Abstract
Phosphoglycerate kinase 1 (PGK1) plays important roles in glycolysis, yet its forward reaction kinetics are unknown, and its role especially in regulating cancer cell glycolysis is unclear. Here, we developed an enzyme assay to measure the kinetic parameters of the PGK1-catalyzed forward reaction. The Km values for 1,3-bisphosphoglyceric acid (1,3-BPG, the forward reaction substrate) were 4.36 μm (yeast PGK1) and 6.86 μm (human PKG1). The Km values for 3-phosphoglycerate (3-PG, the reverse reaction substrate and a serine precursor) were 146 μm (yeast PGK1) and 186 μm (human PGK1). The Vmax of the forward reaction was about 3.5- and 5.8-fold higher than that of the reverse reaction for the human and yeast enzymes, respectively. Consistently, the intracellular steady-state concentrations of 3-PG were between 180 and 550 μm in cancer cells, providing a basis for glycolysis to shuttle 3-PG to the serine synthesis pathway. Using siRNA-mediated PGK1-specific knockdown in five cancer cell lines derived from different tissues, along with titration of PGK1 in a cell-free glycolysis system, we found that the perturbation of PGK1 had no effect or only marginal effects on the glucose consumption and lactate generation. The PGK1 knockdown increased the concentrations of fructose 1,6-bisphosphate, dihydroxyacetone phosphate, glyceraldehyde 3-phosphate, and 1,3-BPG in nearly equal proportions, controlled by the kinetic and thermodynamic states of glycolysis. We conclude that perturbation of PGK1 in cancer cells insignificantly affects the conversion of glucose to lactate in glycolysis.
Keywords: glycolysis, serine, lactic acid, cancer, Warburg effect, cellular metabolism, energy homeostasis, PGK1 forward reaction kinetics, phosphoglycerate kinase
Introduction
The aerobic glycolysis (Warburg effect, WE)2 is a metabolic hallmark of cancer cells. Inhibiting WE is recognized as an approach to treat cancer (1–5). The rate-limiting enzymes along glycolysis are recognized as targets for inhibiting WE (6–12).
Previous studies demonstrated that PGK1 was a rate-limiting enzyme in the glycolysis of cancer cells (13–19). PGK1 catalyzes a step at the middle of glycolysis and produces ATP and 3-PG (a precursor for serine). Given the high glycolytic rate, PGK1 activity accounts for a large part in maintaining the energy homeostasis and serine biosynthesis. Clinically, PGK1 was overexpressed in many types of tumors (20–24). Experimentally, it is found that this enzyme was dynamically modulated in cells. This enzyme was transcriptionally up-regulated by HIF1 (25) but down-regulated by PPAR-γ (26). Hu et al. (13) reported that PGK1 could be acetylated by PCAF and Sirtuin 7, and the acetylation of PGK1 enhanced its activity and promoted glycolysis and liver cancer tumorigenesis. Zhang et al. (14) demonstrated that polarized M2 macrophages secreted IL-6, which enhances PGK1 phosphorylation in tumor cells and promoted glycolysis, and this phosphorylation is associated with malignance and prognosis of human GBM. Li et al. (15) revealed that, apart from its canonical activity, PGK1 could translocate into mitochondria to phosphorylate and to activate PDHK1, which in turn phosphorylated and inhibited PDH, impairing the TCA cycle and enhancing glycolysis. In contrast, Tanner et al. (27) reported that a perturbation of PGK1 did not significantly affect glycolysis.
The mixed reports indicated a vague understanding of the mechanism by which PGK1 regulates glycolysis. As the rate control of glycolysis is fundamentally a question of kinetics and thermodynamics of glycolysis, we sought to investigate the effect of perturbing PGK1 on glycolysis with respect to thermodynamics and related kinetics.
To investigate the above-mentioned questions, we sought to know the kinetics of PGK1. PGK1 catalyzes a reversible reaction, and its reverse-reaction kinetics is known, but its forward reaction kinetics is unknown because there are no available methods for research. Some studies reported that the forward reaction activity of PGK1 could be measured by the production of NADH, with a reaction mixture containing GA3P, β-NAD, and ADP (13, 28). The forward reaction of PGK1 can promote GAPDH to produce NADH (13, 28). This method may tell the difference of the NADH generation rate with or without PGK1, but it cannot accurately measure the activity of PGK1 nor the kinetic parameters. Therefore, we developed a method to accurately measure the forward reaction kinetics of PGK1.
Results
Rationale to measure the forward reaction rate of PGK1
We designed a coupled-enzyme assay to measure the forward-reaction rate of PGK1.
Step 1
In a reaction mixture containing GA3P, NAD, and Pi, by adding excessive amounts of GAPDH, Reaction 1 would rapidly reach equilibrium.
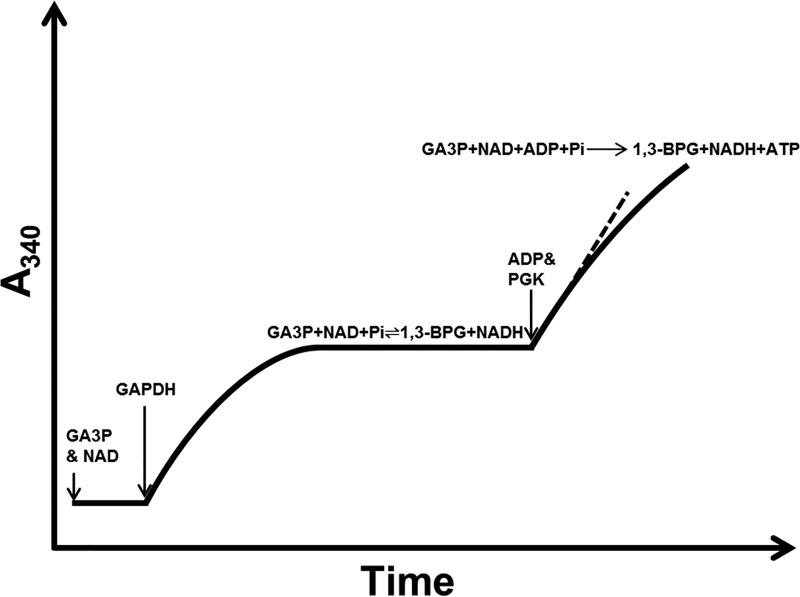
The equilibrium of the reactions is monitored at 340 nm, which increases at first and then remains stable (Fig. 1), which is indicative of the equilibrium state.
Figure 1.
Rationale to measure the forward reaction rate of PGK1. In a reaction mixture containing GA3P, NAD, and Pi, add an excessive amount of GAPDH, bringing the reaction rapidly to equilibrium state, which is monitored at A340. We then added ADP and PGK1 into the reaction mixture to initiate the PGK1-catalyzed forward reaction and recorded the initial rate (see also the text).
Step 2
By adding ADP and PGK1 into the reaction mixture, we initiate Reaction 2 (Fig. 1).
Consumption of 1,3-BPG immediately disrupts the equilibrium state of Reaction 1, thus driving GA3P and NAD to 1,3-BPG and NADH. The rate of NADH generation can be readily spectrophotometrically monitored. According to Reactions 1 and 2, we could derive the following equations: 1) the number of 3-PG molecules generated = the numbers of 1,3-BPG molecules consumed; 2) the number of 1,3-BPG molecules consumed = the numbers of GA3P molecules consumed; 3) the number of GA3P molecules generated = the numbers of NADH molecules generated; and 4) therefore, NADH generation rate = turnover rate of 1,3-BPG to 3-PG catalyzed by PGK1. Therefore, the initial rate of PGK1 could be accurately measured.
Kinetic parameters of PGK1
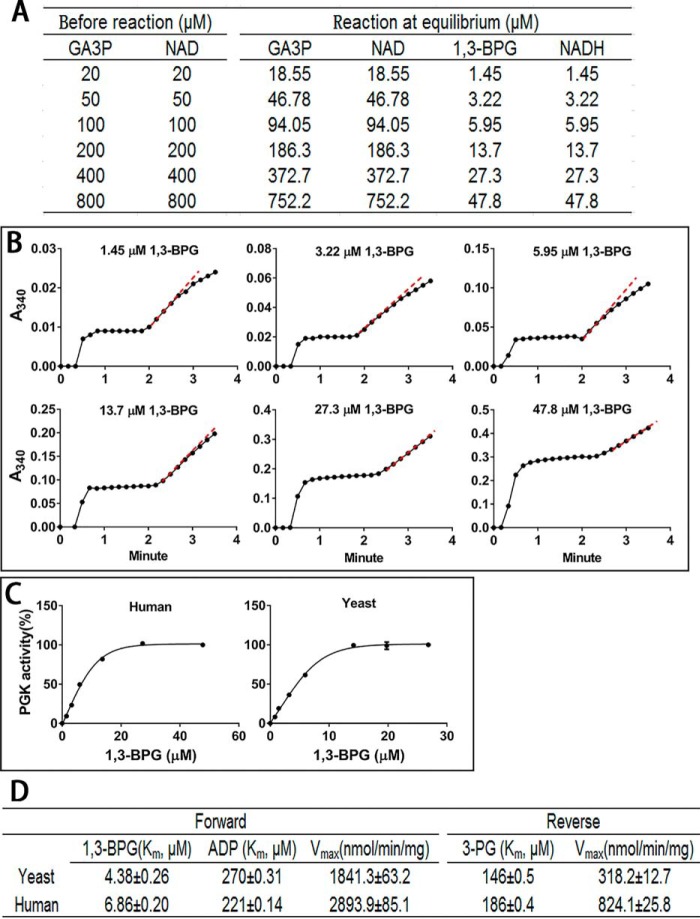
For measurement, based on the equilibrium of Reaction 1, we set serial concentrations of GA3P, which gave rise to the serial concentrations of 1,3-BPG (Fig. 2A). Then we determined the Km and Vmax values of human and yeast PGK1 (Fig. 2, B and C). The Km values for 1,3-BPG were 4.36 μm (yeast) and 6.86 μm (human); the Km values for 3-PG were 146 μm (yeast) and 186 μm (human), and the Vmax of the forward reaction (from 1,3-BPG to 3PG) was about 3.5-fold (human) and 5-fold (yeast) higher than that of the reverse reaction (Fig. 2D), indicating that the enzyme favors the forward reaction. By setting 1,3-BPG at a saturating concentration (about 50 μm) and varying the ADP concentrations (0.02 to 2 mm), we obtained the Km value for ADP (Fig. 2D).
Figure 2.
Measurement of the kinetic parameters of the forward reaction catalyzed by human and yeast PGK1. A, set the initial concentration of GA3P and NAD in the reaction mixture and add 1 unit/ml GAPDH. When the reaction reaches equilibrium, the concentration of 1,3-BPG in the mixture can be readily calculated. B, add 2 mm ADP and 0.0025 units of recombinant human PGK1 or yeast PGK1 to initiate reactions with a serial concentration of 1,3-BPG. C, kinetic curves of PGK1 versus 1,3-BPG. D, Km and Vmax values of PGK1 of the forward and reverse reactions. Data are means ± S.D., n = 3. All the results were repeated by three independent experiments.
Potential physiological implications of the kinetic parameters of the PGK1 for 3-PG shuttle to serine
We sought to explore the physiological meaning of the kinetic parameters of PGK1 in glycolysis. The low Km for 1,3-BPG, the high Km for 3-PG, and the much higher rates of the forward reaction than those of the reverse reaction are the biochemical bases to maintain a low concentration of 1,3-BPG and a high concentration of 3-PG in cells. Indeed, cellular 3-PG concentrations were kept at relatively high concentrations, between 0.18 and 0.55 mm (depending on the cell lines) (Table 1). 3-PG is a precursor for serine synthesis. Phosphoglycerate dehydrogenase (PHGDH), which catalyzes the first step for de novo serine synthesis, has the Km value of 0.26 mm for 3-PG (29). Keeping a high cellular 3-PG concentration is very important for this molecule to shuttle to serine synthesis, because the specific activity (at saturating concentration of 3-PG) of PHGDH in cancer cell is very low. The activity was undetectable even using 0.15 mg of cell lysate protein in our assay system, whereas the HK activity could be accurately determined using 0.03 mg of cell lysate protein.
Table 1.
Intracellular Glc and glycolytic intermediate concentration (mm) in cells with or without PGK1 knockdown
Data are means ± S.D., n = 3. The results were repeated by two independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001. siPGK1-transfected cells versus NC-transfected cells are shown.
| Metabolites (mm) | Glc | G6P | F6P | FBP | DHAP | GA3P | 3PG | 2PG | PEP | Pyr | ADP | ATP | NAD | NADH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HeLa-NC | 5.56 ± 0.037 | 0.56 ± 0.07 | 0.12 ± 0.08 | 0.38 ± 0.035 | 0.75 ± 0.04 | 0.05 ± 0.02 | 0.28 ± 0.04 | 0.09 ± 0.03 | 0.10 ± 0.014 | 0.20 ± 0.03 | 0.71 ± 0.12 | 3.80 ± 0.58 | 0.68 ± 0.11 | 0.033 ± 0.005 |
| HeLa-siPGK1 | 5.52 ± 0.03 | 0.57 ± 0.1 | 0.14 ± 0.11 | 0.65 ± 0.002*** | 1.28 ± 0.08*** | 0.09 ± 0.03* | 0.28 ± 0.03 | 0.07 ± 0.02 | 0.11 ± 0.02 | 0.14 ± 0.03 | 0.61 ± 0.07 | 3.10 ± 0.54 | 0.66 ± 0.12 | 0.035 ± 0.008 |
| MGC80–3-NC | 3.9 ± 0.07 | 0.43 ± 0.04 | 0.12 ± 0.03 | 0.28 ± 0.01 | 0.58 ± 0.05 | 0.05 ± 0.02 | 0.18 ± 0.05 | 0.10 ± 0.066 | 0.15 ± 0.004 | 0.27 ± 0.02 | 0.59 ± 0.014 | 3.74 ± 0.21 | 0.58 ± 0.04 | 0.039 ± 0.004 |
| MGC80–3-siPGK1 | 3.96 ± 0.11 | 0.44 ± 0.05 | 0.11 ± 0.02 | 0.55 ± 0.014*** | 1.17 ± 0.1*** | 0.096 ± 0.04* | 0.14 ± 0.07 | 0.078 ± 0.04 | 0.15 ± 0.001 | 0.21 ± 0.02* | 0.83 ± 0.062 | 4.54 ± 0.8 | 0.78 ± 0.12 | 0.048 ± 0.005 |
| RKO-NC | 5.2 ± 0.47 | 0.31 ± 0.015 | 0.11 ± 0.011 | 0.37 ± 0.034 | 0.47 ± 0.04 | 0.043 ± 0.003 | 0.55 ± 0.07 | 0.066 ± 0.028 | 0.06 ± 0.03 | 0.56 ± 0.04 | 6.7 ± 0.19 | 11.3 ± 0.54 | 1.45 ± 0.03 | 0.16 ± 0.01 |
| RKO-siPGK1 | 5.2 ± 0.14 | 0.29 ± 0.1 | 0.13 ± 0.037 | 0.50 ± 0.04* | 0.77 ± 0.091** | 0.073 ± 0.005** | 0.43 ± 0.12 | 0.085 ± 0.017 | 0.049 ± 0.013 | 0.48 ± 0.07 | 6.9 ± 0.25 | 11.5 ± 0.64 | 1.45 ± 0.06 | 0.16 ± 0.003 |
| SK-HEP-1-NC | 3.88 ± 0.24 | 0.29 ± 0.028 | 0.078 ± 0.022 | 0.24 ± 0.037 | 0.55 ± 0.047 | 0.045 ± 0.006 | 0.49 ± 0.09 | 0.13 ± 0.045 | 0.23 ± 0.064 | 0.51 ± 0.096 | 0.77 ± 0.05 | 9.3 ± 0.07 | 0.93 ± 0.001 | 0.025 ± 0.003 |
| SK-HEP-1-siPGK1 | 3.4 ± 0.18 | 0.35 ± 0.05 | 0.11 ± 0.032 | 0.39 ± 0.035** | 0.8 ± 0.045** | 0.066 ± 0.009* | 0.52 ± 0.11 | 0.13 ± 0.054 | 0.29 ± 0.022 | 0.51 ± 0.10 | 0.77 ± 0.05 | 9.9 ± 0.42 | 0.72 ± 0.55 | 0.029 ± 0.003 |
| A549-NC | 4.2 ± 0.04 | 0.21 ± 0.04 | 0.038 ± 0.003 | 0.14 ± 0.03 | 0.28 ± 0.03 | 0.021 ± 0.002 | 0.22 ± 0.044 | 0.071 ± 0.03 | 0.084 ± 0.04 | 0.64 ± 0.066 | 0.84 ± 0.057 | 10.7 ± 0.25 | 1.78 ± 0.033 | 0.28 ± 0.009 |
| A549-siPGK1 | 3.7 ± 0.05*** | 0.177 ± 0.01 | 0.05 ± 0.013 | 0.20 ± 0.035* | 0.54 ± 0.08*** | 0.042 ± 0.003*** | 0.21 ± 0.052 | 0.069 ± 0.018 | 0.085 ± 0.031 | 0.625 ± 0.014 | 0.9 ± 0.069 | 11.1 ± 0.56 | 1.78 ± 0.038 | 0.28 ± 0.01 |
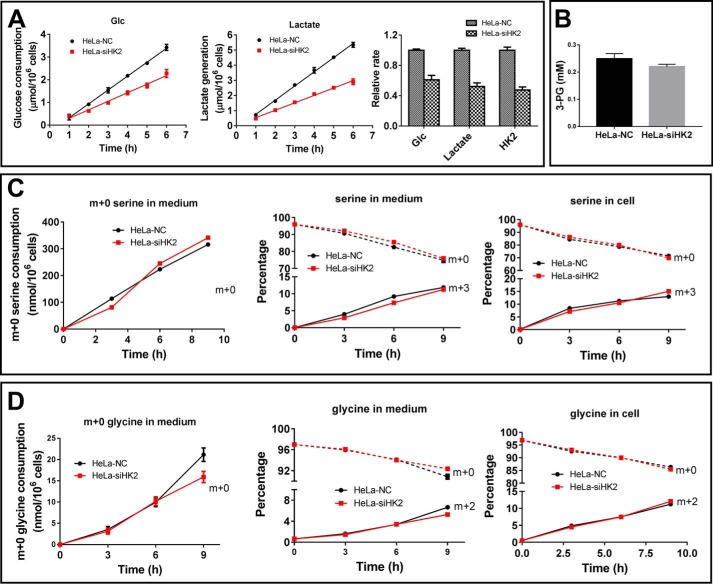
HK2 knockdown reduced HK activity by ∼50%, glucose consumption by ∼40%, and lactate generation by ∼50% (Fig. 3A). However, HK2 knockdown did not significantly reduce the 3-PG concentration (Fig. 3B) and serine synthesis, as manifested by the analysis of the serine isotopologues (Fig. 3C). The m + 0 serine species is provided by the culture medium, and m + 3 serine isotopologue was generated from [13C6]glucose through 3-PG. The consumption rate of m + 0 serine was comparable between control cells and HK2 knockdown cells (Fig. 3C, left panel), and the percentages of extracellular and intracellular m + 0 and m + 3 serine species were also comparable between control cells and HK2 knockdown (Fig. 3C, middle and right panels). A fraction of serine was further used for glycine synthesis. The consumption rate of glycine (m + 0, which is provided by culture medium) is comparable between control and HK2 knockdown cells (Fig. 3D, left panel) and so were the percentages of extracellular and intracellular glycine (m + 2, which is derived from m + 3 serine) (Fig. 3D, middle and right panels). These data support that glucose carbon shuttling to serine and glycine was not significantly affected by HK2 knockdown, and even HK2 knockdown reduced the glycolysis rate to lactate by 50%.
Figure 3.
Effect of HK2 knockdown on glycolysis and serine synthesis in HeLa cells. A, proportional relationship between HK2 activity and glycolysis rate (lactate generation and glucose consumption). B, 3-PG concentrations in HeLa–NC and HeLa–siPGK1 cells. C, glucose carbon shuttle to serine. Cells were incubated in the presence of 6 mm [13C6]glucose. Cells and medium were collected at 3, 6, and 9 h, and serine and glycine isotopologues were analyzed by LC-MS. Left panel, consumption of m + 0 serine provided by the culture medium; middle panel, the percentage of m + 0 and m + 3 serine in medium; right panel, percentage of m + 0 and m + 3 serine in cells. D, left panel, consumption of m + 0 glycine provided by the culture medium; middle panel, percentage of m + 0 and m + 2 glycine in medium; right panel, percentage of m + 0 and m + 2 glycine in cells. Data are mean ± S.D., n = 3. All the results were repeated by two independent experiments.
Taken together, the kinetic parameters of PGK1 are associated with a stable steady-state concentration of 3-PG in cancer cells, which is around the Km value of PHGDH, underlying a sound biochemical basis for glycolysis to shuttle to the de novo serine synthesis pathway.
Characteristics of cell lines
We used five cell lines from different tissue origins (cervical cancer cell line HeLa, gastric cancer cell line MGC80-3, colon cancer cell line RKO, lung cancer cell line A549, and liver cancer cell line SK-HEP-1). These cells were derived from different organs from different patients, representing five types of cancer cell lines. They exhibited widespread mutations (30–33). HeLa cells had WT TP53, but p53 protein was repressed because of overexpression of human papillomavirus type 16 E6 (34, 35). In MGC80-3, decreased expression of TWSG1 was detected compared with normal gastric cells (36). In RKO cells, TP53 was WT, whereas PTEN, KRAS, BRAF, and PIK3CA were mutated (30, 32). WT TP53 was detected in both SK-HEP-1 and A549. BRAF was mutated in SK-HEP-1, whereas CDKN2A and KRAS were mutated in A549 cells (31, 33).
Despite the differences, they shared the same metabolic feature, the Warburg effect. They exhibited a similar pattern of glycolytic enzymes (Fig. 4A), and they converted most incoming glucose to lactate (Fig. 4B). If the glucose consumption rate is expressed by kilograms of glucose/kg of cells per day, the number would be larger than 1.5 kg per kg of cancer cells per day (Fig. 4C). Thus, the data demonstrated that these cells shared the feature of the Warburg effect. This is consistent with the general consensus that the Warburg effect is characteristic of essentially all types of cancer cells (37–39). Mechanistically, the Warburg effect is programmed by a complex signaling network composed of oncogenic activation and tumor suppressor inactivation or insufficiency, including but not limited to Ras, Raf, ERK, JNK, Myc, HIF, p53, PI3K, Akt, etc. (38, 40–47).
Figure 4.
Characteristics of cell lines–Warburg effect. A, glycolytic enzyme activities in five cancer cell lines. B, glycolytic rate expressed by glucose consumption or lactate generation. Data are means ± S.D., n = 3. All the results were repeated by at least five independent experiments. C, estimation of glucose consumption and lactate generation by 1 kg of cancer cells in 24 h of each cell line.
Thermodynamic state of glycolysis and pattern of glycolytic intermediates
For clarity, we list the following: the terms and abbreviations; the specific activity; the enzyme activity assayed at saturating substrate concentrations; the relative enzyme concentration; and as the absolute concentrations of enzymes in cells could not be determined, we used relative concentration, which is based on the specific activity, e.g. the relative PGK1 concentration of HeLa cells is 1, then the relative concentration of the PGK1-knockdown cell is the PGK1-specific activity in knockdown cells divided by the PGK1-specific activity in control cells. So we term [PGK1] the relative concentration of PGK1. In addition, the abbreviations used are as follows: [Glc], the concentration of glucose; [G6P], the concentration of glucose 6-phosphate; [F6P], the concentration of fructose 6-phosphate; [FBP], the concentration of fructose 1,6-bisphosphate; [DHAP], the concentration of dihydroxyacetone phosphate; [GA3P], the concentration of glyceraldehyde 3-phosphate; [1,3-BPG], the concentration of 1,3-bisphosphoglycerate; [3-PG], the concentration of 3-phosphoglycerate; [2-PG], the concentration of 2-phosphoglyerate; [PEP], the concentration of phosphoenolpyruvate; and [Pyr], the concentration of pyruvate.
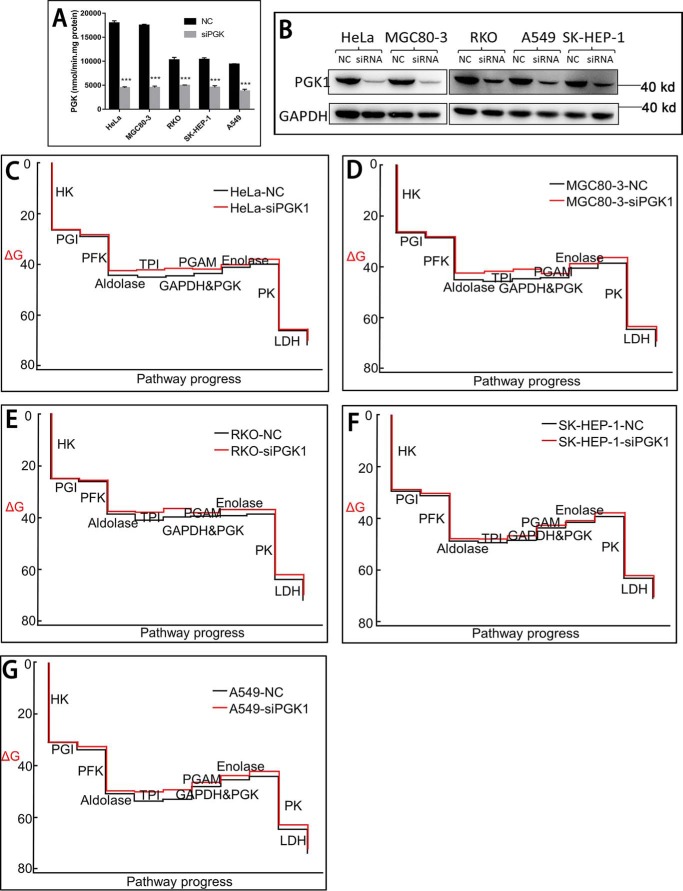
The transient knockdown specifically reduced PGK1 activity without significantly affecting other glycolytic enzymes (Table 2 and Fig. 5, A and B). The knockdown efficiency varied from cell to cell (Table 2), ranging from 70% (HeLa) to 52% (RKO).
Table 2.
Enzyme activity (nanomoles/min·mg of protein) in cells with or without PGK1 knockdown
Data are means ± S.D., n = 3. The results were repeated by two independent experiments. ***, p < 0.001. siPGK1-transfected cells versus NC-transfected cells are shown.
| HK | PGI | PFK | Aldolase | TPI | GAPDH | PGK | PGAM | enolase | PK | LDH | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HeLa-NC | 77.4 ± 4.8 | 3552.5 ± 25.9 | 390.7 ± 7.9 | 244.6 ± 9.8 | 19638.4 ± 605.6 | 3431.5 ± 194.6 | 14789.9 ± 863.7 | 1963.8 ± 93.5 | 1741.2 ± 105.8 | 3752.1 ± 187.1 | 7036.9 ± 486.3 |
| HeLa-siPGK1 | 79.0 ± 2.86 | 3562.2 ± 31.5 | 395.2 ± 22.2 | 259.5 ± 17.9 | 20721.7 ± 442.8 | 3215.4 ± 126.1 | 4476.4 ± 227.3*** | 1866.2 ± 115.6 | 1714.9 ± 75.6 | 3521.9 ± 174.7 | 7220.3 ± 403.7 |
| MGC80–3-NC | 82.5 ± 5.5 | 2450.9 ± 175.9 | 388.9 ± 11.9 | 239.7 ± 7.3 | 20166.3 ± 267.6 | 4952.7 ± 380.9 | 19296.7 ± 1236.7 | 2282.6 ± 77.5 | 774.6 ± 21.9 | 3477.9 ± 233.9 | 7613.1 ± 588.0 |
| MGC80–3-siPGK1 | 81.9 ± 6.2 | 2308.9 ± 154.2 | 409.8 ± 18.7 | 249.4 ± 8.2 | 19697.4 ± 215.2 | 5216.5 ± 226.2 | 5543.6 ± 446.9*** | 2194.9 ± 89.0 | 741.1 ± 24.7 | 3561.6 ± 426.5 | 7962.3 ± 89.0 |
| RKO-NC | 84.6 ± 3.9 | 2665.9 ± 46.8 | 231.3 ± 4.5 | 358.6 ± 7.8 | 29215.6 ± 210.9 | 2416.4 ± 27.0 | 10263.4 ± 553.0 | 935.4 ± 46.8 | 600.2 ± 13.5 | 3741.6 ± 46.8 | 4521.1 ± 71.4 |
| RKO-siPGK1 | 81.3 ± 1.9 | 2650.3 ± 27.0 | 223.5 ± 4.5 | 335.2 ± 23.4 | 28888.2 ± 164.3 | 2400.9 ± 54.0 | 4936.8 ± 119.1*** | 873.0 ± 27.0 | 561.2 ± 23.4 | 3554.5 ± 168.6 | 4458.7 ± 54.0 |
| SK-HEP-1-NC | 88.3 ± 6.1 | 1665.4 ± 28.6 | 336.6 ± 2.4 | 296.8 ± 8.2 | 20842.6 ± 159.0 | 3611.2 ± 49.5 | 10360.8 ± 343.3 | 956.4 ± 28.6 | 766.7 ± 24.7 | 4188.3 ± 75.6 | 5062.2 ± 28.6 |
| SK-HEP-1-siPGK1 | 84.8 ± 3.5 | 1632.4 ± 49.5 | 333.9 ± 10.9 | 288.6 ± 8.2 | 19473.9 ± 804.3 | 3516.5 ± 49.5 | 4562.1 ± 343.3*** | 906.9 ± 28.6 | 709.0 ± 37.8 | 4138.8 ± 75.6 | 4979.8 ± 28.6 |
| A549-NC | 56.6 ± 2.7 | 4523.0 ± 98.2 | 196.5 ± 3.1 | 300.1 ± 28.4 | 18306.5 ± 267.7 | 2508.0 ± 64.3 | 9378.3 ± 92.8 | 1264.7 ± 37.1 | 664.5 ± 18.6 | 6388.0 ± 37.1 | 5894.9 ± 234.5 |
| A549-siPGK1 | 53.2 ± 4.8 | 4242.1 ± 103.4 | 206.8 ± 3.3 | 267.0 ± 23.5 | 18863.9 ± 103.4 | 2594.9 ± 39.1 | 3779.5 ± 390.8*** | 1241.0 ± 39.1 | 654.4 ± 19.5 | 6498.6 ± 67.7 | 5618.5 ± 67.7 |
Figure 5.
PGK1 knockdown does not affect the thermodynamic state of the glycolysis in cancer cells. A and B, PGK1 activity and Western blotting of PGK1 in cells with or without PGK1 knockdown. C–G, thermodynamic state of each reaction along the glycolysis in cancer cells. All the results were repeated by two independent experiments.
We quantified the cellular concentrations of the glycolytic intermediates, including ATP, ADP, NAD, and NADH (Table 1). Using these data, we calculated the mass action ratio (Q) and the actual Gibbs free energy (ΔG) of each reaction along the glycolysis (Table 3). According to Q and ΔG, it is clear that the reactions from the step catalyzed by aldolase to the step catalyzed by enolase were all at near-equilibrium state (Fig. 5, C–G). The reaction catalyzed by the PGI is also at near-equilibrium state. PGK1 knockdown did not significantly affect the thermodynamic state of the glycolysis (Fig. 5, C–G). The reactions catalyzed by HK2, PFK1, and PK were far from equilibrium, generating the driving force for glycolysis.
Table 3.
ΔG values of glycolytic enzymes in cells with or without PGK1 knockdown calculated according to the data in Table 1
| Enzyme | HK | PGI | PFK | Aldolase | TPI | GAPDH and PGK | PGAM | Enolase | PKM | LDH | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Keq | 850 | 0.5 | 310 | 0.000064 | 0.05 | 158.00 | 0.17 | 0.48 | 363000.00 | 26300.00 | |
| ΔG0 (kJ/mol) | −16.7 | 1.7 | −14.2 | 23.8 | 7.50 | −12.50 | 4.40 | 1.70 | −31.40 | −25.10 | |
| HeLa-NC | Q | 0.019 | 0.22 | 0.59 | 0.000100 | 0.07 | 257.26 | 0.31 | 1.14 | 10.75 | 897.00 |
| ΔG (kJ/mol) | −26.66 | −2.62 | −15.75 | −0.94 | 0.73 | 1.28 | 2.70 | 1.50 | −26.33 | −6.10 | |
| HeLa-siPGK1 | Q | 0.020 | 0.25 | 0.91 | 0.000180 | 0.07 | 135.08 | 0.24 | 1.53 | 6.47 | 1448.57 |
| ΔG (kJ/mol) | −26.45 | −2.24 | −14.62 | 0.60 | 0.91 | −0.38 | 2.08 | 2.25 | −27.64 | −4.87 | |
| MGC80-3-NC | Q | 0.017 | 0.27 | 0.37 | 0.000100 | 0.08 | 195.38 | 0.56 | 1.51 | 11.43 | 635.56 |
| ΔG (kJ/mol) | −26.86 | −2.08 | −16.97 | −0.90 | 1.25 | 0.57 | 4.21 | 2.22 | −26.17 | −6.99 | |
| MGC80-3-siPGK1 | Q | 0.020 | 0.26 | 0.92 | 0.000200 | 0.08 | 67.93 | 0.56 | 1.88 | 7.63 | 891.43 |
| ΔG (kJ/mol) | −26.44 | −2.21 | −14.61 | 0.88 | 1.25 | −2.15 | 4.21 | 2.78 | −27.21 | −6.12 | |
| RKO-NC | Q | 0.035 | 0.35 | 1.98 | 0.000055 | 0.09 | 185.85 | 0.12 | 0.91 | 15.87 | 348.21 |
| ΔG (kJ/mol) | −25.05 | −1.37 | −12.63 | −2.50 | 1.53 | 0.45 | 0.24 | 0.91 | −25.32 | −8.54 | |
| RKO-siPGK1 | Q | 0.033 | 0.45 | 2.26 | 0.000112 | 0.09 | 85.59 | 0.20 | 0.58 | 16.65 | 390.00 |
| ΔG (kJ/mol) | −25.22 | −0.76 | −12.29 | −0.64 | 1.62 | −1.55 | 1.53 | −0.26 | −25.20 | −8.25 | |
| SK-HEP-1-NC | Q | 0.006 | 0.27 | 0.25 | 0.000103 | 0.08 | 1126.12 | 0.27 | 1.77 | 26.83 | 382.35 |
| ΔG (kJ/mol) | −29.52 | −2.08 | −17.92 | −0.86 | 1.24 | 5.09 | 2.29 | 2.63 | −23.97 | −8.30 | |
| SK-HEP-1-siPGK1 | Q | 0.008 | 0.31 | 0.28 | 0.000135 | 0.08 | 861.95 | 0.24 | 2.23 | 22.51 | 351.76 |
| ΔG (kJ/mol) | −28.84 | −1.68 | −17.70 | −0.16 | 1.26 | 4.40 | 2.04 | 3.23 | −24.42 | −8.51 | |
| A549-NC | Q | 0.004 | 0.18 | 0.29 | 0.000042 | 0.08 | 1146.11 | 0.32 | 1.18 | 97.52 | 231.56 |
| ΔG (kJ/mol) | −30.71 | −3.10 | −17.61 | −3.18 | 1.02 | 5.13 | 2.79 | 1.59 | −20.65 | −9.59 | |
| A549-siPGK1 | Q | 0.004 | 0.28 | 0.32 | 0.000113 | 0.08 | 529.91 | 0.33 | 1.23 | 91.18 | 224.64 |
| ΔG (kJ/mol) | −30.73 | −1.95 | −17.30 | −0.62 | 1.11 | 3.15 | 2.84 | 1.70 | −20.82 | −9.67 |
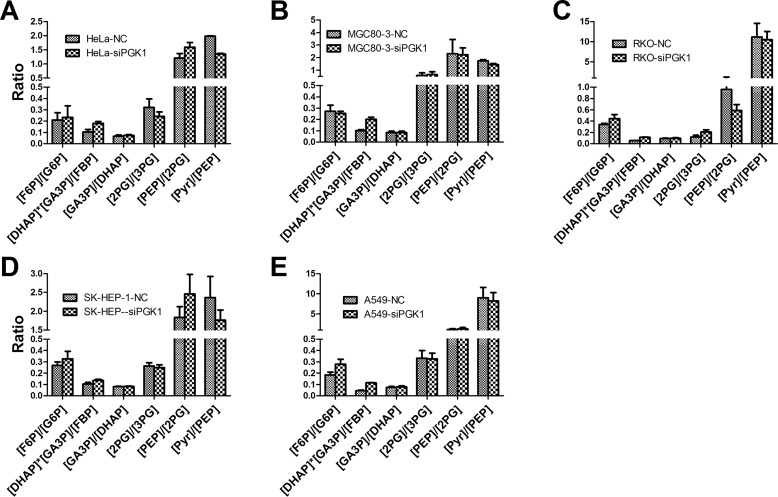
As the reactions catalyzed PGI, and aldolase through enolase were all at near-equilibrium state in the steady-state glycolysis, we could reason out a pattern of the glycolytic intermediates, which is characterized by the [F6P]/[G6P], [DHAP][GA3P]/[FBP], [GA3P]/[DHAP], [2-PG]/[3-PG], and [PEP]/[2-PG]. This pattern was not significantly affected by the perturbation of PGK1 (Fig. 6). In essence, this pattern is determined by the thermodynamic nature of the glycolysis in cancer cells.
Figure 6.
Pattern of the glycolytic intermediates in cancer cells. The data are from Table 1. The unit for [DHAP][GA3P]/([FBP]) is millimolar. Data are means ± S.D., n = 3. All the results were repeated by two independent experiments.
Effect of PGK1 knockdown on the glycolytic intermediates
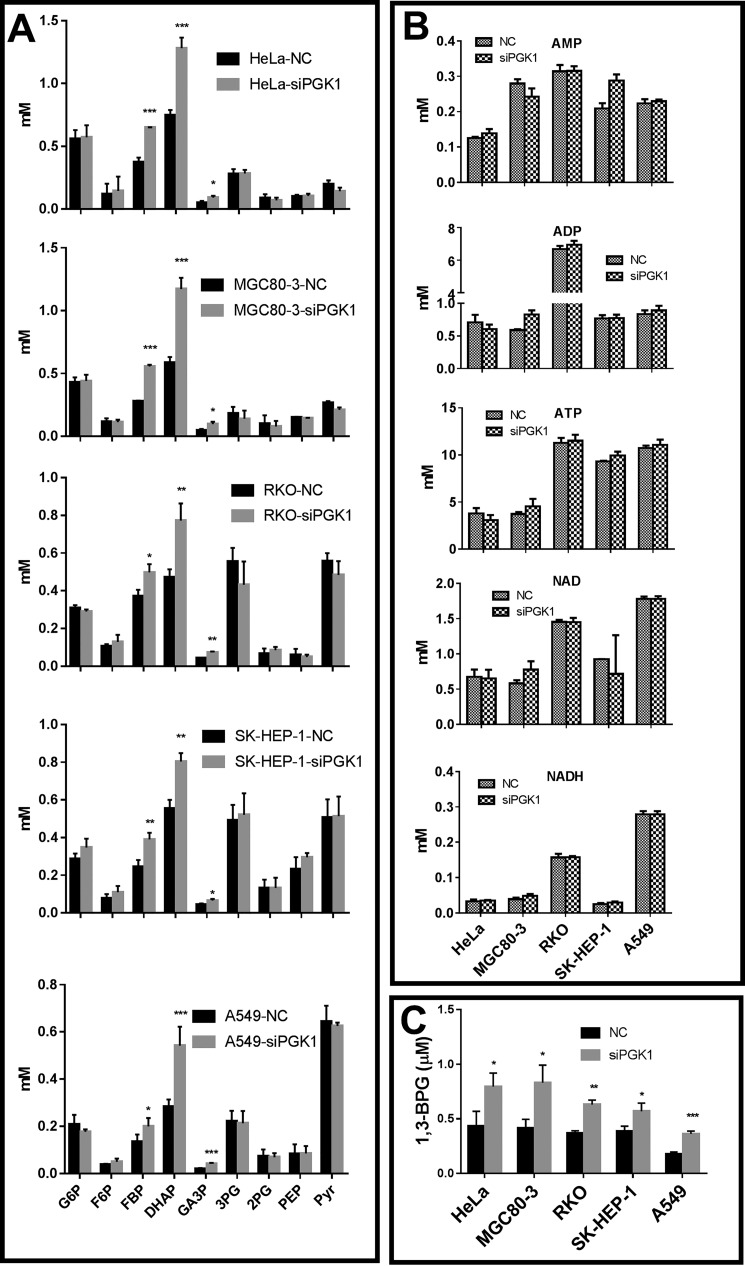
Because the reactions from aldolase to enolase are all at near-equilibrium states, in theory, we could infer that when the PGK1 amount is reduced, its rate would be temporarily reduced, leading to an accumulation of its substrate 1,3-BPG. As the reactions catalyzed by aldolase, TPI, and GAPDH are all at near-equilibrium, the concentrations of FBP, DHAP, GA3P, and 1,3-BPG would all accumulate in the same or a similar proportion according to the thermodynamic state of these reactions. Experimentally, we observed that siRNA knockdown induced an increase of the concentrations of FBP, DHAP, and GA3P (Fig. 7A). Nevertheless, we lack method to quantify the amount of 1,3-BPG. siRNA knockdown did not significantly affect the concentrations of other glycolytic intermediates, including ATP, ADP, NAD, and NADH (Fig. 7B).
Figure 7.
Effect of PGK1 knockdown on the concentrations of the glycolytic intermediates in cancer cells. A, PGK1 knockdown only significantly increased the concentration of FBP, DHAP, and GA3P. B, PGK1 knockdown did not significantly affect AMP, ADP, ATP, NAD, and NADH. C, estimated 1,3-BPG. The calculation is described under “Materials and methods.” Data are mean ± S.D., n = 3. All the results were repeated by two independent experiments.
Estimating the intracellular [1,3-BPG]
As the reactions from aldolase to enolase were at a near-equilibrium state, and as the PGK1 knockdown by siRNA did not significantly alter the thermodynamic state of glycolysis, we could estimate 1,3-BPG according to the mass action ratio of GAPDH, Q = ([1,3-BPG][NADH])/([GA3P][NAD][Pi]), where Q is approximately equal to Keq. The estimated [1,3-BPG] was about 1.48–2.04-fold higher in PGK1-knockdown cells than in control cells (Fig. 7C).
PGK1 knockdown does not significantly affect the glycolysis rate
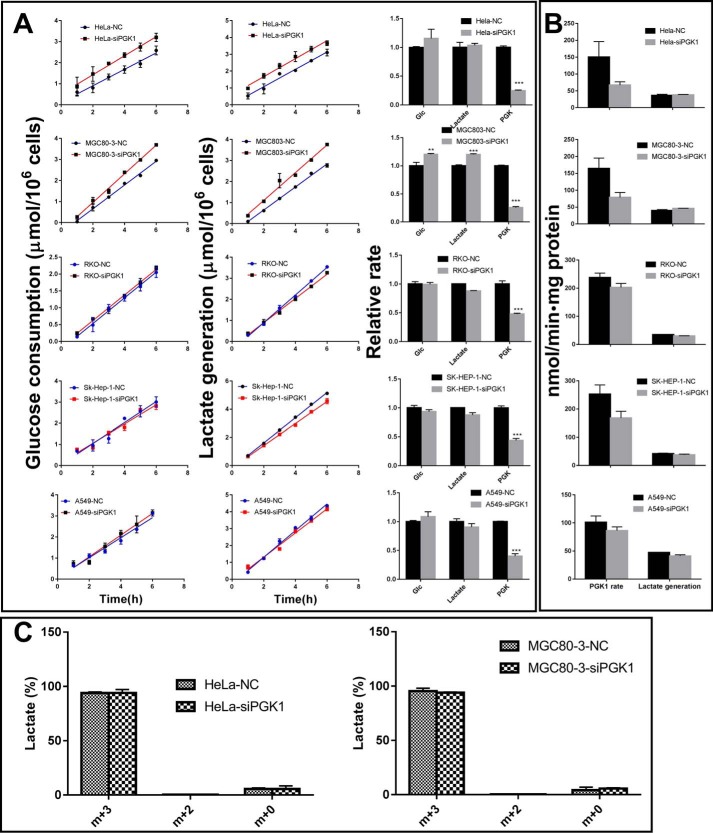
According to the PGK1 kinetic curve and the Km value of PGK1 toward 1,3-BPG (Fig. 2C), intracellular [1,3-BPG] is evidently not saturating PGK1 in both control and PGK1 knockdown cells. Theoretically, the increased [1,3-BPG] can significantly increase the catalytic rate of PGK1 and cancel out or at least attenuate the effect of the siRNA-induced loss of PGK1 amount on its catalytic rate as well as on the glycolytic rate. This was validated by the experiments, which demonstrated that lactate generation and glucose consumption by cancer cells were insignificantly or marginally affected by siRNA knockdown (Fig. 8A). In addition, the following lines of evidence also support that glycolysis rate in cancer cells is not sensitive to PGK1 knockdown:
Figure 8.
PGK1 knockdown insignificantly or marginally affects the glycolysis rate. A, glucose consumption rate, lactate generation rate, and PGK1 activity. B, actual PGK1 activity at the cellular 1,3-BPG concentrations (Fig. 7C) and the cellular lactate generation rate. C, tracing glucose carbon to lactate. Cells were incubated in the presence of 6 mm [13C6]glucose for 6 h. The percentages of the generated lactate isotopologues were measured by LC-MS. Data are means ± S.D., n = 3. All the results were repeated by two independent experiments.
First, The PGK1-specific activities are three orders of magnitude higher than the rate-limiting HK (Fig. 4A). Even though the actual activities of PGK1 in PGK1-knockdown cells were lower than those in control cells, they were all significantly higher than the cellular glycolysis rate (Fig. 8B).
Second, as the reactions from aldolase to enolase were all in a near-equilibrium state, the reaction catalyzed by pyruvate kinase, with the large and negative ΔG (Table 3), is the force to drive the glycolysis flux to pyruvate. Kinetically, the actual activities of pyruvate kinase in control and PGK1-knockdown cells (Table S1) were comparable, and they were 1 to 2 orders of magnitude higher than the glycolytic rate, which is sufficient to drive the upstream intermediates to pyruvate. The results were consistent with our previous study concerning the flux control of glycolysis in cancer cells with respect to the kinetics and thermodynamics of this enzyme (48).
Third, the Q values of the reaction catalyzed by LDH in both control and PGK1-knockdown cells (Table 3) were much smaller than the Keq, generating a sufficiently large and negative ΔG that favors the forward reaction. Kinetically, the actual LDH activities in control and PGK1-knockdown cells were 2 orders of magnitude higher than the glycolytic rate (Table S1).
Finally, we used [13C6]glucose to trace lactate generation, and the results showed that glucose-derived lactate (m + 3) consisted of about 95% of total lactate in control and PGK1 knockdown cells (Fig. 8C). This confirmed that glucose-derived lactate in both control and PGK1-knockdown cells was not significantly different from each other, supporting the notion that glycolysis in cancer cells is not sensitive to perturbation of PGK1.
PGK1 knockdown moderately reduces serine consumption and de novo serine synthesis
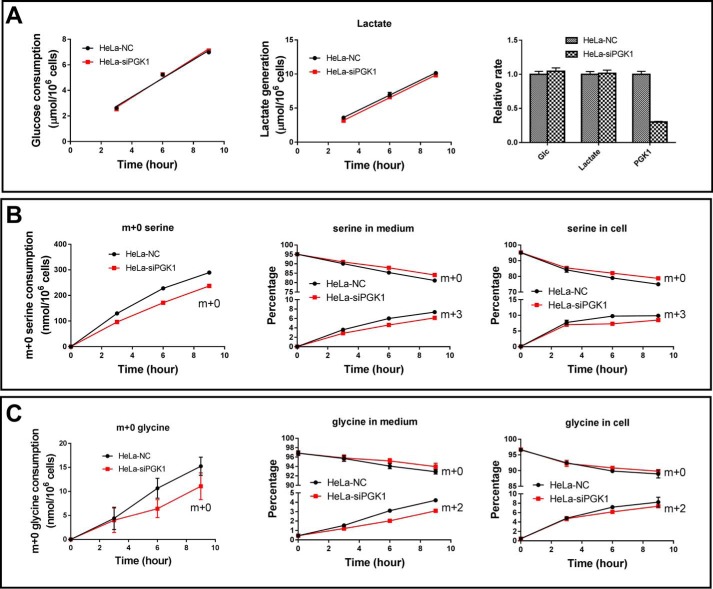
We used [13C6]Glc to trace glucose carbon incorporation into serine. The m + 0 serine species is provided by the culture medium, and the m + 3 serine isotopologue was generated from [13C6]glucose through 3-PG. Although glucose consumption rate and lactate generation rate were comparable between HeLa–NC and HeLa–siPGK1 (Fig. 9A), the consumption rate of m + 0 serine was about 20% lower in HeLa–siPGK1 cells than in control cells (Fig. 9B). HeLa–siPGK1 showed a higher percentage of extracellular and intracellular m + 0 serine than HeLa–NC, whereas the m + 3 species was the reverse (Fig. 9B). Consistently, glycine consumption rate was lower by HeLa–siPGK1 than by HeLa–NC and so were the percentages of glycine isotopologues (Fig. 9C). The data indicate that PGK1 knockdown moderately reduced serine consumption and serine de novo synthesis by HeLa cells.
Figure 9.
PGK1 knockdown moderately reduces serine consumption and de novo serine synthesis. A, glucose consumption and lactate generation by HeLa–siPGK1 and HeLa–NC. B, serine consumption and serine synthesis by HeLa–siPGK1 and HeLa–NC. Left panel, consumption of m + 0 serine provided by the culture medium; middle panel, percentage of m + 0 and m + 3 serine in medium; right panel, percentage of m + 0 and m + 3 serine in cells. C, glycine consumption and glycine synthesis by HeLa–siPGK1 and HeLa–NC. Left panel, consumption of m + 0 glycine provided by the culture medium; middle panel, percentage of m + 0 and m + 2 glycine in medium; right panel, percentage of m + 0 and m + 2 glycine in cells. Data are means ± S.D., n = 3. All the results were repeated by two independent experiments.
Cell-free system
If the above-proposed mechanism is valid, we should observe it in a different model. Glycolysis in living cells is influenced by many factors, e.g. glycolysis is connected to subsidiary metabolic branches (pentose phosphate pathway, serine synthesis pathway, etc.), and glycolytic enzymes are influenced by dynamic regulation, including compartmentation, allosteric regulation, and chemical modification (phosphorylation, acetylation, etc.), all of which could influence the rate and the intermediate concentrations. To demonstrate the effect of perturbing PGK1 on glycolysis, we used a cell-free glycolysis system, as described by us previously (49), to avoid or at least minimize the influences. We used 30-min incubation for the subsequent experiments concerning the cell-free glycolysis.
We prepared HeLa–siPGK1 cell lysate and titrated the PGK activity in the cell lysate. The results derived from the cell-free glycolysis system were fully agreeable with those from the living cells as described below.
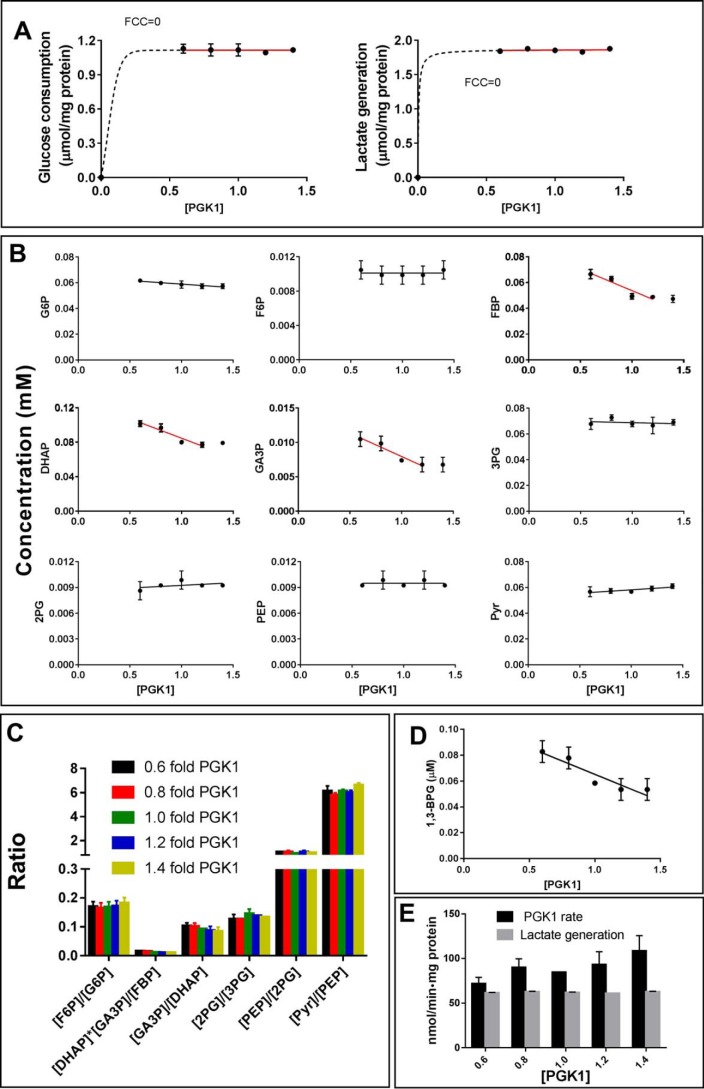
First, we used the PGK1-knockdown cell lysate to measure the flux control coefficient (FCC) values. The FCC, through the unperturbed point (i.e. the relative [PGK1] at 1.0), was nearly zero, based on either the rate of glucose consumption or the rate of lactate generation (Fig. 10A).
Figure 10.
Cell-free glycolysis model for studying the effect of PGK1 titration on glycolysis. A, glucose consumption rate and lactate generation rate in the reaction mixture titrated with PGK1. PGK1 of HeLa cells was knocked down by siPGK1, and the cells were collected, and cell lysate was prepared. The cell lysate was then titrated with pure PGK1. The cell lysate with different amounts of PGK1 was separately added into the reaction mixture to start the reaction, which was terminated at 30 min. Glucose, lactate, and glycolytic intermediates were measured. The flux control coefficient was calculated according to the tangent of the nonlinear regression at the point [PGK1] = 1.0. B, concentrations of glycolytic intermediates in the reaction mixture titrated with PGK1. C, pattern of the glycolytic intermediates. D, concentration of 1,3-BPG versus PGK1. The concentration of 1,3-BPG is described under “Materials and methods.” E, actual PGK activity at the 1,3-BPG concentrations (D) and the lactate generation rate. Data are means ± S.D., n = 3. Results were repeated by two independent experiments.
Second, the [FBP], [DHAP], and [GA3P] were nearly inversely proportional to [PGK1], except the last point ([PGK1] at 1.4), and no significant change of other intermediates (G6P, F6P, 3-PG, 2-PG, PEP, and pyruvate) was detected (Fig. 10B). The ratios of [F6P]/[G6P], [DHAP][GA3P]/[FBP], [GA3P]/[DHAP], [2-PG]/[3-PG], and [PEP]/[2-PG] were not significantly affected by the amount of PGK1 (Fig. 10C). Accordingly, we could estimate the concentration of 1,3-BPG (Fig. 10D).
Third, the actual PGK1 activities in cell-free system with different PGK1 concentrations were all significantly higher than the glycolysis rate (Fig. 10E).
Finally, as the sum of the ΔG of the reaction from aldolase to enolase was between −0.08 and 0.35 kJ/mol ( S2), the forward flux depends on pyruvate kinase, which catalyzed a reaction with a ΔG of −26 kJ/mol (Table S2). Kinetically, its actual activity was much high than the glycolysis rate (Table 3). For LDH-catalyzed reaction, the ΔG favors the forward reaction (Table S2), and the actual LDH activities were much higher than the glycolysis rate (Table S3).
siRNA knockdown off-target concern and data consistency
To exclude the possible off-target effect of siRNA knockdown on the results, we thoroughly checked the data as follows: (a) siRNA specifically knocked down the targeted enzyme, with no significant or marginal effect on other glycolytic enzymes (Table 2); (b) effects of the siRNA knockdown on cells were consistent between different cell lines, as assessed by the lactate production, glucose consumption, glycolytic intermediates, mass action ratios, and ΔG (Tables 1 and 3 and Figs. 6, 7A, and 8A); and (c) effects (lactate production, glucose consumption, FCC, and glycolytic intermediate pattern) of the targeted enzyme modulated by siRNA in living cells and cell-free models are consistent with each other (Figs. 5–8, and 10). Moreover, the data are quantitatively interrelated and consistent with the biochemical principle. In addition, we used siRNA with different sequence to repeat the experiments, and we obtained the consistent results (Figs. S1 and S2 and Table S4).
Glycolysis under different conditions
For these experiments, we had to confirm the quality control, i.e. the siRNA only knocks down PGK1 without significantly affecting other glycolytic enzymes. The quality controls are summarized in Fig. S3A. siRNA specifically knocks down PGK1 by ∼70% in HeLa cells and ∼45% in RKO after 48 h of transfection. When the transfected cells were trypsinized and seeded to plates, knockdown efficiency was kept almost the same (Fig. S3B).
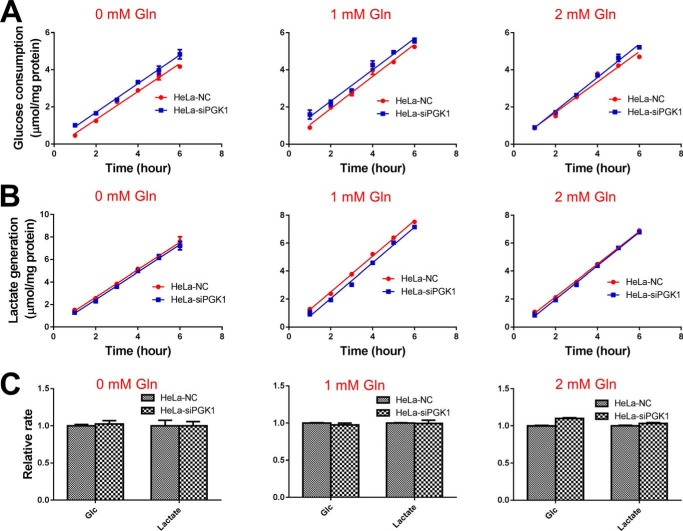
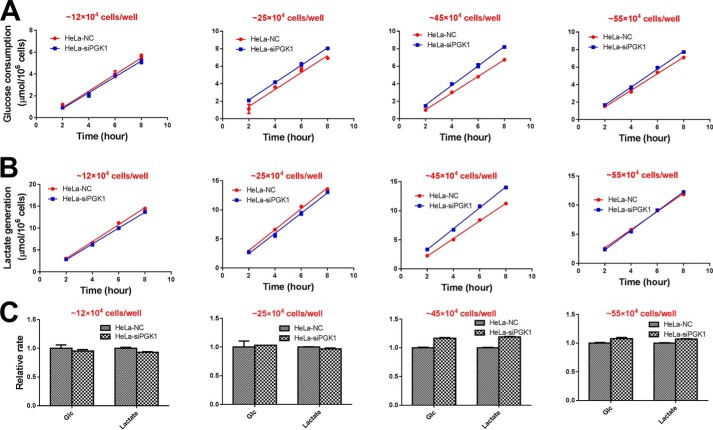
We measured glycolysis rate (glucose consumption and lactate generation) under several conditions, including different glucose concentrations, different glutamine concentrations, and different cell density. In all the above condition, we did not observe a significant difference of glucose consumption and lactate generation rate between PGK1 knockdown and control cells (Figs. 11–13 and Figs. S4–S6).
Figure 11.
Glycolysis rate of HeLa–NC and HeLa–siPGK1 cells under different glucose concentrations. A, glucose and lactate concentrations in culture medium. B, glucose consumption rate under different glucose concentrations. C, lactate generation rate under different glucose concentrations. D, relative rate of glucose consumption and lactate generation. Data are means ± S.D., n = 3. Results were repeated by two independent experiments.
Figure 12.
Glycolysis rate of HeLa–NC and HeLa–siPGK1 cells under different glutamine concentrations. A, glucose consumption rate under different glutamine concentrations. B, lactate generation rate under different glutamine concentrations. C, relative rate of glucose consumption and lactate generation. Data are means ± S.D., n = 3. Results were repeated by two independent experiments.
Figure 13.
Glycolysis rate of HeLa–NC and HeLa–siPGK1 cells under different cell density. A, glucose consumption rate under different cell density. Cells were seeded in 12-well plates. B, lactate generation rate under different cell densities. C, relative rate of glucose consumption and lactate generation. Data are means ± S.D., n = 3. Results were repeated by two independent experiments.
PGK1 knockdown and cell growth
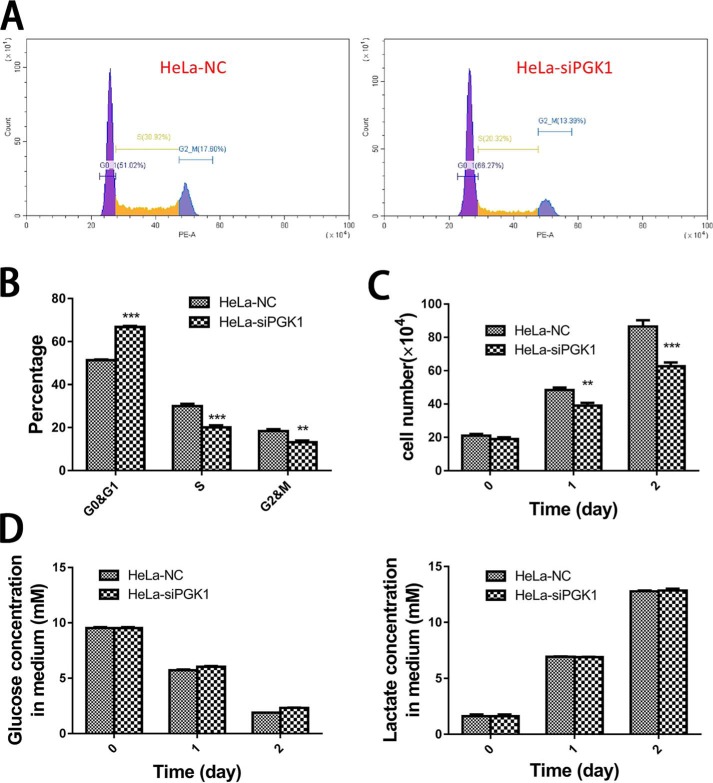
PGK1 knockdown increased HeLa cell percentage at G0 and G1 phases from ∼53 to ∼65% and correspondingly reduced the cell percentage at S and G2 and M phases (Fig. 14, A and B). Consistently, PGK1 knockdown moderately reduced cell growth rate (Fig. 14C). In contrast, glucose consumption and lactate generation were not significantly different between the two groups of cells. However, by taking the cell number into consideration, the rate of glycolysis in cells with PGK1 knockdown would be higher than that in control cells (Fig. 14D).
Figure 14.
Effect of PGK1 knockdown on cell growth. A, representative flow cytometry figures of cell cycle analysis of HeLa–NC and HeLa–siPGK1 cells. B, cell cycle distribution of HeLa–NC and HeLa–siPGK1 cells. C, cell number of HeLa–NC and HeLa–siPGK1 cells at 0, 1, and 2 days after the transfected cells were seeded to new plates. D, glucose and lactate concentrations in culture medium of HeLa–NC or HeLa–siPGK1 cells. Data are means ± S.D., n = 3. Results were repeated by two independent experiments.
RKO cells were different from HeLa cells (Fig. S7). PGK1 knockdown did not inhibit RKO cell growth and lactate generation, but induced about 20% reduction of glucose consumption.
We do not know why PGK1 knockdown exerted different effects on different cells. Presumably, the different effect is related to the different genetic background and tissue origin.
Discussion
In summary, the study investigated three issues. First, we developed a coupled-enzyme assay for measuring the activity of PGK1 with respect to its forward reaction and determined the forward-reaction kinetic parameters. Second, the kinetic parameters of PGK1 are potentially important for glucose carbon to shuttle to serine synthesis. Third, integration of the thermodynamic states, the concentrations of the glycolytic intermediates, and the enzyme kinetics in the system of glycolysis could well-interpret the effect of the perturbation of PGK1 on the glycolysis in cancer cells.
The flux control of aerobic glycolysis is fundamentally a question of kinetics and thermodynamics of the glycolysis as a system. Regarding kinetics, we observed that the [1,3-BPG] is inversely proportional to [PGK1] when glycolysis is at steady state. According to the principle of the enzyme–substrate kinetics, the reasonable interpretation is that PGK1 activity in the glycolysis is flexible, and this flexibility renders PGK1 insensitive to perturbation. Regarding thermodynamics, we observed that the ΔG values of the reactions along the glycolysis remained unchanged in response to perturbation of PGK1, reflecting the rigid thermodynamics of the glycolysis. ΔG values control the Q values of the reactions along the glycolysis and hence determine the concentration of the glycolytic intermediates, including 1,3-BPG. Therefore, the activity of PGK1 in the glycolysis is also controlled by the overall thermodynamic state of the glycolysis. Collectively, the effect of perturbation of PGK1 on glycolysis is related to not only PGK1 itself but also the thermodynamic state of the glycolysis.
Inhibiting aerobic glycolysis is a strategy for treating cancer (5). The way to inhibit aerobic glycolysis is to define the rate-limiting enzymes and target them. The way to define the rate-limiting enzyme is to examine whether inhibition of an enzyme is correlated with an inhibition of overall glycolytic rate or not. This approach is depending on “proof of concept,” but its drawback is also obvious, because it does not answer why the enzyme is rate-limiting. This is probably a major reason why there are many mixed reports of the enzymes, including PFK1 (10, 50, 51), GAPDH (8–12, 27), PGK1 (13–15, 27), PGAM (27, 52), PKM2 (16–19), LDH (27, 53, 54), etc. Revealing the kinetic and thermodynamic aspects concerning the effect of perturbing PGK1 may help to resolve the issue.
PGK1 knockdown may increase the rate to the subsidiary branches of glycolysis, e.g. to methylglyoxal (MG). MG is a metabolite generated from nonenzymatic degradation of DHAP and GA3P (55). MG reacts with arginine, lysine, and cysteine residues in proteins, forming advanced glycation end products (56). PGK1 knockdown induced an increase of concentrations of DHAP and GA3P; hence, it could enhance MG generation. Bollong et al. (57) reported that PGK1 inhibitor CBR-407–1 induced an increase of concentrations of DHAP and GA3P and enhanced generation of MG, which covalently modified KEAP1, leading to a reduced ubiquitination and the accumulation of NRF2. Similarly, as GAPDH knockdown could significantly increase the steady-state concentrations of DHAP and GA3P, GAPDH inhibition may also enhance MG generation and its downstream signaling.
It is important to dissect the effects of PGK1 knockdown on glycolysis and on cell growth. PGK1 knockdown induced a moderate growth inhibition of HeLa cells, but it was irrelevant with glycolysis. Presumably, the growth inhibition induced by PGK1 knockdown might be associated with its nonglycolytic activities. Li et al. (58) reported that EGFR- and ERK-activated casein kinase 2α (CK2α) phosphorylated nuclear PGK1. Phosphorylated PGK1 bound with CDC7, recruited DNA helicase to replication origins, and promoted DNA replication and cell growth. Similarly, other glycolytic enzymes have nonglycolytic functions, which have multiple biological roles, as reviewed by Lu and Wang (59).
There are two isotypes of PGK, PGK1 and PGK2. PGK1 is ubiquitously expressed in all cells, whereas PGK2 is expressed in spermatogenic cells (60). However, a few cancers displayed moderate to strong nuclear and cytoplasmic PGK2 staining, including renal cancer, breast cancer, pancreatic cancer, ovarian cancer, and testis cancer (RRID:SCR_006710, gene name: PGK2) (http://proteinatlas.org/ENSG00000170950-PGK2/pathology)3, indicating that cancer cells could express both PGK1 and PGK2. If so, when PGK1 is perturbed, PGK2 might compensate for the perturbed levels of PGK1.
Materials and methods
Cell lines
Human gastric cancer cell line MGC80-3, cervical cancer cell line HeLa, liver cancer cell line SK-HEP-1, colon cancer cell line RKO, and lung cancer cell line A549 were obtained from Cell Bank of Type Culture Collection of the Chinese Academy of Science (Shanghai, China) and were cultured in RMPI 1640 medium with 10% FBS. Cells were maintained in a humidified incubator at 37 °C with 5% CO2.
Reagents and enzymes
Reagents were from Sigma, including the following: ATP (catalog no. A3377); ADP (catalog no. A5285); NAD (catalog no. N0632); NADH (catalog no. N8129); NADP (catalog no. N8035); NADPH (catalog no. N7505); glucose (catalog no. G8270); G6P (catalog no. G7879); F6P (catalog no. V900924); GA3P (catalog no. G5251); 3-PG (catalog no. P8877); 2-PG (catalog no. 19710); PEP (catalog no. P7001); Pyr (catalog no. V900232); lactic acid (catalog no. L1750); HK (catalog no. H4502); PGI (catalog no. P5381); PFK (catalog no. F0137); aldolase (catalog no. A8811); TPI (catalog no. T6258); GAPDH (catalog no. G2267); PGK (catalog no. P7634); enolase (catalog no. E6126); PK (catalog no. P7768); LDH (catalog no. L2500); G6PDH (catalog no. G8404); and α-GPDH (catalog no. G6751). FBP were purchased from Aladdin (China, catalog no. F111301).
Knockdown of PGK by siRNA
1.5 × 105 cells were seeded into each well of 6-well plates and cultured overnight. Cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's protocol, with either negative control siRNA (NC) or siPGK1 (Ribobio, China). The siRNA sequences were as follows: siPGK1, sense, GCAUCAAAUUCUGCUUGGA dTdT, and antisense, UCCAAGCAGAAUUUGAUGC dTdT; siPGK1-1, sense, GAGTCAATCTGCCACAGAA dTdT, and antisense, UUCUGUGGCAGAUUGACUC dTdT; NC, sense, UUCUCCGAACGUGUCACGU dTdT, and antisense, ACGUGACACGUUCGGAGAA dTdT. 48 h after transfection, cells were washed with PBS, and 2 ml of fresh complete RMPI 1640 medium plus 8 mm glucose were added to each well. Then we collected 10 μl of media at 1–6 h and then determined glucose and lactate. The cells were counted and collected for enzyme activity assay, Western blotting, or intracellular intermediate determination.
Western blotting
Cells were washed with cold PBS and then lysed with M-PERTM mammalian protein extraction reagent (Thermo Fisher Scientific) supplemented with mixture (MedChemExpress) on ice for 30 min. Protein concentration was determined using BCA protein assay kit (Thermo Fisher Scientific). The protein was boiled for 5 min with loading buffer, and 20 μg was subjected to 12% SDS-PAGE, transferred to polyvinylidene difluoride membrane, and incubated with primary body PGK1 (Proteintech, catalog no. 17811-1-AP). GAPDH (Proteintech, catalog no. 60004-1-lg) was used as internal control.
Glucose and lactate determination
We determined glucose and lactate according to the methods described previously with some modification (61). Briefly, 10 μl of collected media were diluted five times by adding 40 μl of water, and then we added 10 μl of mixture or standard solution of glucose/lactate to a 96-well plate, together with 190 μl of reaction buffer. Cell-free system samples were added with 190 μl of reaction buffer directly without dilution. The reaction buffer contained 200 mm HEPES (pH 7.4), 100 mm KCl, 5 mm Na2HPO4, 5 mm MgCl2, 0.5 mm EDTA, 2 mm ATP, 0.2 mm NADP, 0.2 units/ml HK2, and 0.2 units/ml G6PDH for glucose determination or containing 200 mm glycine, 170 mm hydrazine (pH 9.2), 2 mm NAD, and 5 units/ml LDH for lactate determination. 340 nm absorbance was recorded using SpectraMax i3 (Molecular Devices) after a 60-min reaction, and glucose or lactate concentration was calculated according to standard curve.
Glycolytic enzyme activity assay
For determination of PGK1 activity in forward reaction, 1 mm GA3P and 1 mm NAD were added to a cuvette, mixed well with 1 ml of reaction buffer (200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA and 5 mm MgCl2 (pH 7.4)), and the absorbance was recorded at 340 nm. Then, 1 unit/ml GAPDH was added, and waiting until absorbance at 340 nm reached a plateau, we added 2 mm ADP and 1 μg of protein lysate to start the reaction. The initial slope value was used to calculate the enzyme activity.
We determined other enzyme activity at saturating substrate concentrations according to previously reported methods (61) with some modifications. Briefly, 1 ml of reaction buffer was added to the cuvette, and the substrates were added as below. The reaction was started by adding cell lysate and mixed, and then the absorbance at 340 nm was recorded using a spectrophotometer (DU® 700, Beckman Coulter). The following substrates were added: HK, 0.2 mm NADP, 2 mm ATP, 10 mm glucose, 1 unit/ml G6PDH, 30 μg of protein lysate; PGI, 2 mm F6P, 0.2 mm NADP, 1 unit/ml G6PDH, 5 μg of protein lysate; PFK, 0.1 mm ADP, 2 mm ATP, 2 mm F6P, 1 unit/ml aldolase, 0.1 mm NADH, 1 unit/ml α-GPDH, 10 μg of protein lysate; aldolase, 0.1 mm NADH, 1.5 mm FBP, 1 unit/ml α-GPDH, 15 μg of protein lysate; TPI, 2 mm GA3P, 0.1 mm NADH, 1 unit/ml α-GPDH, 1 μg of protein lysate; GAPDH, 2 mm NAD, 2 mm GA3P, 4 μg of protein lysate. The following were also added: PGK in reverse reaction, 2 mm ATP, 2 mm 3PG, 0.1 mm NADH, 1 unit/ml GAPDH, 5 μg of protein lysate; PGAM, 2 mm ADP, 1 mm 3PG, 1 unit/ml enolase, 0.1 mm NADH, 1 unit/ml PK, 1 unit/ml LDH, 10 μg of protein lysate; enolase: 2 mm ADP, 1 mm 2-PG, 1 unit/ml PK, 0.1 mm NADH, 1 unit/ml LDH, 10 μg of protein lysate; PK, 2 mm ADP, 2 mm PEP, 0.1 mm NADH, 5 units/ml LDH, 2 μg of protein lysate; and LDH, 0.1 mm NADH, 2 mm pyruvate, 2 μg of protein lysate.
In vitro cell-free system model for glycolysis
Previously we had described an in vitro cell-free system as a glycolysis model (48, 49). We used the reaction buffer containing 200 mm HEPES, 0.5 mm EDTA, 100 mm KCl, 5 mm MgCl2, 5 mm Na2HPO4, 4 mm ADP, 1.5 mm ATP, 5 mm glucose, 0.1 mm NADH, and 2 mm NAD for this glycolysis system. 70 μl of lysate (8–10 μg/μl protein) was added to 630 μl of reaction buffer, and the mixture was incubated at 37 °C for 30 min. Then we added 600 μl of 1 m HClO4 to the mixture to terminate the reaction and later 100 μl of 3 m K2CO3 was added to neutralize the buffer. The mixture was kept on ice for 20 min further. Supernatant was obtained by 10,000 × g, centrifuged at 4 °C, and used for glycolytic intermediate determination.
Determination of glycolytic intermediates in cell-free system
We used previously reported methods to determine the glycolytic intermediates (61). The reaction buffer in this part contained 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, and 5 mm MgCl2, with pH adjusted to 7.4.
For G6P and F6P, 100 μl of supernatant and 0.2 mm NADP were added to 900 μl of reaction buffer, and the reaction was started by adding 1 unit/ml G6PDH. The first reaction to measure G6P ended when 340 nm absorbance reached a plateau, and then 1 unit/ml PGI was added to measure F6P.
For FBP, DHAP, and GA3P, 100 μl of supernatant and 0.1 mm NADH were added to 900 μl of reaction buffer, and the reaction to measure DHAP was started by adding 1 unit/ml α-GPDH. When the first reaction ended, 1 unit/ml TPI was added to measure GA3P. Finally, 1 unit/ml aldolase was added to measure FBP.
For 3PG, 50 μl of supernatant, 2 mm ATP, 0.1 mm NADH, 1 unit/ml PGK were added to 950 μl of reaction buffer, and the reaction was started by adding 1 unit/ml GAPDH.
For 2-PG, PEP, and Pyr, 100 μl of supernatant, 0.1 mm NADH were added to 900 μl of reaction buffer, and the reaction to measure Pyr was started by adding 1 unit/ml LDH. When the first reaction ended, 2 mm ADP and 1 unit/ml PK were added to measure PEP. Finally, 1 unit/ml enolase was added to measure 2-PG.
Determination of intracellular glycolytic intermediates
Negative control or siPGK1-transfected cells were washed by ice-cold PBS twice, and 600 μl of 1 m pre-cold HClO4 was added to every three wells of a 6-well plate. Cells were collected by a scraper, incubated on ice for 30 min, and neutralized by 100 μl of 3 m K2CO3. Then the supernatant was obtained by 10,000 × g centrifugation at 4 °C. Then, 50 μl of 2 m NaOH was added to the supernatant and kept at 60 °C for 5 min and neutralized again by adding 50 μl of 2 m HCl. These two steps possibly eliminated intracellular NAD(H)/NADP(H). In the meantime, a same 4th well of cells was trypsinized and collected to determine the cell number and cell size by a cell counter (JIMBIO). Through the following reactions, intermediates plus NADP (for G6P, F6P, and glucose) or NADH (for FBP-Pyr) were converted to products and NADPH or NAD; terminating the reaction by NaOH or HCl will conserve the NADPH or NAD, which could be measured by cycling methods (61, 63). For all the reactions, 10 μl supernatant was mixed with 40 μl of reaction buffer. Different reactions differ in reaction buffers as follows: G6P: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 2 mm ATP, 0.5 mm NADP, 0.5 units/ml G6PDH; F6P: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 2 mm ATP, 0.5 mm NADP, 0.5 units/ml G6PDH, plus 0.5 units/ml PGI; Glc: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 2 mm ATP, 0.5 mm NADP, 0.5 units/ml G6PDH, plus 0.5 units/ml HK; DHAP: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 0.5 units/ml α-GPDH; GA3P: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 0.5 units/ml α-GPDH, plus 0.5 units/ml TPI; FBP: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 0.5 units/ml α-GPDH, 0.5 units/ml TPI, plus 0.5 units/ml aldolase; 3PG: 200 mm HEPES, 100 mm KCl, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 1 mm ATP, 0.5 units/ml PGK, plus 0.5 units/ml GAPDH; Pyr: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 1 mm ADP, 0.5 units/ml LDH; PEP: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 1 mm ADP, 0.5 units/ml LDH, plus 0.5 units/ml PK; 2-PG: 200 mm HEPES, 100 mm KCl, 5 mm Na2HPO4, 0.5 mm EDTA, 5 mm MgCl2 (pH 7.4), 0.5 mm NADH, 1 mm ADP, 0.5 units/ml LDH, 0.5 units/ml PK, plus 0.5 units/ml enolase.
For G6P, F6P, and Glc determinations, after 30 min of incubation at 37 °C, the reaction was terminated by adding 10 μl of 2 m NaOH, mixed well, and kept at 60 °C for 5 min to eliminate NADP, then the mixture was neutralized by adding 20 μl of 1 m HCl. For the rest of the intermediates, after 30 min of incubation at 37 °C, the reaction was terminated by adding 20 μl of 1 m HCl, kept at 60 °C for 5 min to eliminate NADH, and then the mixture was neutralized by adding 10 μl of 2 m NaOH. After that, 70 μl of neutralized mixture or standard solution was added to a 96-well plate together with 100 μl of develop buffer (0.4 m Tris-HCl, 0.2 mm G6P, 1 unit/ml G6PDH, 0.1 mm MTS, 0.1 mm PES (pH 7.8)) to cycling NADPH (for G6P, F6P, and glucose). For the rest of the intermediates, the develop buffer contained 0.4 m Tris-HCl (pH 7.8), 5 m ethanol, 2 units/ml alcohol dehydrogenase, 0.1 mm MTS, 0.1 mm PES. After 30 min of incubation with develop buffer at 37 °C, the 490 nm absorbances of 96-well plates were recorded and analyzed.
Determination of isotopic serine and glycine by LC-MS/MS
Forty eight hours after NC or siHK2 transfection, cells were washed with PBS twice and cultured with glucose-free RMPI 1640 medium supplemented with 10% FBS and 7 mm [13C6]glucose for 3, 6, and 9 h. Medium at 0, 3, 6, and 9 h was collected for LC-MS/MS measurements. Meanwhile, cells at 0, 3, 6, 9 h were washed with PBS three times, and intracellular amino acids were extracted by adding 80% pre-cold methanol. Cells were collected by a scraper, and 20,000 × g centrifugation at 4 °C was performed, and the supernatant was evaporated by a vacuum centrifugal concentrator and dissolved in 100 μl of water. 10 μl of dissolved metabolites or collected medium or standard was mixed with 20 μl of AQC (catalog no. ab145409, AbCAM, amino acid derivatization agent) and 120 μl of borate buffer, incubated at 60 °C for 20 min, and the AQC-derivatized sample was used for LC-MS/MS by a Waters Acquity UPLC system coupled to a QTrap 4000 mass spectrometer with ESI probe (Applied Biosystems Inc., Foster City, CA). An AccQ-Tag Ultra RP column was used to perform the liquid chromatography. Mobile phase A was 25 mm ammonia formate with 1% acetonitrile (pH 3.05), and mobile phase B was 100% acetonitrile. The gradient program was as follows: 0–0.54 min, 99.9% A, 0.1% B; 0.54–5.74 min, 99.9% A, 90.9% A; 5.74–7.74 min, 90.9% A, 78.8% A; 7.74–8.04 min, 78.8% A, 40.4% A; 8.04–8.64 min, 40.4% A; 8.64–8.73 min, 40.4% A, 99.9% A; 8.73–9.5 min, 99.9% A. 1-μl sample or standard solution was injected to perform the analysis with a flow rate at 0.7 ml/min. During the performance, the column was kept at 55 °C. The following parameters were optimized and used for MASS analysis: 40 p.s.i. curtain gas, medium collision gas, 5500 V, ion spray voltage, temperature of the ion source 500 °C, 40 p.s.i. ion source gas1, and 40 p.s.i. ion source gas2.
Cell cycle assay
Cell cycle assay was performed using a cell cycle staining kit (catalog no. 70-CCS012, MultiSciences, China) according to the manufacturer's protocol. Briefly, HeLa or RKO cells were transfected with NC or siPGK1 for 48 h, trypsinized, and seeded to a new 6-well plate overnight, then collected and subjected to flow cytometer analysis.
Determination of ATP, ADP, NAD, and NADH in cells
Cells in 6-well plates were washed with ice-cold PBS twice, and 0.6 ml of 80% (v/v) pre-cold (−20 °C) methanol was added per well to extract the intracellular metabolites. Then a scraper was used to collect the cells, and the cell debris was discarded by 20,000 × g centrifugation at 4 °C. The supernatant was evaporated by a vacuum centrifugal concentrator and was dissolved in 100 μl of water following UPLC analysis. Waters ACQUITY UPLC system with an ACQUITY UPLC HSS T3 column was used to perform the liquid chromatography. The mobile phase A was 20 mm Triethylamine in 99/1% water/acetonitrile (pH 6.5) and mobile phase B was 100% acetonitrile. The gradient program was as follows: 0–3 min, 100% A; 3–4 min, 100% A–98.5% A; 4–7 min, 98.5% A–92% A; 7–7.1 min, 92% A–100% A; 7.1–10 min, 100% A. A 10- sample or standard solution was injected to perform the analysis with a flow rate at 0.3 ml/min. During the performance, the column was kept at 40 °C.
Estimation of 1,3-BPG
Assume at steady state that the reaction calculated by GAPDH was at equilibrium. So we can estimate the concentration by equation 1,
| (Eq. 1) |
where Keq is the equilibrium constant of GAPDH; [NAD]/[NADH] was set 78 (48), and [Pi] in the cell was set 1.5 mm (64).
Analysis of isotopic lactate by LC-MS/MS
Isotopic lactate tracing is based on our previously reported method (54). [13C6]Glucose was purchased from Sigma. 48 h after transfection, HeLa–NC and HeLa–siPGK1 were washed with PBS twice and cultured in glucose-free RPMI 1640 medium supplemented with 10% ultrafiltrated FBS and 8 mm [13C6]glucose for 6 h. Then the culture medium was collected and diluted 40 times with 100% acetonitrile and centrifuged at 25,000 × g for 10 min at 4 °C. Supernatant was collected for LS-MS/MS analysis according to methods reported previously by us (54, 65). Briefly, an ACQUITY BEH amide column was used to perform LC and kept at 50 °C during analysis, and the injection volume was 7.5 μl. Mobile phase A was 10 mm ammonium acetate in 85% acetonitrile, 15% water, pH 9.0; and mobile phase B was 10 mm ammonium acetate in 50% acetonitrile, 50% water, pH 9.0. The gradient program was as follows: 0–0.4 min, 100% A; 0.4–2 min, 100–30% A; 2–2.5 min, 30–15% A; 2.5–3 min, 15% A; 3–3.1 min, 15–100% A; 3.1–7.5 min, 100% A. A 4000 QTRAP mass spectrometer (AB SCIEX) equipped with an ESI ion source (Turbospray) operating in negative ion mode was used for MS detection, and the same parameter settings were employed (54).
Calculation of the Gibbs free energy change ΔG of glycolytic reactions
ΔG was calculated according to Equation 2,
| (Eq. 2) |
where ΔG310′0 is the standard transformed Gibbs free energy at 37 °C, and Q was calculated according to intermediate concentrations as listed in the tables. For intracellular metabolite calculations, NAD/NADH was set as 78 according to our previously reported study (48). Take HK of HeLa–NC for example. Q of the HK-catalyzed reaction in cell equals [G6P][ADP]/[ATP][Glc], which was 0.019; however, ΔG310′0 is not available. According to ΔG = ΔH − TΔS, and because the change of ΔH and ΔS is negligible between 37 and 25 °C (66, 67), we deduced the equation to a new form as shown in Equation 3,
| (Eq. 3) |
ΔG298′0 and ΔH298′0 are available in Refs. 68–72. So we get Equation 4,
| (Eq. 4) |
Calculation of FCC
Nonlinear regression of the glucose consumption or lactate generation data was performed using GraphPad7. Then a tangent was obtained at the point of enzymes at 1.0. FCC was calculated by Equation 5 (62),
| (Eq. 5) |
where the slope stands for the tangent (ΔJ/ΔE) obtained at point of enzymes at 1.0, E stands for the enzyme concentration, which is 1.0, and J stands for the lactate generated or glucose consumed at enzyme 1.0.
Statistical analysis
All experiments were repeated at least two times, and all data were analyzed using GraphPad Prism7.
Data availability
All data are contained within the manuscript and supporting information.
Author contributions
C. J., X. Z., H. W., Y. W., and X. H. data curation; C. J., X. Z., H. W., Y. W., and X. H. formal analysis; C. J., X. Z., H. W., Y. W., and X. H. validation; C. J., X. Z., H. W., Y. W., and X. H. investigation; C. J., H. W., Y. W., and X. H. methodology; H. W. and X. H. project administration; X. H. conceptualization; X. H. resources; X. H. supervision; X. H. funding acquisition; X. H. writing-original draft.
Supplementary Material
Acknowledgments
We thank Professor Daming Gao (Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences) for the kind gift of recombinant human PGK1.
This work was supported in part by National Basic Research Program of China (973 Program) Grant 2013CB911303, National Natural Science Foundation of China Project Grant 81470126 and Key Project Grant 2018C03009 funded by Zhejiang Provincial Department of Sciences and Technologies, and Fundamental Research Funds for the Central Universities Grants 2017XZZX001-01 and 2019FZJD009, National Ministry of Education, China (to X. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- WE
- Warburg effect
- G6P
- glucose 6-phosphate
- F6P
- fructose 6-phosphate
- FBP
- fructose 1,6-bisphosphate
- GA3P
- glyceraldehyde 3-phosphate
- 3-PG
- 3-phosphoglycerate
- 2-PG
- 2-phosphoglycerate
- PEP
- phosphoenolpyruvate
- Pyr
- pyruvate
- HK
- hexokinase
- PGI
- phosphohexose isomerase
- PFK
- phosphofructokinase
- TPI
- triose-phosphate isomerase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PGK
- phosphoglycerate kinase
- PK
- pyruvate kinase
- LDH
- lactate dehydrogenase
- FCC
- flux control coefficient
- NC
- negative control
- DHAP
- dihydroxyacetone phosphate
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- 1,3-BPG
- 1,3-bisphosphoglyceric acid
- PHGDH
- phosphoglycerate dehydrogenase
- MG
- methylglyoxal
- FBS
- fetal bovine serum
- G6PDH
- glucose-6-phosphate dehydrogenase
- PES
- phenazine ethosulfate
- AQC
- 6-aminoquinoline N-succinimidyl ester.
References
- 1. Altman B. J., Stine Z. E., and Dang C. V. (2016) From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 10.1038/nrc.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cascone T., McKenzie J. A., Mbofung R. M., Punt S., Wang Z., Xu C., Williams L. J., Wang Z., Bristow C. A., Carugo A., Peoples M. D., Li L., Karpinets T., Huang L., Malu S., et al. (2018) Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987.e4 10.1016/j.cmet.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakazawa M. S., Keith B., and Simon M. C. (2016) Oxygen availability and metabolic adaptations. Nat. Rev. Cancer 16, 663–673 10.1038/nrc.2016.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang M., and Vousden K. H. (2016) Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- 5. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 6. Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R., Hawkins C., and Guha A. (2011) Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 208, 313–326 10.1084/jem.20101470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeWaal D., Nogueira V., Terry A. R., Patra K. C., Jeon S. M., Guzman G., Au J., Long C. P., Antoniewicz M. R., and Hay N. (2018) Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 9, 446 10.1038/s41467-017-02733-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liberti M. V., Dai Z., Wardell S. E., Baccile J. A., Liu X., Gao X., Baldi R., Mehrmohamadi M., Johnson M. O., Madhukar N. S., Shestov A. A., Chio I. I. C., Elemento O., Rathmell J. C., Schroeder F. C., et al. (2017) A predictive model for selective targeting of the Warburg effect through GAPDH inhibition with a natural product. Cell Metab. 26, 648–659.e8 10.1016/j.cmet.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhong X. Y., Yuan X. M., Xu Y. Y., Yin M., Yan W. W., Zou S. W., Wei L. M., Lu H. J., Wang Y. P., and Lei Q. Y. (2018) CARM1 methylates GAPDH to regulate glucose metabolism and is suppressed in liver cancer. Cell Rep. 24, 3207–3223 10.1016/j.celrep.2018.08.066 [DOI] [PubMed] [Google Scholar]
- 10. Shestov A. A., Liu X., Ser Z., Cluntun A. A., Hung Y. P., Huang L., Kim D., Le A., Yellen G., Albeck J. G., and Locasale J. W. (2014) Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife 3, 10.7554/eLife.03342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Locasale J. W. (2018) New concepts in feedback regulation of glucose metabolism. Curr. Opin. Syst. Biol. 8, 32–38 10.1016/j.coisb.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yun J., Mullarky E., Lu C., Bosch K. N., Kavalier A., Rivera K., Roper J., Chio I. I., Giannopoulou E. G., Rago C., Muley A., Asara J. M., Paik J., Elemento O., Chen Z., Pappin D. J., et al. (2015) Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396 10.1126/science.aaa5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu H., Zhu W., Qin J., Chen M., Gong L., Li L., Liu X., Tao Y., Yin H., Zhou H., Zhou L., Ye D., Ye Q., and Gao D. (2017) Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 65, 515–528 10.1002/hep.28887 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Yu G., Chu H., Wang X., Xiong L., Cai G., Liu R., Gao H., Tao B., Li W., Li G., Liang J., and Yang W. (2018) Macrophage-associated PGK1 phosphorylation promotes aerobic glycolysis and tumorigenesis. Mol. Cell 71, 201–215.e7 10.1016/j.molcel.2018.06.023 [DOI] [PubMed] [Google Scholar]
- 15. Li X., Jiang Y., Meisenhelder J., Yang W., Hawke D. H., Zheng Y., Xia Y., Aldape K., He J., Hunter T., Wang L., and Lu Z. (2016) Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell 61, 705–719 10.1016/j.molcel.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., and Cantley L. C. (2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 10.1038/nature06667 [DOI] [PubMed] [Google Scholar]
- 17. Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., Thomas C. J., Vander Heiden M. G., and Cantley L. C. (2011) Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 10.1126/science.1211485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaneton B., Hillmann P., Zheng L., Martin A. C. L., Maddocks O. D. K., Chokkathukalam A., Coyle J. E., Jankevics A., Holding F. P., Vousden K. H., Frezza C., O'Reilly M., and Gottlieb E. (2012) Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462 10.1038/nature11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anastasiou D., Yu Y., Israelsen W. J., Jiang J. K., Boxer M. B., Hong B. S., Tempel W., Dimov S., Shen M., Jha A., Yang H., Mattaini K. R., Metallo C. M., Fiske B. P., Courtney K. D., et al. (2012) Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 8, 839–847 10.1038/nchembio.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hwang T. L., Liang Y., Chien K. Y., and Yu J. S. (2006) Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics 6, 2259–2272 10.1002/pmic.200500345 [DOI] [PubMed] [Google Scholar]
- 21. Yan H., Yang K., Xiao H., Zou Y. J., Zhang W. B., and Liu H. Y. (2012) Over-expression of cofilin-1 and phosphoglycerate kinase 1 in astrocytomas involved in pathogenesis of radioresistance. CNS Neurosci. Ther. 18, 729–736 10.1111/j.1755-5949.2012.00353.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ahmad S. S., Glatzle J., Bajaeifer K., Bühler S., Lehmann T., Königsrainer I., Vollmer J. P., Sipos B., Ahmad S. S., Northoff H., Königsrainer A., and Zieker D. (2013) Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int. J. Oncol. 43, 586–590 10.3892/ijo.2013.1971 [DOI] [PubMed] [Google Scholar]
- 23. Zieker D., Königsrainer I., Tritschler I., Löffler M., Beckert S., Traub F., Nieselt K., Bühler S., Weller M., Gaedcke J., Taichman R. S., Northoff H., Brücher B. L., and Königsrainer A. (2010) Phosphoglycerate kinase 1 a promoting enzyme for peritoneal dissemination in gastric cancer. Int. J. Cancer 126, 1513–1520 10.1002/ijc.24835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ai J., Huang H., Lv X., Tang Z., Chen M., Chen T., Duan W., Sun H., Li Q., Tan R., Liu Y., Duan J., Yang Y., Wei Y., Li Y., and Zhou Q. (2011) FLNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell. Physiol. Biochem. 27, 207–216 10.1159/000327946 [DOI] [PubMed] [Google Scholar]
- 25. Kress S., Stein A., Maurer P., Weber B., Reichert J., Buchmann A., Huppert P., and Schwarz M. (1998) Expression of hypoxia-inducible genes in tumor cells. J. Cancer Res. Clin. Oncol. 124, 315–320 10.1007/s004320050175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shashni B., Sakharkar K. R., Nagasaki Y., and Sakharkar M. K. (2013) Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARγ in breast cancer pathophysiology. J. Drug Target 21, 161–174 10.3109/1061186X.2012.736998 [DOI] [PubMed] [Google Scholar]
- 27. Tanner L. B., Goglia A. G., Wei M. H., Sehgal T., Parsons L. R., Park J. O., White E., Toettcher J. E., and Rabinowitz J. D. (2018) Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49–62.e8 10.1016/j.cels.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen X., Zhao C., Li X., Wang T., Li Y., Cao C., Ding Y., Dong M., Finci L., Wang J. H., Li X., and Liu L. (2015) Terazosin activates Pgk1 and Hsp90 to promote stress resistance. Nat. Chem. Biol. 11, 19–25 10.1038/nchembio.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan J., Teng X., Liu L., Mattaini K. R., Looper R. E., Vander Heiden M. G., and Rabinowitz J. D. (2015) Human phosphoglycerate dehydrogenase produces the oncometabolite d-2-hydroxyglutarate. ACS Chem. Biol. 10, 510–516 10.1021/cb500683c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahmed D., Eide P. W., Eilertsen I. A., Danielsen S. A., Eknæs M., Hektoen M., Lind G. E., and Lothe R. A. (2013) Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71 10.1038/oncsis.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y., Chen Y., Hu Y., Wang J., Xie X., He G., Chen H., Shao Q., Zeng H., and Zhang H. (2018) Genomic alterations across six hepatocellular carcinoma cell lines by panel-based sequencing. Transl. Cancer Res. 7, 231–239 10.21037/tcr.2018.02.14 [DOI] [Google Scholar]
- 32. Berg K. C. G., Eide P. W., Eilertsen I. A., Johannessen B., Bruun J., Danielsen S. A., Bjørnslett M., Meza-Zepeda L. A., Eknaes M., Lind G. E., Myklebost O., Skotheim R. I., Sveen A., and Lothe R. A. (2017) Multi-omics of 34 colorectal cancer cell lines–a resource for biomedical studies. Mol. Cancer 16, 116 10.1186/s12943-017-0691-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blanco R., Iwakawa R., Tang M., Kohno T., Angulo B., Pio R., Montuenga L. M., Minna J. D., Yokota J., and Sanchez-Cespedes M. (2009) A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 30, 1199–1206 10.1002/humu.21028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yaginuma Y., and Westphal H. (1991) Analysis of the p53 gene in human uterine carcinoma cell lines. Cancer Res. 51, 6506–6509 [PubMed] [Google Scholar]
- 35. Hoppe-Seyler F., and Butz K. (1993) Repression of endogenous p53 transactivation function in HeLa cervical carcinoma cells by human papillomavirus type 16 E6, human mdm-2, and mutant p53. J. Virol. 67, 3111–3117 10.1128/JVI.67.6.3111-3117.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan J., Zeng J., Shuai C., and Liu Y. (2018) TWSG1 is a novel tumor suppressor in gastric cancer. DNA Cell Biol. 37, 574–583 10.1089/dna.2018.4188 [DOI] [PubMed] [Google Scholar]
- 37. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu P. P., and Sabatini D. M. (2008) Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 10.1016/j.cell.2008.08.021 [DOI] [PubMed] [Google Scholar]
- 39. Cairns R. A., Harris I. S., and Mak T. W. (2011) Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- 40. Wieman H. L., Wofford J. A., and Rathmell J. C. (2007) Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell 18, 1437–1446 10.1091/mbc.e06-07-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gottlob K., Majewski N., Kennedy S., Kandel E., Robey R. B., and Hay N. (2001) Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 15, 1406–1418 10.1101/gad.889901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones R. G., and Thompson C. B. (2009) Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23, 537–548 10.1101/gad.1756509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dang C. V. (1999) c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19, 1–11 10.1128/MCB.19.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J. K., Markowitz S., Zhou S., Diaz L. A. Jr., Velculescu V. E., Lengauer C., Kinzler K. W., Vogelstein B., and Papadopoulos N. (2009) Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 325, 1555–1559 10.1126/science.1174229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., and Thompson C. B. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 10.1158/0008-5472.CAN-03-2904 [DOI] [PubMed] [Google Scholar]
- 46. Matoba S., Kang J. G., Patino W. D., Wragg A., Boehm M., Gavrilova O., Hurley P. J., Bunz F., and Hwang P. M. (2006) p53 regulates mitochondrial respiration. Science 312, 1650–1653 10.1126/science.1126863 [DOI] [PubMed] [Google Scholar]
- 47. Vousden K. H., and Ryan K. M. (2009) p53 and metabolism. Nat. Rev. Cancer 9, 691–700 10.1038/nrc2715 [DOI] [PubMed] [Google Scholar]
- 48. Xie J., Dai C., and Hu X. (2016) Evidence that does not support pyruvate kinase M2 (PKM2)-catalyzed reaction as a rate-limiting step in cancer cell glycolysis. J. Biol. Chem. 291, 8987–8999 10.1074/jbc.M115.704825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie J., Wu H., Dai C., Pan Q., Ding Z., Hu D., Ji B., Luo Y., and Hu X. (2014) Beyond Warburg effect–dual metabolic nature of cancer cells. Sci. Rep. 4, 4927 10.1038/srep04927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang Y., Ishak Gabra M. B., Hanse E. A., Lowman X. H., Tran T. Q., Li H., Milman N., Liu J., Reid M. A., Locasale J. W., Gil Z., and Kong M. (2019) MiR-135 suppresses glycolysis and promotes pancreatic cancer cell adaptation to metabolic stress by targeting phosphofructokinase-1. Nat. Commun. 10, 809 10.1038/s41467-019-08759-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A. 3rd., Peters E. C., Driggers E. M., and Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 10.1126/science.1222278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hitosugi T., Zhou L., Elf S., Fan J., Kang H. B., Seo J. H., Shan C., Dai Q., Zhang L., Xie J., Gu T. L., Jin P., Alečković M., LeRoy G., Kang Y., et al. (2012) Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 22, 585–600 10.1016/j.ccr.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fantin V. R., St-Pierre J., and Leder P. (2006) Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 10.1016/j.ccr.2006.04.023 [DOI] [PubMed] [Google Scholar]
- 54. Ying M., Guo C., and Hu X. (2019) The quantitative relationship between isotopic and net contributions of lactate and glucose to the TCA cycle. J. Biol. Chem. 294, 9615–9630 10.1074/jbc.RA119.007841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richard J. P. (1993) Mechanism for the formation of methylglyoxal from triosephosphates. Biochem. Soc. Trans. 21, 549–553 10.1042/bst0210549 [DOI] [PubMed] [Google Scholar]
- 56. Shipanova I. N., Glomb M. A., and Nagaraj R. H. (1997) Protein modification by methylglyoxal: chemical nature and synthetic mechanism of a major fluorescent adduct. Arch. Biochem. Biophys. 344, 29–36 10.1006/abbi.1997.0195 [DOI] [PubMed] [Google Scholar]
- 57. Bollong M. J., Lee G., Coukos J. S., Yun H., Zambaldo C., Chang J. W., Chin E. N., Ahmad I., Chatterjee A. K., Lairson L. L., Schultz P. G., and Moellering R. E. (2018) A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature 562, 600–604 10.1038/s41586-018-0622-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X., Qian X., Jiang H., Xia Y., Zheng Y., Li J., Huang B.-J., Fang J., Qian C.-N., Jiang T., Zeng Y.-X., and Lu Z. (2018) Nuclear PGK1 Alleviates ADP-dependent inhibition of CDC7 to promote DNA replication. Mol. Cell 72, 650–660.e8 10.1016/j.molcel.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 59. Lu S., and Wang Y. (2018) Nonmetabolic functions of metabolic enzymes in cancer development. Cancer Commun. 38, 63 10.1186/s40880-018-0336-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chiarelli L. R., Morera S. M., Bianchi P., Fermo E., Zanella A., Galizzi A., and Valentini G. (2012) Molecular insights on pathogenic effects of mutations causing phosphoglycerate kinase deficiency. PloS One 7, e32065 10.1371/journal.pone.0032065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Passonneau J., and Lowry O. H. (1993) Enzymatic Analysis: A Practical Guide. Humana Press Inc., Totowa, NJ [Google Scholar]
- 62. Kacser H., and Burns J. A. (1995) The control of flux. Biochem. Soc. Trans. 23, 341–366 10.1042/bst0230341 [DOI] [PubMed] [Google Scholar]
- 63. Gibon Y., and Larher F. (1997) Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 251, 153–157 10.1006/abio.1997.2283 [DOI] [PubMed] [Google Scholar]
- 64. Giachelli C. M. (2003) Vascular calcification: in vitro evidence for the role of inorganic phosphate. J. Am. Soc. Nephrol. 14, S300–S304 10.1097/01.ASN.0000081663.52165.66 [DOI] [PubMed] [Google Scholar]
- 65. Zhang W., Guo C., Jiang K., Ying M., and Hu X. (2017) Quantification of lactate from various metabolic pathways and quantification issues of lactate isotopologues and isotopomers. Sci. Rep. 7, 8489 10.1038/s41598-017-08277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Méndez E. (2008) Biochemical thermodynamics under near physiological conditions. Biochem. Mol. Biol. Educ. 36, 116–119 10.1002/bmb.20157 [DOI] [PubMed] [Google Scholar]
- 67. Alberty R. A. (2006) Biochemical thermodynamics: applications of Mathematica. Methods Biochem. Anal. 48, 1–458 [PubMed] [Google Scholar]
- 68. Li X., Wu F., Qi F., and Beard D. A. (2011) A database of thermodynamic properties of the reactions of glycolysis, the tricarboxylic acid cycle, and the pentose phosphate pathway. Database 2011, bar005 10.1093/database/bar005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Keleti T., Földi J., Erdei S., and Tro T. Q. (1972) Some thermodynamic data on d-glyceraldehyde-3-phosphate dehydrogenase action under optimal conditions. Biochim. Biophys. Acta 268, 285–291 10.1016/0005-2744(72)90323-3 [DOI] [PubMed] [Google Scholar]
- 70. Donnovan L., Barclay K., Otto K., and Jespersen N. (1975) Thermochemistry of reaction catalyzed by lactate-dehydrogenase. Thermochim Acta 11, 151–156 10.1016/0040-6031(75)80018-9 [DOI] [Google Scholar]
- 71. Varga A., Szabó J., Flachner B., Gugolya Z., Vonderviszt F., Závodszky P., and Vas M. (2009) Thermodynamic analysis of substrate induced domain closure of 3-phosphoglycerate kinase. FEBS Lett. 583, 3660–3664 10.1016/j.febslet.2009.10.048 [DOI] [PubMed] [Google Scholar]
- 72. Rekharsky M. V., Egorov A. M., Galchenko G. L., and Berezin I. V. (1981) Thermodynamics of redox reactions involving nicotinamide adenine-dinucleotide. Thermochim Acta 46, 89–101 10.1016/0040-6031(81)80234-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript and supporting information.