Abstract
Escherichia coli was one of the first species to have its genome sequenced and remains one of the best-characterized model organisms. Thus, it is perhaps surprising that recent studies have shown that a substantial number of genes have been overlooked. Genes encoding more than 140 small proteins, defined as those containing 50 or fewer amino acids, have been identified in E. coli in the past 10 years, and there is substantial evidence indicating that many more remain to be discovered. This review covers the methods that have been successful in identifying small proteins and the short open reading frames that encode them. The small proteins that have been functionally characterized to date in this model organism are also discussed. It is hoped that the review, along with the associated databases of known as well as predicted but undetected small proteins, will aid in and provide a roadmap for the continued identification and characterization of these proteins in E. coli as well as other bacteria.
INTRODUCTION
Since the publication of its full genomic sequence in 1997, Escherichia coli has been widely regarded as one of the best-annotated genomes (1). Multiple organizations, projects, and individual investigators have been, and continue to be, involved in updating its annotation, including the National Center for Biotechnology Information (NCBI), UniProtKB/Swiss-Prot, and EcoCyc, to name a few (2–4). Due to these efforts, E. coli is still regarded as a gold standard for annotation. Nevertheless, some important questions regarding the E. coli genome remain unanswered, including the total number of genes. One difficulty in answering this question is the problem of short genes, including those encoding the smallest proteins (5). There are hundreds of thousands of potential small open reading frames (sORFs) that could encode proteins of fewer than 50 amino acids (aa) (1, 6). Even if only a small fraction of these sORFs encode authentic proteins, inadequate annotation of these genes means that a significant fraction of the gene products of one of the best-studied model organisms has been overlooked.
As more research is conducted into identifying and testing for small protein synthesis, it is becoming clear that there are more small proteins synthesized than originally expected and that a number of them are encoded by sORFs lacking commonly expected characteristics, such as canonical ribosome binding sites and start codons (6–9). This review endeavors to summarize current work regarding the prediction, identification, and confirmation of small protein synthesis in E. coli and providing the first glimpses into the functions of these small proteins. It is hoped that this summary will prompt increased study of this family of proteins.
DEFINITION OF A SMALL PROTEIN
It is difficult to find a consensus in the literature regarding the definition of a small protein. Rather than having a functional definition, the group is delineated by an arbitrary size range, including 15 to 50 aa (10), 33 to 100 aa (11), and less than 25 kDa (12). However, any definition regarding sizes leads to the reasonable question about proteins immediately outside that range. If, for example, small proteins are defined as those which are 50 or fewer amino acids, what about a protein that is 51 amino acids? What is the minimum size for a small protein? The majority of the E. coli proteins identified in the last 5 to 10 years have been those containing 50 or fewer amino acids (Fig. 1), suggesting that this is the range where the most progress needs to be made. Many consider any short chain of amino acids to be a peptide, although the term “peptide” itself is derived from the Greek “digested.” Despite the arbitrary nature of the use of size as a criterion, we suggest one overarching distinction; a protein is encoded by a single ORF, regardless of size, and is not processed from a larger protein.
Figure 1.
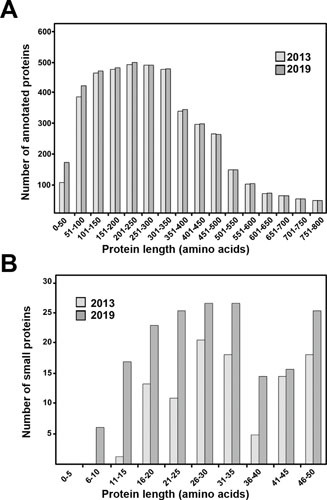
Small protein gene identification in the E. coli genome over the past 6 years. (A) Histogram of currently annotated protein-coding genes in E. coli compared to those identified in 2013. (B) Histogram of currently annotated small protein genes in E. coli compared to those known in 2013. For both (A) and (B), the light gray bars represent small protein genes annotated in 2013, and the dark gray bars represent genes annotated in 2019. Data on annotated genes in 2013 are from E. coli K12 MG1655 genome annotation U00096.2. Data on annotated genes in 2019 were compiled from a combination of annotations from EcoCyc (92) and recent papers identifying new small proteins.
A survey of the literature shows diverse nomenclature, including “small protein,” “short protein,” “miniprotein,” “microprotein,” “μ protein,” and “micropeptide,” while the corresponding genes have been termed “small gene,” “sORF,” “smORF,” and “μORF” (6, 10, 13–16). For the purpose of this review, we will consider “small proteins” to be those containing 50 or fewer amino acids and not derived by processing, and we will denote the corresponding coding sequences “sORFs.”
IDENTIFICATION OF SMALL PROTEIN-ENCODING GENES
The reliable identification of small protein-encoding genes among the thousands of sORFs encoded by the E. coli genome remains a challenge. In addition to the sheer number of possible sORFs, this is due to many factors associated with the limited sequence information present in the sORFs (17). These sequences often lack characteristics commonly used to identify genes, such as evidence of codon adaptation at the nucleotide level and identifiable protein domains at the amino acid level. Additionally, small proteins appear to be more poorly conserved than large proteins, limiting the ability to rely on conservation to identify small proteins (18). The availability of genome-wide transcriptome (RNA-seq) and ribosome binding (ribo-seq or ribosome profiling) datasets is providing valuable information to augment bioinformatic predictions of small protein genes. For example, the detection of a transcript and ribosome binding in a genomic region increases confidence that the sequence might be translated. Ultimately, the application of a combination of parameters usually provides the most accurate prediction of true small protein-encoding genes.
Bioinformatics
When the E. coli genome was originally annotated, the size cutoff for short gene annotation was 153 nucleotides, or 50 amino acids, for the predicted proteins (1). Since that time, traditional gene annotation programs, such as GeneMark, EasyGene, FrameD, and GLIMMER, have repeatedly been found to be less reliable at identifying genes encoding authentic small proteins compared to genes encoding larger proteins (10, 11, 19–21). In an effort to improve predictive capability, several advanced gene screening algorithms have been developed to more accurately identify small protein genes in the genomes of E. coli and other bacteria. As a group, they share a focus on evaluating sequences for characteristics suggestive of translation, transcription, and/or conservation between related bacterial species. Many of these programs take advantage of the large number of sequenced bacterial genomes to perform comparative genomics with a goal of not only identifying new small protein-encoding genes, but also of correcting annotation among the sequences (19, 21, 22). The following section describes bioinformatic methods or programs used to identify putative small protein-encoding genes in E. coli. Table S1 summarizes useful websites for the identification and characterization of small proteins.
Hemm et al. screened intergenic regions of >40 base pairs for conservation using tBLASTn and for sORFs encoded downstream of potential ribosome binding sites (23). sORFs encoding small proteins of 16 to 50 amino acids were considered in this study. Select sORFs were then tested for small protein synthesis, with both strong conservation and the strength of the ribosome binding site found to correlate with the identification of authentic short genes.
Warren et al. performed a broad-range screen for unannotated genes encoding proteins of 33 amino acids or larger in 1,297 bacterial chromosomes and plasmids (11). These sequences were analyzed using comparative genomics, BLASTp, and the gene prediction programs GLIMMER and GeneMark. They identified 1,153 ORFs that they suggested are candidate genes, with the majority encoding proteins of 100 or fewer amino acids.
Goli et al. described an “ensemble method” for short gene identification based on a combination of prominent sequence features, including codon usage bias, GC content at different codon positions, physicochemical and conformational properties of DNA, and amino acid properties (20). They reported that their method was substantially more accurate at identifying known small proteins in E. coli than the gene predictor program FrameD. They found trimer frequency of nucleotides, codon adaptation index, GC content in the first and second position, and nucleotide stacking energy to be the best predictors of short genes.
Óhéigeartaigh et al. reported the development of the program SearchDOGS for identifying missed genes in bacterial genomes (22). The program is based on a comparative genome method that examines nucleotide sequence synteny between related species. Once syntenic relationships between loci have been established, sequences are analyzed for ORFs that may be missed in some genomes. An analysis of nine gammaproteobacterial clades yielded 56 candidate genes encoding proteins of less than 60 amino acids, with 36 of them encoded in E. coli K-12.
Wood et al. published an analysis of 1,474 bacterial genome annotations using comparative genomics (21). Potentially missed genes containing 110 or more nucleotides were identified using GLIMMER and then analyzed using a gene function database called COMBREX. Potential genes were compared to entries in COMBREX using the ComBlast annotation pipeline. Of the 13,602 candidate genes identified in this study, 60% encode proteins containing fewer than 100 amino acids.
Transcription Profiling
Evidence that a genomic region encoding a predicted sORF is transcribed provides support that the corresponding small protein might be translated. Accordingly, publicly available transcription profiling data have been found to correlate with small protein expression (24). To our knowledge, no work has exclusively used transcriptional profiling to specifically identify new small proteins, but this information is valuable, particularly for determining conditions when the proteins might be synthesized.
Ribosome Profiling
Ribosome profiling or Ribo-seq, a deep sequencing approach in which the RNA regions associated with ribosomes are sequenced, has allowed small protein-encoding transcripts to be identified. Due to technical challenges, the resolution of ribosome profiling for bacterial cells is lower than that for eukaryotic cells. This makes identifying short genes more challenging, especially when the sORF analyzed is located close to or overlapping another gene. Nevertheless, the presence of ribosomes on known transcripts is a valuable indicator that translation may be occurring. As more ribosome profiling datasets are being published, these data can be used in support of bioinformatics approaches.
The capabilities of ribosome profiling have been expanded through the use of inhibitors that capture specific states of ribosomal complexes. Tetracycline, long known to inhibit translation in bacteria, enriches samples for ribosomes bound to start codons (86). Two other inhibitors, Onc112 and retapamulin, trap E. coli ribosomes in translation initiation complexes even more efficiently to give even greater enrichment at start codons (6, 25). Comparing the data from experiments with different inhibitors can reveal sites with the highest probability of being true start codons, yielding a more robust method for identifying small protein genes. The ease and low cost of sequencing, combined with the advancements in profiling techniques, will allow for the continued identification of these genes in any bacterium where the inhibitors are effective.
It is worth being cognizant of some limitations of the ribosome profiling approach, including a lack of correlation between ribosome density and amount of protein detected (6). Ultimately, no single identification method developed to date is sufficient to identify all sORFs. For example, predicted ribosome binding sites upstream of known small proteins exhibit a range of binding energies with the 16S rRNA, which can show little correlation with experimental ribosome profiling data (Fig. 2). This lack of correlation could reflect experimental constraints such as the specific conditions of the ribosome profiling experiment or be a consequence of the multitude of factors that might impact translation, including transcript levels, RNA folding, and/or regulatory protein or RNA binding. Ultimately, given the sheer number of potential sORFs combined with their sequence variety, a collection of identification approaches will continue to be the most effective method for identifying new sORFs.
Figure 2.
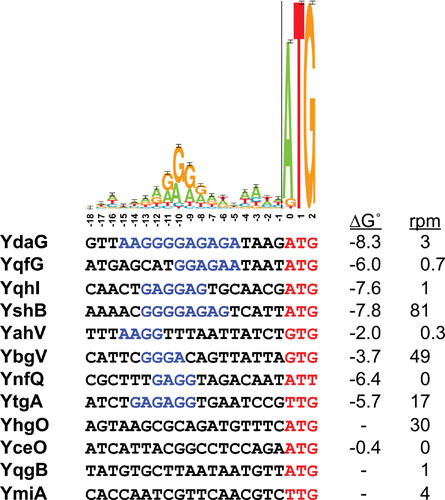
Ribosome binding sites for representative small protein-coding genes. The sequence logo for E. coli ribosome binding sites is reproduced from reference 93. Sequences of 12 small protein genes of unknown function are listed below. Red type corresponds to the predicted start codon, while the blue type indicates stretches of four or more G and A residues. Gibbs free energies (ΔG° in kcal/mol) for the interaction between the sequence shown and the 16S RNA were calculated using IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) (94). No value is given for the three sequences for which no significant interaction was detected. Rpm (reads per million mapped) values for ribosome profiling carried out in the presence of the inhibitor Onc112 are taken from reference 6.
IDENTIFICATION OF SMALL PROTEINS
While computationally predicted ribosome binding sites, conservation, or genome-wide evidence of transcription and ribosome binding can indicate that an sORF is translated, none of these methodologies confirm that the small protein exists as a stable entity in the cell. There are a number of approaches to directly detect small proteins. Given the potential for spurious translation as well as limitations to each of the detection approaches, the strongest evidence that a protein is synthesized and potentially has a function again needs to come from a combination of methods.
Detecting Small Proteins with Mass Spectrometry
Advancements in the sensitivity of mass spectrometers have created an explosion of new proteomic data for bacteria and eukaryotes alike. However, while the use of more sophisticated instrumentation has proven very successful for larger proteins, the identification of small proteins is still hampered by a number of issues. First, many small proteins, particularly hydrophobic, membrane-associated proteins, do not yield more than one potential detectable tryptic peptide (12). Second, even for proteins that can be digested into multiple peptides, it is rare that more than one peptide is observed (12). Third, the low abundance of many small proteins contributes to the failure to observe tryptic peptides, as they may be lost in the background associated with high-sensitivity experiments. Together, these factors make it difficult to identify small proteins unambiguously by mass spectrometry. Overcoming these challenges will allow the more accurate analysis of the small proteomes and raise mass spectrometry to the level of ribosome profiling as a tool for small protein identification.
To achieve better detection, many researchers have utilized approaches to increase the relative abundance of small proteins in their samples. Such approaches, including solvent fractionation (26), molecular weight cutoff filters, and column chromatography, have generally relied on the size of small proteins as the distinguishing trait, as all small proteins share this characteristic. An important consideration is that most of these approaches focus on unbound, soluble proteins. If a small protein is bound to other proteins, the combined molecular weight may exceed the molecular weight cutoff in a small protein screen. Additionally, small, hydrophobic proteins may bind strongly to the materials that compose filters, tubes, or other components used in purification processes. Even with methodological improvements to address these limitations, it is unlikely that a single approach will be possible due to the diversity of the samples being tested and the varied properties of the small proteins.
Although enriching samples for small proteins has the potential to reduce the challenges associated with low abundance, many small proteins still only yield one tryptic peptide during analysis. The observation of multiple, different peptides corresponding to a small protein reduces the number of false positives. Recently, a combination of data from different proteomics experiments was examined to identify new small proteins (27). Although increasing the threshold to two unique tryptic peptides decreased the number of putative small proteins in the experimental data set, it also eliminated all matches from the decoy set. Approaches that add additional criteria such as required fragmentation patterns or external evidence from ribosome profiling beyond a minimal mass spectrometric score (e.g., in MASCOT or SEQUEST) also increase the chance of finding translated proteins. However, as is true for ribosome profiling data, improving the stringency also will lead to an increase in small proteins that are missed.
Quantitative mass spectrometry approaches also have proven useful for identifying small proteins (9, 28). By examining the changes in peptide abundance between samples generated under different growth conditions, some of the aforementioned challenges are circumvented. Although the results are limited, one effective use of this type of quantitative analysis is the identification of small proteins that are significantly more abundant under a specific growth condition. This type of analysis additionally provides useful physiological data that can aid in the characterization of the protein and its function. For example, studies performed in E. coli under heat and cold shock have each revealed new sORFs, including one that is embedded inside another gene (9, 28).
Detecting Small Proteins by N-Terminal Sequencing
The traditional, biochemical method for identifying or verifying protein sequences is Edman degradation. This classic method relies on a chemical reaction that sequentially cleaves the N-terminal residue from a polypeptide chain. Although useful, this method may not be suitable for most small proteins due to the requirements for high sample yield and purity. Thus, this method currently has limited application for proteome-wide small protein identification. In an advance that bypasses some of the limitations, Edman degradation is carried out on proteins transferred to polyvinylidene difluoride (PVDF) membranes after separation by two-dimensional gel electrophoresis (29). By having proteins bound to PVDF, gas-phase protein sequencing can be performed, providing greater sensitivity.
An alternative to traditional N-terminal sequencing relies on the isolation of methionine-containing peptides followed by mass spectrometry. N-terminal methionine-containing peptides can specifically be purified from samples using combined fractional diagonal chromatography (COFRADIC) or similar fractionation methods (30, 31). Most of these studies have only examined known proteins and thus have not led to the discovery of new small proteins. However, 32 proteins of 100 or fewer amino acids were found in the original COFRADIC E. coli dataset (30), and 29 small proteins were identified using a different form of diagonal chromatography (31). One new small protein with a role in stress sensing was detected by COFRADIC in Listeria monocytogenes (16). A challenge imposed by COFRADIC is that each protein is identified by only one peptide, and these peptides may not fragment optimally for mass spectrometric analysis. On the other hand, given that each protein molecule in the cell is represented by only a single N-terminal peptide, the abundance of each signal in the sample should correspond to its stoichiometry in the cell (32).
Confirmation of Small Protein Synthesis Using Epitope-Tagged Alleles
While bioinformatics, transcriptomics, ribosome profiling, mass spectrometry, and N-terminal sequencing are excellent starting points for identifying putative small proteins, comprehensive validation of protein synthesis is key to accurate annotation. The generation of long lists of potential sORFs is enticing, but the challenges to reliable identification mean that the synthesis of small proteins needs to be validated by more than one approach, lest databases become flooded with putative genes. A reliable method of validation is the addition of a tag so that the tagged small protein can be detected by an independent method.
In E. coli and other organisms in which recombination is an established method, a common technique to verify the translation is the construction of chromosomal, translational sORF fusions to epitopes, such as the sequence peptide affinity (SPA) and FLAG tags, which can be detected by immunoblot analysis. This allows for synthesis to be verified (the protein should appear as a specific band by SDS-PAGE) and for steady-state levels of the protein to be observed since the protein is expressed under control of its native promoter and endogenous transcription factors. To date, over 90 E. coli small proteins have been shown to be expressed using an SPA or FLAG epitope tag (6, 10, 24, 33). Another option is chromosomal, translational fusions to fluorescent proteins such as green fluorescent protein (GFP).
Additional options may need to be considered in cases where the putative sORF overlaps an essential gene or when the cells cannot tolerate a fusion at the endogenous chromosomal locus. This includes fusing the putative ORF, its 5′ untranslated region, and its promoter to a reporter, such as GFP or lacZ encoding the assayable enzyme β-galactosidase, at a heterologous location on the chromosome. A number of small proteins have been shown to be expressed using a reporter gene assay (6, 34, 35). A less desirable option, due to the potential for artifacts resulting from overexpression, is fusions to reporters on plasmids.
Beyond showing that a small protein is translated, tests of protein synthesis can provide additional information related to the function of the protein. For example, YnfR, YmcF, and YnfQ were found to be translated upon cold shock (9, 24), suggesting that the functions of these proteins may be important under this stress condition. Tagged proteins also can be useful for additional assays. GFP fusions, for instance, have been used to investigate the localization and orientation of E. coli transmembrane small proteins based on the principle that GFP will not fluoresce if the domain, attached to either the N or C terminus of the small protein, is located in the periplasm (36). Finally, the tagged proteins can be useful for biochemical studies in which the small proteins are purified and/or characterized using the epitope tag.
Small E. coli proteins for which synthesis has been confirmed are given in Table S2, while predicted sORFs in intergenic regions for which no protein has been detected are given in Table S3. Together, the two data sets can serve as useful positive and negative controls for future predictions of small protein genes.
SMALL PROTEINS IN PATHOGENIC E. COLI AND SALMONELLA ENTERICA
Currently, only a few studies have been conducted to identify small proteins in nonlaboratory strains of E. coli or in other Enterobacteria species. One study in S. enterica identified 130 unannotated sORFs and confirmed the synthesis of 25 new small proteins (37). Four of the small proteins, designated Mia-28 (24 aa), Mia-31 (13 aa), Mia-63 (45 aa), and STM14_1499 (35 aa), were induced under low-magnesium stress, a condition that is experienced by S. enterica undergoing phagocytosis by macrophages. Thus, the expression pattern is an indication that the small proteins might be involved in pathogenesis. Separately, the YshB (36 aa) protein in S. enterica was recently shown to be upregulated before phagocytosis of the bacteria by macrophages, and S. enterica mutants lacking YshB exhibited impaired virulence in mouse models (38). In the enteropathogenic E. coli strain O157:H7 Sakai, ribosome profiling provided evidence for the translation of 14 sORFs as well as several slightly longer ORFs for cells grown in rich medium (39). Synthesis of one of the potential small proteins (designated X049, 38 aa) was corroborated by mass spectroscopy. Interestingly, another possible small protein gene (designated X033) was identified in a transposon screen for mutations that lead to decreased colonization of ruminating cattle by O157:H7 (39). It is likely, but remains to be determined, if other small proteins are involved in the pathogenesis of E. coli and other enteric bacteria.
PROPERTIES OF SMALL PROTEIN GENES AND SMALL PROTEINS
Recent studies using ribosome profiling and mass spectroscopy have led to the identification of small proteins encoded by sORFs with diverse, nontraditional sequence characteristics. Small proteins have been detected from sORFs with rare start codons and unrecognizable ribosome binding sites (Fig. 2), as well as those with no detectable ribosome binding in vivo (6). They also have been detected from sORFs located within larger ORFs, in both the sense and antisense strands (6, 25). These results suggest that future screens for small proteins should consider those encoded by both traditional, intergenic sORFs and those with unconventional sequence characteristics and encoded by intragenic sORFs. An exciting avenue of investigation will be characterizing intragenically encoded small proteins and determining if the functions of these proteins intersect with the larger proteins encoded by the parent gene.
While small proteins generally do not have enough amino acids for protein domain determination, one exception is a hydrophobic α helix, which can range from 6 to over 20 amino acids (40). It was initially proposed that hydrophobic α helix-containing small proteins may be the predominant form of this class of proteins (10) and thus could be a predictive factor for identifying new small proteins. However, an equal or greater number of hydrophilic small proteins have been identified in more recent screens for small proteins in E. coli (6, 10, 24, 41). Nevertheless, biochemical experiments have shown that bioinformatic predictions of hydrophobic helices do correlate well with membrane localization, suggesting that a hydrophobic α helix is still a useful indicator of small protein localization and potential function (10, 36).
FUNCTIONS OF sORF TRANSLATION
Before turning to the identified functions of small proteins, it is worth noting that just the process of translating an sORF upstream of or overlapping a downstream ORF may be the critical activity. While the proteins encoded by these regulatory sORFs historically have been referred to as leader peptides in bacteria, we will adopt the nomenclature of uORF (upstream ORF) predominant in the eukaryotic literature. Translation of a uORF can regulate the translation or transcription of the downstream genes. In general, translational pausing versus successful translation of the uORF leads to different mRNA secondary structures or affects the binding of the Rho transcription termination factor, impacting the translation or transcription of downstream genes. These uORFs are commonly located in the leaders of amino acid biosynthetic operons (reviewed in reference 42). In such cases, the small protein usually contains multiple codons for the amino acid synthesized by the enzymes encoded on the mRNA. If the amino acid is abundant, the small protein is synthesized frequently, which in turn inhibits expression of the downstream biosynthetic genes. If the cell is deficient in the amino acid, translation is stalled for the uORF-encoded small protein, allowing expression of the downstream gene to proceed via newly accessible regions of the 5′ untranslated region preceding the next gene. Currently, nine small proteins with these properties have been identified for E. coli operons encoding genes involved in amino acid biosynthesis. HisL (15 aa), IlvL (32 aa), LeuL (28 aa), ThrL (21 aa), and PheL (15 aa) are encoded by uORFs upstream of histidine, isoleucine, leucine, threonine, and phenylalanine operons, respectively. IvbL (32 aa) is encoded at the beginning of an operon encoding a protein involved in both isoleucine and valine biosynthesis, TrpL (14 aa) and TnaC (24 aa) are encoded by uORFs for two tryptophan biosynthetic operons, and PheM (14 aa) is encoded upstream of an operon encoding a phenylalanine tRNA.
Another theme that is emerging among genes preceded by uORFs in both bacteria and in eukaryotic cells is polyamine biosynthesis (43). One example is the speFL ORF encoded upstream of the E. coli and S. enterica speF gene, which encodes ornithine decarboxylase required for polyamine synthesis (44, 45). Again, a block in translation elongation at two consecutive arginine residues in the sORF, in this case due to ornithine binding to the ribosome, impacts Rho-mediated termination and the structure of the mRNA leader, allowing for transcription and translation of the downstream gene.
uORFs upstream of E. coli genes not associated with amino acid or polyamine metabolism include IdlP (27 aa), encoded upstream of the iraD gene and regulated by CsrA (34), MgtL (17 aa), encoded upstream of the mgtA gene and regulated by magnesium levels (46), PyrL (44 aa), encoded upstream of the pyrB gene and regulated by UTP abundance (47), RhoL (33 aa), encoded upstream of the rho gene and thought to be regulated by Rho-dependent transcription termination (48), and Uof (28 aa), which is proposed to be involved in the regulation of Fur protein levels in response to iron abundance (35).
Screens for small proteins suggest that more uORFs, translation of which can either positively or negatively impact the downstream gene (6), remain to be discovered. It is even conceivable that expression of some operons is controlled by more than one uORF. The IlvX small protein (16 aa) was discovered to be encoded downstream of the ilvL uORF of the ilvLXGMEDA operon (10). IlvX accumulates in cells grown in minimal medium, but the role, if any, of this new sORF in isoleucine biosynthesis has not been investigated (23). In addition to further characterizing the regulatory mechanisms, a critical question remaining to be answered is whether the uORF-encoded small proteins have independent functions.
FUNCTIONS OF SMALL PROTEINS
Functional characterization of small proteins is still in its infancy. Multiple factors make it challenging to elucidate the functions. First, sequence comparisons are a less useful tool for small proteins than for their larger counterparts. As already mentioned, the short amino acid sequences make independent domain identification difficult, and the limited number of bacterial small proteins identified also precludes comparisons. In addition, there is increasing evidence that small proteins of similar function can have remarkable sequence flexibility (49, 50), making it difficult to functionally group small proteins beyond predictions for cellular localization.
Experimentally, a number of features also make small protein characterization more challenging. The short amino acid sequences make antibody development more difficult, since there are fewer potential antigens in short sequences. This is particularly true for transmembrane small proteins, which may contain only a single, hydrophobic α helix. Additionally, standard biochemical techniques may not be effective at detecting small proteins. For example, unmodified small proteins can have a molecular weight between 1.5 and 5 kDa, much lower than the resolution available using standard acrylamide gel chromatography. The proteins also are less likely to be detected by standard reagents for staining proteins in gels, as each stain favors certain amino acids and a small protein may have few of these particular residues. Due to these constraints, most of the characterized small proteins have been expressed and purified with epitope tags, such as the SPA, hemagglutinin (HA), hexahistidine (6XHis) tags, and others. However, the size of these tags relative to the small protein means that there is a substantial probability that the small protein function may be altered or eliminated by the addition of the tag. Small proteins serving regulatory roles may also be present at low levels, increasing the difficulty of protein purifications. Finally, small protein genes may be missed in genetic screens due to a lack of sORF annotation and to the lower likelihood that a small protein gene will be mutated.
As a starting point for characterizing the functions of individual small proteins, several studies have focused on large-scale characterization of groups of small proteins. Expression analysis of tagged small proteins under different growth and stress conditions has shown that many accumulate under specific conditions, suggestive of functions under these conditions (9, 23, 24, 28). Other studies have assayed groups of small protein genes for phenotypes associated with their deletions (51). All of these studies have yielded results suggestive of functions for multiple small proteins.
The current challenge in small protein studies in E. coli is moving beyond these generalized approaches to functionally and mechanistically elucidating the roles small proteins play during a particular growth or stress condition. Although relatively few E. coli small proteins have been characterized in this way, they can be organized into some functional groups, which are listed in Table 1 and which we will describe next.
Table 1.
Overview of known small protein functions
| Class | Function | Examples | References |
|---|---|---|---|
| Proteins encoded by uORFs (leader peptides) | Translation regulates expression of downstream gene | HisL, IlvL, LeuL, ThrL, PheL, IvbL, TrpL, TnaC, PheM, SpeFL, IdlP, MgtL, PyrL, RhoL, Uof, IlvX, ToiL, PssL, YoaL, BaxL, ArgL | 6, 34, 35, 42, 44, 46–48 |
| Toxins | Hydrophobic proteins that are toxic to the cell at high levels; in many cases, synthesis is controlled by a regulatory RNA (type I toxin-antitoxin systems) | DinQ, EcnB, HokA, HokB, HokC, HokD, HokE, IsbA, IsbB, IsbC, IsbD, IsbE, LdrA, LdrB, LdrC, LdrD, ShoB, TisB | 52, 53, 90 |
| Ribosomal proteins | May replace homologous ribosomal proteins under specific conditions such as zinc limitation | RpmH, RpmJ, Sra, YkgO | 10, 61–64, 91 |
| Regulators of sensor kinase activity | Provide a mechanism for feedback regulation of two-component systems | MgrB | 66, 67 |
| Transmembrane protein regulators of transporters | Modulate the specificity, activity, or levels of transporters | AcrZ, KdpF, MgtS | 68–72, 85 |
| Cytoplasmic protein regulators of transporters | Modulate the specificity, activity or levels of transporters | SgrT, MntS | 73–75 |
| Components of cytochrome oxidase complexes | Associate with cytochrome oxidases; CydX has been shown to be critical for activity | CydX, CydH/CydY | 60, 80, 81 |
Toxic Small Proteins
E. coli contains more than a dozen small proteins that have been shown to be, or are predicted to be, toxins of the type I toxin-antitoxin family, in which synthesis of the small toxic protein is inhibited by a base pairing antitoxin RNA. Small proteins expressed from type I toxin-antitoxin systems are predicted to contain a hydrophobic helix (reviewed in reference 52). When derepressed, these hydrophobic small proteins are thought to localize to the cell membrane, oligomerize to form pores, and compromise the integrity of the membrane. Consistent with this model, overexpression of several of these small proteins has been shown to result in membrane depolarization, decreased cell density in liquid culture, and decreased colony forming units (53 to 55). For the HokB protein, it was also found that an oxidoreductase can disassemble the HokB pore, leading to degradation of the protein and providing a mechanism to downregulate the effects of the toxin (56). The role of type I toxin proteins at endogenous levels is still under debate. One model is that the toxin-antitoxin systems solely exist as selfish DNA elements, while studies have suggested that toxin-antitoxin systems help cells survive stress conditions by leading to slow growth rather than death (57, 58). One other small protein, the 48-aa lipoprotein EcnB, also has been described as a toxin, but in this case, a divergently encoded 41-aa lipoprotein, EcnA, is reported to be the antitoxin (59).
Intriguingly, there are no obvious differences in the amino acid sequences of the toxic small proteins and hydrophobic membrane proteins that are normally expressed at high levels and have other functions in the cell. For example, overexpression of the hydrophobic small proteins CydX and AcrZ did not have a substantial impact on growth (60). Possibly, toxicity is associated with a propensity to oligomerize, but further research is needed to determine what features distinguish toxic and nontoxic small hydrophobic membrane proteins.
Ribosomal Small Proteins
Three abundant ribosomal proteins in E. coli have fewer than 50 amino acids. RpmH (46 aa) and RpmJ (38 aa) are subunits L34 and L36, respectively, of the 50S ribosomal complex (61, 62). sra encodes the S22 subunit (45 aa) of the 30S ribosomal complex (63, 64). A fourth small protein, YkgO (46 aa), is a paralog of RpmJ and accumulates to high levels in cells after exposure to the chelating agent EDTA (23). YkgO lacks a zinc-binding motif found in RpmJ (10). Thus, it has been proposed that ribosomal proteins like YkgO may have evolved to replace zinc-dependent ribosomal proteins under zinc-limiting conditions (65).
Small Protein Modulator of a Sensor Kinase
The E. coli genome encodes multiple two-component regulatory systems, which control responses to a wide range of environmental stresses. In these systems, the sensor kinase generally is in the membrane. Interestingly, one small transmembrane protein, MgrB (47 aa), whose levels are induced by the PhoQP two-component system in limiting magnesium, plays a role in the regulation of the PhoQ histidine kinase (66, 67) by binding to and inhibiting this kinase. MgrB thus acts as a negative feedback regulator of the PhoQP system (67). It will be interesting to see if other small proteins similarly modulate phosphate transfer in two-component systems.
Small Protein Modulators of Transporters
The largest category of E. coli small proteins characterized to date are those that modulate the activities or levels of transporters. Most of these small proteins are predicted to consist of a single hydrophobic α helix and localize to the cell membrane. This suggests an environment at the E. coli membrane in which large transmembrane complexes are surrounded by, interact with, and are potentially regulated by, small transmembrane proteins. Consistent with this model, AcrZ (49 aa) has been shown to interact with the AcrAB-TolC multidrug efflux pump and is required for the optimal export of certain antibiotics (68). It is thought to therefore regulate the substrate specificity of the transporter. The KdpF (29 aa) small protein interacts with the Kdp-ATPase potassium ion transporter (69, 70). The small protein may stabilize the complex, as it can be replaced by high lipid levels (70). The MgtS (31 aa) protein is a transmembrane small protein that accumulates under multiple stresses, including magnesium deprivation (23, 71). The small protein is required for the accumulation of the magnesium transporter MgtA, and evidence suggests that the small protein inhibits degradation of MgtA by the FtsH protease (71). Surprisingly, MgtS interacts with a second membrane protein, the PitA phosphate symporter (72). This interaction also increases intracellular magnesium levels.
Two small proteins without a detectable hydrophobic α helix also play roles in regulating the activities of larger membrane protein complexes. SgrT (43 aa) is expressed under sugar-phosphate stress and interacts with the PtsG sugar transporter to inhibit transporter activity (33, 73). This interaction promotes recovery of cells grown with toxic glucose analogs such as α-methyl glucoside (73). MntS (42 aa), whose expression is induced by low manganese, is involved in the regulation of cellular manganese levels (74). MntS expression in the presence of high manganese increases intracellular manganese levels phenocopying the effects of reducing MntP manganese exporter activity (75). It is anticipated that a number of other transmembrane proteins as well as potentially cytosolic small proteins modulate the activities or levels of transporters.
Small Proteins Associated with Cytochrome Oxidase Complexes
Perhaps the best-characterized small transmembrane domain proteins found to interact with larger membrane proteins are those associated with cytochrome oxidase complexes in E. coli as well as in other bacteria. While these complexes have been studied for more than 70 years, the fact that small proteins are part of the complexes has only been realized recently. The first hint came from purification of the E. coli cytochrome bd-I oxidase, revealing an unidentified lower-molecular-weight band (76). Next, a small ORF, annotated as ybgT, was identified downstream of the cydAB genes in the cydAB-ybgT-ybgE operon encoding this oxidase (77). YbgT, now renamed CydX (37 aa), was not identified as a subunit of the complex until it was independently characterized as a small protein (60, 78) and shown to be required for oxidase activity (60, 79). Intriguingly, the recent determination of the E. coli cytochrome bd-I oxidase structure by cryo-EM (Fig. 3) shows that there is a second small protein component of the complex (80, 81). This protein, now denoted CydH or CydY (29 aa) but previously annotated as YnhF, was found to bind to the Q-loop, a highly variable portion of the CydA subunit. CydX has been proposed to play a role in folding and stabilizing the complex, given decreased yields and altered redox-difference spectra for the purified complex in the absence of the small protein (79). Recent characterization of CydX in S. enterica showed that the protein is similarly required for cytochrome bd oxidase function and heme orientation, and that the mutants lacking the small protein exhibit sensitivity to nitric oxide, reduced proliferation in macrophages, and increased resistance to select antibiotics (82). The role of the CydH/CydY protein still needs to be determined (80, 81).
Figure 3.
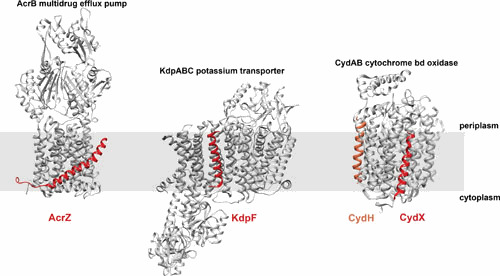
Structures of representative small proteins. Structures of AcrZ, KdpF, CydX, and CydH (red) in association with the AcrB multidrug efflux pump (PDB 4C48 [84]), Kdp potassium transporter (PDB 5MRW [69]), and cytochrome bd-I oxidase (PDB 6RKO [80]), respectively. The approximate position of the membrane is indicated by shading.
In addition to the cydABX-ybgE operon, E. coli and related species encode the paralogous appCBXA operon encoding the cytochrome bd-II oxidase. AppX is the small protein paralog to CydX. The two proteins are thought to play similar roles in their respective complexes, and there is evidence indicating that there may be some cross-interaction between these small proteins and the two complexes (60). Consistent with limited requirements for specific amino acids for functionality, mutational analysis of the CydX small protein did not identify any single amino acid required for small protein function in the complex (50). These data, combined with the growing body of work showing small proteins associated with cytochrome oxidases in other bacteria (49, 78, 83), suggest that much remains to be learned about this class of small proteins.
The structures of the AcrBZ, KdpABCF, and CydABXH complexes (69, 80, 81, 84, 85) (Fig. 3) show the positions of the small proteins relative to the larger subunits of each complex. It is worth noting that in all of these structures, the small protein is located toward the exterior of the complex, interacting with the membrane. This relationship hints that the small proteins may not be constant components of each complex and instead may have the potential to separate from the complex as part of their regulatory functions. It has already been shown that small proteins can bind to more than one protein (60, 72). Given the positions of the small proteins in the complexes, it is also possible that they modulate the lipid environment around the larger proteins. Important directions for future research are further understanding the nature and lifetime of the interactions of small proteins with large membrane protein complexes, which would help elucidate the impact of these small proteins on the structures and activities of the complexes.
MORE SMALL PROTEINS
Results from multiple studies strongly suggest that more small proteins remain to be identified in E. coli. In one recent study, 38 of tested 41 sORFs predicted by ribosome profiling after treatment with retapamulin and Onc112 were found to be expressed, a 92% success rate (6). This success rate, combined with the fact that over 412 sORFs showed a signal above threshold in this study, supports the idea that more small proteins exist. Likewise, over 150 sORFs were predicted in a similar experiment examining tetracycline-stalled ribosome binding (86). Finally, over 260 sORFs were predicted to be preceded by a stronger than average ribosome binding site (10). A comparison of the small proteins predicted by these three screens yields 65 sORFs identified in at least two studies and 12 identified in all three. These approaches do not even consider small proteins encoded by sORFs lacking ribosome binding signals. Thus, it is possible that the E. coli genome encodes hundreds of as-yet-unidentified small proteins encoded in intergenic regions. This accounting also does not consider several other groups of proteins of low molecular weight, including prophage- and pseudogene-encoded proteins and small proteins encoded intragenic to larger proteins. Some of these groups of proteins deserve brief mention. It also is possible that fragments of larger proteins such as signal peptides, which are not covered here, have independent functions.
Prophage-Encoded Small Proteins
The genome of E. coli contains a number of prophage regions that are the result of ancestral phage integration into the genome (1, 87). Given that several bacteriophage small proteins have been identified and, in some cases, characterized (reviewed in 13), it is not surprising that small proteins are encoded in prophage regions. Small proteins have been found to be encoded in the Qin, CP4-6, DLP12, Rac, and CPS-53 prophages as well as the KpLE2 phage-like element. These proteins are often expressed at high levels in cells growing under normal conditions (6, 10, 24), raising the question about possible protein functions outside of phage survival. In intact phages, small proteins have been found to play many roles in the phage life cycle (13). They regulate host protein function, play structural roles in the phage capsids, and promote cell lysis through interactions with the membrane. At this point, it is not known how many small E. coli prophage proteins are synthesized and how many have endogenous functions in the bacterial cell.
Proteins Encoded by Pseudogenes
Pseudogenes are genes that have undergone mutation to the point where a portion of the ORF is noncoding (88). In the case of small proteins, larger genes may undergo a nonsense or frameshift mutation that transforms a larger ORF into one that encodes a small protein. Multiple small proteins in E. coli are expressed from ORFs annotated as pseudogenes (10) such as YmjD, YnfP, and YkgS. Similar to prophage small proteins, it is generally thought that pseudogenes are nonfunctional. It is possible, however, that at least a subset of these proteins have roles in the cell. The degradation of larger genes may represent a common evolutionary mechanism for creating sORFs. These small proteins could then acquire an independent biological function. It remains to be determined if any pseudogene small proteins are functional in E. coli.
Proteins of 50 to 100 Amino Acids
As shown in Fig. 1, the number of proteins identified in E. coli that range from 50 to 100 amino acids also has increased substantially in the past 6 years. Proteins in this range were reported in recent small protein identification papers, and it is very likely that more of these “larger” small proteins will be discovered in the future. Structurally, one distinction between these proteins and the small proteins considered in this review is that a larger number of amino acids facilitates the adoption of more complex protein domains. Proteins of this size can contain multiple hydrophobic α helices and domains such as a zinc ribbon and zinc finger. In fact, many proteins of 51 to 100 amino acids in E. coli are well characterized, including ribosomal proteins, the chaperone protein GroS (97 aa), the carbon metabolism regulator CsrA (61 aa), and the cold- and stress-inducible Csp proteins (69 to 74 aa). Similar to proteins with 50 or fewer amino acids, it is likely that new protein families remain to be identified in this group of proteins. For example, three proteins, the paralogous YmcF (62 aa) and YnfQ (62 aa) proteins and YnfU (56 aa), which are all encoded adjacent to Csp proteins, have structural homology to the zinc-binding domains of larger proteins (6, 9). Ultimately, more than half of the genes encoding these proteins have unknown or only predicted functions, indicating that they also represent an underexplored area of E. coli proteomics.
Proteins Encoded by Dual-Function RNAs
There is increasing evidence that small proteins can be encoded in E. coli from sORFs within larger genes. Previously, this possibility was considered extremely rare outside of viral genomes, but may be more prevalent than expected for chromosomally-encoded genes. The GndA small protein (36 or 54 aa; the N-terminal aa has not been unambiguously identified) was recently identified as a heat shock-induced small protein and is encoded within, but out of frame of, the larger gnd gene (28). In addition, ribosome-profiling with the retapamulin inhibitor revealed a number of examples of ribosomal binding within larger genes. Of these internal sORFs, expression has been confirmed for a small protein encoded in the same ORF as, but at the 3′ end of, the sfsA gene (25).
Another variation of dual function is transcripts that have been shown to act as a regulatory RNA and also encode a small protein. The best-characterized example in E. coli is the dual-function SgrS RNA, which both acts as an RNA regulator of the ptsG RNA and encodes the SgrT small protein, which as mentioned above, inhibits PtsG function (73).
As one additional variation of overlapping coding capacity, some small proteins have been found to be encoded antisense to other genes (6). The existence of intragenically encoded small proteins again illustrates how small protein coding capacity has been underestimated and raises intriguing questions about how the additional coding capacity arises and whether the proteins have antagonistic or synergistic functions.
OUTLOOK
Small proteins represent an exciting area of discovery in bacteria, archaea, and eukaryotes (89). As one of the best-characterized organisms, E. coli is an ideal system for the continued identification and characterization of these enigmatic molecules. Experimental techniques for identifying new sORFs and confirming small protein expression have been well established, and there is substantial evidence that more small proteins remain to be identified. The largest challenge is determining the functional roles for these newly identified proteins. The fact that more than 70% of larger proteins have been characterized in E. coli provides a strong basis for studying these small proteins. Understanding of the functions of AcrZ, CydX, MntS, and other small proteins was possible because the proteins they interact with were already characterized. Conversely, elucidating the roles of the small proteins can provide insights into the physiological roles of the larger proteins with which they interact. For example, the finding that the low-magnesium-induced MgtS protein binds PitA revealed that this phosphate transporter also has an important role in maintaining magnesium homeostasis (72). The resources and information available for E. coli should continue to facilitate small protein discovery and characterization in this organism and provide a gateway for small protein discoveries in other organisms.
SUPPLEMENTARY MATERIAL
ACKNOWLEDGMENTS
We thank A. Burton, M. Orr, L. Waters, and A. Zhong for comments on the review.
Research in the Hemm laboratory is funded by grants from the National Institute of Allergy and Infectious Diseases (1R15AI094548-01) and the National Science Foundation (1149274), as well as by funds to the Jess and Mildred Fisher Endowed Chair of Biological Sciences at Towson University. Research in the Storz laboratory is funded by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development.
No potential conflicts of interest relevant to this review were reported.
Contributor Information
Matthew R. Hemm, Department of Biological Sciences, Towson University, Towson, MD
Jeremy Weaver, Division of Molecular and Cellular Biology, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD; Present address: Thermo Fisher Scientific, Rockford, IL.
Gisela Storz, Division of Molecular and Cellular Biology, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD.
Susan T. Lovett, Brandeis University, Waltham, MA
Deborah Hinton, Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD.
REFERENCES
- 1.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res 33:D334–D337 10.1093/nar/gki108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39(Database):D583–D590 10.1093/nar/gkq1143. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624 10.1093/nar/gkw569. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudd KE, Humphery-Smith I, Wasinger VC, Bairoch A. 1998. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19:536–544 10.1002/elps.1150190413. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Weaver J, Mohammad F, Buskirk AR, Storz G. 2019. Identifying small proteins by ribosome profiling with stalled initiation complexes. MBio 10:e02819-18 10.1128/mBio.02819-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hücker SM, Vanderhaeghen S, Abellan-Schneyder I, Wecko R, Simon S, Scherer S, Neuhaus K. 2018. A novel short l-arginine responsive protein-coding gene (laoB) antiparallel overlapping to a CadC-like transcriptional regulator in Escherichia coli O157:H7 Sakai originated by overprinting. BMC Evol Biol 18:21 10.1186/s12862-018-1134-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hücker SM, Vanderhaeghen S, Abellan-Schneyder I, Scherer S, Neuhaus K. 2018. The novel anaerobiosis-responsive overlapping gene ano is overlapping antisense to the annotated gene ECs2385 of Escherichia coli O157:H7 Sakai. Front Microbiol 9:931 10.3389/fmicb.2018.00931. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Lima NG, Khitun A, Rosenbloom AD, Yuan P, Gassaway BM, Barber KW, Rinehart J, Slavoff SA. 2017. Comparative proteomics enables identification of nonannotated cold shock proteins in E. coli. J Proteome Res 16:3722–3731 10.1021/acs.jproteome.7b00419. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol 70:1487–1501 10.1111/j.1365-2958.2008.06495.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren AS, Archuleta J, Feng WC, Setubal JC. 2010. Missing genes in the annotation of prokaryotic genomes. BMC Bioinformatics 11:131 10.1186/1471-2105-11-131. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller SA, Kohajda T, Findeiss S, Stadler PF, Washietl S, Kellis M, von Bergen M, Kalkhof S. 2010. Optimization of parameters for coverage of low molecular weight proteins. Anal Bioanal Chem 398:2867–2881 10.1007/s00216-010-4093-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiMaio D. 2014. Viral miniproteins. Annu Rev Microbiol 68:21–43 10.1146/annurev-micro-091313-103727. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Short JD, Pfarr CM. 2002. Translational regulation of the JunD messenger RNA. J Biol Chem 277:32697–32705 10.1074/jbc.M204553200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Basrai MA, Hieter P, Boeke JD. 1997. Small open reading frames: beautiful needles in the haystack. Genome Res 7:768–771 10.1101/gr.7.8.768. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Impens F, Rolhion N, Radoshevich L, Bécavin C, Duval M, Mellin J, García Del Portillo F, Pucciarelli MG, Williams AH, Cossart P. 2017. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat Microbiol 2:17005 10.1038/nmicrobiol.2017.5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochman H. 2002. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet 18:335–337 10.1016/S0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- 18.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777 10.1146/annurev-biochem-070611-102400. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samayoa J, Yildiz FH, Karplus K. 2011. Identification of prokaryotic small proteins using a comparative genomic approach. Bioinformatics 27:1765–1771 10.1093/bioinformatics/btr275. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goli B, Nair AS. 2012. The elusive short gene: an ensemble method for recognition for prokaryotic genome. Biochem Biophys Res Commun 422:36–41 10.1016/j.bbrc.2012.04.090. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Wood DE, Lin H, Levy-Moonshine A, Swaminathan R, Chang YC, Anton BP, Osmani L, Steffen M, Kasif S, Salzberg SL. 2012. Thousands of missed genes found in bacterial genomes and their analysis with COMBREX. Biol Direct 7:37 10.1186/1745-6150-7-37. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Óhéigeartaigh SS, Armisén D, Byrne KP, Wolfe KH. 2014. SearchDOGS bacteria, software that provides automated identification of potentially missed genes in annotated bacterial genomes. J Bacteriol 196:2030–2042 10.1128/JB.01368-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemm MR, Paul BJ, Miranda-Ríos J, Zhang A, Soltanzad N, Storz G. 2010. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol 192:46–58 10.1128/JB.00872-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VanOrsdel CE, Kelly JP, Burke BN, Lein CD, Oufiero CE, Sanchez JF, Wimmers LE, Hearn DJ, Abuikhdair FJ, Barnhart KR, Duley ML, Ernst SEG, Kenerson BA, Serafin AJ, Hemm MR. 2018. Identifying new small proteins in Escherichia coli. Proteomics 18:e1700064 10.1002/pmic.201700064. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meydan S, Marks J, Klepacki D, Sharma V, Baranov PV, Firth AE, Margus T, Kefi A, Vázquez-Laslop N, Mankin AS. 2019. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol Cell 74:481–493.e6 10.1016/j.molcel.2019.02.017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan Z, Wang X, Raetz CR. 2011. Identification of a chloroform-soluble membrane miniprotein in Escherichia coli and its homolog in Salmonella typhimurium. Anal Biochem 409:284–289 10.1016/j.ab.2010.10.035. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miravet-Verde S, Ferrar T, Espadas-García G, Mazzolini R, Gharrab A, Sabido E, Serrano L, Lluch-Senar M. 2019. Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol 15:e8290 10.15252/msb.20188290. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan P, D’Lima NG, Slavoff SA. 2018. Comparative membrane proteomics reveals a nonannotated E. coli heat shock protein. Biochemistry 57:56–60 10.1021/acs.biochem.7b00864. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reim DF, Speicher DW. 1994. A method for high-performance sequence analysis using polyvinylidene difluoride membranes with a biphasic reaction column sequencer. Anal Biochem 216:213–222 10.1006/abio.1994.1027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. 2003. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 21:566–569 10.1038/nbt810. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Kramer G, Sprenger RR, Nessen MA, Roseboom W, Speijer D, de Jong L, de Mattos MJ, Back J, de Koster CG. 2010. Proteome-wide alterations in Escherichia coli translation rates upon anaerobiosis. Mol Cell Proteomics 9:2508–2516 10.1074/mcp.M110.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Damme P, Van Damme J, Demol H, Staes A, Vandekerckhove J, Gevaert K. 2009. A review of COFRADIC techniques targeting protein N-terminal acetylation. BMC Proc 3(Suppl 6):S6 10.1186/1753-6561-3-s6-s6. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA 104:20454–20459 10.1073/pnas.0708102104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H, McGibbon LC, Potts AH, Yakhnin H, Romeo T, Babitzke P. 2017. Translational repression of the RpoS antiadapter IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. MBio 8:e01355-17 10.1128/mBio.01355-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecerek B, Moll I, Bläsi U. 2007. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J 26:965–975 10.1038/sj.emboj.7601553. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontaine F, Fuchs RT, Storz G. 2011. Membrane localization of small proteins in Escherichia coli. J Biol Chem 286:32464–32474 10.1074/jbc.M111.245696. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baek J, Lee J, Yoon K, Lee H. 2017. Identification of unannotated small genes in Salmonella. G3 (Bethesda) 7:983–989 10.1534/g3.116.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bomjan R, Zhang M, Zhou D. 2019. YshB promotes intracellular replication and is required for Salmonella virulence. J Bacteriol 201:00314–00319 10.1128/JB.00314-19. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhaus K, Landstorfer R, Fellner L, Simon S, Schafferhans A, Goldberg T, Marx H, Ozoline ON, Rost B, Kuster B, Keim DA, Scherer S. 2016. Translatomics combined with transcriptomics and proteomics reveals novel functional, recently evolved orphan genes in Escherichia coli O157:H7 (EHEC). BMC Genomics 17:133 10.1186/s12864-016-2456-1. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hildebrand PW, Preissner R, Frömmel C. 2004. Structural features of transmembrane helices. FEBS Lett 559:145–151 10.1016/S0014-5793(04)00061-4. [DOI] [PubMed] [Google Scholar]
- 41.Alix E, Blanc-Potard AB. 2009. Hydrophobic peptides: novel regulators within bacterial membrane. Mol Microbiol 72:5–11 10.1111/j.1365-2958.2009.06626.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Kolter R, Yanofsky C. 1982. Attenuation in amino acid biosynthetic operons. Annu Rev Genet 16:113–134 10.1146/annurev.ge.16.120182.000553. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Ivanov IP, Atkins JF, Michael AJ. 2010. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38:353–359 10.1093/nar/gkp1037. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Zvi T, Pushkarev A, Seri H, Elgrably-Weiss M, Papenfort K, Altuvia S. 2019. mRNA dynamics and alternative conformations adopted under low and high arginine concentrations control polyamine biosynthesis in Salmonella. PLoS Genet 15:e1007646 10.1371/journal.pgen.1007646. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrero Del Valle A, Seip B, Cervera-Marzal I, Sacheau G, Seefeldt AC, Innis CA. 2020. Ornithine capture by a translating ribosome controls bacterial polyamine synthesis. Nat Microbiol 5:554–561 10.1038/s41564-020-0669-1. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chadani Y, Niwa T, Izumi T, Sugata N, Nagao A, Suzuki T, Chiba S, Ito K, Taguchi H. 2017. Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing. Mol Cell 68:528–539.e5 10.1016/j.molcel.2017.10.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Levin HL, Schachman HK. 1985. Regulation of aspartate transcarbamoylase synthesis in Escherichia coli: analysis of deletion mutations in the promoter region of the pyrBI operon. Proc Natl Acad Sci USA 82:4643–4647 10.1073/pnas.82.14.4643. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto Y, Shigesada K, Hirano M, Imai M. 1986. Autogenous regulation of the gene for transcription termination factor rho in Escherichia coli: localization and function of its attenuators. J Bacteriol 166:945–958 10.1128/JB.166.3.945-958.1986. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen RJ, Brenner EP, VanOrsdel CE, Hobson JJ, Hearn DJ, Hemm MR. 2014. Conservation analysis of the CydX protein yields insights into small protein identification and evolution. BMC Genomics 15:946 10.1186/1471-2164-15-946. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hobson JJ, Gallegos AS, Atha BW III, Kelly JP, Lein CD, VanOrsdel CE, Weldon JE, Hemm MR. 2018. Investigation of amino acid specificity in the CydX small protein shows sequence plasticity at the functional level. PLoS One 13:e0198699 10.1371/journal.pone.0198699. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hobbs EC, Astarita JL, Storz G. 2010. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J Bacteriol 192:59–67 10.1128/JB.00873-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72:579–589 10.1128/MMBR.00025-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol 70:1076–1093 10.1111/j.1365-2958.2008.06394.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen K, Gerdes K. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol 32:1090–1102 10.1046/j.1365-2958.1999.01431.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Wilmaerts D, Bayoumi M, Dewachter L, Knapen W, Mika JT, Hofkens J, Dedecker P, Maglia G, Verstraeten N, Michiels J. 2018. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. MBio 9:e00744-18 10.1128/mBio.00744-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilmaerts D, Dewachter L, De Loose PJ, Bollen C, Verstraeten N, Michiels J. 2019. HokB monomerization and membrane repolarization control persister awakening. Mol Cell 75:1031–1042.e4 10.1016/j.molcel.2019.06.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317 10.1371/journal.pbio.1000317. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun 391:209–213 10.1016/j.bbrc.2009.11.033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol 280:583–596 10.1006/jmbi.1998.1894. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.VanOrsdel CE, Bhatt S, Allen RJ, Brenner EP, Hobson JJ, Jamil A, Haynes BM, Genson AM, Hemm MR. 2013. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J Bacteriol 195:3640–3650 10.1128/JB.00324-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wada A, Sako T. 1987. Primary structures of and genes for new ribosomal proteins A and B in Escherichia coli. J Biochem 101:817–820 10.1093/jb/101.3.817. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Panagiotidis CA, Canellakis ES. 1984. Comparison of the basic Escherichia coli antizyme 1 and antizyme 2 with the ribosomal proteins S20/L26 and L34. J Biol Chem 259:15025–15027. [PubMed] [Google Scholar]
- 63.Wada A. 1986. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. II. Characterization of four new proteins. J Biochem 100:1595–1605 10.1093/oxfordjournals.jbchem.a121867. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Izutsu K, Wada C, Komine Y, Sako T, Ueguchi C, Nakura S, Wada A. 2001. Escherichia coli ribosome-associated protein SRA, whose copy number increases during stationary phase. J Bacteriol 183:2765–2773 10.1128/JB.183.9.2765-2773.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63:294–307 10.1111/j.1365-2958.2006.05513.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Salazar ME, Podgornaia AI, Laub MT. 2016. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol Microbiol 102:430–445 10.1111/mmi.13471. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Lippa AM, Goulian M. 2009. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet 5:e1000788 10.1371/journal.pgen.1000788. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. 2012. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci USA 109:16696–16701 10.1073/pnas.1210093109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang CS, Pedersen BP, Stokes DL. 2017. Crystal structure of the potassium-importing KdpFABC membrane complex. Nature 546:681–685 10.1038/nature22970. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gassel M, Möllenkamp T, Puppe W, Altendorf K. 1999. The KdpF subunit is part of the K(+)-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J Biol Chem 274:37901–37907 10.1074/jbc.274.53.37901. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, Yin X, Wu Orr M, Dambach M, Curtis R, Storz G. 2017. Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc Natl Acad Sci USA 114:5689–5694 10.1073/pnas.1703415114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin X, Wu Orr M, Wang H, Hobbs EC, Shabalina SA, Storz G. 2019. The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol Microbiol 111:131–144 10.1111/mmi.14143. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lloyd CR, Park S, Fei J, Vanderpool CK. 2017. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J Bacteriol 199:e00869–e00816 10.1128/JB.00869-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waters LS, Sandoval M, Storz G. 2011. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J Bacteriol 193:5887–5897 10.1128/JB.05872-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin JE, Waters LS, Storz G, Imlay JA. 2015. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet 11:e1004977 10.1371/journal.pgen.1004977. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller MJ, Gennis RB. 1983. The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J Biol Chem 258:9159–9165. [PubMed] [Google Scholar]
- 77.Muller MM, Webster RE. 1997. Characterization of the tol-pal and cyd region of Escherichia coli K-12: transcript analysis and identification of two new proteins encoded by the cyd operon. J Bacteriol 179:2077–2080 10.1128/JB.179.6.2077-2080.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolán HG, Tsolis RM. 2012. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol 2:47 10.3389/fcimb.2012.00047. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T. 2014. Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett 588:1537–1541 10.1016/j.febslet.2014.03.036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Safarian S, Hahn A, Mills DJ, Radloff M, Eisinger ML, Nikolaev A, Meier-Credo J, Melin F, Miyoshi H, Gennis RB, Sakamoto J, Langer JD, Hellwig P, Kühlbrandt W, Michel H. 2019. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 366:100–104 10.1126/science.aay0967. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Theßeling A, Rasmussen T, Burschel S, Wohlwend D, Kägi J, Müller R, Böttcher B, Friedrich T. 2019. Homologous bd oxidases share the same architecture but differ in mechanism. Nat Commun 10:5138 10.1038/s41467-019-13122-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duc KM, Kang BG, Lee C, Park HJ, Park YM, Joung YH, Bang IS. 2020. The small protein CydX is required for cytochrome bd quinol oxidase stability and function in Salmonella Typhimurium: a phenotypic study. J Bacteriol 202:00348-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Safarian S, Rajendran C, Müller H, Preu J, Langer JD, Ovchinnikov S, Hirose T, Kusumoto T, Sakamoto J, Michel H. 2016. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 352:583–586 10.1126/science.aaf2477. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515 10.1038/nature13205. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stock C, Hielkema L, Tascón I, Wunnicke D, Oostergetel GT, Azkargorta M, Paulino C, Hänelt I. 2018. Cryo-EM structures of KdpFABC suggest a K+ transport mechanism via two inter-subunit half-channels. Nat Commun 9:4971 10.1038/s41467-018-07319-2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahigashi K, Takai Y, Kimura M, Abe N, Nakayashiki T, Shiwa Y, Yoshikawa H, Wanner BL, Ishihama Y, Mori H. 2016. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res 23:193–201 10.1093/dnares/dsw008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 88.Lerat E, Ochman H. 2005. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res 33:3125–3132 10.1093/nar/gki631. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orr MW, Mao Y, Storz G, Qian SB. 2020. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 48:1029–1042 10.1093/nar/gkz734. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 33:1040–1050 10.1093/nar/gki256. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hansen FG, Hansen EB, Atlung T. 1982. The nucleotide sequence of the dnaA gene promoter and of the adjacent rpmH gene, coding for the ribosomal protein L34, of Escherichia coli. EMBO J 1:1043–1048 10.1002/j.1460-2075.1982.tb01294.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karp PD, Ong WK, Paley S, Billington R, Caspi R, Fulcher C, Kothari A, Krummenacker M, Latendresse M, Midford PE, Subhraveti P, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Santos-Zavaleta A, Mackie A, Collado-Vides J, Keseler IM, Paulsen I. 2018. The EcoCyc database. Ecosal Plus 8: 10.1128/ecosalplus.ESP-0006-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100 10.1093/nar/18.20.6097. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mann M, Wright PR, Backofen R. 2017. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res 45(W1):W435–W439 10.1093/nar/gkx279. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


