Abstract
Background
The randomized phase II INTELLANCE-2/EORTC_1410 trial on EGFR-amplified recurrent glioblastomas showed a trend towards improved overall survival when patients were treated with depatux-m plus temozolomide compared with the control arm of alkylating chemotherapy only. We here performed translational research on material derived from this clinical trial to identify patients that benefit from this treatment.
Methods
Targeted DNA-sequencing and whole transcriptome analysis was performed on clinical trial samples. High-throughput, high-content imaging analysis was done to understand the molecular mechanism underlying the survival benefit.
Results
We first define the tumor genomic landscape in this well-annotated patient population. We find that tumors harboring EGFR single-nucleotide variations (SNVs) have improved outcome in the depatux-m + TMZ combination arm. Such SNVs are common to the extracellular domain of the receptor and functionally result in a receptor that is hypersensitive to low-affinity EGFR ligands. These hypersensitizing SNVs and the ligand-independent EGFRvIII variant are inversely correlated, indicating two distinct modes of evolution to increase EGFR signaling in glioblastomas. Ligand hypersensitivity can explain the therapeutic efficacy of depatux-m as increased ligand-induced activation will result in increased exposure of the epitope to the antibody–drug conjugate. We also identified tumors harboring mutations sensitive to “classical” EGFR tyrosine-kinase inhibitors, providing a potential alternative treatment strategy.
Conclusions
These data can help guide treatment for recurrent glioblastoma patients and increase our understanding into the molecular mechanisms underlying EGFR signaling in these tumors.
Keywords: amphiregulin, depatux-m, EGFR, extracellular domain mutations, ligand hypersensitivity
Key Points.
SNVs in EGFR are correlated with improved survival to depatux-m + temozolomide.
Common SNVs in EGFR increase sensitivity of the receptor to its ligands.
Some gliomas harbor EGFR tyrosine-kinase inhibitor sensitive mutations.
Importance of the Study.
Recurrent glioblastoma patients have a dismal prognosis; the median survival is ~6–8 months and there is no standard of care. Depatux-m is an antibody–drug conjugate with signs of clinical activity in recurrent glioblastomas when given in combination with temozolomide. Here, we used material from a randomized phase II trial and identified patients that have survival benefit from this combination. We show that specific mutations increase sensitivity to EGFR ligands and this “hypersensitivity” can explain the observed treatment benefit. Our results indicate that ligand hypersensitivity and ligand independence are two, inversely correlated, mechanisms to increase EGFR signaling. We also identified tumors harboring mutations sensitive to “classical” EGFR tyrosine-kinase inhibitors, providing a potential alternative treatment strategy. These data can help guide treatment for recurrent glioblastoma patients.
The epidermal growth factor receptor (EGFR) gene is amplified in approximately half of all glioblastoma patients.1,2 Unfortunately, and despite EGFR being a driver mutation in glioblastomas, pharmacological inhibition of the receptor has not been demonstrated to affect patient survival or tumor growth.3–5 Depatuxizumab mafodotin (depatux-m, ABT414) is an antibody–drug conjugate that consists of an antibody directed against EGFR,6,7 conjugated to a toxin (monomethyl auristatin F) that blocks microtubule polymerization. The antibody is specific for tumors that overexpress EGFR and preferentially binds to the active conformation of the receptor and the constitutively active variant, EGFRvIII.6–8 Depatux-m therefore should specifically target glioblastoma cells by using the high expression level of EGFR. Phase I clinical trials have tested the drug for safety and toxicity and showed some encouraging responses especially in recurrent glioblastoma patients with EGFR amplification.9–11
INTELLANCE-2/EORTC_1410 was a randomized phase II trial on EGFR-amplified recurrent glioblastomas that showed a trend towards improved overall survival (HR 0.71, 95% CI [0.50, 1.02], P = .06 in the primary analysis, HR 0.66, 95% CI [0.48, 0.93], P = .024 in follow-up analysis) when patients were treated with depatux-m and temozolomide (TMZ) compared with the control arm of alkylating chemotherapy only. In the present study, we aimed to identify patients that benefit from this combination and to understand the mechanism of increased sensitivity.
Methods
Patient Samples
Recurrent GBM patients were considered eligible for the INTELLANCE-2/EORTC_1410 trial (NCT02343406) if they had been diagnosed with a histologically confirmed, EGFR-amplified glioblastoma at first occurrence. Amplification of the EGFR locus was centrally determined using FISH in one of the three laboratories (Histogenex, Antwerp Belgium, Mosaic, Lake Forest California, Peter MacCallum Cancer Institute Melbourne, Australia) using the Vysis EGFR CDx Assay (Abbott Molecular, Des Plaines, IL; not on market).12 A tumor was considered EGFR-amplified when the EGFR/centromere chromosome 7 (CEP7) ratio was ≥2 in ≥15% recorded nuclei, with 50 nuclei/tumor analyzed. Tumors with polysomy for chromosome 7 (CEP7 copy number > 3) but without focal amplification of the EGFR gene in ≥ 15% nuclei were considered to be EGFR-nonamplified and not included. Two hundred sixty patients were randomized in the trial to receive either i) TMZ or, if progressing within 16 weeks of day 1 of the last temozolomide cycle, CCNU (n = 26 and 60, respectively); ii) depatux-m (n = 86); or iii) TMZ plus depatux-m (n = 88). For this analysis, the database was locked on January 12, 2018 (longer term follow-up data). MGMT promoter methylation status data were previously described and determined using a methylation-specific PCR.13 All patients gave written informed consent for trial participation, pathology review, and molecular testing.
Sequencing
Material, either tissue sections or tissue blocks, were centrally collected at Erasmus MC. Evaluation of the area with highest tumor content was done by the pathologist (J.M.K.) on a hematoxylin and eosin stained section. One to twenty 5μ sections were then sent to Almac Diagnostics (Craigavon, UK) for macro-dissection, DNA and RNA extraction, and sequencing. DNA/RNA extraction was performed using the Allprep DNA/RNA FFPE kit (Qiagen, Venlo, The Netherlands). Sequencing was done on the Trusight Tumor 170 panel (Illumina, Eindhoven, The Netherlands) which uses a combination of DNA and RNA sequencing to interrogate SNVs in ~150 genes, amplification of 59 genes, and fusion and splice-variant expression in 55 genes. SNV, copy-number, fusion-gene, and splice-variant expression calling was done using the Illumina Basespace sequence hub. Very deep sequencing was performed to enable quantification of subclonal EGFR variants. All variants with a variant allele fraction (VAF) > 15% were included in the analysis, except for EGFR, where all VAFs were included as all variants in EGFR are subclonal. SNVs with quality scores <70 and/or present in the Exac database at fractions >0.001 were omitted from the analysis. Splice variants/mutations were calculated as the “spliced-in fraction”; the number of mutant reads as fraction of the total reads over that particular variant. Data were further analyzed in R using ggplot2, survival, and GenVisR packages. Expression values were estimated using featureCounts using gencode-29 as gene annotation. One sample yielding only 707 reads was excluded from further analysis.
Whole transcriptome sequencing of rRNA depleted cDNA was done on the same isolate by GenomeScan (Leiden, The Netherlands) at a depth of 50 million paired-end reads per sample. HTStream was used to remove duplicate reads, fastp for low base quality trimming and adaptor removal and further quality assessment. Alignment to hg38 was done using STAR (2.6.1d). Stranded read-counts were estimated with STARs builtin “--quantMode GeneCounts” option. Samples with read-count <750,000 were excluded from further analysis (29 samples). DESeq2 was used for expression analysis and its VST-normalization for survival analysis using Coxph regression in R.
Data Analysis
For generating the waterfall plot of chromosomal changes, we defined trisomy as whole chromosome copy-number > 2.4 and LOH as copy-number < 1.6. For generating the waterfall plot per gene, we set a threshold for high-copy amplification to > 6 copies per cell, copy number gain (including trisomy) between 3 and 6, and deep (homozygous) deletions at <1 copy per cell. All analyses to define variants associated with survival were done on samples with high copy amplification of EGFR only; samples without such high-level amplification may represent a different molecular entity.
Constructs and Image Analysis
EGFR mutation constructs were generated by in-fusion cloning into a piggybac vector (System Biosciences, Palo Alto, CA) with eGFP cloned 3′ to the transmembrane domain as described.14 This position was chosen to avoid potential interference with ligand binding or receptor internalization–signaling sites. These constructs retain the important physical properties of EGFR with respect to signal transduction and protein–protein interactions.14 Stable HeLa cell-lines (ATCC, Manassas, VA) were created for all constructs. Cells were plated in 96 or 384 well plates for further analysis. Following transfection, we selected for cells that expressed the (mutant) EGF receptor using FACS. As we did not select for a single cell clone, levels of EGFR expression were variable between individual cells. This way the observed responses can be evaluated across a wide range of expression levels. Quantification of EGFR signals intensities shows a high correlation between intensity and mRNA expression levels. This was done by comparing mRNA expression with signal intensities in various lung-cancer cell lines (manuscript in preparation). Using this approach we show that the various mutation constructs had expression levels comparable to the endogenous EGFR expression in these cells (except for the cell lines expressing the EGFR_A289D or EGFRvIII mutation, where the construct is expressed at slightly lower levels; for EGFRvIII this was despite repeated attempts). Moreover, expression levels were highly similar between the various mutation constructs, again except for the cell line expressing the EGFR_A289D or EGFRvIII mutation.
All images were obtained using an Opera Phenix high-throughput high-content confocal microscope (Perkin Elmer, Hamburg, Germany). At least 10 images were obtained per well so that hundreds of individual cells per condition were analyzed ensuring robustness of measurements. Image analysis was performed using Harmony software (Perkin Elmer) using identical settings for all conditions within each experiment and further analyzed using R. Experiments were performed at least in three independent replicates.
EGFR antibody (clone H11, DAKO, Amstelveen, The Netherlands) and a phospho-specific EGFR antibody (AB32430, anti phospho Y1068, Abcam, Cambridge, UK) were used at 1:500 dilution. Secondary antibodies used were alexafluor 647 goat anti-mouse, alexafluor 594 rabbit anti-mouse and alexafluor 488 goat anti-rabbit (A21240, A11062, and A11008, respectively, Invitrogen, Bleiswijk, The Netherlands). Hoechst was used as counterstain to visualize nuclei.
Statistical Analysis
Distribution of frequencies was compared between subtypes using the chi-squared test. A Fisher’s exact test was used in case the assumptions for chi-square distribution were violated as indicated in the respective tables. Kaplan–Meier survival curves were generated using the survival package in R.15 Overall survival was used to identify the molecular markers associated with outcome, with survival defined from the point of randomization until the date of death. If unavailable, the date of day known to be alive was used. The significance of prognostic factors was determined using Cox regression in univariate analysis. Differential gene expression was determined using de DESeq2 bioconductor package. P values less than .05, which were adjusted for a false discovery rate <0.05, were considered significant.
Results
Mutational Landscape of INTELLANCE-2/EORTC_1410 Trial Samples
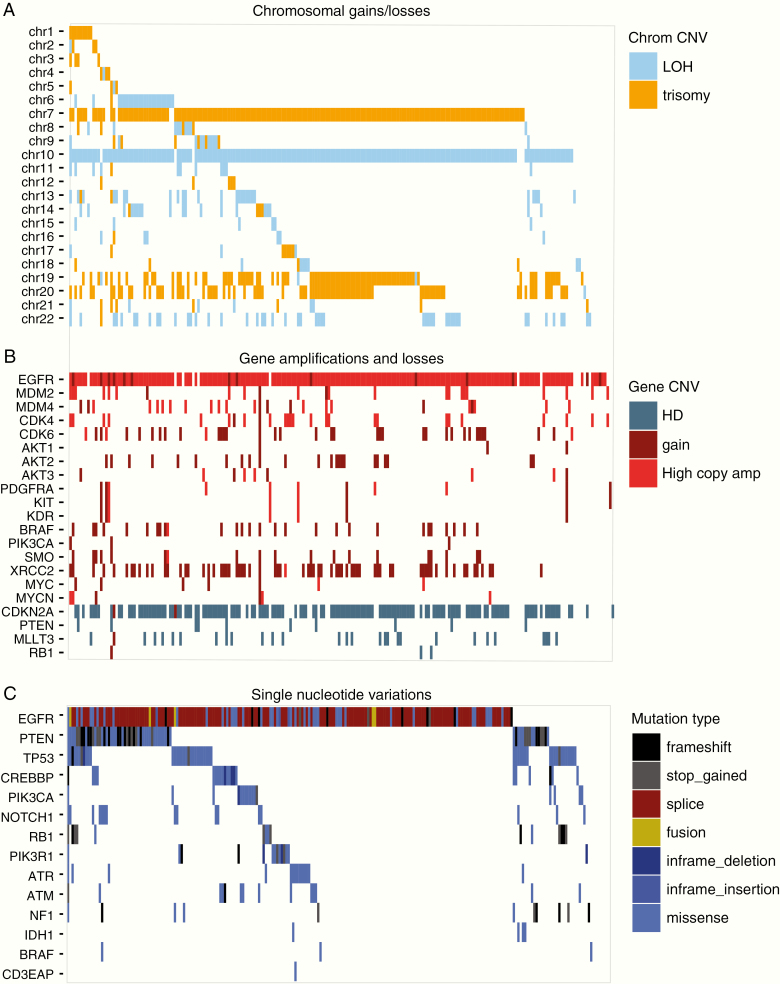
We first analyzed the molecular characteristics of tumors from patients included in the INTELLANCE-2/EORTC_1410 trial. This is important as this allows defining the genomic landscape in tumors of patients that are eligible for clinical trial inclusion. Glioblastomas often have trisomy of chromosome 7 and loss of chromosome 10 and, of the large scale genomic changes, these were indeed the two most commonly found (204 and 184 samples for chromosome 7 and 10, respectively, Figure 1A). Combined trisomy 7, LOH 10 was observed in 176 of 236 samples (75%). Other common large chromosomal changes included gain of chromosomes 19 and 20 which are also frequently observed in glioblastomas.
Figure 1.
Genomic landscape of samples included in the INTELLANCE-2/EORTC_1410 trial. Shown are waterfall plots of chromosomal changes (A), gains and losses of individual genes (B), and SNVs within individual genes (C). The copy number changes, gene amplifications/deletions, and mutations are similar to observed in other (EGFR-amplified) glioblastoma datasets. Patients included in this study therefore were not selected for a specific molecularly subtype. LOH = loss of heterozygosity; HD = homozygous deletion.
On the gene level, most, but not all, samples harbored high copy amplification of the EGFR locus (copy number > 6 in 200 tumors16); this was mainly observed in tumors with trisomy 7 (166/200, Figure 1B). Other high copy amplified genes included MDM2 (n = 20), MDM4 (n = 21), CDK4 (n = 24), and CDK6 (n = 4). Most samples also harbored a homozygous deletion of the CDKN2A locus, and a small population had such deletions in the PTEN locus.
Mutations were identified in driver genes common to GBMs and included EGFR (n = 115), PTEN (n = 62), TP53 (n = 48), CREBBP (n = 21), and PIK3CA (n = 16) (Figure 1C). Mutational hotspots were identified in TP53 (Supplementary Figure 1), PTEN (see below), and EGFR. As may be expected, truncating mutations were common to tumor suppressor genes (PTEN, RB1, and NF1).
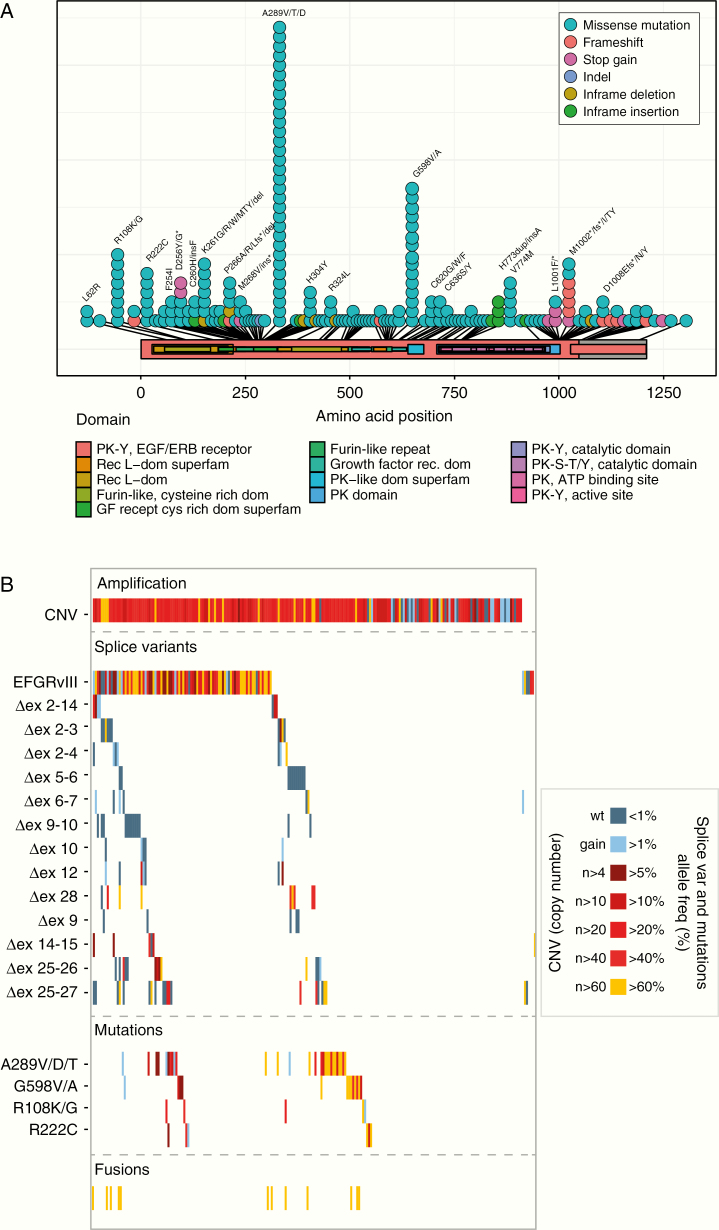
Common missense mutations identified in EGFR clustered on the extracellular domain of the protein and included R108, A289, and G598. Interestingly, we identified a new hotspot at the 3′ (intracellular) end of the gene where truncating mutations tended to cluster (Figure 2A). Such mutations often involved amino acids L1001 and M1002. We also identified three mutations in EGFR that, in lung cancer, are associated with response to EGFR tyrosine-kinase inhibitors (TKIs).17 These mutations, G719A, G719D and S786I (~1.3% of analyzed samples), were present at a relatively high mutant allele fraction (in two samples, the variant allele fraction, VAF, exceeded >20%). Since the type of mutation may predict response to EGFR TKIs, such inhibitors may provide a new treatment option for these patients.18–20
Figure 2.
Genetic changes within the EGFR gene of samples from patients included in the INTELLANCE-2/EORTC_1410 trial. (A) Lolliplot of SNVs identified showing characteristic hotspot mutations in the extracellular domain. (B) Waterfall plots of genetic changes subdivided in copy number gains, splice variant expression, and mutations. The waterfall plot is color coded to represent the level of copy number gain (in the CNV plot) or the percentage of mutant alleles (percentage spliced in).
The most common (oncogenic) splice variant identified was the glioblastoma-specific in-frame deletion of exons 2–7 (EGFRvIII), present in approximately half of all EGFR-amplified samples (n = 101, Figure 2B). Other variants present at significant population frequency included deletions of exons 9–10 (n = 14), exons 25–26 (n = 14), and exons 25–27 (n = 22), the latter two affecting the intracellular domain. EGFR fusion genes were identified in 13 samples, with fusion partners often located in the vicinity of the EGFR gene locus (SEPT14, SEC61G, LANCL2). Similar to previously reported for glioblastomas, the mutations, splice variants, and fusion genes identified in EGFR were almost always subclonal and many samples harbored more than one genetic change (Figure 2B).1 Only 13/200 EGFR-amplified samples did not harbor any genetic change in the EGFR locus.
A289 Missense Mutations and 3′ Truncating Mutations in EGFR Are Associated With Response to Depatux-m + TMZ
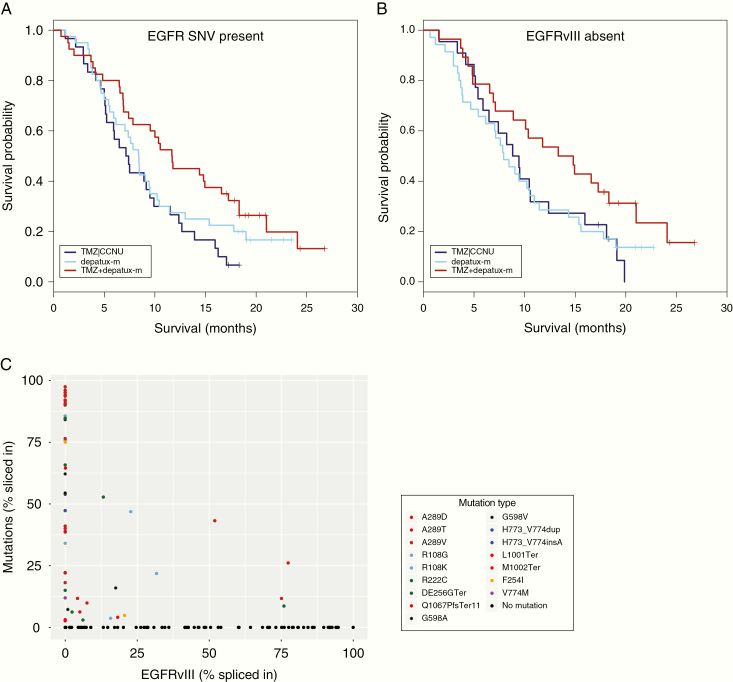
We performed correlative analysis on genetic changes to identify those associated with survival in the depatux-m + TMZ arm. Interestingly, the presence of SNVs (any protein altering SNV within the coding region) in EGFR was associated with survival in univariate analysis: the HR was 0.495 with 95% CI [0.283, 0.865], P = .014 in the combination arm and 0.751, 95% CI [0.444, 1.272], P = 0.287 in the depatux-m monotherapy arm, compared with the CCNU or TMZ control arm (Table 1, Figure 3). Multivariable analysis confirmed that depatux-m + TMZ treatment was associated with survival in samples with EGFR-SNVs, independent of known prognostic factors such as age and MGMT promoter methylation status (HR 0.45, 95% CI [0.26, 0.76], P = 0.003; Supplementary Table 1). No such association was found in the samples without EGFR-SNVs (HR 0.85, 95% CI [0.48, 1.50], P = .57). Since depatux-m specifically targets EGFR, we focused further on individual variants to determine which of these were most associated with survival. A trend was observed in tumors harboring A289 hotspot mutations (HR 0.386, 95% CI [0.138, 1.082], P = .070, Table 1, Figure 4A). No other individual SNV reached such statistical values, but this may be related to the low number of samples harboring individual SNVs and the corresponding limited statistical power.
Table 1.
Genetic changes in EGFR and other genes associated with response to depatux-m + TMZ
| EGFR mutation/Gene | Treatment | HR | 95% CI | Pr(>|z|) | n | events | survival (months) | |
|---|---|---|---|---|---|---|---|---|
| Any mutation | TMZ|CCNU | 28 | 26 | 6.8 | ||||
| depatux-m | 0.751 | 0.444 | 1.272 | 0.287 | 37 | 31 | 8.4 | |
| TMZ+depatux-m | 0.495 | 0.283 | 0.865 | 0.014 | 33 | 26 | 11.7 | |
| Hotspot mutation | TMZ|CCNU | 15 | 14 | 7.1 | ||||
| depatux-m | 1.058 | 0.530 | 2.113 | 0.873 | 21 | 20 | 7.0 | |
| TMZ+depatux-m | 0.510 | 0.247 | 1.055 | 0.070 | 21 | 16 | 10.5 | |
| A289 | TMZ|CCNU | 8 | 8 | 7.5 | ||||
| depatux-m | 0.799 | 0.313 | 2.037 | 0.638 | 11 | 11 | 6.2 | |
| TMZ+depatux-m | 0.386 | 0.138 | 1.082 | 0.070 | 11 | 8 | 10.4 | |
| G598 | TMZ|CCNU | 4 | 4 | 4.2 | ||||
| depatux-m | 1 | 0 | NA | |||||
| TMZ+depatux-m | 0.254 | 0.027 | 2.370 | 0.229 | 2 | 1 | 7.8 | |
| R108 | TMZ|CCNU | 3 | 2 | 5.2 | ||||
| depatux-m | 1.814 | 0.325 | 10.120 | 0.497 | 5 | 5 | 7.8 | |
| TMZ+depatux-m | 1.076 | 0.207 | 5.590 | 0.931 | 6 | 6 | 9.3 | |
| EGFRvIII absent | TMZ|CCNU | 22 | 20 | 9.1 | ||||
| depatux-m | 1.004 | 0.570 | 1.771 | 0.988 | 35 | 30 | 7.9 | |
| TMZ+depatux-m | 0.582 | 0.311 | 1.088 | 0.090 | 28 | 21 | 14.1 | |
| EGFRvIII present | TMZ|CCNU | 43 | 40 | 7.5 | ||||
| depatux-m | 0.894 | 0.553 | 1.446 | 0.649 | 33 | 29 | 7.3 | |
| TMZ+depatux-m | 0.673 | 0.412 | 1.099 | 0.114 | 33 | 27 | 9.8 | |
| ∆ex25-27 | TMZ|CCNU | 11 | 11 | 5.1 | ||||
| depatux-m | 0.285 | 0.087 | 0.937 | 0.039 | 6 | 5 | 9.4 | |
| TMZ+depatux-m | 0.255 | 0.077 | 0.846 | 0.026 | 5 | 4 | 16.9 | |
| ∆ex25-26 | TMZ|CCNU | 5 | 3 | 8.4 | ||||
| depatux-m | 1.691 | 0.399 | 7.166 | 0.476 | 6 | 5 | 5.5 | |
| TMZ+depatux-m | 0.629 | 0.104 | 3.804 | 0.614 | 3 | 2 | 14.4 | |
| ∆ex27 | TMZ|CCNU | 2 | 2 | 6.6 | ||||
| depatux-m | 2.145 | 0.117 | 39.265 | 0.607 | 1 | 1 | 3.9 | |
| TMZ+depatux-m | 0.351 | 0.031 | 3.978 | 0.398 | 2 | 2 | 14.1 | |
| C-term del | TMZ|CCNU | 2 | 2 | 6.8 | ||||
| depatux-m | 1.414 | 0.085 | 23.570 | 0.809 | 1 | 1 | 3.5 | |
| TMZ+depatux-m | 0.000 | 0.000 | Inf | 0.999 | 3 | 1 | NA | |
| C-term SNV | TMZ|CCNU | 3 | 3 | 5.1 | ||||
| depatux-m | 0.107 | 0.009 | 1.278 | 0.077 | 4 | 2 | 17.8 | |
| TMZ+depatux-m | 0.077 | 0.005 | 1.192 | 0.067 | 3 | 1 | NA | |
| all-C-term | TMZ|CCNU | 16 | 14 | 5.1 | ||||
| depatux-m | 0.514 | 0.229 | 1.155 | 0.107 | 15 | 12 | 8.5 | |
| TMZ+depatux-m | 0.309 | 0.130 | 0.735 | 0.008 | 13 | 9 | 16.9 | |
| all C-term trunc | TMZ|CCNU | 13 | 13 | 5.1 | ||||
| depatux-m | 0.480 | 0.181 | 1.271 | 0.140 | 8 | 7 | 8.8 | |
| TMZ+depatux-m | 0.175 | 0.054 | 0.574 | 0.004 | 7 | 4 | 18.3 | |
| PTEN all | TMZ|CCNU | 16 | 16 | 8.4 | ||||
| depatux-m | 0.692 | 0.353 | 1.353 | 0.282 | 23 | 19 | 8.4 | |
| TMZ+depatux-m | 0.499 | 0.241 | 1.034 | 0.061 | 20 | 15 | 10.2 | |
| PTEN HD | TMZ|CCNU | 14 | 13 | 5.4 | ||||
| depatux-m | 1.291 | 0.540 | 3.088 | 0.566 | 9 | 9 | 5.5 | |
| TMZ+depatux-m | 0.392 | 0.125 | 1.229 | 0.108 | 7 | 6 | 8.2 | |
| PTEN SNV | TMZ|CCNU | 13 | 13 | 8.8 | ||||
| depatux-m | 0.876 | 0.636 | 1.207 | 0.417 | 18 | 15 | 8.7 | |
| TMZ+depatux-m | 0.659 | 0.475 | 0.914 | 0.013 | 15 | 11 | 11.0 | |
| ARID1A SNV | TMZ|CCNU | 2 | 2 | 2.6 | ||||
| depatux-m | 0.541 | 0.086 | 3.410 | 0.513 | 5 | 5 | 5.4 | |
| TMZ+depatux-m | 0.139 | 0.015 | 1.258 | 0.079 | 5 | 3 | 16.6 | |
| ARID1A LOH | TMZ|CCNU | 3 | 3 | 10.5 | ||||
| depatux-m | 1 | 0 | NA | |||||
| TMZ+depatux-m | 0.286 | 0.047 | 1.745 | 0.175 | 6 | 5 | 15.1 | |
| AIRD1A SNV+LOH | TMZ|CCNU | 5 | 5 | 4.2 | ||||
| depatux-m | 0.572 | 0.158 | 2.070 | 0.395 | 6 | 5 | 8.2 | |
| TMZ+depatux-m | 0.269 | 0.075 | 0.961 | 0.043 | 11 | 8 | 15.4 | |
| RP11.770J1.4 | TMZ|CCNU | 10 | 10 | 6.7 | ||||
| depatux-m | 0.831 | 0.379 | 1.821 | 0.643 | 19 | 17 | 8.4 | |
| TMZ+depatux-m | 0.322 | 0.100 | 1.036 | 0.057 | 8 | 4 | 14.8 | |
| DHFR | TMZ|CCNU | 28 | 27 | 9.5 | ||||
| depatux-m | 1.003 | 0.599 | 1.677 | 0.992 | 36 | 32 | 7.9 | |
| TMZ+depatux-m | 0.587 | 0.345 | 1.001 | 0.050 | 36 | 28 | 11.7 |
OS = Median overall survival. Hotspot mutations are defined as amino acid changes present in at least four samples and involve amino acids, A289, G598, R222, R108, K261, P266, H304, H773, V774, and/or M1002.
Figure 3.
Genetic changes within the EGFR gene that were associated with prolonged survival with depatux-m + TMZ. (A) Presence of SNVs; (B) absence of EGFRvIII expression. Hazard rates of these changes are listed in Table 1. Both genetic changes are correlated as samples containing SNVs often do not express EGFRvIII and vice versa (C).
Figure 4.
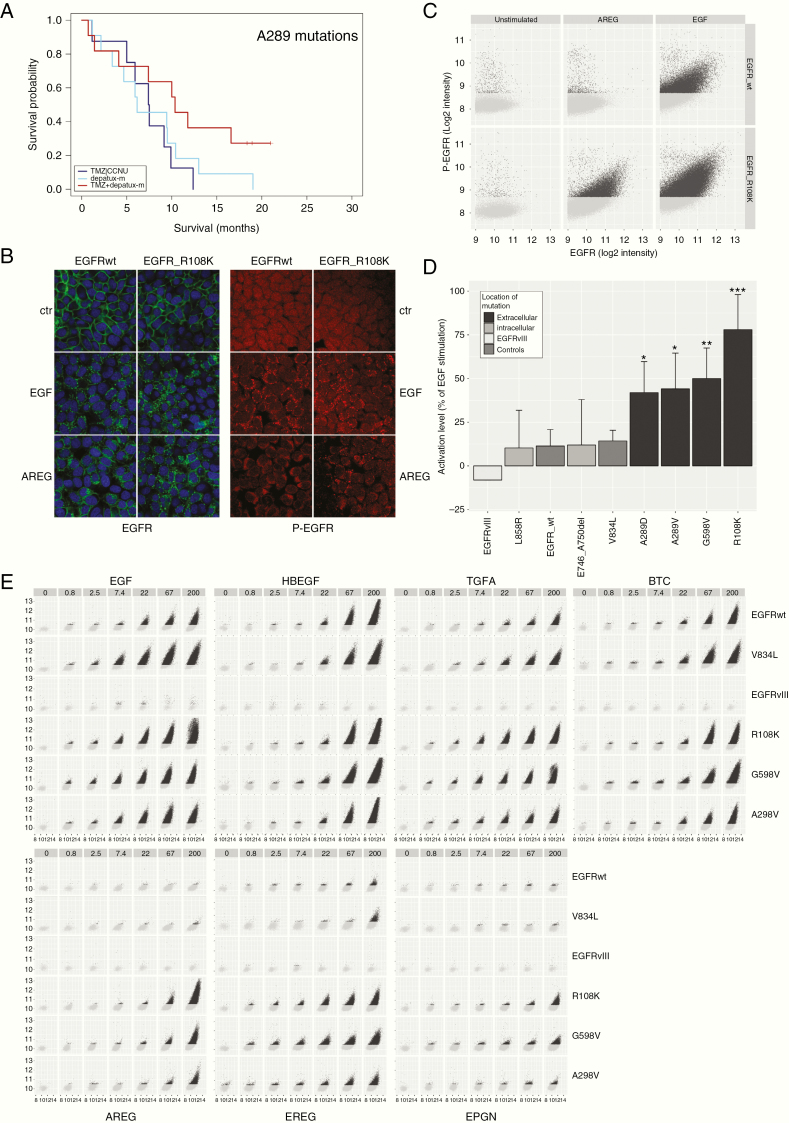
(A) A289x hotspot mutations are associated with response to depatux-m + TMZ. (B) Example of microscopic images of EGFR-wt and EGFRR108K stimulated with either EGF or AREG. Green: EGFR, red: phospho-EGFR, blue: Hoechst. AREG resulted in increased receptor endocytosis and phospho-EGFR signal in EGFRR108K compared to EGFR-wt. (C) Example of analysis depicting the level of phospho-EGFR (y-axis) against total EGFR (x-axis) within individual EGFR positive vesicles. Each dot represents one EGFR-positive submenbranous vesicle. (D) Averages of experiments plotted in (C). (E) Similar to (B) the level of phospho-EGFR (y-axis) is plotted against total EGFR (x-axis) within individual EGFR positive vesicles. As can be seen, apart from EGFRvIII, all constructs responded, dose dependently, to the high affinity ligands EGF, HB-EGF, TGFA, and BTC by an increase in EGFR phosphorylation. Low affinity ligands AREG, EREG, and EPGN were not able to stimulate EGFR-wt constructs, but resulted in a strong activation of extracellular domain mutations EGFRR108K, EGFRA289V, and EGFRG598V. The absence of responses in EGFRvIII-expressing cells is in line with the notion that this mutation is independent of ligand. Images were taken at 40× magnification.
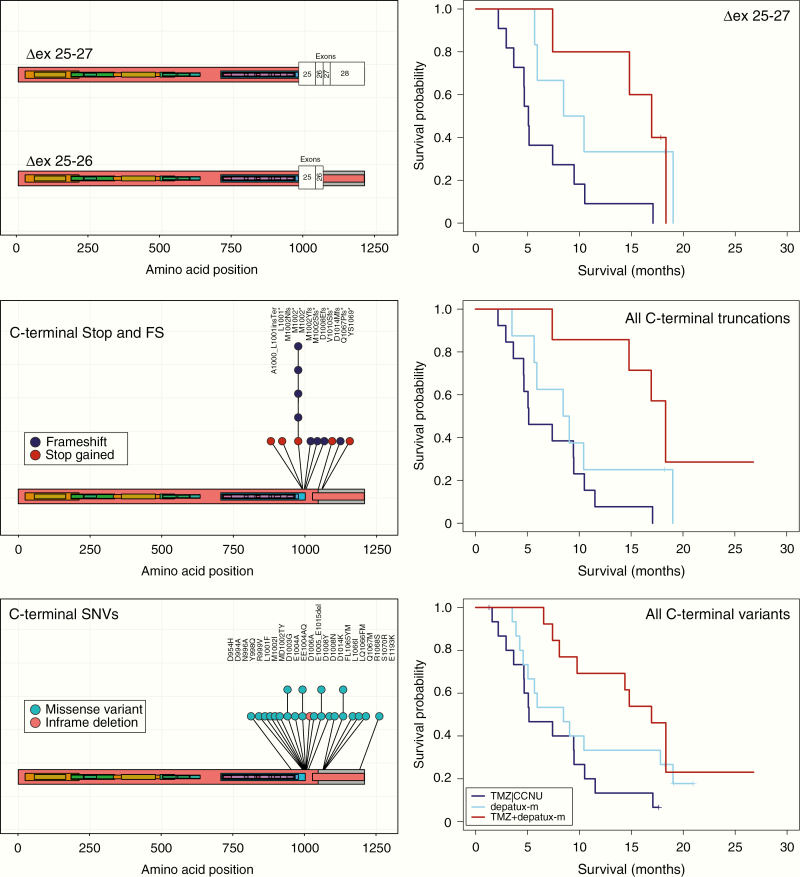
On the splice variant level, we found that a deletion of exons 25–27, affecting the C-terminal intracellular domain, was also associated with survival in the depatux-m + TMZ arm. Although there are relatively few samples (n = 22) with this genetic change, the HR was 0.255 with 95% CI [0.077, 0.846] and P = .026 (Table 1, Figure 5). The ∆ex 25–27 splice variants introduce a frame shift in exon 28 resulting in a deletion of the C-terminal tail (exons 25–28) of EGFR. We therefore included C-terminal truncating mutations (SNVs leading to frameshifts and premature termination codons and fusion genes at the 3′ end of the gene) in this analysis and found that the association remained significant (HR 0.175, 95% CI [0.054, 0.574], P = .004 for the combination of depatux-m +TMZ). The association also remained significant when other 3′ mutations were included in the analysis: 3′ end missense mutations (n = 10) center around two hotspots at amino acids 993–1014 and 1065–1070 and it is possible that they affect a functional domain similar to the domain lost in the truncating mutations (Table 1, Figure 5). Exon deletions overlapping the hotspots ∆ex 25–26 or ∆ex 27, identified in 14 and 5 samples, respectively, may affect a similar domain. When all C-terminal mutations were combined (∆ex 25–28, ∆ex 25–26, ∆ex 27, protein truncation SNVs, and/or other 3’SNVs fusion genes, n = 28 samples), the HR for depatux-m + TMZ was 0.309 [0.130, 0.735], P = .008. Depatux-m monotherapy was not significantly associated with survival (HR 0.514 [0.229, 1.155], P = .107, Table 1, Figure 5). We acknowledge that care should be taken when conducting such post hoc and combinatorial analyses, therefore, the analysis of each variant type is listed separately (Table 1).
Figure 5.
C-terminal truncating mutations are associated with response to depatux-m + TMZ. Deletion of exons 25–27 results in a frame shift of the protein. Such changes are associated with response to depatux-m + TMZ (top right). Nonsense and frameshift mutations show a similar trend (middle panels, left lolliplot of individual mutations, right survival analysis), as do the C-terminal SNVs (bottom panels, left lolliplot of individual mutations, right survival analysis). Hazard rates of individual changes are listed in Table 1.
Expression of EGFRvIII is common in glioblastomas and, since depatux-m also has affinity for this deletion variant, we were particularly interested in association with survival. In contrast to what might be expected, the absence of EGFRvIII expression showed a trend towards association with survival: the HR was 0.582, 95% CI [0.311, 1.088], P = .090 in the combination arm and 1.004, 95% CI [0.570, 1.771], P = .988 in the depatux-m monotherapy arm both compared with the CCNU or TMZ control arm (Table 1, Figure 3, all treatment arms shown in Supplementary Figure 2). Multivariable analysis including MGMT promoter methylation status and age confirmed the trend towards association of depatux-m + TMZ with survival in samples without EGFRvIII expression-SNVs (HR 0.57, 95% CI [0.31, 1.03], P = .064, Supplementary Table 1).
Because MGMT status is predictive for response to TMZ chemotherapy, we stratified the molecular markers associated with survival (EGFR-SNVs and absent EGFRvIII expression) by this factor. Although sample size is relatively small, both depatux-m + TMZ and MGMT promoter methylation status were associated with improved outcome and both associations remained significant in a multivariable analysis containing both factors (Supplementary Figure 3).
Extracellular Missense Mutations Result in a Receptor With Increased Ligand Sensitivity
We next aimed to determine why SNVs in EGFR are associated with response to depatux-m + TMZ. First, the presence of SNVs and absence of EGFRvIII expression identify a similar population: the majority of samples expressing EGFRvIII do not express additional EGFR mutation variants (18/96) and the majority of samples harboring SNVs do not express EGFRvIII (18/57), P = .004 (chi-square test, Figure 3C). The level of EGFRvIII expression is also lower in samples expressing SNVs: The spliced-in fraction (the number of mutant reads as fraction of the total EGFR reads) was 0.12 ± 0.22 vs. 0.29 ± 0.36, P < .001. Conversely, the level of EGFR SNVs was lower in samples expressing EGFRvIII compared with those that do not express EGFRvIII (spliced-in fraction, calculated only using reads covering the affected base, was 0.11 ± 0.22 vs. 0.35 ± 0.38, P < .001). The difference in expression is even larger when only focusing on hotspot mutations (i.e., any mutation occurring in at least four samples) where the spliced in fraction of EGFRvIII was 0.09 ± 0.18 vs. 0.45 ± 0.33 in mutation positive v negative tumors (P < .001). The inverse correlation between expression of EGFRvIII and SNVs was not specific to samples included in this study; glioblastoma samples included in the randomized phase II BELOB trial and in TCGA samples show a near identical and statistically significant inverse correlation (Supplementary Figure 4).1,21 This inverse correlation suggests a divergent evolution where, after the initial amplification of EGFR, glioblastomas develop either EGFRvIII or other EGFR mutations.
To understand why SNVs in EGFR are associated with survival in the depatux-m + TMZ arm, we performed functional analysis on various EGFR-mutation constructs. The majority of hotspot mutations in glioblastomas are localized in the extracellular, ligand binding domain of EGFR, and we hypothesized that they may affect ligand-induced activation. We therefore stimulated various mutation constructs with two different EGFR ligands: one high affinity ligand (EGF) and one low affinity ligand (Amphiregulin, AREG). Stimulation with 200 ng/ml EGF resulted in a strong increase EGFR-phosphorylation in all constructs (except for EGFRvIII; this constitutively active variant is ligand independent22,23) (Figure 4). In contrast, the low affinity EGFR ligand amphiregulin (AREG, 200 ng/ml) only marginally activated the control constructs EGFR-wt and EGFRV834L and did not activate EGFRvIII. Interestingly, AREG stimulation had a strong effect on all constructs harboring extracellular missense mutations found in glioblastomas (EGFRR108K, EGFRG598V, and EGFRA289V). AREG activated these EGFR-mutation constructs at levels 40%–80% of that observed by EGF stimulation, a 3-8 fold increase compared with EGFRwt constructs (P < .02 for any extracellular mutation construct vs. EGFRwt and P < .05 for any of the extracellular mutation constructs vs. EGFRV834L, Figure 4). Mutations commonly found in lung cancer (EGFRL858R and EGFRA746-E750del) responded to EGF and AREG similar to the control constructs.
To determine whether extracellular domain mutations were also more sensitive to other EGFR ligands, we tested all seven known ligands for EGFR activation. For each ligand, we performed a dose–response analysis ranging from 200 ng/ml (maximal stimulation) to 0.8 ng/ml. In the control constructs, all high affinity EGFR ligands (EGF, TGFα, HB-EGF, and BTC) resulted in a strong activation and all low affinity ligands AREG, EREG, and EPGN resulted in a markedly weaker activation (Figure 6). In contrast, however, all extracellular missense mutations, EGFRA298V, EGFRG598V, and EGFRR108K, showed strong activation towards all EGFR ligands, including the low affinity ligands AREG, EPGN, and EREG (Figure 6). EGFRvIII did not respond to any of the ligands. These experiments therefore show that EGFR containing extracellular missense mutations render the receptor more sensitive to stimulation, especially by the weak activators AREG, EREG, and EPGN.
The hypersensitivity of extracellular domain mutations may explain the increased responsiveness to depatux-m: receptors are more easily activated and, as activation leads to receptor internalization, increased internalization with the antibody/drug conjugate. Hypersensitivity likely also leads to an increased exposure of the epitope for the antibody (i.e., the activated conformation of EGFR).24
Our molecular imaging analysis also showed that EGFRvIII mainly has an intracellular localization (Supplementary Figure 5). This is in contrast to control constructs which are mainly localized to the membrane. Other activating mutations such as EGFRL858R also showed a certain degree of increased intracellular localization, but only EGFRvIII showed such prominent intracellular localization. The increased responsiveness in samples without EGFRvIII expression therefore can be explained by the near absence of EGFRvIII on the extracellular membrane: this may prevent effective binding to depatux-m.
Functional analysis indeed confirmed the direct correlation between receptor internalization and EGFR antibody internalization: EGFR receptor internalization still occurred in the presence of either ABT806 or cetuximab, regardless of ligand or mutation present, whereas this internalization could be completely inhibited by erlotinib or lapatinib (respectively, type I and II TKIs). The internalization was accompanied by uptake of EGFR antibodies as demonstrated by staining cells only with secondary antibodies directed at the FC fragment (Supplementary Figure 6).
Additional Genetic Events Associated With Survival in the Depatux-m + TMZ Arm
We also performed correlative analysis to screen for other events associated with patient survival in the depatux-m + TMZ arm. We focused on genetic events (SNVs and CNVs) present in at least nine samples and selected those showing a trend (P < .10) by Cox regression analysis. Of the 149 genes examined, inactivating PTEN mutations were associated with outcome to depatux-m + TMZ; the HR was 0.499 with 95% CI [0.241, 1.034], P = .061 (Table 1, Supplementary Figure 7). For this analysis, we combined samples with homozygous deletion and SNVs (individual mutation types showed a similar trend, Table 1). Combining homozygous deletion with SNVs was warranted as many of the SNVs in PTEN led to premature stop codons and most of the missense mutations are listed in the COSMIC database25 with high pathogenic prediction scores (FATHMM26) (Supplementary Table 2).
A second gene associated with survival was ARID1A, a tumor suppressor gene that is mutated in various cancer types including ovarian, endometrial, and uterine cancer.27 Mutations in this gene are often heterozygous which suggests that inactivation of one allele is sufficient to relieve the tumor suppressive effect of the protein.27 The identified SNVs in ARID1A in our samples were also heterozygous. When combining samples with ARID1A LOH (n = 10) and missense mutations (n = 12), the HR for depatux-m + TMZ was 0.27 (95% CI [0.074, 0.961], P = .04) (Table 1, Supplementary Figure 8; analysis of LOH and SNVs individually is also listed). SNVs in dihydro-folate reductase (DHFR, a gene required for the de novo synthesis of purines) and RP11.770J1.4 (a long intragenic noncoding RNA) were also associated with response to the combination of depatux-m + TMZ (HR 0.587, P = .050 and HR 0.322, P = .057, respectively, Supplementary Figure 8).
Because inactivating alterations in PTEN and TP53 are associated with EGFR kinase inhibitor response in several cancer types (see, e.g., ref. 28), we performed a multivariate analysis including these genes and show that depatux-m + TMZ remained a factor associated with survival (Supplementary Table 3).
Gene Expression Analysis
Whole transcriptomic analysis identified genes associated with survival in each of the three treatment arms (Supplementary Tables 4–6). For each of these gene-lists, several genes were co-expressed which is suggestive for higher-order interactions (Supplementary Figure 9). Gene-set enrichment analysis identified various pathways associated with survival including “cell cycle,” “cell activation,” and “meiotic cell cycle process,” and various pathways associated with immune response including “immune system development” and “lymphocyte activation.” We did not find a correlation between the level of immune infiltration and survival as determined by Immunophenoscore analysis.29 One gene, N-MYC downstream regulated gene 2 (NDRG2), was specifically associated with survival in the depatux-m + TMZ arm and not in the other two arms of the study (Supplementary Figure 9), suggesting that this gene is predictive for response to the combination treatment. Other studies have also shown similar correlation between expression and survival of NDRG2 in glioblastomas.30
The survival curves in both depatux-m treated arms show a tail suggesting more long survivors (>365 days from randomization) when treated with the drug. Gene expression analysis between long and short survivors identified 15 differentially expressed genes, including CDK4 and 6 genes that neighbor it. We find approximately 2.2 times more CDK4-amplified samples in patients with survival < 365 days (P = .004, see also ref. 31). Expression of FOXF1 was significantly higher in short- compared with long survivors (P = .010, Supplementary Figure 10).
Discussion
In this study, we have performed detailed molecular analyses on glioblastomas of patients treated within the INTELLANCE-2/EORTC_1410 randomized phase II clinical trial. Our results suggest that patients harboring tumors with EGFR SNVs may derive more benefit from the combination of depatux-m + TMZ.
Three mechanisms can explain the survival benefit of these variants to depatux-m + TMZ. First, we show that extracellular domain mutations result in a receptor that is hypersensitive to activation by the various EGFR ligands. Since EGFR is internalized after receptor activation,32 the hypersensitivity likely increases internalization and so increase uptake of the antibody–drug conjugate. Second, hypersensitive mutations increase transformation towards the active conformation of the protein. EGFR can switch between inactive (closed) and active (domain II exposed) conformation; the presence of ligand locks the protein in the active conformation.20,33 Hypersensitive mutations may shift the equilibrium towards the active conformation of the protein. Indeed, such mutations have been demonstrated to increase exposure of the epitope for the ABT806 antibody, and so result in increased binding of depatux-m.24 Thirdly, we show that EGFRvIII expression is inversely correlated with the presence of SNVs in EGFR. Since EGFRvIII mainly has an intracellular localization (see also ref. 34), its near absence on the extracellular membrane may prevent binding to depatux-m, samples without EGFRvIII expression (i.e., predominantly those with EGFR SNVs) are more likely to respond to depatux-m. Other mechanisms, however, may also determine sensitivity/resistance.35
Our results are in line with two recent publications both using mice engrafted with cell lines specifically overexpressing either EGFRA289V or EGFRG598V ECD mutations. Both studies showed significant survival benefit from treatment with ABT806 (i.e., the antibody used in depatux-m).24,36 Since our study also suggested benefit when such mutations are present, and given the mechanistic insight of its possible mode of action, further investigation into the efficacy of depatux-m in glioblastoma patients with ECD mutations is warranted.
We also show that intracellular EGFR truncating mutations and splice variants are associated with response to depatux-m + TMZ. Other studies have shown that such truncating mutations result in altered receptor internalization37,38 and this altered internalization therefore may be linked to treatment response and survival benefit. Similarly, bi-allelic inactivation of PTEN also was associated with response to depatux-m + TMZ. Since poly-phosphoinositides are key regulators of membrane trafficking, they also may contribute to altered receptor endocysis. PtdIns(4,5)P2, for example, is required in the progression of early endocytosis39 and PtdIns(4,5)P2 is produced from PTEN substrate PtdIns(3,4,5)P3.
One important caveat of this study is that the correlative analysis are post hoc, and therefore, these observations require confirmation in an independent dataset: our analysis may have incorrectly identified markers associated with survival in the depatux-m + TMZ arm due to multiple testing. Such dataset may be available in the INTELLANCE-1 trial that examined the effect of depatux-m in combination with chemoradiation compared with chemoradiation only in newly diagnosed glioblastoma patients (clinicaltrials.gov identifier NCT02573324). Of note, this trial did not meet its primary endpoint, though trial results have not been published to date. The difference between the two trials may lie in the fact that the patients treated within INTELLANCE-1 received surgery prior to treatment. The remaining tumor cells in INTELLANCE-1 therefore were likely in areas with an intact blood-brain barrier, which may have reduced accessibility to depatux-m. In addition, most of the samples analyzed in the current study were derived from primary tumors (in ~80% of cases) as surgery at recurrence is seldom performed. Various changes can occur during tumor evolution and some specifically affect EGFR variants.40–42EGFRvIII expression, for example, is often lost at tumor recurrence43–45; ~1/3 of EGFR-mutations are also lost at tumor recurrence.46 Nevertheless, the copy number changes, gene amplifications/deletions and mutations are similar to observed in other (EGFR-amplified) glioblastoma datasets, and therefore patients included in the INTELLANCE-2/EORTC_1410 trial did not select for a specific and molecularly defined tumor type.1 Another limitation of our study is the that response is extrapolated from survival data. Correlation of response as seen on MRI with molecular features may identify different genetic markers.
Our data also provide a model for tumor evolution with respect to EGFR-dependency. GBMs require EGFR signaling for growth and EGFR amplification is the first step in GBMs to meet this requirement. After this initial amplification, the tumor evolves to facilitate the need for EGFR signaling by gaining additional and activation mutations. Our data show that there are at least two different modes of evolution in glioblastomas: by becoming independent of ligand (EGFRvIII) or by becoming hypersensitive to ligand (extracellular hotspot mutations). Since both EGFRvIII and extracellular domain mutations have tumorigenic properties,22,47,48 only one of these mutations is required to facilitate the need for EGFR signaling. This explains why, in all the glioblastoma datasets examined, EGFRvIII expression is inversely correlated with presence of EGFR-missense mutations (Figure 3 and Supplementary Figure 4).
In addition to finding associations of specific EGFR variants associated with response to depatux-m + TMZ, we also found rare mutations that, in pulmonary adenocarcinoma patients, respond to EGFR-TKIs. Several lines of evidence suggest that responses to EGFR-TKIs are dependent on the type of mutation and not on the type of tumor. For example, TKI-sensitive mutations in pulmonary adenocarcinomas are also sensitive to these TKIs in other tumor types.49–51 Such responses have also been documented for the mutations identified in this study.18,19 These TKI-sensitive mutations have been identified in other glioblastoma datasets, and although present in only a small minority (~1–2%) of EGFR-amplified GBMs), EGFR-TKIs may prove an interesting treatment option for patients harboring such tumors. However, one complicating factor is that EGFR mutations in glioblastomas almost invariably are subclonal. In addition, they often show high intratumoral (and temporal) heterogeneity.46 It is therefore possible that EGFR-TKIs may only be effective in tumors with a high variant allele fraction of the TKI-sensitive mutation.
Funding
We are grateful to AbbVie, Inc. for supporting this independent EORTC study. This research was sponsored by a grant from the “Westlandse ride.” Our manuscript refers to one unpublished manuscript: The clinical report of the INTELLANCE 2/EORTC 1410 randomized phase 2 clinical trial by Martin van den Bent et al. This manuscript has been submitted to Neuro-Oncology.
Conflict of interest statement:
P.J.F. received research funding by AbbVie; M.J.v.d.B. consultancy for Abbvie, cellgene, boehringer, BMS, AGIOS. P.A., J.L., and E.B. are employees of AbbVie and may own stock. M.W. has received research grants from Abbvie, Adastra, Bayer, Merck, Sharp & Dohme (MSD), Dracen, Merck (EMD), Novocure, OGD2, Piqur und Roche, and honoraria for lectures or advisory board participation or consulting from Abbvie, Basilea, Bristol Meyer Squibb, Celgene, Merck, Sharp & Dohme (MSD), Merck (EMD), Novocure, Orbus, Roche, Teva, and Tocagen. J.M.S. received research funding from Pfizer and Catalysis, consulting or advisory board for Abbvie, Celgene, and Pfizer and travel expenses from Astellas and Abbvie. PC consultancy for Abbvie, Astra Zeneca, BMS, Merck Serono, MSD, Daiichi Sankyo, Vifor, and Leo pharma, and received a research grant from Astra Zeneca. EORTC (MMO, TGO, and VGO) has received research funding from AbbVie.
Authorship
Conceptualization: P.J.F., M.v.d.B.; Methodology: Y.H., W.V., T.G., P.J.F.; Investigation: Y.H., W.V., I.d.H., M.d.W., and J.M.K.; Writing—Original Draft: P.J.F; Writing—Review and Editing: all authors; Funding Acquisition: P.J.F.; Resources: M.E., J.S., A.W., J-.S.F., E.F., P.M.C., M.W., P.A., J.L., and E.B.; Data Curation: M.M., T.G., M.v.R; Supervision:: P.J.F. and M.v.d.B.
Supplementary Material
References
- 1. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lassman AB, Aldape KD, Ansell PJ, et al. . Epidermal growth factor receptor (EGFR) amplification rates observed in screening patients for randomized trials in glioblastoma. J Neurooncol. 2019;144(1):205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van den Bent MJ, Brandes AA, Rampling R, et al. . Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27(8):1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uhm JH, Ballman KV, Wu W, et al. . Phase II evaluation of gefitinib in patients with newly diagnosed Grade 4 astrocytoma: Mayo/North Central Cancer Treatment Group Study N0074. Int J Radiat Oncol Biol Phys. 2011;80(2):347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sepúlveda-Sánchez JM, Vaz MÁ, Balañá C, et al. . Phase II trial of dacomitinib, a pan-human EGFR tyrosine kinase inhibitor, in recurrent glioblastoma patients with EGFR amplification. Neuro Oncol. 2017;19(11):1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johns TG, Stockert E, Ritter G, et al. . Novel monoclonal antibody specific for the de2-7 epidermal growth factor receptor (EGFR) that also recognizes the EGFR expressed in cells containing amplification of the EGFR gene. Int J Cancer. 2002;98(3):398–408. [DOI] [PubMed] [Google Scholar]
- 7. Luwor RB, Johns TG, Murone C, et al. . Monoclonal antibody 806 inhibits the growth of tumor xenografts expressing either the de2-7 or amplified epidermal growth factor receptor (EGFR) but not wild-type EGFR. Cancer Res. 2001;61(14):5355–5361. [PubMed] [Google Scholar]
- 8. Johns TG, Adams TE, Cochran JR, et al. . Identification of the epitope for the epidermal growth factor receptor-specific monoclonal antibody 806 reveals that it preferentially recognizes an untethered form of the receptor. J Biol Chem. 2004;279(29):30375–30384. [DOI] [PubMed] [Google Scholar]
- 9. Lassman AB, van den Bent MJ, Gan HK, et al. . Safety and efficacy of depatuxizumab mafodotin + temozolomide in patients with EGFR-amplified, recurrent glioblastoma: results from an international phase I multicenter trial. Neuro Oncol. 2019;21(1):106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gan HK, Reardon DA, Lassman AB, et al. . Safety, pharmacokinetics, and antitumor response of depatuxizumab mafodotin as monotherapy or in combination with temozolomide in patients with glioblastoma. Neuro Oncol. 2018;20(6):838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Bent M, Gan HK, Lassman AB, et al. . Efficacy of depatuxizumab mafodotin (ABT-414) monotherapy in patients with EGFR-amplified, recurrent glioblastoma: results from a multi-center, international study. Cancer Chemother Pharmacol. 2017;80(6):1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lassman AB, Roberts-Rapp L, Sokolova I, et al. . Comparison of biomarker assays for EGFR: implications for precision medicine in patients with glioblastoma. Clin Cancer Res. 2019;25(11):3259–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 14. Erdem-Eraslan L, Gao Y, Kloosterhof NK, et al. . Mutation specific functions of EGFR result in a mutation-specific downstream pathway activation. Eur J Cancer. 2015;51(7):893–903. [DOI] [PubMed] [Google Scholar]
- 15.Therneau, TM, Grambsch, PM. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 16. French PJ, Eoli M, Sepulveda JM, et al. . Defining EGFR amplification status for clinical trial inclusion. Neuro Oncol. 2019;21(10):1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor AD, Micheel CM, Anderson IA, Levy MA, Lovly CM. The Path(way) less traveled: a pathway-oriented approach to providing information about precision cancer medicine on my cancer genome. Transl Oncol. 2016;9(2):163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voss JS, Holtegaard LM, Kerr SE, et al. . Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44(7):1216–1222. [DOI] [PubMed] [Google Scholar]
- 19. Agatsuma N, Yasuda Y, Ozasa H. Malignant pleural mesothelioma harboring both G719C and S768I mutations of EGFR successfully treated with Afatinib. J Thorac Oncol. 2017;12(9):e141–e143. [DOI] [PubMed] [Google Scholar]
- 20. Gao Y, Vallentgoed WR, French PJ.. Finding the right way to target EGFR in glioblastomas; Lessons from lung adenocarcinomas. Cancers (Basel). 2018;10(12):489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erdem-Eraslan L, van den Bent MJ, Hoogstrate Y, et al. . Identification of patients with recurrent glioblastoma who may benefit from combined bevacizumab and CCNU Therapy: a report from the BELOB trial. Cancer Res. 2016;76(3):525–534. [DOI] [PubMed] [Google Scholar]
- 22. Nishikawa R, Ji XD, Harmon RC, et al. . A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91(16):7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang HS, Nagane M, Klingbeil CK, et al. . The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272(5):2927–2935. [DOI] [PubMed] [Google Scholar]
- 24. Orellana L, Thorne AH, Lema R, et al. . Oncogenic mutations at the EGFR ectodomain structurally converge to remove a steric hindrance on a kinase-coupled cryptic epitope. Proc Natl Acad Sci U S A. 2019;116(20):10009–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Forbes SA, Beare D, Boutselakis H, et al. . COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34(3):511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mellinghoff IK, Wang MY, Vivanco I, et al. . Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. [DOI] [PubMed] [Google Scholar]
- 29. Charoentong P, Finotello F, Angelova M, et al. . Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. [DOI] [PubMed] [Google Scholar]
- 30. Skiriutė D, Steponaitis G, Vaitkienė P, et al. . Glioma malignancy-dependent NDRG2 gene methylation and downregulation correlates with poor patient outcome. J Cancer. 2014;5(6):446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cimino PJ, McFerrin L, Wirsching HG, et al. . Copy number profiling across glioblastoma populations has implications for clinical trial design. Neuro Oncol. 2018;20(10):1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan M, Narasimhan S, de Heus C, et al. . EGFR dynamics change during activation in native membranes as revealed by NMR. Cell. 2016;167(5):1241–1251.e11. [DOI] [PubMed] [Google Scholar]
- 34. Ekstrand AJ, Liu L, He J, et al. . Altered subcellular location of an activated and tumour-associated epidermal growth factor receptor. Oncogene. 1995;10(7):1455–1460. [PubMed] [Google Scholar]
- 35. Scott AM, Roberts-Rapp L, Gan H, et al. . Determinants of responses to ABT-414: results of next generation sequencing. Neuro-Oncology. 2015; 17(Suppl 5): v10. [Google Scholar]
- 36. Binder ZA, Thorne AH, Bakas S, et al. . Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell. 2018;34(1):163–177.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen WS, Lazar CS, Lund KA, et al. . Functional independence of the epidermal growth factor receptor from a domain required for ligand-induced internalization and calcium regulation. Cell. 1989;59(1):33–43. [DOI] [PubMed] [Google Scholar]
- 38. Chang CP, Kao JP, Lazar CS, et al. . Ligand-induced internalization and increased cell calcium are mediated via distinct structural elements in the carboxyl terminus of the epidermal growth factor receptor. J Biol Chem. 1991;266(34):23467–23470. [PubMed] [Google Scholar]
- 39. Bohdanowicz M, Grinstein S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol Rev. 2013;93(1):69–106. [DOI] [PubMed] [Google Scholar]
- 40. Kim H, Zheng S, Amini SS, et al. . Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim J, Lee IH, Cho HJ, et al. . Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Cazzato E, Ladewig E, et al. . Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Felsberg J, Hentschel B, Kaulich K, et al. ; German Glioma Network Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Clin Cancer Res. 2017;23(22):6846–6855. [DOI] [PubMed] [Google Scholar]
- 44. van den Bent MJ, Gao Y, Kerkhof M, et al. . Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol. 2015;17(7):935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weller M, Butowski N, Tran DD, et al. ; ACT IV trial investigators Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 46. Draaisma K, Chatzipli A, Taphoorn M, et al. . Molecular evolution of IDH wild-type glioblastomas treated with standard of care affects survival and design of precision medicine trials: a report from the EORTC 1542 study. J Clin Oncol. 2019; JCO1900367. [DOI] [PubMed] [Google Scholar]
- 47. Lee JC, Vivanco I, Beroukhim R, et al. . Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3(12):e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feng H, Hu B, Vuori K, et al. . EGFRvIII stimulates glioma growth and invasion through PKA-dependent serine phosphorylation of Dock180. Oncogene. 2014;33(19):2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iyevleva AG, Novik AV, Moiseyenko VM, Imyanitov EN. EGFR mutation in kidney carcinoma confers sensitivity to gefitinib treatment: a case report. Urol Oncol. 2009;27(5):548–550. [DOI] [PubMed] [Google Scholar]
- 50. Masago K, Asato R, Fujita S, et al. . Epidermal growth factor receptor gene mutations in papillary thyroid carcinoma. Int J Cancer. 2009;124(11):2744–2749. [DOI] [PubMed] [Google Scholar]
- 51. Ali SM, Alpaugh RK, Buell JK, et al. . Antitumor response of an ERBB2 amplified inflammatory breast carcinoma with EGFR mutation to the EGFR-TKI erlotinib. Clin Breast Cancer. 2014;14(1):e14–e16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.