Abstract
Sperm head shaping is a key event in spermiogenesis and is tightly controlled via the acrosome–manchette network. Linker of nucleoskeleton and cytoskeleton (LINC) complexes consist of Sad1 and UNC84 domain–containing (SUN) and Klarsicht/ANC-1/Syne-1 homology (KASH) domain proteins and form conserved nuclear envelope bridges implicated in transducing mechanical forces from the manchette to sculpt sperm nuclei into a hook-like shape. However, the role of LINC complexes in sperm head shaping is still poorly understood. Here we assessed the role of SUN3, a testis-specific LINC component harboring a conserved SUN domain, in spermiogenesis. We show that CRISPR/Cas9-generated Sun3 knockout male mice are infertile, displaying drastically reduced sperm counts and a globozoospermia-like phenotype, including a missing, mislocalized, or fragmented acrosome, as well as multiple defects in sperm flagella. Further examination revealed that the sperm head abnormalities are apparent at step 9 and that the sperm nuclei fail to elongate because of the absence of manchette microtubules and perinuclear rings. These observations indicate that Sun3 deletion likely impairs the ability of the LINC complex to transduce the cytoskeletal force to the nuclear envelope, required for sperm head elongation. We also found that SUN3 interacts with SUN4 in mouse testes and that the level of SUN4 proteins is drastically reduced in Sun3-null mice. Altogether, our results indicate that SUN3 is essential for sperm head shaping and male fertility, providing molecular clues regarding the underlying pathology of the globozoospermia-like phenotype.
Keywords: spermatogenesis, sperm, gene knockout, reproduction, spermatozoa, infertility, LINC complex, manchette, sperm shaping, SUN3
Introduction
Spermiogenesis involves drastic morphological changes of spermatids, including acrosome formation, elongation and condensation of the nucleus, and disposal of residual cytoplasm, to differentiate into mature spermatozoa consisting of a head and a tail (1, 2). During this complex process, a transient cytoskeletal structure called the manchette appears concurrently with spermatid nucleus elongation. The manchette, which is first evident in step 8 spermatids in mice (3), is composed of parallel arrays of microtubules aligned with the long axis of the nucleus and a belt-like perinuclear ring made of actins where the microtubules are anchored, forming a sleeve-like structure that encircles the spermatid nucleus (4–6). It is believed that manchette microtubules emanate from perinuclear rings, as these microtubules appear from the post-acrosomal region in mouse and bovine spermatids (7). As spermatids differentiate, the microtubular structure and the perinuclear ring move caudally to the posterior pole of spermatid nucleus, sculpting the nucleus into a hook-like morphology.
The important role of the manchette in sperm head shaping has been demonstrated in several studies in which disruption of manchette-related proteins resulted in various forms of sperm head abnormalities and eventually impaired male fertility. For example, loss of functional HOOK1 causes ectopic positioning of the manchette microtubular structure within spermatids, resulting in abnormal sperm head morphologies, such as club-shaped and crescent forms (8). Similarly, disruption of RIM-BP3, a HOOK1-interacting protein located in the manchette, also causes ectopic positioning of manchette and sperm head abnormalities similar to those of Hook1 mutant mice (9). The microtubule plus-end-tracking protein CLIP-170 localizes prominently to the manchette rings during manchette formation, and male mice with knockout of Clip-170 are subfertile because of defective sperm head shaping and abnormal elongation of the manchette tubules (2, 10). Despite the essential role of the manchette in spermiogenesis, how the microtubule manchette is assembled and regulated remains poorly understood.
Sad1/UNC84 (SUN)6 homology proteins are a family of nuclear membrane proteins that share a conserved C terminus, the SUN domain. SUN proteins and Klarsicht/ANC-1/Syne-1 homology (KASH) proteins form a linker of nucleoskeleton and cytoskeleton (LINC) complex that functions like a bridge across the inner and outer nuclear membranes to physically connect the nucleus to the cytoskeleton (11). This complex is responsible for various important cellular functions, such as mechanotransduction, cellular signaling, nuclear anchorage, and positioning. So far, five SUN proteins have been described in mammals and have been demonstrated or suggested to play roles in germ cell development (12–17). SUN1 and SUN2 are ubiquitously expressed in various tissues (12, 13) and mediate meiotic telomere attachment to the nuclear envelope (18–22). SUN3, SUN4, and SUN5 are specifically expressed in the testes (14–17). SUN5 localization is restricted to the head–tail junctions of sperm and is essential for anchoring the sperm head to the tail (23). SUN4 localizes to the spermatid nuclear envelope in close association with manchette microtubules (15, 16). Knockout of Sun4 in mice disrupts the lateral interactions of the manchette to the nucleus, and the nucleus thus fails to elongate, eventually resulting in a globozoospermia-like phenotype and male infertility (15, 16). SUN3 protein expression begins at postnatal day 15, and, similar to SUN4, its localization is closely associated with the manchette in developing spermatids (14). However, because of the lack of animal models, whether Sun3 indeed plays a role in spermiogenesis is not clear.
To investigate the physiological functions of Sun3 during mammalian spermatogenesis, we generated a Sun3 knockout mouse model using CRISPR/Cas9 genome editing technology. We found that Sun3−/− male mice are infertile, displaying abnormal sperm head morphology and irregular acrosome localization, likely resulting from disruption of manchette assembly. These findings demonstrate that Sun3 is essential for sperm head shaping during spermiogenesis.
Results
Generation of Sun3−/− mice by CRISPR/Cas9-mediated genome editing
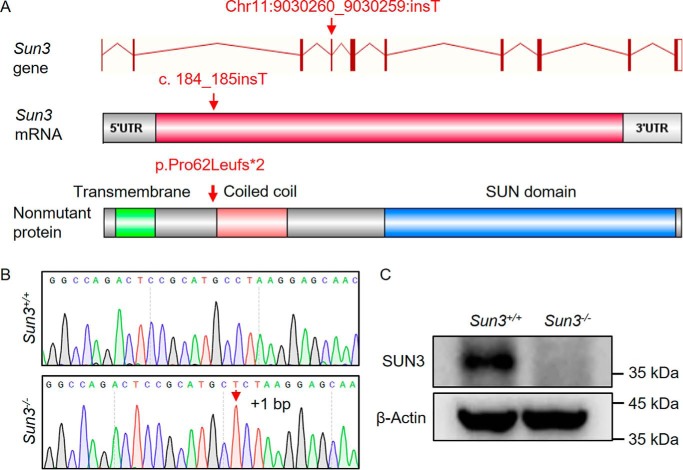
Consistent with previous findings (14, 16), SUN3 was expressed testis-specifically (Fig. S1A) and was detected in spermatids, localizing to the nuclear pole distal to the acrosome and overlapping with α-tubulin and SUN4 (Fig. S1B). To investigate the biological function of SUN3, we generated Sun3-null mice using CRISPR/Cas9 genome editing technology (Fig. 1A). Sanger sequencing of genomic DNA from the mutant mice revealed that a thymidine was inserted between nucleotide position 184 and 185 (c.184_185insT) in exon 4 of Sun3, predicted to result in premature translation termination (p.Pro62Leufs*2) (Fig. 1B). Western blotting analyses further confirmed that full-length SUN3 proteins were absent in the testes of Sun3−/− mice (Fig. 1C and Fig. S2).
Figure 1.
Generation of Sun3−/− mice. A, diagram illustrating the CRISPR/Cas9 targeting strategy to generate Sun3 knockout mice. A thymine deoxyribotide was inserted in exon 4 between nucleotide positions 184 and 185, predicted to cause a frameshift at amino acid position 63 that introduces a premature stop codon at position 64 (p.Pro62Leufs*2). B, representative Sanger sequencing chromatograms showing the thymine deoxyribotide inserted between nucleotide positions 184 and 185, as indicated by the red arrow. C, Western blotting using the rabbit anti-SUN3 antibody confirms the lack of full-length SUN3 proteins in testis lysates from 8-week-old Sun3−/− mice.
Deletion of Sun3 impairs spermatogenesis
Mice lacking Sun3 appeared normal, displaying no obvious abnormalities in development and behavior. Given the testis-restricted expression pattern of SUN3, we studied fertility in Sun3−/− male mice. Mating attempts of Sun3−/− males with WT females did not produce any offspring, indicating that Sun3−/− males are infertile. Sun3−/− females showed no overt abnormalities in fertility.
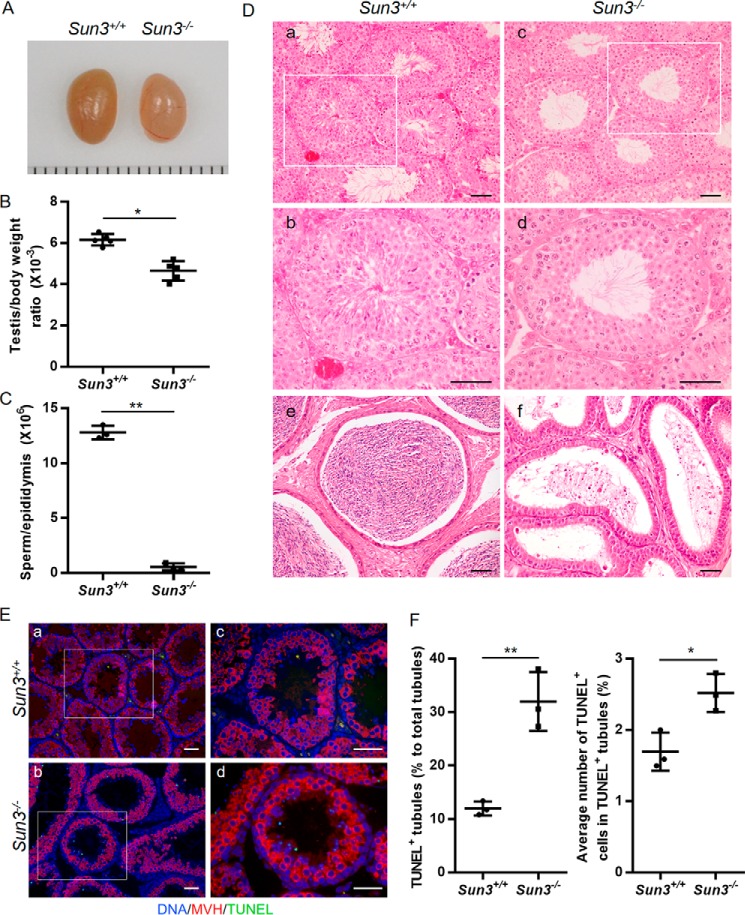
Sun3−/− mice had smaller testes (Fig. 2, A and B) and sharply declined epididymal sperm numbers (0.17 ± 0.03 million/ml) compared with WT mice (12.80 ± 0.36 million/ml) (Fig. 2C). To further characterize the spermatogenic defects in Sun3−/− mice, H&E staining of testis and epididymis sections was performed. All types of spermatogenic cells were present in an orderly way in Sun3+/+ seminiferous tubules and mature spermatozoa, and a canonical hook-shaped head could be seen in the lumen of tubules (Fig. 2D, a and b). However, in Sun3−/− mice, all elongating and elongated spermatids as well as spermatozoa were observed to have a noncanonical round head (Fig. 2D, c and d). Consistent with the result of sperm counting per epididymis, the numbers of these noncanonical spermatids and spermatozoa were apparently lower in seminiferous tubules and in the cauda epididymides of Sun3−/− mice compared with those in Sun3+/+ mice (Fig. 2D, e and f). Furthermore, a TUNEL assay in combination with a germ cell–specific marker (MVH) was performed, and the results indicated apoptosis in cells with a small amorphous nuclear shape, which corresponds to the noncanonical spermatids in Sun3−/− mice (Fig. 2E). The frequency of TUNEL+ tubules (11.99% ± 0.75% versus 32.00% ± 3.17%) and the number of TUNEL+ cells per TUNEL+ tubule (1.70% ± 0.15% versus 2.52% ± 0.15%) in Sun3−/− mice were significantly increased compared with Sun3+/+ mice (Fig. 2F), suggesting that the spermatids of Sun3−/− mice underwent apoptosis. These results indicate that Sun3 is essential for spermatogenesis and particularly for the development of spermatids.
Figure 2.
Deletion of Sun3 impairs spermatogenesis. A, representative images of testes from 8-week-old Sun3+/+ and Sun3−/− mice. Each grid represents 1 mm. B, average testes/body weight ratio of 8-week-old Sun3+/+and Sun3−/− mice (n = 5). C, epididymal sperm number of 8-week-old Sun3+/+ and Sun3−/− mice (n = 3). D, representative images of H&E-stained testicular sections (a–d) and epididymides (e and f) from 8-week-old Sun3+/+ and Sun3−/− mice. b and d, higher-magnification images of the rectangular area outlined with white boxes in a and c, respectively. Scale bars = 50 μm. E, TUNEL assay performed on testicular sections from 8-week-old Sun3+/+ and Sun3−/− mice. c and d, higher-magnification images of the rectangular area outlined with white boxes in a and b, respectively. Scale bars = 50 μm. F, percentage of TUNEL-positive tubules in testis sections from 8-week-old Sun3+/+and Sun3−/− mice (n = 3). F, quantification of TUNEL-positive cells per TUNEL-positive tubule. 298 tubules were examined from three Sun3+/+ mice. 375 tubules were examined from three Sun3−/− mice. Data are presented as mean ± S.D. Student's t test; *, p < 0.05; **, p < 0.01.
Sun3−/− mice produce round-headed spermatozoa
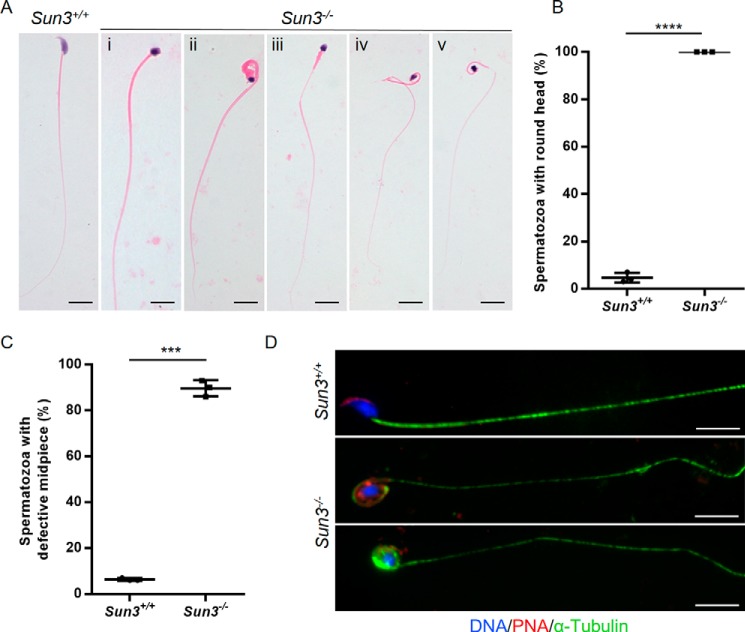
We further analyzed the sperm morphology in the epididymis and found that, in contrast to the typical hook-shaped appearance of sperm heads in Sun3+/+ mice, all sperm heads in Sun3−/− mice were amorphous with a smaller and more rounded shape (Fig. 3, A and B). Additionally, ∼89.67% ± 2.02% of spermatozoa from Sun3−/− mice also displayed various midpiece defects, such as irregular caliber, bending, coiling, and/or cracking (Fig. 3, A and C, and Fig. S3). Immunofluorescence staining of peanut agglutinin (PNA), a marker of acrosomes, was performed on sperm smears and seminiferous tubules. In Sun3+/+ mice, acrosomes with a typical crescent shape were found on top of the nucleus in the anterior dorsal part of the sperm head. However, acrosomes of spermatozoa from Sun3−/− mice were missing, mislocalized, or fragmented (Fig. 3D).
Figure 3.
Morphological abnormalities of spermatozoa from Sun3−/− mice. A, representative images of spermatozoa from 8-week-old Sun3+/+ and Sun3−/− mice. Spermatozoa from Sun3+/+ mice present with typical morphology, whereas spermatozoa from Sun3−/− mice present an amorphous head accompanied by various midpiece defects, such as thickening (i and ii), coiling (ii and v), thinning (iii–v), and cracking (iv). Scale bars = 10 μm. B, quantification of spermatozoa with an amorphous head from 8-week-old Sun3+/+and Sun3−/− mice (n = 3). Student's t test; ****, p < 0.0001. C, quantification of spermatozoa with a defective midpiece from 8-week-old Sun3+/+and Sun3−/− mice (n = 3). Student's t test; ***, p < 0.001. D, representative images of spermatozoa from 8-week-old Sun3+/+and Sun3−/− mice stained for PNA (red) and α-tubulin (green). DNA was counterstained with Hoechst. Scale bars = 10 μm.
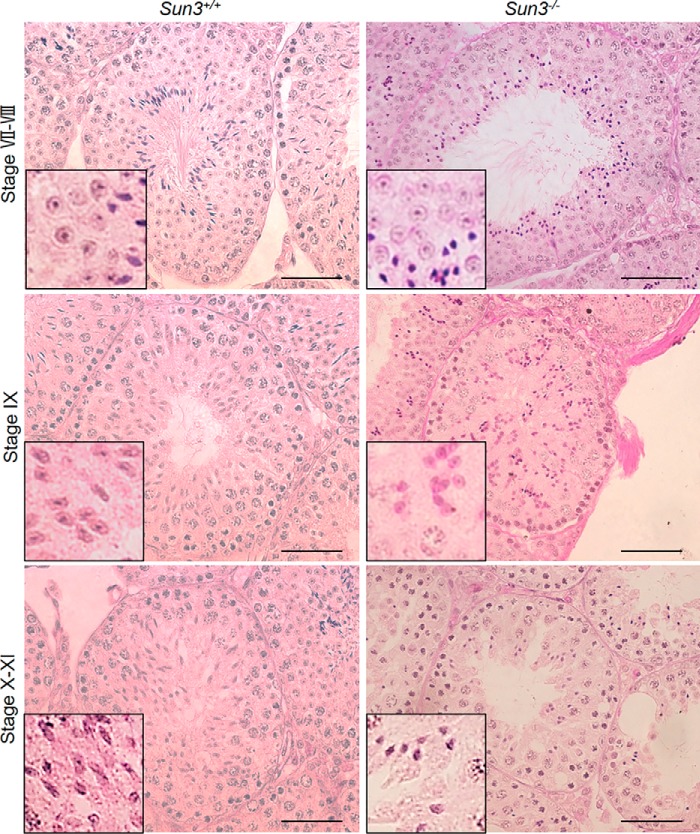
Because failure of sperm head elongation usually leads to a rounded head appearance, to understand the specific step at which head abnormalities occur in Sun3−/− mice, we compared spermatids of Sun3+/+ and Sun3−/− mice at different steps of spermiogenesis. PAS staining revealed that the morphology of spermatids until steps 7 and 8 were comparable in Sun3+/+ and Sun3−/− mice, indicating that the development of round spermatids was normal in Sun3−/− mice (Fig. 4). At step 9, the nuclei of spermatids became flattened and started to elongate along with condensation of chromatin in Sun3+/+ mice, whereas in Sun3−/− mice, spermatids still had a round nucleus although the chromatin had been condensed. At step 10, spermatid nuclei in Sun3+/+ mice became thinner and more elongated; however, the nuclei of spermatids were small and deformed and remained round in Sun3−/− mice (Fig. 4). We also noted that the number of spermatids was markedly reduced from step 10. Altogether, these findings demonstrate that abnormal morphology of Sun3−/− spermatids occurs when spermatids start to elongate.
Figure 4.
Abnormalities of spermatids occur when spermatids start to elongate in Sun3−/− mice. PAS staining of testis sections from 8-week-old Sun3+/+ and Sun3−/− mice revealed abnormal sperm nuclei from step 9 in Sun3−/− mice. Scale bars = 50 μm.
Manchette assembly is disrupted in Sun3−/− spermatids
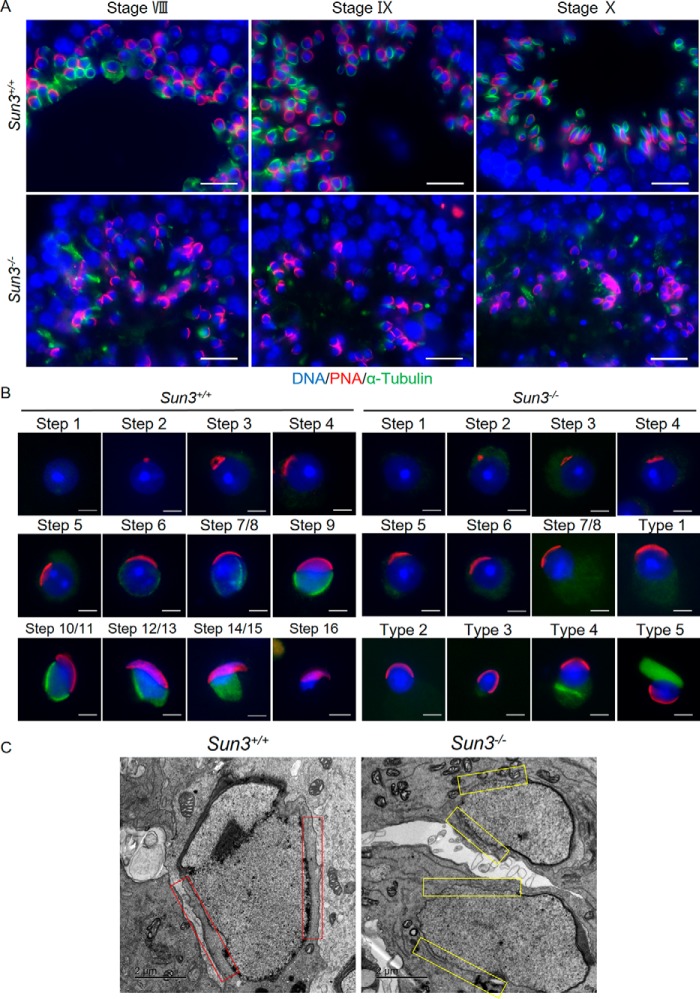
Because the microtubule manchette is essential for elongation of spermatids, and SUN3 localization is closely associated with the manchette (4, 14), we next wanted to find out whether the structure of the manchette was disrupted after Sun3 deletion. Coimmunofluorescence staining of testis sections with antibodies against α-tubulin, a marker of manchette microtubules, and PNA was conducted. In Sun3+/+ mice, manchette microtubules were found to be tightly attached to the nuclear periphery at the caudal region opposite the acrosome of spermatids; however, these structures were not detected in Sun3−/− mice despite some weak and diffuse labeling that was ectopically positioned (Fig. 5A). To confirm loss of manchette microtubules in Sun3 knockout spermatids, immunofluorescence staining on testis cell smears was performed. In Sun3+/+ mice, spermatids from step 1 to step 16 could be distinguished based on the morphology of the acrosome, and the acrosome covering the anterior side of the nucleus as well as the microtubules of the manchette tightly surrounding the caudal region could be seen from step 7/8 to step 14/15 in Sun3+/+ spermatids. In Sun3−/− mice, we did not detect any morphological abnormalities in spermatids until step 7/8, and acrosomes covering the anterior side of the nucleus appeared to be normal in round spermatids. However, the typical microtubule arrays of the manchette were not observed in all spermatids from Sun3−/− mice. Aberrantly polymerized microtubule bundles that were dissociated from the nucleus were observed in about 5% of spermatids (Fig. 5B). Moreover, the perinuclear ring, a belt-like structure surrounding the nucleus where manchette microtubules insert, was also absent in spermatids from Sun3−/− mice (Fig. S4). Transmission EM (TEM) further revealed the presence of manchette microtubule bundles that were closely associated with the nuclear envelope in elongating spermatids from Sun3+/+ testes; however, these recognizable structures were not seen in elongating spermatids from Sun3−/− testes, although some disorganized microtubule bundles that had lost their interaction with the nucleus were observed (Fig. 5C). Thus, we conclude that SUN3 is not only indispensable for manchette formation but also likely required for the organization of manchette during sperm head shaping in mice.
Figure 5.
Manchette formation is disrupted in Sun3−/− mice. A, representative images of testis sections from 8-week-old Sun3+/+ and Sun3−/− mice, stained by anti-PNA (red) and anti-α-tubulin (green) antibodies. DNA was counterstained with Hoechst. Scale bars = 20 μm. B, representative images of spermatids from 8-week-old Sun3+/+ and Sun3−/− mice, stained by anti-PNA (red) and anti-α-tubulin (green) antibodies. DNA was counterstained with Hoechst. Types 1–5 show various abnormal spermatids in Sun3−/− mice. Scale bars = 2 μm. C, manchette microtubule bundles were not observed in Sun3−/− mice by transmission EM. Red rectangles show the presence of manchette microtubule bundles in Sun3+/+ mice, whereas yellow rectangles indicate the absence of such microtubule bundles in Sun3−/− mice. Scale bars = 2 μm.
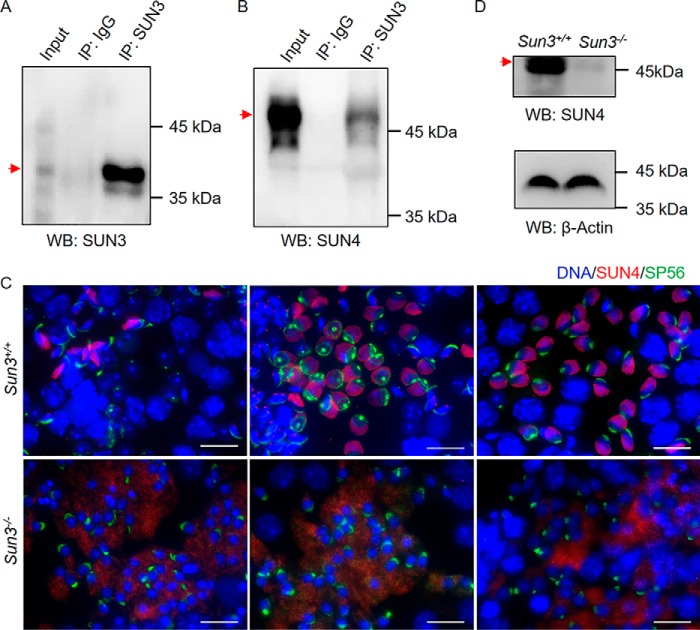
SUN3 interacts with SUN4 in mouse testes
Deletion of SUN4, another SUN protein that associates with the manchette, also leads to defects in acrosome and manchette formation in mice (15), similar to the findings in Sun3-null mice. To explore whether SUN3 interacts with SUN4 in testes, we performed immunoprecipitation using an anti-SUN3 antibody with testis lysates from Sun3+/+ mice. Western blotting detected SUN3 and SUN4 in lysates immunoprecipitated by the anti-SUN3 antibody but not in IgG-immunoprecipitated lysates (Fig. 6, A and B). Immunofluorescence staining in Sun3+/+ and Sun3−/− testicular sections revealed clear signals of SUN4 proteins surrounding the caudal region of the nucleus opposite the acrosome, where the manchette locates in step 7/8 spermatids, elongating spermatids, and elongated spermatids in Sun3+/+ mice, whereas only unspecific diffuse signals were observed in the cytoplasm of spermatids from Sun3−/− mice (Fig. 6C). Western blotting of testicular lysates revealed that the level of SUN4 proteins was drastically reduced in adult Sun3−/− mice compared with Sun3+/+ mice (Fig. 6D). These findings demonstrated that SUN3 and SUN4 interact and that SUN3 is required to maintain the level of SUN4 proteins in vivo.
Figure 6.
SUN3 interacts with SUN4 in mouse testes, and SUN4 protein levels are drastically reduced in Sun3−/− mice. A, Western blotting of immunoprecipitated testis extract showing the presence of SUN3 proteins in lysates immunoprecipitated by rabbit anti-SUN3 antibodies. Rabbit IgG was used as a negative control. B, Western blotting showing the presence of SUN4 proteins in testis lysates immunoprecipitated by guinea pig anti-SUN3 antibodies. Rabbit IgG was used as a negative control. IP, immunoprecipitation; WB, Western blotting. C, immunofluorescence staining of SUN4 (red), SP56 (green), and Hoechst (blue) on testis cell spreads from 8-week-old Sun3+/+ and Sun3−/− mice, showing mislocalization of SUN4 in Sun3−/− spermatids. Scale bars = 20 μm. D, Western blotting showing the levels of SUN4 proteins in Sun3+/+ and Sun3−/− testes. β-Actin was used as a loading control. For A, B, and D, the bands corresponding to the sizes of proteins of interest are indicated by arrows.
Discussion
Sperm head shaping is a key event during spermiogenesis, and misshaping of sperm heads often leads to male infertility (24). In this study, we generated mice lacking Sun3 through CRISPR/Cas9 technology and investigated the function of SUN3 during spermiogenesis. We found that loss of SUN3 leads to a drastic reduction in sperm numbers, a globozoospermia-like phenotype accompanied by multiple sperm tail defects resulting from failure of manchette formation during sperm head shaping, and, ultimately, male infertility. Additionally, we also demonstrated that SUN3 interacts with SUN4 in vivo and is required to maintain the level of SUN4 proteins in testes. We report, for the first time, that Sun3 is indispensable for sperm head shaping and particularly required for manchette formation.
A typical manchette is characterized by highly organized microtubule bundles attached to the perinuclear ring, forming a sleeve-like structure surrounding the posterior part of the spermatid nucleus (5, 25). This transient structure appears when spermatids are going to elongate and disappears when the sperm heads are properly shaped. Several genes have been reported to be implicated in manchette function in mice, including Hook1, Katnb1, Lrguk1, Meig1, Pacrg, and Spef2 (8, 26–29). The manchettes in these mutant mice displayed a highly disorganized structure and/or perturbed disassembly, resulting in spermatozoa with a deformed shape (8, 26–29). However, we found that, in most cases, the canonical manchette structure was not detected in spermatids after Sun3 knockout, and some aberrantly polymerized straight microtubule bundles were observed in a few step 8–16 spermatids (∼5%), but these bundles were completely dissociated from the nucleus. The nuclear shape of the epididymal sperm from Sun3 knockout mice was generally rounder than that observed in other disrupted manchettes. This suggests that SUN3 is essentially required for formation and coupling of the manchette.
SUN3 and SUN4 are exclusively expressed in spermatids and colocalize in close association with the manchette (14, 16, 30), suggesting a cooperative function in coupling of the manchette to the nuclear periphery so that the cytoplasmic forces can be transduced to shape the sperm nuclei. In Sun4−/− mice, manchette microtubules were disorganized, lost their lateral interaction with the nucleus, and, in some cases, were even completely missing, resulting in a globozoospermia-like phenotype (15). Deletion of Sun4 caused SUN3 mislocalization with a tendency to form aggregates in the cytoplasm, suggesting that SUN3 localization depends on SUN4 (15). In this study, manchette tubules were not observed in the majority of spermatids in Sun3−/− mice, although a few spermatids showed some straight microtubule bundles, but these structures were dissociated from nuclei, which closely resembled the phenotype of Sun4−/− mice. Interestingly, the level of SUN4 proteins was drastically reduced in Sun3−/− testes, indicating that SUN3 is also required to maintain the level of SUN4 proteins in vivo. Thus, it indicates that the functions of SUN3 and SUN4 are likely interdependent. It is worth mentioning that we tried many times to optimize the experimental protocols of coimmunoprecipitation using the anti-SUN4 antibody; however, we could not detect SUN3 in lysates immunoprecipitated by anti-SUN4 antibodies, for which the possibilities that anti-SUN4 antibody might not be suitable for coimmunoprecipitation experiments or that the interaction of SUN4 with the anti-SUN4 antibody may interfere with binding of SUN3 with SUN4 should be considered. It would be interesting to conduct further studies to confirm the interaction between SUN3 and SUN4, to investigate how SUN3 interacts with SUN4, and particularly how they coordinate and function in assembly or stabilization of the manchette cytoskeletal structure.
Anchorage and positioning of the cell nucleus plays an important role during diverse developmental processes such as fertilization, cell migration, and establishment of polarity (31–33). In mammalian somatic cells, the LINC complex, composed of SUN domain and KASH domain proteins, forms a protein bridge within the perinuclear space that connects the nucleus with the cytoskeleton (34, 35). Two spermiogenesis-specific LINC complexes have been described; one is SUN1η–NESPRIN3, localized on the anterior acrosomal side, and the other is SUN3–NESPRIN1, situated at the posterior pole of the spermatid nucleus (14). However, unlike the various functions of SUN proteins in spermatogenesis, the known KASH domain proteins appear to be not that important. Mice mutant for KASH domain proteins, Nesprin1 or Nesprin3, were fully fertile, and disruption of genes encoding for other important KASH proteins (Nesprin2, Nesprin4, and Lrmp) also displayed no overt impact on fertility (36, 37). So far, only KASH5 has been shown to be essential for spermatogenesis. KASH5 is predominantly expressed at meiotic stages and aggregated at one nuclear pole in round and elongated spermatids (18). Mice lacking KASH5 manifested meiotic arrest; thus, we currently do not know whether KASH5 plays a role in spermiogenesis as well. However, given the localization pattern of KASH5 in spermatids, it is a poor candidate as a LINC complex partner for SUN3. Thus, we assume that NESPRIN1 is not essentially required for spermiogenesis and that SUN3 may form links with other alternative KASH proteins to physically couple the manchette to the nucleus.
Altogether, our study indicates that Sun3 is required for sperm head shaping during mammalian spermiogenesis. Disruption of Sun3 in mice caused a drastic reduction in sperm numbers and severe sperm head defects because of manchette formation failure, ultimately resulting in male infertility. These results will deepen our understanding of the role of LINC complexes in sperm head shaping and provide new molecular cues regarding the underlying pathology of globozoospermia.
Experimental procedures
Mice
Sun3−/− mice were generated using CRISPR/Cas9 genome editing as described previously (38). To generate Sun3−/− mice, a guide RNA sequence (GGCGTTGCTTAAAGACATGA) targeting exon 4 (5′-GACTCCGCATGCTCTAAGGA-3′) of the Sun3 gene was coinjected with Cas9 mRNAs into B6D2F1 (C57BL/6 × DBA/2) zygotes, which were subsequently transferred to the oviducts of pseudopregnant ICR female mice. Sanger sequencing of tail genomic DNA confirmed a female founder mouse carrying a homozygous 1-nt insertion in exon 4 of Sun3 (5′-GACTCCGCATGCTCTAAGGA-3′, the inserted T nucleotide is underlined). The female founder mouse was crossed to WT C57BL/6 mice. The resulting heterozygous mice were crossed to obtain homozygous mutants. Animal experiments were approved by the Institutional Animal Care Committee of the University of Science and Technology of China. The following sequences of primers were used to confirm the genotype of Sun3 mice: 5′-GGCAGGCTGAGAAAGACACACATG-3′ and 5′-GGCTTCACAGCTGACAATGGCAT-3′.
Western blotting
Testes from adult mice (8–12 weeks old) were homogenized in radioimmune precipitation assay buffer (25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors using a Tissuelyzer and then cleared by centrifugation. Western blotting was carried out as described previously (39, 40). Antibodies included rabbit polyclonal anti-SUN3 (Proteintech, 26434-AP-1, 1:1000), guinea pig polyclonal anti-SUN3 (1:3000) (16), rabbit polyclonal anti-SUN4 (Proteintech, 19721-1-AP, 1:3000) and anti-β-actin (Abcam, ab8227, 1:100).
Sperm counting
Adult male mice were sacrificed by cervical dislocation. Their epididymides were dissected, minced into small pieces in 1 ml of Dulbecco's modified Eagle's medium, and incubated at 37 °C in a 5% CO2 humidified incubator for 1 h to allow sperm release. Sperm counts were determined using a hemocytometer under a microscope (Nikon Eclipse E200).
Histological examination, TUNEL assay, and immunofluorescence staining
Mice were euthanized by cervical dislocation. Testes were removed and fixed overnight in Bouin's solution (for H&E and PAS) or 4% paraformaldehyde (for immunofluorescence staining and the TUNEL assay). Samples were dehydrated through a graded series of ethanol, embedded in paraffin, and sectioned at 5 μm for subsequent H&E and PAS staining. Sperm smears were prepared from epididymis as described previously (41). To prepare testis smears, testes were peeled off the testicular capsule, cut into pieces in PBS, and filtered through a 200-mesh cell strainer. The filtered cell suspensions were spread onto a glass slide.
H&E and PAS staining of tissue sections or sperm smears was performed as described previously (41). The TUNEL assay was performed using the In Situ Cell Death Detection Kit (Roche, 11684795910) according to the manufacturer's instructions. For immunofluorescence, tissue sections, testis smears, or sperm smears were blocked in antibody dilution buffer (10% normal donkey serum, 3% BSA, and 0.05% Triton X-100 in PBS) for 30 min, followed by overnight incubation at 4 °C with primary antibodies against α-tubulin (Sigma, F2168, 1:200), rabbit polyclonal anti-SUN4 (1:3000), and lectin PNA (Invitrogen, L32458, 1:200). After washing with PBST four times, sections were incubated with secondary antibodies (Invitrogen, Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 555 donkey anti-rabbit IgG, 1:200) at 37 °C for 1 h. Finally, sections were mounted with Vectashield mounting medium (Vector Laboratories, H-1000) containing Hoechst 33342 (Invitrogen, H21492). Staining of epididymal sperm mitochondria was performed using MitoTracker probes (Molecular Probes, M-7512). Images were captured using a Nikon Eclipse 80i microscope (Nikon) equipped with a digital camera incorporating a charge-coupled device (Hamamatsu) and analyzed using NIS-Element microscope imaging software (Nikon).
Coimmunoprecipitation
Tissue extracts were prepared using a Dounce homogenizer in cold immunoprecipitation buffer with 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm Na3VO4, 1 mm EDTA, 1 mm EGTA, 50 mm Tris (pH 7.5), and 150 mm NaCl dissolved in 500 ml of PBS supplemented with 1 mm phenylmethylsulfonyl fluoride and protein inhibitor mixture (Roche, 04693116001). All reagents were of analytical grade. Lysates were then centrifuged at 15,000 × g at 4 °C for 15 min, and the supernatant was divided into two aliquots. Each aliquot was incubated with 1.5 μg of rabbit anti-SUN3 antibody (Proteintech, 26434-AP-1) or rabbit IgG nonspecific antibody (CST, 2729S). After incubation at 4 °C overnight with rotation, beads were washed five times with radioimmune precipitation assay buffer. Finally, the beads were resuspended in 3× SDS-sample buffer (120 mm Tris/HCl, 10% SDS, 20% glycerol, 20% 2-mercaptoethanol, and bromphenol blue (pH 6.8)), boiled at 100 °C for 15 min, and subsequently subjected to Western blotting.
Transmission EM
Ultrastructural analysis of Sun3−/− mouse testes was performed as described previously (42). Briefly, testes from Sun3+/+ and Sun3−/− mice were fixed in 4% paraformaldehyde containing 0.05% glutaraldehyde in 0.1 m phosphate buffer and then post-fixed in 1% osmium tetroxide. Dehydration was carried out in ethanol, and the samples were embedded in Epon 812. Ultrathin sections were obtained, counterstained with uranyl acetate and lead citrate, and examined with a JEOL JEM-1230 transmission electron microscope.
Statistical analysis
Testis/body weight ratio, sperm number, sperm morphology, and TUNEL-positive cells in Sun3+/+ and Sun3−/− mice were analyzed statistically by using Student's t test. Results are presented as mean ± S.D. Statistical significance is defined as p < 0.05.
Author contributions
Q. G. and R. K. methodology; Q. G. and R. K. writing-original draft; C. Y. and M. A. resources; C. Y. visualization; M. A., X. J., H. M., and Q. S. writing-review and editing; X. J. and Q. S. conceptualization; H. M. formal analysis; Q. S. funding acquisition.
Supplementary Material
This work was supported by National Natural Science Foundation of China (NSFC) Grant 31890780, 31630050 (to Q. S.), and 31871514 (to X. J.); National Key Research and Developmental Program of China Grants 2018YFC1004700 (to X. J.) and 2016YFC1000600 (to Q. S.); and Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB19000000. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- SUN
- Sad1 and UNC84
- KASH
- Klarsicht/ANC-1/Syne-1 homology
- LINC
- linker of nucleoskeleton and cytoskeleton
- PNA
- peanut agglutinin
- PAS
- periodic acid-Schiff.
References
- 1. Jan S. Z., Hamer G., Repping S., de Rooij D. G., van Pelt A. M., and Vormer T. (2012) Molecular control of rodent spermatogenesis. Biochim. Biophys. Acta 1822, 1838–1850 10.1016/j.bbadis.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 2. Lehti M. S., and Sironen A. (2016) Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 151, R43–R54 10.1530/REP-15-0310 [DOI] [PubMed] [Google Scholar]
- 3. O'Donnell L., and O'Bryan M. K. (2014) Microtubules and spermatogenesis. Semin. Cell Dev. Biol. 30, 45–54 10.1016/j.semcdb.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 4. Kierszenbaum A. L. (2001) Spermatid manchette: plugging proteins to zero into the sperm tail. Mol. Reprod. Dev. 59, 347–349 10.1002/mrd.1040 [DOI] [PubMed] [Google Scholar]
- 5. Courtens J. L., and Loir M. (1981) The spermatid manchette of mammals: formation and relations with the nuclear envelope and the chromatin. Reprod. Nutr. Dev. 21, 467–477 10.1051/rnd:19810312 [DOI] [PubMed] [Google Scholar]
- 6. Kierszenbaum A. L., and Tres L. L. (2004) The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch. Histol. Cytol. 67, 271–284 10.1679/aohc.67.271 [DOI] [PubMed] [Google Scholar]
- 7. Moreno R. D., and Schatten G. (2000) Microtubule configurations and post-translational α-tubulin modifications during mammalian spermatogenesis. Cell Motil. Cytoskeleton 46, 235–246 [DOI] [PubMed] [Google Scholar]
- 8. Mendoza-Lujambio I., Burfeind P., Dixkens C., Meinhardt A., Hoyer-Fender S., Engel W., and Neesen J. (2002) The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 11, 1647–1658 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- 9. Zhou J., Du Y.-R., Qin W.-H., Hu Y.-G., Huang Y.-N., Bao L., Han D., Mansouri A., and Xu G.-L. (2009) RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development 136, 373–382 10.1242/dev.030858 [DOI] [PubMed] [Google Scholar]
- 10. Akhmanova A., Mausset-Bonnefont A. L., van Cappellen W., Keijzer N., Hoogenraad C. C., Stepanova T., Drabek K., van der Wees J., Mommaas M., Onderwater J., van der Meulen H., Tanenbaum M. E., Medema R. H., Hoogerbrugge J., Vreeburg J., et al. (2005) The microtubule plus-end-tracking protein CLIP-170 associates with the spermatid manchette and is essential for spermatogenesis. Gene Dev. 19, 2501–2515 10.1101/gad.344505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tzur Y. B., Wilson K. L., and Gruenbaum Y. (2006) SUN-domain proteins: “velcro” that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 7, 782–788 10.1038/nrm2003 [DOI] [PubMed] [Google Scholar]
- 12. Hodzic D. M., Yeater D. B., Bengtsson L., Otto H., and Stahl P. D. (2004) Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279, 25805–25812 10.1074/jbc.M313157200 [DOI] [PubMed] [Google Scholar]
- 13. Padmakumar V. C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A. A., Gotzmann J., Foisner R., and Karakesisoglou I. (2005) The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118, 3419–3430 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- 14. Göb E., Schmitt J., Benavente R., and Alsheimer M. (2010) Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE 5, e12072 10.1371/journal.pone.0012072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calvi A., Wong A. S., Wright G., Wong E. S., Loo T. H., Stewart C. L., and Burke B. (2015) SUN4 is essential for nuclear remodeling during mammalian spermiogenesis. Dev. Biol. 407, 321–330 10.1016/j.ydbio.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 16. Pasch E., Link J., Beck C., Scheuerle S., and Alsheimer M. (2015) The LINC complex component Sun4 plays a crucial role in sperm head formation and fertility. Biol. Open 4, 1792–1802 10.1242/bio.015768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yassine S., Escoffier J., Abi Nahed R., Pierre V., Karaouzene T., Ray P. F., and Arnoult C. (2015) Dynamics of Sun5 localization during spermatogenesis in wild type and Dpy19l2 knock-out mice indicates that Sun5 is not involved in acrosome attachment to the nuclear envelope. PLoS ONE 10, e0118698 10.1371/journal.pone.0118698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morimoto A., Shibuya H., Zhu X., Kim J., Ishiguro K., Han M., and Watanabe Y. (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198, 165–172 10.1083/jcb.201204085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding X., Xu R., Yu J., Xu T., Zhuang Y., and Han M. (2007) SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 20. Penkner A. M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., and Jantsch V. (2009) Meiotic Chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139, 920–933 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- 21. Lei K., Zhang X., Ding X., Guo X., Chen M., Zhu B., Xu T., Zhuang Y., Xu R., and Han M. (2009) SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 10207–10212 10.1073/pnas.0812037106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Link J., Leubner M., Schmitt J., Göb E., Benavente R., Jeang K.-T., Xu R., and Alsheimer M. (2014) Analysis of meiosis in SUN1 deficient mice reveals a distinct role of SUN2 in mammalian meiotic LINC complex formation and function. PLoS Genet. 10, e1004099 10.1371/journal.pgen.1004099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shang Y., Zhu F., Wang L., Ouyang Y. C., Dong M. Z., Liu C., Zhao H., Cui X., Ma D., Zhang Z., Yang X., Guo Y., Liu F., Yuan L., Gao F., et al. (2017) Essential role for SUN5 in anchoring sperm head to the tail. Elife 6, e28199 10.7554/eLife.28199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan W. (2009) Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol. Cell Endocrinol. 306, 24–32 10.1016/j.mce.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kierszenbaum A. L., Rivkin E., and Tres L. L. (2011) Cytoskeletal track selection during cargo transport in spermatids is relevant to male fertility. Spermatogenesis 1, 221–230 10.4161/spmg.1.3.18018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunleavy J. E. M., Okuda H., O'Connor A. E., Merriner D. J., O'Donnell L., Jamsai D., Bergmann M., and O'Bryan M. K. (2017) Katanin-like 2 (KATNAL2) functions in multiple aspects of haploid male germ cell development in the mouse. PLoS Genet. 13, e1007078 10.1371/journal.pgen.1007078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y., DeBoer K., de Kretser D. M., O'Donnell L., O'Connor A. E., Merriner D. J., Okuda H., Whittle B., Jans D. A., Efthymiadis A., McLachlan R. I., Ormandy C. J., Goodnow C. C., Jamsai D., and O'Bryan M. K. (2015) LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 11, e1005090 10.1371/journal.pgen.1005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lehti M. S., Zhang F.-P., Kotaja N., and Sironen A. (2017) SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development 144, 2683–2693 10.1242/dev.152108 [DOI] [PubMed] [Google Scholar]
- 29. Li W., Tang W., Teves M. E., Zhang Z., Zhang L., Li H., Archer K. J., Peterson D. L., Williams D. C. Jr., Strauss J. F. 3rd, and Zhang Z. (2015) A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development 142, 921–930 10.1242/dev.119834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao X., Tarnasky H. A., Lee J. P., Oko R., and van der Hoorn F. A. (1999) Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 211, 109–123 10.1006/dbio.1999.9297 [DOI] [PubMed] [Google Scholar]
- 31. Fridkin A., Penkner A., Jantsch V., and Gruenbaum Y. (2009) SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol. Life Sci. 66, 1518–1533 10.1007/s00018-008-8713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Starr D. A. (2009) A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122, 577–586 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burke B., and Roux K. J. (2009) Nuclei take a position: managing nuclear location. Dev. Cell 17, 587–597 10.1016/j.devcel.2009.10.018 [DOI] [PubMed] [Google Scholar]
- 34. Sosa B. A., Rothballer A., Kutay U., and Schwartz T. U. (2012) LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047 10.1016/j.cell.2012.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang W., Shi Z., Jiao S., Chen C., Wang H., Liu G., Wang Q., Zhao Y., Greene M. I., and Zhou Z. (2012) Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 22, 1440–1452 10.1038/cr.2012.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horn H. F., Kim D. I., Wright G. D., Wong E. S., Stewart C. L., Burke B., and Roux K. J. (2013) A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol. 202, 1023–1039 10.1083/jcb.201304004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., and Han M. (2007) Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development 134, 901–908 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- 38. Jiang L., Li T., Zhang X., Zhang B., Yu C., Li Y., Fan S., Jiang X., Khan T., Hao Q., Xu P., Nadano D., Huleihel M., Lunenfeld E., Wang P. J., et al. (2017) RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr. Biol. 27, 1498–1505.e6 10.1016/j.cub.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 39. Jiang X., Ma T., Zhang Y., Zhang H., Yin S., Zheng W., Wang L., Wang Z., Khan M., Sheikh S. W., Bukhari I., Iqbal F., Cooke H. J., and Shi Q. (2015) Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol. Reprod. 92, 79 [DOI] [PubMed] [Google Scholar]
- 40. Yin H., Ma H., Hussain S., Zhang H., Xie X., Jiang L., Jiang X., Iqbal F., Bukhari I., Jiang H., Ali A., Zhong L., Li T., Fan S., Zhang B., et al. (2019) A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet. Med. 21, 62 10.1038/s41436-018-0015-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu F., Yan P., Zhang J., Cui Y., Zheng M., Cheng Y., Guo Y., Yang X., Guo X., and Zhu H. (2019) Deficiency of TPPP2, a factor linked to oligoasthenozoospermia, causes subfertility in male mice. J. Cell. Mol. Med. 23, 2583–2594 10.1111/jcmm.14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang B., Ma H., Khan T., Li T., Zhang H., Gao J., Zhou J., Ma A., Li Y., Yu C., Bao J., Ali A., Murtaza G., Yin H., Gao Q., et al. (2020) A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J. Exp. Med. 217, e20182365 10.1084/jem.20182365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.