Abstract
Background
Malignant brain tumors (BT) are among the cancers most frequently associated with constitutional mismatch repair deficiency (CMMRD), a rare childhood cancer predisposition syndrome resulting from biallelic germline mutations in mismatch repair genes. This study analyzed data from the European “Care for CMMRD” (C4CMMRD) database to describe their clinical characteristics, treatments, and outcome with the aim of improving its diagnosis/treatment.
Methods
Retrospective analysis of data on patients with CMMRD and malignant BT from the C4CMMRD database up to July 2017.
Results
Among the 87 registered patients, 49 developed 56 malignant BTs: 50 high-grade gliomas (HGG) (with giant multinucleated cells in 16/21 histologically reviewed tumors) and 6 embryonal tumors. The median age at first BT was 9.2 years [1.1–40.6], with nine patients older than 18. Twenty-seven patients developed multiple malignancies (including16 before the BT). Most patients received standard treatment, and eight patients immunotherapy for relapsed HGG. The 3- and 5-year overall survival (OS) rates were 30% (95% CI: 19–45) and 22% (95% CI: 12–37) after the first BT, with worse prognosis for HGG (3-year OS = 20.5%). Six patients were alive (median follow-up 2.5 years) and 43 dead (38 deaths, 88%, were BT-related). Other CMMRD-specific features were café-au-lait macules (40/41), multiple BTs (5/15), developmental brain anomalies (11/15), and consanguinity (20/38 families).
Conclusions
Several characteristics could help suspecting CMMRD in pediatric malignant BTs: giant cells on histology, previous malignancies, parental consanguinity, café-au-lait macules, multiple BTs, and developmental brain anomalies. The prognosis of CMMRD-associated BT treated with standard therapies is poor requiring new therapeutic up-front approaches.
Keywords: brain tumor, café-au-lait spot, childhood cancer, constitutional mismatch repair deficiency, high-grade glioma, MMR biallelic germline mutation, predisposition
Key Points.
• CMMRD diagnosis is difficult in malignant brain tumors due to phenotypic overlaps.
• Skin or brain anomalies and giant cell glioblastoma can guide towards CMMRD.
• Prognosis of patients with CMMRD and brain tumor is poor and needs to be improved.
Importance of the Study.
Constitutional mismatch repair deficiency (CMMRD) is a cancer predisposition syndrome resulting from biallelic germline mutations in mismatch repair genes, leading to childhood malignancies. As CMMRD diagnosis is challenging in pediatric patients with malignant brain tumors, we analyzed data (personal and familial history, histological characteristics of the tumors, treatments, and outcome) concerning 49 patients with CMMRD and malignant brain tumors from the European C4CMMRD database to identify specific characteristics. This analysis allowed highlighting the following specific features that may guide practitioners: café-au-lait macules, high-grade gliomas (HGG) particularly those with giant multinucleated cells, asynchronous multiple brain tumors, developmental brain anomalies, and high rate of parental consanguinity. Most patients received standard treatments, and eight underwent immunotherapy for HGG at relapse. We also found that the prognosis of patients with CMMRD and brain tumor, especially HGG, is not good. We discussed new treatment and prevention strategies with immunotherapeutic approaches to improve their outcome.
Base substitution and insertion-deletion mismatches generated during DNA replication are corrected by the DNA mismatch repair (MMR) pathway.1 Germline mutations in one of the four MMR genes (MLH1, MSH2, MSH6, and PMS2) cause cancer predisposition. Heterozygous germline mutations in one of the four MMR genes cause Lynch syndrome which is associated with an increased risk of colorectal, gynecological, urinary tract, and other cancers during the fourth and fifth decades of life.2 Constitutional mismatch repair deficiency (CMMRD) (OMIM#276300), first described in 1999,3,4 is a rare autosomal recessive cancer predisposition syndrome caused by homozygous or compound heterozygous germline mutations in one of the four MMR genes. Patients with CMMRD may develop a large variety of neoplasms, including malignant brain, gastrointestinal tract, and hematologic tumors,5 most frequently in childhood and adolescence.
On account of its rarity, international collaborations have been put in place, such as the European “Care for CMMRD” (C4CMMRD) consortium and the International Biallelic Mismatch Repair Deficiency Consortium, to better identify and manage these patients. The European C4CMMRD Consortium was launched in Paris in 2013. It established and published diagnostic criteria and surveillance guidelines.5,6 Since the first reports, more than 200 patients with CMMRD and cancer have been described. International collaborations allowed increasing the knowledge on this syndrome, its phenotype, the pathophysiological mechanisms of tumor development, and potential therapeutic options.5,7–11 However, despite an increasing number of publications, CMMRD is still underdiagnosed. To increase awareness about the characteristics of malignant brain tumors in patients with CMMRD, we analyzed the data on patients with CMMRD and brain tumors collected in the database of the C4CMMRD consortium. In this study, we describe the family history, clinical characteristics, treatments, and outcome of these patients.
Patients and Methods
Inclusion Criteria
The European C4CMMRD consortium includes pediatric oncologic centers in different European countries. Patients who may have CMMRD according to the diagnostic criteria established by the consortium5 are referred to the genetic clinics to perform a germline molecular analysis. All patients gave their informed consent before genetic testing, according to each country’s legislation. Their clinical data and family history are retrospectively and prospectively collected in the C4CMMRD database.
Patients are considered to have CMMRD in the presence of i) biallelic pathogenic germline mutations in any of the four MMR genes, ii) monoallelic pathogenic germline mutation and variant of unknown significance (VUS) on the other allele, or iii) biallelic VUS in any of the four MMR genes. Loss of expression of one MMR protein by immunohistochemistry analysis in non-neoplastic cells, abnormal functional tests,7 and/or microsatellite instability in non-neoplastic tissue12,13 was mandatory for the diagnosis of CMMRD in patients with mono or biallelic VUS. After confirmation of the predisposition syndrome, patients are registered in the European C4CMMRD consortium database by the clinicians.
Data Collected
The study was approved by an institutional review board and was conducted in accordance with the Declaration of Helsinki. Data on all patients with CMMRD and at least one brain tumor were extracted from the C4CMMRD consortium database. Clinical data, such as personal history of neoplasms, clinical features, detailed family history including a pedigree, tumor subtype, and type of treatment and response, were recorded. Histological diagnosis was performed according to the WHO guidelines available at diagnosis time.14,15 A histological review according to the 2016 WHO guidelines14 was performed using the available paraffin-embedded tumor sections, including hematoxylin-eosin-saffron (HES) staining for morphological description and immunohistochemical (IHC) analysis of PMS2, MLH1, MSH2, and MSH6 expression. A central review of the available presurgery brain MRI data was performed whenever possible.
Statistics
Baseline values (i.e., at diagnosis) were expressed as medians and interquartile range for continuous variables, and as numbers and percentages for categorical variables. Continuous variables were compared using the nonparametric Student’s t test, and categorical variables using the nonparametric chi-square test. Overall survival (OS) rates were calculated using the Kaplan–Meier method and compared with the log rank method. OS rates were estimated from the date of diagnosis of the first malignancy or of the first brain tumor to death whatever the cause, or the date of the last follow-up. The 95% confidence interval (CI) values for survival rates were estimated with the Rothman method.
Results
General Characteristics
In total, 87 patients were registered in the C4CMMRD consortium database at the end of July 2017. Among them, 49 patients (56%; 26 females and 23 males) from 10 countries had at least one brain tumor, including 31 patients already described in previous studies.7,13,16–20
Overall, 95 malignancies were diagnosed in these 49 patients: 56 central nervous system (CNS) malignant neoplasms, 21 Lynch syndrome-associated carcinomas, 15 hematological malignancies, and 3 sarcomas. One patient developed a meningioma within the radiation field more than 10 years after craniospinal irradiation. This tumor was most likely not related to the CMMRD syndrome. The median ages at onset of the first tumor and first brain tumor were 7 [1.1–22.6] and 9.2 [1.1–40.6] years, respectively. Nine patients developed their first brain tumor after the age of 18 years.
Twenty-seven patients (55%) had multiple malignancies (median: two tumors per patient), including two patients who developed five sequential cancers. Sixteen patients had 21 malignancies (10 gastrointestinal tumors, 9 hematologic malignancies, 2 sarcomas) before the first brain tumor, and 11 patients had another malignancy after the brain tumor. Seven patients (patients 5, 6, 10, 15, 30, 32, and 34) developed two distinct malignant brain tumors with a median interval of 1.5 years [range 0.4–17.9] between tumors. In this series, only one patient had his brain tumor identified by screening after a first hematological malignancy (patient 19). Table 1 summarizes the clinical data of this series.
Table 1.
Description of the series: patients’ medical history and genetic characteristics
| Previously described | N° | Sex | First tumor (age at onset) | Second tumor (age at onset) | Other tumors (age at onset) | Status (FU from 1st BT) | Gene | Mutation 1 | Mutation 2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | HGG (5y) | Dead (0.9y) | PMS2 | c.2007-2A>G, p.? | c.2007-2A>G, p.? | |||
| (14)1 | 2 | F | ADK (21y) | HGG (33y) | ADK (33y) | Dead (0.7y) | PMS2 | c2531C>A, p.Pro844His (VUS) | c2531C>A, p.Pro844His (VUS) |
| (16)1, C072, P13 | 3 | F | ADK (23y) | ADK (26y) | HGG (35y), ADK (36 and 37y) | Dead (4y) | PMS2 | c.137G>T, p.Ser46Ile | c.137G>T, p.Ser46Ile |
| 4. A | M | HGG (11y) | Dead (0.5y) | MSH6 | c.3725G>A, p.Arg1242His (VUS) | c.3725G>A, p.Arg1242His (VUS) | |||
| 4.B | F | HGG (13y) | Dead (0.5y) | MSH6 | c.3725G>A, p.Arg1242His (VUS) | c.3725G>A, p.Arg1242His (VUS) | |||
| 5 | M | HGG (6y) | HGG (7y) | Dead (2.8y) | PMS2 | c.2007-2A>G, p.? | c.2007-2A>G, p.? | ||
| 6 | M | MB (7y) | ADK (22y) | HGG (25y), meningioma | Dead (18.7y) | MSH6 | c.1196C>T, p.Pro399Leu (VUS) | c.2061T>G, p.Cys687Trp (VUS) | |
| (30)1 | 7.A | F | HGG (5y) | Dead (1.8y) | MSH6 | c.1596_1597dup, p.Glu533Valfs*39 | c.3261del, p.Phe1088Serfs*2 | ||
| C142 | 7.B | F | HGG (17y) | Dead (1.3y) | MSH6 | c.1596_1597dup, p.Glu533Valfs*39 | c.3261del, p.Phe1088Serfs*2 | ||
| (12)1, C192 | 8 | F | Osteosarcoma (11y) | HGG (13y) | Dead (0.4y) | PMS2 | c.161T>C, p. Ile54Thr (VUS) | c.1831dup, p.Ile611Asnfs*2 | |
| P93, ID134 | 9 | M | TALL (2y) | HGG (3y) | Alive (CR, 2.1y) | PMS2 | C.2007-2A>G, p.? | C.2007-2A>G, p.? | |
| ID184 | 10 | F | PNET (6y) | ADK (9y) | HGG (14y) | Dead (7.5y) | PMS2 | c.862C>T p.Gln288* | c.862C>T p.Gln288* |
| (31)1, C232 | 11 | M | TLL (7y) | TLL (11y) | HGG (14y), ADK (14y) | Dead (1.9y) | MSH6 | c.1763_1771dup, p.His588_Pro590dup (VUS) | c.1763_1771dup, p.His588_Pro590dup (VUS) |
| (8)1 | 12.A | F | HGG (12y) | TLL (14y) | ADK (14 and 21y) | Dead (9.7y) | PMS2 | c.24-12_107delinsAAAT, p.? | c.1-?_23+?del, p.? |
| (9)1 | 12.B | M | TLL (6y) | HGG (10y) | Dead (2.7y) | PMS2 | c.24-12_107delinsAAAT, p.? | c.1-?_23+?del, p.? | |
| ID144, Pub5 | 13 | F | MB (1y) | Alive (CR, 2.3y) | MSH6 | c.2426_2428del, p.Val809del (VUS) | c.2426_2428del, p.Val809del (VUS) | ||
| (18)1, C10.12 | 14 | M | HGG (6y) | Dead (1.1y) | PMS2 | c.2007-2A>G, p.? | c.2007-2A>G, p.? | ||
| (6)1 | 15 | M | Sarcoma (3y) | HGG (11y) | Burkitt lymphoma (18y), ADK (19y), HGG (20y) | Dead (9.9y) | PMS2 | c.903G>T, p.Lys301Asn | c.1145-?_2174+?dup, p.? |
| (24)1 | 16 | M | HGG (3y) | Dead (0.6y) | MSH2 | c.1-?_1076+?del, p.? | c.454del, p.Met152Cysfs*22 | ||
| (21)1 | 17.A | F | AML (5y) | MB (6y) | Dead (3.7y) | MLH1 | c.199G>T, p.Gly67Trp | c.199G>T, p.Gly67Trp | |
| 17.B | F | HGG (3y) | Dead (0.2y) | MLH1 | c.199G>T, p.Gly67Trp | c.199G>T, p.Gly67Trp | |||
| (5)1, C182 | 18 | M | ADK (22y) | ADK (32y) | HGG (41y) | Dead (1.5y) | PMS2 | c.(988 + 1_9891)_(1144 + 1_1145-1)del, p.(Glu330_Glu381del) | c.2249G>A, p.Gly750Asp (VUS) |
| (2)1, C09.12 | 19 | M | TLL (4y) | HGG (8y) | Alive (CR, 3y) | PMS2 | c.2007-2A>G, p.? | c.2007-2A>G, p.? | |
| 20 | F | HGG (6y) | Dead (0.2y) | MSH2 | c.508C>T, p.Gln170* | c.508C>T, p.Gln170* | |||
| Pub6 | 21 | F | HGG (18y) | Dead (2.2y) | PMS2 | c.686_687del, p.Ser229Cysfs*19 | c.686_687del, p.Ser229Cysfs*19 | ||
| (17)1, C29.12 | 22 | M | HGG (6y) | Dead (0.8y) | PMS2 | c.(2275 + 1_2276-1)_(*160_?)del p.? | c.(2275 + 1_2276-1)_(*160_?)del p.? | ||
| 23 | M | HGG (3y) | Alive (PD, 2.7y) | MSH6 | c.3386_3388delGTG, p.Cys1129_Val1130delinsLeu (VUS) | c.3386_3388delGTG, p.Cys1129_Val1130delinsLeu (VUS) | |||
| 24 | M | HGG (7y) | Dead (0.8y) | PMS2 | c.(705 + 1_706-1)_(803 + 1_804-1)del, p.(Leu236Hisfs*30) | c.(705 + 1_706-1)_(803 + 1_804-1)del, p.(Leu236Hisfs*30) | |||
| (1)1, C052 | 25 | F | HGG (22y) | ADK (25y) | Osteosarcoma (25y), AML (31y) | Dead (10y) | PMS2 | c.400C>T, p.Arg134* | c.1579del, p.Arg527Glyfs*68 |
| 26 | M | BLL (12y) | HGG (13y) | Dead (0.1y) | PMS2 | c.(2275 + 1_2276-1)_(*160_?)del p.? | c.803 + 2T>G, p.? | ||
| 27 | M | HGG (7y) | TLL (9y) | Dead (1.4y) | PMS2 | c.746_753del, p.Asp249Valfs*2 | c.1738A>T, p.Lys580* | ||
| (10)1, C01.22 | 28 | F | HGG (19y) | ADK (24y) | Dead (5.8y) | PMS2 | c.1730dup, p.Arg578Alafs*3 | c.137G>T, p.Ser46Ile | |
| (26)1, C20.22 | 29.A | F | HGG (6y) | Dead (0.3) | MSH6 | c2216C>A, p.Thr739Lys (VUS) | c2216C>A, p.Thr739Lys (VUS) | ||
| (27)1, C20.12 | 29.B | F | HGG (9y) | Dead (0.3y) | MSH6 | c2216C>A, p.Thr739Lys (VUS) | c2216C>A, p.Thr739Lys (VUS) | ||
| 30 | M | HGG (6y) | HGG (6y) | Dead (1.2y) | MSH6 | c.1800_1813dup, p.Thr605Ilefs*10 | c.1800_1813dup, p.Thr605Ilefs*10 | ||
| P53 | 31 | F | HGG (10y) | Alive (CR, 1.8y) | PMS2 | c.634C>T, p.Gln212* | c.1239del, p.Asp414Thrfs*34 | ||
| 32 | F | HGG (13y) | HGG (13y) | Dead (1.1y) | MSH6 | c.2731C>T,p.Arg911* | c.3013C>T, p.Arg1005* | ||
| (121)7 | 33.A | M | HGG (10y) | Dead (9y) | PMS2 | c.137G>T, p.Ser46Ile | c.804-2A>G; p.? | ||
| 33.B | F | ADK (21y) | HGG (24y) | Dead (2.9y) | PMS2 | c.137G>T, p.Ser46Ile | c.804-2A>G; p.? | ||
| (7)1 | 34 | M | HGG (7y) | TLL (7y) | HGG (8y) | Dead (1.4y) | PMS2 | c.2113G>A, p.Glu705Lys (VUS) | c.706-?_903+?del, p.? |
| 35 | M | ADK (12y) | TLL (16y) | HGG (18y) | Dead (2.4y) | PMS2 | c.862C>T, p.Gln288* | c.862C>T, p.Gln288* | |
| (19)1 | 36.A | F | MB (5y) | AML (7y) | Dead (3.7y) | MLH1 | c.678-7_686del, p.? | c.678-7_686del, p.? | |
| (20)1, C152 | 36.B | M | TLL (5y) | HGG (6y) | Dead (1.6y) | MLH1 | c.678-7_686del, p.? | c.678-7_686del, p.? | |
| 37.A | M | HGG (12y) | Dead (1.4y) | PMS2 | c.137G>T, p.Ser46Ile | c.1145?_2174?dup, p.? | |||
| 37.B | F | MB (5y) | Alive (CR, 15y) | PMS2 | c.137G>T, p.Ser46Ile | c.1145?_2174?dup, p.? | |||
| 38.A | F | HGG (6y) | Dead (0.1y) | MSH2 | c.1667T>C, p.Leu556Ser (VUS) | c.1667T>C, p.Leu556Ser (VUS) | |||
| ID204 | 38.B | F | HGG (18y) | Dead (0.6y) | MSH2 | c.1667T>C, p.Leu556Ser(VUS) | c.1667T>C, p.Leu556Ser (VUS) | ||
| ID234 | 38.C | F | ADK (9y) | HGG (20y) | Dead (0.3y) | MSH2 | c.1667T>C, p.Leu556Ser (VUS) | c.1667T>C, p.Leu556Ser (VUS) | |
| ID194 | 38.D | M | ADK (8y) | ADK (8y) | HGG (13y) | Dead (1y) | MSH2 | c.1667T>C, p.Leu556Ser (VUS) | c.1667T>C, p.Leu556Ser (VUS) |
1Lavoine N et al., J Med Genet 2015.
2Bodo S et al., Gastroenterology. 2015.
3Tesch VK et al., Front Immunol. 2018.
4Gallon R et al., Hum. Mutat. 2019.
5Taeubner J et al., Eur J Hum Genet. 2018.
6Baris HN et al., Ped. Blood Cancer, 2016.
7Giunti L et al., J Hum Genet. 2009.
BT: brain tumor, FU: follow-up, y: years, HGG: High-grade glioma, MB: Medulloblastoma, PNET: Primitive neuro-epithelial tumor, ADK: digestive Adenocarcinoma, TALL: T Acute lymphoblastic leukemia, TLL/BLL: T/B lymphoblastic lymphoma, AML: Acute myeloid leukemia, RC: complete remission, PD: progressive disease.
These patients belonged to 38 nuclear families. Consanguinity was reported in 20/38 (53%) families. In 16 families, siblings developed a CMMRD-associated malignancy, and in nine families more than one child had a brain tumor.
Histological Characteristics
Histological diagnosis of the 56 malignant CNS tumors according to the WHO 2016 guidelines could not be provided in all cases because only 26/56 cancers could be histologically reviewed. According to the histological report at diagnosis, all 56 CNS neoplasms were malignant: high grade gliomas (HGG) (n = 50; 89%) and embryonal tumors (n = 5 medulloblastomas and n = 1 supra-tentorial tumor, formerly named “primitive neuroectodermal tumor”). HGGs were further classified in glioblastoma (n = 40), anaplastic astrocytoma (n = 5), oligodendroglioma (=3), anaplastic pleomorphic xanthoastrocytoma (n = 1), and anaplastic ganglioglioma (n = 1). No low grade lesion was reported, but for one meningioma.
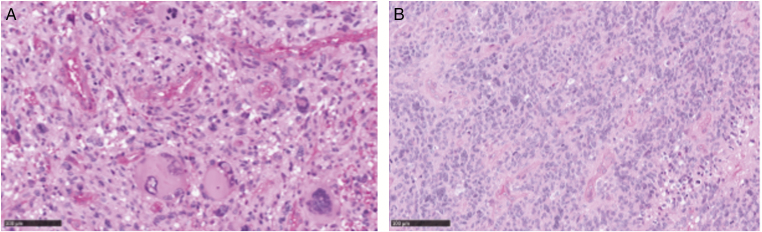
The histological review of the 26 available tumor samples (Table 2) identified them as glioblastoma with wild type IDH except for one (patient 4.B) (n = 21), anaplastic astrocytoma with wild type IDH (n = 3; among which two had unusual angiocentric features), anaplastic pleomorphic xanthoastrocytoma (n = 1), and anaplastic ganglioglioma (n = 1). Most of these gliomas displayed a particular pleomorphic appearance, and five were classified as giant cell glioblastomas (Figure 1A). Eleven glioblastomas included few giant multinucleated cells, but not enough to be considered as classic giant cell glioblastoma (Figure 1B), and only five did not have any giant cell. IHC results on MMR protein expression were available for 23 tumors and showed in all cases complete loss of expression of at least one MMR protein in the tumor and in normal cells that matched the genetic testing results (Table 2).
Table 2.
Histological review of brain tumors in 26 patients with CMMRD
| No. | Mutated gene | Histological review of high-grade glioma | Immunostaining for MMR proteins |
|---|---|---|---|
| 1 | PMS2 | glioblastoma IDH-wildtype (with few giant multinucleated cells) | PMS2- |
| 2 | PMS2 | giant cell glioblastoma | PMS2- |
| 4. A | MSH6 | glioblastoma IDH-wildtype (with few giant multinucleated cells) | MSH6- |
| 4.B | MSH6 | glioblastoma IDH-mutant (with few giant multinucleated cells) | MSH6- |
| 5 | PMS2 | glioblastoma (with few giant multinucleated cells) | PMS2- |
| 8 | PMS2 | giant cell glioblastomas | PMS2- |
| 11 | MSH6 | glioblastoma (with few giant multinucleated cells) | MSH2-/MSH6- |
| 12.A | PMS2 | glioblastoma (without giant cells) | PMS2- |
| 12.B | PMS2 | glioblastoma (without giant cells) | PMS2- |
| 14 | PMS2 | glioblastoma (without giant cells) | MLH1-/PMS2- |
| 15 | PMS2 | giant-cell glioblastoma | MLH1-/PMS2- |
| 16 | MSH2 | giant-cell glioblastoma | * |
| 18 | PMS2 | anaplastic pleomorphic xanthoastrocytoma | MSH2-/PMS2-/MLH1- |
| 19 | PMS2 | anaplastic astrocytoma grade III, IDH-wildtype (with angiocentric features) | PMS2- |
| 22 | PMS2 | giant-cell glioblastoma | PMS2- |
| 23 | MSH6 | glioblastoma (with few giant multinucleated cells) | MSH2-/MSH6- |
| 24 | PMS2 | glioblastoma (with few giant multinucleated cells) | PMS2- |
| 25 | PMS2 | anaplastic ganglioglioma | PMS2- |
| 26 | PMS2 | anaplastic astrocytoma grade III, IDH-wildtype | PMS2- |
| 29.A | MSH6 | glioblastoma (with few giant multinucleated cells) | MSH2-/MSH6- |
| 29.B | MSH6 | anaplastic astrocytoma grade III, IDH-wildtype (with angiocentric features) | MSH2-/MSH6- |
| 30 | MSH6 | glioblastoma (without giant cells) | NA |
| 31 | PMS2 | glioblastoma (with few giant multinucleated cells) | PMS2- |
| 34 | PMS2 | glioblastoma (with few giant multinucleated cells) | MLH1-/PMS2- |
| 36.B | MLH1 | glioblastoma (without giant cells) | MLH1-/PMS2- |
| 37.A | PMS2 | glioblastoma (with few giant multinucleated cells) | NA |
NA: Not available.
*Immunostaining results not interpretable due to technical reasons.
Figure 1.
Images of a giant cell glioblastoma (A) and a glioblastoma with few giant multinucleated cells (B). Tumor sections were stained with HES (scale 100μm).
Radiological Characteristics
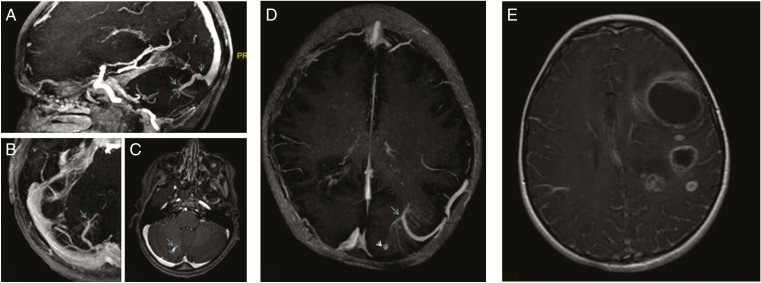
Presurgery brain MRI images were available for 15 patients. Brain developmental vascular anomalies were detected in 11 patients, but none was associated with cavernoma. Vascular abnormalities were visible on the enhanced T1 sequences (Figure 2a–d). They were located in the cerebellum (n = 4 patients) and in the supratentorial region (n = 7 patients), but not adjacent to the brain tumor. The radiological review indicated that five patients presented multiple brain tumors with asynchronous development at diagnosis (Figure 2e). Tumors were located in the parietal (n = 5), frontal (n = 5), temporal (n =4), and occipital (n = 2) brain lobes. No focal area of signal intensity was observed in this series.
Figure 2.
Brain developmental vascular anomalies and multiple tumor lesions in patients with CMMRD. Brain MRI images from three patients 1) MIP images (a and b) and axial T1 slices with gadolinium (c) showing the arteriovenous abnormality in the right cerebellar lobe, connected with the initial part of the right sinus; 2) MIP images (d) showing the left temporal arteriovenous malformation, with an aspect of “jellyfish head,” associated with an arterial aneurysms; and 3) Axial T1 slice with gadolinium (e) showing the presence of multiple tumor lesions.
Other Characteristics of Patients With CMMRD Syndrome
Except for one, all patients with CMMRD and available clinical data (40/41) had café-au-lait macules (CALM) (2 to 10 macules). One patient had a giant hyperpigmented lesion, and 9/21 patients also presented hypopigmented skin macules. A plexiform neurofibroma was reported in 4 of the 18 patients with data on the presence of neurofibroma. Nineteen patients underwent colonoscopic examination at a median age of 16 years [range 9–24] before or after the brain tumor diagnosis, and all had at least one adenoma. Moreover, hepatic adenoma was detected in one patient.
Germline Mutation Screening
Genetic screening for MMR gene variants in all patients with tumors (Table 1) showed the presence of biallelic pathogenic germline mutations (n = 33 patients), monoallelic pathogenic germline mutation and VUS on the other allele (n = 3 patients), and biallelic VUS (n = 13 patients). The diagnosis of CMMRD was confirmed in all patients with VUS by functional tests results and/or loss of expression of the MMR protein in non neoplastic cells (Supplementary Material). The affected MMR genes were: PMS2 (n = 27 patients), MSH6 (n = 12 patients), MSH2 (n = 6 patients), and MLH1 (n = 4 patients). The mutation in one of the MMR genes was homozygous in 29 children with CMMRD (22 families: 20 families with consanguinity and 2 without reported consanguinity) and compound heterozygous in 20 patients (16 families).
Cancer Management and Outcome
Data on tumor treatment were available for 45 patients and 50 tumors. Most patients received standard treatment that combined surgery, whenever possible (47/50), with radiotherapy (39/50) and various chemotherapy protocols (34/49). No unusual treatment-related toxicity was reported.
Moreover, eight patients received immunotherapy with anti-PD1 antibodies for HGG at relapse. Disease progression was observed in seven of these patients within the first 2 months of immunotherapy (1 to 4 injections), and six of them died at 5.2 months [1.8–9.5] after the first injection. Two patients were still alive, one with progressive disease after immunotherapy discontinuation (follow-up: 8.6 months), and the other one was still on treatment.
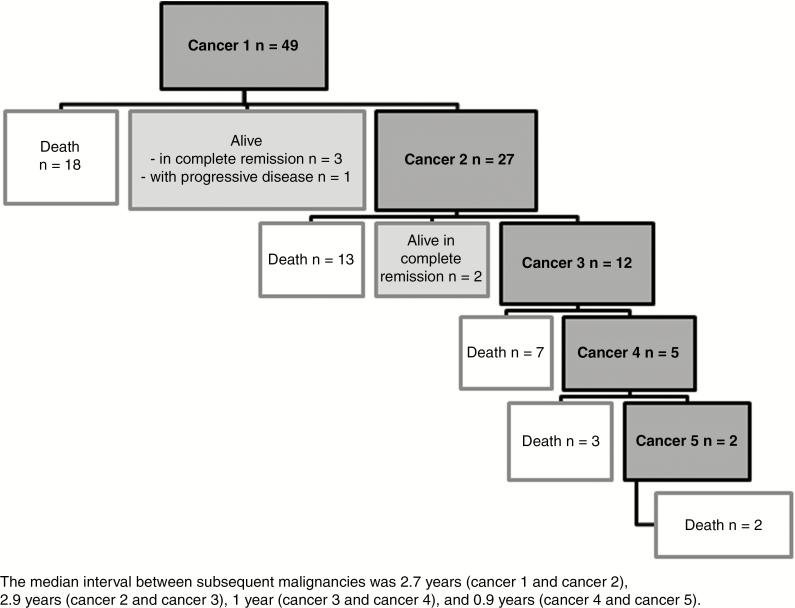
The outcome of the entire series is summarized in Figure 3. The median follow-up from the date of diagnosis of the first brain tumor was 1.5 years [0–18.7]. Only six patients (including patient 19, who had his brain tumor identified by screening) were still alive with a median follow-up of 2.5 years [1.8–15] after the diagnosis of the first brain tumor among whom five were in complete remission (2.3 and 15 years after the diagnosis of medulloblastoma for two patients; 1.8, 2.1, and 3 years after the diagnosis of glioblastoma/anaplastic astrocytoma for the other three). The last patient who developed glioblastoma was still alive but with progressive disease. Overall, 43 (88%) patients died at a median age of 13.2 years [3.5–42]. Death was related to the first brain tumor in 31 patients (median survival of 11 months after diagnosis [2–108]), to a second brain tumor in seven patients, and to another cancer in five (hematological malignancy in three, and digestive adenocarcinoma in two patients).
Figure 3.
Outcome of patients with constitutional mismatch repair deficiency (CMMRD) and brain tumor from the C4CMMRD database (n = 49 patients). The median interval between subsequent malignancies was 2.7 years (cancer 1 and cancer 2), 2.9 years (cancer 2 and cancer 3), 1 year (cancer 3 and cancer 4), and 0.9 years (cancer 4 and cancer 5).
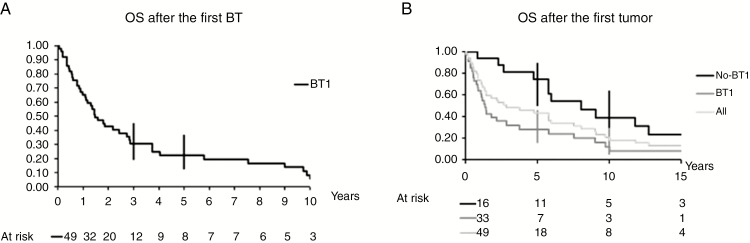
The patients’ outcome was evaluated by assessing the survival of each patient after the first brain tumor (Figure 4A), and after the first tumor (any type) (Figure 4B). Overall, the 3- and 5-year OS rates after the first brain tumor (n = 49) were 30% (95% CI: 19–45) and 22% (95% CI: 12–37) (Figure 4A). The 3- and 5-year OS rates were much lower in patients with HGG [20.5% (95% CI: 11–36) and 17% (95% CI: 8–32), respectively] than in patients with embryonal tumors [5-year OS rate: 60% (95% CI: 38–96)] (log rank: 0.0049). Among the six patients with embryonal tumor, only one child died due to her medulloblastoma at 3.7 years after diagnosis. Three patients died of a subsequent glioblastoma or of hematological malignancy.
Figure 4.
Overall survival (OS) after the first brain tumor (BT) (A), and after the first tumor (any tumor: n = 49 patients; BT as first tumor: n =33; not BT as first tumor: n = 16 patients) (B).
The 5-year OS rate after the first tumor (any type) (n = 49) was 43% (95% CI 30–58). Comparison of the 5-year OS rate according to the type of first tumor (brain tumor versus any other tumor type) showed that it was significantly lower if the first tumor was a brain tumor (n = 33) [5-year OS = 28% (95% CI: 15–46)] compared with any other tumor type (n = 16) [5-year OS = 74.5% (95% CI: 50–90)] (log rank: 0.0092) (Figure 4B).
Discussion
This detailed description of a large series of patients with CMMRD and at least one brain tumor using data collected by an international consortium shows that patients had a wide variety of cancers (brain, gastrointestinal and hematologic malignancies), as previously reported for other CMMRD series.5,21 As expected, malignant brain tumors were mostly HGGs, but we also observed embryonal tumors, including five medulloblastomas. Although medulloblastoma is not a frequent tumor in CMMRD, it represented 17% of all malignant brain tumors in our series. Therefore, it is very striking that a recent study on a large medulloblastoma sample (n = 1022) did not find any patient with biallelic germline mutations in MMR genes.22 As already reported,5,7,16 other rare malignant glial tumors, such as anaplastic astrocytoma, anaplastic pleomorphic xanthoastrocytoma, and anaplastic ganglioglioma, were also observed. The outcome of CMMRD-associated brain tumors was poor (5-year OS = 22%), especially in patients with HGG. The 3-year OS of 20.5% for patients with glioblastoma is worse than the previously reported 3-year OS rate for pediatric glioblastoma without cancer predisposition syndrome (30%).23
Since the first reports on patients with CMMRD, international collaborative research projects have considerably increased the knowledge on tumor characteristics and evolution.8 However, despite the delineation of clinical criteria that should raise the suspicion of CMMRD,5 which should entail molecular genetic diagnosis, CMMRD is still underdiagnosed in pediatric patients with a malignant brain tumor. From this series, we can highlight the following features that could guide the clinician towards the possible presence of CMMRD, thus justifying additional investigations on MMR proteins/genes by IHC and/or appropriate microsatellite instability analysis,12,13 functional assays,7,24 and MMR gene mutation analysis:
CALM: Except one, all patients had CALM that led to the initial misdiagnosis of neurofibromatosis type 1 (NF1) in some cases. In addition, plexiform neurofibroma was reported in four patients. This phenotypic overlap between NF1 and CMMRD, mostly based on skin pigmentation alterations, has been described previously.5,25–27 As malignant brain tumors are rare in NF1, CMMRD is a valid differential diagnosis in children with malignant brain tumor and a phenotype reminiscent of NF1. The correct diagnosis of the underlying disease must be based on the identification of a clearly pathogenic germline NF1 or MMR gene mutation in these children.28
HGG with giant cells: Histological analysis highlighted that many gliomas were characterized by a malignant glial heterogeneous population with giant multinucleated and pleomorphic cells. When pleomorphic cells were abundant, tumors were identified as giant cell glioblastomas, in accordance with the WHO guidelines. This specific feature, already described in malignant HGG with germline MMR gene mutations,10,29 justifies the diagnostic work-up for CMMRD with at least IHC analysis of MMR proteins in the tumor and adjacent normal tissues, especially when found in “ultramutated” glioblastomas. However, we must stress that normal expression of all MMR proteins does not exclude the diagnosis of CMMRD in highly suggestive cases, as reported for some patients with CMMRD and missense MSH6 mutations.18
Radiological findings of metachronous multiple brain lesions and/or brain vascular malformations: Patients with CMMRD may have multiple different tumor lesions (5/15 patients in our series). This feature has already been described at diagnosis in few case reports.30,31 Patients with CMMRD may also have non-neoplastic congenital brain malformations, usually asymptomatic.5,32 In contrast to the low reported incidence of brain developmental vascular anomalies in the general population (2.6%–6.4%), our radiological data on 15 patients with CMMRD and brain tumor highlighted a high rate (73%) of brain vascular anomalies, not adjacent to the brain tumor and not related to the tumor treatment. We suggest that this feature should be added to the current scoring system with the other brain malformations (corpus callosum agenesis and nontherapy-induced cavernoma),5 as indication criteria for CMMRD genetic testing in patients with cancer.
Family history of cancer, affected siblings, and/or consanguinity.
Although our knowledge on CMMRD-related tumor development and sensitivity to treatments has improved,7 specific therapeutic strategies are limited33 and often based on case report studies. In this series, the different therapeutic strategies often combined surgery, radiation therapy, and chemotherapy, in function of the time of diagnosis and tumor extension. Unlike other DNA damage repair syndromes,34 no excessive toxicity after chemoradiation therapies was observed, as already reported. The real impact of temozolomide, frequently used in the standard glioblastoma treatment, cannot be specifically studied. Nevertheless, it has become clear that, due to the strong resistance of MMR-deficient cells to alkylating anti-neoplastic agents,7,35–37 temozolomide is less effective in MMR-deficient tumors, and might even provide a growth advantage to tumor cells.37–40 Therefore, this drug should be avoided in patients with CMMRD. The hypermutated phenotype related to CMMRD8 could offer opportunities for new therapeutic approaches as MMR-deficient tumors are more responsive to PD-1 inhibition than MMR-proficient tumors.41–43 There is growing evidence that immunotherapies, such as immune checkpoint inhibitors, prolong survival in patients with CMMRD with recurrent or refractory glioblastoma9,44 and in patients with MMR-deficient noncolorectal cancer (objective response rate of 71%).42 Information on our small group of eight patients who received immunotherapy at HGG relapse was very limited because most of them were included in a clinical trial, limiting access to data. However, in our patients, immunotherapy efficacy was slightly disappointing compared with the initial publications.9,44 This finding may be related to the advanced disease at the moment of such treatment.
To conclude, we still recommend surgical resection of the malignant brain tumor followed by radiotherapy and chemotherapy not based on temozolomide. However, we think that upfront immunotherapy after the initial resection at the time of radiotherapy might represent a better option for these patients and should be evaluated.
The early identification of CMMRD and tumor prevention are probably the most important elements in the context of a tumor predisposition syndrome with high penetrance. Genetic counseling should give clear information on the risk in siblings and on the Lynch syndrome-associated cancer risk for the wider family. As patients with CMMRD are at high risk of developing multiple cancers, they should undergo regular cancer surveillance according to the guidelines recently published in consensus papers.6,10,45 Indeed, very early detection of asymptomatic tumors can facilitate their complete resection. Nevertheless, cancer surveillance does not guarantee cancer detection at a curable stage, and the development of preventive treatment strategies would be a major step forward. Aspirin and other nonsteroidal anti-inflammatory drugs are considered to be cancer-preventive mainly due to their antiproliferation and apoptosis-inducing activities and have been shown to reduce the colorectal cancer risk in patients with Lynch syndrome.46–48 Although the use of aspirin has been suggested for cancer prevention in patients with CMMRD,49 its efficacy has not been proven and the potential benefit has to be balanced with the hemorrhagic risk, especially in patients with frequent brain developmental vascular anomalies. Immunotherapy with checkpoint inhibitors also could be an attractive preventive strategy in healthy carriers of biallelic MMR gene mutations. It has been hypothesized that combining checkpoint inhibitors with neoantigen-based vaccines could increase the potential of immunotherapies.50 Although the safety and efficacy of such vaccines remain to been proven,50 these approaches are very promising.
Conclusion
CMMRD diagnosis is still challenging in patients with a pediatric malignant brain tumor, also due to the phenotypic overlap between NF1 and CMMRD. Nevertheless, specific features and a family history of cancer often in the context of consanguinity and/or affected siblings may guide the clinician. The prognosis of patients with a CMMRD-related brain tumor (especially glioblastoma) is not as good as originally thought. Therefore, treatment and prevention need to be improved, including immunotherapies and new upfront therapeutic approaches.
Supplementary Material
Funding
Société Française de lutte contre les cancers et leucémies de l’enfant et de l’adolescent (SFCE) and Fédération enfants et santé funded the histological review of brain tumors associated with CMMRD. This study was presented at an oral session of the 18th International Symposium on Pediatric Neuro-Oncology (ISPNO), Denver July 2018 and of the 50th Annual Congress of the International Society of Pediatric Oncology (SIOP), Kyoto November 2018.
Authors’ Contributions
Léa Guerrini-Rousseau, Laurence Brugières, Jacques Grill, Franck Bourdeaut, Karin Dahan, Christine Devalck, Cécile Faure-Conter, Maurizio Genuardi, Yael Goldberg, Michaela Kuhlen, Enrico Opocher, Vanessa Perez-Alonso, Astrid Sehested, Irene Slavc and Sheila Unger provided clinical data. Katharina Wimmer, Chrystelle Colas, and Yael Goldberg contributed to collect and analyze the genetic data. Pascale Varlet and Felipe Andreiuolo provided histological review. Salma Moalla provided radiological review. Laurence Brugières, Jacques Grill, and Léa Guerrini-Rousseau contributed to the study design. Chrystelle Colas and Laurence Brugières were in charge of the C4CMMRD database. All authors contributed to the article writing, reviewing, and editing.
Conflict of interest statement. None declared.
References
- 1. Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. [DOI] [PubMed] [Google Scholar]
- 2. Lynch HT, Lynch PM, Lanspa SJ, et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Q, Lasset C, Desseigne F, et al. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59(2): 294–297. [PubMed] [Google Scholar]
- 4. Ricciardone MD, Ozçelik T, Cevher B, et al. Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res. 1999;59(2):290–293. [PubMed] [Google Scholar]
- 5. Wimmer K, Kratz CP, Vasen HFA, et al. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium “care for CMMRD” (C4CMMRD). J Med Genet. 2014;51(6):355–365. [DOI] [PubMed] [Google Scholar]
- 6. Vasen HFA, Ghorbanoghli Z, Bourdeaut F, et al. Guidelines for surveillance of individuals with constitutional mismatch repair-deficiency proposed by the European Consortium “Care for CMMR-D” (C4CMMR-D). J Med Genet. 2014;51(5):283–293. [DOI] [PubMed] [Google Scholar]
- 7. Bodo S, Colas C, Buhard O, et al. Diagnosis of constitutional mismatch repair-deficiency syndrome based on microsatellite instability and lymphocyte tolerance to methylating agents. Gastroenterology. 2015;149(4):1017–1029. [DOI] [PubMed] [Google Scholar]
- 8. Shlien A, Campbell BB, de Borja R, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. [DOI] [PubMed] [Google Scholar]
- 9. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 10. Tabori U, Hansford JR, Achatz MI, et al. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin Cancer Res. 2017;23(11):e32–e37. [DOI] [PubMed] [Google Scholar]
- 11. Durno C, Boland CR, Cohen S, et al. Recommendations on surveillance and management of biallelic mismatch repair deficiency (BMMRD) syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Gastroenterology. 2017;152(6):1605–1614. [DOI] [PubMed] [Google Scholar]
- 12. Ingham D, Diggle CP, Berry I, et al. Simple detection of germline microsatellite instability for diagnosis of constitutional mismatch repair cancer syndrome. Hum Mutat. 2013;34(6):847–852. [DOI] [PubMed] [Google Scholar]
- 13. Gallon R, Mühlegger B, Wenzel S-S, et al. A sensitive and scalable microsatellite instability assay to diagnose constitutional mismatch repair deficiency by sequencing of peripheral blood leukocytes. Hum Mutat. 2019;40(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of rumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 15. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lavoine N, Colas C, Muleris M, et al. Constitutional mismatch repair deficiency syndrome: clinical description in a French cohort. J Med Genet. 2015;52(11):770–778. [DOI] [PubMed] [Google Scholar]
- 17. Tesch VK, IJspeert H, Raicht A, et al. No overt clinical immunodeficiency despite immune biological abnormalities in patients with constitutional mismatch repair deficiency. Front Immunol. 2018;9:1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taeubner J, Wimmer K, Muleris M, et al. Diagnostic challenges in a child with early onset desmoplastic medulloblastoma and homozygous variants in MSH2 and MSH6. Eur J Hum Genet. 2018;26(3):440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baris HN, Barnes-Kedar I, Toledano H, et al. Constitutional mismatch repair deficiency in Israel: high proportion of founder mutations in MMR genes and consanguinity. Pediatr Blood Cancer. 2016;63(3):418–427. [DOI] [PubMed] [Google Scholar]
- 20. Giunti L, Cetica V, Ricci U, et al. Type A microsatellite instability in pediatric gliomas as an indicator of Turcot syndrome. Eur J Hum Genet. 2009;17(7):919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sijmons RH, Hofstra RMW. Review: Clinical aspects of hereditary DNA Mismatch repair gene mutations. DNA Repair (Amst). 2016;38:155–162. [DOI] [PubMed] [Google Scholar]
- 22. Waszak SM, Northcott PA, Buchhalter I, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018;19(6):785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grill J, Massimino M, Bouffet E, et al. Phase II, open-label, randomized, multicenter trial (HERBY) of bevacizumab in pediatric patients with newly diagnosed high-grade glioma. J Clin Oncol. 2018;36(10):951–958. [DOI] [PubMed] [Google Scholar]
- 24. Shuen AY, Lanni S, Panigrahi GB, et al. Functional repair assay for the diagnosis of constitutional mismatch repair deficiency from non-neoplastic tissue. J Clin Oncol. 2019:JCO1800474. [DOI] [PubMed] [Google Scholar]
- 25. Daou B, Zanello M, Varlet P, et al. An unusual case of constitutional mismatch repair deficiency syndrome with anaplastic ganglioglioma, colonic adenocarcinoma, osteosarcoma, acute myeloid leukemia, and signs of neurofibromatosis type 1: case report. Neurosurgery. 2015;77(1):E145–152; discussion E152. [DOI] [PubMed] [Google Scholar]
- 26. Wimmer K, Rosenbaum T, Messiaen L. Connections between constitutional mismatch repair deficiency syndrome and neurofibromatosis type 1. Clin Genet. 2017;91(4):507–519. [DOI] [PubMed] [Google Scholar]
- 27. Suerink M, Ripperger T, Messiaen L, et al. Constitutional mismatch repair deficiency as a differential diagnosis of neurofibromatosis type 1: consensus guidelines for testing a child without malignancy. J Med Genet. 2019;56(2):53–62. [DOI] [PubMed] [Google Scholar]
- 28. Guerrini-Rousseau L, Suerink M, Grill J, et al. Patients with High-Grade Gliomas and Café-au-Lait Macules: Is Neurofibromatosis Type 1 the Only Diagnosis? AJNR Am J Neuroradiol. 2019;40(6):E30–E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galuppini F, Opocher E, Tabori U, et al. Concomitant IDH wild-type glioblastoma and IDH1-mutant anaplastic astrocytoma in a patient with constitutional mismatch repair deficiency syndrome. Neuropathol Appl Neurobiol. 2018;44(2):233–239. [DOI] [PubMed] [Google Scholar]
- 30. Ilencikova D, Sejnova D, Jindrova J, et al. High-grade brain tumors in siblings with biallelic MSH6 mutations. Pediatr Blood Cancer. 2011;57(6):1067–1070. [DOI] [PubMed] [Google Scholar]
- 31. Amayiri N, Al-Hussaini M, Swaidan M, et al. Synchronous glioblastoma and medulloblastoma in a child with mismatch repair mutation. Childs Nerv Syst. 2016;32(3):553–557. [DOI] [PubMed] [Google Scholar]
- 32. Shiran SI, Ben-Sira L, Elhasid R, et al. Multiple brain developmental venous anomalies as a marker for constitutional mismatch repair deficiency syndrome. AJNR Am J Neuroradiol. 2018;39(10):1943–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abedalthagafi M. Constitutional mismatch repair-deficiency: current problems and emerging therapeutic strategies. Oncotarget. 2018;9(83):35458–35469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuler N, Palm J, Kaiser M, et al. DNA-damage foci to detect and characterize DNA repair alterations in children treated for pediatric malignancies. PLoS ONE. 2014;9(3):e91319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hawn MT, Umar A, Carethers JM, et al. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55(17):3721–3725. [PubMed] [Google Scholar]
- 36. Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22(12):1931–1937. [DOI] [PubMed] [Google Scholar]
- 37. Nagel ZD, Kitange GJ, Gupta SK, et al. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017;77(1):198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fedier A, Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity (review). Int J Oncol. 2004;24(4):1039–1047. [PubMed] [Google Scholar]
- 39. Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7): 2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McFaline-Figueroa JL, Braun CJ, Stanciu M, et al. Minor changes in expression of the mismatch repair protein MSH2 Exert a major impact on glioblastoma response to temozolomide. Cancer Res. 2015;75(15):3127–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelderman S, Schumacher TN, Kvistborg P. Mismatch repair-deficient cancers are targets for anti-PD-1 therapy. Cancer Cell. 2015;28(1):11–13. [DOI] [PubMed] [Google Scholar]
- 43. Nebot-Bral L, Brandao D, Verlingue L, et al. Hypermutated tumours in the era of immunotherapy: the paradigm of personalised medicine. Eur J Cancer. 2017;84:290–303. [DOI] [PubMed] [Google Scholar]
- 44. AlHarbi M, Ali Mobark N, AlMubarak L, et al. Durable response to nivolumab in a pediatric patient with refractory glioblastoma and constitutional biallelic mismatch repair deficiency. Oncologist. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Durno CA, Aronson M, Tabori U, et al. Oncologic surveillance for subjects with biallelic mismatch repair gene mutations: 10 year follow-up of a kindred. Pediatr Blood Cancer. 2012;59(4):652–656. [DOI] [PubMed] [Google Scholar]
- 46. Burn J, Gerdes A-M, Macrae F, et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet. 2011;378(9809):2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chubak J, Kamineni A, Buist DS, et al. Aspirin Use for the Prevention of Colorectal Cancer: An Updated Systematic Evidence Review for the U.S. Preventive Services Task Force. Rockville (MD): Agency for Healthcare Research and Quality (US); 2015. http://www.ncbi.nlm.nih.gov/books/NBK321661/. Accessed August 27, 2018. [PubMed] [Google Scholar]
- 48. Ait Ouakrim D, Dashti SG, Chau R, et al. Aspirin, ibuprofen, and the risk of colorectal cancer in lynch syndrome. J Natl Cancer Inst. 2015;107(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leenders EKSM, Westdorp H, Brüggemann RJ, et al. Cancer prevention by aspirin in children with Constitutional Mismatch Repair Deficiency (CMMRD). Eur J Hum Genet. 2018;26(10):1417–1423.: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Westdorp H, Kolders S, Hoogerbrugge N, et al. Immunotherapy holds the key to cancer treatment and prevention in constitutional mismatch repair deficiency (CMMRD) syndrome. Cancer Lett. 2017;403:159–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.