SUMMARY
Mice engineered for conditional, cell type-specific gene inactivation have dominated the field of mouse genetics because of the high efficiency of Cre-loxP-mediated recombination. Recent advances in CRISPR/Cas9 technologies have provided alternatives for rapid gene mutagenesis for loss-of-function (LOF) analysis. Whether these strategies can be streamlined for rapid genetic analysis with the efficiencies comparable with those of conventional genetic approaches has yet to be established. We show that a single adeno-associated viral (AAV) vector containing a recombinase-dependent Staphylococcus aureus Cas9 (SaCas9) and a single guide RNA (sgRNA) are as efficient as conventional conditional gene knockout and can be adapted for use in either Cre- or Flp-driver mouse lines. The efficacy of this approach is demonstrated for the analysis of GABAergic, glutamatergic, and monoaminergic neurotransmission. Using this strategy, we reveal insight into the role of GABAergic regulation of midbrain GABA-producing neurons in psychomotor activation.
In Brief
Hunker et al. generate single adeno-associated viral vectors for conditional gene mutagenesis in the adult mouse nervous system with efficiencies equivalent to those of conventional gene inactivation strategies. On the basis of this efficacy, they provide a resource of Cre- and Flp-dependent constructs for targeting catecholamine, glutamate, and GABA neurotransmission.
Graphical Abstract

INTRODUCTION
The study of gene function in specific cell types of the central nervous system has relied largely on the generation of mouse lines with targeted alleles that incorporate Cre-recombinase and Cre recognition sites (loxP) flanking a portion of a gene coding region (Gu et al., 1993). Although this method revolutionized cell type-specific analysis of gene function, it is laborious and requires crossing targeted mouse lines to designated Cre-driver mouse lines to achieve cell type-specific gene inactivation.
With the advent of single-cell sequencing, the desire to address the functionality of selectively expressed genes has expanded. CRISPR/Cas9 has emerged as an efficient means to rapidly generate indel mutations that result in loss of function (LOF) (Cong et al., 2013; Deltcheva et al., 2011; Jinek et al., 2012). Viral vector-based delivery of CRISPR/Cas9 has also been demonstrated (Bak et al., 2018; Nishiyama et al., 2017; Swiech et al., 2015), but the large size of Staphylococcus pyogenes Cas9 (SpCas9) requires the generation of separate viral vectors for the delivery of the SpCas9 and corresponding single guide RNA (sgRNA) (Duan et al., 2016; Swiech et al., 2015). Cre-dependent transgenic mouse and rat lines that allow cell type-specific expression of SpCas9 have also been generated that use only a single viral vector (Bä ck et al., 2019; Platt et al., 2014). However, this method requires crossing the SpCas9 conditional lines to Cre-driver lines, which can reduce the speed of genetic analysis. Adding to the complexity, several genetic loci that are used to generate cell type-specific Cre expression are expressed transiently in gametogenesis or during development (Song and Palmiter, 2018), thus careful attention must be paid to ensure that ectopic recombination or mosaic expression of SpCas9 is not observed.
In addition to SpCas9, the smaller SaCas9 has also been characterized and shown to be amenable to expression along with its corresponding sgRNA via adeno-associated virus (AAV) delivery (Ran et al., 2015). Using a single AAV containing a Cre-dependent SaCas9 and a sgRNA, it was recently demonstrated that SaCas9 is capable of mediating cell type-specific mutagenesis (Kumar et al., 2018), though the efficiency using this strategy was quite low. Whether this was the result of targeting an alternatively spliced exon or the inefficiency of the CRISPR/SaCas9 is not clear. It is also unclear whether viral-based delivery of SaCas9 and the sgRNA from a single AAV can be used in different cell types or whether other recombinases can be used to promote conditional expression of SaCas9 for gene mutagenesis. Here, we generated a single AAV containing a recombinase-dependent expression cassette for SaCas9 with a non-conditional sgRNA (AAV-CMV-FLEX-SaCas9-U6-sgRNA) and provide a standardized workflow for assessing mutagenesis in small populations of targeted neurons. We found this approach to be as efficient as conventional gene knockout, and it can be used to target genes in cells that would be otherwise difficult to genetically isolate.
RESULTS
Single Vector CRISPR/SaCas9 Is as Efficient as Conventional Knockout
To establish the efficiency of a Cre-dependent CRISPR/SaCas9 viral-based system, we generated an AAV containing an inverted SaCas9 flanked by two sets of staggered loxP sites for Cre-mediated inversion/excision (FLEX or DIO), in addition to a U6 promoter and sgRNA (AAV-CMV-FLEX-SaCas9-U6-sgRNA; Figure 1A). The optimal size of the insert between the inverted terminal repeats (ITRs) of AAV is 4.7 kb, which is the approximate size of the AAV genome (Grieger and Samulski, 2005; Lusby et al., 1980; Muzyczka et al., 1984; Wu et al., 2010). With the addition of two sets of loxP sites (to permit stable inversion of SaCas9), the overall insert size between ITRs is 4.85 kb, which is just beyond the optimal insert size. To establish whether reducing the size of the insert affects conditional mutagenesis, we created a second construct containing only one set of inverted loxP sites (AAV-CMV-FLEX(1LoxP)-SaCas9-U6-sgRNA; Figure 1B), reducing the size to 4.7 kb. The disadvantage of this strategy is repeated Cre-mediated inversion that results in constitutive on/off/on/off expression of the transgene, which may reduce efficiency.
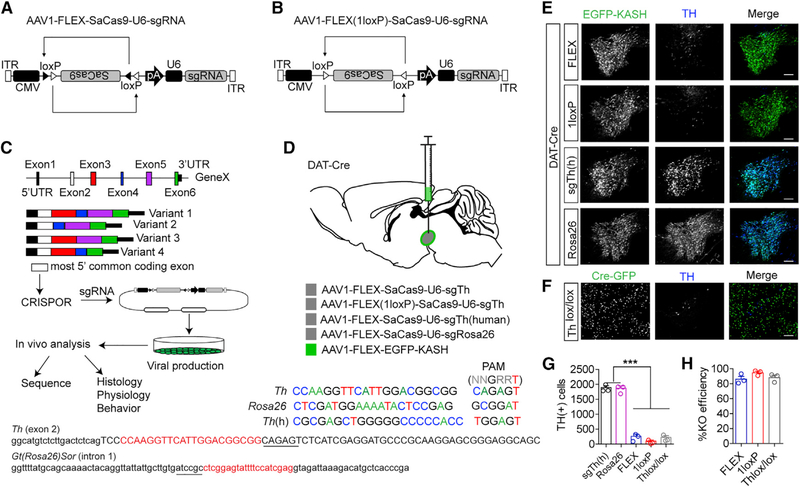
Figure 1. Generation of Single Vector CRISPR/SaCas9.
A) Design of AAV-FLEX-SaCas9-U6-sgRNA.
(B) Design of AAV-FLEX(1LoxP)-SaCas9-U6-sgRNA.
(C) Schematic for designing, generating, and analyzing CRISPR AAVs.
(D) Illustration of viral injections into the VTA and guide sequences for Th, Gt(Rosa26)Sor, and Th(h). Exon 2 of Th containing the sequence of the guide is denoted; note that lowercase is intron sequence and uppercase is exon. The intronic sequence containing the guide for Gt(Rosa26)Sor is also denoted, and PAMs are underlined.
(E) Example images of coronal VTA sections co-stained for EGFP (for identifying virally transduced and Cre-positive neurons using AAV1-FLEX-EGFP-KASH) and tyrosine hydroxylase (TH; a marker for dopamine neurons) showing loss of TH staining in the 1loxP and FLEX strategies compared with controls.
(F) Example images of coronal VTA sections co-stained for GFP and TH in Thlox/lox mice injected with AAV1-CAG-Cre-GFP. Scale bars: 250 mm.
(G) Cell count quantification of VTA TH-positive cells (n = 3 mice/group; one-way ANOVA F[4, 10] = 214.3, p < 0.0001; Tukey’s multiple comparisons test, ***p < 0.001).
(H) Comparison of knockout efficiencies among the FLEX, 1LoxP, and Thlox/lox strategies. Data are presented as mean ± standard error of the mean (SEM).
For the design of sgRNAs, we reasoned that targeting the most 5′ common exon of the gene would provide the most reliable inactivation. We obtained genomic sequence from the online resource, the University of California, Santa Cruz (UCSC), Genome Browser (Waterston et al., 2002) and aligned with reference sequences for all known splice variants obtained from the Mouse Genome Informatics database (Bult et al., 2019; Smith et al., 2019; Figure 1C). Sequence for the designated exon was analyzed using the CRISPOR prediction tool (Concordet and Haeussler, 2018; Haeussler et al., 2016) to identify guides with the highest predicted efficiency and fewest predicted off-targets. Guides were synthesized and subcloned into the AAV shuttle plasmid (Figure 1C) as previously described (Ran et al., 2015). The AAV virus was produced in house (Gore et al., 2013) using a single packaging plasmid that provides high-efficiency viral production (Grimm et al., 2003). AAV-CMVFLEX-SaCas9-U6-sgRNA or AAV-CMV-FLEX(1loxP)-SaCas9-U6-sgRNA were co-injected with a Cre-dependent expression cassette containing the KASH peptide sequence fused to EGFP to integrate EGFP into the nuclear envelope (Swiech et al., 2015). This allowed easy visualization of the injection site and permited subsequent nuclear fluorescence-assisted cell sorting (FACS) for sequencing analysis (Figures 1C and D).
To determine the efficiency of viral SaCas9-mediated mutagenesis relative to conventional gene inactivation using the Cre-loxP strategy in mice, we generated AAVs containing a guide targeted to the Th allele (Figure 1D), which encodes the rate-limiting enzyme in catecholamine production, tyrosine hydroxylase (TH). To ensure that observed changes in TH levels are not the result of general toxicity associated with double-strand breaks introduced by SaCas9, we generated a guide targeting intron 1 of the Gt(Rosa26)Sor allele (Rosa26). This allele has been shown to be ubiquitously expressed and has no observable impact on development (Friedrich and Soriano, 1991; Zambrowicz et al., 1997), and insertions into intron 1 of Gt(Rosa26)Sor are commonly used for generation of transgenic mice (Huang and Zeng, 2013). We also generated a vector to control for SaCas9 expression using a guide for the human Th allele (Th[h]) that is not homologous to mouse. AAV1-CMV-FLEX-SaCas9-U6-sgTh, AAV1-CMV-FLEX(1loxP)-SaCas9-U6-sgTh, AAV1-CMV-FLEX-SaCas9-U6-sgTh(h), or AAV1-CMV-FLEX-SaCas9-U6-sgRosa26 (control) with AAV1-FLEX-EGFP-KASH was injected into the ventral tegmental area (VTA) of mice expressing Cre-recombinase under the control of the endogenous dopamine transporter (DAT) locus Slc6a3 (DAT-Cre; Figure 1D; Zhuang et al., 2005). Viral targeting resulted in high-efficiency transduction that was specific to TH-positive neurons (Figures S1A–S1C). Four weeks after injection, we assessed genetic LOF through Th mutagenesis with viral CRISPR/SaCas9 and compared with Th inactivation using a Thlox/lox mouse line (Darvas and Palmiter, 2010) injected with AAV1-CMV-Cre-GFP. Using immunohistochemistry (IHC) with an antibody specific to TH, we determined that both AAV1-CMV-FLEX-SaCas9-U6-sgTh and AAV1-CMV-FLEX (1loxP)-SaCas9-U6-sgTh were as effective at Th inactivation as conventional gene knockout (Figures 1E–1H).
To determine whether the efficiency of AAV1-CMV-FLEXSaCas9-U6-sgTh and AAV1-CMV-FLEX(1loxP)-SaCas9-U6-sgTh for inactivation of Th is due to the guide selected for mutagenesis, we generated a second guide located just downstream of the original Th guide (Figures S1D–S1F). In addition, to establish whether the observed efficiency is a reflection of the in vivo efficacy of SaCas9, as opposed to SpCas9, we also generated an AAV for the non-conditional expression of SpCas9 with a smaller promoter (hSynp) to allow efficient packaging of the larger SpCas9 insert (4.2 kb versus 3.2 kb for SaCas9) and a second bicistronic AAV for the delivery of the sgRNA driven by the U6 promoter and an expression cassette for EGFP-KASH downstream of hSynp (Swiech et al., 2015; Figures S1D, S1G, and S1H). In both experiments, we observed equally efficient inactivation of Th expression (Figures S1I and S1J).
Analysis of Cell Type-Specific Mutagenesis
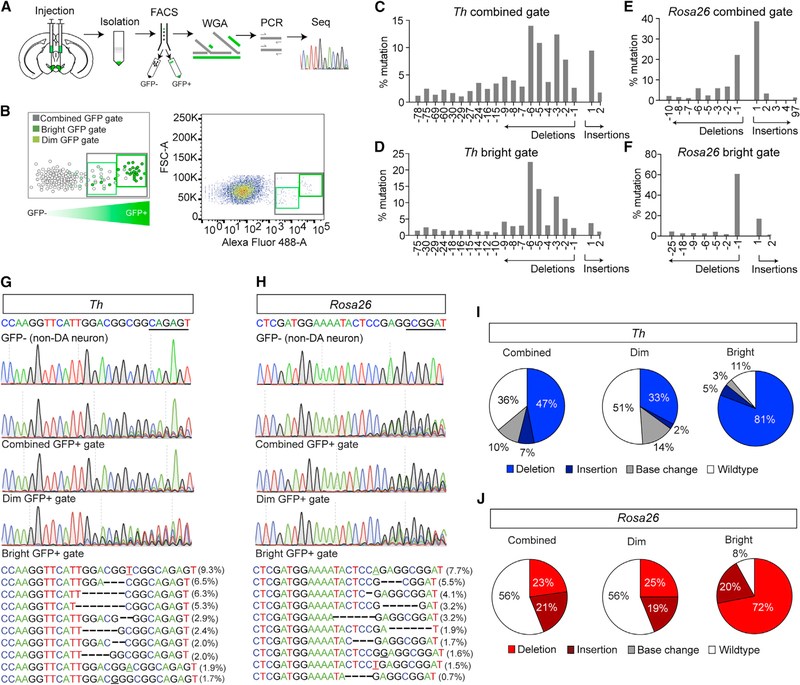
Previous analysis of the efficiency of SaCas9 in vivo indicated a low rate of mutagenesis (Kumar et al., 2018). However, this analysis was performed on isolated genomic DNA harvested from a sample containing both targeted and non-targeted cell types (Kumar et al., 2018). Analysis of isolated cell types following targeted mutagenesis with FACS on isolated nuclei using EGFP-KASH demonstrated a higher rate of mutagenesis with SpCas9 (Swiech et al., 2015). To determine whether the number of detectable insertions and deletions (indels) corresponds to the efficient loss of immunoreactivity of TH, we performed FACS on genetically isolated nuclei from tissue punches of the injection site (Figure 2A), similar to that described previously (Swiech et al., 2015). As an additional step following nuclear isolation, we performed whole-genome amplification (WGA) and sequencing of amplicons from targeted PCR flanking the guide sequence and PAM (Figure 2A), as well as exome sequencing of WGA DNA. EGFP-KASH tagged nuclei were isolated using one of three gates, a “bright” gate containing the brightest fluorescent nuclei, a “dim” gate containing fluorescent nuclei with lower fluorescence and some unlabeled autofluorescent cells, and a combined gate encompassing both the bright and dim gates (Figure 2B). Approximately 1,000 nuclei were sorted per gate, and genomic DNA was extracted and amplified. Sequences flanking the guides targeted to Th and Gt(Rosa26)Sor were amplified by PCR and deep-sequenced.
Figure 2. Analysis of Cell Type-Specific Mutagenesis.
(A) Schematic for isolation, FACS, WGA, PCR, and sequencing (seq) for identifying indels at targeted sites.
(B) Schematic of FACS illustrating designated gates (left) and example FACS (right).
(C and D) Indel frequencies for the combined gate (C) and bright gates (D) for targeted analysis of Th.
(E and F) Indel frequencies for the combined gate (E) and bright gates (F) for targeted analysis of Gt(Rosa26)Sor.
(G and H) Sanger sequencing of FACS sorted GFP-negative (top) and GFP-positive (middle) nuclei of mice co-injected with AAV1-FLEX-SaCas9-U6-sgTh (G) and AAV1-FLEX-EGFP-KASH or AAV1-FLEX-SaCas9-U6-sgGt(Rosa26)Sor (H) and AAV1-FLEX-EGFP-KASH into the VTA. Most common indels (bottom) for Th-targeted (G) and Gt(Rosa26)Sor-targeted (H) nuclei.
(I and J) Percentage of sequence reads with wild-type, insertion, deletion, or base changes for Th-targeted (I) and Gt(Rosa26)Sor-targeted (J) nuclei.
We observed a diversity of indels in GFP-positive nuclei from both Th and Gt(Rosa26)Sor-targeted cells (Figures 2C–2H). Sequence reads from GFP-negative nuclei contained no evidence of mutagenesis, as expected (Figures 2G and 2H). Consistent with previous observations that SaCas9 produces double-strand DNA breaks 3 bases upstream of the PAM (Ran et al., 2015), we observed the majority of indels adjacent to or flanking this position that was evident in the chromatogram and sequence reads (Figures 2G and 2H). The highest proportion of mutagenized sequence was observed in the bright gate, followed by the combined gate, and finally the dim gate (Figures 2I and 2J). For Th, the proportion of mutagenized sequenced in the bright gate most closely resembled the overall loss of immunoreactivity for TH (Figures 1G, 1H, and 2I), although the combined gate identified the greatest diversity of indels (Figures 2C–2F).
To confirm mutagenesis of Th and establish the extent of off-target indels, we acquired whole-exome sequencing (WES) of WGA DNA from GFP-positive and GFP-negative nuclei from Th and Gt(Rosa26)Sor-targeted mice. A total of 763 insertions or deletions were localized to 322 genes following correction for known population-level mutations (Table S1). Of the 322 genes, 47 contained indels at the same position in all four samples, 40 in three, and 60 in two. The remaining 175 were unique to one sample, and we confirmed insertions or deletions in exons using the Integrative Genomics Viewer (Robinson et al., 2011) in 158 of these (Table S2). These unique indels were distributed across all four samples (Rosa26 GFP-positive, 16; Rosa26 GFP-negative, 86; Th GFP-positive, 44; Th GFP-negative, 11). The guide directed toward Gt(Rosa26)Sor was in intron 1 and not identified by WES. However, this method did confirm an indel in exon 2 of the Th locus that was located at the site of the SaCas9 cleavage site within the sgRNA that was unique to GFP-positive nuclei from mice injected with AAV1-CMV-FLEX-SaCas9-U6-sgTh and AAV1-FLEX-EGFP-KASH (Table S2).
Viral CRISPR/SaCas9 Can Avoid Pitfalls of Conventional Genetics
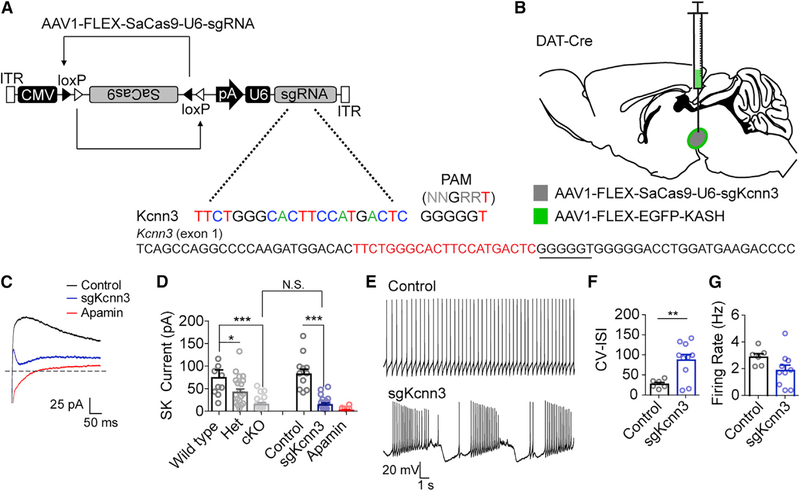
A caveat to the use of conventional Cre-loxP strategies is the observation that many genes can be transiently expressed in germ cells, which can yield mosaic genotypes (Song and Palmiter, 2018). This requires the generation of heterozygous delta (constitutively inactivated) alleles to guard against mosaicism (Zweifel et al., 2008) that may produce haploinsufficiency phenotypes. To establish whether AAV-CMV-FLEX-SaCas9-U6-sgRNA can be used to circumvent this problem, we targeted the gene encoding the calcium-activated, small conductance potassium channel SK3, Kcnn3 (Figures 3A and 3B). SK3 is the predominant ion channel underlying SK-mediated tail currents in dopamine neurons (Sarpal et al., 2004; Wolfart et al., 2001), and Kcnn3 has been conditionally inactivated in previously using conventional methodologies (Deignan et al., 2012).
Figure 3. Alleviating Confounds of Conventional Genetics.
(A) Design of AAV1-FLEX-SaCas9-U6-sgKcnn3. Exon 1 of Kcnn3 is denoted with guide sequence highlighted in red; the PAM is underlined.
(B) Illustration of viral injections into the VTA.
(C) Example traces of tail currents evoked following a 500 ms step from ‒70 to 0 mV.
(D) Quantification of tail current amplitudes following genetic knockout or CRISPR knockout of Kcnn3 or application of apamin (wild-type, n = 10; Het, n = 19; cKO, n = 15; control, n = 11; sgKcnn3, n = 15; apamin, n = 6; one-way ANOVA F[5, 70] = 12.05, p < 0.0001; Bonferroni’s select comparison, *p < 0.05 and ***p < 0.001).
(E) Example traces of action potential firing from control or sgKcnn3-targeted dopamine neurons.
(F and G) Quantification of the coefficient of variation of the inter-spike interval (F) and firing rate (G) in control or sgKcnn3-targeted dopamine neurons (control, n = 6; sgKcnn3, n = 9; Student’s unpaired t test, **p < 0.01). For sgKcnn3 control, mice were injected with AAV1-FLEX-SaCas9-sgTh(h). Data are presented as mean ± SEM.
Because DAT-Cre can result in ectopic recombination in germ cells, we generated a delta allele for Kcnn3 by crossing Kcnn3lox/lox mice to a broadly expressed Cre-driver line, Meox2-Cre, as previously described (Zweifel et al., 2008). Kcnn3⊿ mice were crossed to DAT-Cre mice, and the resulting DAT-Cre::Kcnn3⊿/+ mice were crossed to Kcnn3lox/lox mice to generate wild-type (WT) mice (Slc6a3+/+::Kcnn3+/lox), heterozygous mice (Slc6a3+/+::Kcnn3⊿/lox or Slc6a3Cre/+::Kcnn3+/lox), and knockout mice (Slc6a3Cre/+::Kcnn3⊿/lox). Heterozygous mice displayed a reduced SK-mediated tail current in dopamine neurons that was further reduced by biallelic inactivation (Figure 3D). Importantly, inactivation of Kcnn3 using AAV1-CMV-FLEX-SaCas9-U6-sgKcnn3 was as efficient as conditional gene knockout at reducing SK3-mediated tail currents (Figures 3C and 3D). A small amount of SK-mediated current remains in dopamine neurons following Kcnn3 knockout (Deignan et al., 2012), as demonstrated by a slightly further reduction in SK-mediated tail currents using the SK-specific toxin apamin (Figure 3D). Consistent with previous reports that SK3 channels regulate dopamine neuron activity patterns (Shepard and Bunney, 1991; Soden et al., 2013; Wolfart et al., 2001), Kcnn3 mutagenesis with AAV1-CMV-FLEX-SaCas9-U6-sgKcnn3 significantly increased spike firing irregularity (Figures 3E and 3F) without altering firing frequency (Figure 3G).
Analysis of the combined gate from FACS revealed a diversity of insertions and deletions similar to those observed for Th and Gt(Rosa26)Sor mutagenesis (Figures S2A and S2B). Unlike Th and Gt(Rosa26)Sor, which had predicted off-targets in intragenic regions, the Kcnn3 guide had a predicted off-target in the coding region of Socs6. Sequence analysis of the PCR amplicon flanking the predicted off-target site revealed no insertions of deletions, but did detect a 1% rate of base change, likely reflecting sequencing infidelity (Figures S2C and S2D).
AAV-CRISPR/SaCas9 Can Be Used to Mutate Genes in Difficult-to-Isolate Cell Types
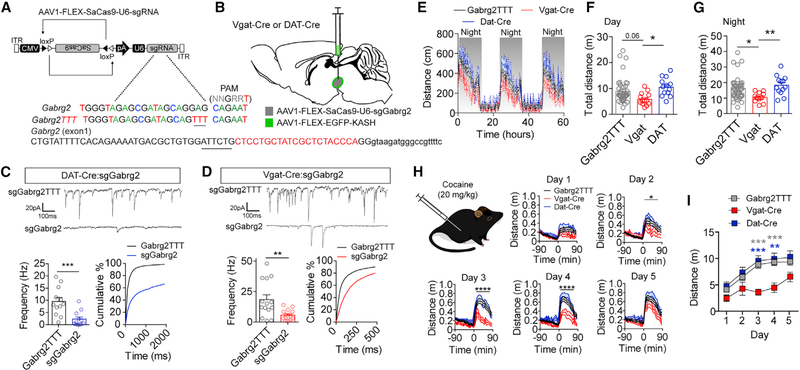
The VTA consists of multiple cell types, including GABAergic, dopaminergic, and glutamatergic populations (Lammel et al., 2014; Morales and Margolis, 2017). Because VTA GABA-producing cells do not have identifiable markers that allow for both neurotransmitter and anatomical specificity, they are more difficult to isolate for selective gene inactivation experiments. To assess the efficiency of mutagenesis in these cells, we targeted a broadly expressed gene that plays a key role in GABAergic synaptic transmission, Gabrg2, which encodes the GABAA receptor subunit γ2. Within the VTA, both dopaminergic and non-dopaminergic neurons express GABAA receptors. The GABAA γ2 subunit is required for proper GABAA receptor formation and insertion into the cell membrane at the synapse (Schweizer et al., 2003). To selectively mutate Gabrg2, we generated AAV1-CMV-FLEX-SaCas9-U6-sgGabrg2 (Figure 4A) for selective mutagenesis in either GABA neurons (Slc32a1-Cre or Vgat-Cre) or dopamine neurons (DAT-Cre) (Figure 4B). Viral labeling of GABA-producing neurons in the VTA did not overlap with TH in the VTA, indicating specificity for viral transduction (Figure S3A).
Figure 4. Targeting Difficult-to-Isolate Cell Types.
(A) Design of AAV1-FLEX-SaCas9-U6-sgGabrg2 and control virus AAV1-FLEX-SaCas9-U6-sgGabrg2TTT. Exon 1 of Gabrg2 is denoted with guide sequence highlighted in red; the PAM is underlined; note that the guide targets the 3′ sequence, so it is written in the reverse complement.
(B) Illustration of viral injections into the VTA.
(C and D) Representative traces and quantification of mIPSC frequency recorded from VTA (C) dopamine (n = 12 cells/group) or (D) GABA (controls, n = 14 cells; sgGabrg2, n = 16 cells) cells. Left: average mIPSC frequency (Student’s t test, **p < 0.01 and ***p < 0.001). Right: cumulative distribution of inter-event intervals.
(E–G) Comparison of locomotion among Vgat-Cre (n = 13), DAT-Cre (n = 13), and control (n = 44) mice (E) across three consecutive days and nights in 15 min time bins, (F) the sum of total distance traveled across 2 consecutive days (one-way ANOVA F[2, 66] = 4.179, p < 0.05; Tukey’s multiple-comparisons test, *p < 0.05), and (G) the sum of total distance traveled across three consecutive nights (one-way ANOVA, F[2, 66] = 5.363, p < 0.001; Tukey’s multiple-comparisons test, *p < 0.05 and **p < 0.01).
(H) Locomotor responses 90 min pre- and post-cocaine (20 mg/kg, s.c. injection) injection across five consecutive days in Vgat-Cre, DAT-Cre, and control mice (day 2: two-way repeated measures ANOVA F[70,1890] = 1.40, *p < 0.05; day 3: two-way repeated measures ANOVA F[70,1890] = 3.17, ****p < 0.0001; day 4: twoway-repeated measures ANOVA F[70,1890] = 2.85, ****p < 0.0001).
(I) Sum of 90 min total distance traveled post-cocaine injection for 5 consecutive days in Vgat-Cre, DAT-Cre, and control mice (two-way repeated-measures ANOVA, effect of genotype, F[2, 54] = 6.259, p < 0.01; Tukey’s multiple-comparisons test, **p < 0.01 and ***p < 0.001). Data are presented as mean ± SEM.
In addition to addressing the role of GABAA receptor signaling in GABAergic neurons of the VTA, we sought to establish an additional control for AAV-CMV-FLEX-SaCas9-U6-sgRNA expression. It has been shown that the bases within the seed region of the sgRNA directly adjacent to the PAM site are crucial for the unwinding of DNA and subsequent mutagenesis by SaCas9 (Jiang et al., 2015; Nishiyama et al., 2017; Sternberg et al., 2014). Therefore, we generated an identical guide for targeting Gabrg2, except three bases in the seed region were mutated (sgGabrg2TTT; Figure 4A). To test whether the control sgRNA prevents mutagenesis of Gabrg2, we co-injected mice with either AAV1-CMV-FLEX-SaCas9-U6-sgGabrg2 and AAV1-FLEX-EGFP-KASH or AAV1-CMV-FLEX-SaCas9-U6-sgGabrg2TTT and AAV1-FLEX-EGFP-KASH and isolated nuclei via FACS.
Deep sequencing of a PCR-amplified fragment of genomic DNA that included the SaCas9-targeted locus demonstrated a variety of insertions and deletions in the sgGabrg2-targeted neurons (Figures S3B and S3C). Analysis of the sgGabrg2TTT control revealed a low level of mutagenesis, including a 1% rate of base change and a 1% rate of deletions (Figures S3B and S3C). The Gabrg2 guide had predicted off-targets in exons of two genes, Paxbpl and Wdfy3. Sequence analysis of these predicted off-target sites revealed a low rate of mutagenesis in Paxbpl, including a 5% base change and a 5% deletion rate. Similarly, Wdfy3 had a 2% base change and 2% deletion rate (Figures S3D and S3E).
To quantify the degree of LOF of Gabrg2 following mutagenesis, we recorded miniature inhibitory postsynaptic currents (mIPSCs) in CRISPR/SaCas9-targeted VTA dopamine and VTA GABA neurons. Mutagenesis of Gabrg2 in VTA dopamine neurons (Figure 4C) and in VTA GABA neurons (Figure 4D) resulted in a significant reduction in mIPSC frequency relative to their corresponding controls. In addition to a reduction in mIPSC frequency, we also observed as significant reduction in mIPSC amplitude in both targeted neuronal cell types relative to controls (Figures S3F and S3G).
Although GABAergic signaling plays an important role in psychomotor activation and behavioral sensitization of psychostimulant drugs such as cocaine (Bocklisch et al., 2013; Cameron and Williams, 1994; Liu et al., 2005; Steffensen et al., 2008) the role of GABAergic signaling in GABA neurons of the VTA is largely unknown. Analysis of basal locomotion revealed that reduced synaptic GABA signaling onto VTA GABA-producing neurons resulted in a significant reduction in distance traveled (Figures 4E–4G). In contrast, reducing synaptic GABA signaling in dopamine neurons had no significant effect compared with controls (Figures 4E–4G). Subcutaneous (s.c.) cocaine injections produce psychomotor activation that increases with subsequent injections (Kalivas and Stewart, 1991). To determine if Gabrg2 mutagenesis in VTA GABA and dopamine subpopulations alters psychomotor sensitization, we performed 5 days of repeated cocaine injections (20 mg/kg s.c., once daily; Figure 4H) and recorded distance traveled pre- and post-injection. Mutagenesis of Gabrg2 in VTA GABA-producing neurons resulted in a significant attenuation of locomotor sensitization relative to controls or mutagenesis of Gabrg2 in VTA dopamine-producing neurons (Figure 4I) demonstrating a key role for GABAergic regulation of VTA GABA-producing neurons in this process.
AAV-CRISPR/SaCas9 Is Effective for Projection-Specific Phenotypic Analysis
We next sought to determine whether this approach can be used for projection-specific phenotypic analysis. It has been previously shown that constitutive global inactivation of the DAT gene, Slc6a3, in mice results in robust hyperlocomotor activity (Giros et al., 1996). It has not been established whether this hyperlocomotor phenotype can be observed through the selective adult inactivation of Slc6a3 in the VTA, or more specifically, in the dopamine projection to the nucleus accumbens (NAc). Here, we performed parallel experiments in which we injected an AAV-CMV-FLEX-SaCas9-U6-sgRNA containing a guide targeted to Slc6a3 (Figure 5A) directly into the VTA of DAT-Cre mice (VTA targeted; Figure 5B) or injected the retrograde transducing virus CAV containing an expression cassette for Cre (CAV2-CMV-Cre) into the NAc and AAV1-CMV-FLEX-SaCas9-Ug-sgSlc6a3 into the VTA (intersect targeted; Figure 5C).
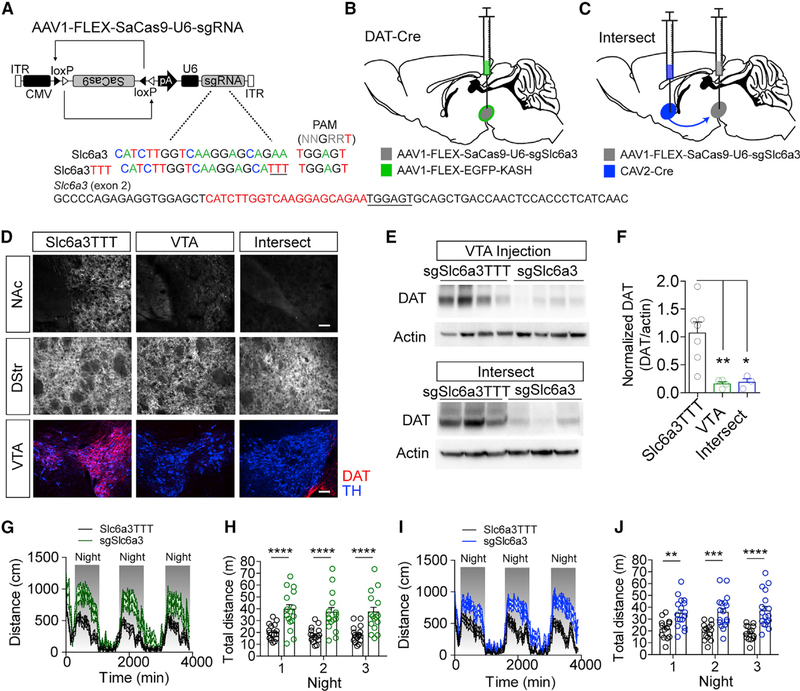
Figure 5. Intersectional Strategy Targeting Slc6a3 Produces Robust DAT Knockout and Hyperactivity.
(A) Design of AAV1-FLEX-SaCas9-U6-sgSlc6a3 and control virus AAV1-FLEX-SaCas9-U6-sgSlc6a3TTT. Exon 2 of Slc6a3 is denoted with guide sequence highlighted in red; the PAM is underlined.
(B) Illustration of viral injections into the VTA.
(C) Illustration of intersectional strategy with injection of CAV2-CMV-Cre into the terminals of VTA dopamine neurons in the nucleus accumbens (NAc) and AAV1-FLEX-SaCas9-U6-sgSlc6a3 into VTA dopamine cell bodies in wild-type mice.
(D) Example IHC images of DAT and TH in the NAc, dorsal striatum (DStr), and VTA in control, VTA-targeted, or intersect-strategy mice. Scale bar: 250 mm.
(E) Western blots from NAc punches probing for DAT and actin comparing the single AAV1-FLEX-SaCas9-U6-sgSlc6a3 VTA injection and the intersect strategy with controls.
(F) Quantification of DAT levels from the western blots (controls, n = 7; VTA sgSlc6a3, n = 4; VTA-NAc sgSlc6a3, n = 3; one-way ANOVA, F[2, 11] = 9.055, p < 0.001; Tukey’s multiple-comparisons test, *p < 0.05 and **p < 0.01).
(G and H) Locomotion measured for AAV1-FLEX-SaCas9-U6-sgSlc6a3 single VTA injection (controls, n = 16; sgSlc6a3, n = 15), across 3 consecutive days and nights in 15 min time bins (G) and comparison of total distance traveled across 3 consecutive nights (H) (two-way repeated-measures ANOVA, effect of genotype, F[1, 29] = 23.66, p < 0.0001; Bonferroni multiple comparisons, ****p < 0.0001).
(I and J) Locomotion measured for the intersectional strategy (controls, n = 14; sgSlc6a3, n = 17), across 3 consecutive days and nights in 15 min time bins (I) and comparison of total distance traveled across three consecutive nights (J) (two-way repeated-measures ANOVA, effect of virus, F[1, 29] = 19.65, p < 0.0001; Bonferroni multiple comparisons, **p < 0.01, ***p < 0.001, and ****p < 0.0001). Data are presented as mean ± SEM.
Reductions in DAT protein in the NAc and VTA were evident by IHC (Figure 5D) and Western blot analysis of DAT protein levels from tissue punches of the NAc to isolate dopamine terminals in both VTA-targeted and intersect-targeted mice (Figures 5E and 5F). We did not observe significant reductions in TH protein levels in the NAc tissue punches of these mice (Figure S4A), indicating that compensatory downregulation of TH is not observed with this genetic strategy (Salvatore et al., 2016). Sequence analysis of the PCR amplicon surrounding the sgSlc6a3 targeting site revealed a number of insertions and deletions, similar to that observed for other targeted genes. Base pair changes were observed in a small number of sequence reads from the same region in the sgSlc6a3TTT control (4%) and the predicted off-target gene Ndn (3%) (Figures S4B and S4C).
Analysis of locomotion revealed significant elevations in both day and night distance traveled in VTA-targeted and intersect-targeted mice (Figures 5G–5J; Figure S4D). In addition to inducing a hyperlocomotor phenotype, congenital inactivation of Slc6a3 globally is also associated with insensitivity to cocaine (Giros et al., 1996). The contribution of mesolimbic dopamine projections from the VTA to the NAc in mediating this cocaine insensitivity is not known. To address this, we assessed locomotor activity in response to 5 days of repeated cocaine injections (20 mg/kg s.c., once daily) in VTA- and intersect-targeted mice (Figure S4E). Both intersect and VTA-targeted mice showed an initial response to cocaine that did not differ significantly from control mice (Figures S4F–S4H). With repeated injections, control mice developed a sensitized response as predicted, but VTA- and intersect-targeted mice displayed a locomotor desensitization that was significantly different from control mice (Figure S4H).
AAV-CRISPR/SaCas9 Can Be Combined with Optogenetic Circuit Analysis
We next sought to determine whether AAV-CRISPR/SaCas9 can be effectively combined with optogenetics for circuit connectivity analysis. To test this, we generated AAV1-FLEX-SaCas9-U6-sgRNA constructs containing guides for targeting the vesicular glutamate transporters Vglut1 (Slc17a7) and Vglut2 (Slc17a6), and the vesicular GABA transporter Vgat (Slc32a1) (Figure 6A). These vectors were co-injected into designated Cre-driver lines with AAV1-FLEX-ChR2-EYFP (Figures 6B–6J).
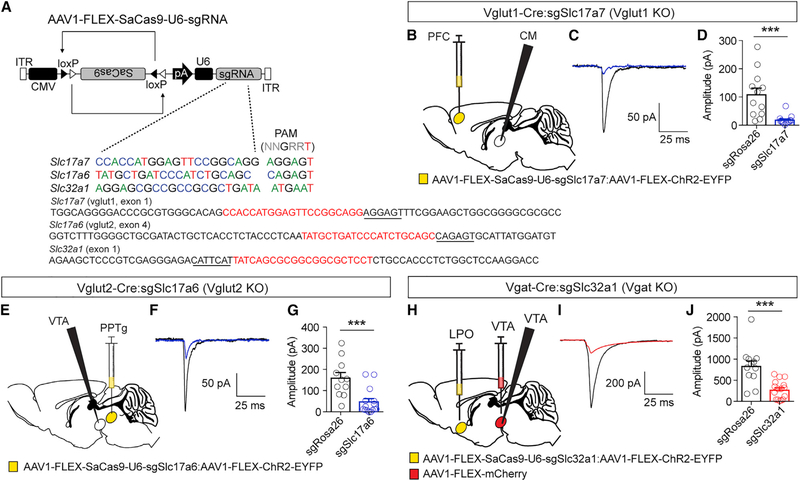
Figure 6. Targeted Mutagenesis of Vesicular Glutamate and GABA Transporters.
(A) Schematic of Cre-dependent AAVs for targeting Vglut1 (Slc17a7), Vglut2 (Slc17a6), and Vgat (Slc32a1). Exons of targeted loci are denoted with guide sequence highlighted in red; the PAM is underlined; note that the Slc32a1 guide targets the 3′ sequence, so it is written in the reverse complement.
(B) Schematic of experimental design for testing efficacy of sgSlc17a7 in Vglut1-Cre mice.
(C) Example light-evoked EPSC in CM neuron from control and AAV1-FLEX-SaCas9-U6-sgSlc17a7-injected mice.
(D) Quantification of light-evoked EPSCs (control n = 12; sgSlc17a7 n = 13; Student’s unpaired t test, ***p < 0.001).
(E) Schematic of experimental design for testing efficacy of sgSlc17a6 in Vglut2-Cre mice.
(F) Example light-evoked EPSC in VTA neuron from control and AAV1-FLEX-SaCas9-U6-sgSlc17a6 mice.
(G) Quantification of light-evoked EPSCs (control n = 11; sgSlc17a6 n = 15; Student’s unpaired t test, ***p < 0.001).
(H) Schematic of experimental design for testing efficacy of sgSlc32a1 in Vgat-Cre mice.
(I) Example light-evoked IPSC in GABA neuron of the VTA from control and AAV1-FLEX-SaCas9-U6-sgSlc32a1 mice.
(J) Quantification of light-evoked IPSCs (control n = 12; sgSlc32a1 n = 16; Student’s unpaired t test, ***p < 0.001). Data are presented as mean ± SEM.
For Vglut1, AAV1-FLEX-SaCas9-U6-sgSlc17a7 was injected with AAV1-FLEX-ChR2-EYFP into the prefrontal cortex (PFC) of Vglut1-Cre mice and recordings were made in the centromedian nucleus (CM) of the thalamus (Figure 6B). Light-evoked excitatory postsynaptic currents (EPSCs) were recorded at a holding potential of ‒60 mV in AAV1-FLEX-SaCas9-U6-sgSlc17a7 or AAV1-FLEX-SaCas9-U6-sgRosa26 (control) mice. CRISPR/SaCas9 mutagenesis resulted in a significant reduction in light-evoked EPSCs (Figures 6C and 6D). FACS using the combined gate followed by targeted deep sequencing of the targeted region confirmed mutagenesis and revealed the presence of indels (Figures S5A and S5B).
For Vglut2, AAV1-FLEX-SaCas9-U6-sgSlc17a6 was injected with AAV1-FLEX-ChR2-EYFP into the pedunculopontine tegmental nucleus (PPTg) of Vglut2-Cre mice, and recordings were made in the VTA (Figure 6E). As above, light-evoked EPSCs were recorded at a holding potential of ‒60 mV in AAV1-FLEX-SaCas9-U6-sgSlc17a6 or AAV1-FLEX-SaCas9-U6-sgRosa26 (control) mice. Consistent with inactivation of Vglut1, inactivation of Vglut2 resulted in a significant reduction in light-evoked EPSCs (Figures 6F and 6G) and the induction of a variety of indels (Figures S5C and S5D).
For Vgat, AAV1-FLEX-SaCas9-U6-sgSlc32a1 was injected with AAV1-FLEX-ChR2-EYFP into the lateral pre-optic area (LPO) of the hypothalamus of Vgat-Cre mice. GABA-releasing neurons of the VTA were isolated by injection of AAV1-FLEX-mCherry into this region, and light-evoked IPSCs were recorded at a holding potential of ‒60 mV (high chloride internal) in AAV1-FLEX-SaCas9-U6-sgSlc32a1 or AAV1-FLEX-SaCas9-U6-sgRosa26 (control) mice. CRISPR/SaCas9 mutagenesis resulted in a significant reduction in light-evoked IPSCs (Figures 6H–6J). FACS using the combined gate followed by targeted deep sequencing of the region flanking the guide confirmed mutagenesis through a variety of indels (Figures S5E and S5F).
A Flp Recombinase-Dependent AAV-CRISPR/SaCas9 Yields Efficient Inactivation
In addition to Cre-driver mouse lines, there is an emergence of mouse lines expressing the optimized yeast recombinase FlpO that utilizes FRT recognition sites (Dymecki, 1996; Sadowski, 1995). To determine whether SaCas9-mediated mutagenesis can be combined with Flp-mediated recombination, we generated AAV-CMV-FLEXfrt-SaCas9-U6-sgRNA. To test the efficiency of AAV-CMV-FLEXfrt-SaCas9-U6-sgRNA, we subcloned a guide for targeting Grin1 (AAV1-CMV-FLEXfrt-SaCas9-U6-sgGrin1; Figures 7A and 7B), the gene encoding the essential NR1 subunit of the NMDA receptor. We also generated AAVs for Flp-dependent mCherry expression to facilitate fluorescence-assisted patch clamp, and a Flp-dependent EGFP-KASH for nuclear FACS.
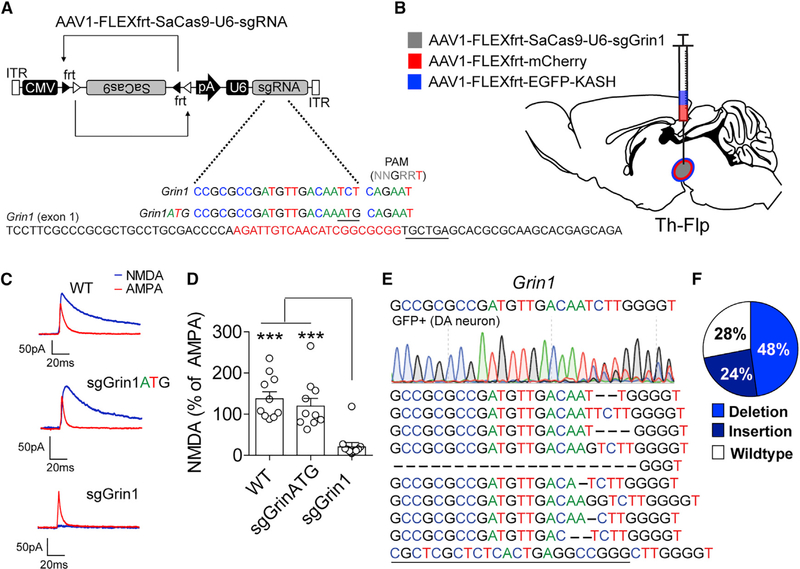
Figure 7. Flp-Dependent CRISPR/SaCas9 Produces Robust Gene Mutagenesis.
(A) Design of AAV1-FLEXfrt-SaCas9-U6-sgGrin1 and control virus AAV1-FLEXfrt-SaCas9-U6-sgGrin1ATG. Exon 1 of Grin1 is denoted with guide sequence highlighted in red; the PAM is underlined.
(B) Illustration of sagittal section with viral injections into the VTA.
(C) Representative traces of NMDA and AMPA currents evoked in dopamine neurons by electrical stimulation in WT mice or mice expressing sgGrin1 or sgGrin1ATG. AMPA current recordings were made in the presence of AP5; NMDA currents were determined by subtracting the AMPA current from the compound current trace recorded in ACSF.
(D) Quantification of the peak NMDA current as a percentage of the peak AMPA current (n = 10 cells/group; one-way ANOVA, F[2, 27] = 15.55, p < 0.0001; Tukey’s multiple-comparisons test, ***p < 0.001).
(E) Sanger sequencing of FACS sorted GFP-positive nuclei from the combined gate of mice co-injected with AAV1-FLEXfrt-SaCas9-U6-sgGrin1 and AAV1-FLEXfrt-EGFP-KASH into the VTA of Th-Flp mice and the most common indels (bottom).
(F) Percentage of targeted deep sequencing reads with wild-type, insertion, deletion, or base changes. Data are presented as mean ± SEM.
Genetic inactivation of Grin1 in dopamine neurons has been previously shown to eliminate NMDA receptor-mediated current in these cells (Engblom et al., 2008; Zweifel et al., 2008). Analysis of electrically evoked EPSCs in fluorescently identified dopamine neurons through co-expression of AAV1-CAG-FLEXfrt-mCherry and AAV1-CMV-FLEXfrt-SaCas9-U6-sgGrin1 in Th-Flp mice (Poulin et al., 2018; Figure 7C) revealed significantly reduced NMDAR-mediated EPSCs relative to AMPA-type glutamate receptor EPSCs in VTA dopamine neurons compared with either WT dopamine neurons or dopamine neurons transduced with the control virus AAV1-CMV-FLEXfrt-SaCas9-U6-sgGrin1ATG (Figures 7C and 7D). Consistent with other genes mutated with Cre-dependent CRISPR/SaCas9, Flp-dependent CRISPR/SaCas9 targeting of Grin1 resulted in a dramatic reduction in NMDA receptor-mediated current. Sequence analysis of the Grin1-targeted region in GFP-labeled nuclei from the combined FACS gate confirmed a diversity of indels consistent with results obtained using Cre-dependent mutagenesis (Figures 7E and 7F). On the basis of the efficiency of FLP-dependent mutagenesis of Grin1, the guides targeting all other genes describe here were subcloned into AAV-FLEXfrt-SaCas9-U6-sgRNA, and both Cre- and Flp-dependent constructs were deposited in the online repository Addgene (addgene.org).
In addition to the detection of indels with targeted deep sequencing, we also sought to establish whether the simple T7 endonuclease 1 (T7E1) mismatch cleavage assay could detect mutations at targeted loci. Following FACS, WGA, and PCR, we detected mismatch cleavage in nine of nine guides tested in GFP-positive nuclei with the bands detected matching the size predicted on the basis of the location of the SaCas9 cleavage site in the amplicon (Figure S6). Thus, the T7E1 mismatch cleavage assay is also an effective means for confirming in vivo mutagenesis.
DISCUSSION
We demonstrate that a single viral vector for the conditional expression of SaCas9 along with a non-conditional sgRNA is sufficient to mediate robust mutagenesis and LOF. A major consideration for the use of AAV-FLEX-SaCas9-U6-sgRNA is the selection of controls to account the expression of SaCas9, the ubiquitous expression of the sgRNA, and potential off-target effects. We considered several approaches to address this, including the design of guides that target noncoding DNA within the mouse genome to control for the expression of SaCas9, injection of AAV-CMV-FLEX-SaCas9-U6-sgRNA into WT mice to control for viral transduction and constitutive expression of the targeting sgRNA, selective targeting of a locus with no known developmental or neuronal function to control for the induction of double-strand DNA breaks and mutating regions of the sgRNA to maintain the same potential off-targets as the chosen guide. On the basis of these considerations, we concluded that the best controls are either the generation of a separate virus containing a 3 base mutation within the important sgRNA seed region or a ubiquitous control that targets Gt(Rosa26)Sor.
In addition to the selection of controls, an equally important consideration is the selection of the sgRNA. Using the simple strategy outlined in Figure 1A in which sgRNAs were targeted to the most 5ʹ common exon, we routinely and consistently achieved efficiency that was equivalent to conventional gene knockout. In 9 of 9 targeted genes characterized here, the first guide we chose on the basis of this criterion was highly effective. In addition to the 9 gene targets described here, we have also successfully targeted 11 other independent genes encoding a variety of ion channels, synaptogenic proteins, and G-protein-coupled receptors using this strategy. In all cases, we observe highly efficient gene mutagenesis and inactivation with the selection of a single sgRNA.
A final major consideration of viral-mediated SaCas9 delivery is the analysis of on- and off-target effects. To assess mutagenesis, we generated a standardized workflow that was applied to all genes investigated. For FACS, we analyzed three distinct gates in our sgTh and sgRosa26 control and determined that sequence analysis of nuclei from “bright” gate best corresponds to the observed LOF; however, because the combined gate provided the broadest range of unique indels we used this for subsequent NGS WES and targeted analysis of the remaining genes. Our analysis of predicted off-targets and WES suggests that the rate of mutagenesis in predicted off-targets is very low. Our WES analysis of GFP-positive and GFP-negative nuclei from AAV1-CMV-FLEX-SaCas9-U6-sgTh and AAV1-CMV-FLEX-SaCas9-U6-sgRosa26 did not reveal high levels of unique insertions and deletions in Cre-positive EGFP-expressing cells. In fact, the highest level of insertions detected were in Rosa26-targeted, GFP-negative nuclei that are non-Cre-expressing neurons. It is also interesting to note that in the deep sequencing analysis of the nine targeted loci described here, the majority of mutations observed were deletions with an average insertion/deletion ratio of 0.15. In contrast, the majority of indels detected by WES were insertions. After population correction for all four samples combined, including those that were unique and non-unique for a given sample was 511 and the number of deletions was 252, yielding an insertion/deletion ratio of 2.02, which is much greater than the insertion/deletion ratio of the targeted loci.
We cannot conclude that SaCas9 does not generate off-target mutations; however, given that nearly half the number of indels detected were present in two or more samples, and the number of indels detected in GFP-positive neurons was not different than the number of indels in GFP-negative neurons it appears that SaCas9 does not generate ubiquitous off-target mutagenesis. Consistent with this, recent trio analysis in mice and monkeys in which whole genomes were sequenced in Cas9 modified offspring and non-modified parents revealed no statistical differences in the number of mutations identified (Iyer et al., 2018; Luo et al., 2019).
Use of AAV-CMV-FLEX-SaCas9-U6-sgRNA has distinct advantages over other methods. Because the SaCas9 and sgRNA are packaged within a single vector, the delivery is not subject to confounds of using viral vectors of different serotypes, or the generation of two independent viral vectors of the same serotype that may differ in effective titer. Conventional gene inactivation requires the generation of a conditional allele, backcrossing, and crossing to a recombinase driver line of interest that can take years before testing. Moreover, the ability to restrict gene mutagenesis and protein loss to adult mice avoids technical confounds such as ectopic germline expression of the recombinase or early developmental expression that is not restricted to the cell line of interest. A disadvantage is the lack of one-to-one assessment of genotype to phenotype. SaCas9 induces a variety of genetic mutations, and in many cases, it is difficult to directly assign the impact of the specific mutation to the phenotypic outcome. However, because most Cas9-induced mutations are non-sense mutations, it is likely to induce non-sense-mediated mRNA decay. Thus, a common outcome would be a reduction in mRNA and subsequent reductions in protein levels. An additional benefit of SaCas9 conditionally expressed by either Cre or Flp recombinase is that cell type-specific Cre and Flp driver lines can be crossed to generate compound heterozygotes, and different genes in different cell types can be inactivated to test for transcellular genetic interactions.
GABAergic signaling has been shown to play an important role in behavioral sensitization to cocaine (Edwards et al., 2017), but the contribution of GABAA receptor signaling in GABAergic neurons of the VTA has been difficult to assess. We were able to show that a significant reduction in synaptic GABAA signaling on GABAergic but not dopaminergic neurons is sufficient to reduce psychomotor sensitization to cocaine. Thus, single vector SaCas9-mediated mutagenesis can be used to establish novel genotype-phenotype interactions with cellular and anatomical specificity.
Viral-mediated CRISPR/SaCas9 provides an efficient means for intersectional targeting of projection populations. Using this strategy, we were able to demonstrate the effectiveness of circuit-level analysis of gene inactivation. It has been previously demonstrated that loss of DAT in mice results in hyperactivity and reduced responsiveness to cocaine (Giros et al., 1996). Whether these effects are mediated by the loss of DAT in mesoaccumbens dopamine neurons or nigrostriatal dopamine neurons was not previously easy to address. By selectively targeting Slc6a3 in VTA dopamine neurons, or in VTA projections to the NAc, we were able to demonstrate that loss of DAT in the mesoaccumbens pathway is sufficient to cause hyperactivity.
On the basis of our collective observations, we conclude that the addition of AAV-FLEX-SaCas9-U6-sgRNA to the genetic toolbox provides a highly reliable and efficient means for generating gene-knockout models, furthering our capacity for genetic and cellular isolation to probe gene function in the regulation of the central nervous system. In addition to the generation of conventional AAV described here, these shuttle vectors can be easily used in combination with packaging systems such as AAV2-Retro (Tervo et al., 2016) or AAV-PHP.eB and AAV-PHP.S, allowing intravenous administration and systemic delivery (Chan et al., 2017). When combining AAV-FLEX-SaCas9-U6-sgRNA with other AAVs, such as those providing expression of specific opsins, it is important to consider AAV serotype. Anecdotally, we have observed that mixed AAV serotypes can result in significantly reduced transduction efficiency by one virus. For example, mixed AAV1 and AAV5 results in diminished transduction by AAV5. When viruses of the same serotype are used, transduction efficiency is not altered in either virus. Thus, although these viral strategies will likely yield significant insights into the genetic regulation of circuit function, appropriate optimizations should be performed to maximize efficiency.
STAR*METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Larry Zweifel (larryz@uw.edu).
All unique stable reagents generated in this study will be made available upon request but we may require a payment and/or Material Transfer Agreement if there is a potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mice are on a C57BL/6J background. The Slc6a3Cre/+ mouse line was generated as described (Zhuang et al., 2005), and the Slc32a1Cre/+, Slc17a7Cre/+, and Slc17a6Cre/+ mouse lines were acquired from Jackson Laboratories (stock numbers 016962, 023527, and 028863, respectively). ThFlp/+ mice were kindly given by Dr. Rajeshwar Awatramani (Poulin et al., 2018). Kcnn3lox/lox mice were generously provided by Dr. John Adelman (Jackson stock number 019083). The Kcnn3⊿ allele was generated by crossing Kcnn3lox/lox mice with a ubiquitously expressed Cre (Meox2Cre/+). Slc6a3Cre/+; Kcnn3⊿/+ mice were crossed with Kcnn3lox/lox mice to generate Slc6a3Cre/+; Kcnn3lox/+ (heterozygous for Kcnn3 in dopamine neurons) and Slc6a3Cre/+; Kcnn3lox/D (Kcnn3 knockout in dopamine neurons).
Mice were group-housed (max 5 mice/cage) on a twelve-hour light/dark cycle with ad libitum access to food and water. All experiments were performed during the light phase in accordance with the Institutional Animal Care and Use Committee at the University of Washington. Equal numbers of male and female mice were used in each experiment. Mice were 3–6 months old for behavioral experiments and immunohistochemistry, 8–10 weeks old for slice electrophysiology, and 3–4 months old for FACS analysis.
METHOD DETAILS
Selection and design of sgRNAs
The full length sequence for every gene was found on the UCSC genome browser database (http://genome.ucsc.edu/) (Kent et al., 2002), and the exons and splice isoforms of each gene were identified and aligned (Mouse Genome Informatics (MGI) database) (Bult et al., 2019). The most 5′ common coding exon (present in all known splice isoforms) was selected and the sequence was uploaded to the CRISPOR website (crispor.tefor.net) to determine possible sgRNAs and PAM sequences. The final sgRNA was chosen based on specificity, probability of frameshift mutations, and location on exon. Each sgRNA was ordered as short oligos (Sigma) with a 5′ CACC- 3′ overhang on the forward primer, and a 5′ -AAAC 3′ overhang on the reverse primer to allow for seamless integration into the AAV1-FLEX-SaCas9-sgRNA vector. The following are oligos used for the generation of the sgRNAs in this study. Th forward: CACCGCCAAGGTTCATTGGACGGCGG, reverse: AAACCCGCCGTCCAATGAACCTTGGC; Gt(Rosa26)Sor forward: CACCGCTCGATGGAAAATACTCCGAG, reverse: AAACCTCGGAGTATTTTCCATCGAGC; Slc6a3 forward: CACCGCA TCTTGGTCAAGGAGCAGAA, reverse: AAACTTCTGCTCCTTGACCAAGATGC; Kcnn3 forward: CACCGTTCTGGGCACTTCCATGA CTC, reverse: AAACGAGTCATGGAAGTGCCCAGAAC; Gabrg2 forward: CACCGTGGGTAGAGCGATAGCAGGAG, reverse: AAACCTCCTGCTATCGCTCTACCCAC; Slc32a1 forward: CACCGAGGAGCGCCGCCGCGCTGATA, reverse: AAACTATCAGCGC GGCGGCGCTCCTC; Slc17a7 forward: CACCGCCACCATGGAGTTCCGGCAGG, reverse: AAACCCTGCCGGAACTCCATGGT GGC; Slc17a6 forward: CACCGTATGCTGATCCCATCTGCAGC, reverse: AAACGCTGCAGATGGGATCAGCATAC; Grin1 forward: CACCGCCGCGCCGATGTTGACAATCT, reverse: AAACAGATTGTCAACATCGGCGCGGC.
Cloning sgRNAs into pAAV1-FLEX-SaCas9-sgRNA
The pAAV1-FLEX-SaCas9-sgRNA plasmid was digested overnight with BsaI-HFv2 and gel purified (Qiaquick Gel Extraction Kit, QIAGEN). The ordered sgRNA oligos were resuspended to a concentration of 100uM. The oligos were phosphorylated at 37°C for 30min using the following reaction: 1uL of each 100uM oligo, 1uL T4 ligase buffer (NEB), 0.5uL phosphonucleotide kinase (PNK, NEB) and 6.5uL H2O. To anneal the oligos, the entire reaction was placed at 100°C for 5 minutes and allowed to slowly return to room temperature. 50ng of digested pAAV-FLEX-SaCas9-sgRNA and 1uL T4 ligase (NEB) were added directly to the reaction and incubated at room temperature for 2 hours. 2uL of the reaction was electroporated using DBH10 βelectrocompetent cells (NEB). Colonies were grown in LB + AMP and minipreps (QIAGEN) were performed to extract DNA. A restriction digest using BsaI-HFv2 and HindIII-HF was performed to screen for positive colonies. One positive colony was selected and the DNA was extracted using a maxiprep kit (Invitrogen). The insertion of the sgRNA was confirmed via Sanger sequencing (Genewiz) using the following primer: 5′ GACTATCATATGCTTACCGT 3′.
Viral production and stereotaxic injections
All viruses were prepared in-house as described previously (Gore et al., 2013). Below is a comprehensive list of all viruses used in this study. AAV1-FLEX-SaCas9-sgTh, AAV1-FLEX(1loxP)-SaCas9-sgTh, AAV1-FLEX-SaCas9-sgGt(Rosa26)Sor, AAV1-FLEXSaCas9-sgKcnn3, AAV1-FLEX-SaCas9-sgTh(2), AAV1-FLEX-SaCas9-sgGabrg2, AAV1-FLEX-SaCas9-sgGabrg2TTT, AAV1-FLEX-SaCas9-sgSlc6a3, AAV1-FLEX-SaCas9-sgSlc6a3TTT, AAV1-FLEX-SaCas9-sgGrin1, AAV1-FLEX-SaCas9-sgGrin1ATG, AAV1-FLEX-SaCas9-sgSlc17a7, AAV1-FLEX-SaCas9-sgSlc17a6, AAV1-FLEX-SaCas9-sgSlc32a1, AAV1-hSyn-SpCas9, AAV1-U6-sgTh(SpCas9)-hSyn-EGFP-KASH, AAV1-FLEX-EGFP-KASH, AAV1-FLEXfrt-EGFP-KASH, AAV1-FLEX-mCherry, AAV1-FLEXfrt-mCherry, AAV1-FLEX-ChR2-EYFP.
All CRISPR/SaCas9 viruses were made using serotype AAV1 packaging vector with AAV2 ITRs. The AAV1-FLEX-EGFP-KASH and AAV1-FLEXfrt-EGFP-KASH virus used for FACS and genetic sequencing were synthesized using an AAV1 packaging vector and AAV1 ITRs. The following coordinates were used for stereotaxic viral injections relative to Bregma: VTA (M-L = 0.5, A-p = 3.25*F, D-V = ‒4.4, F = (Distance in mm from Bregma to Lambda)/4.21), LPO (M-L = 1.0, A-p = 0.2, D-V = ‒5.0), PPTg (M-L = 1.2, A-p = ‒4.5*F, D-V = ‒3.75), and PFC (M-L = 0.3, A-p = 2.1, D-V = ‒2.0).
Basal locomotion and cocaine sensitization
Standard Allentown cages were placed into locomotion chambers (Columbus instruments) that use infrared beam breaks to calculate distance traveled. For basal locomotion, mice were singly housed in clean cages and provided with ad libitum access to food and water. Locomotion was monitored for 72 consecutive hours. For cocaine sensitization, mice were placed in clean cages with no food or water for 3 hours. Subcutaneous injections of either saline (0.2mL, days 1 and 2) or cocaine (20mg/kg, days 3–7, Sigma-Aldrich) were delivered 90 minutes into the recording session each day for 7 consecutive days.
Slice electrophysiology
Mice were allowed 3–4 weeks recovery after surgery to allow for viral expression and gene knockout. Horizontal (200 mm) or coronal (250 mm) brain slices were prepared from 8–10 week old mice in an ice slush solution containing (in mM); 92 NMDG, 2.5 KCl, 1.25 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, 3 Na-pyruvate, 0.5 CaCl2, 10 MgSO4, pH 7.3–7.4 (Ting et al., 2014). Slices recovered for ~12 min in the same solution at 32°C and then were transferred to a room temperature solution containing (in mM): 92 NaCl, 2.5 KCl, 1.25 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 2 thiouria, 5 Na-ascorbate, 3 Na-pyruvate, 2 CaCl2, 2 MgSO4. Slices were allowed to recover for an additional 60 minutes. All solutions were continuously bubbled with O2/CO2. Whole-cell patch clamp recordings were made using an Axopatch 700B amplifier (Molecular Devices) using 4–6 MΩ electrodes. Recordings were made in ACSF containing (in mM): 126 NaCl, 2.5 KCl, 1.2 NaH2PO4, 1.2 MgCl2 11 D-glucose, 18 NaHCO3, 2.4 CaCl2, at 32°C continually perfused over slices at a rate of ~2 ml/min. VTA dopamine or GABA neurons were identified by fluorescence.
Recording of mIPSCs for sgGabrg2 functional validation
AAV1-FLEX-SaCas9-sgGabrg2 and AAV1-FLEX-EGFP-KASH were coinjected into the VTA of DAT-Cre or Vgat-Cre mice. mIPSC recordings were made in the presence of 2 mM Kynurenic acid (Sigma) and 500 nM Tetrodotoxin (Tocris) to block glutamatergic signaling and action potential firing, respectively. Electrodes were filled with an internal solution containing (in mM): 140 CsCl, 0.2 CaCl2, 8 NaCl, 2 EGTA, 10 HEPES, 1 QX-314, 0.5 Na3GTP, 4 MgATP, pH 7.2, 275–285 mOsm. Cells were held at ‒60mV and access resistance was monitored throughout the experiment. The mIPSCs were analyzed using Mini Analysis software (Synaptosoft). The minimum threshold for mIPSC amplitude height was 7pA.
Recording of SK currents for sgKcnn3 functional validation
AAV1-FLEX-SaCas9-sgKcnn3 and AAV1-FLEX-mCherry were co-injected into the VTA of DAT-Cre mice. SK currents were measured as described (Soden et al., 2013). Electrodes were filled with an internal solution containing (in mM): 130 K-gluconate, 10 HEPES, 5 NaCl, 1 EGTA, 5 Mg-ATP, 0.5 Na-GTP, pH 7.3, 280 mOsm. Neurons were held at ‒70 mV in voltage-clamp mode and tail currents were evoked with a 500 ms depolarization to 0 mV. Tail current amplitudes were measured 85 ms after the end of the voltage step, which corresponded with the average time of maximum current in control neurons. Apamin (300 nM, Tocris) was bath applied to a subset of neurons to block SK3 channels.
Recording of AMPA/NMDA currents for sgGrin1 functional validation
AAV1-FLEXfrt-SaCas9-sgGrin1 and AAV1-FLEXfrt-mCherry were co-injected into the VTA of TH-Flp mice. AMPA/NMDA currents were measured as described (Sanford et al., 2017). Picrotoxin (100 uM, Abcam) was included in the bath and electrodes were filled with an internal solution containing (in mM): 120 CsMeSO3, 20 HEPES, 0.4 EGTA, 2.8 NaCl, 2.5 Mg-ATP, 0.25 Na-GTP, 5 QX-314 bromide, pH 7.3, 280 mOsm. Neurons were held at +40 mV while a concentric bipolar electrode placed rostral to the VTA delivered 0.4 ms stimuli at 0.1 Hz to elicit an EPSC containing both AMPA and NMDA components. Approximately 15 traces were averaged per cell, followed by bath application of AP5 (100 μM, Abcam) to isolate the AMPA component. 15 AMPA EPSC traces were averaged and digitally subtracted from the initial recording in order to generate the NMDA trace.
Recording of light-evoked EPSCs/IPSCs for sgSlc17a7. sgSlc17a6, and Slc32a1 functional validation
Slc17a7 (Vglut1): AAV1-FLEX-ChR2-YFP was co-injected into the PFC along with either AAV1-FLEX-SaCas9-sgSlc17a7 or AAV1-FLEX-SaCas9-sgGt(Rosa26)Sor. Recordings were made from areas of dense ChR2 fiber innervation in the centromedian thalamic nucleus. Slc17a6 (Vglut2): AAV1-FLEX-ChR2-YFP was co-injected into the PPTg along with either AAV1-FLEX-SaCas9-sgSlc17a6 or AAV1-FLEX-SaCas9-sgGt(Rosa26)Sor. Recordings were made from the VTA. Slc32a1 (Vgat): AAV1-FLEX-ChR2-YFP was co-injected into the LPO along with either AAV1-FLEX-SaCas9-sgSlc32a1 or AAV1-FLEX-SaCas9-sgGt(Rosa26)Sor. AAV1-FLEXmCherry was injected into the VTA. Recordings were made from mCherry-labeled (Vgat+) neurons in the VTA.
Light-evoked synaptic transmission was induced with 5 ms light pulses delivered at 0.1 Hz from an optic fiber placed directly in the bath. Amplitudes were calculated from an average of at least 10 events. Light-evoked EPSCs were measured while holding at ‒60 mV in the presence of picrotoxin (100 μM), with an internal recording solution containing (in mM): 130 K-gluconate, 10 HEPES, 5 NaCl, 1 EGTA, 5 Mg-ATP, 0.5 Na-GTP, 5 QX-314, pH 7.3, 280 mOsm. Light-evoked IPSCs were measured with holding at ‒60 mV in the presence of kynurenic acid (2 mM) with an internal recording solution containing (in mM): 135 KCl, 12 NaCl, 0.5 EGTA, 10 HEPES, 2.5 Mg-ATP, 0.25 Na-GTP, 5 QX-314, pH 7.3, 280 mOsm.
Immunohistochemistry
Mice were anesthetized with Beuthanasia (Merck Sharp and Dohme Corp) and perfused transcardially with ice-cold phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). The brain was removed and placed in 4% PFA overnight, then transferred to 30% sucrose in PBS for 2–3 days. Brains were mounted in OCT (Fisher) at ‒19°C and frozen sections (30 μm) were cut using a cryostat (Leica). The following primary antibodies were used for protein detection: Tyrosine hydroxylase (mouse monoclonal, 1:2000, Millipore or rabbit polyclonal, 1:2000, Fisher Scientific), GFP (rabbit polyclonal, 1:2000, Invitrogen or chicken polyclonal, 1:6000, Abcam), HA (rabbit polyclonal, 1:2000, Sigma). The slices were first blocked in 1xTBS with 0.3% Triton and 3% normal donkey serum (TBST+NDS). Slices were incubated in primary antibody diluted in TBST+NDS overnight at 4°C overnight. The following day, brain sections were washed 3x in TBST and then incubated with secondary antibodies (Jackson immuno, Alexa Fluor-488; Cy3) diluted 1:250 in TBST+NDS and then washed 3x times and mounted onto slides using Fluoromount (Southern Biotech). Fluorescence was imaged using a Nikon Upright Widefield microscope, and cells were counted using ImageJ.
Protein isolation
Mice were allowed 5–8 weeks for recovery and viral expression after surgery. Tissue punches of the ventral midbrain or ventral striatum were flash frozen in liquid nitrogen and then homogenized in 10mM HEPES pH 7.4 with 0.1mM protease inhibitor cocktail (Sigma) using glass dounces (25x with dounce A, 25x with dounce B). The homogenate was centrifuged at 1000xg for 10min at 4°C. The supernatant was removed and centrifuged at 14000RPM for 20min at 4°C. The protein pellet was vigorously resuspended in RIPA lysis buffer containing (in mM): 50 Tris pH 7.4, 150 NaCl, 0.1% SDS, 0.5% Na-Deoxycholate, 1% NP-40, 0.1mM PIC and rotated at 4°C for 1 hour. The protein suspension was centrifuged at 14000RPM for 15min at 4°C to pellet any insoluble material. The supernatant was resuspended in 6x sample buffer containing: 6.3% SDS, 60% glycerol, 374mM Tris Base, 5.5% b-mercaptoethanol, 0.67% Bromo Blue, and stored at ‒80°C.
Western blot
Protein samples were denatured at 100°C for 5min before loaded in a 10% precast polyacrylamide gel (Biorad) and run at 80mV for ~1.5hours in running buffer containing (in mM): 25 Tris base, 192 glycine and 0.1% SDS. Protein was transferred from the gel to a PVDF membrane via electrophoresis in transfer buffer containing (in mM) 25 Tris base, 192 glycine and 20% methanol. The membranes were incubated in blocking buffer containing (in mM): 10mM Tris pH 7.4, 100mM NaCl, 0.1% Tween 20, and 5% dehydrated milk for 30min at room temperature. The membranes were incubated in the following primary antibodies overnight at 4°C with rotation: Tyrosine Hydroxylase (rabbit polyclonal, 1:10,000, Millipore), Actin (mouse monoclonal, 1:10,000, Abcam), Dopamine Transporter (rat polyclonal, 1:2000, Millipore). The blots were washed 3x in wash buffer containing (in mM): 10mM Tris pH 7.4, 100mM NaCl, and 0.1% Tween 20, and incubated in 1:5000 secondary antibody conjugated to HRP for 2 hours (goat anti-rabbit, goat anti-mouse, goat anti-rat, Abcam). The blots were washed 3x in wash buffer, and incubated for 5min in ECL Western Blotting Substrate (Thermo Fisher) to expose the protein. Protein levels were quantified using ImageJ.
Nuclear isolation and FACS
The tissue punches of individual mice within an experimental or control group were combined (3 mice/group). Tissue punches of the ventral midbrain were flash frozen in liquid nitrogen for later use or directly homogenized in 2mL of homogenization buffer containing (in mM): 320 Sucrose (sterile filtered), 5 CaCl (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 EDTA pH 8 (sterile filtered), 0.1% NP40, 0.1 Protease Inhibitor Cocktail (PIC, Sigma), 1 β-mercaptoethanol according to Swiech et al., 2014. Sterile filtration was performed using 0.45 mm syringe filters (Millipore). The volume was then brought up to 5mL using homogenization buffer and was incubated on ice for 5 minutes. For centrifugation, 5mL of 50% Optiprep density gradient medium (Sigma) containing (in mM): 5 CaCl (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 PIC, 1 β-mercaptoethanol was added to the homogenate and mixed by inversion. The mixture was gently loaded on 10mL of 29% iso-osmolar Optiprep solution in a 1×3 1/2 in Beckman centrifuge tube (SW32 Ti rotor) and spun at 7500 RPM for 30min at 4°C. The cell debris was removed using a KimWipe and the supernatant was gently poured out. The nuclei pellet was vigorously resuspended in sterile 1xPBS and immediately taken to be further processed.
Generation of amplicons
GFP-positive nuclei were sorted using a BD AriaFACS III into a PCR tube strip containing REPLI-g Advanced Single Cell Storage buffer (QIAGEN). Whole genome amplification (WGA) was performed on the samples directly following FACS using the REPLI-g Advanced DNA Single Cell kit (QIAGEN) according to manufacturer’s instructions.
For generation of the specific amplicons, 1ul of WGA DNA was diluted 1:50 and amplified (PCR 1) with Phusion High Fidelity Polymerase (Thermo Fisher) using the following protocol (all temperatures in Celsius). Initial denaturation at 95 degrees for 30 s followed by 34 cycles of denaturation at 95 degrees for 10 s, annealing at 66 degrees for 20 s, and extension at 73 degrees for 10 s. A final extension was performed at 72 degrees for 5 min.
The product of PCR 1 was diluted 1:100 and amplified again (PCR 2) with a second set of primers to decrease non-specific amplification and increase amplified DNA yield using the thermocycler protocol. The amplicons were gel extracted using the MinElute gel extraction kit (QIAGEN), and stored at 20°C or directly used in downstream processing.
T7E1 Assay
The T7 endonuclease 1 (T7E1) assay was performed on amplicons spanning targeted regions. A 10uL reaction including 150ng of amplicons, 1uL of NEB Buffer 2 and water was placed into a thermocycler and run with an initial denaturation of 95 degrees for 5 min, followed by annealing using a ramp of 95–85 degrees at ‒2 deg/s and 85–25 degrees at ‒0.1 deg/s 1uL of T7E1 was added to each sample and placed in 37°C water bath for 15min. Samples were run on a 2% agarose gel.
Sequencing
For whole exome sequencing, 500ng of WGA genomic DNA from GFP+ sorted nuclei was sent to Genewiz on dry ice. Samples were processed using the Genewiz standardized workflow of fragmentation, library prep, hybridization, PCR amplification, and sequencing using Illumina HiSeq 2X150PE. Sequences were referenced to the mouse reference genome Mus musculus mm9. The following barcode sequences were used, Rosa26-GFP(+): AGATGTAC, TH-GFP(+): CCGAATA, Rosa26-GFP(−): CCATCCTC, and TH-GFP(−): TCTTCACA. Total reads after adaptor trimming and percent greater than 20X coverage was, Rosa26-GFP(+): 218,032,474; 82.05%, TH-GFP(+): 215,568,918; 93.94%, Rosa26-GFP(−): 199,822,240; 81.54%, and TH-GFP(−): 192,337,296; 97.67%.
For both targeted deep sequencing (Amplicon EZ) and Sanger sequencing, amplicons were sent to Genewiz. Amplicons ranged in size from ~170bp to ~450bp. Differences in amplicon size was required due to the unique sequence architecture of each gene. All Sanger sequencing was completed using the primer in the same direction as the sgRNA except for sgSlc17a7. A large G-C rich region just upstream of the sgRNA-targeted region inhibited PCR amplification. Therefore, amplification primers were required to be designed downstream of the G-C rich region, leaving only 47 bp between the 5ʹ end of the forward primer and the SaCas9 cleavage site. 47 bp is too close to the modified region to obtain Sanger sequencing results of the mutations. Therefore, the reverse primer was used instead. The following primers were used for sequencing: Primer set 1: Th forward: TTGTGCTCCTACAGTCTCTGG, reverse: ACATATCTTATGCATGCTACC; Gt(Rosa26)Sor forward: AAAGTCGCTCTGAGTTGTTATCAG, reverse: GGAGCGGGAGAAATGGA TATG; Gabrg2 forward: GGCATCAAATCATCAGCATA, reverse: TATGAAACGAGCTTAAGCTC; Slc6a3 forward: CTCTAGACATGTC TACTGAAT, reverse: TGTAGCACAGGTAGGGAAACC; Slc17a7 forward: GGATTGGCAGGGGAC, reverse: ACATTGTGCC TGGGTGTCGCC; Slc17a6 forward: CTTAAATTGACCTGAGGAGGC, reverse: TGCAGTACAGTTAATACCTAC; Slc32a1 forward: CGCAGCAAGCTGACCAATGTG, reverse: GAGCCTGGCAAGGACAGCTCA; Grin1 forward: CAATACGCTTCAGCACCTCGG, reverse: AGGAAGTAGTGGTCACCTGGC; Kcnn3 forward: CAATGCATACTCGAGTGGCAG, reverse: GACAAGCTGGCTCTGGA GTTG; Pxbp1 forward: CTTCTCACCTGGACTCCTCTT, reverse: ACACTAGGCACATCTGAAAGC; Socs6 forward: GACTGAGTTCT CAGATGTGTC, reverse: CCTGACCTGGTCTGCCAAATG; Wdfy3 forward: GAAGGAAGTAGGAAAGAGGAA, reverse: ACGGG CTGCTCCTCTCCATCAA; Ndn forward: CGAAGAACGGATAGAAGATGT, reverse: TCACATAGATGAGGCTCAGGA. Primer set 2: Th forward: TTCCAGTTCTGCCATGGCTCT, reverse: ACATATCTTATGCATGCTACC; Gt(Rosa26)Sor forward: AAAGGCTAA CCTGGTGTGTGG, reverse: GGAGCGGGAGAAATGGATATG; Gabrg2 forward: GGCATCAAATCATCAGCATA, reverse: TCTCAAATCTGGGTGGTCTC; Slc6a3 forward: CTCTAGACATGTCTACTGAAT, reverse: GATGACTGAGAGCAGGAA; Slc17a7 forward: GGATTGGCAGGGGAC, reverse: CCTCTGTCAGTTGCTCTCAGA; Slc17a6 forward: CTTAAATTGACCTGAGGAGGC, reverse: TCAATTGCACCCAAACTTGAC; Slc32a1 forward: CGCAGCAAGCTGACCAATGTG, reverse: ACTCACCTGAATGGCAT; Grin1 forward: CAATACGCTTCAGCACCTCGG, reverse: GCTTGTGGGTGACAGAAGTG; Kcnn3 forward: CAATGCATACTCGAGTGGCAG, reverse: TGCTGCGACTGTTGCTGCTGA; Pxbp1 forward: CTTCTCACCTGGACTCCTCTT, reverse: CCACGTGCTGAGCC; Socs6 forward: GACTGAGTTCTCAGATGTGTC, reverse: ATGTAGAGCTATGACTG; Wdfy3 forward: GAAGGAAGTAGGAAAGAGGAA, reverse: AACATCCCGGAAAGTGAG; Ndn forward: CGAAGAACGGATAGAAGATGT, reverse: GATTGGTCAGCCTCAGGTGCA.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed using GraphPad Prism version 6. For comparison of multiple treatments, a repeated-measures one-way ANOVA with Tukey’s post hoc multiple comparisons test between all groups was used (unless stated otherwise in the text). For analysis of experiments with multiple treatments and time-points, a two-way repeated-measures ANOVA with Bonferonni’s post hoc multiple comparisons test was used. Multiple comparisons correspond to time-points unless otherwise stated. Two-tailed Student’s t test was used. All data were tested for normality and represented as mean ± S.E.M.. All p values and sample sizes can be found in the figure legends.
DATA AND CODE AVAILABILITY
WES data generated from Rosa26-positive, Rosa26-negative, TH-positive and TH-negative cells associated with Figure 2 have been deposited with the National Center for Biotechnology Information Sequence Read Archive, accession number: PRJNA606851.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti Tyrosine hydroxylase | Millipore | Cat#MAB5280; RRID:AB_2201526 |
| Rabbit polyclonal anti Tyrosine hydroxylase | Fisher Scientific | Cat#AB152; RRID:AB_390204 |
| Rabbit polyclonal anti GFP | Invitrogen | Cat#A11122; RRID:AB_221569 |
| Mouse monoclonal anti Actin | Abcam | Cat#ab6276; RRID:AB_2223210 |
| Rat polyclonal anti Dopamine Transporter | Millipore | Cat# MAB369, RRID:AB_2190413 |
| Chicken polyclonal GFP | Abcam | Cat#ab13970; RRID:AB_300798 |
| Rabbit polyclonal HA | Sigma | Cat#H6908; RRID:AB_260070 |
| Bacterial and Virus Strains | ||
| AAV1-FLEX-SaCas9-sgTh | This paper | N/A |
| AAV1-FLEX(1loxP)-SaCas9-sgTh | This paper | N/A |
| AAV1-FLEX-SaCas9-sgGt(Rosa26)Sor | This paper | N/A |
| AAV1-FLEX-SaCas9-sgKcnn3 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgTh(2) | This paper | N/A |
| AAV1-FLEX-SaCas9-sgGabrg2 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgGabrg2TTT | This paper | N/A |
| AAV1-FLEX-SaCas9-sgSlc6a3 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgSlc6a3TTT | This paper | N/A |
| AAV1-FLEX-SaCas9-sgGrin1 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgGrin1ATG | This paper | N/A |
| AAV1-FLEX-SaCas9-sgSlc17a7 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgSlc17a6 | This paper | N/A |
| AAV1-FLEX-SaCas9-sgSlc32a1 | This paper | N/A |
| AAV1-hSyn-SpCas9 | This paper | N/A |
| AAV1-U6-sgTh(SpCas9)-hSyn-EGFP-KASH | This paper | N/A |
| AAV1-FLEX-EGFP-KASH | This paper | N/A |
| AAV1-FLEXfrt-EGFP-KASH | This paper | N/A |
| AAV1-FLEX-mCherry | University of Washington | N/A |
| AAV1-FLEXfrt-mCherry | University of Washington | N/A |
| AAV1-FLEX-ChR2-EYFP | University of Washington | N/A |
| Biological Samples | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Kynurenic acid | Sigma | Cat#K3375-5G |
| Tetrodotoxin | Tocris | Cat#1069 |
| Apamin | Tocris | Cat#1652 |
| Picrotoxin | Abcam | Cat#ab120315 |
| AP5 | Abcam | Cat#ab120003 |
| OptiPrep density gradient medium | Sigma | Cat# D1556-250ML |
| Protease Inhibitor Cocktail | Sigma | Cat# P8340 |
| Critical Commercial Assays | ||
| Qiaquick gel extraction kit | QIAGEN | Cat#28704 |
| ECL Western Blotting Substrate | Thermo Fisher | Cat#PI80196 |
| REPLI-g Advanced DNA Single Cell kit | QIAGEN | Cat#150363 |
| MinElute gel extraction kit | QIAGEN | Cat#28606 |
| Maxiprep DNA kit | Invitrogen | Cat#K210007 |
| Deposited Data | ||
| National Center for Biotechnology Information Sequence Read Archive | This paper | PRJNA606851 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T/17 cells | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Slc6a3Cre/+ | Zhuang et al., 2005 | N/A |
| Mouse: Slc32a1tm2(cre)Lowl | Jackson Laboratories | Stock #: 016962 |
| Mouse: B6;129S-Slc17a7tm1.1(cre)Hze/J | Jackson Laboratories | Stock #: 023527 |
| Mouse: B6J.129S6(FVB)-Slc17a6tm2(cre)Lowl/MwarJ | Jackson Laboratories | Stock #: 028863 |
| Mouse: ThFlp/+ | Poulin et al., 2018 | N/A |
| Mouse: B6.129S6(Cg)-Kcnn3tm2.1Jpad/J | Jackson Laboratories | Stock #: 019083 |
| Oligonucleotides | ||
| Primers for sgRNAs, see Table S1 | This paper | N/A |
| Primer for confirmation of sgRNA subcloning: 5′ GACTATCATATGCTTACCGT 3′ | This paper | N/A |
| Primers for amplification of targeted loci, see Table S2 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | N/A |
| Snapgene software | GSL biotech | Snapgene.com |
| Mini Analysis Software | Synaptosoft | N/A |
| Clampfit | Molecular Devices | N/A |
Highlights.
Single viral vector CRISPR/SaCas9 for efficient in vivo mutagenesis
Cre- and Flp-dependent cell type-specific gene mutagenesis
Combinatorial strategies for elucidating circuit-specific gene function
ACKNOWLEDGMENTS
This work was supported by NIH grants P30-DA048736, R01-MH104450, and R01-DA044315 (L.S.Z.), F31-MH116549 and T32GM007270 (A.C.H.), 5T32AA007455-33 (G.H.), and R01-MH110556 (R.A.); the Brain and Behavior Research Foundation NARSAD Independent Investigator Award (L.S.Z.) and Young Investigator Award (M.E.S.); and the University of Washington Innovator Award (L.S.Z.). We thank Dr. James Allen for assistance in AAV production, Donna Prunkard and Xiaoping Wu for assistance with FACS, and members of the Zweifel lab for thoughtful discussion.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.092.
REFERENCES
- Bä ck S, Necarsulmer J, Whitaker LR, Coke LM, Koivula P, Heathward EJ, Fortuno LV, Zhang Y, Yeh CG, Baldwin HA, et al. (2019). Neuronspecific genome modification in the adult rat brain using CRISPR-Cas9 transgenic rats. Neuron 102, 105–119.e8. [DOI] [PubMed] [Google Scholar]
- Bak RO, Dever DP, and Porteus MH (2018). CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat. Protoc 13, 358–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklisch C, Pascoli V, Wong JC, House DR, Yvon C, de Roo M, Tan KR, and Lüscher C (2013). Cocaine disinhibits dopamine neurons by potentiation of GABA transmission in the ventral tegmental area. Science 341, 1521–1525. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Blake JA, Smith CL, Kadin JA, and Richardson JE; Mouse Genome Database Group (2019). Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47 (D1), D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DL, and Williams JT (1994). Cocaine inhibits GABA release in the VTA through endogenous 5-HT. J. Neurosci 14, 6763–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sá nchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, and Gradinaru V (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, and Haeussler M (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46 (W1), W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvas M, and Palmiter RD (2010). Restricting dopaminergic signaling to either dorsolateral or medial striatum facilitates cognition. J. Neurosci 30, 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deignan J, Luja n, R., Bond C, Riegel A, Watanabe M, Williams JT, Maylie J, and Adelman JP (2012). SK2 and SK3 expression differentially affect firing frequency and precision in dopamine neurons. Neuroscience 217, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, and Charpentier E (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Ma G, Huang X, D’Amore PA, Zhang F, and Lei H (2016). The clustered, regularly interspaced, short palindromic repeats-associated endonuclease 9 (CRISPR/Cas9)-created MDM2 T309G mutation enhances vitreous-induced expression of MDM2 and proliferation and survival of cells. J. Biol. Chem 291, 16339–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM (1996). A modular set of Flp, FRT and lacZ fusion vectors for manipulating genes by site-specific recombination. Gene 171, 197–201. [DOI] [PubMed] [Google Scholar]
- Edwards NJ, Tejeda HA, Pignatelli M, Zhang S, McDevitt RA, Wu J, Bass CE, Bettler B, Morales M, and Bonci A (2017). Corrigendum: circuit specificity in the inhibitory architecture of the VTA regulates cocaine-induced behavior. Nat. Neurosci 20, 1189. [DOI] [PubMed] [Google Scholar]
- Engblom D, Bilbao A, Sanchis-Segura C, Dahan L, Perreau-Lenz S, Balland B, Parkitna JR, Luja n, R., Halbout B, Mameli M, et al. (2008). Glutamate receptors on dopamine neurons control the persistence of cocaine seeking. Neuron 59, 497–508. [DOI] [PubMed] [Google Scholar]
- Friedrich G, and Soriano P (1991). Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5, 1513–1523. [DOI] [PubMed] [Google Scholar]
- Giros B, Jaber M, Jones SR, Wightman RM, and Caron MG (1996). Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature 379, 606–612. [DOI] [PubMed] [Google Scholar]
- Gore BB, Soden ME, and Zweifel LS (2013). Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Curr. Protoc. Neurosci 65, 4.35.1–4.35.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, and Samulski RJ (2005). Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol 79, 9933–9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Kay MA, and Kleinschmidt JA (2003). Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther 7, 839–850. [DOI] [PubMed] [Google Scholar]
- Gu H, Zou YR, and Rajewsky K (1993). Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73, 1155–1164. [DOI] [PubMed] [Google Scholar]
- Haeussler M, Schö nig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, and Zeng H (2013). Genetic approaches to neural circuits in the mouse. Annu. Rev. Neurosci 36, 183–215. [DOI] [PubMed] [Google Scholar]
- Iyer V, Boroviak K, Thomas M, Doe B, Riva L, Ryder E, and Adams DJ (2018). No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 14, e1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, and Doudna JA (2015). Structural biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, and Stewart J (1991). Dopamine transmission in the initiation and expression of drug- and stress-induced sensitization of motor activity. Brain Res. Brain Res. Rev 16, 223–244. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Stanford W, de Solis C, Aradhana, Abraham ND, Dao TJ, Thaseen S, Sairavi A, Gonzalez CU, and Ploski JE (2018). The development of an AAV-based CRISPR SaCas9 genome editing system that can be delivered to neurons in vivo and regulated via doxycycline and Cre-recombinase. Front. Mol. Neurosci 11, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, and Malenka RC (2014). Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology 76 (Pt B), 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QS, Pu L, and Poo MM (2005). Repeated cocaine exposure in vivo facilitates LTP induction in midbrain dopamine neurons. Nature 437, 1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, He Y, Zhang C, He X, Yan L, Li M, Hu T, Hu Y, Jiang J, Meng X, et al. (2019). Trio deep-sequencing does not reveal unexpected off-target and on-target mutations in Cas9-edited rhesus monkeys. Nat. Commun 10, 5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusby E, Fife KH, and Berns KI (1980). Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J. Virol 34, 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, and Margolis EB (2017). Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci 18, 73–85. [DOI] [PubMed] [Google Scholar]
- Muzyczka N, Samulski RJ, Hermonat P, Srivastava A, and Berns KI (1984). The genetics of adeno-associated virus. Adv. Exp. Med. Biol 179, 151–161. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Mikuni T, and Yasuda R (2017). Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron 96, 755–768.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin JF, Caronia G, Hofer C, Cui Q, Helm B, Ramakrishnan C, Chan CS, Dombeck DA, Deisseroth K, and Awatramani R (2018). Mapping projections of molecularly defined dopamine neuron subtypes using intersectional genetic approaches. Nat. Neurosci 21, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdó ttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski PD (1995). The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol 51, 53–91. [PubMed] [Google Scholar]
- Salvatore MF, Calipari ES, and Jones SR (2016). Regulation of tyrosine hydroxylase expression and phosphorylation in dopamine transporter-deficient mice. ACS Chem. Neurosci 7, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford CA, Soden ME, Baird MA, Miller SM, Schulkin J, Palmiter RD, Clark M, and Zweifel LS (2017). A central amygdala CRF circuit facilitates learning about weak threats. Neuron 93, 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal D, Koenig JI, Adelman JP, Brady D, Prendeville LC, and Shepard PD (2004). Regional distribution of SK3 mRNA-containing neurons in the adult and adolescent rat ventral midbrain and their relationship to dopamine-containing cells. Synapse 53, 104–113. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer C, Balsiger S, Bluethmann H, Mansuy IM, Fritschy JM, Mohler H, and Lüscher B (2003). The gamma 2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol. Cell. Neurosci 24, 442–450. [DOI] [PubMed] [Google Scholar]
- Shepard PD, and Bunney BS (1991). Repetitive firing properties of putative dopamine-containing neurons in vitro: regulation by an apamin-sensitive Ca(2+)-activated K+ conductance. Exp. Brain Res 86, 141–150. [DOI] [PubMed] [Google Scholar]
- Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, Baldarelli RM, Beal JS, Campbell J, Corbani LE, et al. (2019). The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 47 (D1), D774–D779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soden ME, Jones GL, Sanford CA, Chung AS, Güler AD, Chavkin C, Lujá n R, and Zweifel LS (2013). Disruption of dopamine neuron activity pattern regulation through selective expression of a human KCNN3 mutation. Neuron 80, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AJ, and Palmiter RD (2018). Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 34, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffensen SC, Taylor SR, Horton ML, Barber EN, Lyle LT, Stobbs SH, and Allison DW (2008). Cocaine disinhibits dopamine neurons in the ventral tegmental area via use-dependent blockade of GABA neuron voltage-sensitive sodium channels. Eur. J. Neurosci 28, 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, and Doudna JA (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, and Zhang F (2015). In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol 33, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Daigle TL, Chen Q, and Feng G (2014). Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol 1183, 221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. ; Mouse Genome Sequencing Consortium (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Wolfart J, Neuhoff H, Franz O, and Roeper J (2001). Differential expression of the small-conductance, calcium-activated potassium channel SK3 is critical for pacemaker control in dopaminergic midbrain neurons. J. Neurosci 21, 3443–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, and Colosi P (2010). Effect of genome size on AAV vector packaging. Mol. Ther 18, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, and Soriano P (1997). Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U S A 94, 3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Masson J, Gingrich JA, Rayport S, and Hen R (2005). Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J. Neurosci. Methods 143, 27–32. [DOI] [PubMed] [Google Scholar]
- Zweifel LS, Argilli E, Bonci A, and Palmiter RD (2008). Role of NMDA receptors in dopamine neurons for plasticity and addictive behaviors. Neuron 59, 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WES data generated from Rosa26-positive, Rosa26-negative, TH-positive and TH-negative cells associated with Figure 2 have been deposited with the National Center for Biotechnology Information Sequence Read Archive, accession number: PRJNA606851.