Abstract
Contact between inflammatory cells and endothelial cells (ECs) is a crucial step in vascular inflammation. Recently, we demonstrated that the cell-surface level of endomucin (EMCN), a heavily O-glycosylated single-transmembrane sialomucin, interferes with the interactions between inflammatory cells and ECs. We have also shown that, in response to an inflammatory stimulus, EMCN is cleared from the cell surface by an unknown mechanism. In this study, using adenovirus-mediated overexpression of a tagged EMCN in human umbilical vein ECs, we found that treatment with tumor necrosis factor α (TNF-α) or the strong oxidant pervanadate leads to loss of cell-surface EMCN and increases the levels of the C-terminal fragment of EMCN 3- to 4-fold. Furthermore, treatment with the broad-spectrum matrix metalloproteinase inhibitor batimastat (BB94) or inhibition of ADAM metallopeptidase domain 10 (ADAM10) and ADAM17 with two small-molecule inhibitors, GW280264X and GI254023X, or with siRNA significantly reduced basal and TNFα-induced cell-surface EMCN cleavage. Release of the C-terminal fragment of EMCN by TNF-α treatment was blocked by chemical inhibition of ADAM10 alone or in combination with ADAM17. These results indicate that cell-surface EMCN undergoes constitutive cleavage and that TNF-α treatment dramatically increases this cleavage, which is mediated predominantly by ADAM10 and ADAM17. As endothelial cell-surface EMCN attenuates leukocyte–EC interactions during inflammation, we propose that EMCN is a potential therapeutic target to manage vascular inflammation.
Keywords: inflammation, endothelial cell, proteolytic enzyme, leukocyte, glycoprotein, endothelium, matrix metalloproteinase (MMP), endomucin, glycocalyx
Introduction
Vascular inflammation, which can be stimulated by infection, oxidative stress and ischemia, has been implicated in a number of disease processes, including sepsis, atherosclerosis, Alzheimer's disease, and diabetes and its complications (1–4). Interactions between inflammatory cells and endothelial cells (ECs),4 a crucial step in vascular inflammation, can result in leukostasis and capillary occlusion (5) as well as migration of inflammatory cells across the endothelium into tissues, leading to tissue damage (6). It has been well documented that this multistep process is mediated by adhesion molecules and their binding partners on inflammatory cells and ECs that are induced by inflammatory stimuli (7).
It has been reported that, coincident with increased expression of adhesion molecules on the luminal endothelial cell surface during inflammation, there is a “thinning” of the glycocalyx, a layer of glycoproteins, proteoglycans, glycosaminoglycans, and associated plasma proteins (8–10). The glycocalyx has been speculated to form a barrier that prevents adhesion of circulating blood cells to the quiescent endothelium layer, presumably because of the repulsive properties of the negatively charged dense sugar epitopes (11). Moreover, thinning of this glycocalyx layer is associated with release of inflammatory cytokines, chemokines, and their receptors and, more importantly, exposure of adhesion molecules on the surface of the endothelium to their binding partners on inflammatory cells (12).
Endomucin (EMCN), a component of the glycocalyx, is a single-transmembrane, heavily O-glycosylated sialomucin expressed by endothelial cells, specifically by capillaries and venous but not most arterial ECs (13, 14). Recently, we demonstrated that the presence of EMCN on the endothelial surface prevents interactions between inflammatory cells and quiescent, noninflamed ECs. Moreover, we have shown that stimulation of ECs with inflammatory cytokines such as tumor necrosis factor α (TNFα), which is known to induce expression of proadhesion molecules on the endothelium, also leads to a significant decrease in apically localized EMCN as well as a decline in EMCN mRNA. Together, these changes facilitate contact between circulating leukocytes and the inflamed endothelium (15). The finding that EMCN overexpression in vitro or in vivo can suppress leukocyte–EC interactions under inflammatory conditions suggests that preservation of cell-surface EMCN may be a novel approach to regulating inflammation. However, the mechanism by which cell-surface EMCN levels are regulated is unknown and, thus, the topic of this investigation.
Matrix metalloproteinase (MMP) superfamily members, including MMPs and a disintegrin and metalloproteinase (ADAM), are secreted or membrane-bound Zn2+-dependent proteinases. One of the most well-studied actions of these enzymes is their ability to catalyze cleavage of the ectodomain of transmembrane proteins and reduce their cell surface level. Here we used overexpression of a tagged EMCN in conjunction with inhibitors against pan-MMPs, inhibitors, or siRNA against ADAM10 and/or ADAM17 to investigate whether EMCN is cleaved and whether these enzymes are involved in its cleavage from the endothelial surface. The results of these studies provide mechanistic insights into the posttranslational regulation of EMCN.
Results
Treatment of EC with TNFα or pervanadate reduced EMCN cell surface levels
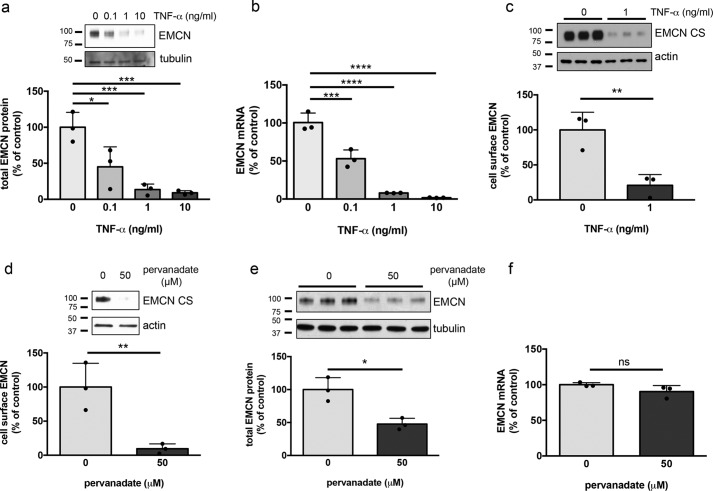
Treatment of HUVECs for 24 h with TNFα, a potent inflammatory cytokine, at a dose as low as 0.1 ng/ml led to a dose-dependent reduction in total EMCN protein and mRNA levels (Fig. 1, a and b). TNFα at 1 ng/ml reduced cell-surface EMCN by ∼85% (Fig. 1c). The concentrations of TNFα here are within the range of plasma levels in patients with vascular inflammation (16, 17), so the results highlight the relevance of cell-surface EMCN reduction in inflammation.
Figure 1.
TNFα or pervanadate treatment leads to reduced cell-surface EMCN and down-regulation of EMCN mRNA in HUVECs. a–f, confluent HUVECs were treated with TNFα at the indicated concentrations for 24 h or with pervanadate (50 μm) for 30 min after overnight serum starvation. Cell-surface protein was isolated by biotinylation, whereas total protein was harvested directly. Total (a and e) and cell surface (c and d) EMCN levels were determined by Western blotting. EMCN mRNA levels were evaluated by Real Time quantitative PCR (b and f). TNFα caused a dose-dependent reduction in total EMCN protein (a) and mRNA (b). TNFα treatment led to a reduction in cell-surface EMCN at 1 ng/ml (c). Pervanadate treatment resulted in decreased cell-surface EMCN (d) and decreased total EMCN protein (e) but did not affect EMCN mRNA levels (f). n = 3, one-way ANOVA or Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant.
Pervanadate, a strong oxidant, has been employed to study the cleavage of glycocalyx components and cell-surface proteins, including syndecan-1, ICAM-1, and occludin, potentially through oxidation and inactivation of protein tyrosine phosphatase (18–20). We therefore examined the effects of pervanadate treatment on EMCN. Pervanadate (50 μm) led to an ∼90% reduction of cell-surface EMCN protein (Fig. 1d) and an approximately 50% decrease of total EMCN protein in HUVECs after 30-min exposure (Fig. 1e). As expected, EMCN mRNA levels were not affected during the short time frame of this treatment (Fig. 1f).
Release of an EMCN C-terminal fragment (EMCN-CTF) by TNFα or pervanadate
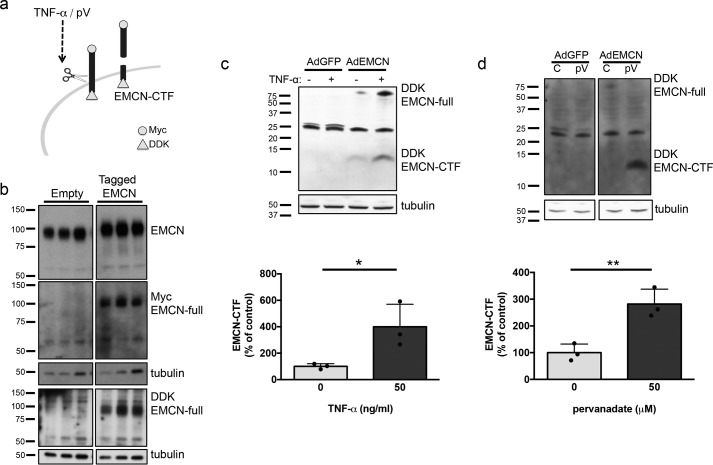
It has been well documented that inflammatory stimuli such as TNFα, interferon-γ, and lipopolysaccharide induce proteolysis and shedding of numerous cell-surface proteins (21–23). The ability of pervanadate to dramatically reduce cell-surface EMCN in 30 min and without affecting mRNA levels strongly suggests proteolytic cleavage of EMCN. To test this possibility, an adenoviral construct was designed to express a tagged EMCN with a myc peptide epitope at the N terminus and a DDK peptide epitope at the C terminus (Fig. 2a). The adenoviral constructs with or without tagged EMCN were transfected into HUVECs, cell lysates were harvested, and expression of the tagged EMCN was evaluated using antibodies against full-length EMCN as well as against the specific tags. Increased total EMCN protein was observed in lysates of cells transfected with and overexpressing the tagged EMCN (Fig. 2b). The anti-myc and anti-DDK antibodies reacted with a protein from cell lysate of HUVECs transfected with the tagged EMCN construct migrating at a molecular mass similar to that of full-length EMCN because of the small size of a Myc tag and a DDK tag (∼1.2 kDa and ∼1 kDa, respectively). This protein was absent from the lysates of HUVECs transfected with the empty construct (Fig. 2b), verifying expression of myc- and DDK-tagged EMCN.
Figure 2.
TNFα or pervanadate treatment induces EMCN cleavage in HUVECs. a, schematic of the tagged EMCN and its cleavage. b, HUVECs were transfected with a construct encoding EMCN tagged with myc on the N terminus and DDK on the C terminus or with an empty construct as a control. Cell lysates were harvested 72 h after transfection for Western blot analysis. c and d, Western blotting confirmed the overexpression of tagged EMCN. HUVECs were transduced with AdEMCN or AdGFP as a control and serum-starved overnight. Cell lysates were harvested after treatment with TNFα (1 ng/ml, 24 h) or pervanadate (50 μm, 30 min) and processed for protein analysis. Western blot analysis for the DDK tag demonstrated the presence of EMCN-CTF in AdEMCN-transduced groups, which was absent in the AdGFP group. Treatment with TNFα or pervanadate led to an increase in EMCN-CTF. n = 3, Student's t test. *, p < 0.05; **, p < 0.01.
To evaluate the potential cleavage of EMCN, an adenovirus was used to overexpress the tagged EMCN (AdEMCN) in cells that were then treated with TNFα or pervanadate. A fragment of ∼12 kDa that corresponded to the size of the EMCN-CTF was detected with the DDK antibody in cell lysates from HUVECs transduced with AdEMCN; this fragment was not detectable in lysates from cells transduced with a control adenovirus expressing GFP (AdGFP) (Fig. 2c). The levels of the 12-kDa EMCN-CTF fragment were increased to 400% following treatment with TNFα (1 ng/ml, 24 h) (Fig. 2c) or to 280% with pervanadate (50 μm, 30 min) (Fig. 2d) compared with untreated controls. The presence of EMCN-CTF in the overexpression system without stimuli suggests a low level of constitutive EMCN cleavage, and the increased EMCN-CTF with treatment by TNFα or pervanadate indicates induced cleavage of EMCN.
Pan-MMP inhibition partially rescued cell-surface EMCN reduction by TNFα
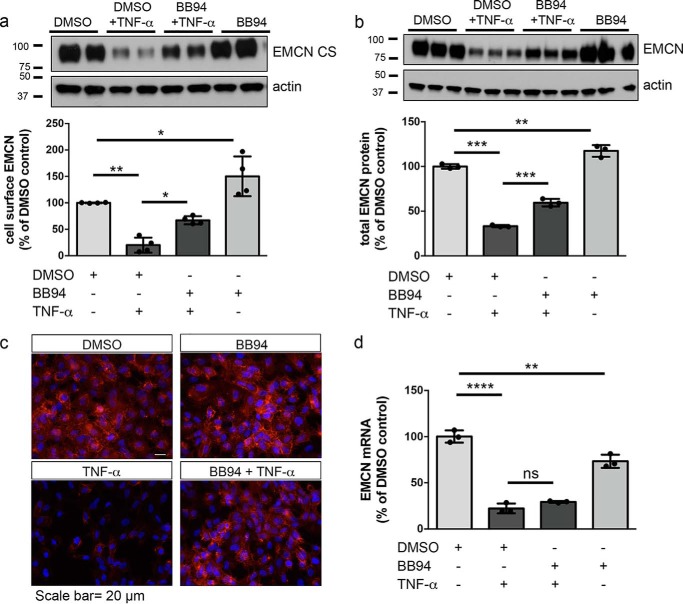
The observation that TNFα treatment of HUVECs led to elevated levels of EMCN-CTF supported the hypothesis that inflammation induces cleavage of EMCN. The involvement of MMP family members in proteolytic cleavage of cell-surface proteins during inflammation has been well studied (12, 24). To elucidate the mechanism of EMCN cleavage, we first examined the effect of a broad-spectrum MMP inhibitor, BB94 (batimastat), a hydroxamate-type inhibitor of MMPs that binds competitively to the active sites of MMPs and ADAMs.
In the absence of stimulation, pretreatment of HUVECs with BB94 increased cell-surface EMCN to 150% (Fig. 3a) and total EMCN protein to 110% (Fig. 3b) of the DMSO vehicle control, respectively. Considering that BB94 alone reduced EMCN mRNA to 70% of the vehicle control (Fig. 3d), BB94 most likely preserves cell-surface EMCN by attenuating constitutive EMCN protein cleavage. With TNFα stimulation, pretreatment of HUVECs with BB94 preserved cell-surface EMCN to 67% of the DMSO control (Fig. 3a) and total EMCN protein to 60% of the DMSO control (Fig. 3b) compared with 20% and 33%, respectively, without BB94 pretreatment. Inhibition of TNFα-induced cell-surface EMCN reduction by BB94 pretreatment was also observed by immunostaining of cell-surface EMCN on nonpermeabilized samples (Fig. 3c). Treatment with GM6001, another broad-spectrum inhibitor of MMPs, provided similar preservation of total EMCN protein compared with the TNFα-alone group (Fig. S1). Given that BB94 had no effect on the TNFα-induced reduction in EMCN mRNA (Fig. 3d), the effects of BB94 were likely posttranslational. BB94 pretreatment also completely blocked the pervanadate-induced release of EMCN-CTF and reduction of total EMCN protein (Fig. S2). Together, these results support a role of members of the MMP family in not only constitutive cleavage of cell-surface EMCN but also under inflammatory conditions.
Figure 3.
Pan-MMP inhibition blocks the TNFα-induced reduction in cell-surface and total EMCN protein. a–d, after overnight serum starvation, confluent HUVECs were pretreated with the pan-MMP inhibitor BB94 (5 μm) or DMSO as a vehicle control for 30 min prior to TNFα treatment (1 ng/ml, 24 h). Cell-surface protein (a) and total cell lysates (b) were harvested for protein analysis. Cells were fixed and stained for EMCN (c, red) or mRNA was harvested for gene expression analysis (d). BB94 blocked TNFα-induced reduction of cell surface EMCN (a) as well as total EMCN protein (b). This was confirmed by immunostaining of EMCN (c, red). (d) BB94 pretreatment did not alter TNFα-induced reduction in EMCN mRNA (d). Scale bar = 50 μm. n = 3, one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant.
Inhibition of ADAM10 and ADAM17 using small-molecule inhibitors prevented loss of cell-surface EMCN
ADAMs, a family of transmembrane MMPs, have been heavily implicated in shedding/cleavage of adhesion molecules and chemokines during inflammation (24). In particular, ADAM10 and ADAM17 have been shown to mediate proteolytic cleavage of CX3CL1, VCAM-1, ICAM-1, and JAM-A in ECs (25–29). In our work, we noted a significant but transient increase in ADAM17 mRNA levels in TNFα-treated HUVECs after 2 h (Fig. S3), suggesting potential involvement of ADAM17 in TNFα-induced EMCN cleavage.
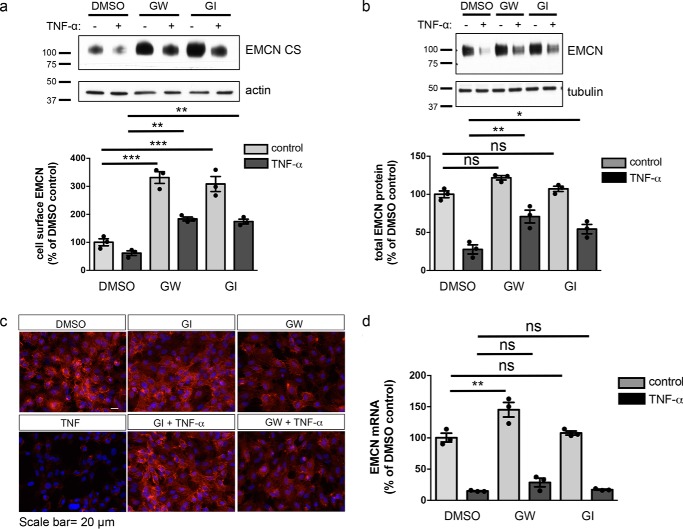
To determine whether these members of the ADAM family were involved in preserving cell-surface EMCN, GW280264X (GW) and GI254023X (GI) were used to discriminate the activities of ADAM10 and ADAM17 (30). GW inhibits ADAM10 and ADAM17, whereas GI preferentially inhibits ADAM10 (IC50, 5.3 nm) compared with ADAM17 (IC50, 541 nm). In these studies, the cells were pretreated for 30 min with GW, GI, or a DMSO control following treatment with or without TNFα.
In the absence of TNFα stimulation, pretreatment with GW or GI significantly increased cell-surface EMCN protein to 231% and 208% of the vehicle control, respectively. Given that neither GW nor GI significantly altered total EMCN protein levels posttranslationally (Fig. 4b), this result supports a critical role of at least ADAM10 in preserving cell surface levels of EMCN (Fig. 4a). In the presence of TNFα, although pretreatment with GW or GI did not influence the TNFα-induced reduction in EMCN mRNA, it did lead to attenuated reduction in total EMCN protein levels from 28% in the TNFα-alone group to 71% and 54% of the vehicle control, respectively (Fig. 4b). More importantly, under TNFα treatment, GW and GI normalized the levels of cell-surface EMCN from 61% in TNFα-alone group to 183% and 174% of the vehicle control, respectively (Fig. 4a), which was corroborated by immunostaining of cell-surface EMCN on nonpermeabilized samples (Fig. 4c). Those results suggest that inhibition of at least ADAM10 preserves cell-surface EMCN at basal levels and under inflammatory conditions.
Figure 4.
Inhibition of ADAM10 and ADAM17 using small-molecule inhibitors blocks loss of cell-surface EMCN. a–d, following overnight serum starvation, confluent HUVECs were treated with GW (10 μm), GI (5 μm), or DMSO as a vehicle control for 30 min before TNFα treatment (1 ng/ml, 24 h). Cell-surface protein (a) and total cell lysates (b) were harvested for protein analysis. Cells were fixed and stained for EMCN protein expression (c, red), or mRNA was harvested for gene expression analysis (d). GW and GI blocked TNFα-induced reduction of cell-surface EMCN protein (a) and total EMCN protein (b), which was also evident by immunostaining of EMCN (c, red). Neither GW nor GI treatment affected TNFα-induced reduction of EMCN mRNA (d). Scale bar = 50 μm. n = 3, one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
To determine whether the effects of GW and GI on preserving cell-surface EMCN were due to EMCN cleavage, levels of EMCN-CTF were determined in TNFα-stimulated HUVECs overexpressing tagged EMCN with and without GW or GI pretreatment. In HUVECs overexpressing tagged EMCN, GI completely blocked TNFα-induced release of EMCN-CTF, whereas GW significantly reduced TNFα-induced release of EMCN-CTF from 338% to 182% of the vehicle control (Fig. S4). Although not statistically significant, without TNFα stimulation, there was a trend toward reduction of constitutive cleavage of EMCN by both inhibitors as well (Fig. S4). This result supports a critical role of at least ADAM10 in constitutive and TNFα-induced cleavage of EMCN.
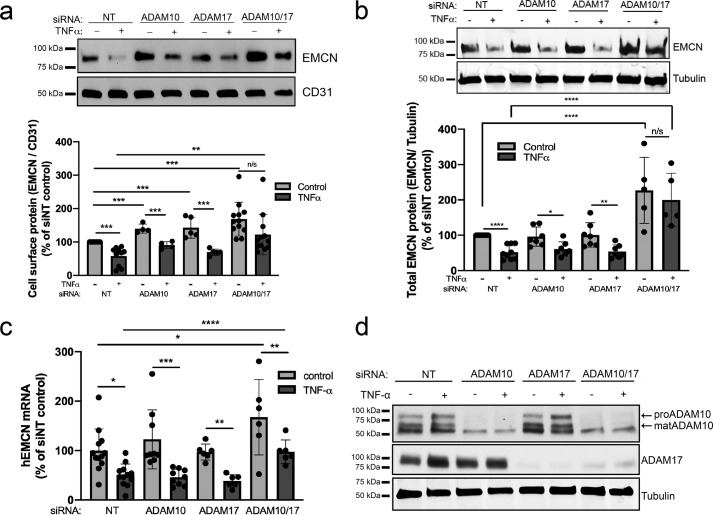
Inhibition of ADAM10 and ADAM17 using siRNA prevented loss of cell-surface EMCN
To define the requirement for ADAM10 and ADAM17 in TNFα-induced EMCN ectodomain cleavage, HUVECs were transfected with siRNA (50 nm) against ADAM10 and ADAM17 alone or in combination. Successful knockdown of ADAM10 and ADAM17 was verified by mRNA and protein 24 h after transfection; combination siRNA transfection did not impact ADAM knockdown (Fig. S5, a–d). Representative Western blot images of ADAM10 and ADAM17 knockdown are shown in Fig. 5d.
Figure 5.
siRNA knockdown of ADAM10 and ADAM17 blocks the TNFα-induced reduction in total and cell-surface EMCN protein. a–d, following overnight serum starvation, HUVECs at 70% confluency were treated with siRNA targeting ADAM10 (50 nm), ADAM17 (50 nm), or a combination of ADAM10 and ADAM17 (50 nm) overnight before TNFα treatment (1 ng/ml, 24 h). Cell-surface (a) and total (b) protein lysate was harvested for analysis using Western blotting. Total RNA was collected in RNA-Bee for quantitative PCR analysis (c). Representative Western blotting of ADAM10 and ADAM17 knockdown was performed using siRNA alone and in combination (d). Combination treatment with siADAM10 and siADAM17 blocked the TNFα-induced reduction of total and cell-surface EMCN protein. n = 7–12, one-way ANOVA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant.
Although TNFα stimulation reduced cell-surface EMCN levels in HUVECs transfected with siNT and siADAM17, only siADAM10 alone (28% increase) or in combination with siADAM10/siADAM17 (63% increase) resulted in significantly higher levels of surface EMCN following TNFα stimulation compared with siNT controls. These results support a role of ADAM10, alone or in combination with ADAM17, in regulating TNFα-induced EMCN shedding. Furthermore, when looking at the role of ADAM10 and ADAM17 in constitutive EMCN shedding, knockdown of ADAM10 (46%) and ADAM17 (35%) alone and in combination (68%) resulted in elevated EMCN surface expression (Fig. 5a). This suggests that, although ADAM10 plays a prominent role in TNFα-induced EMCN surface shedding, constitutive shedding of surface EMCN appears to be regulated by ADAM10 and ADAM17 alone and in combination.
When looking at the role of ADAM10 and ADAM17 in regulating total EMCN protein following TNFα stimulation, only combined knockdown of ADAM10 and ADAM17 (148%) increased total EMCN protein expression compared with siNT controls. The finding that ADAM10 alone prevented only TNFα-induced loss of cell-surface but not total EMCN protein suggests a role of ADAM10 in cleaving EMCN's cell-surface ectodomain under proinflammatory conditions. Accordingly, the results showed that TNFα stimulation reduced EMCN protein levels in HUVECs transfected with siNT (48% decrease), siADAM10 (34% decrease), and siADAM17 (46% decrease). Interestingly, combined knockdown of ADAM10/ADAM17 increased EMCN total protein expression in the absence of TNFα (126% increase). The finding that only combination knockdown of ADAM10 and ADAM17 increased EMCN total protein levels in the presence and absence of TNFα suggests a dual role of ADAM10 and ADAM17 in posttranscriptional regulation of EMCN expression (Fig. 5b).
TNFα stimulation reduced EMCN mRNA in HUVECs transfected with siNT (51% reduction), siADAM10 (76% reduction), siADAM17 (58% reduction), and siADAM10/siADAM17 (69% reduction). Given that neither siADAM10 nor siADAM17 alone had any effect on TNFα-induced reduction in EMCN mRNA (Fig. 5c), ADAM10 and ADAM17 likely regulate EMCN at a posttranslational level. These results further confirm a critical role of ADAM10 and ADAM17 in constitutive as well as TNFα-induced ectodomain shedding of EMCN and provide molecular insights into cleavage of EMCN under inflammatory conditions.
Discussion
Proteolytic cleavage is an important posttranslational modification involved in regulating biological functions of cell-surface proteins. EMCN, an EC-specific cell-surface sialomucin and part of the glycocalyx, plays a critical role in regulating the interactions between ECs and inflammatory cells (15). This study provides evidence that EMCN undergoes both constitutive and induced cleavage. The detection of the C-terminal fragment of EMCN (i.e. the transmembrane and intracellular domains) at low levels in cell lysate of unstimulated cultured ECs and significant increased levels in the presence of TNFα or pervanadate indicate constitutive cleavage of EMCN under quiescent conditions and induced cleavage under inflammatory conditions, suggesting that EMCN could be regulated posttranslationally through cleavage. CD43 (leukosialin), the major membrane sialoprotein on leukocytes, is also proteolytically cleaved during inflammatory activation (31, 32). Given the repulsive barrier function of sialomucins, shedding of the extracellular domain of EMCN and CD43 from the cell surface would facilitate EC–leukocyte interactions during inflammation.
Heparan sulfate proteoglycans, such as syndecans, another major component of the endothelial glycocalyx, are similarly cleaved during inflammation. Mice lacking syndecan-1 have increased leukocyte–EC interactions, suggesting a role of syndecan-1 in suppressing these interaction under normal conditions (33, 34). At the same time, the abundant heparan sulfate in syndecan-1 also binds to proinflammatory cytokines and chemokines. Inhibition of syndecan-1 cleavage has been shown to impede removal of CXC chemokines bound to syndecan-1 in lipopolysaccharide-challenged mice and to exacerbate neutrophilic inflammation (35).
Given that the estimated molecular mass of EMCN-CTF released by TNFα or pervanadate treatment is about 12 kDa and the intracellular and transmembrane domains of EMCN combined are about 8 kDa, the cleavage site for EMCN-CTF is predicted to be in the extracellular domain. However, we have been unable to detect the N-terminal fragment of EMCN in cell lysate or conditioned medium, leaving the fate of the cleaved EMCN extracellular N-terminal fragment undetermined. It is possible that an additional cleavage event removes the myc tag from the N-terminal fragment during proteolytic processing of EMCN, that the extracellular fragment of EMCN undergoes degradation upon initial cleavage, and/or that the fragment is internalized and degraded; any of these events would render the N-terminal fragment nondetectable using our current methods. Many substrates of ADAMs, including Notch and amyloid precursor protein, undergo more than one cleavage event (24). Further studies using EMCN with the tag positioned at various locations of its extracellular domain in the presence of protease inhibitors may circumvent this limitation and provide more information about EMCN extracellular fragment.
MMPs and ADAMs have been shown to mediate shedding of the EC glycocalyx under inflammatory and ischemia conditions (8, 36, 37). Use of pan-MMP inhibitors pointed to involvement of the MMP superfamily in EMCN cleavage. Then, motivated by prior observations of the roles of specific ADAM family members in shedding of cell-surface proteins (24), we further employed small-molecule inhibitors as well as siRNA against ADAM10 and ADAM17 alone and in combination to dissect their relative contributions in EMCN shedding. The results indicate a critical role of ADAM10 and ADAM17 in constitutive shedding of cell-surface EMCN. For induced shedding of cell-surface EMCN, however, although both ADAM10 and 17 appear to be involved in pervanadate-induced cell-surface EMCN reduction (Fig. S6), only ADAM10 or ADAM10 in combination with ADAM17 is responsible for TNFα-induced shedding (Figs. 4 and 5), suggesting that the mediators of EMCN shedding could be stimulus/context-dependent.
Interestingly, differential regulation by ADAMs has been observed for the cleavage of TNFα itself. Whereas constitutive cleavage of TNFα in macrophages, NIH3T3 cells and ECs was found to be mediated by both ADAM10 and ADAM17 (38), phorbol ester or pervanadate stimulation of TNFα shedding in mouse embryonic fibroblasts was primarily mediated by ADAM17 (39). In addition to EMCN as a substrate in ECs, it has been shown that ADAM10 or ADAM17 or both cleave EC junction proteins, VE-cadherin (40) and JAM-A (29), as well as, adhesion molecules between inflammatory cells and ECs, vascular cell adhesion molecule-1 also known as VCAM-1 (27) and intracellular adhesion molecule-1 or ICAM-1 (28). Furthermore, in a murine model of acute pulmonary inflammation induced by lipopolysaccharide leukocyte-specific knockout of ADAM10 (41) or endothelial-specific deletion of ADAM17 (22) prevents leukocyte recruitment, supporting the critical role of ADAM10 and 17 in regulating inflammation. Further investigation of the effects of ADAM10 and ADAM17 deletion on their substrates in vivo could provide valuable information regarding their temporal and cell-type modulation to maximize the beneficial effects.
This study also defines a role of ADAM family members (ADAM10 and ADAM17) in regulating EMCN transcription in addition to transcriptional control of EMCN by GATA2, which has been reported previously (42). This is based on our data, in which cells depleted of both ADAM10 and ADAM17 showed an increase in EMCN mRNA in the presence and absence of 24 h of TNFα stimulation. These findings suggest that the ADAM family members ADAM10 and ADAM17 may regulate EMCN transcription under constitutive and TNFα-stimulated conditions.
Because of its role as a critical contributor to the pathogenesis of numerous diseases, vascular inflammation has been the focus of much research. Onset of inflammation includes increased soluble proinflammatory mediators and increased adhesion molecules on circulating inflammatory cells as well as endothelial cells at sites of inflammation. So far, the mainstay of inflammation-targeting drugs has been nonsteroidal anti-inflammatory drugs and proinflammatory cytokine-neutralizing antibodies (for example, TNFα), which focus on soluble mediators in inflammation. Although they alleviate the symptoms of some inflammatory diseases, they also systematically suppress critical components of the host immune response and increase the possibility of adverse effects (43). EMCN, an EC-specific cell-surface protein, is a more specific target, and our results suggest that regulation of cell-surface EMCN and its cleavage may be an attractive therapeutic approach.
Experimental procedures
Reagents
TNFα was purchased from PeproTech (Rocky Hill, NJ), reconstituted in sterile water with 0.2% BSA, and stored at −80 °C. Pervanadate was made freshly before each use by mixing equal amount of H2O2 and sodium orthovanadate solution (pH 10). The broad-spectrum MMP inhibitor BB94 (batimastat) was purchased from Tocris and GM6001 from Calbiochem (Billerica, MA). The ADAM10 and ADAM17 inhibitor GW280264X and the ADAM10 inhibitor GI254023X were purchased from AOBIOUS (Gloucester, MA). All inhibitors were reconstituted with DMSO, stored at −80 °C, and diluted freshly and added 30 min before inflammatory stimuli. siRNA against ADAM10 (L-004503-00-0005), ADAM17 (L-003453-00-0005), or siRNA nontargeting control (D-001810-01-20) were purchased from Dharmacon (Lafayette, CO).
Expression vector and adenovirus for tagged endomucin
A cDNA fragment encoding a double-tagged human EMCN protein (Myc tag at the N-terminal of the full-length human EMCN after the signal peptide sequence and DDK tag at its C terminus) was synthesized by Genscript (Piscataway, NJ). This cDNA was then cloned into the pcDNA 4 plasmid with a pCMV promoter via EcoRV and NotI cloning sites as the expression vector. The adenovirus expressing tagged EMCN was generated and titered by Vector Biolabs (Malvern, PA). Briefly, this cDNA was inserted into a shuttle vector with a pCMV promoter via EcoRV and NotI, and then the expression cassette was transferred into an adenoviral vector (Ad5 with E1/E3 deletion) by homologous recombination. The adenoviral vector was linearized and transfected into 239 cells to generate the adenovirus. An adenovirus expressing GFP under the same pCMV promoter was used as the control.
Antibodies
The following primary antibodies were used: rat anti-human EMCN for detection of endogenous EMCN unless stated otherwise (ab45771, Abcam, Cambridge, MA), rabbit anti-Myc tag (2278, Cell Signaling, Danvers, MA), goat anti-DDK (ab1257, Abcam), rabbit anti-EMCN against the C-terminal region of endogenous EMCN used in Fig. S2 (NBP1-71558, Novus, Littleton, CO), mouse anti-tubulin (CP06, Calbiochem), mouse anti-actin (sc-58673, Santa Cruz), rabbit anti-GAPDH (sc-25778, Santa Cruz, Dallas, TX) and mouse anti-CD31 (ab7388, Abcam). Primary antibodies were used at 1:2000 for Western blotting and 1:300 for immunostaining unless indicated otherwise. The following secondary antibodies were used: HRP-linked sheep anti-mouse IgG (NA931, GE Healthcare), HRP-linked donkey anti-rabbit IgG (NA934, GE Healthcare), HRP-linked goat anti-rat IgG (NA935, GE Healthcare), HRP-linked rabbit anti-goat IgG (sc-2768, Santa Cruz), and DyLight550 goat anti-rat IgG (ab98387, Abcam). Secondary antibodies were used at 1:5000 for Western blotting and 1:300 for immunostaining unless indicated otherwise.
Cell culture
HUVECs (provided by the Center for Excellence in Vascular Biology, Brigham and Women's Hospital, Boston, MA) were cultured on 0.2% gelatin–coated T-75 flasks in EGM-2 medium (EBM-2 with the SingleQuot kit, Lonza, Walkersville, MD) supplemented with 20% FBS and l-glutamine and maintained at 37 °C with 5% CO2. For all experiments, HUVECs between passages 4 and 6 were used. Prior to inflammatory stimulus treatments, confluent HUVECs that were confluent for less than 24 h were incubated overnight in low-serum medium (EBM-2 with 0.5% FBS and l-glutamine).
Plasmid transfection and adenoviral transduction
To confirm the expression of tagged EMCN, pcDNA plasmids with or without tagged EMCN cDNA were introduced into HUVECs using Amaxa HCAEC Nucleofactor Kit (Lonza). After trypsinization, 1.25 × 106 cells were resuspended in 100 μl of Nucleofactor transfection reagent with 2 μg of the pcDNA vector, including the tagged EMCN or empty vector. The cells were then electroporated with the program S-005, recovered in EGM-2, and distributed into 12-well plates coated with 0.2% gelatin. Twenty-four hours after plating, the cells were incubated in low-serum medium for 48 h before cell lysates were harvested.
For the remainder of the experiments, tagged EMCN was delivered into HUVECs via the adenovirus. AdEMCN or AdGFP was added to HUVECs at a multiplicity of infection of 30 24 h after the cells were plated. Another 24 h later, the cells were incubated overnight in low-serum medium before treatment.
Protein isolation and protein expression analysis
Cell-surface protein isolation by biotinylation
To determine the presence and relative protein levels of EMCN on HUVEC cell membranes, the Pierce Cell Surface Protein Isolation Kit (Thermo Fisher Scientific, Waltham, MA) was used according to the manufacturer's instruction. Briefly, cell-surface protein was biotinylated, purified from cell lysates using a NeutrAvidin-agarose affinity column, and subjected to Western blot analysis.
Total protein isolation
After the designated treatments, cells were collected, lysed in cell lysis buffer (Cell Signaling) containing protease and phosphatase inhibitors (Roche) with high-frequency ultrasonication, and clarified at 14,000 rpm for 20 min at 4 °C. Total protein was then quantified by Pierce BCA assay (Thermo Fisher Scientific) and subjected to Western blot analysis.
Western blot analysis
Equal amounts of total protein or equal volumes of biotinylated protein were separated by SDS-PAGE under reducing conditions and transferred to PVDF membranes (Immobilon-P, Millipore, Billerica, MA). For proteins with a molecular mass below 20 kDa, equal amounts of protein (40 μg) were separated by SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes (N8017, Sigma). Membranes were blocked in TBS containing 0.1% Tween (TBS-T) and 5% nonfat milk for 1 h at room temperature, washed with TBS-T, and then probed with a primary antibody in TBS-T with 1% BSA overnight at 4 °C. Secondary antibody incubations were performed with the corresponding horseradish peroxidase–conjugated antibody at room temperature for 1 h with thorough washes before and after. Proteins were detected by chemiluminescence with SuperSignal West Pico or Dura (Thermo Scientific). α-Tubulin was used as the loading control for total protein samples. For cell-surface assays, α-tubulin from the flow-through or CD31 from the biotinylated membrane fraction was used as a loading controls.
mRNA isolation and gene expression analysis
After treatment, total mRNA was collected from HUVECs in RNA-Bee reagent (Amsbio, Cambridge, MA) and extracted according to the manufacturer's instructions. First-strand cDNA was reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad), following the manufacturer's protocol, from 1 μg of total mRNA in a 20-μl reaction. For real-time PCR, reactions were performed on a LightCycler 480 II (Roche, Indianapolis, IN) using 5.2 μl of diluted (1:10) cDNA reactions, 0.33 μl of a mixture of 10 μm forward and reverse primers, and 5.5 μl of Faststart Universal SYBR Green Master Mix (Roche) according to the manufacturer's instructions. The primers for each gene were as follows: EMCN forward (5′-ATTTGTTCTGGTGGGTTTGT-3′) and reverse (5′-TGCAGGACTTTCTCCTTTTC-3′), ADAM10 forward (5′-TCCCCTTGCAACGATTTTAG-3′) and reverse (5′-AATACTGCCCACCAATGAGC-3′), ADAM17 forward (5′-GTCTGAGAGCAAAGAATCAAGC-3′) and reverse (5′-TCTCCTATTCCTGACCAGCG-3′), and hypoxanthine-guanine phosphoribosyltransferase forward (5′-CCTGGCGTCGTGATTAGTGAT-3′) and reverse (5′-AGACGTTCAGTCCTGTCCATAA-3′).
PCR cycling was performed at 95 °C for 10 min, followed by 95 °C for 15 s and 60 °C for 1 min for a total of 40 cycles. The cycle threshold values corresponding to the PCR cycle number at which fluorescence emission in real-time reaches a threshold above the baseline emission were automatically calculated by a LightCycler 480 II (Roche) according to the manufacturer's instructions. PCR reactions where cDNA templates were replaced with RNase-free water were used as negative controls. Amplification of HPRT was performed on each sample as the housekeeping gene for normalization. Each biological sample was assayed in triplicate for each set of primers, and the averaged cycle threshold value of the triplicate was used to represent each biological sample for further analysis.
siRNA knockdown
HUVECs were seeded at 70% confluence 1 day prior to siRNA transfection. siADAM10 (50 nm, L004503-00-0005), siADAM17 (50 nm, L-003453-00-0005), or siNT (50 nm, D-001810-01-05) was incubated with Dharmafect 1 transfection reagent in Opti-MEM (Life Technologies, 51985034) at room temperature for 30 min to allow complex formation. siRNA was added in EGM-2 complete medium supplemented with 10% FBS in the absence of penicillin–streptomycin. The following day, culture media were changed.
Immunostaining and fluorescence microscopy analysis
For localization of EMCN on cultured cells, HUVECs grown on fibronectin-treated glass coverslips were fixed in 1% formalin overnight at 4 °C, washed three times for 5 min in PBS, and blocked with 3% secondary serum species and 1% BSA in PBS for 1 h at room temperature. Coverslips were then incubated in rat anti-human endomucin at 4 °C overnight, followed by three 5-min washes in PBS. The cells were then incubated in DyLight 550 goat anti-rat secondary antibody for 1 h at room temperature and washed in PBS. DAPI was used to stain cell nuclei. Fluorescent images were taken with a Zeiss fluorescence microscope (Axioskop 2 MOT Plus, ×40 objective).
Statistical analysis
Statistical analysis was carried out with Prism 4.0b for Macintosh. All data are shown as mean ± S.E. The mean values from each group were compared by Student's t test between two groups or one-way analysis of variance (ANOVA) with Tukey's post hoc test for more than two groups, except for dose–response (Fig. 1, a and b) and time point experiments (Fig. S3) with Dunnett's post hoc test. In all tests, p < 0.05 was considered statistically significant.
Data availability
All data are contained within the article.
Author contributions
J. Y., M. S.-G., Y.-S. N., and P. A. D. conceptualization; J. Y., M. E. L., I. C., K. L. S.-T., Y.-S. N., and P .A. D. data curation; J. Y., M. E. L., and M. S.-G. formal analysis; J. Y. validation; J. Y., M. E. L., I. C., K. L. S.-T., M. S.-G., Y.-S. N., and P. A. D. investigation; J. Y., M. E. L., Y.-S. N., and P. A. D. methodology; J. Y. and P. A. D. writing-original draft; J. Y., M. E. L., I. C., K. L. S.-T., M. S.-G., Y.-S. N., and P. A. D. writing-review and editing; M. S.-G., Y.-S. N., and P. A. D. supervision; P. A. D. resources; P. A. D. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Arboleda-Velasquez for advice regarding the design of the tagged EMCN construct and Ashley Woodward for technical assistance with the cell surface protein biotinylation assay.
This work was supported by National Institutes of Health Grants P30 EY03790 and R01EY26539 (to P. A. D.) and F32 EY028405 (to M. E. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
- EC
- endothelial cell
- EMCN
- endomucin
- TNF
- tumor necrosis factor
- MMP
- matrix metalloproteinase
- ADAM
- a disintegrin and metalloproteinase
- HUVEC
- human umbilical vein endothelial cells
- CTF
- C-terminal fragment
- GW
- GW280264X
- GI
- GI254023X
- cDNA
- complementary DNA
- ANOVA
- analysis of variance
- siNT
- non-target siRNA.
References
- 1. Libby P. (2002) Inflammation in atherosclerosis. Nature 420, 868–874 10.1038/nature01323 [DOI] [PubMed] [Google Scholar]
- 2. Hartge M. M., Unger T., and Kintscher U. (2007) The endothelium and vascular inflammation in diabetes. Diab. Vasc. Dis. Res. 4, 84–88 10.3132/dvdr.2007.025 [DOI] [PubMed] [Google Scholar]
- 3. Schouten M., Wiersinga W. J., Levi M., and van der Poll T. (2008) Inflammation, endothelium, and coagulation in sepsis. J. Leukocyte Biol. 83, 536–545 10.1189/jlb.0607373 [DOI] [PubMed] [Google Scholar]
- 4. Zlokovic B. V. (2011) Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat. Rev. Neurosci. 12, 723–738 10.1038/nrn3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kern T. S. (2007) Contributions of inflammatory processes to the development of the early stages of diabetic retinopathy. Exp. Diabetes Res. 2007, 95103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Langer H. F., and Chavakis T. (2009) Leukocyte-endothelial interactions in inflammation. J. Cell. Mol. Med. 13, 1211–1220 10.1111/j.1582-4934.2009.00811.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muller W. A. (2002) Leukocyte-endothelial cell interactions in the inflammatory response. Lab. Invest. 82, 521–533 10.1038/labinvest.3780446 [DOI] [PubMed] [Google Scholar]
- 8. Mulivor A. W., and Lipowsky H. H. (2004) Inflammation- and ischemia-induced shedding of venular glycocalyx. Am. J. Physiol. Heart Circ. Physiol. 286, H1672–1680 10.1152/ajpheart.00832.2003 [DOI] [PubMed] [Google Scholar]
- 9. Rehm M., Bruegger D., Christ F., Conzen P., Thiel M., Jacob M., Chappell D., Stoeckelhuber M., Welsch U., Reichart B., Peter K., and Becker B. F. (2007) Shedding of the endothelial glycocalyx in patients undergoing major vascular surgery with global and regional ischemia. Circulation 116, 1896–1906 10.1161/CIRCULATIONAHA.106.684852 [DOI] [PubMed] [Google Scholar]
- 10. Lipowsky H. H., Gao L., and Lescanic A. (2011) Shedding of the endothelial glycocalyx in arterioles, capillaries, and venules and its effect on capillary hemodynamics during inflammation. Am. J. Physiol. Heart Circ. Physiol. 301, H2235–H2245 10.1152/ajpheart.00803.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varki A. (2008) Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 10.1016/j.molmed.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garton K. J., Gough P. J., and Raines E. W. (2006) Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J. Leukocyte Biol. 79, 1105–1116 10.1189/jlb.0106038 [DOI] [PubMed] [Google Scholar]
- 13. Morgan S. M., Samulowitz U., Darley L., Simmons D. L., and Vestweber D. (1999) Biochemical characterization and molecular cloning of a novel endothelial-specific sialomucin. Blood 93, 165–175 10.1182/blood.V93.1.165 [DOI] [PubMed] [Google Scholar]
- 14. Kuhn A., Brachtendorf G., Kurth F., Sonntag M., Samulowitz U., Metze D., and Vestweber D. (2002) Expression of endomucin, a novel endothelial sialomucin, in normal and diseased human skin. J. Invest. Dermatol. 119, 1388–1393 10.1046/j.1523-1747.2002.19647.x [DOI] [PubMed] [Google Scholar]
- 15. Zahr A., Alcaide P., Yang J., Jones A., Gregory M., dela Paz N. G., Patel-Hett S., Nevers T., Koirala A., Luscinskas F. W., Saint-Geniez M., Ksander B., D'Amore P. A., and Argüeso P. (2016) Endomucin prevents leukocyte-endothelial cell adhesion and has a critical role under resting and inflammatory conditions. Nat. Commun. 7, 10363 10.1038/ncomms10363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gårdlund B., Sjölin J., Nilsson A., Roll M., Wickerts C. J., and Wretlind B. (1995) Plasma levels of cytokines in primary septic shock in humans: correlation with disease severity. J. Infect. Dis. 172, 296–301 10.1093/infdis/172.1.296 [DOI] [PubMed] [Google Scholar]
- 17. Silveira R. C., and Procianoy R. S. (2003) Interleukin-6 and tumor necrosis factor-α levels in plasma and cerebrospinal fluid of term newborn infants with hypoxic-ischemic encephalopathy. J. Pediatr. 143, 625–629 10.1067/S0022-3476(03)00531-6 [DOI] [PubMed] [Google Scholar]
- 18. Reiland J., Ott V. L., Lebakken C. S., Yeaman C., McCarthy J., and Rapraeger A. C. (1996) Pervanadate activation of intracellular kinases leads to tyrosine phosphorylation and shedding of syndecan-1. Biochem. J. 319, 39–47 10.1042/bj3190039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wachtel M., Frei K., Ehler E., Fontana A., Winterhalter K., and Gloor S. M. (1999) Occludin proteolysis and increased permeability in endothelial cells through tyrosine phosphatase inhibition. J. Cell Sci. 112, 4347–4356 [DOI] [PubMed] [Google Scholar]
- 20. Essick E., Sithu S., Dean W., and D'Souza S. (2008) Pervanadate-induced shedding of the intercellular adhesion molecule (ICAM)-1 ectodomain is mediated by membrane type-1 matrix metalloproteinase (MT1-MMP). Mol. Cell. Biochem. 314, 151–159 10.1007/s11010-008-9776-7 [DOI] [PubMed] [Google Scholar]
- 21. Pruessmeyer J., Martin C., Hess F. M., Schwarz N., Schmidt S., Kogel T., Hoettecke N., Schmidt B., Sechi A., Uhlig S., and Ludwig A. (2010) A disintegrin and metalloproteinase 17 (ADAM17) mediates inflammation-induced shedding of syndecan-1 and -4 by lung epithelial cells. J. Biol. Chem. 285, 555–564 10.1074/jbc.M109.059394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dreymueller D., Martin C., Kogel T., Pruessmeyer J., Hess F. M., Horiuchi K., Uhlig S., and Ludwig A. (2012) Lung endothelial ADAM17 regulates the acute inflammatory response to lipopolysaccharide. EMBO Mol. Med. 4, 412–423 10.1002/emmm.201200217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramnath R., Foster R. R., Qiu Y., Cope G., Butler M. J., Salmon A. H., Mathieson P. W., Coward R. J., Welsh G. I., and Satchell S. C. (2014) Matrix metalloproteinase 9-mediated shedding of syndecan 4 in response to tumor necrosis factor α: a contributor to endothelial cell glycocalyx dysfunction. FASEB J. 28, 4686–4699 10.1096/fj.14-252221 [DOI] [PubMed] [Google Scholar]
- 24. Dreymueller D., Pruessmeyer J., Groth E., and Ludwig A. (2012) The role of ADAM-mediated shedding in vascular biology. Eur. J. Cell Biol. 91, 472–485 10.1016/j.ejcb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 25. Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K. J., Rose-John S., and Ludwig A. (2003) The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 102, 1186–1195 10.1182/blood-2002-12-3775 [DOI] [PubMed] [Google Scholar]
- 26. Garton K. J., Gough P. J., Blobel C. P., Murphy G., Greaves D. R., Dempsey P. J., and Raines E. W. (2001) Tumor necrosis factor-α-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1). J. Biol. Chem. 276, 37993–38001 [DOI] [PubMed] [Google Scholar]
- 27. Garton K. J., Gough P. J., Philalay J., Wille P. T., Blobel C. P., Whitehead R. H., Dempsey P. J., and Raines E. W. (2003) Stimulated shedding of vascular cell adhesion molecule 1 (VCAM-1) is mediated by tumor necrosis factor-α-converting enzyme (ADAM 17). J. Biol. Chem. 278, 37459–37464 10.1074/jbc.M305877200 [DOI] [PubMed] [Google Scholar]
- 28. Tsakadze N. L., Sithu S. D., Sen U., English W. R., Murphy G., and D'Souza S. E. (2006) Tumor necrosis factor-α-converting enzyme (TACE/ADAM-17) mediates the ectodomain cleavage of intercellular adhesion molecule-1 (ICAM-1). J. Biol. Chem. 281, 3157–3164 10.1074/jbc.M510797200 [DOI] [PubMed] [Google Scholar]
- 29. Koenen R. R., Pruessmeyer J., Soehnlein O., Fraemohs L., Zernecke A., Schwarz N., Reiss K., Sarabi A., Lindbom L., Hackeng T. M., Weber C., and Ludwig A. (2009) Regulated release and functional modulation of junctional adhesion molecule A by disintegrin metalloproteinases. Blood 113, 4799–4809 10.1182/blood-2008-04-152330 [DOI] [PubMed] [Google Scholar]
- 30. Scholz F., Schulte A., Adamski F., Hundhausen C., Mittag J., Schwarz A., Kruse M. L., Proksch E., and Ludwig A. (2007) Constitutive expression and regulated release of the transmembrane chemokine CXCL16 in human and murine skin. J. Invest. Dermatol. 127, 1444–1455 10.1038/sj.jid.5700751 [DOI] [PubMed] [Google Scholar]
- 31. Rieu P., Porteu F., Bessou G., Lesavre P., and Halbwachs-Mecarelli L. (1992) Human neutrophils release their major membrane sialoprotein, leukosialin (CD43), during cell activation. Eur. J. Immunol. 22, 3021–3026 10.1002/eji.1830221138 [DOI] [PubMed] [Google Scholar]
- 32. Bazil V., and Strominger J. L. (1993) CD43, the major sialoglycoprotein of human leukocytes, is proteolytically cleaved from the surface of stimulated lymphocytes and granulocytes. Proc. Natl. Acad. Sci. U.S.A. 90, 3792–3796 10.1073/pnas.90.9.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Götte M., Joussen A. M., Klein C., Andre P., Wagner D. D., Hinkes M. T., Kirchhof B., Adamis A. P., and Bernfield M. (2002) Role of syndecan-1 in leukocyte-endothelial interactions in the ocular vasculature. Invest. Ophthalmol. Vis. Sci. 43, 1135–1141 [PubMed] [Google Scholar]
- 34. Kharabi Masouleh B., Ten Dam G. B., Wild M. K., Seelige R., van der Vlag J., Rops A. L., Echtermeyer F. G., Vestweber D., van Kuppevelt T. H., Kiesel L., and Götte M. (2009) Role of the heparan sulfate proteoglycan syndecan-1 (CD138) in delayed-type hypersensitivity. J. Immunol. 182, 4985–4993 10.4049/jimmunol.0800574 [DOI] [PubMed] [Google Scholar]
- 35. Hayashida K., Parks W. C., and Park P. W. (2009) Syndecan-1 shedding facilitates the resolution of neutrophilic inflammation by removing sequestered CXC chemokines. Blood 114, 3033–3043 10.1182/blood.V114.22.3033.3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mulivor A. W., and Lipowsky H. H. (2002) Role of glycocalyx in leukocyte-endothelial cell adhesion. Am. J. Physiol. Heart Circ. Physiol. 283, H1282–1291 10.1152/ajpheart.00117.2002 [DOI] [PubMed] [Google Scholar]
- 37. Mulivor A. W., and Lipowsky H. H. (2009) Inhibition of glycan shedding and leukocyte-endothelial adhesion in postcapillary venules by suppression of matrix metalloprotease activity with doxycycline. Microcirculation 16, 657–666 10.3109/10739680903133714 [DOI] [PubMed] [Google Scholar]
- 38. Hikita A., Tanaka N., Yamane S., Ikeda Y., Furukawa H., Tohma S., Suzuki R., Tanaka S., Mitomi H., and Fukui N. (2009) Involvement of a disintegrin and metalloproteinase 10 and 17 in shedding of tumor necrosis factor-α. Biochem. Cell Biol. 87, 581–593 10.1139/O09-015 [DOI] [PubMed] [Google Scholar]
- 39. Zheng Y., Saftig P., Hartmann D., and Blobel C. (2004) Evaluation of the contribution of different ADAMs to tumor necrosis factor α (TNFα) shedding and of the function of the TNFα ectodomain in ensuring selective stimulated shedding by the TNFα convertase (TACE/ADAM17). J. Biol. Chem. 279, 42898–42906 10.1074/jbc.M403193200 [DOI] [PubMed] [Google Scholar]
- 40. Schulz B., Pruessmeyer J., Maretzky T., Ludwig A., Blobel C. P., Saftig P., and Reiss K. (2008) ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circ. Res. 102, 1192–1201 10.1161/CIRCRESAHA.107.169805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pruessmeyer J., Hess F. M., Alert H., Groth E., Pasqualon T., Schwarz N., Nyamoya S., Kollert J., van der Vorst E., Donners M., Martin C., Uhlig S., Saftig P., Dreymueller D., and Ludwig A. (2014) Leukocytes require ADAM10 but not ADAM17 for their migration and inflammatory recruitment into the alveolar space. Blood 123, 4077–4088 10.1182/blood-2013-09-511543 [DOI] [PubMed] [Google Scholar]
- 42. Kanki Y., Kohro T., Jiang S., Tsutsumi S., Mimura I., Suehiro J., Wada Y., Ohta Y., Ihara S., Iwanari H., Naito M., Hamakubo T., Aburatani H., Kodama T., and Minami T. (2011) Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J. 30, 2582–2595 10.1038/emboj.2011.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kreuger J., and Phillipson M. (2016) Targeting vascular and leukocyte communication in angiogenesis, inflammation and fibrosis. Nat. Rev. Drug Discov. 15, 125–142 10.1038/nrd.2015.2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.