Abstract
Diapause in response to seasonality is an important model for rapid evolutionary adaptation that is highly genetically variable, and experiences strong natural selection. Forward genetic methods using various genomic and transcriptomic approaches have begun to characterize the genetic architecture and candidate genes underlying diapause evolution. Largely in parallel, reverse genetic studies have identified functional roles for candidate genes that may or may not be genetically variable. We illustrate the disconnect between the evolutionary and physiological literature using a suite of studies of the role of the circadian clock in diapause regulation. These extensive studies in two different disciplines provide excellent opportunities for integration, which should facilitate rapid progress in understanding both the regulation and evolution of diapause.
Introduction
Diapause, a form of dormancy in insects and other arthropods, is an adaptive and plastic phenotype that allows insects to persist in seasonally variable environments. Insects enter diapause in advance of unfavorable conditions and in response to predictive environmental cues (Box 1). Because it allows insects to persist and adapt to new environments, diapause has been a powerful model for understanding evolution by natural selection [1]. Moreover, natural populations often harbor ample genetic variation affecting both the capacity for and the timing of diapause (Box 1; [2,3]). This combination of strong selection and segregating genetic variation allows diapause to rapidly evolve over contemporary timescales, including in response to changing climates [4], developing agricultural practices [5], and during biological invasions and range expansions [6]. Thus, genetics, selection and evolution of diapause informs several basic and applied topics, including the genetic architecture of rapid adaptation, responses to climate change, evolutionary physiology, and ecological genetics, thereby offering opportunities to uncover novel targets for pest control (Fig. 1).
Box 1.
Diapause concepts and terms
Diapause:
A physiologically-dynamic and hormonally-controlled state of decelerated or arrested morphological development that allows insects to survive unfavorable conditions. Diapause is typically induced/terminated by environmental stimuli (e.g., photoperiod or temperature), though some diapause responses may appear functionally obligate in the field.
Phases of diapause:
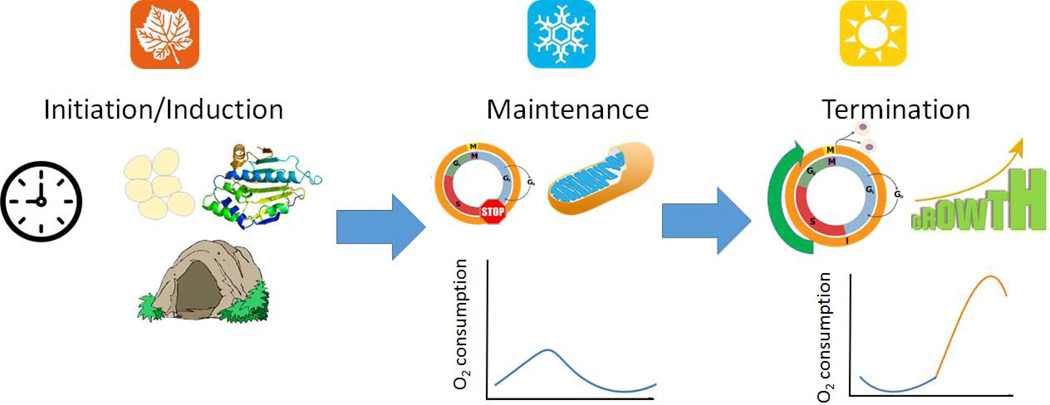
Different eco-physiological states through which diapausing organisms progress (Fig. B1). They are typically described as:
1. Initiation/Induction:
The period before an insect enters diapause, characterized by some combination (but not always all) of:
assessing environmental conditions which may be mediated through measuring daylength via the circadian clock
regulating energetic resources (increased feeding, decreased insulin signaling/PI3K and upregulating fatty acid synthesis)
upregulating stress responses (e.g., HSPs [pictured], FOXO activity, cryoprotectants, immune responses)
regulating hormone levels, e.g., prothoracicotropic hormone (PTTH; larval and pupal diapause), Ecdysone (20HE; pupal diapause) or Juvenile Hormone (JH; adult diapause)
seeking out protected overwintering sites
2. Maintenance:
The period during which the organism is unresponsive to changes in the environment. This usually characterized by cell cycle arrest and decreases in transcription, cellular respiration and metabolism, allowing organisms to conserve energy reserves.
3. Termination:
The period during which the organism becomes competent to resume normal growth, development and activity in response to favorable environmental conditions. This is generally characterized by increases in transcription, cellular respiration, metabolism, and hormonal signaling.
Population-level diapause metrics:
Genetic, physiological and ecological studies generally focus on the photoperiodic initiation (more common) or termination (less common) of diapause because it is closely-tied to seasonal timing and phenology. Two common metrics include:
1. Diapause incidence:
The proportion of individuals that enter diapause under unambiguous, diapause-inducing conditions (e.g., short days and/or low temperatures).
2. Diapause timing:
The seasonal timing of diapause initiation or termination.
Timing is also inferred by measuring Critical photoperiod (CPP) in insects with photoperiodic diapause: The number of hours of light in a 24 hour Light/Dark cycle that will cause 50% of the population to enter or terminate diapause (may be inadequate in some cases, see [1])
Both diapause incidence and timing vary across latitudinal and altitudinal clines, e.g., CPP and diapause incidence are typically positively correlated with latitude and altitude.
1. Saunders DS: Insect clocks: Elsevier; 2002.
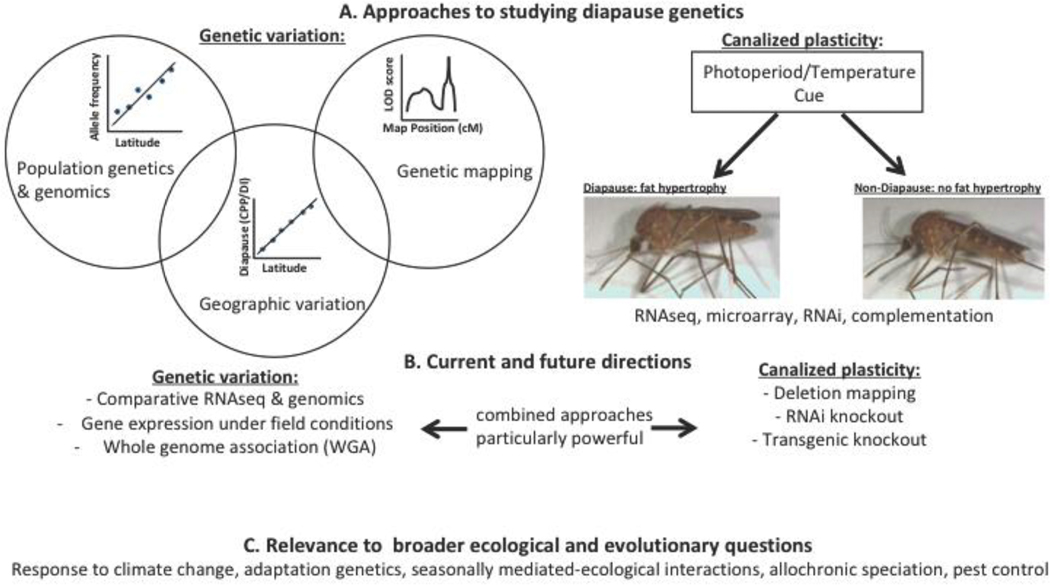
Figure 1.
Overview of experimental approaches investigating diapause genetics and evolution (A), potential future directions (B), and relevance of diapause genetics and evolution to broader ecological and evolutionary questions (C). Panel A highlights many ways in which genetic variation and plasticity are evaluated. Studies analyze genetic variation in diapause by first documenting geographic variation in diapause phenotypes. Crossing populations with divergent phenotypes can then allow researchers to identify the genetic variants that underlie the observed phenotypic variation. Additionally, population genetics & genomics can be used to identify alleles that show coincident patterns of geographic variation. Alternatively, many studies explore the genetic regulation of canalized plasticity in the diapause response by exposing experimental cohorts of a single population to diapause-inducing and diapause-averting cues, and measuring differences in gene expression (RNAseq/microarrays) or determining how manipulating the level of a transcript of interest (RNAi) or the genetic background (complementation) influences the diapause response. Panel B highlights potential methods that are or can be used to identify genetic factors affecting diapause among and within populations, with the suggestion that combining approaches provides a particularly strong basis for identifying causal genetic factors. Panel C highlights the potential of understanding the regulation and evolution of diapause to inform research relating to several critical biological topics.
Studying diapause has provided many important insights into adaptation to variable climates and how physiological plasticity is regulated in animals. Recently, rapid advances in “-omics” technologies have led to exciting progress in understanding the genetic and physiological mechanisms of diapause regulation and evolution. Previous reviews discuss the physiological stages of diapause progression [7], the hormonal regulation of diapause [8], epigenetic regulation of diapause [9], and diapause energetics [10]. Here, we focus on recent studies of genetic variation in diapause, emphasizing the progress achieved by “-omics” approaches. We also point out a continuing disconnect between forward genetic methods and reverse, functional genetics, using the role of the circadian clock in regulating diapause as an example. Combining these tools provides a powerful approach to better understand how diapause evolves, and to better leverage genetic variation to elucidate how this complex phenotype is regulated.
Genetic variation for diapause phenotypes over space and time
A rich history of studying genetic differentiation across latitudinal gradients (i.e., “clines”) provides an expansive view of how insects adapt to spatial environmental heterogeneity [11,12]. Clines in diapause incidence and/or annual timing (Box 1) have been described in hundreds of species and are among the most robust biogeographic trends in animals [3,13]. These clines are formed as geographic populations adapt to local conditions along seasonal gradients. Geographic variation in diapause phenotypes (Box 1) has often been leveraged to infer genetic architecture by crossing laboratory strains derived from different geographic populations [14,15]. Early studies often identified simple, nearly Mendelian factors underlying diapause variation. The advent of DNA sequencing and genetic tools enabled finer, molecular genetic dissection, particularly for a few well-studied and experimentally tractable organisms (Table 1). For example, Williams et al. [16] crossed geographically-derived lines of Drosophila melanogaster and identified a locus of major effect for diapause incidence. Deletion mapping further resolved the locus to variants in the insulin-regulated phosphatidylinositol 3-kinase (PI3-kinase) gene, Dp110, which also contributed to diapause differences among geographically disparate populations. Subsequent tests for associations between the variants and gene expression were equivocal, but the identification of Dp110 was exciting because it supported years of comparative endocrinology suggesting a role for insulin signaling in diapause. Paaby et al. [17] found similar clines in alleles of the insulin-like receptor (InR) among D. melanogaster populations from both North America and Australia, though alleles were only indirectly related to diapause.
Table 1.
List of studies by species that apply various approaches that fall under the umbrellas of forward genetic approaches (unbiased screens for genetic variation) and reverse genetic or targeted approaches (focusing on a candidate gene or genes). Organisms studied using four or more approaches are listed in bold. With the exception of D. melanogaster and C. pipiens, most species have been studied using only one or two approaches. Moreover, single-species transcriptomics has been the most liberally applied approach. *Reference numbers refer to references listed in supplemental table S1, not to references in the main text.
| Class of Approach | Forward (untargeted) | Genetic | Reverse Genetic or targeted | ||||
|---|---|---|---|---|---|---|---|
| Approach | GENETIC MAPPING (line crossing) | GENETIC ASSOCIATION (outbred) | SINGLE POPULATION TRANSCRIPTOMICS | COMPARATIVE TRANSCRIPTOMICS | REGULATION GENOMICS | MANIPULATIVE FUNCTIONAL GENETICS | TARGET GENE VARIATION |
| Questions/Goals | candidate genes; genetic architecture | candidate genes; genetic architecture | Transcriptional basis | Genetic variation in transcription | regulatory regions or molecules | target gene functional roles | targeted associations with phenotype or geography |
| Study organism | D. Melanogaster | D. melanogaster | D. melanogaster | R. pomonella | C. pipiens | D. melanogaster | D. melanogaster |
| C. pipiens | R. pomone11a | C. pipiens | O. nubilalis | S. crassipalpis | C. pipiens | O. nubilalis | |
| O. nubilalis | P. aegeria | R. pomonella | W. smithii | C. costata | N. vitripennis | ||
| W. smithii | Ae. albopictus | S. crassipalpis | |||||
| B. minax | R. pedestris | ||||||
| D. antiqua | M. siamensis | ||||||
| S. crassipalis | |||||||
| D. montana | |||||||
| C. costata | |||||||
| M. rotunda | |||||||
| A. gifuensis | |||||||
| D. antiqua | |||||||
| B. mori | |||||||
| T. diversipes | |||||||
| H. cunea | |||||||
| References* | 1 – 4 | 5 – 7 | 8 – 22 | 23 – 25 | 26 – 27 | 28 – 32 | 33 – 35 |
Schmidt et al. [18] also leveraged naturally segregating geographic variation, performing Quantitative Trait Locus (QTL) analysis and fine-mapping by crossing lines of D. melanogaster derived from geographic populations that differed in diapause incidence. Their experiments identified a locus of major effect, couch potato (cpo), which encodes an RNA binding protein that is highly expressed in the ring gland, the primary endocrine tissue of D. melanogaster. Polymorphisms at multiple SNPs were correlated with latitude, which also predicts diapause incidence in North American D. melanogaster. Furthermore, polymorphisms in cpo also change seasonally in synchrony with diapause expression [19]. Thus, the frequency of cpo polymorphisms is associated with diapause in both space (clinal variation) and time (seasonal variation). Higher levels of cpo expression are also associated with diapause maintenance in the Northern house mosquito, Culex pipiens, though this is the opposite relationship to that observed in D. melanogaster [20].
Associations between diapause and polymorphisms in genes involved in the circadian clock (Fig. 2) have also been uncovered using both targeted and untargeted approaches. Again studying different populations of D. melanogaster, Tauber et al. [21] identified genetic variants of timeless in isofemale lines varying in diapause incidence. They identified a recently derived allele (ls-tim) encoding an additional 23 N-terminal amino acids relative to the ancestral (s-tim) allele. The derived ls-tim allele has a weaker physical interaction with the circadian light receptor Cryptochrome1 (CRY1) than the ancestral timeless allele (s-tim). Thus, the ls-tim allele is predicted to attenuate photosensitivy and promote entry into diapause even under long photoperiods [22]. The allele exhibits a latitudinal cline across North America coincident with the cline in diapause incidence [23,24]. Surprisingly, in Europe, the ls-tim allele frequency decreases with increasing latitude, but this is likely due to the recent spread of the allele from Italy to Scandinavia [25]. Other taxa exhibit geographic variation in tim sequence polymorphism [26] and expression levels [27]. Furthermore, polymorphism in another core circadian clock gene, period, also varies with latitude and is associated with diapause variation in D. melanogaster [28], the European corn borer [29], the parasitic wasp Nasonia vitripennis [30], and the speckled wood butterfly, Pararge aegeria [26]. Thus, studies of geographic variation provide substantial evidence for a link between clock gene polymorphisms and diapause (see “The role of the circadian clock” section below).
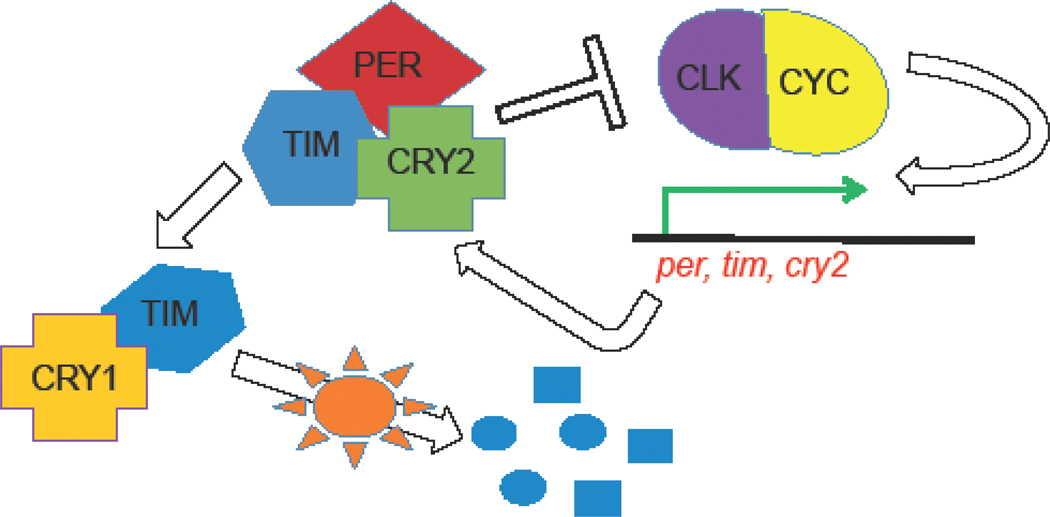
Figure 2.
The structure of the insect circadian clock in mosquitoes and other insects. The positive transcription factors CLOCK (CLK) and CYCLE (CYC) regulate the expression of the core clock genes, period (per), timeless (tim) and cryptochrome2 (cry2), whose respective proteins inhibit the action of CLK and CYC, thereby repressing their own transcription. The TIM protein is degraded by CRY1 in the presence of light (Adapted from [53]).
Quantitative trait locus (QTL) analyses of crosses between seasonal populations of the European corn borer, Ostrinia nubilalis [31], geographic populations of the pitcher plant mosquito, Wyeomyia smithii [32], and members of the Culex pipiens complex [33] also identified loci of major effect on diapause termination and diapause incidence. These results suggest that major effect loci may often segregate in natural populations. However, QTL analyses are typically unable to detect and quantify variation accounted for by loci of small effect. Furthermore, crosses between isogenic lines explore only a subset of variation segregating in natural populations, and thus may fail to detect polygenic variation. Recent studies using genome-wide association techniques suggest that segregating, polygenic variation may indeed be abundant (see Table 1). For example, a whole genome resequening study in the speckled wood butterfly [26] used Genome Wide Association (GWA) analysis to show that many loci of small effect must contribute to population differences in diapause incidence alongside several loci of major effect. A study of temperature-sensitive diapause termination in the apple maggot fly, Rhagoletis pomonella, also used GWA with reduced representation genome resequencing [34]. Despite finding clear evidence for heritable genetic variation, there was no evidence for loci of major effect contributing to that variation, thus supporting a polygenic model. As diapause phenotypes are highly variable in natural populations [3], it seems likely that loci of minor effect play a critical role in the evolution of diapause phenotypes, although the relative influence of loci of major effect may vary across different species.
Transcriptomic variation
Many studies in a broad range of insects and other invertebrates have compared the transcriptomes of diapausing and nondiapausing individuals, but typically in a single population of a single species (Table 1) [35,36]. Observations of similar patterns of gene expression during diapause across species, first through studies on single genes [37], then through transcriptomics, led to the proposal that particular genes [38] or signaling pathways [39] may represent a common “genetic toolkit” for diapause due to evolutionary convergence. A meta-analysis of single-species diapause transcriptomic studies indeed revealed that a common set of transcripts (including circadian clock genes) are differentially regulated during diapause, consistent with convergent evolution rather than shared evolutionary history [35]. Mechanisms that regulate gene expression (DNA methylation, histone modifications and/or individual miRNAs) have also been implicated in diapause regulation in various insects [9], but again, most of this work focuses on single populations of diapausing and nondiapausing species, and there is limited evidence that the same mechanisms are consistently used to regulate diapause across species.
In contrast to the large number of single population studies, only three studies have compared the diapause transcriptomes of genetically distinct populations of the same species. The first compared strains of the European corn borer (Ostrinia nubilalis) [40]; the second compared apple-infesting and hawthorn-infesting populations of R. pomonella [41]; and the third compared the diapause transcriptome of southern and northern populations of Wyeomyia smithii [42]. The goal of all three studies was to identify transcriptional changes contributing to population-level differences in diapause timing (specifically, diapause termination), and to link those changes to genetic variants.
An important advantage of comparative transcriptomic studies is the opportunity to simultaneously examine genetic variation in transcript expression and transcript sequence. Combining transcriptome comparisons with QTL studies is also particularly powerful. For example, all three comparative transcriptome studies identified polymorphisms or gene expression differences potentially related to differences in diapause termination. In the European corn borer, 48 transcripts with either fixed amino acid differences or differential expression during diapause termination among strains mapped to a chromosomal rearrangement previously identified as a major genetic factor influencing diapause termination [40]. Several of these genes are involved in insulin signaling and the circadian clock, which have previously been implicated in diapause regulation as discussed above. Meyers et al. [41] found between-population expression differences in insulin and Wnt signaling, suggesting that these pathways contribute to the early spring emergence of apple-infesting flies relative to the hawthorn-infesting flies. Finally, Emerson et al. [42] identified a transcript designated WsPpdrg1 that was differentially expressed among geographic populations during diapause termination. WsPpdrg1 maps to a major QTL affecting critical photoperiod (CPP; Box 1) and is hypothesized to be involved in photoreception or signal transduction based on its similarity to D. melanogaster proteins. A cautionary note relevant to all of these studies is that diapause transcriptomes may differ substantially between laboratory and ecologically realistic field conditions [36].
The intersection of evolutionary genetics and functional studies: the role of the circadian clock
Over eighty years ago the German botanist Erwin Bünning hypothesized that the same mechanism that organisms use to measure daily (circadian) time might also be used to measure seasonal time (photoperiodic) and thereby initiate responses such as diapause [43]. With the advent of modern molecular genetics, evidence supporting a role for circadian clock genes in the evolution of photoperiodism comes from: 1) screens of geographic or population-level genetic variation, 2) gene expression assays, and 3) genetic knockdown to evaluate functional consequences. In insects, the circadian clock is composed of multiple feedback loops controlled by cycling levels of key signaling proteins and transcription factors (Fig. 2). Studies of genetic variation, including those detailed above, suggest that diapause is associated with variants of the core clock genes period and timeless [21,26]. Variation in clock gene expression has been linked to natural variation in diapause [27,40]. Furthermore, knocking down clock gene transcripts with RNA interference (dsRNAi) also suggests a functional link between the clock and photoperiodic diapause. For example, suppressing period causes multiple species of insects to either delay or avert diapause [44–47].
Nevertheless, the mechanistic link between the clock and the photoperiodic timer remains unresolved. Clock genes have been linked to a range of physiological processes including metabolism and hormonal signaling (Juvenile hormone in adult insects; [45,47,48]). However, techniques such as transcriptome-wide screens have not identified pathways from photoperiod perception to the generation of the diapause phenotype. Additionally, unbiased, genome-wide methods often identify variants with no experimentally determined connections to the circadian clock e.g., [18,26,42]. Emerson et al. [49] noted that it is difficult to determine whether core circadian genes influence seasonal responses via their role in the circadian clock and perception of daylength or whether they pleiotropically regulate genes outside of the clock to generate diapause phenotypes. However, in the intervening decade since that review, we have yet to determine how clock gene variants mechanistically influence diapause (but see [50], though the photoperiodic phenotype is not diapause).
Opportunities for future progress
We suggest that greater integration among what have previously been largely parallel efforts in functional genetics, transcriptomics, and evolutionary genomics would enable more rapid progress towards identifying the molecular regulators of diapause and the genetic basis of diapause evolution. Table 1 illustrates that with a few exceptions, most ‘-omics’ studies of diapause have been carried out in separate species. Transcriptomics of single populations of a single species are by far the most common. These single population transcriptomic comparisons (diapause to nondiapause) have uncovered similar molecular regulators across species. However, comparative transcriptomic approaches leveraging well-characterized intra-specific variation in diapause phenotypes (e.g., [40–42]) provide a stronger connection between candidate genes and diapause phenotypes.
Likewise, forward genetic methods such as GWAS or QTL analysis followed by reverse genetics (e.g., generation of null mutants or RNAi) provide more robust evidence for the roles of candidate genes in diapause regulation. The studies employing genetic screens followed by deletion mapping described above illustrate the power of these combined approaches in D. melanogaster [16,18]. In principle, reverse genetics (e.g., creating loss of function variants) followed by transcriptomics could also identify novel mechanisms. Genome sequencing and de novo assembly are increasingly accessible (e.g., [26]), RNAi has now been implemented in many species, and transgenic approaches show great promise in non-model systems (e.g., [51]) and are now becoming the preferred approach in some well-established study species [52]. We anticipate that these advances will enable powerful integrative approaches to rapidly advance our understanding of how diapause is regulated and evolves in species with genetically variable diapause responses. These advances will contribute significantly to broader issues such as determining the genetic architecture of rapid adaptation, evolutionary responses to climate change, and the identification of novel targets for pest control (Fig. 1).
Supplementary Material
Figure B1.
Phases of diapause development. In temperate environments diapause initiation, maintenance, and termination typically take place in the fall, winter, and spring, respectively.
Highlights.
Insect diapause is an important model for evolution by natural selection.
Genomics-enabled methods are now elucidating genetic architecture and candidate genes.
Forward and reverse genetic studies of diapause are typically applied in parallel.
Combining these approaches will facilitate both evolutionary and physiological studies.
Parallel and integrative efforts are illustrated using studies of the circadian clock.
Acknowledgements
The authors wish to thank Richard Clark and one anonymous reviewer for useful comments on an earlier version of the manuscript. This review was supported by NSF DEB 1638951 to GJR, NIH 1R01AI132409–01A1 to PAA, and an Interdisciplinary Seeds Grant from the OSU Infectious Diseases Institute to MEM.
Footnotes
Competing interests
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bradshaw WE, Lounibos LP: Evolution of dormancy and its photoperiodic control in pitcher plant mosquitos. Evolution 1977, 31:546–567. [DOI] [PubMed] [Google Scholar]
- 2.Danks HV: Insect dormancy: an ecological perspective, vol 1: Biological Survey of Canada (Terrestrial Artropods) Ottawa, ON; 1987. [Google Scholar]
- 3.Tauber MJ, Tauber CA, Masaki S: Seasonal adaptations of insects. New York: Oxford University Press; 1986. [Google Scholar]
- 4.Bradshaw WE, Holzapfel CM: Genetic shift in photoperiodic response correlated with global warming. Proceedings of the National Academy of Sciences of the United States of America 2001, 98:14509–14511.• This study demonstrates rapid evolution of diapause timing (CPP) in response to global warming.
- 5.Landis D, Levine E, Haas M, Meints V: Detection of prolonged diapause of northern corn rootworm in Michigan (Coleoptera: Chrysomelidae). The Great Lakes Entomologist 2017, 25:6. [Google Scholar]
- 6.Armbruster PA: Photoperiodic diapause and the establishment of Aedes albopictus (Diptera: Culicidae) in North America. Journal of medical entomology 2016, 53:1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostal V: Eco-physiological phases of insect diapause. Journal of Insect Physiology 2006, 52:113–127. [DOI] [PubMed] [Google Scholar]
- 8.Denlinger D, Yocum G, Rinehart J, Gilbert L: Hormonal control of diapause. Comprehensive Insect Molecular Science 2004, 3:615–650. [Google Scholar]
- 9.Reynolds JA: Epigenetic influences on diapause In Advances in Insect Physiology. Edited by: Elsevier; 2017:115–144. vol 53.] [Google Scholar]
- 10.Hahn DA, Denlinger DL: Energetics of insect diapause. Annual review of entomology 2011, 56:103–121. [DOI] [PubMed] [Google Scholar]
- 11.Endler JA: Geographic variation, speciation, and clines. Princeton, NJ: Princeton University Press; 1977. [PubMed] [Google Scholar]
- 12.Hoffmann AA, Weeks AR: Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the termperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 2007, 129:133–147. [DOI] [PubMed] [Google Scholar]
- 13.Danilevsky AS: Photoperiodism and seasonal development in insects. Edinburgh, U.K.: Oliver and Boyd; 1965. [Google Scholar]
- 14.Lumme J, Lakovaara S, Oikarinen A, Lokki J: Genetics of photoperiodic diapause in Drosophila littoralis. Hereditas 1975, 79:143–148. [DOI] [PubMed] [Google Scholar]
- 15.Riihimaa A, Kimura MT, Lumme J, Lakovaara S: Geographical variation in the larval diapause of Chymomyza costata (Diptera; Drosophilidae). Hereditas 1996, 124:151–163. [Google Scholar]
- 16.Williams KD, Busto M, Suster ML, So AKC, Ben-Shahar Y, Leevers SJ, Sokolowski MB: Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proceedings of the National Academy of Sciences of the United States of America 2006, 103:15911–15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS: Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Molecular Ecology 2010, 19:760–774. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF: An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 2008, 105:16207–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cogni R, Kuczynski C, Koury S, Lavington E, Behrman EL, O’Brien KR, Schmidt PS, Eanes WF: The intensity of selection acting on the couch potato gene-spatial-temporal variation in a diapause cline. Evolution 2014, 68:538–548. [DOI] [PubMed] [Google Scholar]
- 20.Zhang QR, Denlinger DL: Elevated couch potato transcripts associated with adult diapause in the mosquito Culex pipiens. J. Insect Physiol. 2011, 57:620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, et al. : Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 2007, 316:1895–1898. [DOI] [PubMed] [Google Scholar]
- 22.Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, Landskron J, Stanewsky R, Piccin A, Rosato E, Zordan M, et al. : A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 2007, 316:1898–1900. [DOI] [PubMed] [Google Scholar]
- 23.Pegoraro M, Zonato V, Tyler ER, Fedele G, Kyriacou CP, Tauber E: Geographical analysis of diapause inducibility in European Drosophila melanogaster populations. Journal of Insect Physiology 2017, 98:238–244. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt PS, Matzkin L, Ippolito M, Eanes WF: Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 2005, 59:1721–1732. [PubMed] [Google Scholar]
- 25.Zonato V, Vanin S, Costa R, Tauber E, Kyriacou CP: Inverse European Latitudinal Cline at the timeless Locus of Drosophila melanogaster Reveals Selection on a Clock Gene: Population Genetics of ls-tim. Journal of Biological Rhythms 2018, 33:15–23. [DOI] [PubMed] [Google Scholar]
- 26.Pruisscher P, Nylin S, Gotthard K, Wheat CW: Genetic variation underlying local adaptation of diapause induction along a cline in a butterfly. Molecular Ecology 2018, 27:3613–3626.•• This study combined whole-genome sequencing, genome-wide association and analysis of allele-frequency clines to identify genomic regions affecting diapause induction and to infer alleles of both large and small effect contributing to evolutionary divergence.
- 27.Mathias D, Jacky L, Bradshaw WE, Holzapfel CM: Geographic and developmental variation in expression of the circadian rhythm gene, timeless, in the pitcher-plant mosquito, Wyeomyia smithii. J. Insect Physiol. 2005, 51:661–667. [DOI] [PubMed] [Google Scholar]
- 28.Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP: Natural variation in a Drosophila clock gene and temperature compensation. Science 1997, 278:2117–2120. [DOI] [PubMed] [Google Scholar]
- 29.Levy RC, Kozak GM, Wadsworth CB, Coates BS, Dopman EB: Explaining the sawtooth: latitudinal periodicity in a circadian gene correlates with shifts in generation number. Journal of Evolutionary Biology 2015, 28:40–53. [DOI] [PubMed] [Google Scholar]
- 30.Paolucci S, Salis L, Vermeulen CJ, Beukeboom LW, van de Zande L: QTL analysis of the photoperiodic response and clinal distribution of period alleles in Nasonia vitripennis. Molecular Ecology 2016, 25:4805–4817. [DOI] [PubMed] [Google Scholar]
- 31.Dopman EB, Pérez L, Bogdanowicz SM, Harrison RG: Consequences of reproductive barriers for genealogical discordance in the European corn borer. Proceedings of the National Academy of Sciences 2005, 102:14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradshaw WE, Emerson KJ, Catchen JM, Cresko WA, Holzapfel CM: Footprints in time: comparative quantitative trait loci mapping of the pitcher-plant mosquito, Wyeomyia smithii. Proceedings of the Royal Society B-Biological Sciences 2012, 279:4551–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori A, Romero‐Severson J, Severson D: Genetic basis for reproductive diapause is correlated with life history traits within the Culex pipiens complex. Insect molecular biology 2007, 16:515–524. [DOI] [PubMed] [Google Scholar]
- 34.Ragland GJ, Doellman MM, Meyers PJ, Hood GR, Egan SP, Powell THQ, Hahn DA, Nosil P, Feder JL: A test of genomic modularity among life-history adaptations promoting speciation with gene flow. Molecular Ecology 2017, 26:3926–3942. [DOI] [PubMed] [Google Scholar]
- 35.Ragland GJ, Keep E: Comparative transcriptomics support evolutionary convergence of diapause responses across Insecta. Physiological Entomology 2017, 42:246–256. [Google Scholar]
- 36.Yocum GD, Childers AK, Rinehart JP, Rajamohan A, Pitts-Singer TL, Greenlee KJ, Bowsher JH: Environmental history impacts gene expression during diapause development in the alfalfa leafcutting bee, Megachile rotundata. Journal of Experimental Biology 2018, 221:12.•• The authors examined gene expression differences in bees exposed to laboratory and field conditions and found that the environmental history of the insects had a profound impact on gene expression such that the greatest differential expression was seen between lab and field population late in diapause.
- 37.Denlinger DL: Regulation of diapause. Annual Review of Entomology 2002, 47:93–122. [DOI] [PubMed] [Google Scholar]
- 38.Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA: Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. Proceedings of the Royal Society B-Biological Sciences 2013, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostal V, Stetina T, Poupardin R, Korbelova J, Bruce AW: Conceptual framework of the eco-physiological phases of insect diapause development justified by transcriptomic profiling. Proceedings of the National Academy of Sciences of the United States of America 2017, 114:8532–8537.• The authors compared gene expression differences in the drosophilid fly, Chymomzya costata, throughout diapause development and uncovered specific transcriptional differences during each phase of diapause.
- 40.Wadsworth CB, Dopman EB: Transcriptome profiling reveals mechanisms for the evolution of insect seasonality. Journal of Experimental Biology 2015, 218:3611–3622.•• This study used comparative transcriptomics and analysis of transcript divergence, combined with previous QTL results, to identify candidate genes in the insulin signaling and circadian rhythm pathways affecting diapause termination.
- 41.Meyers PJ, Powell THQ, Walden KKO, Schieferecke AJ, Feder JL, Hahn DA, Robertson HM, Berlocher SH, Ragland GJ: Divergence of the diapause transcriptome in apple maggot flies: winter regulation and post-winter transcriptional repression. Journal of Experimental Biology 2016, 219:2613–2622. [DOI] [PubMed] [Google Scholar]
- 42.Emerson KJ, Bradshaw WE, Holzapfel CM: Microarrays reveal early transcriptional events during the termination of larval diapause in natural populations of the mosquito, Wyeomyia smithii. PloS One 2010, 5:e9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunning E: Die endonome Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber. Deut. Bot. Ges. 1937, 54:590–607. [Google Scholar]
- 44.Dalla Benetta E, Beukeboom LW, van de Zande L: Adaptive Differences in Circadian Clock Gene Expression Patterns and Photoperiodic Diapause Induction in Nasonia vitripennis. The American Naturalist 2019, 193:881–896. [DOI] [PubMed] [Google Scholar]
- 45.Meuti ME, Stone M, Ikeno T, Denlinger DL: Functional circadian clock genes are essential for the overwintering diapause of the Northern house mosquito, Culex pipiens. Journal of Experimental Biology 2015, 218:412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukai A, Goto SG: The clock gene period is essential for the photoperiodic response in the jewel wasp Nasonia vitripennis (Hymenoptera: Pteromalidae). Applied Entomology and Zoology 2016, 51:185–194. [Google Scholar]
- 47.Omura S, Numata H, Goto SG: Circadian clock regulates photoperiodic responses governed by distinct output pathways in the bean bug, Riptortus pedestris. Biological Rhythm Research 2016, 47:937–945. [Google Scholar]
- 48.Eckel-Mahan K, Sassone-Corsi P: Metabolism and the circadian clock converge. Physiological reviews 2013, 93:107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emerson KJ, Bradshaw WE, Holzapfel CM: Complications of complexity: integrating environmental, genetic and hormonal control of insect diapause. Trends in Genetics 2009, 25:217–225.• In this mini-review the authors raise the important concern that it is difficult to distinguish whether the circadian clock as a functional unit or individual clock genes are involved in measuring daylength, regulating hormone production and/or generating phenotypes associated with diapause.
- 50.Pegoraro M, Gesto JS, Kyriacou CP, Tauber E: Role for Circadian Clock Genes in Seasonal Timing: Testing the Bunning Hypothesis. Plos Genetics 2014, 10.•• The authors found that null mutations in the clock genes period, timeless and Clock abolished seasonal differences in chill coma recovery time in females of D. melanogaster, suggesting that the circadian clock regulates seasonal responses.
- 51.Markert MJ, Zhang Y, Enuameh MS, Reppert SM, Wolfe SA, Merlin C: Genomic Access to Monarch Migration Using TALEN and CRISPR/Cas9-Mediated Targeted Mutagenesis. G3-Genes Genomes Genetics 2016, 6:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Xu X, Zhan S, Huang Y: Genome editing in insects: current status and challenges. National Science Review 2019, 6:399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meuti ME, Denlinger DL: Evolutionary links between circadian clocks and photoperiodic diapause in insects. Edited by: Oxford University Press; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.