Abstract
Analyzing RNA-protein complexes is central to the understanding of the molecular circuitry governing cellular processes. In the last years, several proteome-wide studies were dedicated to the identification of RNA-binding proteins. Here, we describe in detail R-DeeP, an approach built on RNA dependence, defined as the ability of a protein to engage in protein complexes only in presence of RNA, with direct or indirect interaction with RNA. This approach provides for the first time quantitative information on the fraction of a protein associated with RNA-protein complexes. R-DeeP is also independent of any potentially biased purification procedures. It is based on cellular lysate fractionation by density gradient ultracentrifugation and subsequent analysis by proteome-wide mass spectrometry or by individual western blotting. The comparison of lysates with and without previous RNase treatment allows the identification of differences in the apparent molecular weight, and hence the size of the complexes. In combination with information from databases of protein-protein complexes, R-DeeP allows the computational reconstruction of protein complexes from proteins migrating in the same fraction. In addition, we computed a pipeline for the statistical analysis of the mass spectrometry dataset to automatically identify RNA-dependent proteins, i.e. proteins whose interactome depends on RNA. With the provided protocol, the individual analysis of selected proteins of interest by western blot can be completed within one to two weeks. For proteome-wide studies, additional time is needed for the integration of the proteomic and statistical analysis. In the future, R-DeeP can also be extended to other fractionation techniques like chromatography.
Keywords: RNA dependence, RNA-protein interaction, RNA-protein complex, RNA-binding protein, ultracentrifugation, density gradient, mass spectrometry, proteome-wide, statistical analysis, quantitative analysis, R-DeeP
Introduction
RNA is a versatile molecule that not only serves as a template for the production of proteins but also associates with proteins to form RNA-protein complexes which play key regulatory roles in various cellular processes, including RNA metabolism and the regulation of gene expression1. Hence, the specific identification of RNA-protein complexes poses a highly relevant scientific challenge of broad interest. Within this context, the importance of RNA-binding proteins (RBPs) has been recently recognized, leading to the development of several strategies to comprehensively identify the cellular pool of RBPs.
RBPs have been compiled by manual curation of the literature for known RBPs2,3 or by the analysis of conserved, canonical RNA-binding domains4. Experimental approaches emerged based on the affinity purification of polyadenylated (poly(A)) RNA species, like study of the mRNA-bound proteome5, RIC6–9 (RNA interactome capture) or serIC10 (serial interactome capture). These studies revealed that RBPs do not necessarily contain conserved RNA-binding domains. Interestingly, RBPs often contain intrinsically disordered regions, which can also bind to RNA6. Since poly(A) pulldown approaches would not identify RPBs binding exclusively to non-polyadenylated RNA species, alternative strategies like RBR-ID11 (identification of RNA-binding regions), RICK12 (RNA interactome using click chemistry) and CARIC13 (click chemistry-assisted RNA interactome capture) were developed, including the use of modified nucleotides combined with UV crosslinking of the interacting proteins. More recently, approaches based on protease digestion allowed the characterization and precise mapping of RNA-binding domains14,15. Novel methods like XRNAX16 (protein-crosslinked RNA extraction), OOPS17 (orthogonal organic phase separation) and PTex18 (phenol toluol extraction) emerged using phenol-chloroform extraction19 for separation of cellular components with different physicochemical properties. Orthogonal to these experimental approaches, in silico methodologies like SONAR20 (support vector machine obtained from neighborhood associated RBPs) successfully supported the identification of new RBPs in various species.
Motivated by the aim of developing a proteome-wide approach, quantitative and free of a potential enrichment bias, we developed a specific method, orthogonal to the established approaches we described above2–18,20–23. Our approach is based on the concept of RNA dependence, which defines a protein as RNA dependent if its interactome and hence probably its function depends on the presence of RNA. In other words, RNA-dependent proteins are proteins engaged in RNA-dependent interactions. RNA-dependent proteins differ from the more strictly defined RBPs as RNA-dependent proteins comprise both proteins interacting directly (RBPs) and indirectly (RBP-interacting proteins) with RNA. The concept of RNA dependence can be directly translated into a screen to quantitatively, specifically, automatically and without enrichment bias identify RNA-dependent proteins, which we called R-DeeP24. It does not rely (i) on the incorporation of modified nucleotides, (ii) nor on the efficiency of RNA-protein crosslinking through UV light, (iii) nor on the efficiency of affinity purification using antisense oligonucleotides or specific antibodies, (iv) nor on any distinct physicochemical differences between RBPs and non-RBPs for purification. Instead, R-DeeP takes advantage of the separation of protein-protein (RNA-independent) and RNA-protein-protein (RNA-dependent) complexes on sucrose density gradients to identify both the RNA-independent and RNA-dependent proteins, as well as the fraction of a protein that is dependent on RNA. Subsequent reconstitution of RNA-dependent and RNA-independent complexes can be achieved by integrating information from protein-protein interactions25 or protein-protein complex26 databases with our data24. Briefly, lysates are fractionated after ultracentrifugation of sucrose density gradients to compare untreated lysates to lysates pre-treated with a cocktail of RNases. The RNA dependence of proteins or their complexes can then be detected by a shift in their apparent molecular weight in the gradient. The present protocol describes the R-DeeP approach step by step from the cell lysis, gradient generation, fractionation to the proteomic or protein-specific detection and the bioinformatical statistical analysis for proteome-wide R-DeeP studies to automatically extract relevant information from the mass spectrometry (MS) results (Fig. 1). R-DeeP significantly identified more than 500 human RNA-dependent proteins, which had not been previously linked to RNA24.
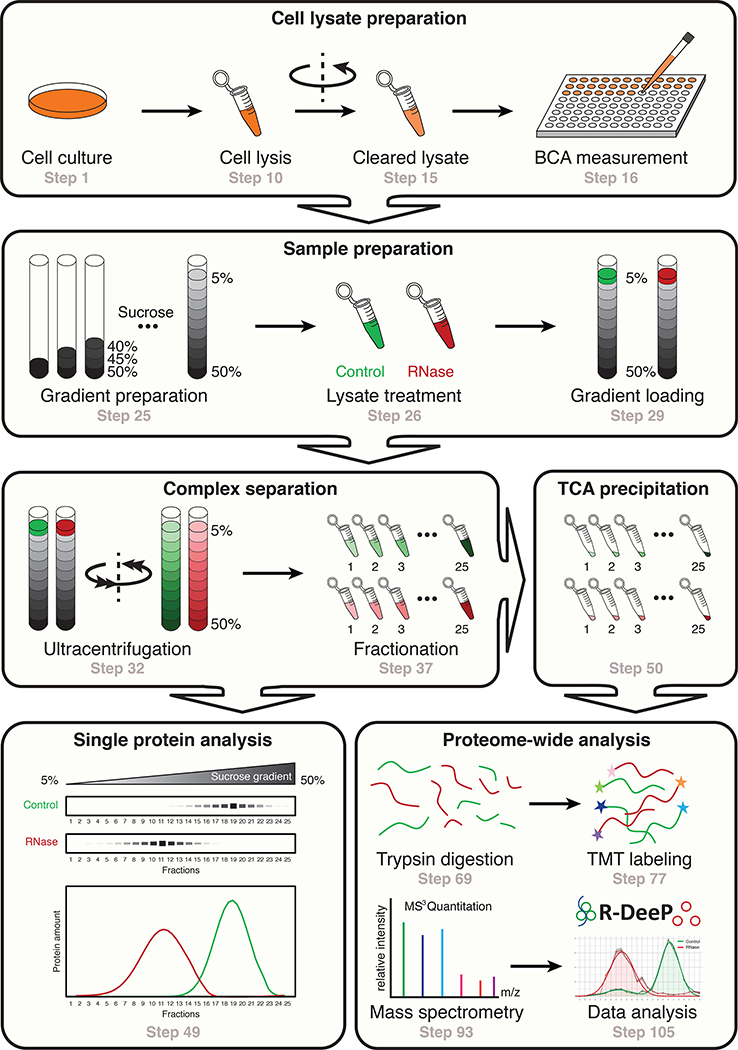
Fig. 1 |. Overview of the R-Deep procedure.
R-Deep is a method to analyze the RNA dependence of protein complexes. First, cells are lysed and lysates are kept untreated (Control) or treated (RNase) with an RNase cocktail (steps 1–26). Second, the protein complexes are separated according to their density in a sucrose density gradient and further fractionated after ultracentrifugation (steps 27–37). The obtained fractions can finally be analyzed at the single protein level (western blot, steps 38–49) or at the proteome-wide level (mass spectrometry, steps 50–105).
Together with the R-DeeP screen, we have compiled the results from 19 different publications - original studies as well as compendia and databases identifying mammalian RBPs proteome-wide - in one central resource available at http://R-DeeP.dkfz.de. This platform offers various search options and can be used for example to search for specific proteins or protein lists. It also indicates in which studies proteins have been identified as RNA-binding and integrate the dataset of a protein-protein complexes database26 to help investigating interaction partners of protein of interest.
Applications of the method
Methods based on size fractionation27 or density gradient ultracentrifugation28–32 have been intensively used in the past to investigate various cellular components and protein-protein complexes. Here, we extend the use of density gradient ultracentrifugation to develop a proteome-wide approach, yet maintaining the option of single protein analysis by western blot if needed.
The R-DeeP approach is highly versatile for individual proteins or proteome-wide studies and can be applied to virtually any species or model system, as long as a cellular/tissue protein extract can be prepared and loaded on the top of the gradient. Since it does not require the incorporation of modified nucleotides, the method is also suitable for cell types which are difficult to transfect or even for application to primary cells or primary tissue samples.
Although we present here its application to human whole cell lysate, alternatively, cells could be fractionated33 and lysates of specific cell compartments (e.g. cytoplasm, cell nucleus, chromatin) could be instead loaded on the sucrose density gradients for further and deeper single proteomic or proteome-wide analysis.
In addition, prior to protein lysate preparation, cells could be pre-treated e.g. with drugs, inhibitors or small molecules that should be tested for their effect on protein-RNA interactions. As an alternative to the present protocol, a single RNase digest could be used instead of a cocktail of different enzymes in order to precisely differentiate and target specific RNA species (e.g. single-stranded, double-stranded or DNA-RNA hybrids). Similarly, a treatment with siRNA could be taken into consideration to target a specific RNA transcript or a specific RNA sequence (e.g. a repetitive sequence).
In its individual protein analysis version, the protocol could be applied to the analysis of a single protein of interest, in its wild-type state as compared to mutant variants (e.g. protein with mutations in the putative RNA-binding site). The R-DeeP analysis is suitable for endogenous protein expression as well as exogenous overexpression of proteins after transfection.
While R-DeeP was developed for the analysis of RNA-dependent proteins (whose interactome is dependent on interactions with RNA), the protocol could be as well applied to investigate DNA-dependent proteins (whose interactome is dependent on interactions with DNA). In principle, treatment with RNases can be omitted and replaced by treatment with DNase I in the treated samples, as compared to an untreated control sample. Alternatively, an RNase-treated lysate could be further treated or not with DNase I to test the DNA dependence of proteins, at the single protein or proteome-wide level. For such an experiment, it would be preferable to work with isolated cell nuclei as starting material.
Alternatively, the R-DeeP approach could be used completely independently of the RNA or DNA dependence concepts, simply to investigate changes and modifications of the interaction state of proteins depending on differences in cellular conditions, which can range from different cell cycle states to various pre-treatments of the cells (starvation, drug treatment, inhibitors of specific cellular activities, etc.). Any condition that would induce a shift of protein(s) in the sucrose density gradient would reveal changes in the interactome of the protein(s).
Thus, this protocol enables the broad application of R-DeeP for individual proteins or the entire proteome across species and for a plethora of different research questions comparing RNA-protein complexes in all types of developmental, physiological or pathological states. We anticipate that this protocol will foster the detection of a number of non-expected protein-RNA interactions, and pave the way to the discovery of new RNA functions in the cell.
Comparison with other methods and limitations
The first methods established to identify RBPs on a proteome-wide level were based on the affinity purification of proteins associated to poly(A) RNA species after UV crosslinking at 254 nm of the RNAs and proteins (RIC, serIC)5,6,10. Alternatively, RNA molecules were labeled in the cell with modified nucleotides like 4-SU (4-thiouridine) prior to photo-crosslinking at 365 nm, a method also called PAR-CLIP34 (photoactivable-ribonucleoside-enhanced crosslinking). These methods have been successfully applied to different species including human6, mouse7 and Drosophila35. However, such approaches strongly rely on efficient crosslinking as well as efficient and specific affinity purification. Moreover, they are suitable for but limited to poly(A) RNA transcripts, whereas a number of noncoding RNA species, emerging as potentially important regulatory molecules, are not polyadenylated36. Therefore, RBPs interacting with noncoding RNAs are excluded from the analysis. To overcome such limitations, strategies emerged taking advantage of the incorporation of 5-ethynyluridine (EU) and click chemistry to attach biotin to EU-labeled RNAs and pulldown associated proteins after 254 nm UV crosslinking using streptavidin-coated beads (RICK)12. In addition to EU treatment, cells can also be incubated with 4-SU before crosslinking at 365 nm (CARIC)13. While such approaches allow in theory the analysis of RBPs interacting with all RNA classes, the studies are dependent on the efficient transcription-dependent incorporation of the modified nucleotides and are only applicable to suitable systems as the nucleotides are usually added to the cell culture medium and diffuse into the cell. This excludes for example, studies in bacterial systems.
More recently, new methods have been developed based on phenol-chloroform extraction to separate cellular components with different physicochemical properties16–18. Such approaches rely on the migration of the UV-crosslinked RNA-bound proteins to the interphase between the aqueous phase (at the top) and the organic phase (at the bottom) following centrifugation. Therefore, these protocols require efficient UV-crosslinking and efficient phase separation to specifically purify the fragile interphase.
All methods established so far to investigate RBPs have their advantages and limitations. Therefore, complementary and orthogonal techniques that do not rely on UV light crosslinking or affinity purification efficiencies will substantially support the discovery and specificity of new RNA-dependent proteins, either by confirming the ability of the protein to bind RNA or by revealing their previously unknown relation to RNA. R-DeeP is unique in two of its properties: first, it does not require any enrichment procedure and hence is independent of differences in affinity or physicochemical properties of the cellular components. It is also independent of the RNA and protein sequences of the binding sites involved in the interactions, which can also affect the efficiency of other methods. Second, it provides quantitative information on the fraction of each RNA-dependent protein that is actually bound to RNA. This includes important quantitative information for known RBPs, as shown for the transcription factor CTCF for which RNA was revealed as a key factor for the recruitment of CTCF to the chromatin24. Additionally, R-DeeP allows the estimation of the size of the complex and the reconstruction of RNA-dependently shifting complexes. A comprehensive comparison of the results of the various techniques in mammalian systems can be taken from the original R-DeeP publication24 and is available as an RBP resource online through the R-DeeP Database (http://R-DeeP.dkfz.de).
The time needed to complete our protocol is comparable to the time needed for example to complete the RIC protocol6,37,38. Similarly, the analysis of the results at the individual protein level by western blotting can be achieved in a couple of days, while a proteome-wide analysis involving a quantitative analysis of MS measurements will take approximatively two weeks.
Limitations of R-DeeP are that the approach does not identify the specific binding site of the RNA on the protein and it does not differentiate between direct and indirect interactions with RNA. But once the protein of interest has been identified, R-DeeP can easily be combined with orthogonal methods which include this property like for example crosslinking and immunoprecipitation (CLIP) followed by T4 polynucleotide kinase (PNK) radioactive assay24. In addition, R-DeeP relies on the preparation of protein extract using detergent-containing lysis buffers. In such conditions, artificial interactions between component of different cellular compartments could occur, which would not exist in intact cells. Although such interactions cannot be excluded, there is so far no indication that this would be the case since these would need to be dependent on RNA in order to give rise to a false-positive shift. Indeed, the reconstitution through R-DeeP of RNA-dependent and RNA-independent control protein-protein complexes returned the expected co-segregation of the protein subunits26. Since R-DeeP is based on ultracentrifugation of cell lysate on a sucrose density gradient, weak or short-lived interactions could be lost during the 18 h-long centrifugation step. This could lead to enrichment in strong interactions.
Experimental design
Here, we describe a set of techniques to identify RNA-dependent proteins in a cellular protein lysate. The method is based on the comparison of two conditions, one being the untreated control, and the other one being the RNase-treated sample. After lysate treatment, protein complexes are separated on a sucrose density gradient and will migrate into different fractions of the gradient, depending on the size, shape and density of the protein-protein or RNA-protein-protein complexes. RNA-dependent complexes are expected to dissociate or partially dissociate after RNase treatment, which will result in the migration of the protein subunits into different fractions in the control as compared to the RNase-treated condition. Such so-called “shifts” can subsequently be detected either by individual western blot analysis or by proteome-wide MS-based analysis.
Controls
The success of the analysis relies on the presence of the appropriate control condition for the experiment. Since we assess directly the RNA dependence of proteins by treatment with a cocktail of different RNases, the appropriate control is the untreated condition (optionally treated with RNase inhibitors if RNA degradation would be observed, depending on the sample). For most other applications (such as other model organisms, other cell types, drug or small molecule treatments), the untreated sample will be the suitable control. However, depending on the aim of the experiment, the control may need to be adapted. For example, in case of treatment with siRNA oligonucleotides, the control sample should be treated with a corresponding non-targeting control siRNA. Since R-DeeP potentially has a broad range of applications, it is recommended to thoroughly think about the experimental design before starting with the experiments. Prior to pursuing the protocol in its proteome-wide version, we recommend to perform an individual western blot analysis for selected known RNA-dependent (e.g. HNRNPU, RPS3) and RNA-independent (e.g. ASNS, MCM7) control proteins, for which antibodies are available and already successfully tested for western blot application24.
Cell lysate preparation
For whole cell lysate preparation, starting with e.g. 2 × 107 cells, it is important to efficiently break the cell walls and to recover as many proteins as possible. The procedure is optimized for loading 1 to 2 mg of protein lysate on the gradient. The buffer conditions are adapted to HeLa cells. The amount and type of detergent can be adapted but should not be increased to higher concentrations to avoid denaturation of protein-containing complexes. Therefore, we introduce a step with several freeze/thaw cycles. Alternatively, a dounce homogenizer could be used to open the cells. For the preparation of lysate from various cell compartments, we recommend to follow the previously published protocol33, with which we obtained clean subcellular fractions24.
Gradient preparation
To successfully complete the protocol, it is essential to prepare the sucrose density gradients in a highly reproducible way. Therefore, we recommend preparing gradients for several replicates in one step, using the same sucrose solutions for all the gradients. Any variations in the content of the solutions or in the volume of the sucrose layers will have repercussions on the migration behavior of the protein complexes at the end. Since control and treated conditions are run in different tubes, reproducibility is essential. The freezing procedure that we describe here is convenient and ensures a high reproducibility in the preparation of the gradients (see the Procedure section, Step 25 and the Supplementary Video 1). The use of the −80 °C freezer or liquid nitrogen to freeze the sucrose layers will only affect the time scale of the preparation but not the quality of the gradients. The sucrose solutions for the gradient preparation range from 5% to 50% sucrose. These concentrations can be taken as a starting point, but the protocol can be optimized as needed and the concentrations of the sucrose solutions can be adapted to the needs. For example, a specific sub-range of the gradient (e.g. 15% to 25%) could be specifically prepared and further fractionated into 25 fractions, which would in principle result in a “zoom in” in the gradient and increase the resolution of this specific part of the gradient to help clarify whether a shift is significant or not.
Ultracentrifugation
Ultracentrifugation is one of the critical steps of the protocol. It is important to ensure access to an ultracentrifuge in the vicinity of the lab to avoid transporting the gradients over longer distances or longer time. So far, we have tested the protocol for two types of rotors (SW40 Ti and SW41 Ti) without any visible effect on the distribution of the tested proteins. Potentially, alternative gradients could be used to perform the R-DeeP protocols. If using a different type of rotor, the size of the sucrose layers in the gradient should be adapted as well as the size of the fractions. In order to avoid disturbing the gradient during the centrifugation step, it is important to adjust the settings to low acceleration and low deceleration. For other types of ultracentrifuges than the one described here, the instruction manual for density gradient procedures should be consulted. Since we experienced a reduction of the whole gradient volume due to evaporation in some centrifuge buckets, we recommend testing the material with test gradients (not loaded with any sample) prior to performing the experiment. Marking the level of the gradient in the tube before centrifugation and comparison to its level after centrifugation allows the estimation of evaporation losses. Such buckets should be accordingly labeled and not further used with gradient tubes but rather left empty during the run, keeping in mind to appropriately balance all the buckets in the centrifuge.
Fractionation
The fractionation procedure also belongs to the critical steps of the protocol and can strongly affect the final results. Therefore, it is essential to maintain a high reproducibility of the fractionation procedure. If possible, the same person equipped with the same pipet should proceed to the fractionation of all gradients of one experiment (see the Procedure section, Steps 34–37 and the Supplementary Video 2). Alternatively, commercially automated gradient fractionators are available, but the automated procedure involves an intensive wash of the machine between each gradient and can lead to the complete loss of the gradient in case of leaking incidents, which can occur. We have good experience with fractionating by hand. We recommend to practice the fractionation procedure on a test gradient in advance (which does not need to be centrifuged or kept at 4 °C). We have set the number of fractions to 25, but any other number of fractions can be taken. For the optimization of this number, a certain number of parameters should be taken into account: (i) the separation of the shifting proteins should remain significant (we recommend at least a shift of one fraction), (ii) the number of fractions to handle, which can become demanding throughout the protocol (a set of two conditions in triplicate, with 25 fractions per gradient leads to 150 fractions, which all need to be reproducibly processed through several steps of the protocol).
Western blot analysis
Analysis of individual proteins by western blotting is extremely helpful to establish the R-DeeP protocol and to test control proteins (in our case, RNA-dependent as well as RNA-independent proteins). In order to detect individual proteins by western blot, it is important to load at least 1 mg of protein lysate onto the sucrose density gradient to yield a sufficient concentration in the individual fractions. Prior to the experiment, we highly recommend testing each antibody of interest directly on a cellular lysate of interest, including various concentrations of the primary antibodies and various amounts of protein lysate (e.g. ranging from 20 ng to 5 ng in 20 μl). The user should use the western blot protocol best established in their laboratory, but optimization of the western blot procedure could include overnight transfer and/or overnight incubation with the primary antibody. The use of an enhanced detection reagent (Amersham ECL Prime Western Blotting Detection Reagent, GE Healthcare, cat. no. RPN2232) could also be helpful to improve the quality of the signal for less abundant proteins or weak antibodies.
TCA precipitation and mass spectrometry analysis
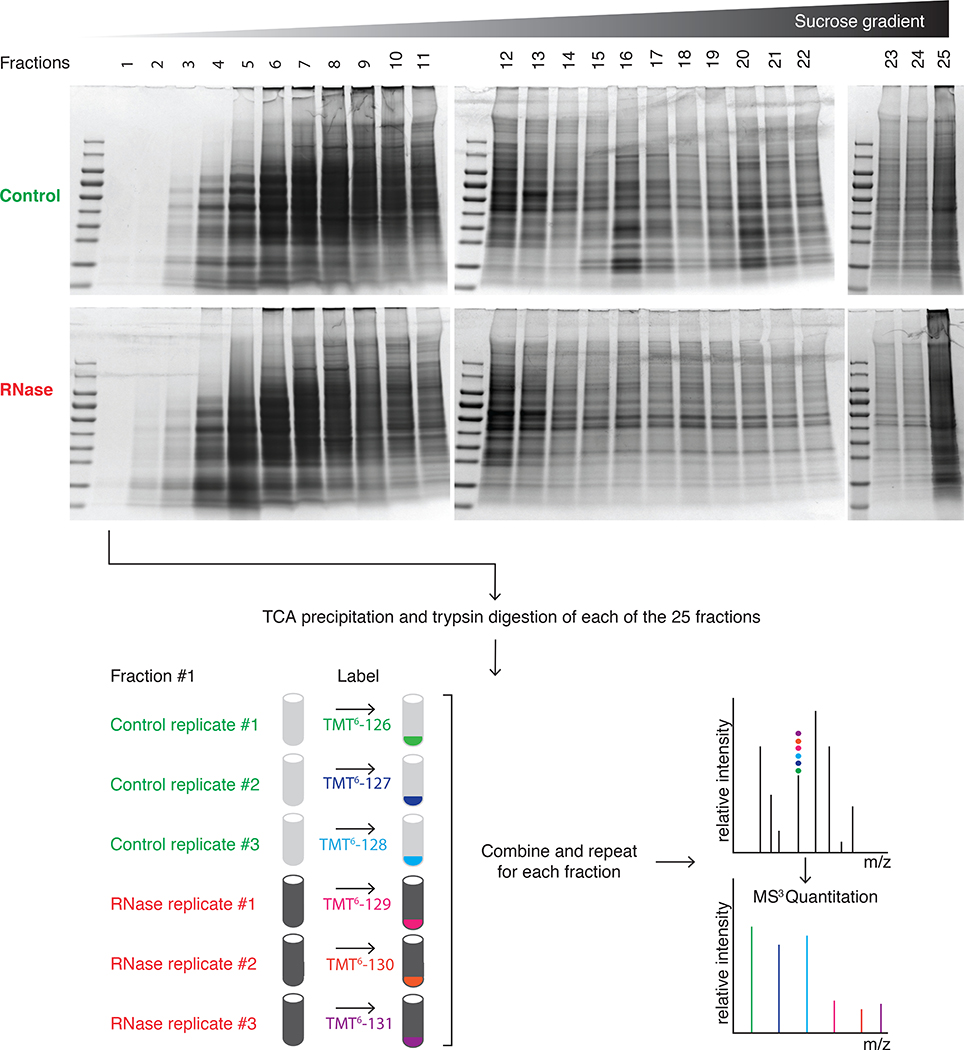
Before protein digestion and TMT labeling for MS, it is essential to fully remove the sucrose solutions. This is achieved by TCA precipitation of the fractions. A checklist for TCA precipitation is provided in Supplementary Table 1 to ensure reproducibility.
Statistical analysis
The statistical analysis procedure performed in this protocol is adapted for the analysis of two conditions in triplicate (3x control treatment and 3x RNase treatment). The pipeline (MS_Statistical_Analysis.R) as provided in the Supplementary Data along with a test dataset (Mass_Spec_RawData_Sample.csv) can be run in RStudio. We recommend the use of RStudio, as it is convenient to visualize the structure of the pipeline. In principle, the pipeline can be applied to the test dataset with only very little previous knowledge in R, provided the user has carefully read the “README.txt” file as provided in the Supplementary Data. For further adaptation of the pipeline to custom-made variants of the protocol, advanced knowledge in R is required. The pipeline offers two functions to visualize the MS data on an individual protein basis. Once loaded into the R environment, the functions can be easily called and will result in a user-friendly graphical representation of the data (see instructions in the “README.txt” file provided in the Supplementary Data along with two examples “HNRPU_HUMAN_FIT.pdf” and “HNRPU_HUMAN_ALL.pdf”).
In brief, the detailed pipeline containing many inline comments to explain each step is divided into seven parts, as follows:
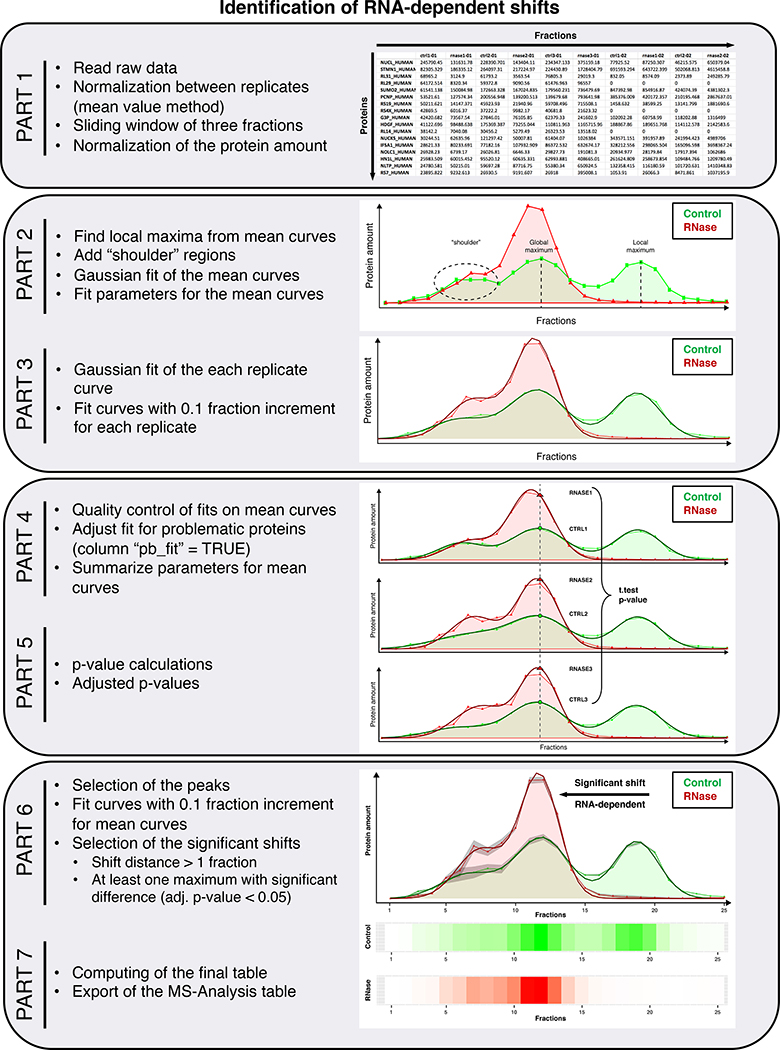
PART 1: a fraction-wise normalization step between the replicates in each condition using the mean value method (median and quantile methods would not work because there are many fractions without protein, value = 0), a moving average of three fractions and a normalization step for each protein to 100 representing the total amount of the protein in all fractions of one replicate.
PART 2: an automatic search for the maxima of the amount of protein and the addition of the so-called shoulder regions, which are not automatically recognized as maxima but cannot be ignored because they still contain a large amount of protein. PART 2 computes the starting parameters (amplitude at the maxima, position/fraction of the maxima and standard deviation) in order to fit all the mean distribution profiles using Gaussian curves and returns for each protein the optimal parameters for each protein profile in the control and RNase-treated conditions (function “optim”, method “BFGS”).
PART 3: a Gaussian fit for each individual replicate, both at the 1 fraction and 0.1 fraction precision.
PART 4: a quality control of the fit with subsequent correction if needed.
PART 5: the assessment of the p-value (Student’s t-test) and adjusted p-values (“FDR” method) for each maximum to evaluate whether the amount of protein in the control sample (in the three replicates) is significantly different from the amount of protein in the RNase-treated sample (in the three replicates) at each maximum respectively.
PART 6: the evaluation of the shifts and subsequent selection of the significant shifts (distance > one fraction and a p-value < 0.05 at one of the maxima in the control or RNase-treated condition).
PART 7: the production and export of a final file for further analysis (an example of such a file can be found as “MS_Analysis_Shifts.csv” as provided in the Supplementary Data) containing information for each protein and specific information for proteins with significant shifts such as:
the average amount of protein in each fraction
the positions of the maxima in the control and RNase samples
the distance between the maxima in the control and the RNase-treated profile
the amount of protein associated with each maximum
the loss and gain of protein at the maxima in the control and RNase-treated protein profile
the significance (FDR-adjusted p-value) of the difference of amplitude between the control and the RNase-treated profile for each maximum
whether the protein presents a right shift (a rare shift toward higher sucrose fractions, that would happen for example when a protein is gaining interaction partners in absence of RNA)
whether the Gaussian fit needed a correction
Significant shifts are detected according to specific selection criteria which include a shift larger than one fraction. To assess the stringency of this criterion, we repeated the analysis with modified parameters allowing shifts for a minimum distance of 0.7 fractions24. This modification added 185 proteins (10%) to the list of 1784 RNA-dependent proteins, showing a minor effect on the final result and demonstrating that the setting of a minimum of one fraction for a shift is a meaningful choice.
Materials
BIOLOGICAL MATERIALS
! CAUTION The authenticity of the cell lines as well as the absence of mycoplasma contamination should be regularly tested.
Cell lines used to prepare lysate: HeLa S3 (ATCC: cat. no. CCL-2.2, RRID: CVCL_0058) and HeLa (ATCC: cat. no. CCL-2, RRID: CVCL_0030) cervical cancer cells and A549 (ATCC: cat. no. CCL-185, RRID:CVCL_0023) lung cancer cells
REAGENTS
▲ CRITICAL Solutions containing organic solvents need to be prepared in glass ware since they can attack plastic material and these substances have an impact on later MS analysis (increasing background signal). In addition, do not use serological pipettes because the labeling will be removed by organic solvent and will be then in the prepared dilution.
▲ CRITICAL Use original tubes and tips from Eppendorf for the preparation of samples for MS - or other plasticware specifically tested for MS analysis. Some plastics are more easily attacked by organic solvents which lead to more background signal in the later MS analysis.
▲ CRITICAL For MS analysis, we recommend to always use ultrapure reagents (purity > 99%) to reduce background signal and contamination as much as possible.
Ham’s F-12K (Kaighn’s) medium (Thermo Fisher Scientific, cat. no. 21127–022)
Dulbecco’s modified Eagle’s medium (DMEM-High Glucose, Sigma-Aldrich, cat. no. D5796)
Fetal bovine serum (FBS), (Thermo Fisher Scientific, cat. no. 10270–106)
Dulbecco’s Phosphate Buffered Saline (PBS), (Sigma-Aldrich, cat. no. D8537)
0.05% Trypsin, EDTA (1x), (Thermo Fisher Scientific, cat. no. 25300)
Bicinchoninic Acid (BCA) solution (Sigma-Aldrich, cat. no. B9643) ! CAUTION BCA is very toxic to aquatic life with long lasting effects, so collect spillage and avoid release to the environment. Dispose of contents and container to an approved waste disposal place.
Pierce BCA Protein Assay Reagent B-25 mL (Thermo Fisher Scientific, cat. no. 23224).
Bovine serum albumin (BSA), (Sigma-Aldrich, cat. no. A1470)
DTT (1.4-Dithiothreitol), (Roche, cat. no. 10197777001) ! CAUTION DTT is harmful if swallowed. Avoid contact with skin and eyes. Wear protective gloves / eye protection / face protection. DTT is very toxic to aquatic life with long lasting effects so collect spillage and avoid release to the environment. Dispose of contents and container to an approved waste disposal place.
Tris-HCl (Acros Organics, cat. no. 167620010)
Sodium chloride, NaCl (Sigma-Aldrich, cat. no. 31434)
Potassium chloride, KCl (Carl Roth, cat. no. 6781.1)
Magnesium chloride, MgCl2 (Carl Roth, cat. no. 2189.1)
Calcium chloride, CaCl2 (Carl Roth, cat. no. 5239.1) ! CAUTION May be harmful if swallowed and cause serious eye irritation. Wear eye protection / face protection.
EDTA disodium salt (Gebru Biotechnik, cat. no. 1034.1000)
Sodium fluoride, NaF (Sigma-Aldrich, cat. no. S7920) ! CAUTION NaF is toxic if swallowed and can cause serious skin and eye irritations. Handle it under the fume hood to avoid inhalation. Wear protective gloves / eye protection / face protection. NaF is very toxic to aquatic life with long lasting effects so collect spillage and avoid release to the environment. Dispose of contents and container to an approved waste disposal place.
NP-40 (PanReac AppliChem, cat. no. A1694,0250) ! CAUTION NP-40 is hazardous. Avoid contact with skin and eyes. Wear protective gloves / eye protection / face protection. NP-40 is very toxic to aquatic life with long lasting effects so collect spillage and avoid release to the environment. Dispose of contents and container to an approved waste disposal place.
Na2HPO4 (Carl Roth, cat. no. P030.1)
Glycerol (Sigma-Aldrich, cat. no. 15523-M)
RNase A (Sigma-Aldrich, cat. no. R4875)
RNase H, recombinant (NEB, cat. no. M0297)
RNase I, 10 U/μl, 1000 U (Thermo Fisher Scientific, cat. no. EN0601)
RNase III, 250 U (Thermo Fisher Scientific, cat. no. AM2290)
RNase T1, 1000 U/μl (Thermo Fisher Scientific, cat. no. EN0542)
DNase I recombinant and 10x DNase I buffer, RNase-free solution (Roche, cat. no. 4716728001)
Sucrose (Sigma-Aldrich, cat. no. 84097–250G)
Liquid nitrogen ! CAUTION Liquid nitrogen has a temperature of −196 °C. Wear protective goggles and appropriate protective clothing.
UltraPure DNase/RNase-free distilled water (Thermo Fisher Scientific, cat. no. 10977049)
SuperSignal West Pico Plus (ThermoFischer Scientific, cat. no. 34578)
Trichloroacetic acid (TCA) (Sigma-Aldrich, cat. no. 91228) ! CAUTION TCA is dangerous and corrosive, always wear safety glasses. ▲ CRITICAL For MS analysis, always use ultrapure TCA.
Tris base (PanReac AppliChem, cat. no. A1086)
Sodium dodecyl sulfate (SDS) (Carl Roth, cat. no. CN30.2) ! CAUTION SDS is dangerous, highly flammable and corrosive. Wear safety glasses and handle with care.
Bromphenol blue-Na-salt (Serva, cat. no. 15375.01)
Glycine (PanReac AppliChem, cat. no. A1067)
Methanol (Carl Roth, cat. no. 7583.1 or EMD, cat. no. MX0475–1) ! CAUTION Methanol is highly toxic and highly flammable. Avoid inhalation and avoid contact with skin and eyes. Wear protecting clothing. Neoprene gloves give better protection than nitrile gloves.
Tween 20 (Fisher Scientific, cat. no. 10485733)
Skim milk powder (Sigma-Aldrich, cat. no. 70166)
Ammonium persulfate (APS), (Sigma-Aldrich, cat. no. A3678) ! CAUTION APS is dangerous. Handle with care.
TEMED (Carl Roth, cat. no. 2367) ! CAUTION It is corrosive, dangerous and highly flammable. Use protective clothing.
Rotiphorese®Gel 30 (37.5:1), (Carl Roth, cat. no. 3029.1) ! CAUTION Causes skin irritation, serious eye irritation and may cause genetic defects. Handle it under the fume hood to avoid inhalation. Wear protective gloves.
Anti ASNS: asparagine synthetase mouse mAb, Santa Cruz, cat. no. sc-365809. RRID: AB_10843357
Anti CTCF: CTCF XP rabbit mAb, Cell Signaling, cat. no. 3418. RRID: AB_2086791
Anti hnRNP U: hnRNP U mouse mAb, Santa Cruz, cat. no. sc-32315. RRID: AB_627741
Anti MCM7: MCM7 (D10A11) XP rabbit mAb, Cell Signaling, cat. no. 3735. RRID: AB_2142705
Anti NCL: C23 (H-250) rabbit pAb, Santa Cruz, cat. no. sc-13057. RRID: AB_2229696 (DISCONTINUED, now replaced by C23 antibody MS-3, Santa Cruz, cat. No. sc-8031. RRID: AB_370271).
Sequencing-grade trypsin (Promega, cat. no. V5111)
HEPES (500 mM solution, pH 8.5), (Sigma-Aldrich, cat. no. H3375)
Ammonium bicarbonate (AmBic, 500 mM solution), (Sigma-Aldrich, cat. no. 09830) ! CAUTION AmBic is a health hazard and cause skin and eye irritation upon contact. Wear protective gloves and eye protection.
cOmplete™, EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, cat. no. 5056489001)
! CAUTION Causes severe skin burns and eye damage. Wear protective gloves and eye protection.
Hydrochloric acid, HCl (Sigma-Aldrich, cat. no. 30721-M) ! CAUTION HCl causes severe skin burns and eye damage. HCl may cause respiratory irritation. Handle it under the fume hood to avoid inhalation. Wear protective gloves / eye protection.
Potassium hydroxide pellets, KOH (PanReac AppliChem, cat. no. 141515.1210) ! CAUTION KOH is harmful if swallowed and causes severe skin burns and eye damage. Wear protective gloves and eye protection.
Acetone, HPLC-MS grade (Fisher Scientific, cat. no. A929–1) ! CAUTION Acetone is highly flammable, handle with care and store it exclusively in explosion-proof fridges. ▲ CRITICAL Always use MS grade acetone.
Water, HPLC-MS grade (Honeywell Burdick & Jackson, cat. no. 365–4). ▲ CRITICAL Always use MS grade water.
Trifluoroacetic acid, TFA (Honeywell Burdick & Jackson, cat. no. BB360P050) ! CAUTION TFA is a corrosive chemical and health hazard level 3 compound. Avoid breathing and any contact with skin and eyes. Wear protective gloves and eye protection. ▲ CRITICAL Always use ultrapure TFA for MS or HPLC.
Acetonitrile, ACN, HPLC-MS grade (Honeywell Burdick & Jackson, cat. no. LC015) ! CAUTION ACN is a flammable toxic health level 4 compound. Inhalation and any contact with skin and/or eyes should be avoided. Wear protective gloves and eye protection. ▲ CRITICAL Always use MS grade ACN.
High-purity formic acid, FA (Merck, cat. no. 111670) ! CAUTION FA is a flammable liquid and health hazard level 4 compound. It can cause severe skin burns and eye damage and is toxic if inhaled. Handle it under the fume hood. Wear protective gloves and eye protection. ▲ CRITICAL Always use ultrapure FA.
Thermo Scientific™ Pierce™ Amine-Reactive Tandem Mass Tag Reagents (TMT-6 plex reagent kit), (Fisher Scientific, cat. no. 90066)
EQUIPMENT
Cell culture
Cell culture incubator (5% CO2, humidified at 37 °C)
Cell culture hood (e.g. CELLGARD ES CLASS II BIOLOGICAL SAFTEY CABINET)
Cell culture vacuum pump (e.g. Integra, Vacusafe)
Laborfuge 400 (Heraeus)
Inverted microscope
Counting chamber Neubauer improved (Roth, cat. no. T729.1)
1.5 ml Eppendorf Safe-Lock Tubes (Eppendorf, cat. no. 003012086)
Cell culture flasks T175 for adherent cells (Greiner Bio-One, cat. no. 660175)
Cell culture flasks T175 for suspension cells (Greiner Bio-One, cat. no. 661195)
25 ml, 10 ml and 5 ml serological pipettes (Nerbe Plus, cat. no. 12–481-9102, 12–461-9108, 12–441-9105)
50 ml centrifuge tubes conical base (TPP, cat. no. 91050)
Nitrile gloves (Ansell, Microflex Xceed)
Cell lysate preparation
Refrigerated tabletop centrifuge (Heraeus, Fresco 17)
1 ml BD Luer slip syringe (VWR, cat. no. BDAM300328)
Syringe needles, 20G x 1 ½ BD Microlance (VWR, cat. no. 613–3937)
Syringe needles, 27G x ¾ BD Microlance (VWR, cat. no. 613–3834)
Plate reader (Labsystems Multiskan MS)
Vortexer (neoLab, cat. no. 7–2020)
Gradient preparation
Tube, Thinwall, Ultra-clear, 13,2 ml, 14×89 mm (Beckmann Coulter, cat. no. 344059) or Tube, Thinwall, Ultra-clear, 14 ml, 14×95 mm (Beckmann Coulter, cat. no. 344060)
neoLab rotator with vortexer plus combi rack for tubes (neoLab, cat. no. 7–0045, 7–0050)
Parafilm 38 m x 10 cm (Sigma-Aldrich, cat. no. P7793)
Liquid nitrogen container with lid (neoLab, cat. no. 1–0042)
Tweezer (neoLab, cat. no. E1358)
RNase treatment
ThermoMixer® F1.5 heat block (Eppendorf, cat. no. 5384000012)
Ultracentrifugation
Optima XE-90 Ultracentrifuge (Beckman Coulter, cat. no. A84471)
SW40 Ti Swinging-Bucket rotor (Beckman Coulter, cat. no. 331302) or SW41 Ti Swinging-Bucket rotor (Beckman Coulter, cat. no. 331362)
Screwdriver to close the centrifuge buckets
Western blot analysis
Protran® Western blotting membranes, nitrocellulose (GE Healthcare, cat. no. 10600002)
Ahlstrom Blotting Paper (neoLab, cat. no. 2–4327)
Immobilon-P PVDF membrane (Sigma-Aldrich, cat. no. IPVH00005)
Tetra vertical electrophoresis cell (Bio-Rad, cat. no. 1658004)
PowerPacTM HC power supply (Bio-Rad, cat. no. 1645052)
BenchRocker 2D (Benchmark Scientific, cat. no. BR2000)
Continuous Operation Rocking and Rolling Action Roller Shakers (IKA, cat. no. 000401000)
ECL Chemocam imager (Intas)
TCA precipitation
Eppendorf tubes 1.5 ml, Protein LoBind Tubes (Eppendorf, cat. no. 30108116)
Pipet tips (Eppendorf, cat. no. 0030073428 (200 μl tips), 0030073460 (1 ml tips))
Refrigerated microfuge (Micro Star 17R, VWR, cat. no. 521–1647)
Centrifuge (Micro Star 17, VWR, cat. no. 521–1646)
Vortex-Genie 1 (Scientific Industries, cat. no. UD-04724–05)
Mass spectrometry (MS)
Bench-top centrifuge
Vortexer
Vacuum centrifuge
Microcentrifuge
Microcentrifuge tubes (Eppendorf)
SOLAμ HRP desalting plate (Thermo Fisher Scientific, 2 mg/1 mL 96-well plate, cat. no. 60209–001)
Oasis HLB 30 μm desalting plate (Waters, 10 mg 96-well plate, cat. no. 186000128)
Vacuum manifold desalting station
9 mm Autosampler minimum volume Inserts (Thermo Fisher Scientific, cat. no. C4010–630P)
Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with an EASY-nLC 1000 Liquid Chromatograph (Thermo Fisher Scientific) and nanospray source (Thermo Fisher Scientific)
Qual Browser software
Image analysis software
ImageJ (http://imagej.net/Downloads)
Statistical analysis of protein amounts per fraction
Open source edition of the integrated development environment RStudio available at: https://www.rstudio.com/products/rstudio/download/
MS_Statistical_Analysis.R (provided in the Supplementary Data): This software provides the custom-made analysis pipeline to automatically identify RNA-dependent proteins.
“README.txt” file (provided in the Supplementary Data), which provides all necessary instructions to successfully run the pipeline “MS_Statistical_Analysis.R” on the test dataset “Mass_Spec_RawData_Sample.csv” (provided in the Supplementary Data).
Training dataset “Mass_Spec_RawData_Sample.csv” (provided in the Supplementary Data), on which the pipeline “MS_Statistical_Analysis.R” can be applied.
REAGENT SETUP
Complete cell culture medium for HeLa S3
Cell culture medium is 500 ml of Ham’s F-12K (Kaighn’s) medium supplemented with 10% (vol/vol) FBS (50 ml). Store at 4 °C for up to 12 months from date of manufacture.
Complete cell culture medium for HeLa
Cell culture medium is 500 ml of DMEM-high glucose medium supplemented with 10% (vol/vol) FBS (50 ml). Store at 4 °C for up to 12 months from date of manufacture.
Complete cell culture medium for A549
Cell culture medium is 500 ml of RPMI medium (Gibco) supplemented with 10% (vol/vol) FBS (50 ml). Store medium at 4 °C for up to 12 months from date of manufacture.
Tris-HCl, 1 M, pH 7.5
To prepare 1 M Tris-HCl, pH7.5, dissolve 30.29 g of Tris in 200 ml water, adjust the pH to 7.5 with HCl, and bring to a final volume of 250 ml with water, filter-sterilize or autoclave and store at room temperature (RT; 15 °C to 25 °C) indefinitely.
NaCl, 5 M
To prepare 5 M NaCl, dissolve 37.86 g of NaCl in water to a final volume of 250 ml, filter-sterilize or autoclave and store at RT indefinitely.
KCl, 2.5 M
To prepare 2.5 M KCl, dissolve 46.59 g of KCl in water to a final volume of 250 ml, filter-sterilize or autoclave and store at RT indefinitely.
MgCl2, 1 M
To prepare 1 M MgCl2, dissolve 50.83 g of MgCl2 in water to a final volume of 250 ml, filter-sterilize or autoclave and store at RT indefinitely.
CaCl2, 1 M
To prepare 1 M CaCl2, dissolve 36.76 g of CaCl2 in water to a final volume of 250 ml, filter-sterilize or autoclave and store at RT indefinitely.
EDTA, 0.5 M, pH 8.0
To prepare 0.5 M EDTA, pH 8, dissolve 36.5 g of EDTA in 200 ml water, adjust the pH to 8.0 with KOH (otherwise the EDTA will not dissolve), and bring to a final volume of 250 ml with water, filter-sterilize or autoclave and store at RT for several months.
NaF, 0.5 M
To prepare 0.5 M NaF, dissolve 1.05 g of NaF in water to a final volume of 50 ml, filter-sterilize and store at RT indefinitely.
DTT, 1 M
To prepare 1 M DTT, dissolve 1.85 g of DTT in water to a final volume of 12 ml, aliquot and store for up to three months at −20 °C.
NP-40, 10%
To prepare 10% (vol/vol) NP-40, mix 5 ml of Triton X-100 with 45 ml of water. Filter-sterilize and store at RT for up to 12 months.
Na2HPO4, 0.5 M
To prepare 0.5 M Na2HPO4, dissolve 17.75 g of Na2HPO4 in water to a final volume of 250 ml, filter-sterilize and store at RT indefinitely.
cOmplete™, EDTA-free protease inhibitor cocktail, 25x
To prepare 25x EDTA-free Protease Inhibitor Cocktail, dissolve 1 tablet in 2 ml water. Can be stored at −20 °C for at least 12 weeks.
Lysis buffer
To prepare 10 ml lysis buffer, mix 250 μl of 1 M Tris-HCl (pH 7.5), 600 μl of 2.5 M KCl, 500 μl of 10% NP-40, 40 μl of 0.5 M EDTA, 20 μl of 0.5 M NaF, 5 μl of 1 M DTT, 400 μl of a 25x cOmplete™, EDTA-free Protease Inhibitor Cocktail stock with 8435 μl water. Store at 4 °C for up to 12 months.
▲ CRITICAL 25x EDTA-free Protease Inhibitor Cocktail has to be added fresh before use.
The final concentrations are 25 mM Tris-HCl (pH 7.5), 150 mM KCl, 0.5% (vol/vol) NP-40, 2 mM EDTA, 1 mM NaF, 0.5 mM DTT, 1x cOmplete™, EDTA-free Protease Inhibitor Cocktail.
iT buffer-glycerol
To prepare 10 ml of iT buffer-glycerol, mix 250 μl of 1 M Tris-HCl (pH 7.5), 274 μl of 5 M NaCl, 20 μl of 2.5 M KCl, 5 μl of 1 M MgCl2, 7 μl of 1 M CaCl2, 6 μl of 0.5 M Na2HPO4, 5 ml of Glycerol, 400 μl of 25x cOmplete™, EDTA-free Protease Inhibitor Cocktail with 4038 μl water. Store at 4 °C for up to 12 months.
▲ CRITICAL 25x EDTA-free Protease Inhibitor Cocktail has to be added fresh before use.
The final concentrations are 25 mM Tris-HCl (pH 7.5), 137 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 0.7 mM CaCl2, 0.3 mM Na2HPO4, 50% Glycerol (vol/vol), 1x cOmplete™, EDTA-free Protease Inhibitor Cocktail.
iT buffer
To prepare 10 ml of iT buffer, mix 250 μl of 1 M Tris-HCl (pH 7.5), 274 μl of 5 M NaCl, 20 μl of 2.5 M KCl, 5 μl of 1 M MgCl2, 7 μl of 1 M CaCl2, 6 μl of 0.5 M Na2HPO4, 400 μl of 25× 1x cOmplete™, EDTA-free Protease Inhibitor Cocktail with 9038 μl water. Store at 4 °C for up to 12 months.
▲ CRITICAL 25x EDTA-free Protease Inhibitor Cocktail has to be added fresh before use.
The final concentrations are 25 mM Tris-HCl (pH 7.5), 137 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 0.7 mM CaCl2, 0.3 mM Na2HPO4, 1x cOmplete™, EDTA-free Protease Inhibitor Cocktail.
BCA-mix
To prepare 2.2 ml BCA-mix, prepare a 1:50 dilution by mixing 44 μl Pierce BCA Protein Assay Reagent B with 2156 μl Bicinchoninic Acid solution. Prepare fresh, do not store.
Sucrose solution buffer
The sucrose solution buffer is 100 mM NaCl, 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA (pH 8). Prepare 500 ml using 10 ml of 5 M NaCl solution, 5 ml of 1 M Tris-HCl (pH 7.4) solution and 500 μl of 1 M EDTA solution; fill up with UltraPure distilled water to 500 ml. Store at 4 °C for up to one year.
5% to 50% (wt/vol) sucrose solutions
The ten sucrose solutions are 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45% and 50% (wt/vol) sucrose in sucrose solution buffer. Prepare 50 ml of each solution by weighing 2.5 g, 5 g, 7.5 g, 10 g, 12.5 g, 15 g, 17.5 g, 20 g, 22.5 g and 25 g of sucrose in ten 50 ml centrifugation tubes. Fill then up to 50 ml with the sucrose solution buffer. Store at 4 °C; prolonged storage is not recommended. Discard if the buffer changes color.
▲ CRITICAL Do not add 50 ml of sucrose solution buffer in each tube; strictly fill up to 50 ml as changing the final volume will change the final sucrose concentration.
Tris base, 2 M, pH 6.8
To prepare 2 M Tris base, pH 6.8, dissolve 60.6 g of Tris base in 200 ml water, adjust pH to 6.8 with HCl and bring to a final volume of 250 ml with water. Store at RT indefinitely.
6x SDS loading dye
For 15 ml 6x SDS loading dye use 3.75 ml of the 2 M Tris base pH 6.8, 4.5 g Glycerol, 1.5 g SDS, 9 ml of the 1 M DTT and 1.5 mg Bromphenol blue. Make aliquots and store them at −20 °C. The aliquots can be stored for several years.
APS, 10 % (wt/vol)
Dissolve 1 g APS in 10 ml water; prepare aliquots and store them at −20 °C for up to one year.
10x SDS running buffer
To prepare 1 l 10x SDS running buffer use 30.3 g Tris base, 144 g Glycine and 10 g SDS. Dissolve everything in 800 ml water and subsequently adjust to 1 l with water; store at RT for up to six months.
1x SDS running buffer
Mix 100 ml 10x SDS buffer with 900 ml water; make fresh and store at RT until use.
10x western blot transfer buffer (Towbin buffer / for wet blot)
To prepare 1 l 10x western blot transfer buffer dissolve 30.3 g Tris base and 144 g Glycine in 800 ml water and subsequently adjust to 1 l with water; store at RT for up to six months.
The final concentrations are 250 mM Tris-HCl, 1.92 M Glycin.
1x transfer buffer (Towbin buffer / for wet blot)
To prepare 1 l 1x western blot transfer buffer mix 100 ml 10x western blot transfer buffer, 200 ml methanol, 700 ml water; store at 4 °C for up to one month or discard after three runs. ▲ CRITICAL The transfer buffer might turn yellow with time. If this happens, discard the transfer buffer in an appropriate liquid waste container. ! CAUTION Transfer buffer contains methanol that is highly toxic. Please discard accordingly in the appropriate liquid waste container.
10x TBS buffer
To prepare 1 l 10x TBS buffer use 30 g Tris base, 80 g NaCl and 2 g KCl. Dissolve everything in 800 ml water and adjust pH to 7.4 with HCl and subsequently adjust to 1 l with water; store at RT for up to three months.
1x TBST
Mix 100 ml 10x TBS buffer, 1 ml Tween-20, 899 ml water; store at RT for up to three months.
Blocking buffer
Dissolve 5% (wt/vol) milk powder in 1x TBST; storage at 4 °C for up to one week; longer storage is not recommended.
Trichloroacetic acid (TCA), 100 %
Dissolve 10 g TCA in 10 ml HPLC-MS grade water; store at 4 °C for up to one year.
! CAUTION TCA is corrosive; wear safety glasses.
▲ CRITICAL Do not store 100% TCA in plastic tubes. It will affect the plastic.
TCA, 10 %
Dilute 100% TCA by using 1 ml 100% TCA and 9 ml MS-grade water. Prepare fresh. Do not store.
▲ CRITICAL Prepare this solution always freshly before use and store on ice.
500 mM Ammonium Bicarbonate (AmBic)
To make 500 mM ammonium bicarbonate stock, add 1.6 g of ammonium bicarbonate to 40 mL of MS-grade water. Store at RT for up to three months.
Trypsin digestion buffer
To prepare 100 μl trypsin digest buffer, add 10 μL of 500 mM AmBic stock solution in 90 μl of MS-grade water. Add 1 μl of sequencing grade trypsin to the buffer.
▲ CRITICAL Prepare fresh when setting up digestion of the protein samples.
Trypsin digest quenching buffer
To prepare the quenching buffer, add HPLC-MS-grade acetonitrile 50% (vol/vol) with 5% (vol/vol) high purity formic acid in 45% (vol/vol) MS-grade water. Store at RT for up to six months.
TFA, 20% (vol/vol)
To make 20% (vol/vol) TFA, add 20% (vol/vol) high purity trifluoroacetic acid to 80% (vol/vol) MS-grade water. Store at RT for up to one year.
TFA, 0.1% (vol/vol)
Dilute 500 μl of 20% (vol/vol) TFA solution in 99.5 ml MS-grade water to make 100 ml of 0.1% (vol/vol) TFA solution. Store at RT for up to one year.
MeOH, 60% (vol/vol)
Add HPLC-MS-grade 60% (vol/vol) methanol in 40% (vol/vol) MS-grade water to make a 60% (vol/vol) MeOH solution. Store at RT for up to one year.
HEPES, 500 mM pH 8.5 stock solution
Dissolve 5.95 g high purity HEPES (free acid) in 45 ml MS-grade water. Titrate the solution to pH 8.5 using NaOH. Make up the final volume of the solution to 50 ml using MS-grade water. Store at RT for up to six months.
HEPES, 166 mM pH 8.5
Dilute 300 μl of 500 mM HEPES, pH 8.5 stock solution in 600 μl MS-grade water to make 166 mM HEPES, pH 8.5 buffer solution.
▲ CRITICAL Prepare fresh for TMT labeling.
Desalting wash buffer
To make desalting wash buffer, add 7% (vol/vol) HPLC-MS-grade methanol and 0.1% (vol/vol) high purity TFA in MS-grade water. Store at RT for up to six months.
MS Loading buffer
Add 7% (vol/vol) HPLC-MS-grade methanol to 1.5% (vol/vol) high-purity formic acid in MS-grade water. Store at RT for up to one year.
EQUIPMENT SETUP
Statistical data analysis
For the statistical analysis of the protein amount per fraction as inferred from the MS data, install the software RStudio and open the provided script “MS_Statistical_Analysis.R” as provided in the Supplementary Data using the “open file” function from the menu “File”. For more detailed information, refer to the “README.txt” file as provided in the Supplementary Data along with the script “MS_Statistical_Analysis.R” and the test dataset “Mass_Spec_RawData_Sample.csv”.
Western blot quantification
For the quantification of western blots, install the “Image J” software (http://imagej.net/Downloads).
Orbitrap Fusion LC-MS/MS instrument method
For TMT 6-plex runs, configure the Orbitrap Fusion to run in data-dependent, SPS-MS3 quantification mode. In this mode, an Orbitrap MS1 scan was taken [scan range, 350 to 1500 mass-to-charge ratio (m/z); resolution (R), 120K; automatic gain control (AGC) target, 2.5 × 105; max ion injection time, 100 ms], followed by ion trap MS2 scans on the most abundant precursors for 4 s [max speed mode; quadrupole isolation, 0.6 m/z; AGC target, 4 × 103; scan rate, rapid; max ion injection time, 60 ms; minimum MS1 scan signal, 5 × 105 normalized units; charge states 2, 3, and 4 included; collision-induced dissociation (CID) energy, 33%] and Orbitrap MS3 scans for quantification [R, 15K; AGC target, 2 × 104; max ion injection time, 125 ms; higher-energy collisional dissociation (HCD) energy, 48%; scan range, 120 to 140 m/z; synchronous precursors selected, 10].
Procedure
Cell culture ● Timing 1 d
▲ CRITICAL Prior to the experiment, the cells of interest should be expanded and passaged 1–2 times, which can take several days depending on the cell line. The procedure described here starts once the cells are ready to be expanded into a large cell culture flask (T175).
-
1
For growing HeLa S3 (suspension) cells, follow option A. For growing HeLa or other adherent cells, follow option B.
(A). Growing HeLa S3 (suspension cells)
Grow cells in cell culture medium for HeLa S3 cells (see Reagent setup) and seed cells 1.5–2.5 × 105/ml into a T175 flask (for suspension cell lines). Grow the cells up to 3–5 × 105 cells/ml at 37° C supplied with 5% CO2.
(B). Growing HeLa, A549 (adherent cells)
Grow cells in the appropriate cell culture medium (see Reagent setup) and seed 6 × 106 cells into a T175 flask containing 100 ml medium (for adherent cell lines). Grow them overnight at 37° C supplied with 5% CO2. Cells should be 80–90% confluent the next day.
▲ CRITICAL STEP At the day of lysate preparation, you should have 2 × 107 cells to obtain enough cell lysate for the gradient. In our hands, this amount of HeLa S3/HeLa cells leads to 4–6 mg of total protein.
Collect cells for cell lysate preparation ● Timing 30 min
-
2
For harvesting HeLa S3 suspension cells, follow option A. For harvesting adherent cells, follow option B.
(A). Harvesting HeLa S3 (suspension cells)
Count cells using a Neubauer cell counting chamber.
-
Take out the appropriate number of cells.
▲ CRITICAL STEP We recommend to start with 2 × 107 cells to obtain enough cell lysate for the gradient.
Transfer cells to 50 ml centrifuge tubes.
Centrifuge cells at 1,200 g for 5 min at RT.
Discard medium by aspiration.
Wash cells by gently resuspending them in 10 ml ice-cold PBS.
Centrifuge at 800 g for 5 min at 4 °C.
Repeat Steps 2A(v) to 2A(vii) one more time.
-
After the last wash, continue with the cell lysis procedure in Step 3, or pause here.
∎ PAUSE POINT Cells can be resuspended in 1 ml iT buffer-glycerol per 2 × 107 cells and stored at −80 °C for up to six months.
(B). Harvesting HeLa, A549 (adherent cells)
Discard medium by aspiration.
Rinse cell monolayer gently with 20 ml warm (37 °C) PBS per T175 flask.
Discard PBS by aspiration.
Add 5 ml warm 0.05% Trypsin, EDTA (1x) per T175 flask and incubate for up to 2 min in the cell culture incubator, checking from time to time under the microscope whether the cells have detached.
When cells have detached from the plastic surface of the flask, add 10 ml 37° C pre-warmed medium per T175 flask.
Resuspend cells by pipetting and transfer cells to a 50 ml centrifuge tube.
Count cells using a Neubauer counting chamber.
-
Take out an appropriate number of cells and transfer them into a new 50 ml centrifuge tube.
▲ CRITICAL STEP We recommend to start with 2 × 107 cells to obtain enough cell lysate for the gradient.
Centrifuge cells at 1,200 g for 5 min at RT.
Discard medium by aspiration.
Wash cells pellet by gently resuspending them in 10 ml ice-cold PBS.
Centrifuge at 800 g for 5 min at 4 °C.
Repeat Steps 2B(x) and 2B(xii) one more time.
-
After the last wash continue with cell lysis procedure in Step 3, or pause.
∎ PAUSE POINT Cells can be resuspended in 1 ml iT buffer-glycerol per 2 × 107 cells and stored at −80 °C for up to six months.
Cell lysis ● Timing ~ 2 h
▲ CRITICAL We recommend to use lysis buffer containing NP-40 or a similar detergent like Triton X-100 but to avoid any detergent causing protein or protein complex denaturation (e.g. SDS).
▲ CRITICAL If you have stored the cells in iT buffer-glycerol, carry out Steps 3–7, otherwise skip these steps and proceed to Step 8 immediately.
-
3
If you have stored the cells in iT buffer-glycerol, firstly centrifuge at 800 g for 5 min at 4 °C.
-
4
Discard supernatant.
-
5
Resuspend cells in 1 ml ice-cold iT buffer without glycerol.
-
6
Centrifuge at 800 g for 5 min at 4 °C.
-
7
Repeat Steps 4–6 one more time.
-
8
Discard the supernatant.
-
9
Lyse cells by adding 100 μl lysis buffer per 2 × 107 cells.
-
10
Incubate cell suspension for 30 min on ice.
-
11
Resuspend by vortexing every 5 min.
-
12
After incubation, flash freeze suspension in liquid nitrogen and thaw suspension again immediately and homogenize with a thin needle (preferentially 27G x ¾). Use needle size 20G x 1½ if the lysate is too viscous.
-
13
Repeat freeze/thaw cycle two more times.
! CAUTION Liquid nitrogen has a temperature of −196 °C. Wear protective goggles and appropriate protective clothing. Be careful when using the needle to avoid stab wounds.
-
14
Centrifuge the suspension at 17,000 g for 10 min at 4 °C.
-
15
Transfer supernatant (cell lysate) to a new tube.
∎ PAUSE POINT The lysate can be flash frozen in liquid nitrogen and stored at −80 °C for up to 12 months.
BCA assay ● Timing ~ 1 h
-
16
▲ CRITICAL STEP We propose here (Steps 16 to 24) an alternative to commercially available kits. Alternatively, protein concentration can be measured using a Qubit™ and associated Qubit™ Protein Assay Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. In order to measure the protein concentration from the lysate, perform a BCA assay (= bicinchoninic acid assay).
-
17
Firstly, prepare the following BSA samples for a standard curve: 0.2, 0.4, 0.5, 0.6, 0.8, 1.0, 1.2 and 1.4 mg/ml of BSA in lysis buffer.
-
18
Prepare a 1:2 dilution of your cell lysate (from Step 15) in lysis buffer (10 μl lysate + 10 μl lysis buffer) in a 1.5 ml tube.
-
19
Transfer 10 μl of each sample and of each BSA standard into a well of a 96-well plate. Alternatively, individual samples can be measured in a photometer using a cuvette.
-
20
Add one well with 10 μl lysis buffer only.
-
21
Add 200 μl BCA-mix per well.
-
22
Incubate plate at 37 °C for 30 min.
-
23
Measure absorption at 540 nm with a plate reader. Subtract the absorption value obtained for the lysis buffer alone.
-
24
Calculate a standard curve using the BSA samples with known concentrations and their corresponding absorption values.
-
25
Determine the cell lysate concentration according to the standard curve.
Gradient preparation
-
26
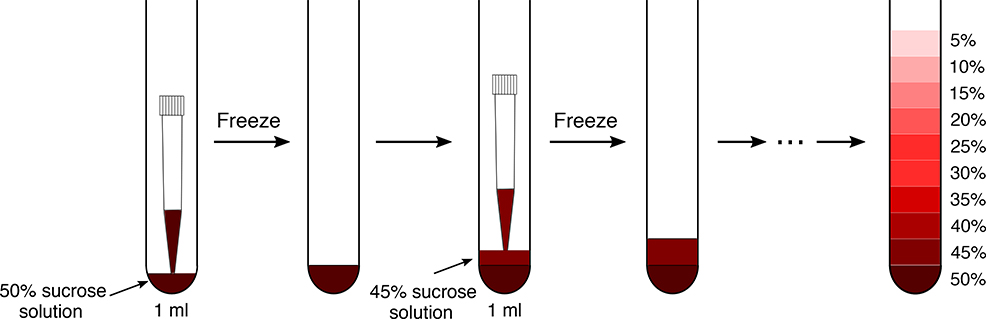
Prepare gradients by freezing the different sucrose layers in liquid nitrogen (option A) or at −80 °C (option B) as depicted Fig. 2. The ultracentrifugation can be performed with different rotors, so the volume of the sucrose gradient will depend on the size of the tubes that are used. We recommend to prepare several sucrose gradients for all the replicates at once for efficiency and reproducibility.
Fig. 2 |. Gradient preparation procedure.
Illustration of the preparation and freezing of a sucrose gradient. A P1000 pipet is used equipped with a filter tip. Be careful to vertically hold the tube while freezing to ensure that each layer is flat and even. Always ensure that the last layer is well frozen before casting the next one on top of it. The whole procedure for sucrose gradient preparation can be followed in Supplementary Video 1.
(A). Gradient preparation by freezing the layers in liquid nitrogen ● Timing ~ 1 h for 10 tubes
▲CRITICAL A detailed explanation of the sucrose density gradient preparation by freezing the different layers in liquid nitrogen is available as Supplementary Video 1.
-
Transfer 1 ml (for 13.2 ml centrifugation tubes) or 1.2 ml (for 14 ml centrifugation tubes) of 50% sucrose solution to the bottom of the tube.
▲CRITICAL STEP Make sure not to release the solution on the walls of the tube but right in the middle.
-
Freeze the solution in the tube in liquid nitrogen. From now on, keep the tubes on ice. The sucrose solution turns white when frozen.
! CAUTION Liquid nitrogen has a temperature of −196 °C. Wear protective goggles and appropriate protective clothing. Use a tweezer to hold the tubes in the liquid nitrogen.
▲CRITICAL STEP Make sure to keep the tube vertical while freezing to ensure the sucrose layer is flat and even.
Ensure the previous layer is well frozen (use for example a pipet with a clean tip and ensure that the sucrose layer is hard) and add the same volume of the 45% sucrose solution on the top of the frozen layer.
Freeze the tube in liquid nitrogen as in Step 25Aii.
Repeat Steps 25Aiii and iv for each of the other sucrose solutions, from the most concentrated sucrose solution (40%) and decreasing in 5% sucrose density steps to the least concentrated sucrose solution (5%). In total, the sucrose density gradient is made up of 10 layers (50%, 45%, 40%, 35%, 30%, 35%, 20%, 15%, 10% and 5% sucrose solutions).
(B). Preparing gradients by freezing the layers at −80 °C ● Timing ~ 3 h for 10 tubes
-
Repeat Step 25Ai
▲CRITICAL STEP Make sure not to release the solution on the walls of the tube but right in the middle.
-
Keep the tube at −80 °C for 15 min.
▲CRITICAL STEP Make sure to keep the tube vertical while freezing to ensure the sucrose layer is flat and even.
▲CRITICAL STEP Cover the tube with parafilm to prevent anything falling inside the tube while it is stored in the −80 °C freezer.
Ensure the previous layer is well frozen and add the same volume of the 45% sucrose solution on top of it.
Keep the tube at −80 °C for 15 min.
-
Repeat Steps 25Biii and iv for each of the other sucrose solutions, starting from the most concentrated sucrose solution (40%) and decreasing in 5% sucrose density steps to the least concentrated sucrose solution (5%). In total, the sucrose density gradient is made up of 10 layers (50%, 45%, 40%, 35%, 30%, 35%, 20%, 15%, 10% and 5% sucrose solutions).
∎ PAUSE POINT The frozen sucrose gradients can be kept at −20 °C for several months, covered with Parafilm to prevent evaporation and/or contamination.
Lysate treatment ● Timing ~ 1 h
-
27
Treat the lysate from Step 15 (at least 1 mg of protein) with five different RNases following option A. Alternatively, and to ensure that the shifts are related only to RNA digestion, use option B to additionally treat all lysates (including the control sample) with DNase I. The RNases that we used have different activities, as depicted in Table 1. Preliminary experiments show that RNase treatment of cell lysate with individual RNases differentially affects the RNA-dependent protein hnRNP U, a well-known RBP (Supplementary Fig. 1), which may reflect differences in specificity as well as activity of the RNases.
Table 1 |.
RNases and their targeted RNA species
| RNase | Target RNA species |
|---|---|
| RNase A | RNase A is an endoribonuclease that attacks at the 3’phosphate of a pyrimidine nucleotide. The sequence of pG-pG-pC-pA-pG will be cleaved to give pG-pG-pCp and A-pG. |
| RNase H | RNase H (Ribonuclease H) is an endoribonuclease that specifically hydrolyzes the phosphodiester bonds of RNA, when hybridized to DNA. |
| RNase I | RNase I is an endoribonuclease that preferentially hydrolyzes single-stranded RNA to nucleoside 3’-monophosphates via nucleoside 2’, 3’-cyclic monophosphate intermediates. |
| RNase III | RNase III is a double-stranded RNA (dsRNA) specific endoribonuclease. In E. coli, RNase III cleaves ribosomal RNA (rRNA) precursors during maturation of rRNA. The enzyme cleaves dsRNA into 12–15 bp dsRNA fragments with 2 to 3 nucleotide 3’ overhangs, and 5’ phosphate and 3’ hydroxyl termini. The termini and overhangs of RNase III cleavage products are thus the same as those produced by Dicer in the eukaryotic RNAi pathway. |
| RNase T1 | RNase T1 is an endoribonuclease that specifically degrades single-stranded RNA at G residues. It cleaves the phosphodiester bond between the 3’-guanylic residue and the 5’-OH residue of adjacent nucleotides with the formation of corresponding intermediate 2’, 3’-cyclic phosphate. The reaction products are 3’-GMP and oligonucleotides with a terminal 3’-GMP. |
(A). Treatment with RNases
-
For each sample to be treated, add 1 μl of each RNase (RNase A, RNase H, RNase I, RNase III, RNase T1) to 100 μl of lysate from Step 15.
▲CRITICAL STEP Use elongated pipet tips to take out the RNase without contaminating the pipet, and thus avoid contaminating control samples.
For the control samples, add the same volume of lysis buffer as the total volume of RNases added to the RNase-treated samples.
-
Incubate the samples at 4 °C for 1 h on a slowly rotating wheel (10 rpm).
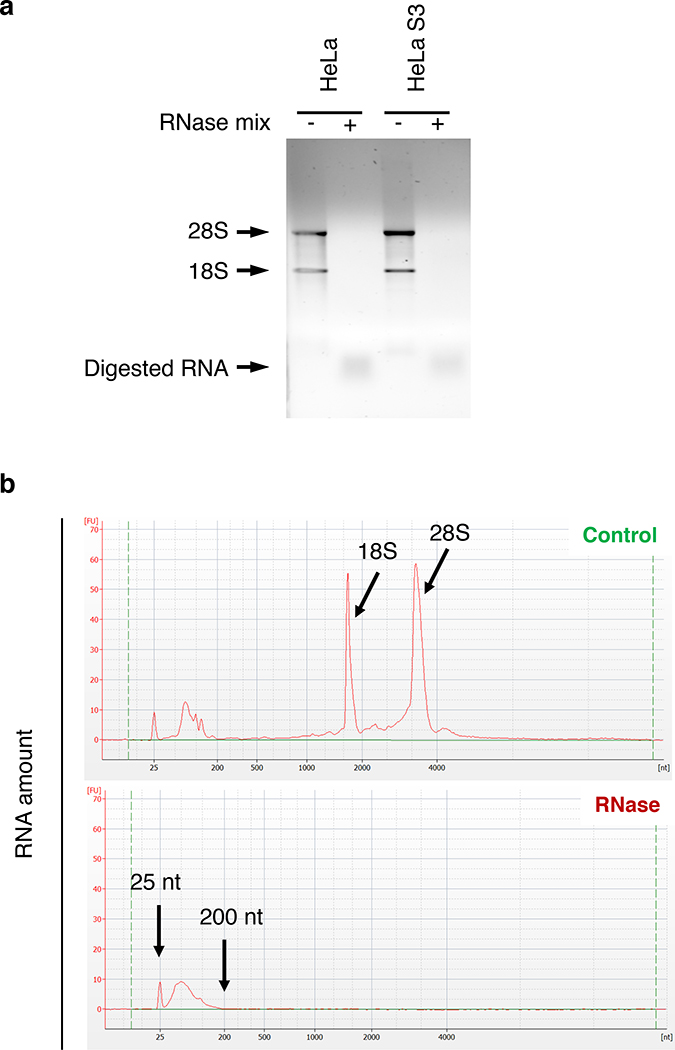
▲CRITICAL STEP Assess the quality of the RNases digestion by electrophoresis of total RNA isolated from control and RNase-treated cell lysates (Fig. 3).
Fig. 3 |. RNase treatment of cleared lysates.
Examples of how to test the efficiency of the RNase treatment. Addition of an RNase mix induces the complete digestion of the two prominent ribosomal RNA species (18S and 28S), as illustrated by the absence of the corresponding bands/peaks. a, Cleared lysates of HeLa and HeLaS3 cells were processed with (+) or without (−) addition of an RNase mix. The respective total RNA samples were loaded onto a 1% agarose gel. b, Total RNA samples from HeLa S3 cells, untreated (Control) or treated with an RNase mix (RNase) were analyzed on a Bioanalyzer to monitor the length of the RNA fragments.
(B). Treatment with RNases and DNase I
-
For each sample to be treated, add 1 μl of each RNase (RNase A, RNase H, RNase I, RNase III, RNase T1) for 100 μl of lysate from Step 15.
▲CRITICAL STEP Use elongated pipet tips to take out the RNase and the DNase without contaminating the pipet, and thus avoid contaminating control samples.
For the control sample, add the same volume of lysis buffer as the total volume of RNases added to the RNase-treated samples.
Add to all samples (both control and RNase-treated) 1/10 of the sample volume of the provided 10x DNase I buffer and 100 U of DNase I for 1 mg of total protein in the lysate.
Incubate the samples at RT for 1 h on a slowly rotating wheel (10 rpm).
Ultracentrifugation ● Timing ~ 19 h
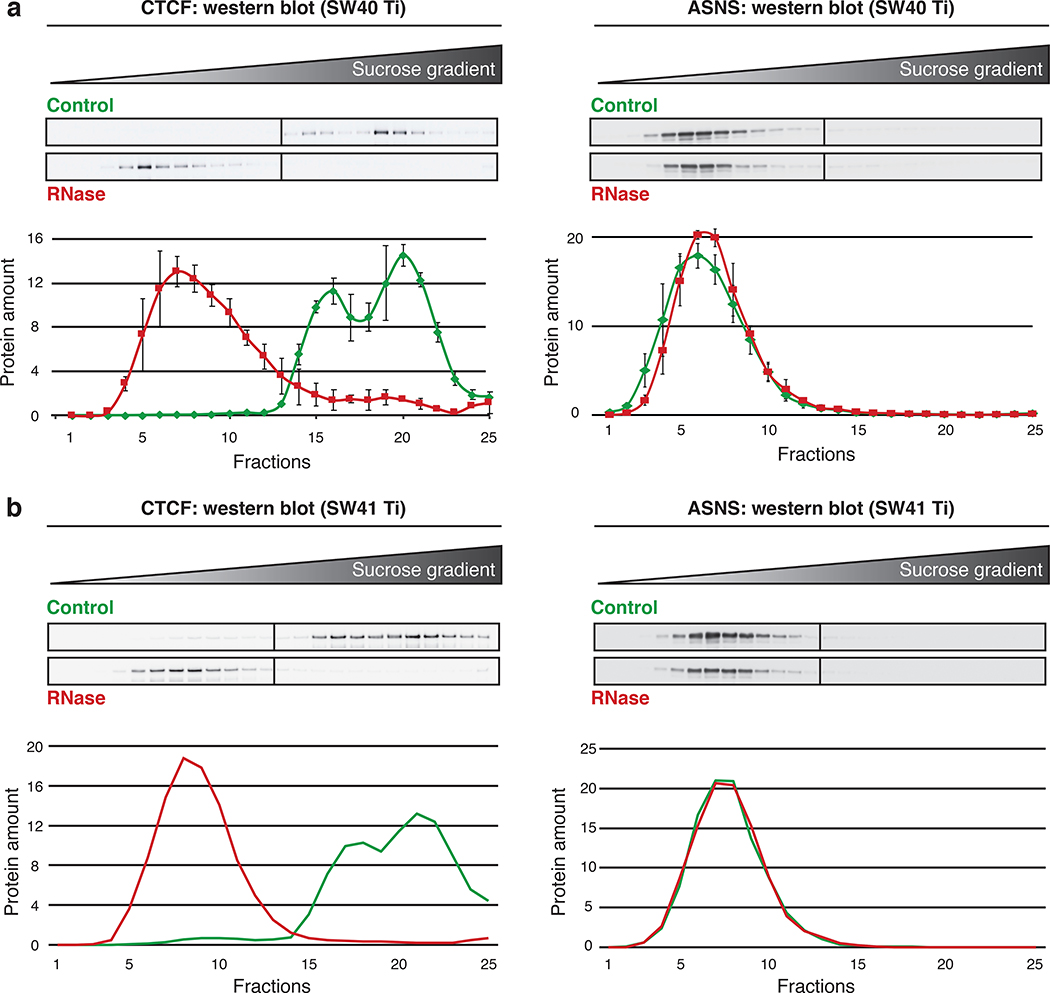
▲CRITICAL The ultracentrifugation can be performed with a SW40 Ti or a SW41 Ti rotor. The following procedure is adapted for both rotors (Fig. 4).
Fig. 4 |. Comparison of gradients from different rotors.
The use of different rotors (SW40 Ti and SW41 Ti) in similar conditions led to the same protein distribution through the 25 fractions of 5% to 50% sucrose density gradients. a, top panel: western blot analysis for RNA-dependent protein CTCF and RNA-independent protein ASNS in 25 fractions of representative control and RNase-treated samples after ultracentrifugation in a SW40 Ti rotor (30,000 rpm/114,000 g for 18 h at 4° C). Bottom panel: graph of the quantitative analysis of three replicates (N = 3) depicting the mean of three experiments with standard error of the mean (SEM). Adapted from Caudron-Herger et al., Mol Cell (2019) (ref. 24). b, Same as in a, with a SW41 Ti rotor (N = 1) (30,000 rpm/111,000 g for 18 h at 4° C). For all western blots, fractions 1 to 25 were loaded onto two membranes (1: fractions 1 to 13 and 2: fractions 14 to 25).
▲CRITICAL The ultracentrifugation must be performed right after the lysate treatment.
▲CRITICAL The centrifugation duration is critical since too short centrifugation time will impede the proper separation of the protein complexes. Depending on the experiment requirements, the centrifugation duration can vary from 2 h to 20 h30–32,39. To the same extent, sucrose concentrations, rotors and centrifugation speed can be adjusted. As depicted in Supplementary Fig. 2, long centrifugation time (here 18 h) at 110,000–115,000 g is necessary in this protocol to obtain a deep migration of large complexes into the gradient. High glucose concentrations (up to 50%) are better adapted for the separation of large complexes30.
-
28
Store the frozen sucrose gradients to 4 °C for at least 1 h prior to the ultracentrifugation to allow them to thaw. Place the rotor and the buckets at 4 °C for at least 1 h prior to centrifugation to cool them down. Turn on the ultracentrifuge, set it to 4 °C and turn on the vacuum to cool it down as well.
-
29
Once the gradients have thawed, equilibrate their exact mass pairwise by adjusting the level of liquid. To do so, only remove some of the top layer.
▲CRITICAL STEP Do not add anything to the thawed gradients. Label the tubes according to the sample they will contain. From now on, avoid any movement that might shake the gradients and disturb the sucrose layers.
-
30
Once the lysate RNase/DNase treatment has finished, carefully pipet the digested lysate to the surface of the sucrose gradient.
▲CRITICAL STEP Use a cut tip to pipet the lysate onto the gradient. Place the end of the tip directly in contact with the surface of the top layer of the sucrose gradient and slowly pipet out the lysate to not disturb the gradient. Try to release the lysate slowly in a circular movement, starting from the tube walls and moving towards the center of the tube (or the other way around, i.e. moving circularly from the center towards the walls). Never pipet back from the gradient, never pipet air bubbles into it.
▲CRITICAL STEP Carefully equilibrate the tubes to avoid imbalance during ultracentrifugation. Usually, ultracentrifuges are equipped with imbalance detector which will turn the centrifuge off to avoid any damage.
-
31
Slowly place the tubes into the buckets, and close them firmly with the screwdriver to prevent any evaporation.
-
32
Carefully place the buckets with respect to their numbering into the rotor and place the rotor into the ultracentrifuge (here, Optima XE-90 from Beckman Coulter).
-
33
Centrifuge the gradients at 30,000 rpm for 18 h, at 4 °C, with deceleration and acceleration parameters set to 9.
▲CRITICAL STEP Set the acceleration and deceleration to the minimum, to avoid any mixing of the gradient. Set these parameters according to the manual instructions. Depending on the centrifuge manufacturer, high numbers do not necessarily mean stronger acceleration/deceleration.
-
34
Prepare in advance 25× 1.5 ml-tubes for each gradient, and label them according to your experiment.
Fractionation ● Timing ~ 15 min per gradient
▲CRITICAL The fractionation process is performed by hand in this protocol, but alternatively, automated fractionation systems are commercially available. A detailed explanation of the sucrose density gradient fractionation is available as Supplementary Video 2.
▲CRITICAL All samples should remain cold during the fractionation. The samples can be kept on ice while fractionating at RT or the whole fractionation procedure can take place in a cold room.
-
35
Carefully take the rotor out of the ultracentrifuge and the tubes out of the buckets using a tweezer without disturbing the gradient.
-
36
To fractionate, hold the tube at approximately 30° with one hand. With a P1000 pipet, place the pipet tip approximately 2 mm under the surface of the solution. Place the tip against the lower wall of the tube and carefully take out 460 μl (for 14 ml tubes) or 392 μl (for 13.2 ml tubes) of the gradient. Be careful to follow down the position of the surface with the tip while pipetting (Fig. 5).
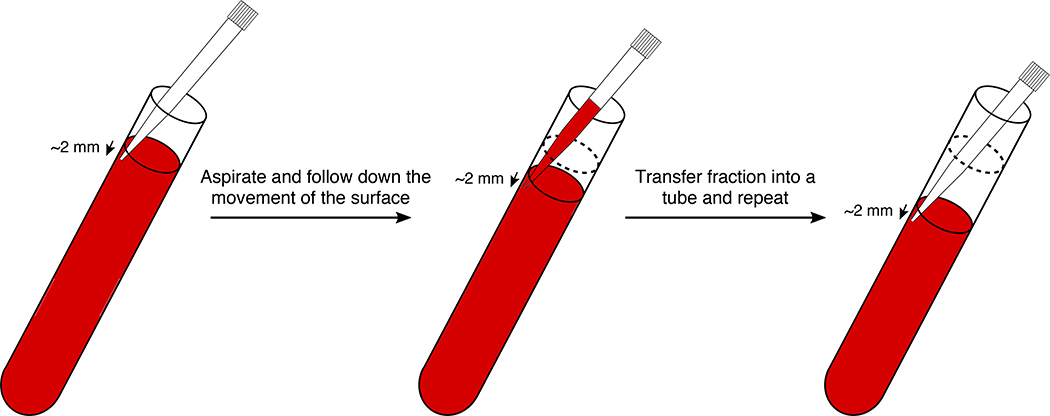
▲CRITICAL STEP It is important to fractionate the gradient slowly and carefully to avoid any mixing. Never pipet back a fraction into the gradient. Be also careful not to pipet any air. This step can introduce some variability in the number of fractions recovered. Different pipet sets and different users might introduce variability in the fraction volumes at the end.
? TROUBLESHOOTING
-
37
Release the fraction in an Eppendorf tube from Step 33 and close it.
-
38
Repeat Steps 35–36 until you have collected the 25 fractions. Repeat the same process for each gradient.
∎ PAUSE POINT The fractions can be kept for several hours at 4 °C or kept frozen for several months at −80 °C.
Fig. 5 |. Gradient fractionation procedure.
Graphical representation of the fractionation process of sucrose density gradients done by hand. A P1000 pipet is used equipped with a filter pipet tip. Pipet the fraction by placing the pipet tip about 2 mm below the surface of the gradient. While pipetting the fraction volume of 460 μl (for 14 ml tubes) or 392 μl (for 13.2 ml tubes), follow down the surface with the tip. Be careful not to aspirate any air during the process. Never pipet back the fractions into the gradient as it would mix the sucrose layers. The whole procedure for sucrose gradient fractionation can be followed in Supplementary Video 2.
Western blot analysis and evaluation ● Timing 1.5 d
▲CRITICAL Next to MS to investigate the whole proteome, single proteins can be analyzed using SDS gels and western blot (Fig. 4 and Supplementary Fig. 3). To directly start with sample preparation for MS, continue with Step 50. The following part, Steps 38–49, will focus on steps that are critical for the R-Deep method. General western blot protocols have been described elsewhere24,40.
-
39
38 Prepare 20 μl of each fraction by mixing with 6x SDS loading dye (1x final concentration).
▲ CRITICAL STEP A protein precipitation beforehand is not needed.
∎ PAUSE POINT Fractions without loading dye can be stored at −80 °C for up to six months, aliquots mixed with loading dye can be stored at −20 °C for up to six months.
-
40
Load 20 μl of each fraction (including loading dye) to an SDS gel.
▲ CRITICAL STEP In contrast to normal western blot analysis, it is important to load the same volumes of each fraction (NOT same protein amounts after quantifying their concentration).
-
41
Run the SDS gel according to the established conditions in your lab.
▲ CRITICAL STEP Since the running can be influenced by many factors, it is important to split the 25 fractions per condition in as few gels as possible (optimal two gels which have each 15 wells; e.g. first gel: fraction 1 to 13, second gel: fraction 14–25) and run them in the same chamber. If possible, the four gels for control and RNase-treated fractions should be run in the same tetracell chamber.
-
42
There are several options for western blotting: semi-dry blotting vs. wet tank blotting and nitrocellulose vs. PVDF vs. nylon membranes. Choose the options that you have the best experience with, but we recommend wet tank blotting onto PVDF membranes. Please note that the loaded protein concentrations are lower than in typical western blot applications, hence, an effective and clear transfer is needed for the later detection.
▲ CRITICAL STEP All fractions from one sample (control or RNase-treated) should run in the same blotting machine to ensure better comparability. After transferring the proteins to the membrane, block the membranes according to your established protocol and continue with antibody incubation.
∎ PAUSE POINT We recommend to do the incubation with the first antibody overnight at 4 °C.
-
43
Detect the signal using chemiluminescence and image.
▲ CRITICAL STEP It is important to develop all fractions of one condition at the same time and under the same conditions because the sum of all signals for a protein will be used to normalize the protein amount in the quantification (see Step 49). A workflow for the analysis of the western blot images (Steps 43–49 below) is provided in Supplementary Fig. 4.
? TROUBLESHOOTING
-
44
Open your western blot image in the ImageJ software.
▲ CRITICAL STEP The ImageJ software uses the following formats: TIFF, GIF, JPEG, DICOM, BMP, PGM or FITS.
-
45
Use the rectangle tool to draw a rectangle around the band with the strongest signal.
▲ CRITICAL STEP This rectangle size is also used for all the other bands of the same protein.
-
46
Move the rectangle from Step 44 around the band of fraction 1 and press “ALT” and “M”. A new window will open showing the measured intensity for this first band (column “IntDen” for “integrated Density” or column “RawIntDen” for “Raw Integrated Density”, which have the same value in absence of image calibration) as well as the area of the rectangle (column “Area” indicating the number of pixels inside the rectangle).
-
47
Move the rectangle to the next band (fraction 2) and press the same buttons as in Step 45.
-
48
Repeat Steps 45–46 until all bands (from the 25 fractions) are measured.
-
49
Transfer the table with the measured values to an excel sheet using copy and paste functions.
▲ CRITICAL STEP Calculation will be done using the measured intensity calculated by ImageJ.
▲ CRITICAL STEP Depending on your settings, the measured intensities can be inverted meaning that stronger bands give lower intensity values. In this case, the measured value needs to be subtracted from the maximum intensity that can be reached within the selected area (the rectangle as defined in Step 44). The maximum intensity can be calculated by multiplying the area (the number of pixels in the rectangle, see Step 45) by 255 for 8-bit images or 35535 for 16-bit images. The type of image is indicated in ImageJ at the top left corner of each image.
-
50
Calculate the normalized protein amount (in %) in each fraction by multiplying the measured intensity by 100 and divide this by the sum of all measured intensities from the 25 fractions of this protein.
? TROUBLESHOOTING
TCA precipitation ● Timing 1–2 d for 50 samples
▲ CRITICAL To prepare samples for MS analysis, it is recommended to use Eppendorf tubes that have low protein binding (see equipment) resulting in an improved final concentration. Plastics from other companies might increase the background signal in the later measurements.
▲ CRITICAL A high reproducibility is very important meaning that all samples should be prepared by the same person and by using the same incubation/centrifugation times for all samples. For 50 samples (25 control fractions + 25 RNase treated fractions), the number of samples should be split into groups and processed one after each other. We recommend no more than 10 samples per group (always keep the same fraction in the same group meaning fraction 1 control and fraction 1 RNase-treated lysates should be processed together): e.g. for 1 replicate, 2× 25 fractions (50 tubes) need to be processed, which can be divided into 5 groups of 8 tubes (4 fractions control + 4 fractions RNase-treated each) and 1 group of 10 tubes (5 fractions control + 5 fractions RNase-treated each). A checklist (Supplementary Table 1) can be very helpful to maintain reproducibility. In case of experiment settings with triplicates, we recommend to TCA-precipitate the replicates of each fraction together: for example, the three controls and the three RNase-treatments from fraction 1 together and so on for each of the 25 fractions.
▲ CRITICAL Wear safety glasses. TCA and acetone are hazardous substances.
-
51
50 Thaw the fractionated samples from Step 37 on ice and transfer 200 μl of each fraction to a fresh tube for precipitation.
∎ PAUSE POINT The leftover of the fractions can be kept frozen for several months at −80° C.
-
52
Add 200 μl HPLC-grade water to the 200 μl taken from each fraction.
-
53
Add 100 μl of 100% TCA to each sample and vortex them.
-
54
Incubate the samples for 15 min on ice.
-
55
Centrifuge the samples for 15 min at 4 °C and 17,000 g.
-
56
Remove all supernatants carefully with a pipet and discard them. The pellets contain the precipitated proteins.
▲ CRITICAL STEP Pellets will be very small or hardly visible. Make sure that the pellets do not get lost.
-
57
To wash the pellets, add 1 ml 10% TCA.
▲ CRITICAL STEP Prepare the 10% TCA solution freshly and keep it on ice.
-
58
Vortex the samples at maximum speed for 5 s.
-
59
Centrifuge the samples for 10 min at 4 °C and 17,000 g.
-
60
Remove the supernatants carefully with a pipet and discard them.
▲ CRITICAL STEP Again, the pellet will be very small or not visible.
-
61
Repeat the washing Steps (56–59) once.
-
62
Add 1 ml cold acetone to the pellets.
▲ CRITICAL STEP Acetone must be stored in explosion-proof fridges.
▲ CRITICAL STEP Acetone needs to be cold because cold acetone washes fewer proteins away compared to warm acetone. If storage in an explosion-proof fridge is not possible, acetone can be left on ice for two hours prior to use to cool it down.
-
63
Vortex the samples at maximum speed for 5 s.
-
64
Centrifuge the samples for 10 min at 4 °C and 17,000 g.
-
65
Remove the acetone carefully with a pipet without losing the pellet.
▲ CRITICAL STEP Again, the pellet will be very small or not visible.
-
66
Centrifuge the samples briefly at 17,000 g for 15 s at 4° C to spin down the remaining acetone.
-
67
Remove as much as possible of the remaining acetone with a pipet.
-
68
Repeat the washing steps with cold acetone (Steps 61–66) two more times.
-
69
Air dry the pellets for 5 to 10 min by leaving them sideway and open on the bench.
▲ CRITICAL STEP The pellets need to be dry before continuing to exclude a negative impact on the later procedure, but an overdried pellet is hard to dissolve in the next step. Close the tube and store the pellet or continue with the protocol once the pellet has dried.
∎ PAUSE POINT Dried pellets can be stored at −80 °C for up to one month.
Digestion and TMT labeling of peptides ● Timing 10 min + overnight digestion + 2 h
▲ CRITICAL A workflow of the whole TMT labeling procedure is provided in Fig. 6.
Fig. 6 |. TMT workflow for sucrose gradient fractions.
Proteins in the 25 sucrose density gradient fractions from control or RNase-treated samples were TCA precipitated and digested into peptides using trypsin. For each fraction, three biological replicates of the control sample and three biological replicates of the RNase-treated sample were labeled with individual tandem mass reporter tags (TMT, here TMT-6 plex), combined on a per-fraction basis and analyzed by LC-MS/MS. Fraction 1 is shown as an example here. The procedure is repeated for fractions 2 to 25.
-
70
Make 1:100 dilution (vol/vol) of sequencing grade trypsin in 50 mM ammonium bicarbonate (AmBic) in MS grade water. For digestion into peptides, add 20 μl of trypsin/AmBic solution per TCA precipitated sample from Step 68 and incubate overnight at 37 °C.
-
71
Quench the digests with 20 μl quenching buffer. Dry the peptides using a vacuum centrifuge (until dried, speed vacs only have one speed setting).
-
72
Desalt digests using SOLAμ HRP 96-well desalting plate (2 mg / 1 ml cartridge size, 1 well per test sample) and vacuum manifold. Condition each well of the desalting plate being used by washing with 2 X 500 μl of 100% (vol/vol) MeOH.
-
73
Equilibrate the desalting plate well with 2 X 150 μl of 0.1% (vol/vol) TFA.
-
74
Resuspend the digests in 100 μl 0.1% (vol/vol) TFA and load them onto the desalting plate well.
-
75
Wash the well with 2 X 150 μl 0.1% (vol/vol) TFA.
-
76
Elute peptides with 125 μl of 60% (vol/vol) MeOH and dry them using vacuum centrifuge.
∎ PAUSE POINT The dried sample can be stored at −80 °C for up to one month.
-
77
For tandem mass tag (TMT) labeling, prepare 166 mM HEPES, pH 8.5 using MS grade water. Vortex to mix.
-
78
Solubilize one aliquot of TMT reagent (0.8 mg) in 85 μl ACN.
-
79
Add 40 μl of 166 mM HEPES solution to the dried peptide sample from Step 75 (approximately 50 μg peptides per sample based on the initial protein amount used per sample for fractionation), vortex well at maximum speed in a vortexer for 5 s to solubilize, and spin the tube in a benchtop microcentrifuge for 30 s at RT to collect all the solution at the bottom of the tube.
-
80
Add 10 μl of TMT reagent in ACN from Step 77 to peptides from Step 78. Vortex to mix at maximum speed in a vortexer for 5 s and spin the tube in a benchtop microcentrifuge for 30 s at RT to collect all the solution at the bottom of the tube.
-
81
Repeat Steps 78 and 79 for each sample to be labeled (a total of six TMT reagent channels per fraction of the sucrose gradient: three biological replicates of the control sample and three biological replicates of the RNase-treated sample). For 25 sucrose gradient fractions collected, there is a total of 25 TMT 6-plexes, which will give in total 6*25 = 150 samples, as depicted in Fig. 6.
-
82
Let the reaction stand at RT to incubate for 1 h.
-
83
After 1 h, remove a small aliquot (1 μl) from each TMT channel to test labeling efficiency (test sample aliquots).
▲ CRITICAL STEP Freeze the rest of the samples remaining after the removal of the test sample aliquots at −80 °C until labeling efficiency has been tested and analyzed in the subsequent steps (Steps 83–88).
▲ CRITICAL STEP Do not quench the remaining sample (see Step 83) until labeling incorporation within each sample has been tested and analyzed in the combined test sample aliquots taken in this step.
∎ PAUSE POINT Peptides in 166 mM HEPES solution with TMT labeling reagent can be stored at −80 °C for up to two weeks.
-
84
Combine the six test sample aliquots for each TMT 6-plex, and quench at RT for 10 min using 1 μl of 500 mM ammonium bicarbonate.
-
85
Acidify the combined test sample by adding 100 μl of 0.1% (vol/vol) TFA in MS grade water.
▲CRITICAL STEP Check pH of the sample using pH-indicator strips and adjust the pH to 3 using 20% (vol/vol) TFA if necessary.
-
86
Desalt the test sample as described in Steps 71–74. Replace 0.1% (vol/vol) TFA with 7% (vol/vol) MeOH/0.1% (vol/vol) TFA to remove unreacted TMT reagent.
-
87
Elute the sample from the plate with 125 μl of 60% (vol/vol) MeOH and dry the sample using a vacuum centrifuge.
∎ PAUSE POINT The dried sample can be stored at −80 °C for up to one month
Mass spectrometric analysis of the test sample ● Timing 1 h
-
88
Reconstitute the dried test sample in 5 μl loading buffer and transfer to a minimum volume insert. Inject 2 μl of the test sample onto the column to be analyzed by LC-MS/MS using parameters described in the ‘Equipment set up’ section.
-
89
To determine the labeling efficiency, the database search is configured to search for TMT6 reporter ions at peptide N-termini and lysines as dynamic modifications.
Calculate the labeling efficiency percentage by: Labeling efficiency percentage = (Fully labeled PSMs/Total PSMs) * 100.
▲ CRITICAL STEP A minimum of 95% labeling efficiency by PSM identifications is critical for TMT labeling.
? TROUBLESHOOTING
▲ CRITICAL STEP Total reporter ion intensities for each replicate should be within 25% of each other.
Quenching and desalting TMT sample ● Timing 2 h
-
90
After achieving > 95% labeling efficiency, quench each of the TMT labeling reactions from Step 82 by adding 5 μl of 500 mM ammonium bicarbonate followed by incubation at RT for 10 min.
-
91
Combine the six TMT-labeled reactions for one fraction into one tube and acidify the sample by the addition of 500 μl of 0.1% (vol/vol) TFA in MS grade water.
▲ CRITICAL STEP Check pH of the sample using pH-indicator strips and adjust the pH to 3 using 20% (vol/vol) TFA if necessary.
-
92
Vacuum centrifuge the sample to dryness.
-
93
Desalt the sample using Oasis HLB 30 μm 96-well desalting plate (10 mg cartridge size, 1 well per TMT 6-plex) as described in steps 71–75. Replace 0.1% (vol/vol) TFA with 7% (vol/vol) MeOH/0.1% (vol/vol) TFA to remove unreacted TMT reagent.
▲ CRITICAL STEP It is important to use a desalting plate with appropriate cartridge size to achieve maximum recovery of the peptides after desalting.
∎ PAUSE POINT The dried sample can be stored at −80 °C for up to one year.
Mass spectrometry and data analysis ● Timing 4 h per multiplex
-
94
Reconstitute the dried sample from Step 92 in 5 μl loading buffer and analyze by LC-MS/MS platform using the configuration described in the Equipment setup section.
-
95Analyze the resulting data files using available software programs for TMT 6-plex experiments and an appropriate database, for instance a target decoy version of the human proteome FASTA (UniProt; www.uniprot.org) using the search parameters described in the table below.
Parameter Value Static modification Carbamidomethyl (C) +57.02146 Da TMT-6 (peptide N-term, K) +229.162932 Da Dynamic modification Oxidation (M) Enzyme Trypsin Maximum missed cleavages 4 Peptide mass tolerance (Da) 1.00 Peptide FDR (%) 1 -
96
Determine protein abundances by aggregating peptide TMT reporter ion intensities on a per protein, TMT channel, and sucrose gradient fraction basis using available software programs like for example MaxQuant41 or Andromeda42.
▲ CRITICAL STEP For inclusion into further analysis, quantification results per protein are required to be based on at least two unique peptide sequences.
? TROUBLESHOOTING
Statistical analysis of the mass spectrometry data ● Timing 1 h
▲ CRITICAL The analysis of the quantitative results per protein per fraction (see for example the provided test dataset “Mass_Spec_RawData_Sample.csv” as provided in the Supplementary Data) can be done using the free software “R”. A very convenient way to use R is to download RStudio, an integrated development environment (for more detailed instruction, refer to the “README.txt” file provided in the Supplementary Data). A complete workflow of the statistical analysis is provided in Fig. 7.
Fig. 7 |. Workflow of the statistical analysis.
The pipeline is divided into seven parts, which include: a normalization step between the replicates and overall normalization (PART 1), a search for the starting parameters in order to fit all the distribution profiles using Gaussian curves (PART 2 and 3), a quality control of the fit with subsequent correction if needed (PART 4), the assessment of the p-value and adjusted p-values (adj. p-value, PART 5), and the evaluation of the shifts and subsequent selection of the significant shifts (PART 6). Lastly, a final table is computed and exported for further analysis (PART 7).
▲ CRITICAL The complete script “MS_Statistical_Analysis.R” as provided in the Supplementary Data is adapted for the analysis of control and RNase-treated gradients (25 fractions each) in triplicate.
▲ CRITICAL Run first the script on the test dataset “Mass_Spec_RawData_Sample.csv” as provided in the Supplementary Data to familiarize yourself with the analysis. Format and organise your MS data exactly like it is in the test dataset to run the script on your own dataset.
-
97
Follow the instructions as found in the “README.txt” file provided in the Supplementary Data to proceed to the installation of RStudio.
-
98
Create a specific folder dedicated to the analysis of the MS dataset and copy the pipeline “MS_Statistical_Analysis.R” and the test dataset “Mass_Spec_RawData_Sample.csv” both provided in the Supplementary Data into that folder.
-
99
Set the folder in RStudio as working directory. Execute the pipeline using the test dataset from Step 97 at once using the command source(“MS_Statistical_Analysis.R”) or step by step as described in the following procedure steps.
▲ CRITICAL STEP The pipeline comprises seven parts as described in the “Experimental design”. The parts are dependent on each other and need to be processed all at once as in Step 98 or one after another in the order PART 1 to PART 7 as explained in the “README.txt” file provided in the Supplementary Data.
-
100
Use PART 1 to read the raw MS data, to proceed to the normalization between replicates using the mean value method, to apply the moving average of three fractions, to normalize the amount of all fractions for each protein to 100 for the total amount of the protein and to calculate average values for each protein in each fraction.
-
101
Use PART 2 of the pipeline to automatically find the starting parameters (amplitude at the maxima, position of the maxima and standard deviation) for the fitting process and to compute a Gaussian fit for each protein in the control and RNase-treated conditions.
-
102
Use PART 3 to compute a Gaussian fit for each protein in each individual replicate.
-
103
Use PART 4 to assess the quality of each fit and adjust the fit if needed.
-
104
Use PART 5 to compute the p-values (Student’s t-test) and the FDR-adjusted p-values for each maximum.
-
105
Use PART 6 to evaluate the shifts based on the selection criteria (shift distance > 1 fraction and at least one of the maxima associated with an adjusted p-value < 0.05).
-
106
Use PART 7 to compute and export the final data containing shift information for further analysis.
-
107
Format and organize your dataset (see “README.txt” file provided in the Supplementary Data) before running the script on it by repeating Steps 99–105.
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 35 | Different volumes in tubes after centrifugation | Evaporation during ultracentrifugation | Test all buckets with test gradients without lysate, marking the level of liquid in the tubes. Avoid using buckets for which evaporation is observed. |
| Variability in number of fractions recovered | Differences between pipet sets or users | Try to use always the same pipet set for every replicate. The replicates should also be performed by the same user. | |
| 42 | Difficulty to detect protein bands | Less than 1 mg lysate was loaded onto gradient; low sensitivity of antibody; low expression of protein | Load 1–2 mg lysate onto gradient; try an alternative antibody; test control proteins to verify western blot procedure; check protein expression in lysate. |
| 49 | Graph is inverted | The ImageJ software gave back inverted mean values | Subtract the inverted mean from 255 (for 8-bit images). |
| Negative protein amounts | Excel mixed up decimal separator “comma” and “point” | Check all mean values again; values cannot be higher than 255 (for 8-bit images). | |
| 88 | TMT label incorporation is <95% | Poor labeling efficiency | Repeat labeling (steps 76–82) for all channels: add another aliquot of TMT reagent to each sample for 1 h and proceed with testing and quenching as described (Steps 83–88). |
| 95 | No proteins / peptides left | Pellet got lost in one of the TCA washing steps | Tubes should be placed into the centrifuge always with the same orientation so that it is clear where the pellet is located. |
| Low protein / peptide concentration | Warm acetone was used for washing steps during TCA precipitation; pellets were overdried and could not be dissolved in the next steps | Acetone needs to be cold to prevent washing away proteins; decrease time for drying, or incubation in an ultrasonic bath with ice can help to re-dissolve the pellet. |
Timing
Step 1, cell culture: 1–3 d
Step 2, collect cells for cell lysate preparation: 30 min
Step 3–15, cell lysis: 2 h
Steps 16–24, BCA assay: 1 h
Step 25, gradient preparation: 1–3 h for 10 tubes
Step 26, lysate treatment: 1 h
Steps 27–33, ultracentrifugation: 19 h
Steps 34–37, fractionation: 15 min per gradient
Steps 38–49, western blot analysis and evaluation: 1.5 d
Steps 50–68, TCA precipitation 1–2 d for 50 samples
Steps 69–86, digestion and TMT labeling of peptides: 1.5 d
Steps 87–88, mass spectrometry analysis of the test sample: 1 h
Steps 89–92, quenching and desalting TMT sample: 2 h
Steps 93–95, mass spectrometry and data analysis: 4 h per multiplex
Steps 96–106, statistical analysis of the mass spectrometry data: 1 h
Anticipated results
This protocol describes how to identify RNA-dependent proteins by sucrose density gradient ultracentrifugation and subsequent western blot analysis for individual proteins of interest or quantitative MS measurement for the whole proteome. We provide a pipeline “MS_Statistical_Analysis.R” in the Supplementary Data for the automated statistical analysis of proteome-wide datasets along with a test dataset “Mass_Spec_RawData_Sample.csv” as depicted in Fig. 1.
In order to be able to detect individual proteins by western blot or MS analysis, it is important to load at least 1 mg of protein lysate on the 13.2 ml or 14 ml sucrose density gradient for centrifugation with SW40 Ti or SW41 Ti rotors respectively. Such an amount of protein lysate can be easily reached with HeLa cells when starting with 2 × 107 cells. In our protocol, based on the comparison of the untreated against the RNase-treated sample, the success of the whole experiment relies on an efficient digestion of the RNA molecules in the lysate. Fig. 3 illustrates how to control that the RNase digestion worked properly. The complete digestion of the two characteristic ribosomal RNA bands (18S rRNA and 28S rRNA) results in the absence of signal in the RNase-treated samples (Fig. 3a). Analysis of the control and RNase-digested samples with a Bioanalyzer should also show the complete disappearance of the two rRNA peaks and the presence of a small amount of RNA molecules below 200 nt length (Fig. 3b). Alternatively, RT-qPCR for selected RNA transcripts or a Bioanalyzer measurement of the isolated RNA molecules could be applied, as shown in the original paper24. An alternative to the present protocol could be treatment with individual RNases. Different effects of the various RNases would be expected since these have different substrate specificities (Table 1) and efficiencies, resulting in shifts of various magnitude and amplitude as illustrated in Supplementary Fig. 1.
The proper preparation of the sucrose density gradients is difficult to control, but at least, if the procedure is followed as shown in the Supplementary Movie 1, all the gradients should have at the end the same level of sucrose solution in the tube. If this is not the case, the deviating tubes should be discarded and the reproducibility of the pipetting should be verified.
The successful separation of the protein-protein and RNA-protein complexes is expected to lead to protein-specific distribution profiles in the fractions of the control and RNase-treated samples. Fig. 4a illustrates the complete RNA-dependent shift of the protein CTCF and the absence of shift for the RNA-independent protein ASNS. Additional expected results are shown in Supplementary Fig. 3a,b with an RNA-dependent protein NCL and an RNA-independent protein MCM7. Moreover, Supplementary Fig. 3c,f illustrate the reproducibility of the protocol in another cell line beside HeLa cells, confirming the RNA dependence of hnRNP U (a well-known RBP and expected to present an RNA-dependent shift) and the RNA independence of ASNS in A549 cells. Finally, Fig. 4b shows that the same results can be obtained in similar conditions with different rotors: SW40 Ti with 30,000 rpm/114,000 g and SW41 Ti with 30,000 rpm/111,000 g, both centrifuging for 18 h at 4 °C.
RNA-dependent proteins can shift to lower and/or to higher density sucrose fractions – what we respectively called left shifts and right shifts. A shift to the lower sucrose density fractions, a left shift, indicates a loss of binding partner(s). A shift towards the higher sucrose density fractions, a right shift, indicates a gain of interaction partner(s). Right shifts are much less abundant as shown in the original paper24 and also illustrated in the file “MS_Analysis_Shifts.csv” as provided in the Supplementary Data. The strong underrepresentation of right shifts can also be taken as a quality measure of the R-DeeP method as more losses than gains of interactions are expected upon RNA digestion. Right shifts are associated to positive shift distances, while left shifts are associated to negative shift distances (MS_Analysis_Shifts.csv). Some proteins can be enriched in multiple fractions, presenting profiles with multiple peaks. Such proteins are part of multiple complexes. Some are represented in the test dataset (“Mass_Spec_RawData_Sample.csv” as provided in the Supplementary Data) with up to four peaks in the control profile and up to three peaks in the RNase profile (MS_Analysis_Shifts.csv).
Fig. 6 depicts the workflow for the TMT labeling of the fractions. In the top panel, the Coomassie staining of the fractions shows already some differences between the control and the RNase-treated sample. This would also be a good indication that RNA-dependent proteins successfully shifted after RNase treatment.
Following the statistical analysis pipeline as depicted in Fig. 7, the final summary file (MS_Analysis_Shifts.csv) is exported to the working directory that was set at the beginning of the pipeline. This file contains information for each protein and can be further analyzed using table-based software to retrieve protein-specific information. The results can be compared to results from previous studies or integrated with results from protein-protein interactions databases25 or protein-protein complex databases26 as illustrated in the original paper24. A further analysis of the results could include analysis of GO43 terms or analysis of the protein sequences. In addition, the pipeline is associated with two functions producing graphical representations of the distribution of a protein of interest among the 25 fractions of the control and RNase-treated gradients. Examples of these graphical representations are illustrated in the file “HNRPU_HUMAN_FIT.pdf” as provided in the Supplementary Data (mean distributions with Gaussian fit curves) along with the file “HNRPU_HUMAN_ALL.pdf” (distribution of the protein in three replicates of the gradients). These graphical representations are helpful to obtain a quick and quantitative overview of the extent of the RNA dependence (or lack thereof) of a protein.
Altogether, the R-DeeP protocol shows a good overlap with the results of previous studies on RBPs24. In addition, our approach does not only target proteins binding directly to RNA but also proteins involved indirectly in interactions with RNA in complexes. The quantitative aspect of the sucrose density gradient ultracentrifugation coupled to the concept of RNA dependence is so far unique and allows the proportion of a protein that is RNA dependent to be quantified, as well as identifying a great number of proteins that had not been linked to RNA before24. We expect that the application of R-DeeP in other model systems will facilitate the detection of a number of non-expected protein-RNA interactions, and will promote the discovery of new RNA functions in the cell.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
A test dataset (Mass_Spec_RawData_Sample.csv), a result summary file (MS_Analysis_Shifts.csv) and two examples of graphics (“HNRPU_HUMAN_FIT.pdf” and “HNRPU_HUMAN_FIT.pdf”) are available in the Supplementary Data in this manuscript.
Code Availability
An analysis software pipeline is provided as “MS_Statistical_Analysis.R” in the Supplementary Data, along with an instruction file (README.txt). The whole analysis package including the examples can be directly downloaded from the documentation section of our database at http://R-DeeP.dkfz.de. The software is distributed as open source software under a 3-clause BSD license.
The analysis of the whole HeLa cell line dataset is available online as a user-friendly database at http://R-DeeP.dkfz.de.
Supplementary Material
Supplementary Video 1: Gradient_Preparation.mp4
Supplementary Video 2: Gradient_Fractionation.mp4
Supplementary Table 1 | Checklist for reproducible TCA precipitation.
Supplementary Fig. 1 | Analysis of samples treated with individual RNases. Western blot analysis for hnRNP U (RNA-dependent protein) in fractions of control and individual RNase-treated samples as indicated (RNase A, RNase I, RNase T1, RNase III or RNase H) in HeLa cells (N = 1). For all western blots, the fractions were loaded onto two membranes (1: fractions 1 to 13 and 2: fractions 14 to 24).
Supplementary Fig. 2 | Effect of various centrifugation times. Western blot analysis for hnRNP U (RNA-dependent protein) in fractions 12 to 25 of control (untreated) samples loaded onto 5% to 50% sucrose density gradients and centrifuged for 18 h, 6 h or 2 h, as indicated. One representative replicate out of two replicates is shown.
Supplementary Fig. 3 | Western blot analysis of individual proteins. a, Western blot analysis for Nucleolin (NCL, RNA-dependent protein) in 25 fractions of representative control and RNase-treated samples in HeLa cells. b, Same as in a for MCM7 (RNA-independent protein). c, same as in a for hnRNP U (RNA-dependent protein) but in A549 cells. d, same as in a for ASNS (RNA-independent protein) but in A549 cells. For all western blots, fractions 1 to 25 were loaded onto two membranes (1: fractions 1 to 13 and 2: fractions 14 to 25).
Supplementary Fig. 4 | Schematic overview of western blot quantification. The single steps for western blot quantification with the software ImageJ are shown (Steps 43-49). The generated intensity table can be copied and pasted to a program (e.g. Microsoft Excel) for further data processing and production of a graphical output. The intensities of each experiment are normalized as follows: for the fraction x.
Acknowledgements
This research was partially supported by the DKFZ NCT3.0 Integrative Project in Cancer Research (NCT3.0_2015.54 DysregPT) grant (to S.D.) and the grants R35GM119455 and P20GM113132 from the National Institute of General Medicine (to A.N.K.). We thank all members of the Diederichs and Kettenbach labs for their comments and suggestions on the method.
Footnotes
Competing financial interest statement
S.D. is co-owner of the siTOOLs Biotech GmbH, Martinsried, Germany, without any relation to this study or competing financial interests. The other authors declare no competing interests.
Supplementary information
Supplementary information is available for this paper.
Supplementary Data: Supplementary_Data.zip containing the following files
MS_Statistical_Analysis.R
README.txt
Mass_Spec_RawData_Sample.csv
MS_Analysis_Shifts.csv
HNRPU_HUMAN_FIT.pdf
HNRPU_HUMAN_ALL.pdf
References
- 1.Kishore S, Luber S & Zavolan M Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics 9, 391–404, doi: 10.1093/bfgp/elq028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook KB, Kazan H, Zuberi K, Morris Q & Hughes TR RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39, D301–308, doi: 10.1093/nar/gkq1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstberger S, Hafner M & Tuschl T A census of human RNA-binding proteins. Nat Rev Genet 15, 829–845, doi: 10.1038/nrg3813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray D et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177, doi: 10.1038/nature12311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltz AG et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46, 674–690, doi: 10.1016/j.molcel.2012.05.021 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Castello A et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406, doi: 10.1016/j.cell.2012.04.031 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kwon SC et al. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20, 1122–1130, doi: 10.1038/nsmb.2638 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Beckmann BM et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun 6, 10127, doi: 10.1038/ncomms10127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Perri JI et al. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat Commun 9, 4408, doi: 10.1038/s41467-018-06557-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad T et al. Serial interactome capture of the human cell nucleus. Nat Commun 7, 11212, doi: 10.1038/ncomms11212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He C et al. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell 64, 416–430, doi: 10.1016/j.molcel.2016.09.034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao X et al. Capturing the interactome of newly transcribed RNA. Nat Methods 15, 213–220, doi: 10.1038/nmeth.4595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R, Han M, Meng L & Chen X Capture and Identification of RNA-binding Proteins by Using Click Chemistry-assisted RNA-interactome Capture (CARIC) Strategy. J Vis Exp, doi: 10.3791/58580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castello A et al. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol Cell 63, 696–710, doi: 10.1016/j.molcel.2016.06.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullari M, Lyon D, Jensen LJ & Nielsen ML Specifying RNA-Binding Regions in Proteins by Peptide Cross-Linking and Affinity Purification. J Proteome Res 16, 2762–2772, doi: 10.1021/acs.jproteome.7b00042 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Trendel J et al. The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell 176, 391–403 e319, doi: 10.1016/j.cell.2018.11.004 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Queiroz RML et al. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat Biotechnol 37, 169–178, doi: 10.1038/s41587-018-0001-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urdaneta EC et al. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat Commun 10, 990, doi: 10.1038/s41467-019-08942-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomczynski P & Sacchi N Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162, 156–159, doi: 10.1006/abio.1987.9999 (1987). [DOI] [PubMed] [Google Scholar]
- 20.Brannan KW et al. SONAR Discovers RNA-Binding Proteins from Analysis of Large-Scale Protein-Protein Interactomes. Mol Cell 64, 282–293, doi: 10.1016/j.molcel.2016.09.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treiber T et al. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol Cell 66, 270–284 e213, doi: 10.1016/j.molcel.2017.03.014 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Sundararaman B et al. Resources for the Comprehensive Discovery of Functional RNA Elements. Mol Cell 61, 903–913, doi: 10.1016/j.molcel.2016.02.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrickson DG, Kelley DR, Tenen D, Bernstein B & Rinn JL Widespread RNA binding by chromatin-associated proteins. Genome Biol 17, 28, doi: 10.1186/s13059-016-0878-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caudron-Herger M et al. R-DeeP: Proteome-wide and Quantitative Identification of RNA-Dependent Proteins by Density Gradient Ultracentrifugation. Mol Cell 75, 184–199 e110, doi: 10.1016/j.molcel.2019.04.018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452, doi: 10.1093/nar/gku1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruepp A et al. CORUM: the comprehensive resource of mammalian protein complexes−−2009. Nucleic Acids Res 38, D497–501, doi: 10.1093/nar/gkp914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirkwood KJ, Ahmad Y, Larance M & Lamond AI Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Mol Cell Proteomics 12, 3851–3873, doi: 10.1074/mcp.M113.032367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hock J et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8, 1052–1060, doi: 10.1038/sj.embor.7401088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smirnov A et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113, 11591–11596, doi: 10.1073/pnas.1609981113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Martinez J, LaCava J & Rout MP Density Gradient Ultracentrifugation to Isolate Endogenous Protein Complexes after Affinity Capture. Cold Spring Harb Protoc 2016, doi: 10.1101/pdb.prot087957 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Behal RH & Cole DG Analysis of interactions between intraflagellar transport proteins. Methods Enzymol 524, 171–194, doi: 10.1016/B978-0-12-397945-2.00010-X (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kalthoff C, Alves J, Urbanke C, Knorr R & Ungewickell EJ Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem 277, 8209–8216, doi: 10.1074/jbc.M111587200 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Gagnon KT, Li L, Janowski BA & Corey DR Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc 9, 2045–2060, doi: 10.1038/nprot.2014.135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hafner M et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141, doi: 10.1016/j.cell.2010.03.009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sysoev VO et al. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun 7, 12128, doi: 10.1038/ncomms12128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn JL & Chang HY Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166, doi: 10.1146/annurev-biochem-051410-092902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castello A et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc 8, 491–500, doi: 10.1038/nprot.2013.020 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Castello A et al. Identification of RNA-binding domains of RNA-binding proteins in cultured cells on a system-wide scale with RBDmap. Nat Protoc 12, 2447–2464, doi: 10.1038/nprot.2017.106 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Pillai RS et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576, doi: 10.1126/science.1115079 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Li Q & Chen XZ Detecting protein-protein interactions by Far western blotting. Nat Protoc 2, 3278–3284, doi: 10.1038/nprot.2007.459 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Tyanova S, Temu T & Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319, doi: 10.1038/nprot.2016.136 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Cox J et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805, doi: 10.1021/pr101065j (2011). [DOI] [PubMed] [Google Scholar]
- 43.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1: Gradient_Preparation.mp4
Supplementary Video 2: Gradient_Fractionation.mp4
Supplementary Table 1 | Checklist for reproducible TCA precipitation.
Supplementary Fig. 1 | Analysis of samples treated with individual RNases. Western blot analysis for hnRNP U (RNA-dependent protein) in fractions of control and individual RNase-treated samples as indicated (RNase A, RNase I, RNase T1, RNase III or RNase H) in HeLa cells (N = 1). For all western blots, the fractions were loaded onto two membranes (1: fractions 1 to 13 and 2: fractions 14 to 24).
Supplementary Fig. 2 | Effect of various centrifugation times. Western blot analysis for hnRNP U (RNA-dependent protein) in fractions 12 to 25 of control (untreated) samples loaded onto 5% to 50% sucrose density gradients and centrifuged for 18 h, 6 h or 2 h, as indicated. One representative replicate out of two replicates is shown.
Supplementary Fig. 3 | Western blot analysis of individual proteins. a, Western blot analysis for Nucleolin (NCL, RNA-dependent protein) in 25 fractions of representative control and RNase-treated samples in HeLa cells. b, Same as in a for MCM7 (RNA-independent protein). c, same as in a for hnRNP U (RNA-dependent protein) but in A549 cells. d, same as in a for ASNS (RNA-independent protein) but in A549 cells. For all western blots, fractions 1 to 25 were loaded onto two membranes (1: fractions 1 to 13 and 2: fractions 14 to 25).
Supplementary Fig. 4 | Schematic overview of western blot quantification. The single steps for western blot quantification with the software ImageJ are shown (Steps 43-49). The generated intensity table can be copied and pasted to a program (e.g. Microsoft Excel) for further data processing and production of a graphical output. The intensities of each experiment are normalized as follows: for the fraction x.
Data Availability Statement
A test dataset (Mass_Spec_RawData_Sample.csv), a result summary file (MS_Analysis_Shifts.csv) and two examples of graphics (“HNRPU_HUMAN_FIT.pdf” and “HNRPU_HUMAN_FIT.pdf”) are available in the Supplementary Data in this manuscript.