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Estes P. S., O'Shea M., Clasen S., and Zarnescu D. C. (2008) Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 39, 170–179 10.1016/j.mcn.2008.06.012 [DOI] [PubMed] [Google Scholar]
- 73. Bianco A., Dienstbier M., Salter H. K., Gatto G., and Bullock S. L. (2010) Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr. Biol. 20, 1487–1492 10.1016/j.cub.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kural C., Kim H., Syed S., Goshima G., Gelfand V. I., and Selvin P. R. (2005) Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science 308, 1469–1472 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- 75. Bramham C. R., Alme M. N., Bittins M., Kuipers S. D., Nair R. R., Pai B., Panja D., Schubert M., Soule J., Tiron A., and Wibrand K. (2010) The Arc of synaptic memory. Exp. Brain Res. 200, 125–140 10.1007/s00221-009-1959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kanhema T., Dagestad G., Panja D., Tiron A., Messaoudi E., Håvik B., Ying S. W., Nairn A. C., Sonenberg N., and Bramham C. R. (2006) Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J. Neurochem. 99, 1328–1337 10.1111/j.1471-4159.2006.04158.x [DOI] [PubMed] [Google Scholar]
- 77. Tanaka J., Horiike Y., Matsuzaki M., Miyazaki T., Ellis-Davies G. C., and Kasai H. (2008) Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science 319, 1683–1687 10.1126/science.1152864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liao L., Pilotte J., Xu T., Wong C. C., Edelman G. M., Vanderklish P., and Yates J. R. 3rd. (2007) BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: an analysis using high-throughput proteomics. J. Proteome Res. 6, 1059–1071 10.1021/pr060358f [DOI] [PubMed] [Google Scholar]
- 79. Smart F. M., Edelman G. M., and Vanderklish P. W. (2003) BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proc. Natl. Acad. Sci. U.S.A. 100, 14403–14408 10.1073/pnas.2436349100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Iacoangeli A., Rozhdestvensky T. S., Dolzhanskaya N., Tournier B., Schütt J., Brosius J., Denman R. B., Khandjian E. W., Kindler S., and Tiedge H. (2008) On BC1 RNA and the fragile X mental retardation protein. Proc. Natl. Acad. Sci. U.S.A. 105, 734–739 10.1073/pnas.0710991105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang F., Chotiner J. K., and Steward O. (2007) Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J. Neurosci. 27, 9054–9067 10.1523/JNEUROSCI.2410-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bonneau A. M., Darveau A., and Sonenberg N. (1985) Effect of viral infection on host protein synthesis and mRNA association with the cytoplasmic cytoskeletal structure. J. Cell Biol. 100, 1209–1218 10.1083/jcb.100.4.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hesketh J. (1994) Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol. Biol. Rep. 19, 233–243 10.1007/BF00986965 [DOI] [PubMed] [Google Scholar]
- 84. Dunaevsky A., Tashiro A., Majewska A., Mason C., and Yuste R. (1999) Developmental regulation of spine motility in the mammalian central nervous system. Proc. Natl. Acad. Sci. U.S.A. 96, 13438–13443 10.1073/pnas.96.23.13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ji Y., Lu Y., Yang F., Shen W., Tang T. T., Feng L., Duan S., and Lu B. (2010) Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons. Nat. Neurosci. 13, 302–309 10.1038/nn.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Luikart B. W., Nef S., Virmani T., Lush M. E., Liu Y., Kavalali E. T., and Parada L. F. (2005) TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J. Neurosci. 25, 3774–3786 10.1523/JNEUROSCI.0041-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Boylan K. L., Mische S., Li M., Marqués G., Morin X., Chia W., and Hays T. S. (2008) Motility screen identifies Drosophila IGF-II mRNA-binding protein–zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet. 4, e36 10.1371/journal.pgen.0040036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Deleted in proof.
- 89. Swanger S. A., and Bassell G. J. (2011) Making and breaking synapses through local mRNA regulation. Curr. Opin. Genet. Dev. 21, 414–421 10.1016/j.gde.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bassell G. J., Zhang H., Byrd A. L., Femino A. M., Singer R. H., Taneja K. L., Lifshitz L. M., Herman I. M., and Kosik K. S. (1998) Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 18, 251–265 10.1523/JNEUROSCI.18-01-00251.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Antar L. N., Afroz R., Dictenberg J. B., Carroll R. C., and Bassell G. J. (2004) Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 24, 2648–2655 10.1523/JNEUROSCI.0099-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.