Abstract
Background
Gliomas are the most common type of primary brain tumor and one of many cancers where males are diagnosed with greater frequency than females. However, little is known about the sex-based molecular differences in glioblastomas (GBMs) or lower grade glioma (non-GBM) subtypes. DNA methylation is an epigenetic mechanism involved in regulating gene transcription. In glioma and other cancers, hypermethylation of specific gene promoters downregulates transcription and may have a profound effect on patient outcome. The purpose of this study was to determine if sex-based methylation differences exist in different glioma subtypes.
Methods
Molecular and clinical data from glioma patients were obtained from The Cancer Genome Atlas and grouped according to tumor grade and molecular subtype (IDH1/2 mutation and 1p/19q chromosomal deletion). Sex-specific differentially methylated probes (DMPs) were identified in each subtype and further analyzed to determine if they were part of differentially methylated regions (DMRs) or associated with differentially methylated DNA transcription regulatory binding motifs.
Results
Analysis of methylation data in 4 glioma subtypes revealed unique sets of both sex-specific DMPs and DMRs in each subtype. Motif analysis based on DMP position also identified distinct sex-based sets of DNA-binding motifs that varied according to glioma subtype. Downstream targets of 2 of the GBM-specific transcription binding sites, NFAT5 and KLF6, showed differential gene expression consistent with increased methylation mediating downregulation.
Conclusion
DNA methylation differences between males and females in 4 glioma molecular subtypes suggest an important, sex-specific role for DNA methylation in epigenetic regulation of gliomagenesis.
Keywords: DNA methylation, glioblastoma, glioma, sex-specific differences, survival
Importance of the Study.
Significantly more males than females are diagnosed with gliomas, and this sex-based bias occurs throughout life. Furthermore, the overall survival of males is significantly shorter compared to females with GBM. However, little is known about the molecular differences that may explain the sex-based differences in incidence and survival. Here we have investigated this discordance by examining epigenetic differences between male and female glioma patients, stratified by molecular subtype. We show that when analyzing DNA methylation data by sex and molecular subtype, distinct DNA methylation patterns emerge. This work highlights the importance of parallel, but separate analyses of male and female data. Improved insights into the molecular differences between male and female glioma patients will be vital for understanding the discrepancies in gliomagenesis rates and prognosis, and may lead to improved treatments for both sexes.
Key Points.
Male and female glioma patients exhibit genome-wide DNA methylation differences.
Glioma molecular subtypes display distinct sex-based methylation patterns.
Sex-based methylation differences may influence gliomagenesis and prognosis.
Gliomas represent about 81% of the malignant brain and central nervous system (CNS) tumors in the United States with more than 100 000 cases diagnosed between 2011 and 2015.1 Classification of these heterogeneous tumors is based on World Health Organization (WHO) criteria and, beginning in 2016, incorporates both molecular as well as histological characteristics.2 More than 55% of gliomas are categorized as WHO grade IV glioblastoma (GBM), which have the worst prognosis, with a median survival of less than 2 years.1,3 Gliomas in WHO grades I–III categories are considered lower grade gliomas (non-GBM) and vary in terms of overall survival based on molecular subtype.1,2 Of key importance in predicting outcome for these non-GBM patients, is the presence of mutations in the isocitrate dehydrogenase genes 1 or 2 (IDH1/2) which correspond to longer survival. IDH1/2 mutations may also be found in GBM, although strictly in instances of clear tumor progression from lower grades, and in only about 10% of GBMs.2 Non-GBM prognosis is also greatly influenced by deletions in the short arm of chromosome 1 and the long arm of chromosome 19 (1p/19q co-deletion) which further prolong survival.4,5 While other genes play a role in response to treatment and outcome,4–6IDH1/2 mutation and 1p/19q co-deletion are hallmark genetic features in the 2016 CNS WHO and “trump” discordant histological assessment.2
As with many other cancers,1,7–10 the incidence of GBMs and non-GBMs is higher in males than females. In GBMs the incidence rate is 1.6 times higher in males compared to females.1,11 Furthermore, the age-adjusted median survival of males is 17.5 months compared to 20.4 months in females with GBM.3 For patients with non-GBMs in the United States, males also have a higher incidence rate than females, about 1.3 times higher for several types of astrocytomas and oligodendrogliomas,1 although these differences in incidence rates are not mirrored by differences in survival time.3 The sex differences in gliomas are evident in all age groups, ruling out the explanation that they are solely the result of sex hormones.1,10,12
Several studies have shown an association between specific DNA methylation patterns and patient outcome, both in gliomas and in other cancers.13–16 Cancer genomes in general are hypermethylated relative to normal cells, particularly in the promoter regions of protein-coding genes.17–19 A working hypothesis is that hypermethylation in the promoter regions of tumor suppressor genes leads to downregulation of proteins needed to maintain proper growth control. The role of methylation in epigenetic regulation of the genome has focused primarily on cytosine–guanine dinucleotides (CpGs). In general, CpGs appear at low frequency throughout the genome, but are enriched in CpG Islands. CpG islands are defined as regions of the genome of at least 500 base pairs with more than 55% GC content19,20; nearly 70% of annotated gene promoter regions are vested with CpG islands.21 In addition to CpG islands, other parts of the genome have been characterized as shores (flanking regions of up to 2 kb), shelves (2–4 kb regions farther out from the islands), and openseas (yet more distant regions with isolated CpGs).19,20,22 Recent studies have indicated that in addition to methylation of CpG islands, methylation of CpG shores plays a key role in gene regulation.13
The importance of epigenetic methylation in cancer progression is illustrated by the CpG Island Methylator Phenotype (CIMP), a hypermethylation pattern detected in the glioma field and in other cancers.14–16,23,24 In glioma patients, the G-CIMP pattern is most common in younger, non-GBM patients with IDH1/2 mutant, non-co-deleted 1p/19q chromosomes.14,25 Patients with CIMP-positive tumors, “G-CIMP-positive,” have significantly improved survival compared to G-CIMP-negative patients.5,14,15,25
Based on these studies, we hypothesized that epigenetic methylation differences in males and females contribute to the differing rates of overall glioma occurrence, treatment responses, and survival differences among gliomas based on molecular subtype. Thus, we investigated DNA methylation patterns in molecular subtypes of gliomas by sex to determine if any common characteristics could explain the known sex-based differences in gliomas.
Materials and Methods
Patient Data
Clinical and molecular data from tumor resections and biopsies were collected from the NIH Genomic Data Commons for 587 glioma participants from The Cancer Genome Atlas (TCGA) PanCancer Atlas Cohort (https://www.cancer.gov/tcga). The TCGA data were collected prior to the 2016 CNS WHO Classification system and include histologic phenotypes, such as oligoastrocytomas, no longer favored. (The historical histologic types entered into TCGA for each of the 3 non-GBM subtypes are shown in Supplementary Table 1.) For this study, we relied on IDH1/2 mutation and 1p/19q co-deletion status to sort non-GBM glioma samples into 3 groups for consistency with the molecular descriptions of the main glioma categories described in the 2016 CNS WHO revision.2 Data were limited to N = 75 untreated primary IDH1/2 wild-type (IDHwt) GBM patients and N = 512 non-GBM patients for which complete Illumina Human Methylation 450K and clinical data were available. Two GBM patients with IDH1/2 mutations and 3 with unknown IDH1/2 status were excluded from this study due to the small sample size. Ninety-four of the non-GBM were IDHwt, 172 were IDH1/2 mutant and co-deleted for 1p/19q (IDHmut-codel), and 246 were IDH1/2 mutant and did not have the 1p/19q co-deletion (IDHmut-non-codel).
PanCancer TCGA clinical data, DNA methylation level 3 PanCancer TCGA data, and RNA sequencing level 3 PanCancer TCGA are described in the work of Malta et al.26 Previously published updates to TCGA glioma patient survival data were also used.15 The DNA methylation data utilized for this study were normalized and batch corrected level 3 data that underwent preprocessing as part of the rigorous PanCancer TCGA analyses as previously described.26 β-values range from 0 to 1, with 0 indicating no DNA methylation and 1 indicating complete DNA methylation. RNA sequencing level 3 TCGA data have been batch corrected, processed, and normalized as previously published.26
Data Analysis
Data were analyzed using R v3.5.3 by molecular subtype and sex. Clinical data were summarized using the “tableone” package. Survival analyses were performed using the “survival” and “survminer” packages in R. The Kaplan–Meier method was used for generating unadjusted survival curves, and the contribution of age to overall survival was assessed using Cox proportional hazards regression models. In all cases, log-rank P-values <.05 were considered statistically significant.
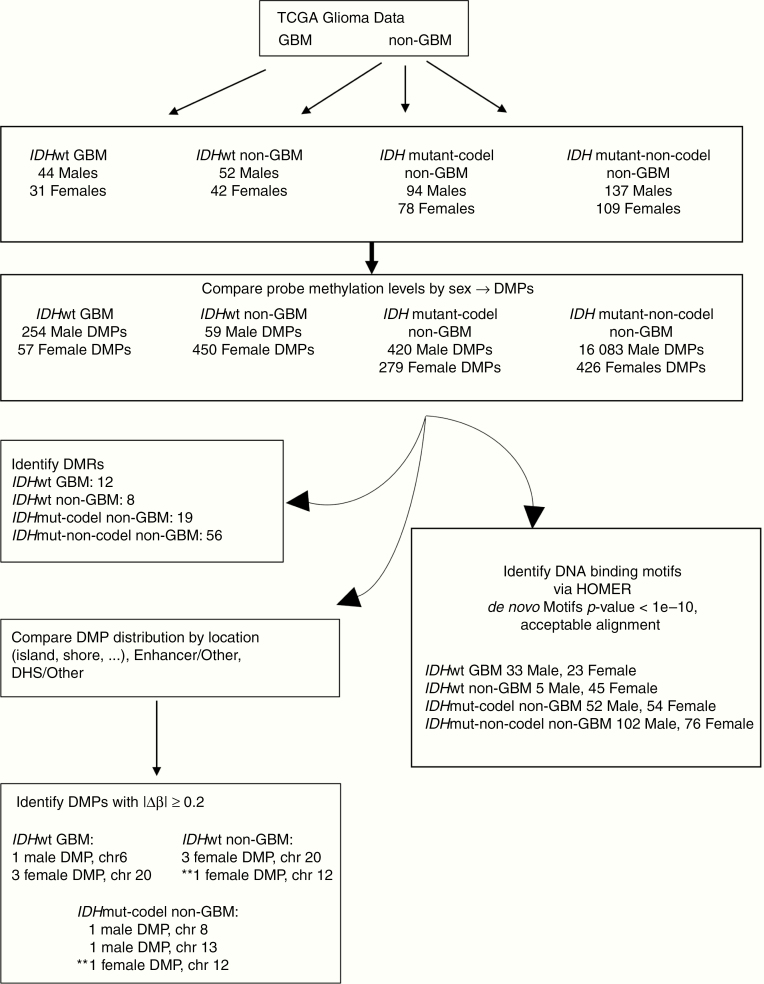
Differentially methylated probes (DMPs) and differentially methylated regions (DMRs) were identified using the R package “ChAMP” v2.12.4.27 DMPs with False Discovery Rate (FDR) adjusted P-values <.05 were considered statistically significant. DMPs hypermethylated in males relative to females are referred to as HyperMale, and DMPs hypermethylated in females compared to males are referred to as HyperFemale. DMRs were identified using the Bumphunter method in ChAMP using the default parameters. ChAMP uses the Benjamini and Hochberg method for FDR estimation to identify differentially methylated sites between males and females. Autosomal probes and regions with FDR corrected P-values <.05 were considered statistically significant. Probes were annotated using the Infinium HumanMethylation450 Bead Chip annotation file, which provides information regarding a probe’s location in known enhancer or DNase I hypersensitive site (DHS) based on experimental data from ENCODE. The reported sensitivity of the Infinium HumanMethylation450 Bead Chip assay is 0.20,20 thus probes with a Δβ magnitude 0.20 or greater were considered unlikely to be false-positives. Hypergeometric Optimization of Motif Enrichment (HOMER) v4.9 (http://homer.ucsd.edu/homer/) was employed to search for known and de novo DNA binding sequences (8–20 bp motifs) with the perl script findMotifGenome.pl using the following criteria: hg19 genome, 200 bp upstream and downstream from each CpG site, and with expected genome-wide distribution of 450K probes as background. HOMER results for “de novo” motifs with P-values <1e−10 were compared to previously published versions to determine those that represented differentially methylated motifs. HOMER input files and raw outputs reported in this paper can be found at https://data.mendeley.com/datasets/rm588t26dp/1. Venn diagrams were generated using an online tool available at http://bioinformatics.psb.ugent.be/webtools/Venn/. Figure 1 is a schematic of the analysis workflow.
Figure 1.
Summary of workflow and key results. Glioma data were obtained from TCGA and divided as indicated into 4 subtypes for analysis. Methylation levels at CpG probes in males and females were compared and those with adjusted P-values <.05 were considered significant. Probes hypermethylated in males compared to females are referred to as “HyperMale DMPs” while probes hypermethylated in females relative to males were termed “HyperFemale DMPs.” Some probes, including some annotated to genes, were found in all 4 glioma subtypes, while others were unique to a given subtype. Significant DMPs in each subtype were compared to each other in terms of distribution throughout the genome (island, shore, shelf, opensea), correspondence to enhancer regions, and localization to DHSs. DMP locations were also used to identify differentially methylated regions throughout the genomes of each subtype. DMPs with |Δβ| ≥ 0.20 were further examined. ** Denotes DMP on chromosome 12 shared by IDHwt non-GBM and IDHmut-codel non-GBM subtypes. Potential transcription factor binding motifs were determined using HOMER. Those de novo motifs with P-values <1e−10 and that align well with known binding sites for a given transcription factor merit further investigation. DHS, DNase I hypersensitive site; DMP, differentially methylated probes; TCGA, The Cancer Genome Atlas.
Results
Samples and Clinical Data
Clinical data are summarized in Table 1. After adjusting for age, median overall survival times were significantly longer in the IDHwt GBM females (N = 31; 8.3 months, 95% CI: 6.6–15.3) compared to IDHwt GBM males (N = 44; 5.4 months, 95% CI 4.7–12.0) (log-rank P-value = .01). Non-GBM patients were divided into 3 groups based on IDH1/2 mutation and 1p/19q co-deletion status. Female IDHwt non-GBM patients (N = 42) had an age-adjusted median overall survival of 21.2 months (95% CI 16.8–60.0) versus 21.0 months (95% CI 16.4–27.2) for IDHwt non-GBM males (N = 52, Table 1). Although the age-adjusted median overall survival times were similar for these 2 groups, survival rates diverge at later times resulting in significantly different age-adjusted overall survival rates (log-rank P-value = .01) (Supplementary Figure 1). One hundred seventy-two patients had an IDH1/2 mutation and were co-deleted for 1p/19q (IDHmut-codel). Due to the low number of deaths during the follow-up period, an age-adjusted median overall survival time could not be determined for either IDHmut-codel females (N = 78) or males (N = 94) (Table 1, Supplementary Figure 1). The largest group of non-GBMs (N = 246) were IDHmut and did not have the 1p/19q co-deletion (IDHmut-non-codel). Female IDHmut-non-codel non-GBM patients (N = 109) had an age-adjusted median overall survival of 60 months (95% CI 60–NA) compared to 368 months (95% CI 60–NA) for males (N = 137) in this subtype. Inspection of age-adjusted Kaplan–Meier overall survival plots revealed male and female survival plots crossing multiple times throughout the course of the disease (Supplementary Figure 1). Despite the apparent difference in median overall survival, there was no significant sex-specific survival difference between female and male IDHmut-non-codel non-GBM patients (log-rank P-value = .41) in this subtype (Supplementary Figure 1).
Table 1.
Summary of Patient Data by Glioma Molecular Subtype and Sex
| Female | Male | P-value | |||
|---|---|---|---|---|---|
| IDHwt GBM | N | 31 | 44 | ||
| Mean age in yrs (SD) | 65.26 (11.89) | 63.00 (8.75) | .35a | ||
| Age range | 39–85 | 47–83 | |||
| Adjusted median overall survivalc (months) [95% CI] | 8.3 [6.6–15.3] | 5.4 [4.7–12.0] | .01b | ||
| Living (%) | No | 31 (100.00) | 44 (100.00) | NA | |
| IDHwt non-GBM | N | 42 | 52 | ||
| Mean age in yrs (SD) | 49.18 (14.31) | 53.83 (14.89) | .14a | ||
| Age range | 24–74 | 21–87 | |||
| Adjusted median overall survivalc (months) [95% CI] | 21.2 [16.8–60.0] | 21.0 [16.4–27.2] | .01b | ||
| Living (%) | No | 22 (52.4) | 32 (61.5) | .50d | |
| Yes | 20 (47.6) | 20 (38.5) | |||
| IDHmut-codel non-GBM | N | 78 | 94 | ||
| Mean age in yrs (SD) | 46.44 (13.06) | 44.12 (12.50) | .24a | ||
| Age range | 20–74 | 17–75 | |||
| Adjusted median overall survivalc (months) [95% CI] | NA [NA–NA] | NA [68.4–NA] | <.01b | ||
| Living (%) | No | 7 (9.0) | 10 (10.6) | .91d | |
| Yes | 71 (91.0) | 84 (89.4) | |||
| IDHmut-non-codel non- GBM | N | 109 | 137 | ||
| Mean age in yrs (SD) | 38.65 (10.95) | 37.23 (10.88) | .31a | ||
| Age range | 18–73 | 14–70 | |||
| Adjusted median overall survivalc (months) [95% CI] | 60 [60–NA] | 368 [60–NA] | .40b | ||
| Living (%) | No | 22 (20.2) | 21 (15.3) | .41d | |
| Yes | 87 (79.8) | 116 (84.7) |
Two IDHwt GBM patients, 1 male and 1 female, were missing 1p/19q co-deletion status data. Age data were missing for 3 females and 4 males in the IDHwt non-GBM subtype.
a P-value from t test.
b P-value from log-rank test.
cAdjusted for by age.
d P-value from chi-square test.
DMPs by Glioma Molecular Subtype and Sex
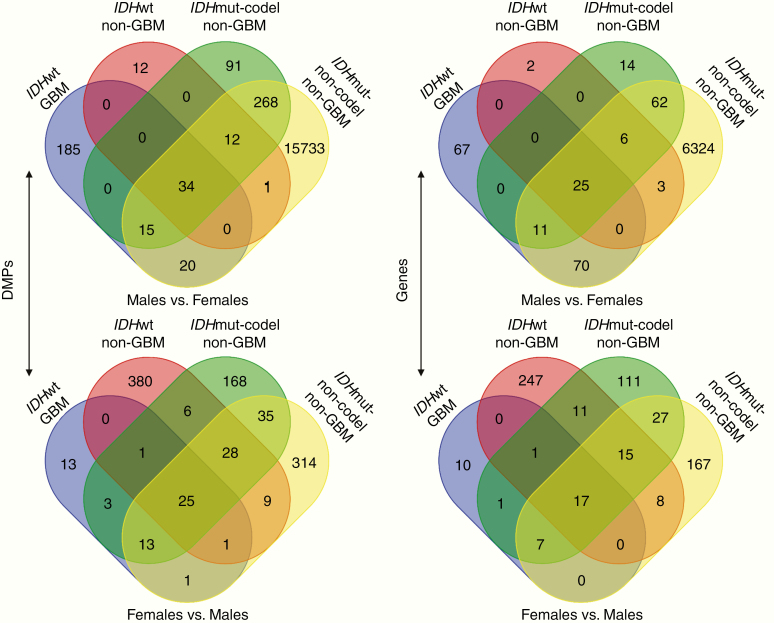
The number of DMPs varied for each glioma subtype (Figure 2). In IDHwt GBMs, 254 DMPs were hypermethylated in males compared to females (HyperMale DMP) while only 57 DMPs were hypermethylated in females relative to males (HyperFemale DMP). In non-GBMs, 59 HyperMale DMPs and 450 HyperFemale DMPs were found in the IDHwt subtype, 420 HyperMale DMPs and 279 HyperFemale DMPs were found in the IDHmut-codel subtype, and 16 083 HyperMale DMPs and 426 HyperFemale DMPs were found in the IDHmut-non-codel subtype (Supplementary File 1). The IDHmut-non-codel tumors were notable for an extremely large number of HyperMale DMPs. Of the 16 083 HyperMale DMPs found in IDHmut-non-codel patients, more than 80% had │Δβ│ values less than 0.05 which may indicate increased heterogeneity for these patients relative to the other 3 groups.
Figure 2.
Intersection of differentially methylated probes (DMPs) and genes in each of the 4 glioma subtypes. DMPs hypermethylated in males relative to females (top left) and DMPs hypermethylated in females relative to males (bottom left). All probes with a significant FDR adjusted P-value (<.05) were compared to determine those shared between different glioma subtypes and those unique to a particular subtype. Distribution of common and unique genes assigned to DMPs hypermethylated in males relative to females (top right) or in females relative to males (bottom right).
The majority of DMPs hypermethylated in males relative to females were unique to a particular glioma subtype, although 34 probes were significantly hypermethylated in males in all 4 glioma subtypes (Figure 2). Similarly in females, each subtype was characterized by a primarily unique set of probes hypermethylated in females compared to males, but all 4 glioma subtypes shared 25 probes in common (Figure 2). Using the Illumina 450K probe annotation package, we found 25 genes that were associated with the DMPs hypermethylated in males and 17 genes associated with DMPs hypermethylated in females in all 4 glioma subtypes (Supplementary Table 2).28 The genes associated with HyperMale DMPs in all glioma subtypes were enriched for cell cycle phase transition genes (TFPD1, RAD21, TUBB4A, ATAD5, and FOXN3). Genes associated with HyperFemale DMPs common to all glioma subtypes were enriched for transcriptional regulators (TLE1, POUF3F2, CDK6, ARID1B, PRDM4, POLDIP3, and DACH1).
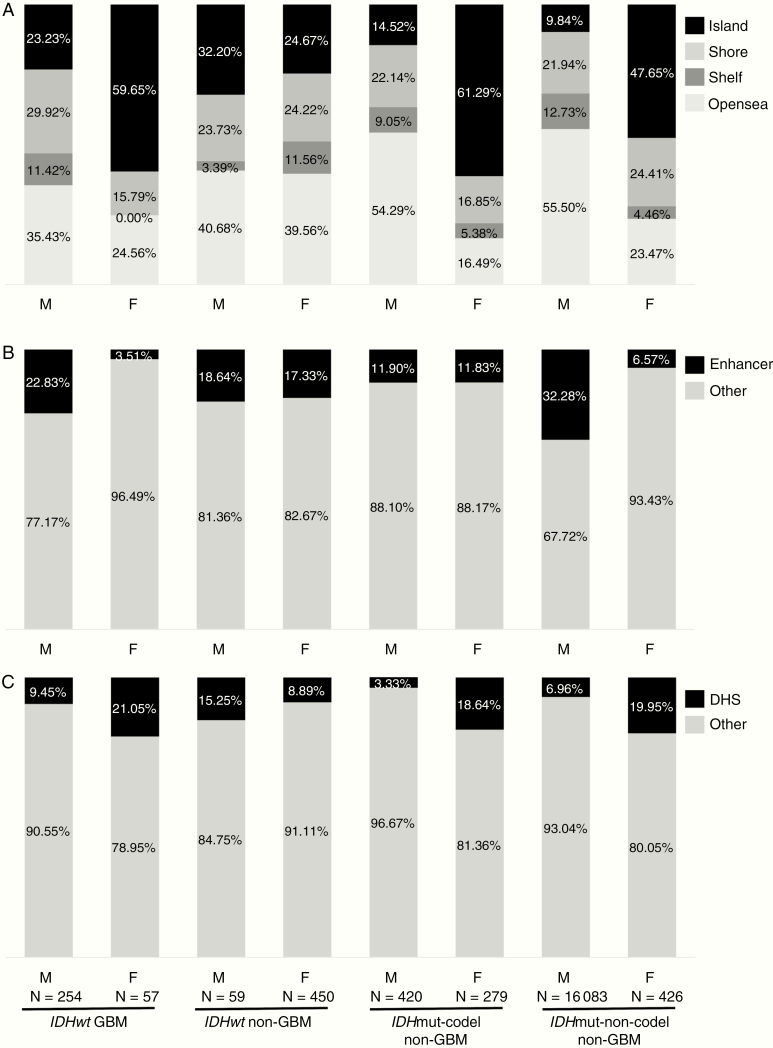
We also assessed DMPs for location in the genome (Figure 3). DMPs hypermethylated in females were primarily localized to CpG islands across the glioma subtypes: 60% in IDHwt GBM, 25% in IDHwt non-GBMs, 61% in IDHmut-codel non-GBMs, and 48% in IDHmut-non-codel non-GBMs. In contrast, probes differentially methylated in males were primarily located in openseas regions (>4 kb from CpG islands): 35% in IDHwt GBM, 41% in IDHwt non-GBMs, 54% in IDHmut-codel non-GBMs, and 56% in IDHmut-non-codel non-GBMs. We assessed also the number of DMPs mapped to known enhancer regions and to DNase I hypersensitive sites (DHS) to probe potential functional consequences of sex-specific differential methylation (Figure 3). Across all glioma subtypes, a higher percentage of probes hypermethylated in males were located in enhancer regions compared to probes hypermethylated in females. In contrast, with the exception of IDHwt non-GBMs, probes hypermethylated in females mapped to known DHS at a higher rate than those hypermethylated in males.
Figure 3.
Distribution of DMPs hypermethylated in males relative to females (“M”), and in females relative to males (“F”) in the 4 glioma subtypes. (A) Distribution of probes characterized as belonging to island, shore, shelf, and opensea regions in the genome. (B) Percentages of probes mapped to enhancer regions or elsewhere in the genome. (C) Percentages of probes corresponding to DNase I hypersensitive sites (DHS) or elsewhere in the genome. The number of DMPs in each category is indicated.
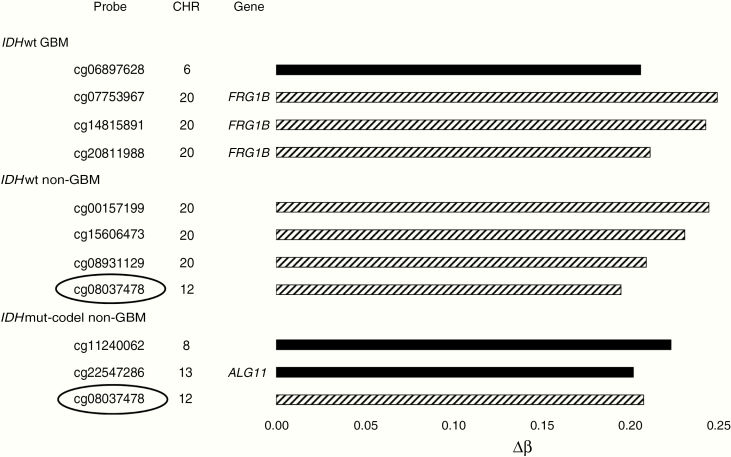
Several DMPs were identified where methylation differed at least 20% between males and females (|Δβ| ≥ 0.2) (Figure 4, Supplementary Table 3). Four DMPs met this criterion in the IDHwt GBM and non-GBM subtypes, and 3 DMPs were found in the IDHmut-codel subtype. None of the DMPs detected in the IDHmut-non-codel category differed by this magnitude. The 3 HyperFemale DMPs in the IDHwt GBM group all mapped to the pseudogene FRG1B on chromosome 20, including 1 (cg07753967) located within 1500 bases of the putative transcriptional start site. The male IDHwt GBM DMP, cg06897628, was located in an opensea on chromosome 6 at position 28762544. Three additional HyperFemale DMPs were found in the IDHwt non-GBMs on chromosome 20, along with 1 HyperFemale DMP, cg08037478, on chromosome 12. The IDHmut-codel non-GBMs had 2 notable HyperMale DMPs, 1 on chromosome 8 and 1 on chromosome 13 which maps to the 3′UTR of a gene for alpha-1,2-mannosyltransferase (ALG11). In addition, IDHmut-codel non-GBMs share the HyperFemale DMP cg08037478 with IDHwt non-GBMs.
Figure 4.
DMPs found for each glioma subtype that differ by a magnitude of 0.2 or more. Solid bars indicate probes hypermethylated in males relative to females. Hatched bars indicate probes hypermethylated in females relative to males. The chromosomal location along with available information regarding correlated genes is shown for each. One probe, cg08037478, was common to both the IDHwt non-GBM and IDHmut-codel non-GBM subtypes (circled). DMP, differentially methylated probe; GBM, glioblastoma.
DMRs by Glioma Molecular Subtype and Sex
We considered the overall genomic context of methylation in each subtype by examining DMRs. DMRs ranging in length from 37 to over 1500 bases were found throughout the genome among the 4 subtypes (Supplementary File 2). The number of DMRs detected for the 4 subtypes included 12 DMRs on 8 different chromosomes for IDHwt GBMs, 8 DMRs across 6 different chromosomes for IDHwt non-GBM, 19 DMRs over 10 different chromosomes in the IDHmut-codel non-GBM, and 56 DMRs spanning 18 chromosomes in the IDHmut-non-codel non-GBM group (Supplementary File 2). While the functional consequences of the identified DMRs cannot be ascertained without additional transcriptomic data analysis, they provide further evidence that each glioma subtype is distinct. Most DMRs were unique to a particular glioma subtype but DMRs common to 2 or 3 subtypes were identified. For instance, IDHwt GBMs, IDHmut-codel non-GBMs, and IDHmut-non-codel non-GBMs all shared a DMR on chromosome 6 that lies in close proximity to 3 closely related members of the heat shock protein 70 family (Hsp70): HSPA1A, HSPA1B, and HSPA1L (Supplementary File 2). IDHwt GBM and IDHwt non-GBMs shared a DMR in common on chromosome 13 that coincides with the middle of a noncoding RNA, RNF219-AS1. DMRs common to both IDHmut-non-codel and IDHwt non-GBMs were found on chromosome 2, mapping to the PAX8 gene, and on chromosome 6 corresponding to pseudogene RP11-373N24.2 (Supplementary File 2). Both IDHmut non-GBM subtypes also shared a DMR on chromosome 20, overlapping a central portion of non-protein coding gene FRG1B.
In addition to genes mentioned above, the DMRs specific to the IDHwt GBM subtype mapped to FAM163A, neuroblastoma-derived secretory protein, on chromosome 1 and 3 separate members of the zinc finger domain containing family: ZDHHC20, ZSCAN1, and ZNF135 on chromosomes 13, 19, and 19, respectively (Supplementary File 2). Two microRNAs, MIR96 and MIR183, previously found to be upregulated in glioma,29 were in close proximity to a DMR on chromosome 7. MIR183 is reported to regulate apoptosis in several cancers including IDHmut glioma,30–32 although the role for MIR96 is less clear. β2-Spectrin, a protein encoded by the SPTBN1 gene on chromosome 2 near another GBM DMR, is a component of TGF-β signaling pathways that serves as a transcriptional cofactor and adaptor protein. Recent studies have suggested a role for β2-Spectrin in DNA repair.33
Analysis of Motifs by Glioma Molecular Subtype and Sex
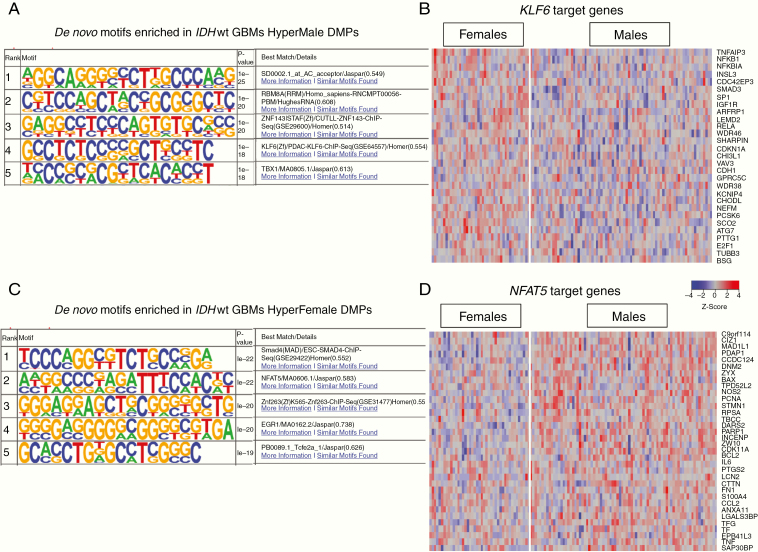
In-depth motif analysis was performed to identify sequence-specific transcription factor (TF) binding sites near DMPs. We first identified binding motifs significantly enriched around HyperFemale DMPs (https://data.mendeley.com/datasets/rm588t26dp/1, Figure 5). Across all glioma subtypes, the motif signature corresponding to the RNA polymerase II apparatus was enriched (geometric test, P < 1e−10) near HyperFemale DMPs. Across all non-GBMs, the motif signature for the TF E2F1 binding site was significantly enriched (geometric test, P < 1e−11) in areas proximal to HyperFemale DMPs. E2F1 plays a crucial role in control of cell cycle and the action of tumor suppressor proteins.34 The motif associated with TF NRF1 was enriched in both IDHwt non-GBMs and IDHwt GBMs (geometric test, P < 1e−12). NRF1 activates the expression of key metabolic genes that regulate cellular growth and nuclear genes required for mitochondrial DNA transcription and replication.35 Finally, we found a number of motifs significantly enriched in HyperFemale DMPs from IDHwt GBM patients only. These motifs included those associated with the TF NFAT5 (geometric test, P < 1e−22), SMAD4 (geometric test, P < 1e−22), FOXH1 (geometric test, P < 1e−12), proto-oncogene SPI1 (geometric test, P < 1e−15), and CEBP (geometric test, P < 1e−12). NFAT5 plays a critical role in integrin-induced cell migration, SMAD4 and FOXH1 are downstream mediators of TGF-β signaling and regulate TGF-β target gene transcription, and CEBP plays a role in immune and inflammatory response signaling.36,37
Figure 5.
Binding motifs and downstream targets for KLF6 and NFAT5, 2 GBM motifs identified by HOMER. (A) Top 5 binding motifs identified by HOMER based on HyperMale DMPs in IDHwt GBM. (B) Transcriptional differences in KLF6 targets for female and male IDHwt GBM patients. (C) Top 5 binding motifs identified by HOMER based on HyperFemale DMPs in IDHwt GBM. (D) Transcriptional differences in NFAT5 targets for female and male IDHwt GBM patients. DMP, differentially methylated probes; GBM, glioblastoma.
We next identified binding motifs significantly enriched near HyperMale DMPs (https://data.mendeley.com/datasets/rm588t26dp/1, Figure 5). No binding motifs shared by all 4 glioma or non-GBM subtypes were found. Motifs associated with TF p53, encoded by TP53, (geometric test, P < 1e−15) and TCF7 (geometric test, P < 1e−18) were significantly enriched in locations near HyperMale DMPs in IDHmut non-GBMs. In IDHwt GBMs, a number of significantly enriched motifs were identified close to HyperMale DMPs. These motifs included the binding sequence associated with KLF6 (geometric test, P < 1e−18) and MAC-1/ITGAM (geometric test, P < 1e−11). KLF6 has been implicated as a tumor suppressor in multiple cancers and regulates p21 signaling. MAC-1/ITGAM encodes the leukocyte integrin subunit α M/CD11b that has been identified as an important regulator of apoptosis especially during brain development via the production of microglial superoxide ions.38,39
We explored the potential effects of TF binding sites impacted by methylation in GBMs by examining the downstream target genes of KLF6 and NFAT5. KLF6 target genes (ATG7, CDKN1A, and SMAD3), involved in apoptotic signaling, were significantly downregulated in male IDHwt GBM patients relative to females (Figure 5). Conversely, the target genes of NFAT5 (CTTN and DNM2 which are involved in cell migration) were significantly downregulated in female as compared to male GBM patients (Figure 5). These results point to a protective effect in females via hypermethylation of NFAT5 binding sites downregulating cell migration genes. In males, however, hypermethylation of the binding sites for KLF6 appears to a deleterious effect via the downregulation of proapoptotic genes.
Discussion
Recent efforts in glioma research have focused on understanding the epigenetic mechanisms that lead to gliomagenesis and that may influence treatment response.5,11,14,15 As with many cancers, males appear more prone to develop both low- and high-grade gliomas, and with GBM, the median survival times in males are significantly shorter than in females.1,11 This sex-based bias has been appreciated for some time, with sex-based differences in cell cycle regulation, mitotic checkpoint signaling, and metabolism previously described.7,40 There is growing interest in understanding the biological mechanisms that can explain these sex differences. For gliomas, sex-based differences occur throughout life, suggesting that sex hormones alone cannot account for these differences.1,11 Using data available from TCGA, we analyzed sex-based differential methylation in all glioma molecular subtypes. This analysis represents the first large-scale analysis of DNA methylation in gliomas where separate but parallel analyses were performed on male and female samples. This approach is critical to uncovering molecular sex-based differences that influence incidence, treatment, and outcome.
We have shown previously that sex-specific responses to the loss of p53 function make male astrocytes more vulnerable to malignant transformation relative to female astrocytes.41 We have also shown using magnetic resonance imaging analysis that standard treatment of temozolomide plus concurrent radiation is more effective in female as compared to male GBM patients.7 Additionally, our previously performed parallel analysis of transcriptomic data from male and female GBM patients revealed sex differences that corresponded to survival.7 In particular we showed that survival in male and female GBM patients may be dependent on different mechanisms with better outcome in females associated with reduced integrin signaling and better outcome in males associated with lower activity in factors that promote cell cycle progression.
Here we have extended our previous work to examine the epigenetic differences between male and female glioma patients, and how those differences impact downstream gene expression. We found that across glioma subtypes the DMPs hypermethylated in females were primarily located in CpG islands, which are enriched in gene promoter regions. Additionally, DMPs hypermethylated in females mapped to known DHS at a higher rate than those hypermethylated in males. DNA methylation in gene promoters and at DHS is associated with lower transcription.42 Taken together these findings indicate an overall downregulation of transcription in female glioma patients as compared to males. In contrast, we found that DMPs hypermethylated in males were primarily located in openseas regions far from gene promoters. We also found a higher percentage of methylation in gene enhancers in males as compared to females. The impact of methylation in enhancer regions is not fully understood.42 These results support our previous work that identified different, sex-specific transcriptomic mechanisms linked to survival in GBM patients.
In addition to examining individual CpGs for differential methylation, we determined whether each glioma subtype possessed genomic regions with sex-specific differential methylation that could lead to differences in gene expression. Determining the functional consequences of these DMRs requires additional study, but they do suggest intriguing candidate genes to include in future investigations. Interestingly, each molecular subtype was characterized by a distinct set of DMRs, although some DMRs were shared by one or more glioma subtype. The paired box domain-containing TF PAX8 on chromosome 2 mapped to DMRs detected in both the IDHwt non-GBM and IDHmut-non-codel non-GBM subtype groups. Previous studies in glioma biopsies and cell lines have shown that PAX8 can regulate telomerase activity.43 Our results suggest that, in these 2 molecular subtypes, telomerase activity might be regulated differently in males and females and that telomerase-directed therapeutic strategies are likely to benefit either male or female patients, but not both sexes. Perhaps more intriguing than PAX8 is the cluster of 3 Hsp70 family members, HSPA1A, HSPA1B, and HSPA1L, that lie in close proximity to a DMR on chromosome 6 shared by the IDHwt GBM, IDHmut-codel, and IDHmut-non-codel molecular subtypes. Several groups have found Hsp70 proteins to be highly expressed in high-grade gliomas,44,45 where they may aid tumor survival by facilitating resistance to radiation46 or chemotherapy.47 Our results indicate that targeting this chaperone may benefit one sex preferentially.
The sex-specific survival disparity is well known in GBMs so we focused on identifying possible DNA binding motifs associated with dysregulated methylation that may explain downstream transcriptomic differences. We found the binding motif for the TF NFAT5 significantly enriched in areas associated with DMPs hypermethylated in females. NFAT5 is a member of the Rel family of TFs, a family that also includes NF-κB. Acting through the aquaporin-4 (AQP4) channel expressed in astrocytes, NFAT5 plays an important role in maintaining osmotic balance in the brain.48,49 Consistent with our previous studies implicating reduced integrin signaling with improved survival in female GBM patients, downstream targets of NFAT5 include genes for cytoskeletal proteins that mediate integrin-induced migration. Two of these target genes, CTTN and DNM2, were downregulated in female GBM patients relative to male GBM patients. This finding supports our hypothesis that the relative hypermethylation in females of the NFAT5 binding site reduces expression of pro-migratory genes and confers a survival advantage to female GBM patients. In males, we found hypermethylation of TF binding sites such as KLF6 and the downregulation of the proapoptotic KLF6 targets ATG7 and CDKN1A.
These results are compelling and represent the first un-merged sex-based analysis of glioma methylation data. However, several limitations should be noted. First, the sample sizes are small and we may not have accounted for all molecular heterogeneity within each glioma subtype, due in part to the availability of molecular features data. In addition, there are no significantly sized and publically available independent verification datasets to computationally validate this work. Finally, detecting DMPs and DMRs using 450K data has limitations due to its reduced CpG representation as compared to whole genome bisulfite sequencing. This can result in false-positive DMRs in the areas in which the 450K array is sparse. We have mitigated this by using Bumphunter which does not call DMRs where CpG coverage is sparse, while this reduces the likelihood of false positives as compared to other methods (ProbeLasso and DMRcate) we have likely missed some true DMRs. Future work includes validation of our epigenetic findings in an independent set of patients and assessing the impact of histological subtypes on epigenetic differences. As part of this study, we identified sex-specific DMRs and differentially methylated TF binding motifs. To fully evaluate the functional consequences of these DMRs and motifs, it will be necessary to assess transcript levels of the implicated RNAs as well as subsequent protein and cellular effects. Nonetheless, this work has identified many avenues to pursue to better understand sex-specific differences in glioma biology.
Funding
This study was supported by pilot funding from the Case Comprehensive Cancer Center [NCI P30CA0437093], the Computational Genomic Epidemiology of Cancer (COGEC) Training Program Fellowship [NCI 5T32CA094186-17 to L.S.], and NCI R01CA174737 to J.B.R. This work made use of the High-Performance Computing Resources in the Core Facility for Advanced Research Computing at Case Western Reserve University.
Conflict of interest statement. There are no conflicts of interest to report.
Authorship Statement:
Experimental design: L.C.S, J.R.C., J.L., J.B.R., M.E.B., and J.S.B-S. Implementation: L.C.S., J.S.B-S., J.B.R., and M.E.B. Data analysis: M.L.J., L.C.S., and V.V. Interpretation: M.L.J., L.C.S, V.V., K.W., J.R.C., J.L., J.B.R., M.E.B., and J.S.B-S. Manuscript draft and revision: M.L.J., L.C.S., V.V., K.W., J.R.C., J.L., J.B.R., M.E.B., and J.S.B-S. Funding: J.S.B-S.
Supplementary Material
Acknowledgments
The authors are grateful to the glioma patients who contributed to the TCGA study.
References
- 1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl 4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Gittleman H, Ostrom QT, Stetson LC, et al. Sex is an important prognostic factor for glioblastoma but not for nonglioblastoma. Neurooncol Pract. 2019;6(6):451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang W, Warrington NM, Taylor SJ, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med. 2019;11(473):eaao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan Y, Liu L, Chen H, et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell. 2016;29(5):711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Radkiewicz C, Johansson ALV, Dickman PW, Lambe M, Edgren G. Sex differences in cancer risk and survival: a Swedish cohort study. Eur J Cancer. 2017;84:130–140. [DOI] [PubMed] [Google Scholar]
- 10. Dorak MT, Karpuzoglu E. Gender differences in cancer susceptibility: an inadequately addressed issue. Front Genet. 2012;3:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thakkar JP, Dolecek TA, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23(10):1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun T, Plutynski A, Ward S, Rubin JB. An integrative view on sex differences in brain tumors. Cell Mol Life Sci. 2015;72(17):3323–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noushmehr H, Weisenberger DJ, Diefes K, et al. ; Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Souza CF, Sabedot TS, Malta TM, et al. A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 2018;23(2):637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kretzmer H, Bernhart SH, Wang W, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. [DOI] [PubMed] [Google Scholar]
- 18. Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014;30(2):75–84. [DOI] [PubMed] [Google Scholar]
- 20. Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. [DOI] [PubMed] [Google Scholar]
- 21. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. [DOI] [PubMed] [Google Scholar]
- 23. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller BF, Sanchez-Vega F, Elnitski L. The emergence of pan-cancer CIMP and its elusive interpretation. Biomolecules. 2016;6(4):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malta TM, de Souza CF, Sabedot TS, et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20(5):608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malta TM, Sokolov A, Gentles AJ, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338–354.e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Butcher L. Illumina450ProbeVariants.db: Annotation Package combining variant data from 1000 Genomes Project for Illumina Human Methylation 450 Bead Chip probes. R package version 1.22.0. 2019. [Google Scholar]
- 29. Lavon I, Zrihan D, Granit A, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12(5):422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bian W, Zhang H, Tang M, et al. Potential role of microRNA-183 as a tumor suppressor in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(5):2065–2072. [DOI] [PubMed] [Google Scholar]
- 31. Zhou J, Zhang C, Zhou B, Jiang D. miR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int J Mol Med. 2019;43(4):1734–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Pusch S, Innes J, et al. Mutant IDH sensitizes gliomas to endoplasmic reticulum stress and triggers apoptosis via miR-183-mediated inhibition of semaphorin 3E. Cancer Res. 2019;79(19):4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horikoshi N, Pandita RK, Mujoo K, et al. β2-spectrin depletion impairs DNA damage repair. Oncotarget. 2016;7(23):33557–33570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72(1):13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang C, Li Z, Lu Y, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103(31):11567–11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hata A, Chen YG. TGF-beta signaling from receptors to smads. Cold Spring Harb Perspect Biol. 2016;8(9):a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jauliac S, López-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4(7):540–544. [DOI] [PubMed] [Google Scholar]
- 38. Narla G, Kremer-Tal S, Matsumoto N, et al. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26(30):4428–4434. [DOI] [PubMed] [Google Scholar]
- 39. Reid DM, Perry VH, Andersson PB, Gordon S. Mitosis and apoptosis of microglia in vivo induced by an anti-CR3 antibody which crosses the blood-brain barrier. Neuroscience. 1993;56(3):529–533. [DOI] [PubMed] [Google Scholar]
- 40. Kfoury N, Sun T, Yu K, et al. Cooperative p16 and p21 action protects female astrocytes from transformation. Acta Neuropathol Commun. 2018;6(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun T, Warrington NM, Luo J, et al. Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J Clin Invest. 2014;124(9):4123–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sharifi-Zarchi A, Gerovska D, Adachi K, et al. DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism. BMC Genomics. 2017;18(1):964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hung N, Chen YJ, Taha A, et al. Increased paired box transcription factor 8 has a survival function in glioma. BMC Cancer. 2014;14:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beaman GM, Dennison SR, Chatfield LK, Phoenix DA. Reliability of HSP70 (HSPA) expression as a prognostic marker in glioma. Mol Cell Biochem. 2014;393(1–2):301–307. [DOI] [PubMed] [Google Scholar]
- 45. Thorsteinsdottir J, Stangl S, Fu P, et al. Overexpression of cytosolic, plasma membrane bound and extracellular heat shock protein 70 (Hsp70) in primary glioblastomas. J Neurooncol. 2017;135(3):443–452. [DOI] [PubMed] [Google Scholar]
- 46. Brondani Da Rocha A, Regner A, Grivicich I, et al. Radioresistance is associated to increased Hsp70 content in human glioblastoma cell lines. Int J Oncol. 2004;25(3):777–785. [PubMed] [Google Scholar]
- 47. Castro GN, Cayado-Gutiérrez N, Zoppino FC, et al. Effects of temozolomide (TMZ) on the expression and interaction of heat shock proteins (HSPs) and DNA repair proteins in human malignant glioma cells. Cell Stress Chaperones. 2015;20(2):253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang XL, Wang X, Peng BW. NFAT5 has a job in the brain. Dev Neurosci. 2018;40(4):289–300. [DOI] [PubMed] [Google Scholar]
- 49. Yi MH, Lee YS, Kang JW, et al. NFAT5-dependent expression of AQP4 in astrocytes. Cell Mol Neurobiol. 2013;33(2):223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.