Abstract
Tissue clearing and light-sheet microscopy have over a 100-year-long history, yet only recently have these fields been combined to facilitate novel experiments and measurements in neuroscience. Since tissue-clearing methods were first combined with modernized light-sheet microscopy a decade ago, the performance of both technologies have rapidly improved, broadening their applications. Here we review the state-of-the-art of tissue clearing methods and light-sheet microscopy and discuss applications of these techniques in profiling cells and circuits in mice. We examine outstanding challenges and future opportunities for expanding these techniques to achieve brain-wide profiling of cells and circuits in primates and humans. Such integration will help provide a systems level understanding of physiology and pathology of our central nervous system.
Tissue clearing and light-sheet microscopy form powerful synergies for neuroscience. In this issue of Neuron, Ueda et al. (2020) review how these technologies enable brain-wide profiling of cells and circuits for a systems-level understanding of physiology and pathology of our nervous system.
Introduction: Brief History of Integration of Tissue Clearing with Light-Sheet Microscopy
The beginning of the 20th century saw the birth of two technologies - tissue clearing and light-sheet microscopy. The earliest report on tissue clearing of opaque biomedical samples appeared in 1911 by the German anatomist, Walter Spalteholz in Leipzig. He tried to make human hearts transparent to study their vascular system (Spalteholz, 1911). Using hydrophobic tissue-clearing reagents (“Wintergrünöl”) such as methyl salicylate and benzyl benzoate on dehydrated specimens, he succeeded in visualizing macroscopic structures in transparent samples for the first time. However, without a technology like light-sheet microscopy, it was not possible to quantify his findings. So even this key advance only brought some qualitative insights into human anatomy. Not so far away from Leipzig, an Austrian chemist, Richard A. Zsigmondy and a German physicist, Henry Siedentopf working in Jena developed the first light-sheet microscope, the “Ultramicroscope” (Siedentopf, 1903). Unlike Spalteholz, Zsigmondy was looking for very small things (“Ultramikronen”), colloidal particles in solution, which he tried to quantify. In principle, it would have been possible to integrate these two technologies over one hundred years ago, but at that time it would not have led very far. Light sheets traversing cleared specimens might create optical sections if one looks at the specimen at the correct angle, but these images still had to be recorded. Most importantly, a three-dimensional model of the specimen must be reconstructed digitally. Without concurrent inventions of electronic cameras and computers, even a hypothetical encounter of Spalteholtz and Zsigmondy would not have produced the same impact that these methods have recently achieved.
Approximately 90 years after these seminal works, the first relevant step towards integration of tissue clearing and light-sheet microscopy was made by Arno Voie and colleagues(Voie et al., 1993) who designed a modern version of light-sheet microscopy (orthogonal-plane fluorescence optical sectioning microscopy; OPFOS), based on lasers and digital camera technology. After clearing the bony structure of the inner ear using EDTA and Spalteholz’s hydrophobic tissue-clearing reagents (methyl salicylate and benzyl benzoate) Voie performed the first fluorescence optical imaging of a tissue-cleared biological specimen, an excised guinea-pig cochlea labeled with fluorescein(Voie et al., 1993). He recorded the images with a CCD camera and successfully reconstructed the spiral in the inner ear with rudimentary homemade 3D reconstruction software. Voie’s approach unfortunately remained largely unnoticed by the broader scientific community for nearly two decades. Light-sheet microscopy rapidly gained momentum in biological imaging in the early 21st century, with applications in diverse fields including microbial oceanography, developmental biology, and neuroscience(Dodt et al., 2007; Fuchs et al., 2002; Holekamp et al., 2008; Huisken et al., 2004). In particular, the work by Ernst Stelzer and colleagues sparked a renewed interest in light-sheet microscopy (Huisken et al., 2004). They reconstructed largely transparent living biological samples, including Medaka fish embryos. Despite the success of this approach for developmental biology, it remained restricted to naturally transparent samples. The first use of light-sheet imaging on neural tissues artificially rendered transparent was made by Hans-Ulrich Dodt and colleagues (Dodt et al., 2007). They took advantage of hydrophobic tissue-clearing reagents, Benzyl Alcohol and Benzyl Benzoate (BABB), that were originally developed by Andrew Murray and Marc Kirschner around 1989 and first applied to fluorescent- and peroxidase-based whole-mount immunocytochemistry of Xenopus oocytes and embryos (Dent et al., 1989). Combining this clearing method with ultramicroscopy and image processing enabled the visualization of neuronal networks at resolution of neural dendrites in whole mouse brains (Dodt et al., 2007). A different set of organic solvents - DBE (dibenzyl ether) and/or THF (tetrahydrofuran) and dichloromethane (DCM) - were combined in 3DISCO to better preserve the fluorescence of GFP during the clearing process (Becker et al., 2014; Becker et al., 2012; Ertürk et al., 2012; Ertürk et al., 2014). Since then, various other hydrophobic tissue-clearing reagents with different attributes have been developed to support various applications, including ultimate DISCO (uDISCO) (Pan et al., 2016), immunolabeling-enabled DISCO (iDISCO) (Belle et al., 2014; Renier et al., 2014), iDISCO+ (Renier et al., 2016), FluoClearBABB (Schwarz et al., 2015), Ethyl Cinnamate (ECi) (Klingberg et al., 2017), Free-of-Acrylamide SDS-based Tissue Clearing (FASTClear) (Perbellini et al., 2017), PolyEthylene Glycol (PEG)-associated solvent system (PEGASOS) (Jing et al., 2018), variable domain of heavy chain antibodies (nanobodies) DISCO (vDISCO) (Cai et al., 2018), Multiscale Architectonic Staining of Human cortex (MASH) (Hildebrand et al., 2018), and stabilised DISCO (sDISCO) (Hahn et al., 2019).
Clearing methods that use water-soluble reagents (hydrophilic tissue-clearing methods) are better at preserving the fluorescence of fluorescent proteins and are also less toxic compared to approaches that use organic reagents. Pioneering works on hydrophilic tissue-clearing methods were conducted by a Russian Physicist, Valery V. Tuchin and colleagues (Bakutkin et al., 1995; Tuchin and Tuchin, 2007). They found that aqueous solutions of a variety of hydrophilic chemicals had high refractive indices (RI), a property believed to be important for tissue clarity. The RI of a certain material is defined as the ratio of the speed of light in a vacuum to that in the material. These high-RI solutions included x-ray contrast agents (Trazograph), a series of alcohols (polyethylene glycol, glycerol, propylene glycol), sugars (glucose and dextrans), and dimethyl sulfoxide (DMSO)(Bakutkin et al., 1995; Bashkatov et al., 1999; Tuchin et al., 1999; Tuchin et al., 1997; Tuchin et al., 2002; Xu et al., 2003; Zimnyakov et al., 1996). Around the same time, Chance and colleagues discovered the RI matching effect of a series of sugars (mannitol, fructose, sucrose and glucose) and alcohols (propanediol and methanol) (Chance et al., 1995; Liu et al., 1996). A different cocktail -FocusClear, which contains another x-ray contrast agent (diatrizoate acid) and a detergent (Tween 20) was used by Chiang and colleagues for whole-brain imaging of cockroach using a confocal microscope (Chiang et al., 2001). In 2011, Atsushi Miyawaki and colleagues developed hydrophilic tissue-clearing methods Scale, which hyperhydrates and delipidates mouse brains with urea-based reagents and a detergent, respectively, resulting in semi-transparent mouse brains (Hama et al., 2011). This clearing approach substantially enhanced the effective imaging depth of two-photon microscopy in adult brain tissue but provided insufficient clearing for effective light-sheet microscopy. This problem was then addressed in 2014 by Hiroki Ueda and colleagues, who developed Clear, Unobstructed Brain/Body Imaging Cocktails and Computational analysis (CUBIC), an efficient hydrophilic tissue-clearing method (Susaki et al., 2014). They used a systematic chemical screening strategy and found a series of aminoalcohols, which can be used to delipidate and decolorize mammalian brains. CUBIC protocols for whole-body clearing also seem to permit light sheet imaging of heart, lung, liver, kidney, pancreas and other organs (Susaki et al., 2014; Susaki et al., 2015; Tainaka et al., 2014) although whole-body imaging still has not been rigorously demonstrated because of the lack of an objective definition of “whole-body imaging”. One of the possible definitions for whole-body/organ imaging would be the successful detection of cells or cellular nuclei in a whole body or a whole organ with more than a certain level of accuracy (e.g. 95% accuracy).
Further chemical screening and profiling led to the development of a series of CUBIC reagents (CUBIC-L or CUBIC-HL for delipidation, CUBIC-R+ for RI-matching, CUBIC-B for decalcification of bone, CUBIC-P for decolorization and CUBIC-X for tissue expansion)(Kubota et al., 2017; Murakami et al., 2018; Tainaka et al., 2018). Parellel efforts revealed the chemical principles of each tissue-clearing processes(Susaki and Ueda, 2016; Tainaka et al., 2016; Tainaka et al., 2018). CUBIC-based clearing was also extended to the fruit fly D.melanogaster. With this approach (FlyClear) endogenous fluorescence in whole undissected animals was preserved across developmental stages from larva to the adult fly (Pende et al., 2018). In addition to these CUBIC-related reagents, other hydrophilic tissue-clearing reagents with different purposes have been independently developed, including thiodiethanol (TDE) immersion (Aoyagi et al., 2015; Costantini et al., 2015; Hasegawa et al., 2016; Staudt et al., 2007); See Deep Brain (SeeDB) (Ke et al., 2013); ScaleS (sorbitol-based Scale) (Hama et al., 2015); FRUIT (a cocktail of fructose and urea) (Hou et al., 2015); Urea-Based Amino-Sugar Mixture (UBasM) (Chen et al., 2017); clearing-enhanced 3D (Ce3D) (Li et al., 2017); See Deep Brain 2 (SeeDB2) (Ke et al., 2016); Clear T (formamide) / Clear T2(a cocktail of formamide and polyethylene glycol) (Kuwajima et al., 2013); Warner’s method (Warner et al., 2014); ClearSee (Kurihara et al., 2015); Rapid clearing method based on Triethanolamine and Formamide (RTF) (Yu et al., 2018); ilLUminate Cleared organs to IDentify target molecules (LUCID) (Mizutani et al., 2018); a series of sugars, sorbitol (Hirshburg et al., 2007) and sucrose (Chance et al., 1995; Tsai et al., 2009); and a series of X-ray contrast, Histodenz in Refractive Index Matched solution (RIMS) protocol (Yang et al., 2014) and iodixanol in System-Wide control of Interaction Time and kinetics of CHemicals (SWITCH) protocol (Murray et al., 2015).
In parallel with the development of hydrophobic and hydrophilic tissue-clearing methods, a hydrogel-based tissue-clearing method called Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/immunostaining/in situ-hybridization-compatible Tissue hYdrogel (CLARITY) was also developed in 2013 by Karl Deisseroth and Kwanghun Chung. In this process, lipids are removed by perfusion of sodium dodecyl sulfate (SDS), a strong detergent and tissue is subsequently transformed into a clear acrylamide gel retaining biological elements (Chung et al., 2013). CLARITY employs an electrophoresis step to accelerate tissue clearing and is therefore more complex than many of the other hydrophobic and hydrophilic tissue-clearing methods that rely on passive diffusion. Following SDS perfusion, proteins and nucleic acids are retained, presumably due to their stabilization by the exogenous acrylamide gel. Using CLARITY, it may be possible to visualize endogenous fluorescent proteins and to label epitopes with fluorescent antibodies (Chung et al., 2013). Although reliable fluorescence preservation and immunolabeling remains challenging, intact CLARITY-processed brains have been successfully imaged at high resolution (NA = 1.0) with light-sheet microscopy(Tomer et al., 2014). Tomer and colleagues developed Passive CLARITY Technique (PACT) by decreasing gel density to improve tissue permeability and probe penetration (Tomer et al., 2014). Gradinaru and colleagues combined PACT with PARS (Perfusion-assisted Agent Release in situ) to render rodent bodies transparent (Treweek et al., 2015; Yang et al., 2014). Other variations of CLARITY have been applied to bones (Greenbaum et al., 2017a) and for the detection of RNA (Greenbaum et al., 2017b; Yang et al., 2014). Sensitive methods for fluorescence in situ hybridization have been used to effectively visualize single RNA molecules within tissue (Shah et al., 2016).
Hydrogel-based methods have been further exploited to achieve super-resolution imaging. Expansion Microscopy, developed by Ed Boyden and colleagues uses a swellable hydrogel to isotropically expand tissues. Expanded structures within tissue may enables imaging at effective resolution higher than that achievable using conventional microscopy (Chen et al., 2015). Chung and colleagues developed another hydrogel-based expansion microscopy method, Magnified Analysis of Proteome (MAP), in which they omitted the protein digestion process to preserve proteins in the expanded brain (Ku et al., 2016), and recently introduced Stabilization under Harsh conditions via Intramolecular Epoxide Linkages to prevent Degradation (SHIELD), in which they used polyepoxy chemicals to protect florescence, antigenicity, transcripts, and tissue architecture (Park et al., 2018). Similar to hydrophobic and hydrophilic tissue-clearing methods, variations of hydrogel-based tissue-clearing reagents with different purposes have been developed, including optimized CLARITY (Lee et al., 2014), CLARITY-TDE (Costantini et al., 2015), Plant-Enzyme-Assisted (PEA)-CLARITY (Palmer et al., 2015), simplified CLARITY (Lai et al., 2016), Active Clarity Technique-Pressure Related Efficient and Stable Transfer of macromolecules into organs (ACT-PREST) (Lee et al., 2016), and Fast Free-of-Acrylamide Clearing Tissue (FACT) (Xu et al., 2017).
Since the earliest combined use of hydrophobic(Dodt et al., 2007), hydrophilic(Susaki et al., 2014) or hydrogel-based(Tomer et al., 2014) tissue-clearing methods and modernized light-sheet microscopy, the performance of both tissue-clearing approaches and light-sheet microscopy have been continuously improved, broadening their utility(Ariel, 2017; Azaripour et al., 2016; Chakraborty et al., 2019; Fu et al., 2016; Gradinaru et al., 2018; Hörl et al., 2018; Migliori et al., 2018; Richardson and Lichtman, 2015; Silvestri et al., 2016; Susaki and Ueda, 2016; Tainaka et al., 2016; Voigt et al., 2019)(Figure 1). Indeed, advanced tissue-labeling/staining techniques, tissue-clearing methods and light-sheet microscopy, whole-brain profiling of cells can be performed in a comprehensive, efficient manner (Belle et al., 2014; Murakami et al., 2018; Renier et al., 2016; Renier et al., 2014; Susaki et al., 2014; Ye et al., 2016). Integration of tissue-clearing and light-sheet microscopy also promises to transform methods for interrogating the structure and connectivity of neural circuits spanning the brain (Ogawa and Watabe-Uchida, 2018) (Economo et al., 2016; Economo et al., 2018; Winnubst et al., 2019).
Figure 1.
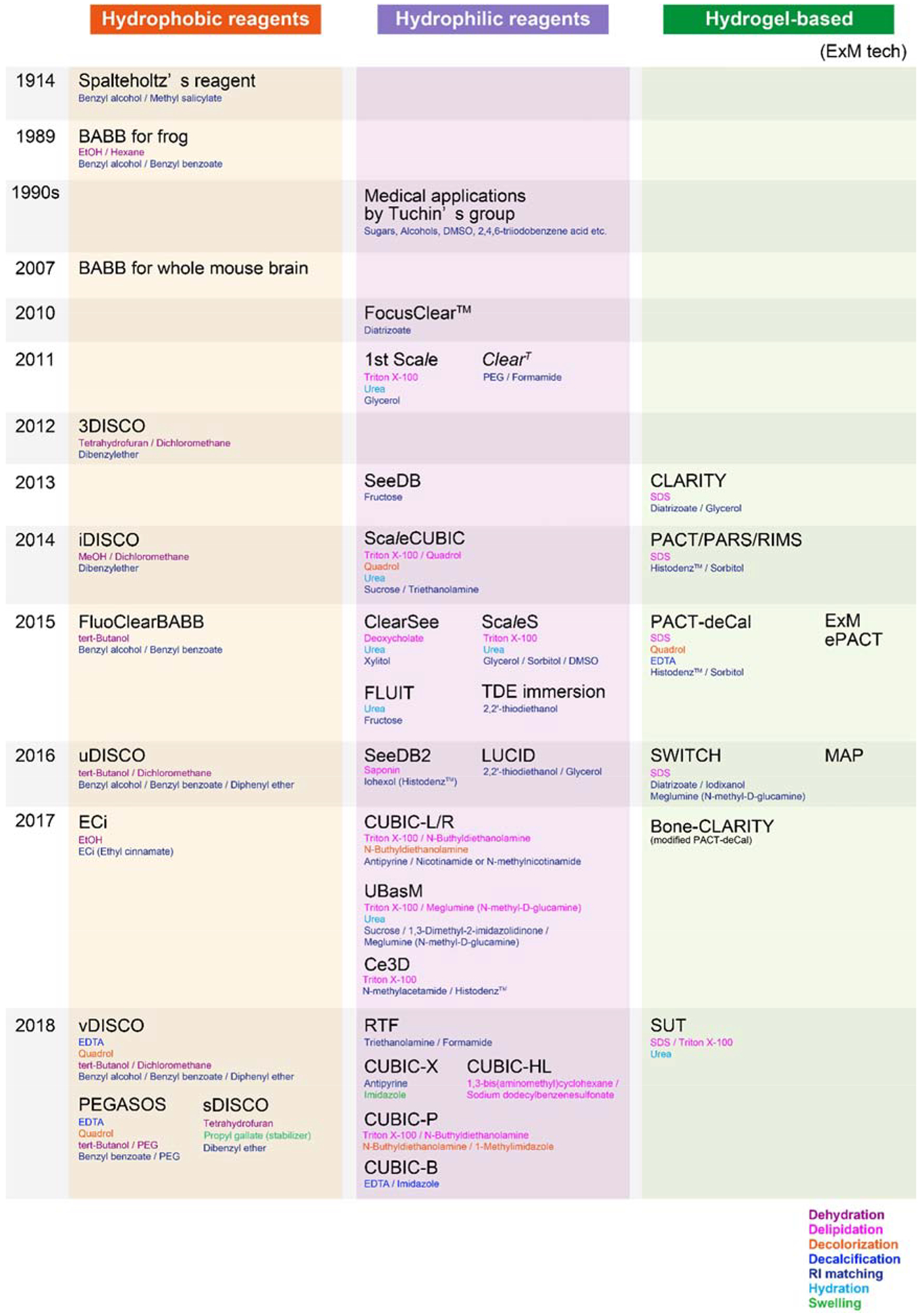
Timeline of Advances in Tissue Clearing Methods.
The Present State of the Art and Challenges in Tissue Clearing
Tissue-clearing methods provide a powerful approach for optical imaging deep within biological specimens. Methods for tissue clearing are under active development so that they may be applied to larger samples and to improve their compatibility with an expanded set of techniques for labeling and visualizing structural and biochemical features of tissues. The underlying physical and chemical principles that underlie tissue clearing processes are also of great interest (Figure 2) and enhanced understanding of these processes continues to drive the development of new tissue-clearing methods.
Figure 2. Physical and Chemical Principles of Tissue Clearing.
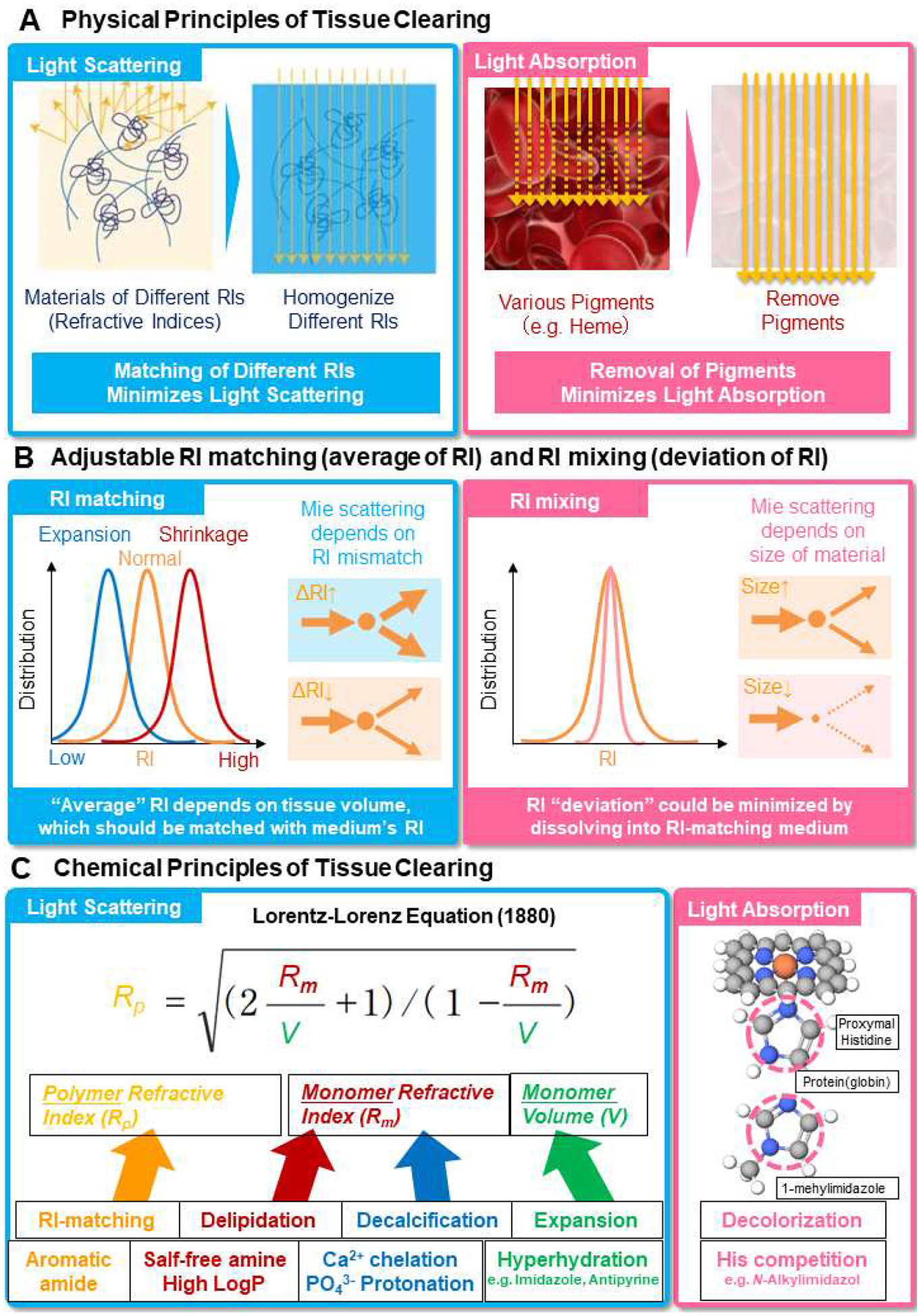
(A) Classical Physical principles of tissue clearing proposed in 1911 by Spalteholz. Light scattering can be minimized by homogenization of RIs of materials whereas light absorption can be minimized by removing pigments. (B) adjustable RI matching (matching the tissue-size-dependent average RI of biological samples with RI-matching medium) and RI mixing (dissolving light-scattering materials into RI-matching medium to minimize RI deviation) proposed in this review. The “average” RI of biological samples depends on the size of tissues, which often expands in hydrophilic and hydrogel-based tissue-clearing methods and shrinks in hydrophobic tissue-clearing methods, and should be matched by RI-matching medium because Mie scattering depends on the RI mismatch between those of light-scattering materials and medium. For adjustable setpoint of average RI, see also Lorentz-Lorenz equation in Figure 2C. RI-deviation problem could be solved by “RI-mixing” by dissolving light-scattering materials (i.e. materials of deviation of RIs from the average RI) into the RI-matching medium since light scattering since Mie scattering depends also on the “size” of light-scattering materials. (C) Chemical principles of tissue clearing. Delipidation, decalcification and expansion processes contribute to the composite RI of biological samples (calculated by Lorentz-Lorenz Equation), which should be matched by RI-matching medium. Decolorization of heme can be achieved by competitive binding of 1-methylimidazole or other amines to iron-containing heme instead of histidine in globin. Each tissue-clearing chemical process are associated with characteristic chemical nature of tissue-clearing chemicals.
Improvements in tissue clearing methods have relied upon both the identification and application of more effective tissue-clearing chemicals (Pan et al., 2016; Tainaka et al., 2014; Tainaka et al., 2018) and enhanced fixation procedures that permit the use of harsher reagents and procedures (e.g. strong detergents and high temperature)(Murray et al., 2015; Park et al., 2019; Yang et al., 2014). The identified potent tissue-clearing chemicals or conditions have been applied not only to entire bodies of rodents (Pan et al., 2016; Tainaka et al., 2014; Yang et al., 2014) but also to entire human organs including a lymph node(Nojima et al., 2017), a kidney(Tainaka et al., 2018; Zhao et al., 2019), an eye ball and even a human brain(Zhao et al., 2019).
Tissue clearing methods, whether hydrophobic, hydrophilic and hydrogel-based, attempt to minimize light scattering – especially Mie scattering(Tuchin, 2015) – caused by mismatches in the RIs of different components of biological materials (e.g. lipids and proteins) with the RI of the medium (e.g. water) (Figure 2A, Light scattering). Among biological materials, water usually exhibits the lowest RI (~1.33), dried lipids exhibit intermediate RIs (1.46–1.48)(Kienle et al., 2014; Pusterla et al., 2017), dried proteins and DNA exhibit higher RIs (1.540–1.598 for dried proteins(Bashkatov et al., 2018) and ~1.58 for dried DNA (Inagaki et al., 1974), respectively) while hydroxyapatite extracted from bone tissue exhibits the highest RI (1.600–1.604)(Antonio, 1949; Ascenzi and Fabry, 1959). In addition to reducing light scattering caused by inhomogeneity of RIs, tissue-clearing methods also minimize light absorption caused by pigments such as heme (Figure 2A, Light absorption).
Practically, most tissue-clearing methods aim to preserve proteins and/or nucleic acids while removing other components of tissue. Effective tissue-clearing methods usually achieve the following five chemical processes; 1) removing lipids (“delipidation”) and 2) hydroxyapatite (“decalcification”) to reduce the inhomogeneity of RIs in biological samples, 3) removing pigments (“decolorization”) to reduce light absorption, and 4) embedding in a hydrophobic solvent or aqueous medium with an RI matching the remaining components of the tissue. In addition, 5) expansion of biological samples sometimes contributes to RI matching (Figure 2B, RI matching). Interestingly, the RIs of RI-matching media differ across hydrophilic (1.52 for CUBIC-R+), hydrogel-based (1.45–1.49 for RIMS), and hydrophobic (1.56 for 3DISCO) methods, likely reflecting commensurate sample expansion and shrinkage (Kubota et al., 2017; Tainaka et al., 2016; Tainaka et al., 2018).
The optimal RIs of different tissue-clearing methods have been explained by considering a physical property of natural biological tissue itself (Murakami et al., 2018). As predicted from a pioneering study on cross-linked gels of extracted natural polymers such as DNA (polynucleotide), protein (polypeptide) and agarose (polysaccharide) by Toyoichi Tanaka and colleague in 1987(Amiya and Tanaka, 1987), a recent study has revealed that fixed and delipidated natural organs behave as an electrolyte polymer gel even without exogenous polymers(Murakami et al., 2018). If a biological tissue acts as a polymer gel, the average RI (Rp) of biological tissue (“polymer gel”) could be described by average RI (Rm) and volume Vm of its components (“monomer”) according to the Lorentz-Lorenz equation (Figure 2C, Light scattering) (L. Lorenz, 1880; Lorentz, 1880), where the average RI of biological tissue Rp is an increasing function of Rm/Vm. Therefore, the expansion and shrinkage of biological tissues lead to the decrease and increase of the average RIs, respectively, which should be matched by RI-matching mediums (Figure 2B, RI matching).
As described above, the “average” RI of biological tissues can be matched by RI-matching medium. However, there is still a remaining issue in delipidated, decalcificated and decolorized biological samples (i.e. cross-linked gels of natural polymers such as proteins and nucleic acids) of the localized deviation of the RIs of these polymers from the average RI of the sample (and hence from the RI-matching medium). One possible solution to address this RI-deviation problem would be “RI-mixing” to dissolve such light-scattering materials (i.e. materials of deviation of RIs from the average RI) into the RI-matching medium since light scattering (Mie scattering) is dependent not only on RI mismatch but also on the “size” of light-scattering materials (Figure 2B, RI mixing). If the effective size of light-scattering materials (e.g. proteins with a μm-scale structure) could become sufficiently smaller than the wavelength of visible lights (0.38–0.75 μm) by dissolving them into the medium, the RIs of materials can be mixed together with those of medium and/or other dissolved light-scattering materials nearby within the wavelength of visible lights. Hence, this “RI-mixing” process would effectively cancel out RI deviation of different light-scattering materials. Importantly, pursuits of RI-matching reagents have already identified a couple of chemicals that might have this RI-mixing property. For example, chemical screening of more than 1,600 hydrophilic chemicals identified effective RI-matching reagents (Tainaka et al., 2018), some of which (e.g. imidazole and antipyrine) also exhibit strong interaction with cross-linked natural polymers such as a gelatin gel(Murakami et al., 2018). Interestingly, known RI-matching reagents (e.g. DMSO) can interact strongly with cross-linked natural polymers even without water, implying that this “RI-mixing” concept might be applicable to hydrophobic reagents. A unified understanding of the physical properties of biological materials and chemical properties of RI-matching and RI-mixing reagents will lead to the efficient engineering of average and deviation of RIs (“RI engineering”) in biological tissues to achieve optimal transparency of larger samples. We also note that the RI-mixing concept built upon Mie scattering theory and our model of RI matching through the Lorentz-Lorenz equation, both of which are proposed in this review, might represent substantial conceptual advancements in the physical principles of tissue clearing since Walter Spalteholz first proposed RI-matching concept in 1911.
In addition to physical principles of RI-matching and RI-mixing in biological tissues, the chemical understanding of other tissue-clearing processes such as delipidation, decolorization and decalcification have also advanced (Figure 2C, Light scattering). Delipidation removes lipids to reduce the inhomogeneity of RIs in biological samples, and also allows other reagents to diffuse deep into biological samples. Therefore, delipidation has already become a common component of hydrophobic, hydrophilic and hydrogel-based tissue-clearing methods. In hydrophobic tissue-clearing methods such as BABB, 3DISCO, uDISCO iDISCO+, and sDISCO, polar solvents including tetrahydrofuran (THF) (3DISCO) (Becker et al., 2014; Becker et al., 2012; Ertürk et al., 2012; Ertürk et al., 2014; Hahn et al., 2019) and alcohols such as ethanol (BABB) (Dent et al., 1989), tert-butanol (FluoBABB, uDISCO)(Pan et al., 2016; Schwarz et al., 2015) and methanol (iDISCO+)(Belle et al., 2014; Renier et al., 2016; Renier et al., 2014) are used as the first step to remove water and partially remove lipids from biological samples. As a second step, hexane (BABB)(Dent et al., 1989) and dichloromethane (DCM) (3DISCO, uDISCO and iDISCO+)(Becker et al., 2014; Becker et al., 2012; Ertürk et al., 2012; Ertürk et al., 2014) are used to extensively remove lipids. In hydrophilic tissue-clearing methods, the nonionic detergent Triton X-100 (Scale, CUBIC-1)(Hama et al., 2011; Susaki et al., 2014; Susaki and Ueda, 2016) and aminoalchohols, including Quadrol (CUBIC-1)(Susaki et al., 2014; Susaki and Ueda, 2016), triethanolamine (CUBIC-2)(Susaki et al., 2014; Susaki and Ueda, 2016) and N-Butyldiethanolamine (CUBIC-L)(Kubota et al., 2017; Tainaka et al., 2018), are used as potent delipidation chemicals. In hydrogel-based tissue-clearing methods such as CLARITY and PACT, the ionic detergent Sodium dodecyl sulfate (SDS) (Chung et al., 2013; Treweek et al., 2015; Yang et al., 2014) is mostly used for delipidation. Improved delipidation procedures have led to the development of more efficient tissue-clearing protocols. The ionic detergent sodium dodecylbenzenesulfonate (CUBIC-HL) (Tainaka et al., 2018), non-ionic detergent 3-[(3-CHolamidopropyl)dimethylAmmonio]-1-PropaneSulfonate (CHAPS) (Zhao et al., 2019), aliphatic amines such as 1,3-bis(aminomethyl)cyclohexane (CUBIC-HL) (Tainaka et al., 2018), and hexanediol (Inoue et al., 2019; Tainaka et al., 2018) were shown to be effective reagents for delipidation of human tissue. Because there is a trade-off between the strength of tissue clearing, especially delipidation, and the preservation of molecular and cellular organization in tissues, we should carefully choose the stringency of tissue clearing according to the required transparency for the imaging of the samples.
Decolorization removes pigments to reduce light absorption in biological samples. Heme in red blood cells can be eluted by aminoalcohols such as Quadrol in mild chemical conditions(Tainaka et al., 2014). The hypothesis for decolorization of heme was proposed that a basic nitrogen in aminoalcohols may competitively bind to iron-containing heme instead of histidine in globin (Figure 2C, Light absorption) (Tainaka et al., 2014; Tainaka et al., 2016). This competitive binding hypothesis was strongly supported by the identification of a more potent decolorization compound, 1-methylimidazole, from more than 1,600 chemicals (Tainaka et al., 2018), which almost completely mimics the functional residue of histidine. We also note that SDS in CLARITY and PACT also successfully decolorize heme, likely as a result of the denaturation of the heme-hemoglobin holoenzyme by SDS(Epp et al., 2015; Lee et al., 2014; Treweek et al., 2015).
Decalcification removes calcium phosphate to reduce light scattering in biological samples containing bone. The inorganic bone mineral, Hydroxyapatite (HAp), was used for chemical screening of 1,600 reagents by measuring the OD600 of a chemically treated HAp suspension. In addition to EDTA, which has been known as a strong calcium chelator, potent decalcification-enhancing compounds were identified. Imidazole was found to facilitate EDTA-based decalcification, perhaps due to the protonation of phosphate ions(Tainaka et al., 2018). This enhanced decalcification may be the result of both chelation of calcium ions by EDTA and protonation of phosphate ions by Imidazole (Tainaka et al., 2018).
In addition to the empirical findings of efficient tissue-clearing reagents and/or conditions, we note that a more unified understanding of tissue-clearing chemistry has been gradually obtained by chemical profiling of potent reagents (Tainaka et al., 2014; Tainaka et al., 2018). In particular, the chemical profiling of more than 1,600 chemicals have been conducted for delipidation, decolorization, decalcification, tissue expansion and RI-matching, respectively (Murakami et al., 2018; Susaki and Ueda, 2016; Tainaka et al., 2016; Tainaka et al., 2018). Based on these screens, we have hypothesized that delipidation is enhanced by salt-free amines with high logP (octanol-water partition coefficient). Similarly, decolorization appears to be associated with competitive binding to iron-containing heme against histidine by N-Alkylimidazole and decalcification a result of chelation of Ca2+ by EDTA and protonating of PO42− by an organic base. Tissue expansion is facilitated by hyperhydration and RI matching by electron enrichment of an RI-matching medium by an aromatic amide (Figure 2C, Light scattering and Light absorption) (Murakami et al., 2018; Tainaka et al., 2018). Taken together, our advancing understanding of the principles underlying tissue clearing provide a foundation for new approaches suitable for specific biological applications.
Another important challenge in tissue clearing is labelling of tissues with antibody. Since 1980s, tissue clearing followed by whole-mount antibody staining was attempted for comprehensive imaging of whole organs and body, especially for nervous systems of insect and shrimp and the Xenopus embryo(Beltz and Kravitz, 1983; Bishop and O’shea, 1982; Dent et al., 1989). Recently, tissue clearing and staining was further applied to three-dimensional observation of murine and human embryos(Belle et al., 2017; Hsueh et al., 2017; Renier et al., 2014), various animal organs and bodies(Cai et al., 2018; Chung et al., 2013; Coutu et al., 2018; Gleave et al., 2013; Hama et al., 2015; Hasegawa et al., 2019; Kubota et al., 2017; Kumar et al., 2010; Renier et al., 2016; Sillitoe and Hawkes, 2002; Susaki et al., 2014; Tainaka et al., 2014; Yang et al., 2014), and pathological samples of human tissues(Chung et al., 2013; Hildebrand et al., 2018; Lai et al., 2018; Liu et al., 2016; Nojima et al., 2017; Tanaka et al., 2017; Zhao et al., 2019). In order to improve the penetration speed of antibodies in a large tissue sample, several approaches have been adopted. First, stringent permeabilization by several methods have been attempted to increase the pore size of fixed specimens and hence enhance the penetration of antibodies. These methods include 1) delipidation (Chung et al., 2013; Duve et al., 1983; Hama et al., 2015; Lai et al., 2018; Susaki et al., 2014; Yang et al., 2014; Zhao et al., 2019), 2) dehydration(Belle et al., 2014; Dent et al., 1989; Duve et al., 1983; Gleave et al., 2013; Renier et al., 2016; Renier et al., 2014; Sillitoe and Hawkes, 2002; Zhao et al., 2019), 3) weaker fixation(Gleave et al., 2013) and 4) partial protein digestion with proteases(Gleave et al., 2013; Kumar et al., 2010; Sillitoe and Hawkes, 2002). Second, weakening non-specific interactions between antibodies and fixed tissues by using chemicals such as Urea (Hama et al., 2015) or SDS (Murray et al., 2015) have been also attempted to inhibit trapping of antibodies and hence accelerate the diffusion of antibodies deep into the tissues. Third, active transports by electrophoresis (Kim et al., 2015a; Lee et al., 2016; Wang et al., 2018) or pressurization such as trans-cardinal perfusion (Cai et al., 2018; Yang et al., 2014) were also tested on tissue samples to enhance the penetration of antibodies. Finally, increasing the amountsof antibodies by their iterative supplies (Chung et al., 2013; Lai et al., 2018) or reducing the size of antibodies by using nanobodies (Cai et al., 2019) were also effective to increase the penetration rate of antibodies. At present, these attempts to increase the penetration rate of antibody successfully can label relatively small and thin tissues, partially dissected tissues, or embryonic tissues with the lesser extracellular matrix. These attempts also successfully can stain an adult mouse brain or a dissected human specimen with relatively sparse epitopes such as c-Fos, amyloid plaques or a microglia marker (Belle et al., 2014; Liebmann et al., 2016; Renier et al., 2016; Renier et al., 2014; Zhao et al., 2019). However, homogeneous antibody staining of adult mouse brains against high-density epitopes such as NeuN and neurofilament has not yet been adequately demonstrated, and therefore remains to be a challenge in tissue clearing.
The Present State of the Art and Challenges in Light-Sheet Fluorescence Microscopy
Light-sheet fluorescence microscopy is a powerful technique for rapid and minimally phototoxic volumetric imaging of biological specimens. The high imaging speed, high signal-to-noise ratio and high light efficiency of this method are the direct result of the central design principle in light-sheet imaging: a thin volume section of the specimen is selectively illuminated by a sheet of laser light, and fluorescence emitted from within this volume section is imaged onto a camera positioned at a right angle to the light sheet. The illuminated volume section and the detection focal plane are coplanar, and thus (1) no out-of-focus regions are exposed to laser light and (2) an image of the entire, thin volume section can be acquired simultaneously. The first feature ensures that the specimen’s “photon budget” is used efficiently and dramatically reduces photo-bleaching and photo-toxic effects compared to conventional and confocal fluorescence microscopy. The second feature provides high imaging speeds limited only by camera performance. This design facilitates rapid three-dimensional imaging simply by moving the light sheet and detection focal plane through the specimen and acquiring a series of images across the volume. Because of these strengths, light-sheet microscopy has found widespread use throughout the life sciences over the course of the past decade and is employed routinely in the fields of developmental biology (Huisken et al., 2004; Keller et al., 2008; McDole et al., 2018; Rozbicki et al., 2015), cell biology (Chen et al., 2014; Liu et al., 2018; Nixon-Abell et al., 2016; Reichmann et al., 2018; Valm et al., 2017), and neuroscience (Ahrens et al., 2013; Chhetri et al., 2015; Dodt et al., 2007; Holekamp et al., 2008; Kawashima et al., 2016; Lemon et al., 2015; Panier et al., 2013; Wolf et al., 2015; Wan et al., 2019).
While even the most basic implementations of light-sheet microscopy already offer key benefits, advances in microscope design have added further strengths and synergies (Figure 3). These improvements have allowed microscopists to address many commonly encountered challenges in biomedical imaging, including fundamental limitations in spatial resolution, limitations in image quality arising from light scattering and aberrations, and limitations in the rapid imaging of large specimens.
Figure 3.
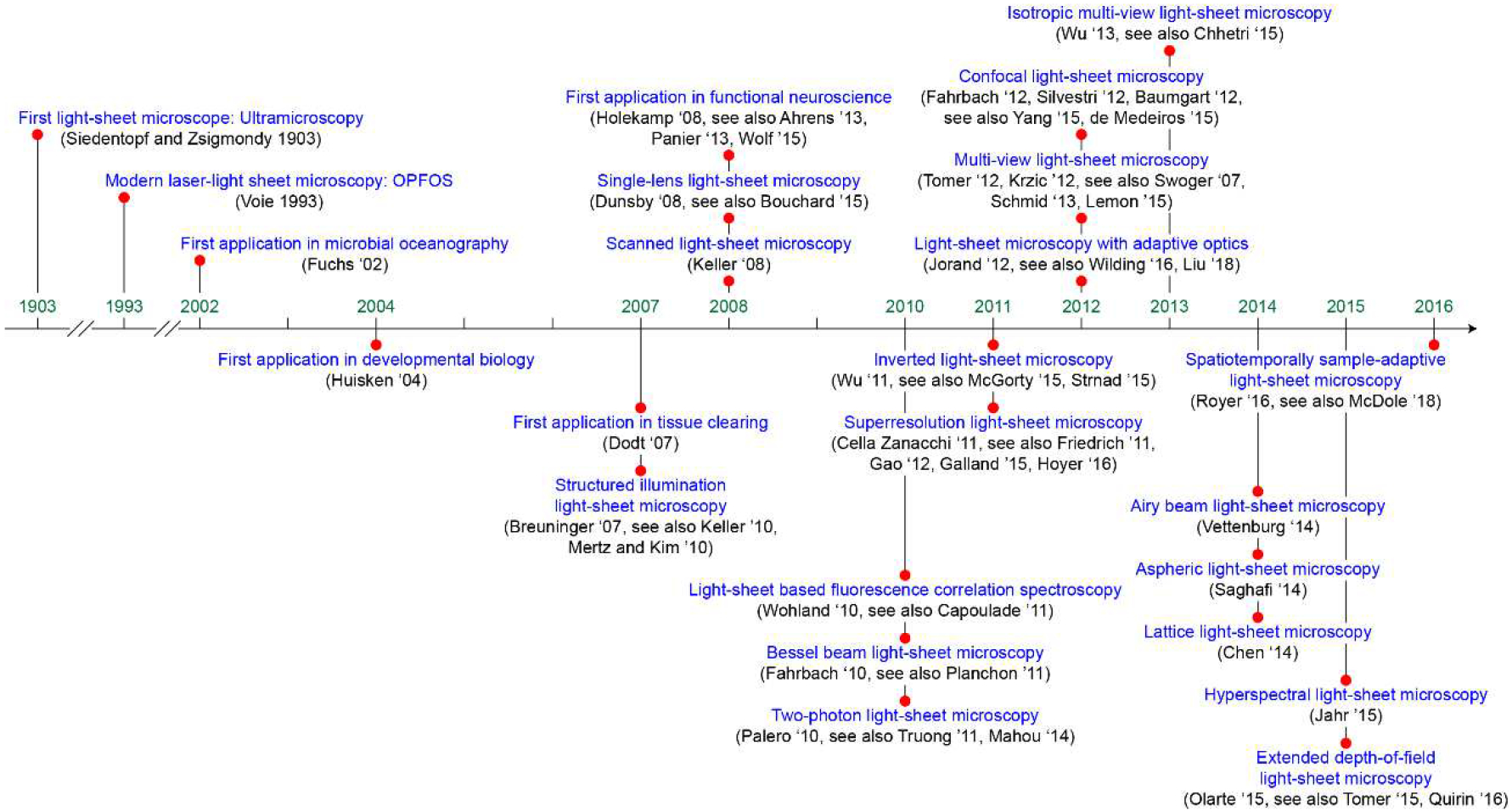
Timeline of Advances in Light-Sheet Microscopy.
Light scattering is a common problem in biological specimens with limited transparency, which includes most living, multi-cellular organisms. By introducing structured illumination (Breuninger et al., 2007; Keller et al., 2010) or line-confocal detection (Baumgart and Kubitscheck, 2012; de Medeiros et al., 2015; Fahrbach and Rohrbach, 2012; Silvestri et al., 2012) in light-sheet microscopy, the contribution of scattered light to image formation can be greatly reduced, thus improving contrast and resolution in deeper regions of the specimen. A complementary strategy for improving depth penetration (and reducing scattering in biological tissues) involves the use of longer-wavelength light for fluorescence excitation and relies on the principle of multi-photon excitation to allow a molecule to enter an excited state through concurrent absorption of multiple lower-energy photons. The use of this concept in light-sheet microscopy was first demonstrated for static light sheets (Palero et al., 2010) and then further improved through integration in beam-scanning light-sheet microscopes (Mahou et al., 2014; Truong et al., 2011). However, even with structured illumination, confocal detection and multi-photon excitation, many biological specimens are still too large to be imaged in high resolution in their entirety, owing primarily to light scattering and absorption. To address this issue, light-sheet microscopy has been further enhanced by multi-view imaging capabilities (Krzic et al., 2012; Lemon et al., 2015; Pende et al., 2018; Schmid et al., 2013; Swoger et al., 2007; Tomer et al., 2012). In such implementations, the specimen is illuminated by more than one light sheet (typically two light sheets from opposite directions)(Huisken and Stainier, 2007) and imaged by more than one camera (typically two cameras from opposing views), which improves physical coverage of large specimens without compromising temporal resolution or temporal continuity across the volume. Finally, scaling up light-sheet imaging to very large transparent specimens is feasible through the use of modified objective arrangements as well as tiling strategies in which sample translation can help overcome the limited field-of-view of the camera and detection optics(Hörl et al., 2018; Migliori et al., 2018; Voigt et al., 2019).
Improvements in spatial resolution have become possible by using beam shaping to create thinner light sheets (Chen et al., 2014; Fahrbach and Rohrbach, 2010; Pende et al., 2018; Planchon et al., 2011; Saghafi et al., 2014; Sheppard, 2013; Vettenburg et al., 2014) as well as by using orthogonal multi-view imaging (Chhetri et al., 2015; Wu et al., 2013). In conventional light-sheet microscopes, axial resolution is typically substantially lower than lateral resolution – often by up to a factor of 5–10 when imaging a large field-of-view. Since axial resolution in the detection system can only be increased up to a limit imposed by the numerical aperture of the detection objective, one option for further improving overall system resolution is the reduction of light sheet thickness. Light sheets constructed from Bessel beams and optical lattices (Chen et al., 2014; Fahrbach and Rohrbach, 2010; Planchon et al., 2011) can be made thin enough to improve overall axial resolution to around 300–400 nm, which is comparable to the lateral resolution limit. However, the field-of-view of such microscopes is typically limited to around 100 μm along the illumination axis and thus other solutions are needed for rapid, high-resolution imaging of larger specimens. By acquiring orthogonal views of a sample, followed by registration and multi-view deconvolution of these multiple views, images with near-isotropic spatial resolution on the order of 300–400 nm can be obtained without the need for constructing thin light sheets (and thus, without the need to limit imaging to a small field-of-view)(Chhetri et al., 2015; Wu et al., 2013). In addition, complementary efforts are underway to investigate the design of light sheets that maintain a narrow beam waist over long spatial distances(Pende et al., 2018; Saghafi et al., 2014). Alternatively, the relationship between size of field-of-view and light sheet waist thickness can be decoupled by tiling/scanning the light sheet waist across the field-of-view(Dean et al., 2015; Fu et al., 2016) or translating the sample relative to the light sheet waist(Migliori et al., 2018), which provides more uniform illumination across the field-of-view.
Although the techniques described above offer high system resolution, the effective spatial resolution in a real biological sample can be substantially lower as a result of optical aberrations. Unfortunately, most biological specimens, even those that are relatively transparent, introduce significant aberrations that alter the path and shape of the light sheet and distort the shape of the detection focal plane. When limiting the observation to sufficiently small regions in the sample, thus ensuring that wavefront errors are relatively uniform across the small field-of-view, light-sheet microscopy can be combined with adaptive optics to efficiently compensate for most, if not all, of these distortions(Jorand et al., 2012; Liu et al., 2018; Wilding et al., 2016). However, conventional adaptive optics are typically unsuited to light-sheet applications that require rapid live imaging of large, optically heterogeneous specimens with a field-of-view exceeding 100 μm. In this latter scenario, geometrical mismatches between light sheets and detection focal planes can be rapidly mapped across the specimen (and over time) by real-time image processing and approximately corrected for through optical defocus and light-sheet tip/tilt manipulations (McDole et al., 2018; Royer et al., 2016). With this complementary approach, diffraction limited performance of the light-sheet microscope can be at least partially recovered in specimens as large and complex as zebrafish, fruit fly or mouse embryos, which typically provides a 2–5 fold improvement in spatial resolution.
Temporal resolution in light-sheet microscopy is generally limited by camera speed. To preserve the high frame rates offered, for example, by state-of-the-art sCMOS cameras in a volumetric imaging setting, the microscope’s detection objective can be moved in synchrony with the light sheet by fast piezo positioners (Ahrens et al., 2013; Lemon et al., 2015). For example, high-performance piezos are capable of moving detection objectives at rates of several tens to hundreds of Hz for travel ranges of up to several hundred μm(Greer and Holy, 2019; Piezosystems_Jena, 2019; Wan et al., 2019). The advantage of this approach is that it ensures optimal image quality by always acquiring images at the native focal plane of the objective; however, the cost of a good piezo is not insignificant. Less expensive alternative solutions include the use of remote focusing with electric tunable lenses (Fahrbach et al., 2013) or extended depth-of-field detection (Olarte et al., 2015; Quirin et al., 2016; Tomer et al., 2015). The reduced cost and ability to keep the objective stationary come at the expense of a reduction in resolution and image quality, which result from imaging away from the native focal plane and, in the latter case, from the use of an elongated detection point-spread function.
Research in recent years has also produced a range of useful, alternative designs of light-sheet microscopy, such as implementations that are compatible with biological preparations on cover slips and other horizontal surfaces (McGorty et al., 2015; Strnad et al., 2016; Wu et al., 2011) or designs that use a single objective for illumination and detection (Bouchard et al., 2015; Dunsby, 2008). The latter concept sacrifices some optical performance by reducing the effective numerical aperture and acquiring images away from the native focal plane, but it also enables light-sheet imaging of samples with limited optical access and supports applications that constrain the placement and orientation of microscope optics.
Whole-Brain Profiling of Cells
Developments in microscopy have helped to bring about a methods-driven renaissance in neuroanatomy that is distinguished by a focus on large-scale projects generating unprecedented amounts of anatomical data. Quantitative, whole-brain profiling of the spatial distribution of cells, their molecular features, and their connectivity represents a powerful application of modernized tissue-clearing and light-sheet microscopy in neuroscience. Comprehensively mapping the distributions of neuronal and glial types across the brain allows brain regions to be delineated with unprecedented precision, and their components to be defined, leading to an enhanced understanding of brain structure and facilitating comparisons across individuals and across species.
Until recent advances in genetic labeling, tissue processing, and light microscopy, comprehensively mapping cells throughout the brain has only been possible in simple organisms, such as the nematode Caenorhabditis elegans (White et al., 1986). The size and complexity of the mammalian brain present barriers to progress in understanding the organization of the nervous system of higher organisms. Accordingly, the classical anatomical literature of the mammalian brain has been piecemeal, with a given study typically examining only one or a few brain regions and one or a few cell types at a time, typically in the male brain. For example, the use of classical stereological methods has led to quantification of interneuron distribution in sensory cortices (Gonchar et al., 2007; Pfeffer et al., 2013; Pronneke et al., 2015; Rudy et al., 2011; Xu et al., 2010), but this approach has proven to be too laborious to be applied to whole-brain cell type analyses (Glaser and Glaser, 2000; Schmitz and Hof, 2005; Williams and Rakic, 1988). A simple method, called isotropic fractionator, was developed for counting cell nuclei in suspension by flow cytometry after tissue dissociation and successfully applied to comparative studies of neuronal and glial cell counts in the mouse brain and across a range of different species (Herculano-Houzel et al., 2015a; Herculano-Houzel et al., 2015b; Herculano-Houzel et al., 2013; Kverkova et al., 2018; Marhounova et al., 2019; Olkowicz et al., 2016). However, since the mammalian brain comprises many brain areas (e.g. mouse brain has >400 unique areas) that would need to be precisely dissected, this method is not appropriate for comprehensive analyses, nor is it easily adaptable for analysis of specific cell types.
Automated block-face imaging methods combining top-view light microscopy and integrated tissue sectioning enable high-quality, high-resolution brain-wide imaging. This approach substantially improved upon histological methods in which thin tissue sections are manually cut, mounted on slides, and imaged individually. The enhanced throughput of automated block-face imaging represents a critical factor for large-scale neuroanatomical projects. Imaging in a block-face configuration also provides enhanced reliability and minimal distortion, producing complete datasets that may be registered to standardized anatomical reference atlases – critical for integrating anatomical data from multiple experiments. A number of block-face imaging methods have been developed. Serial two-photon tomography (STPT) takes advantage of two-photon excitation to provide optical sectioning in standard paraformaldehyde-fixed tissues. STPT can be used to generate a series of two-dimensional images spanning all regions of the brain (Kim et al., 2017; Osten and Margrie, 2013; Ragan et al., 2012) or full three-dimensional image volumes spanning the brain (Economo et al., 2016). Knife-edge scanning microscopy (KESM) (Mayerich et al., 2008) and micro-optical sectioning tomography (fMOST) (Gong et al., 2016; Li et al., 2010; Zheng et al., 2013)can also generate complete, three-dimensional whole-brain imagine volumes using micron-scale sectioning and single-photon imaging of resin-embedded tissue. Block-face serial microscopy tomography (FAST)(Seiriki et al., 2017) uses a spinning-disk confocal to achieve high imaging rates in a block-face configuration.
One application of these methods is brain-wide mapping of genetically defined cell types. For example, Osten and colleagues developed an automated, quantitative brain-wide cell profiling platform (qBrain) that can be used to map the brain-wide distribution of genetically defined cell types across regions in the mouse brain. qBrain combines recombinase-based knock-in “driver” mouse lines that label genetically defined cell types (Huang, 2014; Huang and Zeng, 2013; Madisen et al., 2012), automated brain imaging at single-cell resolution by STPT, and computational analyses that include cell detection by convolutional neural networks (Kim et al., 2015b; Kim et al., 2017; Ragan et al., 2012). The application of this platform to cell type anatomy has immediately demonstrated the power of unbiased whole-brain anatomical mapping in uncovering novel principles of mammalian brain organization. First a comparison of quantitative distribution of three major inhibitory neuron cell types, those expressing somatostatin (SST), parvalbumin (PV) and vasoactive intestinal peptide (VIP), across the mouse brain isocortex revealed a hierarchical organization of the neocortex, as sensory-motor areas were found to be dominated by output-modulating parvalbumin-positive interneurons, whereas association areas comprised large numbers of input-modulating somatostatin-positive interneurons. Another perhaps even more surprising finding from this study was the identification of nine brain regions with different distribution of these cell types between male and female mouse brains, with eight regions containing more VIP+ or SST+ neurons in the female brain, despite the female brain being overall smaller than the male brain. Most regions with sexually dimorphic cell type distributions were from the structures of the vomeronasal system, which regulate reproductive and social behaviors. This finding provides direct, quantitative evidence that anatomical differences underlie sexually dimorphic behaviors (Bayless and Shah, 2016; Simerly, 2002). Given the unexpected results derived from analyses of only 3 cell types by using STPT, it seems likely that further quantitative atlasing of cell type distributions will provide additional insights into how the cellular composition of different brain areas may contribute to their functions.
Block-face imaging methods remain attractive due to their high resolution and compatibility with large tissue samples. However, due to many of the technological advances reviewed here, large-volume imaging using tissue clearing and light sheet microscopy are ‘closing the gap’ and provide several key advantages over block-face methods. Block-face imaging requires specialized instruments that may not be readily accessible to many researchers, while light sheet microscopes are becoming increasingly common fixtures in microscope facilities. Furthermore, light-sheet imaging can achieve acquisition rates for volumetric data that are several orders of magnitude faster than block-face imaging methods. Still, to realize whole-organ cell profiling, clearing methods must achieve high transparency of samples so that the light sheet is not degraded by tissue-induced scattering, aberration, and absorption. In addition, light-sheet imaging methods must maintain consistent quality across large fields of view. Increasingly, these constraints can be met by state of the art of tissue-clearing methods and light-sheet fluorescent microscopy. As a result, this approach is becoming an appealing strategy for whole-brain profiling of cells in intact mammalian brains (Liebmann et al., 2016; Murakami et al., 2018; Renier et al., 2016; Renier et al., 2014; Susaki et al., 2014; Susaki et al., 2015; Sylwestrak et al., 2016; Tatsuki et al., 2016; Tomer et al., 2014). For example, a tissue-clearing/expansion method, CUBIC-X, in combination with custom-made light-sheet fluorescent microscopy with a 10x objective lens (NA = 0.6, working distance = 8 mm), allowed 1.3 million images covering the entire mouse brain to be successfully obtained.
Whole-brain datasets generated by these instruments range from ~100 GB to ~30 TB of data per single mouse brain, necessitating the development of new computational tools for analyses. 3D reconstructions, anatomical registration and signal detection and quantification, for example, have been demonstrated using supervised machine learning algorithms trained on expert annotated ground truth data (Hawrylycz et al., 2011; Kim et al., 2015b; Kuan et al., 2015; Ng et al., 2007; Ragan et al., 2012). In datasets collected using CUBIC-X, GPU-based image analysis was used to extract the three-dimensional coordinates of all cells in the adult mouse brain. Leveraging existing anatomical segmentations of the mouse brain (Dong, 2008; Lein et al., 2007), this approach yielded a whole-brain atlas with single-cell resolution (CUBIC-Atlas) (Murakami et al., 2018). Further development of the cell detection algorithm improved the accuracy and speed of this analysis pipeline so that more than 90 % of cells in the mouse brain could be identified in several hours (up to 2 TB/h) (Matsumoto et al., 2019). Using this cell-nucleus detection algorithm, an updated CUBIC-Atlas 1.2 of 8-week-old C57BL/6J mouse brain was constructed that contains the spatial coordinates and brain regions associated with more than 108 cells (Matsumoto et al., 2019). The improved throughput offered by these protocols allows the analysis of numerous (>100) samples, providing a highly versatile platform for biomedical research, including comparative analyses across a range of mammalian and vertebrate species, and opening a new and exciting frontier in neuroanatomy (Figure 4).
Figure 4. Whole-Brain Profiling of Cells by Light-Sheet Fluorescent Microscopy.
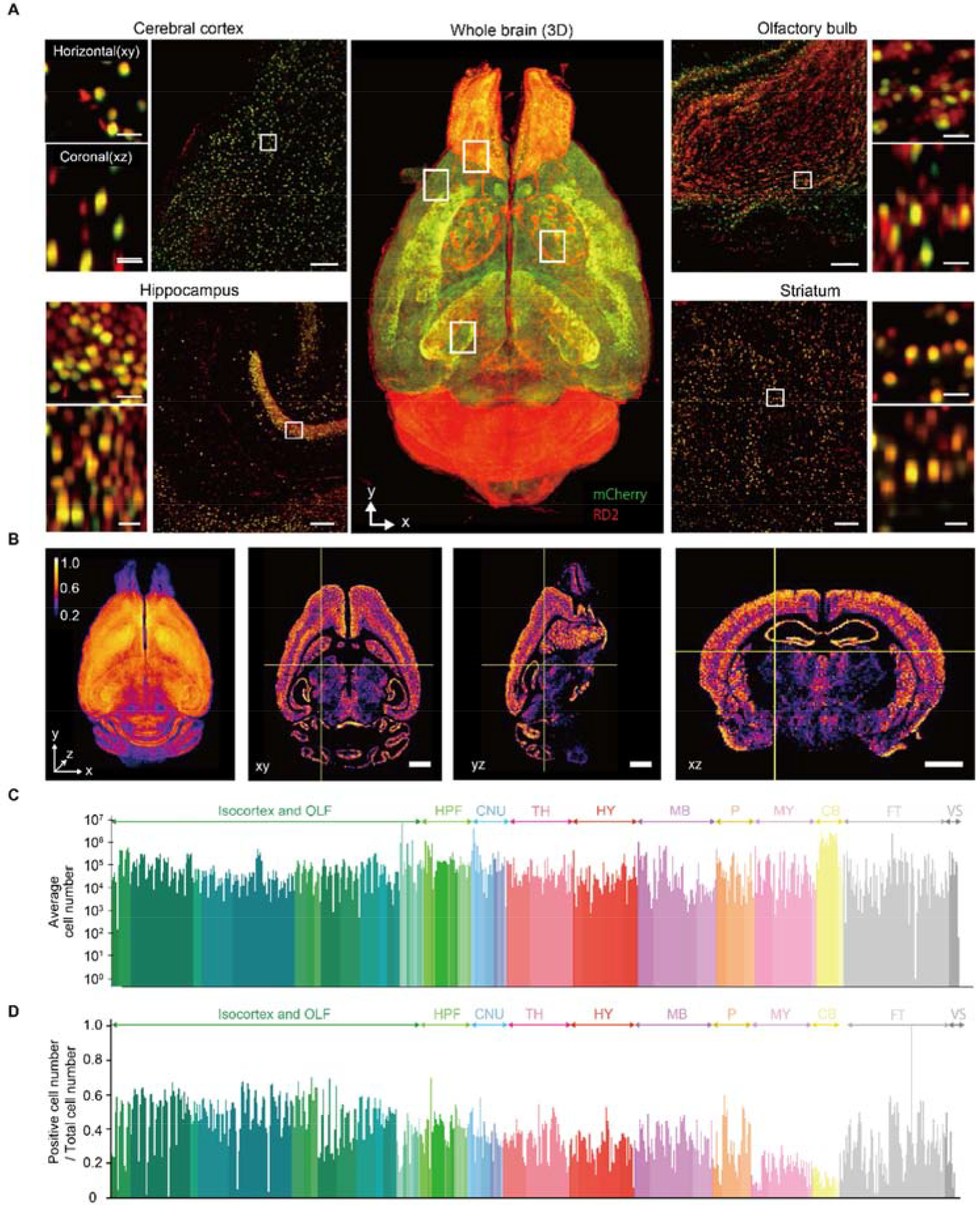
(A) Volume-rendered and single plane images of a brain transduced with AAV-PHP.eB:NSE-H2B-mCherry (mCherry, green) counterstained by RD2 (red) which is cleared by CUBIC-L/R+. Overlapped signals are shown in yellow. A volume-rendered image is shown in the center. Single plane and magnified images are shown for cerebral cortex, hippocampus, olfactory bulb and striatum. Both horizontal (x-y) and coronal (x-z) views are also shown. Scale bar, 200 μm (single plane image) and 25μm (magnified image). (B) 3D and cross-section images of the positive cell number ratio map of AAV-PHP.eB (NSE-H2B-mCherry) infected whole mouse brain. Voxel size, 80 μm. Scale bar, 2 mm. Scale bar, 50 μm. (C) Average cell number of all anatomical region in three 8-week-old C57BL/6N mouse brain. Only the edge region (i.e. having no child region) are shown. OLF, olfactory areas; HPF, hippocampal formation; CNU, cerebral nuclei; HY, Hypothalamus; MB, midbrain; P, pons; MY, medulla; CB, cerebellum; FT, fiber tract; VS, ventricular system. (D) Positive cell number ratio of each anatomical region in AAV-PHP.eB (NSE-H2B-mCherry) infected mouse brain.
The molecular features of cells in the brain – their transcriptomes and proteomes – provide valuable information extending beyond their morphology - both about their type and about cellular processes related to function. Classic approaches for immunofluorescence and fluorescence in situ hybridization for labeling proteins and mRNAs, respectively, have been most frequently applied within thin tissue sections through which fluorophore-conjugated macromolecules can quickly diffuse. In addition to reducing the scattering and absorption of light, processes such as delipidation and hydrogel embedding increase the diffusivity of proteins and oligonucleotides within tissue. For this reason, there is a natural synergy between techniques for clearing tissue samples and for labeling the mRNAs and proteins within them. Nevertheless, devising methods that permit the penetration of antibodies (for immunofluorescence) and oligonucleotides (for in situ hybridization) into tissue samples as large as intact organs has remained challenging. Penetration of macromolecules into tissues cleared using a variety of clearing techniques has been demonstrated, for example, by CLARITY (Chung et al., 2013), iDISCO+ (Renier et al., 2016), vDISCO (Cai et al., 2018), CUBIC-L/R+ and CUBIC-X (Matsumoto et al., 2019; Murakami et al., 2018). Nevertheless, uniformly staining thick samples – particularly intact, adult mammalian brains – may require protocol and/or probe optimization to effectively label different molecular targets. Also, molecular labeling may be time consuming, requiring several weeks or months for probes to thoroughly penetrate large samples even following delipidation and/or hydrogel embedding. Active processes, such as stochastic electrotransport may accelerate macromolecule diffusion(Kim et al., 2015a; Lee et al., 2016). Ongoing methodological development aims to increase the speed, reliability, and set of molecular targets that may be labeled in cleared tissue.
One powerful application of whole-brain tissue clearing and molecular labeling is efficient, brain-wide identification of cells expressing immediate early genes (IEGs). The expression of IEGs such as c-Fos, Arc, Egr-1, FosB, and Npas4 marks transcriptionally activated neurons and that can be used as an indirect measure of cells that have been recently active. Whole-brain mapping of IEG-expressing neurons using tissue clearing and light sheet imaging has been demonstrated using iDisco+(Renier et al., 2016) to delineate the brain-wide set of neural circuits engaged by parenting behavior. Identifying neural circuits using this approach allows the neural circuits engaged by neural processes to be mapped efficiently and comprehensively across the brain.
Whole-Brain Profiling of Circuits
The recent proliferation of neuroanatomical methods has also enabled whole-brain profiling of connectivity in the brain. Connectivity determines how information flows through neural circuits, giving rise to the diversity of mammalian behaviors from the simple startle response of defensive behaviors to the complex neuronal computations during cognitive and emotive processing. ‘Mesoscale connectivity, which describes the long-range projections of neural populations controls which brain areas are connected (Mitra, 2014). At the microscale, mapping the brain-wide connectivity of single neurons provides a fine-scale description of how signals are routed between brain areas. Mapping connectivity using light microscopy at both the mesoscale and microscale involves more stringent requirements for resolution and contrast than mapping cells, as axonal processes with diameters much smaller than neuronal somata must be detected with high fidelity. For this reason, block-face imaging methods have remained the dominant approach, and have been deployed successfully for mapping connectivity. At the mesoscale, STPT has been used to map the long-range connectivity of thalamic projections, connections between the cortex and striatum, and perhaps most notably, region-to-region connectivity as part of the Allen Institute for Brain Science Mouse Connectivity Project. This project has assayed the brain-wide projections of genetically identified populations of cells in hundreds of brain regions(Oh et al., 2014).
While mapping mesoscale connectivity effectively identifies the set of brain regions to which a neural population projects, microscale connectivity controls how those connections are structured at the cellular level. This information is crucial for identifying cell types and defining how information is represented and communicated between brain areas (Figure 5). To determining long-range connectivity in the brain with single-neuron resolution. Economo, Chandrashekar and colleagues extended STPT to full, volumetric imaging of the mouse brain by increasing the acquisition rate by 20-fold and combining it with tissue clearing (Economo et al., 2016) and sparse neuronal labeling. In doing so, they were able to reconstruct the axonal projections of single neurons in their entirety. Using whole-brain imaging to determine axonal connectivity increased the efficiency of this process by several orders of magnitude compared to existing histological approaches (Economo et al., 2019). Efficient, single-neuron reconstruction has also been demonstrated using fMOST(Li et al., 2018). These methods have enabled the brain-wide projections of more than one thousand neurons to be reconstructed in their entirety (Winnubst et al., 2019). Mapping connectivity at the microscale has been instrumental for revealing the heterogeneity of projection neurons and the structure of the projection pathways they comprise in the motor (Economo et al., 2018), somatosensory (Hooks et al., 2018), and visual cortices (Han et al., 2018), the hippocampus (Cembrowski et al., 2018), and claustrum (Wang et al., 2019). Despite the success of projects using block-face imaging to map long-range connectivity in the brain, instruments designed for this purpose are not widely available to most researchers. Advances in light sheet microscopy (Chhetri et al., 2015; Keller and Dodt, 2012; Verveer et al., 2007) promise to substantially reduce the time necessary for brain-wide imaging with sufficient resolution to resolve and reconstruct axons, increasing the efficiency of this process further, enabling comprehensive atlasing of long-range connectivity patterns in the mammalian brain (Economo et al., 2019).
Figure 5. Whole-Brain Profiling of Circuits.
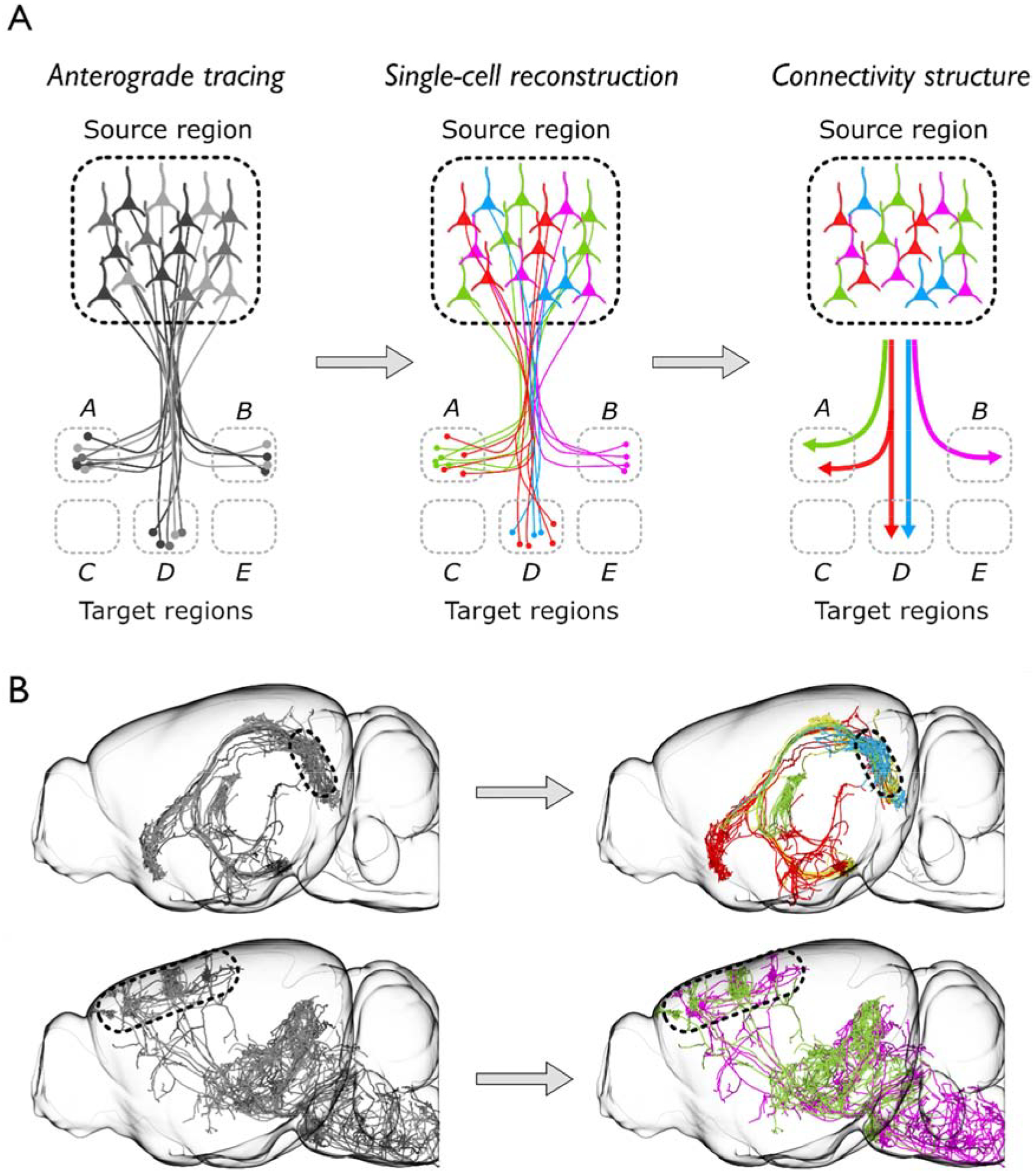
Single-neuron reconstruction reveals structured connectivity patterns. (A) Left, Anterograde tracing of a population of neurons in a source brain region can reveal regions of the brain to which the source connects (i.e. regions A, B, and D, but not C and E). Middle, single-cell axonal reconstruction reveals where individual neurons connect. Right, From single-neuron reconstructions, classes of neurons with similar connectivity can be identified and structured patterns of connectivity across a population can be determined. (B) Top left, Single-neuron reconstructions of projection neurons in the subiculum (boxed region). Top right, Axonal reconstructions reveal distinct brain-wide patterns of connectivity (color-coded) (Cembrowski et al., 2018; Winnubst et al., 2019). Bottom left, Single-neuron reconstructions of pyramidal tract neurons in the motor cortex (boxed region). Bottom right, Axonal reconstructions reveal two distinct types of pyramidal tract neurons based on their brain-wide connectivity (green, magenta) (Economo et al., 2018).
Future Perspective
Seamless integration of tissue-clearing methods and light-sheet microscopy will continue to enable key applications in neuroscience, including whole-brain profiling of cells and circuits in rodents. In the near future, further improvement of clearing, staining, imaging and image informatics will expand the spectrum of applications to whole-brain profiling of cells in primates and humans, one of the grand challenges in neuroscience. Isotropic imaging of cleared and 5-fold expanded entire mouse brains at 1.3 μm × 1.3 μm × 1.3 μm resolution gives rise to ~2.5 TB of data per brain (~0.5 TB × 5-fold volume expansion), and is estimated to be sufficient for extracting cell-nucleus information and constructing three-dimensional single-cell-resolution whole-brain atlases. Since marmoset (~7.6 g) and human brains (~1,330 g) are approximately 15- and 2,660-fold larger, respectively, than mouse brains (~0.5 g)(Stephan et al., 1981), whole-brain profiling of marmoset and human brains will give rise to ~38 TB and 6.65 PB of data, respectively, and provide single-cell-resolution whole-brain atlases comprising ~1.5 × 109 and ~2.7 × 1011 cells, respectively. To achieve whole-brain profiling of cells in primates and humans, several remaining issues need to be addressed. One of the remaining issues toward this goal is more stringent delipidation. Although the chemistry of the delipidation process in tissue clearing has been greatly advanced by chemical screening and profiling(Inoue et al., 2019; Tainaka et al., 2018; Zhao et al., 2019) and by developing stringent fixation protocols(Park et al., 2019), delipidation of the lipid-rich brains of marmosets and humans may require further improvements in delipidation chemistry, in particular for the white matter of the brain. Another remaining issue is auto-fluorescence, in particular human brain specimens. Since the human brain is usually fixed in formaldehyde for a long period of time, there is a need for minimizing auto-fluorescence arising from long-term fixation. The third issue on the path towards whole-brain profiling of cells in such large brains is the need for development of optimal microscopy for marmoset and human brains. Since a marmoset brain is 15 times larger than a mouse brain, the development of new, large-aperture optics with the long working distance required for covering the entire marmoset brain will be necessary. As far as human brains are concerned, an imaging strategy based on physical sectioning of the brain will likely present the most promising approach. Even in this case, further advancements in microscopy will be essential for rapid, high-resolution imaging of the resulting thick slices (e.g. ~5 mm thickness) with a cross-section area of ~100 cm2. Especially for such large tissue samples that may even comprise human tumors (Dodt et al., 2015), light sheets with an extended Rayleigh range (Saghafi et al., 2014) could be particularly helpful. In addition, more powerful image informatics are needed for accurately and rapidly extracting information from terabyte- and petabyte-scale datasets. Although these challenges may seem daunting at present, we believe that they will likely be solved in the near future, enabling the construction of three-dimensional single-cell-resolution whole-brain atlases that serve as a foundation upon which to integrate additional information, such as cell types, cell states (e.g. c-Fos expression) and cellular connectivity.
In the realm of structural connectivity, whole-brain profiling of circuits at a single-neuron level presents another grand challenge for mammalian neuroscience. As discussed above, a recent study characterized the long-range axonal projections of over 1,000 neurons from a few key areas in the mouse brain (Winnubst et al., 2019). However, a detailed brain-wide classification of neurons into ‘morphological cell-types’ in the mouse brain would likely require a sampling of at least 100,000 neurons (~0.1 % of all cells in the mouse brain; since it is estimated that there are ~1,000 brain regions in the mouse brain and likely more than 10 cell types per region)(Winnubst et al., 2019). Further, a more complete description of neuronal cell-types would require establishing correspondences between morphology, gene expression and function at a single neuron level. Faster imaging afforded by novel light-sheet approaches together with an increased throughput in the reconstruction process (a 10–20 fold improvement over current state of the art) due to further automation could make this goal achievable in a few years. Obtaining such comprehensive information on the structural organization of the brain will provide foundational information necessary for functional insights into the mammalian brain in health and disease.
Acknowledgement
We thank E.A. Susaki for helping to construct Figure 1 for advanced tissue-clearing methods, K. Matsumoto for helping to construct Figure 4 and Johan Winnubst for help with renderings in Figure 5. We also gratefully acknowledge grant support from Brain/MINDS JP20dm0207049, Science and Technology Platform Program for Advanced Biological Medicine JP20am0401011, AMED-CREST JP20gm0610006 (AMED/MEXT) (to H.R.U.), Grant-in-Aid for Scientific Research (S) JP25221004 (JSPS KAKENHI) (to H.R.U.), HFSP Research Grant Program (HFSP RGP0019/2018) (to H.R.U.), as well as funding from the Austrian Science Fund (FWF) and the Vienna Science Fund (WWTF) (to H.-U.D.), the Howard Hughes Medical Institute (to P.J.K., M.N.E., and J.C.), and NIMH U01 MH105971 and U01 MH114824 (to P.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
H.R.U. is a coinventor on patent applications covering the CUBIC reagents and a co-founder of CUBICStars Inc. P.J.K. is a coinventor on patents and patent applications covering multi-view and adaptive light-sheet microscopy. P.O. is a co-founder of Certerra Inc. and Certego Therapeutics Inc.
References
- Ahrens MB, Orger MB, Robson DN, Li JM, and Keller PJ (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 10, 413–420. [DOI] [PubMed] [Google Scholar]
- Amiya T, and Tanaka T (1987). Phase transitions in crosslinked gels of natural polymers. Macromolecules 20, 1162–1164. [Google Scholar]
- Antonio A (1949). Quantitative researches on the optical properties of human bone. Nature 163, 604. [DOI] [PubMed] [Google Scholar]
- Aoyagi Y, Kawakami R, Osanai H, Hibi T, and Nemoto T (2015). A rapid optical clearing protocol using 2,2’-thiodiethanol for microscopic observation of fixed mouse brain. PLoS ONE 10, e0116280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel P (2017). A beginner’s guide to tissue clearing. The international journal of biochemistry & cell biology 84, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi A, and Fabry C (1959). Technique for dissection and measurement of refractive index of osteones. J Biophys Biochem Cytol 6, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaripour A, Lagerweij T, Scharfbillig C, Jadczak AE, Willershausen B, and Van Noorden CJ (2016). A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Progress in histochemistry and cytochemistry 51, 9–23. [DOI] [PubMed] [Google Scholar]
- Bakutkin VV, Maksimova IL, Semyonova TN, Tuchin VV, and Kon IL (1995). Controlling optical properties of sclera Paper presented at: Ophthalmic Technologies V (International Society for Optics and Photonics; ). [Google Scholar]
- Bashkatov AN, Berezin KV, Dvoretskiy KN, Chernavina ML, Genina EA, Genin VD, Kochubey VI, Lazareva EN, Pravdin AB, Shvachkina ME, et al. (2018). Measurement of tissue optical properties in the context of tissue optical clearing. J Biomed Opt 23, 1–31, 31. [DOI] [PubMed] [Google Scholar]
- Bashkatov AN, Tuchin VV, Genina EA, Sinichkin YP, Lakodina NA, and Kochubey VI (1999). Human sclera dynamic spectra: in-vitro and in-vivo measurements Paper presented at: Ophthalmic Technologies IX (International Society for Optics and Photonics; ). [Google Scholar]
- Baumgart E, and Kubitscheck U (2012). Scanned light sheet microscopy with confocal slit detection. Opt Express 20, 21805–21814. [DOI] [PubMed] [Google Scholar]
- Bayless DW, and Shah NM (2016). Genetic dissection of neural circuits underlying sexually dimorphic social behaviours. Philos Trans R Soc Lond B Biol Sci 371, 20150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Hahn CM, Saghafi S, Jahrling N, Wanis M, and Dodt HU (2014). Reduction of photo bleaching and long term archiving of chemically cleared GFP-expressing mouse brains. PLoS ONE 9, e114149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Jährling N, Saghafi S, Weiler R, and Dodt HU (2012). Chemical clearing and dehydration of GFP expressing mouse brains. PLoS ONE 7, e33916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, and Chedotal A (2017). Tridimensional Visualization and Analysis of Early Human Development. Cell 169, 161–173 e112. [DOI] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Dominici C, Heitz-Marchaland C, Zelina P, Hellal F, Bradke F, and Chedotal A (2014). A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep 9, 1191–1201. [DOI] [PubMed] [Google Scholar]
- Beltz BS, and Kravitz EA (1983). Mapping of serotonin-like immunoreactivity in the lobster nervous system. The Journal of neuroscience : the official journal of the Society for Neuroscience 3, 585–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CA, and O’shea M (1982). Neuropeptide proctolin (H-Arg-Try-Leu-Pro-Thr-Oh): Immunocytochemical mapping of neurons in the central nervous system of the cockroach. J Comp Neurol 207, 223–238. [DOI] [PubMed] [Google Scholar]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, and Hillman EM (2015). Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat Photonics 9, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger T, Greger K, and Stelzer EH (2007). Lateral modulation boosts image quality in single plane illumination fluorescence microscopy. Opt Lett 32, 1938–1940. [DOI] [PubMed] [Google Scholar]
- Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Foerstera B, Zhao S, Bhatia HS, Mrowka L, Theodorou D, Rempfler M, et al. (2018). Panoptic vDISCO imaging reveals neuronal connectivity, remote trauma effects and meningeal vessels in intact transparent mice. bioRxiv, 374785. [Google Scholar]
- Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, Bhatia HS, Parra-Damas A, Mrowka L, Theodorou D, et al. (2019). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat Neurosci 22, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Phillips MG, DiLisio SF, Shields BC, Winnubst J, Chandrashekar J, Bas E, and Spruston N (2018). Dissociable Structural and Functional Hippocampal Outputs via Distinct Subiculum Cell Classes. Cell 173, 1280–1292.e1218. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Driscoll M, Murphy M, Roudot P, Chang B-J, Vora S, Wong WM, Nielson C, Zhang H, Zhemkov V, et al. (2019). Light-sheet microscopy with isotropic, sub-micron resolution and solvent-independent large-scale imaging. bioRxiv, 605493. [Google Scholar]
- Chance B, Liu H, Kitai T, and Zhang Y (1995). Effects of solutes on optical properties of biological materials: models, cells, and tissues. Anal Biochem 227, 351–362. [DOI] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XFS, Hammer JA, Liu Z, et al. (2014). Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 439–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, and Boyden ES (2015). Expansion microscopy. Science 347, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li G, Li Y, Li Y, Zhu H, Tang L, French P, McGinty J, and Ruan S (2017). UbasM: An effective balanced optical clearing method for intact biomedical imaging. Scientific reports 7, 12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri RK, Amat F, Wan Y, Hockendorf B, Lemon WC, and Keller PJ (2015). Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods 12, 1171–1178. [DOI] [PubMed] [Google Scholar]
- Chiang AS, Liu YC, Chiu SL, Hu SH, Huang CY, and Hsieh CH (2001). Three-dimensional mapping of brain neuropils in the cockroach, Diploptera punctata. J Comp Neurol 440, 1–11. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini I, Ghobril JP, Di Giovanna AP, Mascaro AL, Silvestri L, Mullenbroich MC, Onofri L, Conti V, Vanzi F, Sacconi L, et al. (2015). A versatile clearing agent for multi-modal brain imaging. Sci Rep 5, 9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu DL, Kokkaliaris KD, Kunz L, and Schroeder T (2018). Multicolor quantitative confocal imaging cytometry. Nat Methods 15, 39–46. [DOI] [PubMed] [Google Scholar]
- de Medeiros G, Norlin N, Gunther S, Albert M, Panavaite L, Fiuza UM, Peri F, Hiiragi T, Krzic U, and Hufnagel L (2015). Confocal multiview light-sheet microscopy. Nat Commun 6, 8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean KM, Roudot P, Welf ES, Danuser G, and Fiolka R (2015). Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys J 108, 2807–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Polson AG, and Klymkowsky MW (1989). A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 105, 61–74. [DOI] [PubMed] [Google Scholar]
- Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, and Becker K (2007). Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4, 331–336. [DOI] [PubMed] [Google Scholar]
- Dodt HU, Saghafi S, Becker K, Jahrling N, Hahn C, Pende M, Wanis M, and Niendorf A (2015). Ultramicroscopy: development and outlook. Neurophotonics 2, 041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW (2008). Allen Reference Atlas A Digital Color Brain Atlas of the C57BL/6J Male Mouse (John Wiley & Sons; ). [Google Scholar]
- Dunsby C (2008). Optically sectioned imaging by oblique plane microscopy. Opt Express 16, 20306–20316. [DOI] [PubMed] [Google Scholar]
- Duve H, Thorpe A, and Strausfeld NJ (1983). Cobalt-immunocytochemical identification of peptidergic neurons in Calliphora innervating central and peripheral targets. J Neurocytol 12, 847–861. [DOI] [PubMed] [Google Scholar]
- Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, and Chandrashekar J (2016). A platform for brain-wide imaging and reconstruction of individual neurons. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo MN, Viswanathan S, Tasic B, Bas E, Winnubst J, Menon V, Graybuck LT, Nguyen TN, Smith KA, Yao Z, et al. (2018). Distinct descending motor cortex pathways and their roles in movement. Nature 563, 79–84. [DOI] [PubMed] [Google Scholar]
- Economo MN, Winnubst J, Bas E, Ferreira TA, and Chandrashekar J (2019). Single-neuron axonal reconstruction: The search for a wiring diagram of the brain. J Comp Neurol 527, 2190–2199. [DOI] [PubMed] [Google Scholar]
- Epp JR, Niibori Y, Liz Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, and Frankland PW (2015). Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs(1,2,3). eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, and Dodt HU (2012). Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7, 1983–1995. [DOI] [PubMed] [Google Scholar]
- Ertürk A, Lafkas D, and Chalouni C (2014). Imaging cleared intact biological systems at a cellular level by 3DISCO. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach FO, and Rohrbach A (2010). A line scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt Express 18, 24229–24244. [DOI] [PubMed] [Google Scholar]
- Fahrbach FO, and Rohrbach A (2012). Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media. Nat Commun 3. [DOI] [PubMed] [Google Scholar]
- Fahrbach FO, Voigt FF, Schmid B, Helmchen F, and Huisken J (2013). Rapid 3D light-sheet microscopy with a tunable lens. Opt Express 21, 21010–21026. [DOI] [PubMed] [Google Scholar]
- Fu Q, Martin BL, Matus DQ, and Gao L (2016). Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat Commun 7, 11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Jaffe JS, Long RA, and Azam F (2002). Thin laser light sheet microscope for microbial oceanography. Opt Express 10, 145–154. [DOI] [PubMed] [Google Scholar]
- Glaser JR, and Glaser EM (2000). Stereology, morphometry, and mapping: the whole is greater than the sum of its parts. Journal of chemical neuroanatomy 20, 115–126. [DOI] [PubMed] [Google Scholar]
- Gleave JA, Lerch JP, Henkelman RM, and Nieman BJ (2013). A method for 3D immunostaining and optical imaging of the mouse brain demonstrated in neural progenitor cells. PLoS ONE 8, e72039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, and Burkhalter A (2007). Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Xu D, Yuan J, Li X, Guo C, Peng J, Li Y, Schwarz LA, Li A, and Hu B (2016). High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Treweek J, Overton K, and Deisseroth K (2018). Hydrogel-tissue chemistry: Principles and applications. Annual review of biophysics 47, 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Chan KY, Dobreva T, Brown D, Balani DH, Boyce R, Kronenberg HM, McBride HJ, and Gradinaru V (2017a). Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Science translational medicine 9, eaah6518. [DOI] [PubMed] [Google Scholar]
- Greenbaum A, Jang MJ, Challis C, and Gradinaru V (2017b). Q&A: How can advances in tissue clearing and optogenetics contribute to our understanding of normal and diseased biology? BMC biology 15, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CJ, and Holy TE (2019). Fast objective coupled planar illumination microscopy. Nat Commun 10, 4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Becker K, Saghafi S, Pende M, Avdibašić A, Foroughipour M, Heinz DE, Wotjak CT, and Dodt H-U (2019). High-resolution imaging of fluorescent whole mouse brains using stabilised organic media (sDISCO). Journal of Biophotonics 0, e201800368. [DOI] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, et al. (2015). ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, and Miyawaki A (2011). Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14, 1481–1488. [DOI] [PubMed] [Google Scholar]
- Han Y, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018). The logic of single-cell projections from visual cortex. Nature 556, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Sakamoto Y, Nakagami S, Aida M, Sawa S, and Matsunaga S (2016). Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant and Cell Physiology 57, 462–472. [DOI] [PubMed] [Google Scholar]
- Hasegawa S, Susaki EA, Tanaka T, Komaba H, Wada T, Fukagawa M, Ueda HR, and Nangaku M (2019). Comprehensive three-dimensional analysis (CUBIC-kidney) visualizes abnormal renal sympathetic nerves after ischemia/reperfusion injury. Kidney int 96, 129–138. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M, Baldock RA, Burger A, Hashikawa T, Johnson GA, Martone M, Ng L, Lau C, Larsen SD, and Nissanov J (2011). Digital atlasing and standardization in the mouse brain. PLoS computational biology 7, e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Catania K, Manger PR, and Kaas JH (2015a). Mammalian Brains Are Made of These: A Dataset of the Numbers and Densities of Neuronal and Nonneuronal Cells in the Brain of Glires, Primates, Scandentia, Eulipotyphlans, Afrotherians and Artiodactyls, and Their Relationship with Body Mass. Brain Behavior and Evolution 86, 145–163. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, von Bartheld CS, Miller DJ, and Kaas JH (2015b). How to count cells: the advantages and disadvantages of the isotropic fractionator compared with stereology. Cell and tissue research 360, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Watson C, and Paxinos G (2013). Distribution of neurons in functional areas of the mouse cerebral cortex reveals quantitatively different cortical zones. Front Neuroanat 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand S, Schueth A, Herrler A, Galuske R, and Roebroeck A (2018). Scalable cytoarchitectonic characterization of large intact human neocortex samples. bioRxiv, 274985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshburg J, Choi B, Nelson JS, and Yeh AT (2007). Correlation between collagen solubility and skin optical clearing using sugars. Lasers Surg Med 39, 140–144. [DOI] [PubMed] [Google Scholar]
- Holekamp TF, Turaga D, and Holy TE (2008). Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron 57, 661–672. [DOI] [PubMed] [Google Scholar]
- Hooks BM, Papale AE, Paletzki RF, Feroze MW, Eastwood BS, Couey JJ, Winnubst J, Chandrashekar J, and Gerfen CR (2018). Topographic precision in sensory and motor corticostriatal projections varies across cell type and cortical area. Nat Commun 9, 3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörl D, Rojas Rusak F, Preusser F, Tillberg P, Randel N, Chhetri RK, Cardona A, Keller PJ, Harz H, Leonhardt H, et al. (2018). BigStitcher: Reconstructing high-resolution image datasets of cleared and expanded samples. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, Wang J, Zhang X, Liu B, Fan L, et al. (2015). Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh B, Burns VM, Pauerstein P, Holzem K, Ye L, Engberg K, Wang AC, Gu X, Chakravarthy H, Arda HE, et al. (2017). Pathways to clinical CLARITY: volumetric analysis of irregular, soft, and heterogeneous tissues in development and disease. Sci Rep 7, 5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ (2014). Toward a genetic dissection of cortical circuits in the mouse. Neuron 83, 1284–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, and Zeng H (2013). Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci 36, 183–215. [DOI] [PubMed] [Google Scholar]
- Huisken J, and Stainier DY (2007). Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt Lett 32, 2608–2610. [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, and Stelzer EH (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Hamm RN, Arakawa ET, and Painter LR (1974). Optical and dielectric properties of DNA in the extreme ultraviolet. The Journal of Chemical Physics 61, 4246–4250. [Google Scholar]
- Inoue M, Saito R, Kakita A, and Tainaka K (2019). Rapid chemical clearing of white matter in the post-mortem human brain by 1,2-hexanediol delipidation. Bioorganic & Medicinal Chemistry Letters 29, 1886–1890. [DOI] [PubMed] [Google Scholar]
- Jing D, Zhang S, Luo W, Gao X, Men Y, Ma C, Liu X, Yi Y, Bugde A, Zhou BO, et al. (2018). Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res 28, 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorand R, Le Corre G, Andilla J, Maandhui A, Frongia C, Lobjois V, Ducommun B, and Lorenzo C (2012). Deep and clear optical imaging of thick inhomogeneous samples. PLoS ONE 7, e35795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Zwart MF, Yang CT, Mensh BD, and Ahrens MB (2016). The Serotonergic System Tracks the Outcomes of Actions to Mediate Short-Term Motor Learning. Cell 167, 933–946.e920. [DOI] [PubMed] [Google Scholar]
- Ke M-T, Fujimoto S, and Imai T (2013). SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Ke M-T, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, Sato M, and Imai T (2016). Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep 14, 2718–2732. [DOI] [PubMed] [Google Scholar]
- Keller PJ, and Dodt HU (2012). Light sheet microscopy of living or cleared specimens. Curr Opin Neurobiol 22, 138–143. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Santella A, Khairy K, Bao ZR, Wittbrodt J, and Stelzer EHK (2010). Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods 7, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, and Stelzer EHK (2008). Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069. [DOI] [PubMed] [Google Scholar]
- Kienle DF, de Souza JV, Watkins EB, and Kuhl TL (2014). Thickness and refractive index of DPPC and DPPE monolayers by multiple-beam interferometry. Analytical and Bioanalytical Chemistry 406, 4725–4733. [DOI] [PubMed] [Google Scholar]
- Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, Ruelas L, Hubbert A, McCue M, Vassallo SL, et al. (2015a). Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci USA 112, E6274–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. (2015b). Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep 10, 292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yang GR, Pradhan K, Venkataraju KU, Bota M, Garcia Del Molino LC, Fitzgerald G, Ram K, He M, Levine JM, et al. (2017). Brain-wide Maps Reveal Stereotyped Cell-Type-Based Cortical Architecture and Subcortical Sexual Dimorphism. Cell 171, 456–469 e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingberg A, Hasenberg A, Ludwig-Portugall I, Medyukhina A, Männ L, Brenzel A, Engel DR, Figge MT, Kurts C, and Gunzer M (2017). Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. Journal of the American Society of Nephrology 28, 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzic U, Gunther S, Saunders TE, Streichan SJ, and Hufnagel L (2012). Multiview light-sheet microscope for rapid in toto imaging. Nat Methods 9, 730–733. [DOI] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park JY, Albanese A, Murray E, Cho JH, Park YG, Mangena V, Chen J, and Chung K (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol 34, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, and Ng L (2015). Neuroinformatics of the allen mouse brain connectivity atlas. Methods 73, 4–17. [DOI] [PubMed] [Google Scholar]
- Kubota SI, Takahashi K, Nishida J, Morishita Y, Ehata S, Tainaka K, Miyazono K, and Ueda HR (2017). Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep 20, 236–250. [DOI] [PubMed] [Google Scholar]
- Kumar V, Scandella E, Danuser R, Onder L, Nitschke M, Fukui Y, Halin C, Ludewig B, and Stein JV (2010). Global lymphoid tissue remodeling during a viral infection is orchestrated by a B cell-lymphotoxin-dependent pathway. Blood 115, 4725–4733. [DOI] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, and Higashiyama T (2015). ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, and Mason C (2013). ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kverkova K, Belikova T, Olkowicz S, Pavelkova Z, O’Riain MJ, Sumbera R, Burda H, Bennett NC, and Nemec P (2018). Sociality does not drive the evolution of large brains in eusocial African mole-rats. Sci Rep 8, 9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz L (1880). Über die Refractionsconstante. Ann Phys 11, 70–103. [Google Scholar]
- Lai HM, Liu AKL, Ng HHM, Goldfinger MH, Chau TW, DeFelice J, Tilley BS, Wong WM, Wu W, and Gentleman SM (2018). Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat Commun 9, 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HM, Liu AKL, Ng W-L, DeFelice J, Lee WS, Li H, Li W, Ng HM, Chang RC-C, and Lin B (2016). Rationalisation and validation of an acrylamide-free procedure in three-dimensional histological imaging. PLoS ONE 11, e0158628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, Kim SY, Lee H-J, Cho K, and Jung N (2016). ACT-PRESTO: Rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Scientific reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Park JH, Seo I, Park SH, and Kim S (2014). Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev Biol 14, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Lemon WC, Pulver SR, Hockendorf B, McDole K, Branson K, Freeman J, and Keller PJ (2015). Whole-central nervous system functional imaging in larval Drosophila. Nat Commun 6, 7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AA, Gong H, Zhang B, Wang QD, Yan C, Wu JP, Liu QA, Zeng SQ, and Luo QM (2010). Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408. [DOI] [PubMed] [Google Scholar]
- Li W, Germain RN, and Gerner MY (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proceedings of the National Academy of Sciences 114, E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu B, Sun Q, Zhang Y, Ren M, Zhang X, Li A, Yuan J, Madisen L, Luo Q, et al. (2018). Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc Natl Acad Sci USA 115, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann T, Renier N, Bettayeb K, Greengard P, Tessier-Lavigne M, and Flajolet M (2016). Three-dimensional study of Alzheimer’s disease hallmarks using the idisco clearing method. Cell Rep 16, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AKL, Hurry ME, Ng OT-W, DeFelice J, Lai HM, Pearce R, Wong GTC, Chang RCC, and Gentleman SM (2016). Bringing CLARITY to the human brain: visualization of Lewy pathology in three dimensions. Neuropathology and applied neurobiology 42, 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Beauvoit B, Kimura M, and Chance B (1996). Dependence of tissue optical properties on solute-induced changes in refractive index and osmolarity. J Biomed Opt 1, 200–211. [DOI] [PubMed] [Google Scholar]
- Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, et al. (2018). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz HA (1880). Über die Beziehungzwischen der Fortpflanzungsgeschwindigkeit des Lichtes der Körperdichte. Ann Phys 9, 641–665. [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ 3rd, Gu X, Zanella S, et al. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahou P, Vermot J, Beaurepaire E, and Supatto W (2014). Multicolor two-photon light-sheet microscopy. Nat Methods 11, 600–601. [DOI] [PubMed] [Google Scholar]
- Marhounova L, Kotrschal A, Kverkova K, Kolm N, and Nemec P (2019). Artificial selection on brain size leads to matching changes in overall number of neurons. Evolution; international journal of organic evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Mitani TT, Horiguchi SA, Kaneshiro J, Murakami TC, Mano T, Fujishima H, Konno A, Watanabe TM, Hirai H, et al. (2019). Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat Protoc. [DOI] [PubMed] [Google Scholar]
- Mayerich D, Abbott L, and McCormick B (2008). Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J Microsc 231, 134–143. [DOI] [PubMed] [Google Scholar]
- McDole K, Guignard L, Amat F, Berger A, Malandain G, Royer LA, Turaga SC, Branson K, and Keller PJ (2018). In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level. Cell 175, 859–876.e833. [DOI] [PubMed] [Google Scholar]
- McGorty R, Liu H, Kamiyama D, Dong Z, Guo S, and Huang B (2015). Open-top selective plane illumination microscope for conventionally mounted specimens. Opt Express 23, 16142–16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori B, Datta MS, Dupre C, Apak MC, Asano S, Gao R, Boyden ES, Hermanson O, Yuste R, and Tomer R (2018). Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol 16, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra PP (2014). The circuit architecture of whole brains at the mesoscopic scale. Neuron 83, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani H, Ono S, Ushiku T, Kudo Y, Ikemura M, Kageyama N, Yamamichi N, Fujishiro M, Someya T, and Fukayama M (2018). Transparency-enhancing technology allows three-dimensional assessment of gastrointestinal mucosa: a porcine model. Pathology international 68, 102–108. [DOI] [PubMed] [Google Scholar]
- Murakami TC, Mano T, Saikawa S, Horiguchi SA, Shigeta D, Baba K, Sekiya H, Shimizu Y, Tanaka KF, Kiyonari H, et al. (2018). A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat Neurosci 21, 625–637. [DOI] [PubMed] [Google Scholar]
- Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, et al. (2015). Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell 163, 1500–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A, Dang C, Avants B, Yushkevich P, Gee JC, et al. (2007). Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM transactions on computational biology and bioinformatics 4, 382–393. [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al. (2016). Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima S, Susaki EA, Yoshida K, Takemoto H, Tsujimura N, Iijima S, Takachi K, Nakahara Y, Tahara S, and Ohshima K (2017). CUBIC pathology: three-dimensional imaging for pathological diagnosis. Scientific reports 7, 9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa SK, and Watabe-Uchida M (2018). Organization of dopamine and serotonin system: Anatomical and functional mapping of monosynaptic inputs using rabies virus. Pharmacology Biochemistry and Behavior 174, 9–22. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte OE, Andilla J, Artigas D, and Loza-Alvarez P (2015). Decoupled illumination detection in light sheet microscopy for fast volumetric imaging. Optica 2, 702–705. [Google Scholar]
- Olkowicz S, Kocourek M, Lucan RK, Portes M, Fitch WT, Herculano-Houzel S, and Nemec P (2016). Birds have primate-like numbers of neurons in the forebrain. Proc Natl Acad Sci USA 113, 7255–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osten P, and Margrie TW (2013). Mapping brain circuitry with a light microscope. Nat Methods 10, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palero J, Santos SI, Artigas D, and Loza-Alvarez P (2010). A simple scanless two-photon fluorescence microscope using selective plane illumination. Opt Express 18, 8491–8498. [DOI] [PubMed] [Google Scholar]
- Palmer WM, Martin AP, Flynn JR, Reed SL, White RG, Furbank RT, and Grof CPL (2015). PEA-CLARITY: 3D molecular imaging of whole plant organs. Scientific Reports 5, 13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, and Erturk A (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 13, 859–867. [DOI] [PubMed] [Google Scholar]
- Panier T, Romano SA, Olive R, Pietri T, Sumbre G, Candelier R, and Debregeas G (2013). Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front Neural Circuits 7, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-G, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, et al. (2019). Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol 37, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Becker K, Wanis M, Saghafi S, Kaur R, Hahn C, Pende N, Foroughipour M, Hummel T, and Dodt H-U (2018). High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat Commun 9, 4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbellini F, Liu AK, Watson SA, Bardi I, Rothery SM, and Terracciano CM (2017). Free-of-Acrylamide SDS-based Tissue Clearing (FASTClear) for three dimensional visualization of myocardial tissue. Scientific reports 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, and Scanziani M (2013). Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci 16, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piezosystems_Jena (2019). nanoX 200/400 and nanoSX 400/800. wwwpiezosystemcom https://www.piezosystem.com/products/piezo_actuators/nanox/. [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, and Betzig E (2011). Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods 8, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronneke A, Scheuer B, Wagener RJ, Mock M, Witte M, and Staiger JF (2015). Characterizing VIP Neurons in the Barrel Cortex of VIPcre/tdTomato Mice Reveals Layer-Specific Differences. Cereb Cortex 25, 4854–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusterla JM, Malfatti-Gasperini AA, Puentes-Martinez XE, Cavalcanti LP, and Oliveira RG (2017). Refractive index and thickness determination in Langmuir monolayers of myelin lipids. Biochimica et Biophysica Acta (BBA) - Biomembranes 1859, 924–930. [DOI] [PubMed] [Google Scholar]
- Quirin S, Vladimirov N, Yang C-T, Peterka DS, Yuste R, and Ahrens MB (2016). Calcium imaging of neural circuits with extended depth-of-field light-sheet microscopy. Opt Lett 41, 855–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, and Osten P (2012). Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods 9, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J, Nijmeijer B, Hossain MJ, Eguren M, Schneider I, Politi AZ, Roberti MJ, Hufnagel L, Hiiragi T, and Ellenberg J (2018). Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 361, 189–193. [DOI] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Venkataraju KU, and Zhou Y (2016). Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. [DOI] [PubMed] [Google Scholar]
- Richardson DS, and Lichtman JW (2015). Clarifying Tissue Clearing. Cell 162, 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW, and Keller PJ (2016). Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol 34, 1267. [DOI] [PubMed] [Google Scholar]
- Rozbicki E, Chuai M, Karjalainen AI, Song F, Sang HM, Martin R, Knolker HJ, MacDonald MP, and Weijer CJ (2015). Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nature cell biology 17, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, and Hjerling-Leffler J (2011). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Developmental neurobiology 71, 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghafi S, Becker K, Hahn C, and Dodt HU (2014). 3D-ultramicroscopy utilizing aspheric optics. J Biophotonics 7, 117–125. [DOI] [PubMed] [Google Scholar]
- Schmid B, Shah G, Scherf N, Weber M, Thierbach K, Campos CP, Roeder I, Aanstad P, and Huisken J (2013). High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat Commun 4, 2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, and Hof PR (2005). Design-based stereology in neuroscience. Neuroscience 130, 813–831. [DOI] [PubMed] [Google Scholar]
- Schwarz MK, Scherbarth A, Sprengel R, Engelhardt J, Theer P, and Giese G (2015). Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS ONE 10, e0124650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiriki K, Kasai A, Hashimoto T, Schulze W, Niu M, Yamaguchi S, Nakazawa T, Inoue KI, Uezono S, Takada M, et al. (2017). High-Speed and Scalable Whole-Brain Imaging in Rodents and Primates. Neuron 94, 1085–1100.e1086. [DOI] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Schwarzkopf M, He T-F, Greenbaum A, ho Sohn C, Lignell A, Choi HM, Gradinaru V, and Pierce NA (2016). Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development, dev. 138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard CJR (2013). Pupil filters for generation of light sheets. Opt Express 21, 6339–6345. [DOI] [PubMed] [Google Scholar]
- Siedentopf H, Zsigmondy R (1903). Über Sichtbarmachung und Größenbestimmung ultramikroskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Ann Phys 315, 1–39. [Google Scholar]
- Sillitoe RV, and Hawkes R (2002). Whole-mount immunohistochemistry: a high-throughput screen for patterning defects in the mouse cerebellum. J Histochem Cytochem 50, 235–244. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Bria A, Sacconi L, Iannello G, and Pavone FS (2012). Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt Express 20, 20582–20598. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Costantini I, Sacconi L, and Pavone FS (2016). Clearing of fixed tissue: a review from a microscopist’s perspective. J Biomed Opt 21, 081205–081205. [DOI] [PubMed] [Google Scholar]
- Simerly RB (2002). Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu Rev Neurosci 25, 507–536. [DOI] [PubMed] [Google Scholar]
- Spalteholz W (1911). Über das Durchsichtigmachen von menschlichen und tierischen Präparaten (Hirzel S, Leipzig: ). [Google Scholar]
- Staudt T, Lang MC, Medda R, Engelhardt J, and Hell SW (2007). 2,2’-Thiodiethanol: A new water soluble mounting medium for high resolution optical microscopy. Microsc Res Tech 70, 1–9. [DOI] [PubMed] [Google Scholar]
- Stephan H, Frahm H, and Baron G (1981). New and revised data on volumes of brain structures in insectivores and primates. Folia primatologica; international journal of primatology 35, 1–29. [DOI] [PubMed] [Google Scholar]
- Strnad P, Gunther S, Reichmann J, Krzic U, Balazs B, de Medeiros G, Norlin N, Hiiragi T, Hufnagel L, and Ellenberg J (2016). Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat Methods 13, 139–142. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, and Ueda HR (2015). Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc 10, 1709–1727. [DOI] [PubMed] [Google Scholar]
- Susaki EA, and Ueda HR (2016). Whole-body and Whole-Organ Clearing and Imaging Techniques with Single-Cell Resolution: Toward Organism-Level Systems Biology in Mammals. Cell Chem Biol 23, 137–157. [DOI] [PubMed] [Google Scholar]
- Swoger J, Verveer P, Greger K, Huisken J, and Stelzer EH (2007). Multi-view image fusion improves resolution in three-dimensional microscopy. Opt Express 15, 8029–8042. [DOI] [PubMed] [Google Scholar]
- Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, and Deisseroth K (2016). Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell 164, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, and Ueda HR (2014). Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924. [DOI] [PubMed] [Google Scholar]
- Tainaka K, Kuno A, Kubota SI, Murakami T, and Ueda HR (2016). Chemical Principles in Tissue Clearing and Staining Protocols for Whole-Body Cell Profiling. Annu Rev Cell Dev Biol 32, 713–741. [DOI] [PubMed] [Google Scholar]
- Tainaka K, Murakami TC, Susaki EA, Shimizu C, Saito R, Takahashi K, Hayashi-Takagi A, Sekiya H, Arima Y, Nojima S, et al. (2018). Chemical Landscape for Tissue Clearing based on Hydrophilic Reagents. Cell Rep 24, 2196–2210. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Kanatani S, Tomer R, Sahlgren C, Kronqvist P, Kaczynska D, Louhivuori L, Kis L, Lindh C, and Mitura P (2017). Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nature Biomedical Engineering 1, 796. [DOI] [PubMed] [Google Scholar]
- Tatsuki F, Sunagawa GA, Shi S, Susaki EA, Yukinaga H, Perrin D, Sumiyama K, Ukai-Tadenuma M, Fujishima H, Ohno R, et al. (2016). Involvement of Ca(2+)-Dependent Hyperpolarization in Sleep Duration in Mammals. Neuron 90, 70–85. [DOI] [PubMed] [Google Scholar]
- Tomer R, Khairy K, Amat F, and Keller PJ (2012). Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods 9, 755–763. [DOI] [PubMed] [Google Scholar]
- Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, Sankaran S, Grosenick L, Broxton M, Yang S, and Deisseroth K (2015). SPED Light Sheet Microscopy: Fast Mapping of Biological System Structure and Function. Cell 163, 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, and Deisseroth K (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9, 1682–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10, 1860–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong TV, Supatto W, Koos DS, Choi JM, and Fraser SE (2011). Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods 8, 757–760. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, Lyden PD, and Kleinfeld D (2009). Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci 29, 14553–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchin VV (2015). Tissue optics and photonics: light-tissue interaction. J Biomed Photonics Eng 1, 98–134. [Google Scholar]
- Tuchin VV, Bashkatov AN, Genina EA, Kochubey VI, Lakodina NA, Simonenko GV, Sinichkin YP, Proshina YM, and Razumikhina NA (1999). Optics of living tissues with controlled scattering properties. Paper presented at: Proc SPIE. [Google Scholar]
- Tuchin VV, Maksimova IL, Zimnyakov DA, Kon IL, Mavlyutov AH, and Mishin AA (1997). Light propagation in tissues with controlled optical properties. J Biomed Opt 2, 401–417. [DOI] [PubMed] [Google Scholar]
- Tuchin VV, and Tuchin V (2007). Tissue optics: light scattering methods and instruments for medical diagnosis, Vol 13 (SPIE press; Bellingham: ). [Google Scholar]
- Tuchin VV, Xu X, and Wang RK (2002). Dynamic optical coherence tomography in studies of optical clearing, sedimentation, and aggregation of immersed blood. Appl Opt 41, 258–271. [DOI] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, and Lippincott-Schwartz J (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, and Stelzer EH (2007). High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods 4, 311–313. [DOI] [PubMed] [Google Scholar]
- Vettenburg T, Dalgarno HI, Nylk J, Coll-Llado C, Ferrier DE, Cizmar T, Gunn-Moore FJ, and Dholakia K (2014). Light-sheet microscopy using an Airy beam. Nat Methods 11, 541–544. [DOI] [PubMed] [Google Scholar]
- Voie AH, Burns DH, and Spelman FA (1993). Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J Microsc 170, 229–236. [DOI] [PubMed] [Google Scholar]
- Voigt FF, Kirschenbaum D, Platonova E, Pages S, Campbell RAA, Kastli R, Schaettin M, Egolf L, van der Bourg A, Bethge P, et al. (2019). The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat Methods 16, 1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Wei Z, Looger LL, Koyama M, Druckmann S, Keller PJ, (2019). Single-Cell Reconstruction of Emerging Population Activity in an Entire Developing Circuit. Cell 179, 355–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xie P, Gong H, Zhou Z, Kuang X, Wang Y, Li A. a., Li Y, Liu L, Veldman MB, et al. (2019). Complete single neuron reconstruction reveals morphological diversity in molecularly defined claustral and cortical neuron types. bioRxiv, 675280. [Google Scholar]
- Wang Z, Zhang J, Fan G, Zhao H, Wang X, Zhang J, Zhang P, and Wang W (2018). Imaging transparent intact cardiac tissue with single-cell resolution. Biomedical optics express 9, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner CA, Biedrzycki M, Jacobs SS, Wisser RJ, Caplan JL, and Sherrier DJ (2014). An optical clearing technique for plant tissues allowing deep imaging and compatible with fluorescence microscopy. Plant physiology, pp. 114.244673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans: the mind of a worm. Phil Trans R Soc Lond 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Wilding D, Pozzi P, Soloviev O, Vdovin G, and Verhaegen M (2016). Adaptive illumination based on direct wavefront sensing in a light-sheet fluorescence microscope. Opt Express 24, 24896–24906. [DOI] [PubMed] [Google Scholar]
- Williams RW, and Rakic P (1988). Three-dimensional counting: an accurate and direct method to estimate numbers of cells in sectioned material. J Comp Neurol 278, 344–352. [DOI] [PubMed] [Google Scholar]
- Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, Edson P, Arthur BJ, Bruns C, Rokicki K, Schauder D, et al. (2019). Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain. bioRxiv, 537233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Supatto W, Debregeas G, Mahou P, Kruglik SG, Sintes JM, Beaurepaire E, and Candelier R (2015). Whole-brain functional imaging with two-photon light-sheet microscopy. Nat Methods 12, 379–380. [DOI] [PubMed] [Google Scholar]
- Wu YC, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Bao ZR, Colon-Ramos D, and Shroff H (2011). Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci USA 108, 17708–17713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A, York AG, Winter PW, Waterman CM, Bao ZR, et al. (2013). Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol 31, 1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tamadon A, Liu Y, Ma T, Leak RK, Chen J, Gao Y, and Feng Y (2017). Fast free-of-acrylamide clearing tissue (FACT)—an optimized new protocol for rapid, high-resolution imaging of three-dimensional brain tissue. Scientific reports 7, 9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, and Callaway EM (2010). Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol 518, 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang RK, Elder JB, and Tuchin VV (2003). Effect of dextran-induced changes in refractive index and aggregation on optical properties of whole blood. Physics in Medicine & Biology 48, 1205. [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, and Gradinaru V (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, et al. (2016). Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell 165, 1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Zhu J, Li Y, Ma Y, Wang J, Cheng X, Jin S, Sun Q, Li X, and Gong H (2018). RTF: a rapid and versatile tissue optical clearing method. Scientific reports 8, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Todorov MI, Cai R, Steinke H, Kemter E, Wolf E, Lipfert J, Bechmann I, and Ertürk A (2019). Cellular and Molecular Probing of Intact Transparent Human Organs. bioRxiv, 643908. [Google Scholar]
- Zheng T, Yang ZQ, Li AA, Lv XH, Zhou ZQ, Wang XJ, Qi XL, Li SW, Luo QM, Gong H, et al. (2013). Visualization of brain circuits using two-photon fluorescence micro-optical sectioning tomography. Opt Express 21, 9839–9850. [DOI] [PubMed] [Google Scholar]
- Zimnyakov DA, Tuchin VV, Michin AA, Kon IL, and Serov AN (1996). In-vitro human sclera structure analysis using tissue optical immersion effect Paper presented at: Ophthalmic Technologies VI (International Society for Optics and Photonics; ). [Google Scholar]


