Abstract
The increasing use of advanced nucleic acid sequencing technologies for clinical diagnostics and therapeutics has made vital understanding the costs of performing these procedures and their value to patients, providers, and payers. The Association for Molecular Pathology invested in a cost and value analysis of specific genomic sequencing procedures (GSPs) newly coded by the American Medical Association Current Procedural Terminology Editorial Panel. Cost data and work effort, including the development and use of data analysis pipelines, were gathered from representative laboratories currently performing these GSPs. Results were aggregated to generate representative cost ranges given the complexity and variability of performing the tests. Cost-impact models for three clinical scenarios were generated with assistance from key opinion leaders: impact of using a targeted gene panel in optimizing care for patients with advanced non–small-cell lung cancer, use of a targeted gene panel in the diagnosis and management of patients with sensorineural hearing loss, and exome sequencing in the diagnosis and management of children with neurodevelopmental disorders of unknown genetic etiology. Each model demonstrated value by either reducing health care costs or identifying appropriate care pathways. The templates generated will aid laboratories in assessing their individual costs, considering the value structure in their own patient populations, and contributing their data to the ongoing dialogue regarding the impact of GSPs on improving patient care.
CME Accreditation Statement: This activity (“JMD 2016 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2016 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Genomic sequencing procedures (GSPs) are well suited for the diagnosis and management of inherited disease, particularly when there is significant overlap in clinical presentation or a condition may be caused by any of a large number of genes.1, 2 They are also increasingly valuable for the molecular characterization of a growing number of biomarkers central to the clinical management and treatment of patients with advanced cancer and, where available, tumor material is frequently limited and mutational profiles complex.3, 4 This information is changing the way clinicians are able to diagnose and manage hereditary diseases and the delivery of oncology care.
The American Medical Association Current Procedural Terminology (CPT) Editorial Panel recognized the clinical use of these procedures by introducing several new CPT codes in 2015 to describe DNA or RNA sequence analysis methods that simultaneously assay multiple genes or genetic regions associated with specific clinical conditions. These include codes for an array of multiple gene panels, exome, and genome sequencing. In October 2014, the Center for Medicare and Medicaid Services (CMS) announced that the new GSP CPT codes would be placed on the Clinical Laboratory Fee Schedule and that payment rates would be determined by the gap-fill process. In this process, individual Medicare Administrative Contractors collect data from laboratories and assign rates to new codes. These regional payment rates are then aggregated to inform the new national rate, effective January 1, 2016.
The Association for Molecular Pathology (AMP) Economic Affairs Committee and Board of Directors determined that AMP should support laboratories in collecting cost data to ensure the fair and accurate valuation of GSP codes. The goal of this project was to develop transparent cost data for representative procedures by collecting and analyzing the technical, analytical, postanalytical, and interpretation costs. For a comprehensive analysis of the full costs of performing these GSPs, we included the development, validation, maintenance, quality control, and overhead costs from laboratories providing clinical testing. AMP also recognized that the economic value of these procedures should be assessed and contrasted with the costs of current procedures and processes in different clinical care applications. Three independent payer cost-impact analyses were conducted, modeling the overall cost of current mix of treatments or interventions and the expected mix after the introduction of the new GSPs.
Materials and Methods
Microcosting Models
Five GSPs—CPT codes 81430, 81470, 81445, 81455, and 81415—were selected as representative applications of GSPs that reflected the spectrum of technology and data analysis (Table 1). These included multigene tumor panels, targeted inherited disease panels for hearing loss and X-linked intellectual disability, and exome sequencing. Laboratories performing these types of tests were identified and evaluated as to whether they met the criteria for a representative laboratory. Any laboratory that had been conducting GSPs, including the technical sequencing and clinical interpretation and reporting, for at least 6 months and running at least one in-house batch of ≥five samples per week for one or more of the procedures was eligible for participation. We were assured by each participating laboratory that it performed the necessary validation processes to ensure analytic accuracy and sufficient diagnostic yield, but we did not perform an independent evaluation of test quality. Boston Healthcare Associates and Tynan Consulting were retained to collect data from the laboratories and generate tools, which could be used to share the information.
Table 1.
GSP CPT Codes Targeted for Microcosting Analysis
| CPT code | Description |
|---|---|
| 81430 | Hearing loss (eg, nonsyndromic hearing loss, Usher syndrome, Pendred syndrome); genomic sequence analysis panel, must include sequencing of at least 60 genes, including CDH23, CLRN1, GJB2, GPR98,∗MTRNR1,∗MYO7A, MYO15A, OTOF, PCDH15, SLC26A4, TMC1, TMPRSS3, USH1C, USH1G, USH2A, and WFS1 |
| 81470 | XLID (eg, syndromic and nonsyndromic XLID); genomic sequence analysis panel, must include sequencing of at least 60 genes, including ARX, ATRX, CDKL5, FGD1, FMR1, HUWE1, IL1RAPL1, KDM5C, L1CAM, MECP2, MED12, MID1, OCRL, RPS6KA3, and SLC16AZ∗ |
| 81445 | Targeted genomic sequence analysis panel, solid organ neoplasm, DNA analysis, 5-50 genes (eg, ALK, BRAF, CDKN2A, EGFR, ERBB2, KIT, KRAS, NRAS, MET, PDGFRA, PDGFRB, PGR, PIK3CA, PTEN, RET), interrogation for sequence variants and copy number variants or rearrangements, if performed |
| 81455 | Targeted genomic sequence analysis panel, solid organ or hematolymphoid neoplasm, DNA and RNA analysis when performed, 51 or greater genes (eg, ALK, BRAF, CDKN2A, CEBPA, DNMT3A, EGFR, ERBB2, EZH2, FLT3, IDH1, IDH2, JAK2, KIT, KRAS, MLL,∗NPM1, NRAS, MET, NOTCH1, PDGFRA, PDGFRB, PGR, PIK3CA, PTEN, RET), interrogation for sequence variants and copy number variants or rearrangements, if performed |
| 81415 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis |
CPT copyright 2014 American Medical Association. All rights reserved. CPT is a registered trademark of the American Medical Association.
CPT, Current Procedural Terminology; GSP, genomic sequencing procedure; XLID, X-linked intellectual disability.
Human Gene Nomenclature Committee recommended nomenclature: ADGRV1 (GRP98), KMT2A (MLL), MT-RNR1 (MTRNR1), SLC16A2 (SLC16AZ).
Laboratories shared their standard operating procedures for the entire GSP, including the bioinformatics, interpretation, and clinical reporting. A subset of laboratories allowed on-site visits and physical observation, including timing of specific protocol steps. In total, 65 laboratories were contacted, with 36 willing to discuss potential participation in the project and 9 agreeing to participate and share protocols. Overall, 13 unique test protocols were provided by the nine laboratories. These laboratories represented both small and large academic medical centers as well as commercial reference laboratories. Protocols were parsed into spreadsheets to capture the individual steps and process modules, including DNA extraction, library preparation, sequencing, quality control, and data analysis. The components required to execute each module were identified and documented in the spreadsheet, including the cost of reagents and consumables, cost of and time for equipment use, and the personnel hands-on time. The professional time of M.D. and Ph.D. laboratory directors to interpret and sign out these cases was also included in the data collection and analysis.
Where possible, components were priced using the CMS database for costs of common supplies and equipment. If the items were not identified in the CMS database, reagent and consumable prices as set by the manufacturer or wholesale suppliers were obtained and used in the analysis. Reagent and consumables costs were reduced to a per-unit cost to match the protocol (eg, cost per milliliter or cost per gram). For each step, the necessary item cost per unit was listed and multiplied by total quantity and batch size to calculate a cost per step. Equipment costs were calculated by assigning a useful life to each piece of equipment and then amortizing the price of that equipment to a per-minute rate, assuming a piece of equipment is used 50% of the time for the particular protocol. For each step, the necessary equipment cost per minute was multiplied by the time each piece of equipment was used. Costs were examined per batch but then divided by batch size to enable comparisons on a per-sample basis.
The average salary for each personnel job class was also obtained from the CMS database. If job classifications could not be obtained from the CMS database, average salary was determined through survey results from professional associations. The per-minute rate was calculated on the basis of the assumption of a 40-hour week, 50 weeks per year, and multiplied by the hands-on time per step.
In some cases, laboratories perform confirmatory testing or testing of genetic regions missed by their GSP using traditional chain termination or Sanger sequencing to complete the assay. The per-unit cost of Sanger sequencing was obtained from individual laboratories and multiplied by the average number of Sanger sequencing tests typically needed per patient. Laboratories may also include other complementary methods, such as triplet repeat PCR-based assay for fragile X syndrome in X-linked intellectual disability and PCR-based allele-specific assay for GJB6 gene for hearing loss. Microcosting analysis of Sanger sequencing and other complementary methods was not performed.
An important and unique component of this analysis was capturing the costs of bioinformatics and pipeline development. The pipeline was accounted for as capital equipment. Interviews were conducted with bioinformatics personnel and laboratory directors to assess the time involved in developing a bioinformatics pipeline multiplied by the personnel classification pay scales. The costs of pipeline development and validation, commercial software if purchased, and the computing hardware were amortized over the lifetime of the pipeline (estimated as 1 year).
The costs of analyzing and reporting results from the review of the FASTQ or BAM files to assess the quality of the run to the clinical reporting of results were included. The laboratory personnel and computing time was captured and multiplied by the personnel classification involved (eg, bioinformatician) with per-unit cost of that individual. The total personnel time and software costs involved in reviewing the individual sample results through either individual or group reviews (by fellows, laboratory supervisors, laboratory directors, and/or pathologists) and the costs of the equipment used to store data both for short-term analysis needs and long-term record-keeping were included and amortized on the basis of the estimated total number of samples to be processed and kept.
Finally, the individual cost information from each laboratory procedure for the components of DNA extraction, library preparation, sequencing, bioinformatics, and reporting was blinded and aggregated for side-by-side comparison.
Health Economic Models
To enable the assessment of the value of GSPs in patient management, we developed payer cost-impact models. The goal was to develop the tools to estimate the impact of incorporating genomic testing into patient management compared with current approaches to care. Three intended use populations were chosen as examples: i) patients with advanced non–small-cell lung cancer (NSCLC) in need of treatment optimization, ii) patients being evaluated for syndromic sensorineural hearing loss, and iii) children experiencing neurodevelopmental disorders. The key elements of each health economic model were to generate an estimate of the size of the eligible population (specific to the intended use of the test), the current mix of tests and treatments or interventions, and the anticipated result after the introduction of testing using the relevant GSP. The primary end point of these models was the cost per diagnosis, management, treatment, or intervention mix before and after GSP testing.
The models were on the basis of best practices outlined in an International Society for Pharmacoeconomics and Outcomes Research task force report on budget impact analysis.5 Given that the models are intended to demonstrate to payers the utility of paying for testing, each was framed from the perspective of a typical commercial insurer. These insurers typically focus on the near-term direct costs associated with testing and potential direct cost offsets, so we used a short time frame of 6 months to 1 year. They were built using a mixture of clinical and health economic literature, patient-level data from laboratories or institutions currently conducting GSPs, and key opinion leader inputs or assumptions. All costs were adjusted to 2014 US dollars, when necessary, to account for inflation changes, by using the Consumer Price Index inflation calculator. To determine the patient populations involved, we used epidemiological data obtained from the US Census Bureau, the National Cancer Institute, and published literature.6, 7, 8, 9
For the model of GSP testing in patients with NSCLC, the traditional care pathway was adapted from the National Comprehensive Cancer Network guidelines.10 The model focuses on first-line treatment for advanced or metastatic (stage IIIB/IV) NSCLC patients with a time horizon of 6 months from diagnosis. For the hereditary conditions (sensorineural hearing loss and neurodevelopmental disorders), we relied on literature and information from academic medical centers that see these types of patients to estimate the current mix of diagnostic procedures and their use levels. The data analysis models herein assumed 100% of patients will receive mutational analysis using a GSP and providers and patients will rely on these results to inform their decisions concerning medical care. Key opinion leaders reviewed all of the models to objectively assess and validate the clinical and economic impact of these procedures.
Advanced NSCLC in Need of Treatment Optimization
The cost-impact model identified four treatment pathways: targeted therapy, nontargeted therapy, clinical trial, and hospice care. In the current care pathway, positive results from EGFR or ALK testing were used to direct a patient to the corresponding targeted therapy. Patients who tested negative for EGFR and ALK mutations were directed to traditional chemotherapy regimens or enrolled in clinical trials or hospice care in line with national averages. In the GSP-driven care pathway, targeted genomic sequencing with a multigene panel was used to direct and refine the optimal treatment pathway. Recent National Comprehensive Cancer Network guidelines have endorsed the examination of eight different genetic alterations to help assess next steps with individual patients.10 This list of genetic alterations and mutation frequencies was used for the basis of the GSP-driven model. The targeted therapy determination or enrollment in a clinical trial was guided by the literature and in consult with key opinion leaders (Table 2). Patients without these alterations are directed toward either nontargeted therapy or hospice care. The percentage of patients sent to hospice care was on the basis of data provided by key opinion leaders who have conducted research on implications of next-generation sequencing.
Table 2.
The Specific Mutations Included and the Associated Clinical Actions
| Gene | Type of genetic alteration | Mutation frequency, % | Base case treatment scenario | Alternative treatment scenario |
|---|---|---|---|---|
| EGFR | Mutation | 11 | Erlotinib, gefitinib, or afatanib | Erlotinib, gefitinib, or afatanib |
| ALK | Translocation | 1 | Crizotinib | Crizotinib |
| BRAF | Mutation | 7 | Trial | Vemurafenib or dabrafenib (off-label use) |
| RET | Translocation | 1 | Trial | Cabozantinib (off-label use) |
| ROS1 | Translocation | 2 | Trial | Crizotinib (off-label use) |
| ERBB2 | Mutation | 1 | Trial | Trastuzumab or afatanib (off-label use) |
| MET | Amplification | 7 | Trial | Crizotinib (off-label use) |
| KRAS | Mutation | 32 | Trial | Trial |
| HRAS | Mutation | 0 | Trial | Trial |
| NRAS | Mutation | 0 | Trial | Trial |
| PIK3CA | Mutation | 4 | Trial | Trial |
Baseline mutation frequencies in lung adenocarcinoma were on the basis of the Cancer Genome Research Network's published data.11
For each pathway, the total cost of treatment was established. The components included cost of drug therapy, drug administration, and adverse events (https://www.mskcc.org/research-areas/programs-centers/health-policy-outcomes/cost-drugs; last accessed October 13, 2015). Nontargeted (chemotherapy) drug costs, adverse event treatment costs, and adverse event rates were derived from published literature.12, 13, 14, 15, 16, 17, 18, 19 Although the adverse event rates for targeted versus nontargeted therapy are different, the treatment costs are the same for each. For each adverse event, the cost is multiplied by the rate to determine an average cost and these are summed to determine the total average adverse event cost. Costs for EGFR and ALK testing are derived from the Medicare fee schedule rates (http://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/ClinicalLabFeeSched/index.html; last accessed October 13, 2015). The cost of a NSCLC-specific GSP panel was obtained from our microcosting analyses described herein.
The model assumes payers will not be required to reimburse investigational or experimental drug costs, 50% of clinical trial patients will be randomized to investigational drug arm, and 50% will be randomized to the control drug arm, and clinical trial adverse event treatment costs will be reimbursed regardless of the trial arm. The hospice care cost was sourced from published literature20 and included hospice services provided during a 6-month period.
To determine the total treatment cost for each pathway, the model multiplies the number of patients directed down a particular pathway by that pathway's average cost. The summation of the treatment costs for these four pathways amounts to the total treatment cost for the corresponding care. Diagnostic services costs are added in to determine the total cost of care. The tumor model also estimates two clinical outcomes: total number of adverse events and total months of progression-free survival. Progression-free survival data for each of the targeted therapies was derived from published literature.21, 22
Syndromic Sensorineural Hearing Loss of Unknown Etiology (Multigene Panel) and Neurodevelopmental Disorders in Pediatric Cases of Unknown Etiology (Exome Sequencing)
The Health Economic models for sensorineural hearing loss and neurodevelopmental disorders were developed comparing the overall diagnostic cost of a traditional care pathway with that of one guided by results from performing a GSP. Traditional care pathways for sensorineural hearing loss and neurodevelopmental disorders consisted of radiology, electrocardiography, laboratory tests, physical examinations (genetics and neurology) and consultations (eg, ophthalmology), single-gene tests or limited syndrome-specific panels, cytogenetic testing, and chromosomal microarray (CMA) testing.
In the sensorineural hearing loss model, the GSP-guided care pathway begins with a GJB2/GJB6-directed test, because of its high diagnostic yield of 20% and low cost, on the basis of data from key opinion leaders. This is followed by a comprehensive sensorineural hearing loss–specific multigene GSP panel in patients who are GJB2/GJB6 negative. The diagnostic yield of the GSP panel was set to be 20%, on the basis of data from key opinion leaders (this is a model input, and can be updated by model users depending on the panel used at respective laboratories). The model assumes that patients who do not end up with a genetic diagnosis via GSP will be directed to tests that are used in the traditional care pathway described above.
In the neurodevelopmental disorders model, two GSP-guided care pathways were analyzed depending on relative placement of CMA, fragile X, and biochemical testing, and exome sequencing in the care pathway (CMA + fragile X testing as first line, followed by GSP as second line, or GSP as first line, followed by CMA + fragile X tests as second line). There is ongoing debate in the field regarding the diagnostic yields of CMA compared with exome sequencing, and we wanted model users to determine the most appropriate relative placement of GSP, CMA, and fragile X tests, on the basis of the costs and the diagnostic yields at their institutions and the referral patient population.
All possible diagnostic procedures for a typical patient with sensorineural hearing loss or neurodevelopmental disorders were identified to compare the total costs of the traditional and GSP-guided pathways for these two indications. Use data of these tests for these two indications were obtained from published sources or on the basis of clinical experiences by key opinion leaders.6, 23, 24, 25, 26, 27, 28 We supplemented these data with test use data from collaborators at academic medical centers or large health systems.
Reimbursement rates for the diagnostic procedures were derived from the 2014 Medicare Fee Schedule, where available. Some single-gene tests or syndrome-specific panels whose CPT codes were not listed in the Medicare Fee Schedule required using suggested reimbursement rates, which we obtained from Cahaba GBA, a regional Medicare Administrator Contractor. For CMA and some single-gene tests that were not listed in either of these databases, cost inputs were obtained from key opinion leaders or list prices published online. For GSPs, we used the cost values obtained from our microcosting analyses described herein. We realize that these cost values may not reflect the actual costs of diagnostic procedures in pediatric populations, or those that are covered by commercial insurance. To overcome this limitation, functionality was provided for users to add their own cost inputs manually, or allow them to use a multiplier that will automatically multiply Medicare rates by a fixed number.
Test use data were multiplied with unit cost of each test and number of patients in the health plan, and aggregated to calculate the total cost of the diagnostic approaches described above. The final result summary graphically displays the total costs of the two care pathways in each case.
Results
Microcost Analysis
Detailed microcosting analyses were performed on 13 protocols from nine laboratories performing clinical testing for one or more of the five CPT-based procedures. These laboratories represented both small and large academic medical centers and commercial reference laboratories so as to capture the array of testing methods and approaches to the bioinformatic analyses. One challenge in performing cost analyses for methods with multiple technology platforms and assay steps is the difficulty in determining a representative sample. To address this challenge, several laboratories performing clinical testing that met our definition of a representative laboratory were selected. All costs related to performing these procedures, including the direct costs of performing, analyzing, and reporting patient samples, the expense of developing and validating the technical protocols, the development, validation, quality control, and maintenance of the informatics pipelines, data storage, and the institutional overhead, were combined to calculate a total per sample per laboratory test cost. Individual laboratories were deidentified, and the findings were aggregated for comparison (Table 3) and have been made publicly available [http://www.amp.org/committees/economics/NGSPricingProject.cfm (registration required); last accessed November 12, 2015].
Table 3.
GSP Microcost Summary Data
| Protocol |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Procedure | 5-50 gene tumor panel | >50 gene tumor | XLID panel∗ | Hearing loss panel∗ | Hearing loss panel | Exome sequencing | ||||||||
| Variable | Average batch size | 5 | 5 | 6 | 7 | 8 | 6 | 8 | 9 | 8 | 8 | 10 | 8 | 5 |
| Total preanalytics/analytics consumables cost | DNA extraction | 6 | 12 | 10 | 8 | 5 | 10 | 6 | 6 | 5 | 8 | 3 | 8 | 3 |
| Library preparation | 208 | 217 | 182 | 159 | 163 | 477 | 466 | 196 | 158 | 181 | 420 | 276 | 432 | |
| Sequencing | 85 | 92 | 76 | 137 | 180 | 279 | 124 | 365 | 788 | 985 | 315 | 989 | 806 | |
| Total preanalytics/analytics equipment cost | DNA extraction | 0 | 0 | 0 | 0 | 0 | 4 | 10 | 3 | 1 | 0 | 3 | 0 | 10 |
| Library preparation | 3 | 2 | 10 | 1 | 8 | 13 | 2 | 2 | 3 | 9 | 1 | 17 | 2 | |
| Sequencing | 6 | 8 | 7 | 18 | 21 | 109 | 14 | 113 | 102 | 94 | 136 | 104 | 64 | |
| Total preanalytics/analytics labor cost | DNA extraction | 4 | 6 | 13 | 14 | 3 | 10 | 5 | 3 | 1 | 4 | 3 | 4 | 7 |
| Library preparation | 9 | 8 | 23 | 18 | 7 | 30 | 28 | 11 | 12 | 0 | 38 | 22 | 45 | |
| Sequencing | 4 | 20 | 7 | 18 | 2 | 19 | 1 | 5 | 2 | 1 | 5 | 0 | 2 | |
| Total bioinformatics/data analysis/reporting cost | 86 | 243 | 66 | 110 | 131 | 699 | 160 | 66 | 671 | 256 | 163 | 1670 | 659 | |
| Total validation maintenance overhead cost | 287 | 300 | 195 | 198 | 56 | 298 | 99 | 280 | 207 | 354 | 410 | 300 | 398 | |
| Total assay cost, per sample | 699 | 908 | 589 | 682 | 578 | 1948 | 914 | 1048 | 1949 | 1890 | 1499 | 3388 | 2428 | |
GSP, genomic sequencing procedure; XLID, X-linked intellectual disability.
All costs reported in US dollars. 0 values indicate costs <$1.
As part of a consolidated genetic panel workflow.
For targeted genomic sequence analysis of DNA from solid tumor specimens, the results from five representative laboratories fulfilling the criteria for CPT code 81445 demonstrated costs ranging from $577.99 to $907.82 (Table 3). Only one laboratory participated that fulfilled criteria for CPT code 81455, a tumor panel with >50 genes ($1948). Cost varied with platform, investment in laboratory-developed or commercial bioinformatics, and validation expenses. Assays were mostly on the basis of commercial hotspot mutation panels [from Thermo Fisher Scientific (Waltham, MA) or Illumina (San Diego, CA)], and methods did not typically include duplication/deletion, copy number variation, or translocation testing. Some laboratories performed paired normal tissue testing for germline mutation determination, but those costs were not included in this analysis.
Cost data from two laboratories performing hearing loss genomic sequence analysis that fulfilled the criteria for CPT code 81430 were calculated ($1048 and $1949) (Table 3). Their gene lists had a similar set of genes, and the largest variance in cost was because of the bioinformatic analysis and clinical interpretation by group review. Notably, duplication/deletion testing was typically assessed via another technology (microarray, PCR, or fluorescence in situ hybridization) and, therefore, was not included in microcosting. Cost analyses of complementary assays were not performed.
Costs for exome sequence analysis (81415, single exome) ranged from $1499.32 to $3388.18, with data aggregated from three participating laboratories (Table 3). The laboratory with the lowest cost was performing a medical or clinical exome analysis, whereas the other two were evaluating the full exome. Cost variability was observed for the technical sequencing and variant interpretation even with the same platform and method and appeared to be related to the extent of subsequent analysis (eg, through group review and interpretation versus individual laboratory director review and sign out).
Health Economic Models
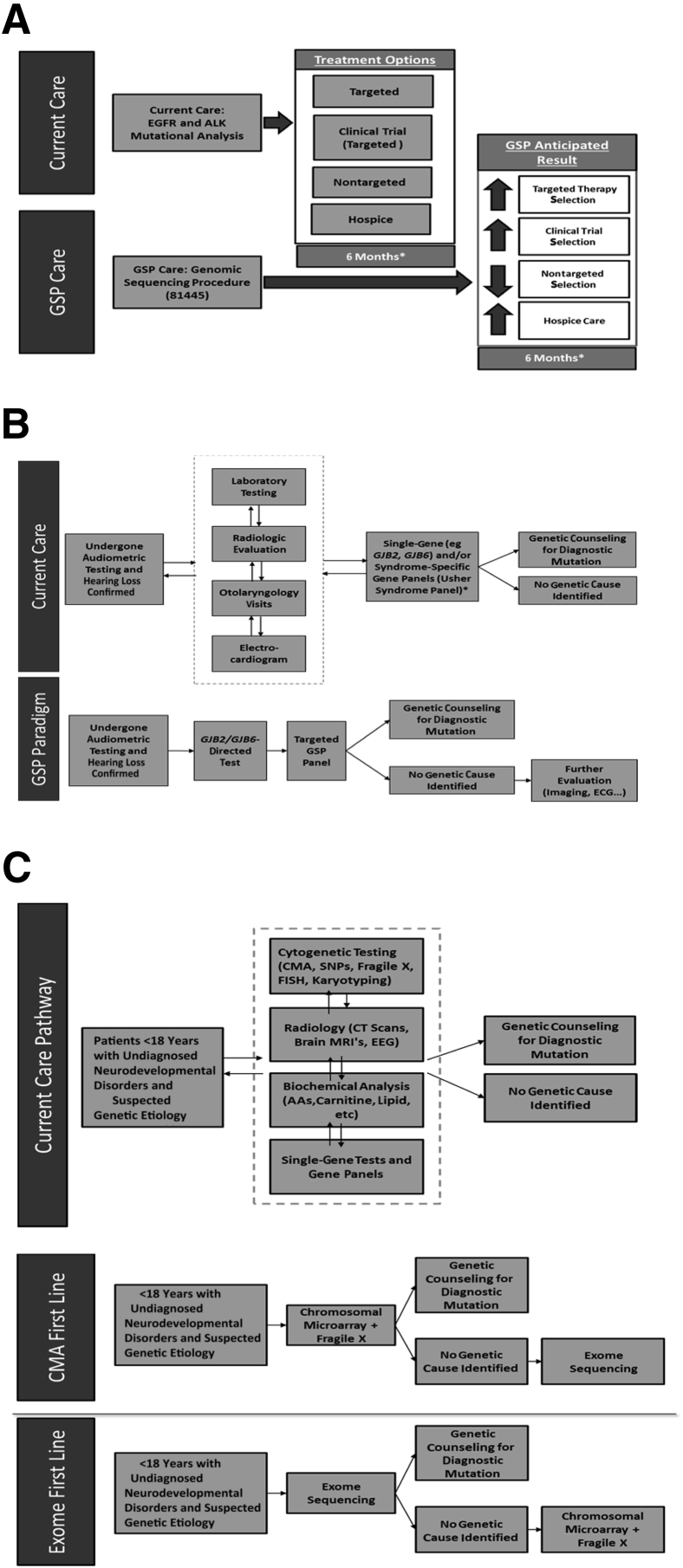
Three independent payer cost-impact analyses were conducted, modeling the overall cost of the current mix of treatments/interventions and the expected mix after the introduction of the new GSPs (Figure 1). All of the models were reviewed by key opinion leaders to objectively assess and validate the clinical and economic impact of these procedures.
Figure 1.
Three independent payer cost-impact analyses. The models present the overall cost of current mix of treatments/interventions and the expected mix after the introduction of the new genomic sequencing procedures (GSPs). A–C: Budget impact models. A: Care pathways for non–small-cell lung cancer (NSCLC). The NSCLC model compares the use of nontargeted therapies, targeted therapies, clinical trials, and hospice care in a care pathway with and without the use of a GSP. B: Care pathways for sensorineural hearing loss. The hearing loss model compares a current care pathway involving an array of laboratory tests and diagnostic procedures, followed by single-gene tests, with an approach where a GSP is used after initial GJB2/GJB6 testing to comprehensively evaluate genetic causes, thus guiding further evaluation efforts. C: Care pathways for neurodevelopmental disorders in patients <18 years. The exome sequencing model compares the use of an array of cytogenetic, radiological, and laboratory/genetic tests with an exome sequencing approach [used in combination with chromosomal microarray (CMA) and fragile X testing]. CT, computed tomography; EEG, electroencephalography; FISH, fluorescence in situ hybridization; MRI, magnetic resonance imaging; SNP, single-nucleotide polymorphism.
Budget Impact of Adopting GSP in Tumor Profiling in NSCLC
Many laboratories currently use multiple platforms or approaches to optimize detection of mutations, copy number alterations, and translocations. In our health economic model, we included the alterations listed in Table 2.
The results of this impact analysis in the care of patients with advanced-stage NSCLC suggest that the use of GSPs results in a theoretical increase in the use of targeted therapy (6% to 13%). This was complemented by a dramatic decrease in use of nontargeted therapy (83% to 20%). This results in a decrease in the total number of adverse events from 207 to 138 (for a plan size of 1 million members). In addition, identification of investigational genetic alterations greatly increases the percentage of patients who could be enrolled in clinical trials from 4% to 54%. Finally, genomic profiling also helped with physicians' and patients' decisions with regard to hospice care for those patients lacking targetable variants, increasing the percentage of patients who entered hospice care from 7% to 13%.
From a cost perspective for a health plan with 1 million covered lives, the cost of targeted therapy increases by the largest amount ($1.1 million to $2.3 million), but the cost of nontargeted therapy decreases by approximately $6.6 million ($8.8 million to $2.2 million). The cost of clinical trials and hospice care also increase by $2.7 million and $60,000, respectively. These increases are welcome, however, because they are less costly than more expensive regimen-based treatments. In sum, the total cost of treatment decreases by $2.7 million (from $10.2 million to $7.5 million). The total cost of genetic testing increases by $0.13 million, when the cost of a GSP is set to $700, which is the average cost of a 5-50 gene tumor panel, according to our microcosting analysis. Moreover, there are additional clinical benefits associated with this care pathway, with a net increase in average progression-free survival and decrease in total number of adverse events.
Budget Impact Model of Adopting GSP in the Diagnosis of Sensorineural Hearing Loss
The goal of this model was to evaluate the clinical and economic (direct cost to a US payer) impact of adopting a GSP in the diagnosis of sensorineural hearing loss of unknown genetic etiology in patients aged <18 years. Earlier care pathways (without the use of GSPs) were compared with implementing GSPs in the algorithm for evaluating patients (Figure 1B). Medicare reimbursement rates were not available for some molecular pathology procedure codes for germline testing, and institutional list prices were used. The results of this analysis revealed both an increase in diagnostic yield and cost savings. For a plan size of 1 million members, a cost savings of $0.24 million and an increase in diagnostic yield from 25% to 36% were demonstrated on incorporation of GSPs into the diagnostic approach, using an average cost of $1499, as per our microcosting analysis. The diagnostic yield of hearing loss GSP was assumed to be 20%. We also used the minimum and maximum cost of hearing loss GSP from our microcosting analysis in the budget-impact model. At a GSP cost of $1048 (minimum), the cost savings from diagnostic workup increased to $0.32 million; and at a GSP cost of $1949 (maximum), the cost savings reduced to $0.16 million.
Budget Impact Model of Adopting Exome Sequencing in Pediatric Neurodevelopmental Disorders
The goal of this model was to evaluate clinical and economic (direct cost to a US payer) impact of incorporating exome sequencing (GSP) in the diagnosis of children with neurodevelopmental disorders of unknown etiology. Traditional care pathways (without the use of exome sequencing) were compared with two alternate approaches, which implement exome sequencing in the algorithm. In one, we compared the traditional pathway with a new one that had 100% of patients receiving CMA and fragile X testing as first line, followed by exome sequencing on patients without a diagnosis. In the other, we compared the traditional pathway with one that had 100% of patients receiving exome sequencing as first line, followed by CMA and fragile X testing as second line on patients without a diagnosis (Figure 1C).
In the first approach with CMA and fragile X testing, followed by exome sequencing, there was a cost savings of $1.33 million for a plan size of 1 million members, and an increase in diagnostic yield from 30% to 40%, using the average cost of exome sequencing determined in this project ($2439). The diagnostic yield of exome sequencing was assumed to be 20%. We also used the minimum and maximum cost of exome sequencing from our microcosting analysis in the budget-impact model. At the exome cost of $1499 (minimum), the cost savings from diagnostic workup increased to $10.1 million, whereas at the exome cost of $3388 (maximum), there was a cost increase of $7.5 million. In the second approach with exome sequencing first, followed by CMA and fragile X testing, there was a cost increase of $0.89 million for a plan size of 1 million members, and an increase in diagnostic yield from 30% to 40%, using the average cost of $2439. At the lowest cost of $1499, there was a cost savings of $10.8 million, whereas at the higher cost of $3388, there was a cost increase of $12.7 million. This analysis suggests that the selective use of exome sequencing can demonstrate possible cost savings.
Discussion
AMP endeavored to gather accurate and transparent cost data for tests fulfilling the criteria for five of the new GSP CPT codes and to construct three payer cost-impact models. Laboratories should be able to use these models to help articulate the cost and value of the GSP services they provide to payers (both Medicare/Medicaid and commercial payers) and colleagues. These models necessarily represent the current landscape in a rapidly evolving field.
The microcosting models demonstrate that there is considerable variability in the costs of GSPs and the depth and breadth of these high-complexity tests offered by various laboratories. This variability reflects the current dynamic nature of the field with multiple platforms, panel designs, analysis pipelines, and practices with regard to results review and reporting. Each laboratory's protocol was unique, and cost comparisons by individual steps were challenging. However, across laboratories, key drivers of higher costs included kit reagents, sequencing equipment, data analysis and reporting, and technical and bioinformatics personnel time. One factor that could reduce cost was the ability to multiplex patients up to a full batch using DNA barcode technology. Institutions using group reviews of the results also tended to have higher personnel costs than the results reviews performed mainly by pipeline and reviewed by individual laboratory directors. We also incorporated validation, maintenance, and overhead costs into the model. This component was challenging to clearly identify and, in some instances, only rough estimates could be provided by the laboratory. List pricing for many of these tests was often two to five times the microcost basis. This is and will continue to be subject to several market forces, including the percentage of nonpayment for test claims and/or institutional overhead variances.
Of particular importance was the development of a tool that laboratories could use to assess their own costs for these or other GSP CPT codes. A blank template similar to the one used in this project is available on the AMP website and includes detailed instructions [http://www.amp.org/committees/economics/NGSPricingProject.cfm (registration required); last accessed November 12, 2015]. A frequently asked questions sheet and instructional video are also available. The template is readily adaptable to the changing procedures and costs involved in providing a variety of GSPs and, over time, can reflect innovations in the technology.
A common theme in discussions regarding advanced technologies in health care is the value of the results of the technology to the beneficiary. To address this issue, three independent health economic analyses were conducted, modeling the overall cost of current mix of treatments or interventions and the anticipated benefit of incorporating GSPs in the diagnosis and treatment determination. For cancer patients, use of GSPs is becoming increasingly important for the identification of potentially targetable genetic changes and the optimization of therapy selection. This technology provides a platform for the simultaneous identification of multiple genes or genetic regions known to harbor tumor hotspot mutations. This approach is especially useful in cases where limited material is available that is not sufficient for performing multiple analyses; it can improve both the diagnostic yield and time to result. Many molecular pathologists and oncologists believe that genomic profiling of a patient's tumor should allow more informed therapy decisions for several tumor types, including lung cancer and melanoma. In our cost-impact analysis, we chose to focus on metastatic NSCLC, where recent National Comprehensive Cancer Network guidelines support the use of multigene panels. We presumed patients with potentially targetable alterations in these genes would be offered the opportunity to enroll in clinical trials. Compared with EGFR and ALK single-gene tests, the use of a multigene GSP that included these emerging biomarkers in conjunction with clinical trial participation reduces costs for the payer and resulted in a reduction in adverse events and improved progression-free survival. This cost reduction is related to the fact that payers are not expected to pay for the use of investigational drugs. If we used the presumption of off-label use of US Food and Drug Administration–approved targeted therapies for other tumor types, the model shows a modest increase in cost related to increased use of targeted therapies. This cost increase was offset by improved outcomes, such as a reduced incidence of adverse events and improved progression-free survival. It is likely that institutions use a combination of both strategies. Individual institutions can adjust these models to fit their own patient populations and group practices.
There is an evolving body of evidence to support the cost-effectiveness of GSPs in the evaluation of heterogeneous cancers. Li et al29 reported the cost-effectiveness of sequencing 34 cancer-associated genes as an aid to treatment selection in patients with metastatic melanoma, resulting in an annual saving of $79.6 million (and a gain of 155 quality-adjusted life years) if their base case were applied to the 8900 patients diagnosed with metastatic melanoma each year in the United States. In conjunction with the 2015 American Society of Clinical Oncology meeting, Intermountain Health presented a study of an internal retrospective health economic analysis of 72 patients, 36 of whom received treatment matched to a GSP molecular profile and 36 matched-control patients who received chemotherapy.30 The progression-free survival was 22.9 weeks for the precision medicine treatment group and 12 weeks for the standard chemotherapy control group. Costs per week were $3204 per week in the targeted group and $3501 in the control cohort. In addition, the costs of treatment-related morbidities were significantly lower for patients receiving GSP-based care compared with standard chemotherapy approaches.
Many payers and some providers have questioned the value of a diagnosis for patients with a genetic disorder of unknown or heterogeneous etiology. We did not attempt to solve this question herein, although the American College of Medical Genetics recently issued a position statement addressing the clinical utility of a diagnosis for patients.31 Rather, we focused on current clinical practice for what is typically referred to as a diagnostic odyssey case and attempted to determine the cost savings and impact of current clinical practice versus the use of a GSP. In both hereditary disease models, the GSP care path would appear to provide an efficient and economical approach to arriving at a definitive diagnosis and thus serve to save significant health care dollars. Williams et al32 reported that in Geisinger's experience, if more than three single genes are considered for analysis, even more complex whole-genome sequencing becomes an economically viable alternative, even when confirmatory testing costs are included, suggesting the economic viability of exome sequencing. Shashi et al33 propose that a 50% success rate for GSPs in undiagnosed genetic disorders could result in a higher rate of genetic diagnosis and a considerable cost savings, especially if used after the initial clinical visit. Soden et al34 evaluated 100 families with 119 children affected by neurodevelopmental disorders with various levels of acuity. With >1000 loci involved in the etiology of neurodevelopmental disorders, whole-genome or whole-exome sequencing was the preferred diagnostic modality. Diagnostic yields ranged from 73% in acutely ill children to 40% in nonacute cases when parent-child trios were sequenced. The cost of prior negative tests in nonacute patients averaged $19,100 per family and if GSPs were performed at symptom onset, diagnoses may have been made 77 months earlier than occurred in the study. As with the model presented herein, this study suggests that cost savings because of the use of GSPs is sensitive to the cost of the procedure and at which point in the diagnostic algorithm it is incorporated.
We anticipate that sequencing technology and costs will change with time and that modeling the cost-impact will only become more refined as we gain more knowledge about genetic changes and therapeutic options. We hope that laboratories will use these templates to assess their individual costs, to consider the value structure in their own patient populations, and to contribute their data to the ongoing dialogue regarding the impact of GSPs on improving patient care.
Acknowledgments
We thank Marc Williams (Geisinger Health System), Wendy Chung (Columbia University), Lisa Schimmenti (University of Minnesota), Katherine Phillips (University of California San Francisco), Heidi Rehm (Partners HealthCare), Douglas Johnson (Vanderbilt University), and Leon Cossler (Albany College of Pharmacy and Health Sciences) for their valuable input and expertise and for sharing their experiences and data from their practices. We also thank the institutions, laboratories, laboratory directors, and staff for sharing their confidential protocols, pipeline development, price information, and time studies. We especially thank the laboratories that allowed site visits and interviews.
Footnotes
Supported in part by unrestricted donations from Agilent Technologies, BD Biosciences, BioReference Laboratories, and Roche.
Disclosures: C.M., D.P., and S.D. of Boston Healthcare Associates were contracted by the Association for Molecular Pathology to perform this study. K.T. of Tynan Consulting was retained as project manager by the Association for Molecular Pathology.
The Genomic Sequencing Procedures Pricing Project Oversight Committee is a Working Group of the Association for Molecular Pathology Economic Affairs Committee. The Genomic Sequencing Procedures Pricing Project Oversight Committee members consisted of Linda M. Sabatini (Chair), Aaron D. Bossler, Jill Hagenkord, Madhuri Hegde, Janina Longtine, Ester Stein, Katherine Tynan (Project Manager), and Vivianna Van Deerlin. The 2014 and 2015 Economic Affairs Committee consisted of Dara Aisner, Aaron D. Bossler (2015 Chair), Samuel Caughron, Pranil Chandra, Jill Hagenkord, Elaine Lyon, Jan Nowak (2014 Cochair), Richard Press, Linda Sabatini, Michele Schoonmaker, and Ester Stein.
Disclaimer
Standard of practice is not being defined, and there may be alternatives. The Association for Molecular Pathology (AMP) makes no warranty, express or implied, regarding the Report and specifically excludes any warranties of merchantability and fitness for a particular use or purpose. AMP shall not be liable for direct, indirect, special, incidental, or consequential damages related to the use of the information contained herein.
References
- 1.Schrijver I., Aziz N., Farkas D.H., Furtado M., Gonzalez A.F., Greiner T.C., Grody W.W., Hambuch T., Kalman L., Kant J.A., Klein R.D., Leonard D.G., Lubin I.M., Mao R., Nagan N., Pratt V.M., Sobel M.E., Voelkerding K.V., Gibson J.S. Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the Association for Molecular Pathology. J Mol Diagn. 2012;14:525–540. doi: 10.1016/j.jmoldx.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankala A., da Silva C., Gualandi F., Ferlini A., Bean L.J., Collins C., Tanner A.K., Hegde M.R. A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann Neurol. 2015;77:206–214. doi: 10.1002/ana.24303. [DOI] [PubMed] [Google Scholar]
- 3.Sholl L.M., Aisner D.L., Varella-Garcia M., Berry L.D., Dias-Santagata D., Wistuba I.I., Chen H., Fujimoto J., Kugler K., Franklin W.A., Iafrate A.J., Ladanyi M., Kris M.G., Johnson B.E., Bunn P.A., Minna J.D., Kwiatkowski D.J. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the lung cancer mutation consortium experience. J Thorac Oncol. 2015;10:768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson D.B., Dahlman K.H., Knol J., Gilbert J., Puzanov I., Means-Powell J., Balko J.M., Lovly C.M., Murphy B.A., Goff L.W., Abramson V.G., Crispens M.A., Mayer I.A., Berlin J.D., Horn L., Keedy V.L., Reddy N.M., Arteaga C.L., Sosman J.A., Pao W. Enabling a genetically informed approach to cancer medicine: a retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. Oncologist. 2014;19:616–622. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan S.D., Mauskopf J.A., Augustovski F., Jaime Caro J., Lee K.M., Minchin M., Orlewska E., Penna P., Rodriguez Barrios J.M., Shau W.Y. Budget impact analysis-principles of good practice: report of the ISPOR 2012 Budget Impact Analysis Good Practice II Task Force. Value Health. 2014;17:5–14. doi: 10.1016/j.jval.2013.08.2291. [DOI] [PubMed] [Google Scholar]
- 6.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., Faucett W.A., Feuk L., Friedman J.M., Hamosh A., Jackson L., Kaminsky E.B., Kok K., Krantz I.D., Kuhn R.M., Lee C., Ostell J.M., Rosenberg C., Scherer S.W., Spinner N.B., Stavropoulos D.J., Tepperberg J.H., Thorland E.C., Vermeesch J.R., Waggoner D.J., Watson M.S., Martin C.L., Ledbetter D.H. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchfield B.B., Feldman J.J., Dunbar J.L., Gardner E.N. The severely to profoundly hearing-impaired population in the United States: prevalence estimates and demographics. J Am Acad Audiol. 2001;12:183–189. [PubMed] [Google Scholar]
- 8.Wisnivesky J.P., Yankelevitz D., Henschke C.I. Stage of lung cancer in relation to its size: part 2. Evidence. Chest. 2005;127:1136–1139. doi: 10.1378/chest.127.4.1136. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. Morb Mortal Wkly Rep Surveill Summ. 2014;63:1–21. [PubMed] [Google Scholar]
- 10.Ettinger D.S., Wood D.E., Akerley W., Bazhenova L.A., Borghaei H., Camidge D.R. Non-small cell lung cancer, version 6.2015. J Natl Compr Canc Netw. 2015;13:515–524. doi: 10.6004/jnccn.2015.0071. [DOI] [PubMed] [Google Scholar]
- 11.Collisson E.A., Campbell J.D., Brooks A.N., Berger A.H., Lee W., Chmielecki J. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vera-Llonch M., Weycker D., Glass A., Gao S., Borker R., Barber B., Oster G. Healthcare costs in patients with metastatic lung cancer receiving chemotherapy. BMC Health Serv Res. 2011;11:305. doi: 10.1186/1472-6963-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shepherd F.A., Rodrigues Pereira J., Ciuleanu T., Tan E.H., Hirsh V., Thongprasert S., Campos D., Maoleekoonpiroj S., Smylie M., Martins R., van Kooten M., Dediu M., Findlay B., Tu D., Johnston D., Bezjak A., Clark G., Santabárbara P., Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 14.Solomon B.J., Mok T., Kim D.W., Wu Y.L., Nakagawa K., Mekhail T., Felip E., Cappuzzo F., Paolini J., Usari T., Iyer S., Reisman A., Wilner K.D., Tursi J., Blackhall F. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 15.Hanna N., Shepherd F.A., Fossella F.V., Pereira J.R., De Marinis F., von Pawel J., Gatzemeier U., Tsao T.C., Pless M., Muller T., Lim H.L., Desch C., Szondy K., Gervais R., Shaharyar, Manegold C., Paul S., Paoletti P., Einhorn L., Bunn P.A. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J Clin Oncol. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 16.Stokes M.E., Muehlenbein C.E., Marciniak M.D., Faries D.E., Motabar S., Gillespie T.W., Lipscomb J., Knopf K.B., Buesching D.P. Neutropenia-related costs in patients treated with first-line chemotherapy for advanced non-small cell lung cancer. J Manag Care Pharm. 2009;15:669–682. doi: 10.18553/jmcp.2009.15.8.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuderer N.M., Dale D.C., Crawford J., Cosler L.E., Lyman G.H. Mortality, morbidity, and cost associated with febrile neutropenia in adult cancer patients. Cancer. 2006;106:2258–2266. doi: 10.1002/cncr.21847. [DOI] [PubMed] [Google Scholar]
- 18.Burke T.A., Wisniewski T., Ernst F.R. Resource utilization and costs associated with chemotherapy-induced nausea and vomiting (CINV) following highly or moderately emetogenic chemotherapy administered in the US outpatient hospital setting. Support Care Cancer. 2011;19:131–140. doi: 10.1007/s00520-009-0797-x. [DOI] [PubMed] [Google Scholar]
- 19.Dranitsaris G., Maroun J., Shah A. Severe chemotherapy-induced diarrhea in patients with colorectal cancer: a cost of illness analysis. Support Care Cancer. 2005;13:318–324. doi: 10.1007/s00520-004-0738-7. [DOI] [PubMed] [Google Scholar]
- 20.Brooks G.A., Li L., Uno H., Hassett M.J., Landon B.E., Schrag D. Acute hospital care is the chief driver of regional spending variation in Medicare patients with advanced cancer. Health Aff (Millwood) 2014;33:1793–1800. doi: 10.1377/hlthaff.2014.0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 22.Shaw A.T., Kim D.W., Nakagawa K., Seto T., Crinó L., Ahn M.J., De Pas T., Besse B., Solomon B.J., Blackhall F., Wu Y.L., Thomas M., O'Byrne K.J., Moro-Sibilot D., Camidge D.R., Mok T., Hirsh V., Riely G.J., Iyer S., Tassell V., Polli A., Wilner K.D., Jänne P.A. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 23.Mafong D.D., Shin E.J., Lalwani A.K. Use of laboratory evaluation and radiologic imaging in the diagnostic evaluation of children with sensorineural hearing loss. Laryngoscope. 2002;112:1–7. doi: 10.1097/00005537-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Lin J.W., Chowdhury N., Mody A., Tonini R., Emery C., Haymond J., Oghalai J.S. Comprehensive diagnostic battery for evaluating sensorineural hearing loss in children. Otol Neurotol. 2011;32:259–264. doi: 10.1097/MAO.0b013e31820160fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preciado D.A., Lawson L., Madden C., Myer D., Ngo C., Bradshaw J.K., Choo D.I., Greinwald J.H. Improved diagnostic effectiveness with a sequential diagnostic paradigm in idiopathic pediatric sensorineural hearing loss. Otol Neurotol. 2005;26:610–615. doi: 10.1097/01.mao.0000178133.89353.1d. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A., Ruscetta M.N., Chi D.H. Ophthalmologic findings in children with sensorineural hearing loss. Arch Otolaryngol Head Neck Surg. 2009;135:119–123. doi: 10.1001/archoto.2008.546. [DOI] [PubMed] [Google Scholar]
- 27.Joint Committee on Infant Hearing Year 2007 position statement: principles and guidelines for early hearing detection and intervention programs. Pediatrics. 2007;120:898–921. doi: 10.1542/peds.2007-2333. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer G.B., Mendelsohn N.J. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Bare L.A., Bender R.A., Sninsky J.J., Wilson L.S., Devlin J.J., Waldman F.M. Cost effectiveness of sequencing 34 cancer-associated genes as an aid for treatment selection in patients with metastatic melanoma. Mol Diagn Ther. 2015;19:169–177. doi: 10.1007/s40291-015-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadauld L., Van Norman S.B., Fulde G., Dermott J.G., Newman D., Butler A.M., Tudor B.P., Gilbert H., Yin Lin K., Stone G., Ford J.M., Haslem D.S. Precision medicine to improve survival without increasing costs in advanced cancer patients. American Society of Clinical Oncology Meeting. J Clin Oncol. 2015;33 Suppl Abstract e17641. [Google Scholar]
- 31.ACMG Board of Directors Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015;17:505–507. doi: 10.1038/gim.2015.41. [DOI] [PubMed] [Google Scholar]
- 32.Williams M, Bock J, Faucett A, Smith-Packard B, Wagner M, Williams J: Quantification of the diagnostic odyssey in patients referred for whole genome sequencing. 2013 American College of Medical Genetics and Genomics Annual Clinical Genetics Meeting. 2013, Abstract 260.
- 33.Shashi V., McConkie-Rosell A., Rosell B., Schoch K., Vellore K., McDonald M., Jiang Y.H., Xie P., Need A., Goldstein D.B., Goldstein D.G. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 34.Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L., Twist G., Noll A., Heese B.A., Zellmer L., Atherton A.M., Abdelmoity A.T., Safina N., Nyp S.S., Zuccarelli B., Larson I.A., Modrcin A., Herd S., Creed M., Ye Z., Yuan X., Brodsky R.A., Kingsmore S.F. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]