Abstract
The mammalian genome comprises nuclear DNA (nDNA) derived from both parents and mitochondrial DNA (mtDNA) that is maternally inherited and encodes essential proteins required for oxidative phosphorylation. Thousands of copies of the circular mtDNA are present in most cell types that are packaged by TFAM into higher-order structures called nucleoids1. Mitochondria are also platforms for antiviral signalling2 and, due to their bacterial origin, mtDNA and other mitochondrial components trigger innate immune responses and inflammatory pathology2,3. We showed previously that instability and cytoplasmic release of mtDNA activates the cGAS-STING-TBK1 pathway resulting in interferon stimulated gene (ISG) expression that promotes antiviral immunity4. Here, we find that persistent mtDNA stress is not associated with basally activated NF-κB signalling or interferon gene expression typical of an acute antiviral response. Instead, a specific subset of ISGs, that includes Parp9, remains activated by the unphosphorylated form of ISGF3 (U-ISGF3) that enhances nDNA damage and repair responses. In cultured primary fibroblasts and cancer cells, the chemotherapeutic drug doxorubicin causes mtDNA damage and release, which leads to cGAS-STING-dependent ISG activation. In addition, mtDNA stress in TFAM-deficient mouse melanoma cells produces tumours that are more resistant to doxorubicin in vivo. Finally, Tfam+/− mice exposed to ionizing radiation exhibit enhanced nDNA repair responses in spleen. Therefore, we propose that damage to and subsequent release of mtDNA elicits a protective signalling response that enhances nDNA repair in cells and tissues, suggesting mtDNA is a genotoxic stress sentinel.
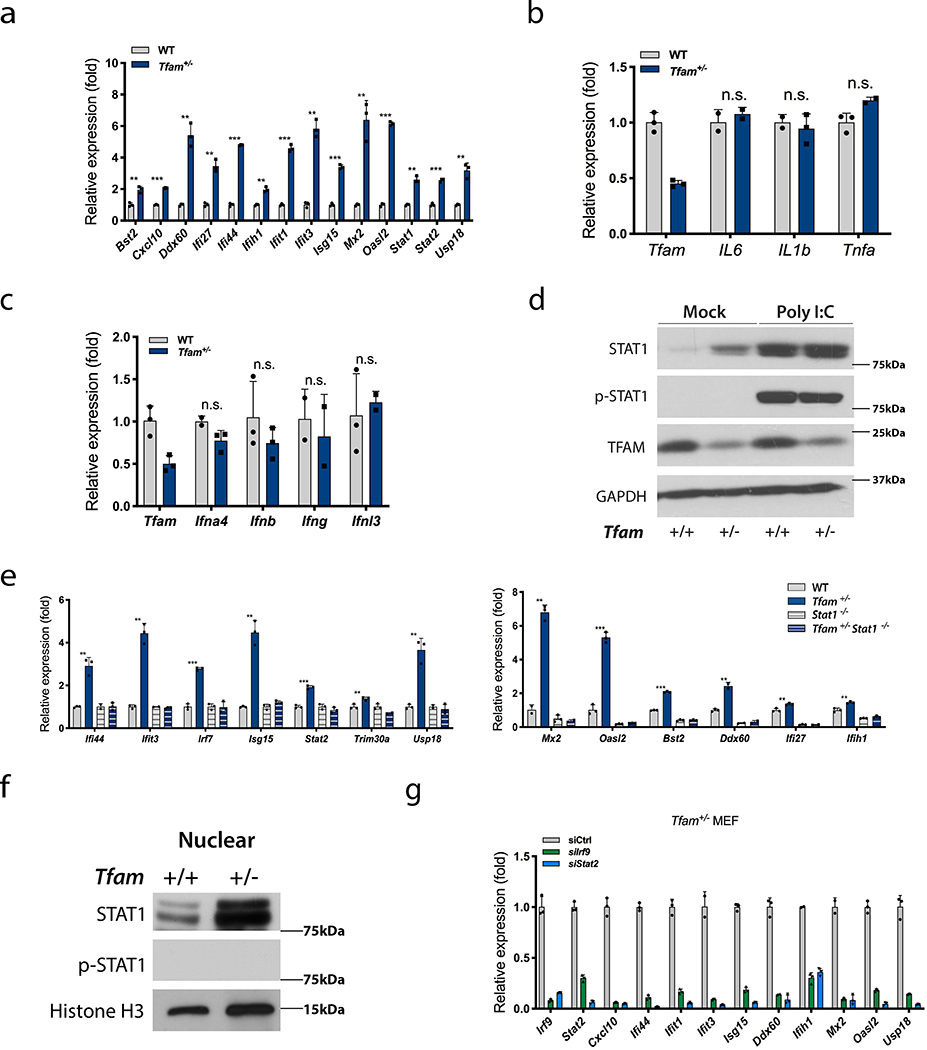
In this study, we endeavoured to better understand the nature and downstream consequences of innate immune signalling due to endogenous mtDNA stress. We showed previously that reduced expression of the mtDNA-binding protein TFAM (i.e. in cells from heterozygous Tfam+/− mice) causes elongation of mitochondria, enlarged nucleoids, and enhanced basal release of mtDNA into the cytoplasm that primes a cGAS-STING-dependent antiviral response4. Innate immune signalling due to cGAS-STING activation by released mtDNA has now been observed in many other cell types and conditions3,5–7. Acute antiviral responses usually engage both NF-κB-dependent activation of proinflammatory cytokines and IRF3/7-mediated induction of type I interferons2. However, we observed that, despite chronic ISG activation in Tfam+/− or siTfam MEFs (Fig. 1a, Extended Data Fig. 1a, b), there was no basal elevation of the NF-κB pathway target genes or protein components (Fig. 1b, Extended Data Figs. 1a and c, e) or type I, II or III interferon genes (Fig. 1c, Extended Data Figs. 1a, d). This led us to probe whether Tfam+/− cells are basally producing interferon (IFN). While treatment of wild-type (WT) MEFs with the viral RNA mimetic poly(I:C) resulted in robust activation of IFNβ (positive control, Extended Data Fig. 1f), conditioned media from Tfam+/− cells failed to stimulate an ISG response when added to WT cells (Extended Data Fig. 1g), consistent with little, if any, IFN being produced basally. In line with this, Tfam+/− cells have minimal, if any phosphorylated STAT1 (Y701) (p-STAT1), despite expressing more unphosphorylated STAT1 (U-STAT1) basally (Fig. 1d). This was not due to an inability to detect p-STAT1, as MEFs stimulated with poly(I:C) displayed strong phosphorylation of STAT1 as expected (Fig. 1d). These results indicate that chronic mtDNA-dependent ISG activation in MEFs is not occurring through canonical type I interferon-mediated JAK-STAT pathway activation, during which p-STAT1 and p-STAT2 form a protein complex with IRF9 called Interferon-Stimulated Gene Factor 3 (ISGF3)8. To test this more rigorously, we crossed Tfam+/− to Stat1−/− mice (which cannot form ISGF3) and analysed MEFs derived from them. MEFs from Stat1−/− mice retain normal mtDNA copy number, mitochondrial mass and membrane potential (Extended Data Figs. 2a–d). However, expression of most ISGs in Tfam+/− Stat1−/− MEFs were at baseline levels (Fig. 1e), demonstrating that mtDNA-induced ISG activation is STAT1 dependent. This was not due to reversal of the mtDNA stress phenotypes of Tfam+/− cells4, as Tfam+/− Stat1−/− cells retained elongated mitochondria and larger nucleoids (Extended Data Fig. 2e). The few ISGs that were STAT1-independent were dependent on IRF3 (Extended Data 2f–h), which is consistent with activated IRF3 driving expression of certain ISGs before signalling through the type I interferon-mediated JAK-STAT pathway9,10.
Figure 1. Innate immune signalling by chronic mtDNA stress requires U-ISGF3 but is not associated with NF-κB or interferon gene activation.
a-c, qRT-PCR analysis of (a) the indicated ISGs, (b) NF-κB target genes and (c) IFN genes in WT and Tfam+/− littermate MEFs. d, Western blot of STAT1, p-STAT1 (Y701), TFAM and GAPDH (loading control) in WT and Tfam+/− littermate MEFs transfected with 2μg Poly I:C or lipofectamine only (Mock) for 12 hours. (n=3 independent experiments) e, qRT-PCR analysis of the indicated ISGs in WT, Tfam+/−, Stat1−/− and Tfam+/− Stat1−/− littermate MEFs. f, Western blot of STAT1, p-STAT1 (Y701) and histone H3 (nuclear marker and loading control) in purified nuclear extracts from WT and Tfam+/− littermate MEFs. (n=3 independent experiments). g, qRT-PCR analysis of the indicated ISGs in Tfam+/− MEFs transfected with siRNA against Irf9 or Stat2 for 72 hours. Data were normalized to cells transfected with a siRNA control (siCtrl), which was given a value of 1.0. The data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates are provided as Supplementary Figures. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: **p < 0.01, ***p < 0.001, n.s. not significant (p > 0.05).
Activation of the JAK-STAT pathway by type I IFN leads to phosphorylation of STAT1 and STAT2, which with IRF9 form the trimeric ISGF3 transcription factor that activates ISGs in the nucleus8. However, unphosphorylated STAT1 and STAT2 also form a complex with IRF9 known as unphosphorylated ISGF3 (U-ISGF3)11–13. U-ISGF3 activates a subset of ISGs whose promoters harbour a specific type of interferon-stimulated response element11. Since mtDNA-induced ISG activation was dependent on U-STAT1 (Fig. 1e), we hypothesized that U-ISGF3 was driving this response. Consistent with this, we observed more U-STAT1 in the nuclei of Tfam+/− cells (Fig. 1f). Also, ISG activation in Tfam+/− cells was dependent on STAT2 and IRF9 in a manner that is not additive (i.e. epistatic) with STAT1, indicating they work together in the same pathway/complex (Fig. 1g, Extended Data Figs. 2i–k). Finally, 66% of the reported U-ISGF3-induced ISGs11 are upregulated in Tfam+/− MEFs (Extended Data Fig. 3a).
In addition to the critical functions of ISGs in antiviral responses8, Minn and colleagues have shown that cancer cells resistant to adjuvant chemotherapy, and radiation therapy exhibit increased expression of a unique subset of ISGs14,15. This interferon-related DNA damage resistance signature (IRDS)14 includes U-ISGF3-regulated ISGs (e.g. Ifit1, Ifit3, and Isg15) that are involved in the DNA damage resistance phenotype11. Since the U-ISGF3-regulated ISGs in Tfam+/− MEFs overlap significantly with the IRDS (Extended Data Fig. 3a), we asked if they confer resistance to chemotherapeutic DNA-damaging agents. Compared to WT controls, Tfam+/− MEFs are resistant to cell death induced by doxorubicin as judged visually (Fig. 2a) or with assays of cell viability and apoptosis (Figs. 2b–d). This resistance phenotype was not observed in Tfam+/− Stat1−/− cells that do not have ISG expression (Extended Data Figs. 3b–d). To gain insight into the mechanism of this resistance, we assessed nDNA damage responses in Tfam+/− MEFs. First, to examine the rate of nDNA repair, we challenged cells with doxorubicin for 12 hours, washed the cells with fresh medium, and then analysed the time course of repair in the absence of the drug. We found that nuclear phosphorylated Tumour Suppressor P53 Binding Protein 1 (p-53BP1) and phosphorylated histone H2A family member X (γH2A.X) DNA damage foci disappeared faster in Tfam+/− MEFs indicative of an enhanced rate of nDNA repair (Figs. 2e, f). Similar results were obtained in Tfam+/− MEFs after exposure to ionizing radiation (IR) (Extended Data Figs. 3e, f).
Figure 2. mtDNA-stress signalling enhances nuclear DNA damage and repair responses.
WT and Tfam+/− MEFs were treated with 1μM doxorubicin (Dox) for 48 hours and analysed for cell death parameters. a, direct visualization of phase images collected using a 4x objective (scale bar represents 200 μm, phase images represent n=3 biological replicates); b, Cell viability analysis using alamarBlue. Error bars indicate means ± SD of n=4 (1μM Dox) or n=3 (1.5μM Dox) biological replicates. c, apoptosis analysis by flow cytometry using Annexin V and Propidium Iodide (PI) followed by flow cytometry analysis. (Contour plots represent n=3 biological replicates.) d, Quantification of flow cytometry analysis in c. for live cells (Annexin V and PI low), apoptotic cells (Annexin V high and PI low) and dead cells (Annexin V and PI high). Error bars indicate means ± SD of n=3 biological replicates. b, e and f, Analysis of nDNA repair rate (i.e. the rate of γH2A.X and p-53BP1 foci resolution during recovery after 12 hours of doxorubicin-mediated damage) in WT and Tfam+/− littermate MEFs. Nuclei are labelled with DAPI (blue), while γH2A.X (magenta) and p-53BP1 (green) were detected by immunofluorescence. Plotted to the right of the images is the average number of foci per nucleus at the indicated times after doxorubicin was removed. g and h, Analysis of the rate of induction of the nDNA damage response (i.e. the rate of γH2A.X and p-53BP1 foci formation) in WT and Tfam+/− littermate MEFs. Cells were analysed at the indicated time points after addition of 1μM doxorubicin. Imaging was done as described in e, f. Plotted to the right is the percentage of positive cells (>20 γH2A.X foci per nucleus or >15 p-53BP1 foci per nucleus) as a function of the time after doxorubicin addition. i and j, Analysis of the rate of induction of the nDNA damage response as described in g and h in Tfam+/− littermate MEFs transfected with two independent Parp9 siRNAs (#1 or #2) or a control (Ctrl) siRNA for 48 hours. e-j, Error bars indicate means ± SD of n=3 biological replicates in which 50 nuclei were quantified. k and l, Analysis of nDNA repair in vivo. WT and Tfam+/− mice with (IR) or without (Ctrl) exposure to 10Gy IR were sacrificed 24 hours post-exposure. Mouse spleens (n=3 per group) were fixed and stained to quantify nuclear γH2A.X and p-53BP1 foci. Plotted to the right is the number of “foci positive” splenocytes defined as DAPI-positive nuclei (blue) with more than 5 γH2A.X (magenta) or 11 p-53BP1 (green) foci. Scale bars represent 20 μm. Error bars indicate means ± SD (n=3 biological replicates, >400 nuclei each). All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * p < 0.05, ** < 0.01, *** p < 0.001.
Given the faster nDNA repair kinetics in Tfam+/− cells, we inspected our prior gene expression profiling results4 for known DNA repair factors and found that several PARP enzymes (PARP9, PARP10, PARP12 and PARP14) were upregulated (Extended Data Fig. 4a). While there was no upregulation of Parp1 and Parp2 (Extended Data Fig. 4a), which have well-documented roles in nDNA repair, Parp9 is an ISG involved in the PARP1-mediated DNA repair pathway16–19. Accordingly, we confirmed that Parp9 is upregulated in Tfam+/− MEFs in a STAT1-dependent fashion19 (Extended Data Fig. 4b). To determine if the increase in PARP9 contributes to the DNA damage resistance phenotype, we knocked down PARP9 in Tfam+/− cells. PARP9 knockdown dampened both the resistance to doxorubicin (Extended Data Figs. 4c, d) and the enhanced rate of p-53BP1 and γH2A.X foci disappearance (Extended Data Figs 4e–h). These data demonstrate that increased PARP9 expression contributes to enhanced nDNA repair observed in Tfam+/− cells. During PARP1-mediated DNA repair, PARP9 recognizes ribosylated histone proteins and recruits the ubiquitin ligase DTX3L to DNA damage sites to ubiquitinate histone H417,18. The ubiquitination of histone H4 enables SET8 to methylate histone H4 for subsequent recruitment of p-53BP118. Therefore, we hypothesized that Tfam+/− MEFs, which have basally increased PARP9, would more quickly induce nDNA damage response in addition to exhibiting enhanced nDNA repair kinetics. To address this, we altered our strategy to assess the rate of DNA repair foci formation immediately after exposure of cells to DNA damage. First, to visualize nDNA damage foci formation very shortly after DNA damage, we fixed the cells at 7.5 and 15 minutes after IR, and observed more p-53BP1 and γH2A.X foci in Tfam+/− MEFs at these time points (Extended Data Figs. 3g, h). Similar results were obtained with 1-hour doxorubicin treatment, where Tfam+/− MEFs induced significantly more p-53BP1 and γH2A.X foci than WT control cells (Figs. 2g, h). As was the case for the enhanced nDNA repair rate, the enhanced induction of DNA damage response was blunted by knocking down PARP9 in Tfam+/− MEFs (Figs. 2i, j). From these DNA-damage response induction/repair studies, we conclude that mtDNA stress primes cells for a faster nDNA damage response and more efficient nDNA repair (i.e. a leftward shift in the entire nDNA damage response and repair curve) and that this is mediated, in part, through upregulation of PARP9. Finally, to determine if mtDNA stress enhances the DNA-damage induction/repair response in vivo, we exposed WT and Tfam+/− mice to IR. Analysis of spleens 24 hours after IR revealed fewer nuclear γH2A.X and p-53BP1 foci in the Tfam+/− mice compared to WT controls, consistent with a more robust nDNA repair response in the Tfam+/− mice (Figs. 2k, l; compare small magenta or green foci inside blue splenocyte nuclei).
We next asked if mtDNA is involved in the response to DNA damaging agents in WT (i.e. Tfam+/+) cells, which are not experiencing mtDNA stress basally. Doxorubicin treatment robustly induced ISG expression in a STAT1-dependent manner, as well as some STAT1 Y701 phosphorylation (Fig 3a, and Extended Data Figs. 5a, b). Remarkably, this was accompanied by elongated mitochondria and large nucleoids (Extended Data Fig. 5c), mirroring the basal mtDNA-stress phenotypes in Tfam+/− MEFs. Additionally, doxorubicin increased mitochondrial reactive oxygen species (ROS) (Fig 3b), mtDNA damage (Fig 3c), and release of mtDNA into the cytoplasm (Figs. 3d, e). To determine if mtDNA is directly involved in ISG induction by doxorubicin, WT MEFs were exposed to ethidium bromide or 2’,3’-dideoxycytidine (ddC) to deplete mtDNA (Extended Figs. 5d, f). The ability of doxorubicin to induce ISGs in these mtDNA-depleted cells was significantly blunted (Fig. 3f and Extended Data Fig. 5e). However, depletion of mtDNA in MEFs did not inhibit ISG activation in response to the innate immune receptor agonist dsDNA9020 (Extended Data Fig 5g), demonstrating that loss of OXPHOS capacity or other metabolic consequences downstream of mtDNA depletion does not compromise the ability of these cells to mount a full ISG signalling response. This further implicated mtDNA release per se as the trigger for ISG induction. Consistent with this interpretation, we found mitochondria-targeted doxorubicin (mitoDOX), which causes direct mtDNA damage21, is sufficient to induce an ISG response in WT MEFs (Fig. 3g). In addition, we also saw robust activation of ISGs with doxorubicin or mitoDOX in mouse LMTK− cells (Fig. 3h and Extended Data Fig. 5i) but not in their ρ° counterparts, which completely lack mtDNA (Fig. 3i and Extended Data Figs. 5h, i). Since ρ° cells not only lack mtDNA, but also mtDNA-encoded proteins, we treated cells with chloramphenicol to inhibit mitochondrial translation (as evidenced by loss of the mtDNA-encoded cytochrome oxidase 1 subunit; Extended Data Fig. 5j), but leave the mtDNA intact. We observed similar induction of ISGs under these conditions (Extended Data Fig. 5k), discounting mtDNA-encoded proteins and further implicating mtDNA itself as a major trigger for doxorubicin-induced ISG activation. Finally, in mouse MC-38 colon cancer cells, we also observed doxorubicin-mediated ISG activation (Extended Data Fig. 6a) that was dependent on mtDNA (Extended Data Figs. 6c–f) and accompanied by phosphorylation of STAT1 (Extended Data Fig. 6b). Furthermore, mitoDOX was sufficient for ISG induction in these cells (Extended Data Fig. 6g). Using CRISPR-Cas9 knock-out cell pools (Extended Data Fig. 6h), we demonstrated that the ISG response to doxorubicin in MC-38 cells was dependent on the cGAS-STING-TBK1 DNA-sensing pathway but not the MAVS pathway for RNA-mediated innate immune signalling (Extended Data Figs. 6i–k). From these results, we conclude that doxorubicin not only damages nDNA, but also directly damages mtDNA, leading to cytoplasmic release and ISG activation in primary MEFs and MC-38 cancer cells. We assume that the residual ISG activation observed in mtDNA-depleted cells treated with doxorubicin (Fig. 3f and Extended Data Figs. 5e, 6e, f) is caused by nDNA damage and release of cytoplasmic chromatin fragments or micronuclei.
Figure 3. Damage and release of mtDNA mediates ISG expression.
WT MEFs were treated with (+Dox) or without (-Dox) 500 nM doxorubicin for 24 hours and subjected to a, qRT-qPCR analysis of the indicated ISGs; b, flow cytometry analysis using MitoSox. The mean fluorescence intensity (MFI) is plotted; c, mtDNA damage analysis by a long-PCR assay. Error bars indicate means ± SD of three biological replicates. d, Western blot analysis of whole cell extract (WCE), Pellet (Pel) and cytoplasmic (Cyt) fractions (see Methods section) derived from WT MEFs treated with (+) or without (−) 500nM doxorubicin (Dox) for 24 hours. GAPDH was used to mark the cytoplasmic (Cyt) fraction, in which the lack of mitochondrial and nuclear contamination was assessed by probing for HSP60 and histone H3. γH2A.X was probed as a marker of DNA damage. (n=3 independent experiments). e, Analysis of mtDNA present in purified cytoplasmic fractions (described in d) by qPCR (three different mtDNA primers were used: Dloop, Dloop2 and ND4). f, WT MEFs with (EtBr) or without (Ctrl) prior ethidium bromide treatment to deplete mtDNA were treated with (+Dox) or without (-Dox) 500nM doxorubicin for 24 hours and analysed by qRT-PCR for expression of the ISGs Ifit1 and Ifit3. g, qRT-PCR analysis of the indicated ISGs in WT MEFs treated with DMSO (Mock) or 3μM mitochondria-targeted doxorubicin (mitoDox) for 24 hours. h and i, qRT-PCR analysis of the indicated ISGs in LMTK− cells with (ρ+) or without (ρ˚) mtDNA that were treated with DMSO (Mock) or 3μM mitoDox for 48 hours. a, e-i, The data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates and the FACS gating strategy are provided as Supplementary Figures. b and c, Error bars indicate means ± SD of n=3 biological replicates. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, n.s. not significant, (p > 0.05).
We next examined how mtDNA-stress-mediated enhancement of nDNA repair might affect chemotherapy drug responses in cancer cells. Using CRISPR-Cas9, we generated TFAM-deficient (TFD) versions of mouse MC-38 colon cancer and mouse melanoma (YUMMER1.7) cells22. In both cases, we observed the stereotypical mtDNA-stress phenotypes we defined initially in MEFs, including mtDNA depletion and enlarged nucleoids (Figs. 4a, 4d, and Extended Data Figs. 7a–d and 8a), which was accompanied by cGAS-STING-TBK1 pathway-mediated basal activation of ISGs (Figs. 4b,e and Extended Data Figs. 7e–g and 8c). Like Tfam+/− MEFs, TFD YUMMER1.7 and MC-38 cells with an enhanced ISG response exhibit resistance to doxorubicin (Fig. 4c and Extended Data Figs. 7h and 8b) that is dependent on the cGAS-STING-TBK1 pathway (Fig. 4f). These results are consistent with mtDNA-stress-induced ISGs mediating an enhanced nDNA damage response in cancer cells in vitro. Therefore, we next tested if this was also the case in an in vivo setting. After confirming that the mtDNA-stress phenotypes in TFD YUMMER1.7 cells could be complemented by ectopically replenishing TFAM (Extended Data Figs. 8d–g), we transplanted WT and TFD YUMMER1.7 cells into nude mice and measured tumour growth responses in the presence and absence of doxorubicin (Fig. 4g). Doxorubicin significantly inhibited the tumour growth rate of WT YUMMER1.7 cells but did not have a significant effect on TFD YUMMER1.7 cells (Figs. 4h, i), leading us to conclude that mtDNA-stress also promotes resistance to DNA damaging agents in vivo.
Figure 4. Analysis of mtDNA-stress phenotypes in TFAM-deficient mouse melanoma cells and associated chemoresistance in vivo.
TFAM-deficient (TFD) YUMMER1.7 mouse melanoma cells were generated by CRISPR-Cas9 editing and analysed compared to unedited (WT) control cells. a, mtDNA abundance (relative mtDNA copy number) by qPCR with Dloop primers. b, qRT-PCR analysis of the indicated ISGs. c, analysis of cell viability in response to 48 hours of 500nM doxorubicin using an alamarBlue assay. (n=6 biological replicates.) d, Immunofluorescence of mitochondria (anti-HSP60, magenta) and mtDNA nucleoids (anti-DNA, cyan). Images are Z-stack projections and scale bar represents 10 μm. (n=3 independent experiments) e, qRT-PCR analysis of the indicated ISGs in WT or TFD YUMMER1.7 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (scr) gRNA control. a, b and e, The data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates are provided as Supplementary Figures. f, The same cell lines as in e were treated with 500nM doxorubicin for 48 hours and then subjected to cell viability analysis using the alamarBlue assay. (n=4 biological replicates). g, Schematic of protocol used for assessment of doxorubicin chemoresistance in vivo using WT and TFD YUMMER1.7 cells. h, Growth measurements of WT and TFD YUMMER1.7 tumours in mice with (Dox) or without (Ctrl) doxorubicin treatment. Tumour volume is plotted as a function of days after cell transplant and the day of Dox treatment is indicated. Error bars indicate means ± SD of n=8 biological replicates. i, Tumour volume (mm3) of WT or TFD tumours on day 8 (WT) and day 18 (TFD) are plotted, which are the days when the control (-Dox) tumours in reached 1000 mm3 (h and i, n=8 mice per group). All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, *** P < 0.001, n.s. not significant (P > 0.05).
Finally, we conducted bioinformatics analysis of Tfam and ISG expression patterns in over 1000 cancer cell lines using data from the Cancer Cell Line Encyclopedia23. We partitioned cells into four quartiles based on Tfam RNA expression. Comparing expression of ISGs in the upper (top 25%) and lower (bottom 25%) quartiles revealed significantly higher ISG expression in cells with lower Tfam expression (Extended Data Fig. 9a). This correlation was not observed with IFN genes (IFNA1, IFNB1 and IFNG; Extended Data Fig. 9a). An inverse relationship to ISG expression was also found with other genes associated with mtDNA metabolism (Lig3, Nthl1 and Polg; Extended Data Fig. 9b), but not with mitochondrial genes in general (e.g. SOD2, SDHD, and VDAC1; Extended Data Fig. 9c). These data are consistent with mtDNA-associated ISG activation, without parallel IFN activation in human cancer cells.
While it is well-established that mtDNA is a direct target of DNA-damaging chemotherapeutic agents24, we show here that mtDNA release is a salient downstream consequence of this damage, resulting in cGAS-STING activation and enhanced nDNA repair responses. While this likely initially involves canonical IFN-JAK-STAT signalling and subsequent ISG induction by ISGF3, our results show that mtDNA can lead to sustained activation of a specific class of ISGs by U-ISGF3 that significantly overlaps with the IRDS subset11,14. Importantly, this form of mtDNA stress enhances nDNA repair capacity, with PARP9 playing a key role based on our results. However, we emphasize that PARP9 is merely one new factor implicated in the response and that it likely collaborates with other ISGs to produce this phenotype14. Furthermore, other forms of mtDNA stress can have deleterious, rather than beneficial, effects on nDNA replication and repair (e.g. through effects on cellular dNTP pools)25. Thus, how different types of mtDNA stress ultimately affect nDNA stability under specific physiological and pathological contexts is worthy of more study. Lastly, we speculate that mtDNA is a sensor of genotoxic stress that mediates a novel mitochondria-to-nucleus, stress-signalling pathway26 to prime nDNA damage and repair responses that is beneficial under normal circumstances. That mtDNA has fewer DNA repair pathways than nDNA27,28 may ensure that it is more prone to damage than nDNA, which allows it to serve as a genotoxic stress sentinel. However, based on our results, we speculate that chronic mtDNA-stress signalling of this type in cancer cells could contribute to chemotherapy drug resistance. If so, inhibition of mtDNA damage and release pathways might be a potential avenue to prevent chemoresistance in human cancer patients.
METHODS
Antibodies and other reagents
The following antibodies were obtained commercially: STAT1 (Cell Signaling Technology, 9172S), p-STAT1 (Tyr 701) (D4A7) (Cell Signaling Technology, 7649), STAT2 (Cell Signaling Technology, 4597S), NF-κB, p-NF-κB, IKKβ, IκB (Cell Signaling Technology, 9936), HSP60 (Cell Signaling Technology, 12165T), cGAS (D3O8O) (Cell Signaling Technology, 31659S), STING (D2P2F) (Cell Signaling Technology, 13647S), TBK1 (D1B4) (Cell Signaling Technology, 3504), MAVS (Cell Signaling Technology, 4983S), anti-DNA (Millipore CBL186), γH2A.X (Ser139, EMD Millipore 05–636), Histone H3 (Abcam ab1791), GAPDH (Ambion, AM4300), VDAC (abcam, ab15895), actin (Santa Cruz, sc47778), IRF9 (ProteinTech, 14167–1-AP), p-53BP1 (Cell Signaling Technology, 2675S) and Vinculin (Sigma, V9131), OXPHOS antibody Cocktail (abcam, ab110413). Rabbit anti-mouse Tfam polyclonal anti-sera was previously described.4,29
The following reagents were obtained commercially: SYBR Green Master Mix (ThermoFisher, #4364346), RNeasy plus RNA extraction kit (QIAGEN, #74136), alamarBlue (Invitrogen, DAL1025), DC™ protein assay kit II (Bio-Rad, 5000112), doxorubicin (Sigma, #44583), ISD Control/LyoVec (Invivogen, tlrl-isdcc) and ISD/LyoVec (Invivogen, tlrl-isdc), uridine, (Sigma, U3750). sodium pyruvate (Invitrogen, #11360070), 2’3’-dideoxycitidine (ddC) (Sigma, D5782–100MG), ethidium bromide (Sigma, E1510–10ML), Lipofectamine2000 (Invitrogen, #11668027), Lipofectamine 3000 (Invitrogen, L3000075), RNAiMAX (Invitrogen, #1847641), MitoTracker Green (Thermo Fisher, #M7514), MitoTracker Deep Red (Thermo Fisher, #M22426). MitoSOX, (Invitrogen, M36008), fibronectin (Sigma, F1141–5MG), Prolong-Antifade (Invitrogen, P36935), DAPI (Thermo Fisher, #1816957), DAKO fluorescence mounting medium (Dako NA Inc, S3023), annexin V binding buffer (Biolegend, #42220), Pacific Blue annexin V (Biolegend, #640918) and propidium iodide (MP Biomedicals, #195458), crystal violet (Fisher Scientific, #C581–25), PicoGreen (Invitrogen, #P7581), formalin (Anatech Ltd. #174), DNA/Hind III marker (Fisher Scientific, SM0101), Chloramphenicol (Sigma, R4408), High Capacity cDNA RT Kit (Thermo Fisher, #4368814), GeneJET genomic DNA purification kit (Thermo Scientific, #K0721) and LongAmp Tag PCR kit (New England Biolabs, #E5200S).
PCR primers (Supplemental Table 1) siRNAs (Supplemental Table 2) and gRNAs (Supplemental Table 3) were synthesized by IDT. The forward (F) and reverse (R) dsDNA90 oligos (Supplemental Table. 4) were synthesized by Sigma and annealed as follows: 5 μL of 3 μg/μL dsDNA-F and dsDNA-R were added to 5 μL of annealing buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 50 nM NaCl) and 35 μL endotoxin-free water and heated and cooled (37˚C for 30 min, 95˚C for 5 min, and cooled to 25˚C at a rate of 5˚C/min).
The following reagents were obtained from the indicated investigators. LPS and Poly I:C, Dr. Akiko Iwasaki (Yale University); mitochondria-targeted doxorubicin (mitoDox), Dr. Shanna Kelly (University of Toronto); pSpCas9(BB)-2A-GFP (pX458) vector, Dr. Feng Zhang (Addgene, # 48138); LentiCRISPRv2 vector, Dr. Qin Yan (Yale University) and pCDH-EF1-FHC vector, Dr. Richard Wood (Addgene, #64874).
Animal strains and cell lines
The Tfam+/− mice were originally derived from Tfamflox mice obtained from Dr. Navdeep Chandel (Northwestern University) and generated as described previously4,29. The Stat1−/− mice30 and female athymic nu/nu mice (Hsd:Athymic Nude-Foxn1nu, Envigo, Huntingdon, UK) were purchased from Jackson Labs (Stock No.012606). All animal husbandry and procedures were IACUC approved by the animal care and use committees at Yale University or the Salk Institute for Biological Studies.
MC-38 colon cancer cells were purchased from ATCC. Mouse YUMMER1.7 melanoma cells were obtained from Dr. Marcus Bosenberg’s (Yale University). Mouse LM thymidine kinase- (LMTK-) cells were obtained from David Clayton (Stanford University). 293FT cells were purchased from ATCC and Lenti-x 293T cells were purchased from Takara CloneTech.
Cell culture and media-exchange experiments
WT and Tfam+/− MEFs were generated from E12.5–14.5 mouse embryos and cultured in DMEM (Invitrogen), supplemented with 10% FBS (Atlanta Biological). All experiments were performed in MEFs passaged five or fewer times, except in the case of cells depleted of mtDNA by ddC or ethidium bromide, which required additional passages. YUMMER1.7 cells were cultured in DMEM/F12 (Gibco) supplemented with 10% FBS (Gibco). MC-38 and LMTK- cells were cultured in DMEM supplemented with 10% FBS (Gibco). For LMTK- ρ˚ cells, the 50 ng/mL uridine and 1 mM sodium pyruvate were added to the growth media. Transfection of poly I:C and dsDNA-90 were performed using Lipofectamine 2000 at a ratio of 2:1 (lipofectamine 2000:μg nucleic acid). Transfection of siRNAs into MEFs was achieved using Lipofectamine RNAiMax according to manufacturer’s instructions. All siRNAs were transfected at a final concentration of 25 nM.
To generate TFAM-deficient (TFD) YUMMER1.7 and MC-38 cells, Tfam gRNA was cloned in pSpCas9(BB)-2A-GFP (pX458) vector and transfected at a ratio of 2:1 (lipofectamine 3000:μg nucleic acid). GFP-positive cells were sorted one cell/well into 96-well plates using a Becton-Dickinson Influx™ cytometer 48 hours post-transfection. The colonies were genotyped using Tfam-deletion primers (mTfam Del, Supplemental Table 1). To generate CRISPR-knockout cell and TFAM-overexpressing cell lines, LentiCRISPRv2 vectors with gRNA31 or pCDH-EF1-FHC vectors32 with or without HA-Flag-TFAM were transfected into 293FT or Lenti-x 293T cells with 2:1 (psPAX2:pMD2G) plasmids using lipofectamine 2000 to produce lentivirus. Lentivirus-containing media were collected 48 hours post-transfection and filtered using a 0.45 μm membrane. YUMMER1.7 cells and MC-38 cells were infected with lentivirus for 24 hours and then selected for antibiotic resistance in 1μg/mL or 5μg/mL puromycin, respectively, for 2–3 weeks.
Media-exchange experiments were performed to assay for IFN production by Tfam+/− cells. WT and Tfam+/− MEFs were seeded into 6-well plates to achieve confluency after 24 hours, at which point the cell-free media from each were collected. For the positive control, WT MEFs were transfected with 2 μg of poly I:C for two hours, washed 3x with PBS, replenished with fresh media, and then collected after 7 hours. Negative controls were plain media as well as conditioned media from unstimulated WT MEFs. All conditioned media were filtered using a 0.45 μm membrane and then added to fresh WT MEFs. After overnight culture with the conditioned or control media, RNA was extracted from cells for qRT-PCR analysis of ISGs.
Quantitative PCR
To quantify mRNA transcript abundance, RNA was extracted using the RNeasy plus RNA extraction kit followed by reverse transcription using High Capacity cDNA RT Kit. Equal amounts of cDNA and the indicated primers (Supplemental Table 1) were used for qPCR using Fast SYBR Green Master Mix. For each biological sample three technical replicates were performed and normalized against the GAPDH Ct value. Relative expression was analysed using the 2−ΔΔCt method and the relative fold change was plotted with the control samples given a value of 1.0. To quantify mtDNA, cells were suspended in 50mM NaOH and boiled for 30 min by neutralization by 1/10 volume of 1M Tris-HCl (pH 8.0). DNA samples were diluted to 10ng/μL and subjected to qPCR analysis using Dloop (two Dloop and ND4 primers, Supplemental Table. 1) and Tert primers (Supplemental Table. 1) to amply mtDNA and nuclear DNA, respectively. Three technical replicates were performed for each biological sample and normalized against nuclear Tert Ct value. Relative copy number was analysed using a 2−ΔΔCt method and the control mtDNA abundance was given a value of 100%.
Nuclear fractionation and cytosolic mtDNA analysis
Nuclear fractionation was performed as described4. In brief, after washing with PBS, cells were resuspended in cold RSB buffer (10 mM NaCl, 1.5 mM CaCl2, 10 mM Tris-HCl pH 7.5) and incubated on ice for 10 min. A motorized Teflon pestle was used to homogenize the cells followed by centrifugation at 980xg for 10 min. Pellets were washed 5 times with PBS, solubilized in 1% SDS, boiled for 5 min, sonicated to shear the DNA and then protein concentrations were determined using DC™ protein assay kit II.
To assay cytosolic mtDNA, 1 × 107 MEFs treated with or without 500 nM doxorubicin for 24 hours were collected for cytoplasmic fractionation exactly as we described previously5. Quantitative PCR was performed on DNA samples from the whole cell and cytosolic fractions using the mtDNA and nuclear DNA primers in Supplemental Table 1. The nuclear DNA amplification value in the whole cell lysate was used for normalizing cell numbers.
Flow cytometry
To assess mitochondrial mass, membrane potential and ROS, cells were stained with 50nM MitoTracker Green and 20nM MitoTracker Deep Red or 5μM MitoSOX, for 30 min. Cells were then washed with PBS, pelleted, and resuspended in PBS with 2% FBS prior to flow cytometry analysis. For apoptosis analysis, cells were challenged with 1 μM doxorubicin for 48 hours. All live and dead cells were collected, washed with PBS, pelleted, re-suspended in 60–100μL Annexin V-binding buffer containing Pacific Blue Annexin V (1:20 dilution) and 1 μg/mL propidium iodide (PI) and subsequently incubated for 15–20 minutes prior to flow cytometry analysis. For mitochondrial mass, membrane potential and ROS assessment, see FACS gating strategy (Supplementary Fig. 11). For the analysis of apoptosis, all of the cells were analysed. All flow cytometry data were analysed using FlowJo software.
Immunofluorescence microscopy
To assess mitochondrial and nucleoid morphology, cells were seeded on coverslips coated with 10μg/ml fibronectin in PBS for 1 hour at 37˚C. Cells were fixed in a pre-warmed (37°C) solution of 4% paraformaldehyde in PBS for 15 minutes at room temperature, permeabilized with 0.1% (v/v) Triton X-100 in PBS for 10 minutes at room temperature, and blocked using filtered PBS containing 1% (w/v) BSA for an hour at room temperature. Coverslips were then incubated with antibodies against DNA (1:150) and HSP60 (1:1000), diluted in filtered PBS containing 1% BSA at 4°C overnight, followed by 4 × 5-minute washes in PBS containing 5mM EDTA. Secondary antibodies (1:500, Alexa fluorophores 488 and 546, Invitrogen) were incubated in PBS containing 1% BSA for 1 hour at room temperature and removed by 4 × 5-minute washes in PBS. Coverslips were then mounted onto slides using Prolong Antifade. Cells were imaged using (For Extended Data Fig. 2e) a Leica SP5 confocal microscope (100x objective) or (For Fig. 4d and Extended Data Figs. 5c, 7d and 8g) a Zeiss LSM 880 Rear Port Laser Scanning Confocal and Airyscan FAST microscope (63x objective), with Z stacks acquired. Airyscan images were processed using Zen Black software.
For γH2A.X and p-53BP1 foci analyses in MEFs, cells were grown in chamber slides coated with 20μg/mL in PBS for 2 hours at 37˚C (154534, Lab-Tek) prior to the indicated treatment. Cells were fixed/permeabilized in 3% PFA/0.5% Triton-X100/2% sucrose for 20 min at the indicated time points. Blocking was performed with 5% fetal bovine serum and 5% normal goat serum in PBS overnight at 4°C followed by overnight incubation with primary antibodies against γH2A.X antibody (1:400) or p-53BP1 antibody (1:300), diluted in the blocking solution at 4˚C. After three times of wash with PBS containing 0.5% Triton-X100, cells were incubated with secondary antibodies (anti-mouse AF Plus 555, anti-rabbit AF Plus 488 (1:400; A32727, A32731, Thermo Fisher)) diluted in the blocking solution for 90 min at room temperature. DNA was stained with 2.5μg/mL DAPI for 15 min at room temperature followed by 3 times of wash. Chambers were removed from the slides and the slides were covered with coverslips using DAKO Fluorescence Mounting Medium. Images were analysed using Nikon Eclipse Ti fluorescence microscope with a Plan Apo 60X/1.40 Oil DIC h objective, a CSU-W1 confocal scanning unit with an iXon Ultra camera (Andor Technology), MLC 400B laser unit (Agilent Technologies) and NIS-Elements 4.30 software (Nikon Corporation).
For γH2A.X and p53BP1 foci analyses in spleen, 8-month-old WT and Tfam+/− littermate mice were subjected to 10 Gy whole body IR. Both control mice (that did not receive IR) and irradiated mice were sacrificed 24 hours after treatment. Spleens were harvested and immediately fixed in aqueous buffered zinc formalin for 24 hours, then transferred to 70% ethanol. Fixed spleens were sent to the UCSD histology core for sectioning. Deparaffinization and rehydration was achieved by immersing the slides in Xylene three times for 5 min and twice each in 100% Ethanol, 95% Ethanol, 70% Ethanol, 50% Ethanol, and deionized water for 10 min. Antigen retrieval was performed by boiling the slides for 20 min in sodium citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0), followed by a 5 min wash in deionized water. Subsequently, slides were submerged in 1% normal goat serum in PBS with 0.5% Triton X-100 and 8% sucrose for 20 min and incubated for 2 hours at room temperature in blocking solution (5% normal goat serum, 5% fetal bovine serum, 0.2% Triton X-100 in PBS) prior to anti-γH2A.X, anti-p-53BP1 and DAPI DNA staining as described above. Images were taken using Nikon Eclipse Ti fluorescence microscope as described above for MEFs, using a Plan Apo 100X/1.40 Oil DIC h objective.
Foci were analysed using the Focinator v2–31 software as previously described33,34. In spleen samples, DAPI DNA staining was used to exclude the high auto-fluorescent red blood cells from the counting process. All representative images were processed using ImageJ.
Cell viability analysis
WT and Tfam+/− littermate MEFs were seeded into 6 well plates at 2,000 cells/well. The next day, the media was replenished with or without 1μM doxorubicin. Light images were taken using an EVOS phase microscope (4x objective) 2 days post-treatment. All live and dead cells were subjected to flow cytometry analysis (see Flow Cytometry Analysis). Cell viability after doxorubicin treatment was measured by fluorescent detection of alamarBlue reagent, according to the manufacturer’s instructions. The number of cells seeded and the incubation time with alamarBlue reagent was adjusted for each cell line: MEFs, 5000 cells, 2.5 hours; MC-38, 2000 cells, 1 hour; YUMMER1.7, 3000 cells, 2 hours. For colony-formation assays, YUMMER1.7 cells were seeded in 6-well plates at 5000 cells/well. The next day, cells were challenged with indicated doses of doxorubicin for 4 hours. Cells were then washed with PBS three times and replenished with fresh warm media. After visible cell colonies formed, cells were fixed with ice-cold methanol on ice for 5 min and stained with 0.05% crystal violet in 10% ethanol at room temperature for 20 min. Cells were washed with water and imaged using ChemiDoc™ MP imaging system (Bio-Rad). Colonies were counted using “analyse particle” command in Image J.
Analysis of mtDNA damage
A long-PCR assay was used to quantify mtDNA damage35 using total cellular DNA extracted with GeneJET Genomic DNA purification Kit from MEFs treated with or without doxorubicin. DNA concentration was measured using PicoGreen. A standard curve for DNA measurement was generated using lambda DNA/Hind III marker. DNA (15 ng) from each sample was subjected to long-PCR analysis using LongAmp Tag PCR kit and mtDNA copy number analysis (Described in Quantitative PCR). The long-PCR products were quantified using PicoGreen and the relative amplification (Rel. Amp) was normalized to mtDNA copy number in each sample.
Tumour cell growth and doxorubicin resistance in vivo
Seven-week-old female athymic nu/nu mice (Hsd:Athymic Nude-Foxn1nu, Envigo, Huntingdon, UK) were used for the in vivo tumour studies. Mice were quarantined for 2 weeks before the experiment. WT or TFD YUMMER1.7 cells were implanted subcutaneously (2 × 106 cells in 100 μL PBS) in the right flank (16 mice per group). Mice were randomized into treatment and non-treatment groups when tumours reached 100 mm3 (8 mice per group), and then received a single intraperitoneal injection of PBS or 10 mg/kg doxorubicin in PBS, respectively. Mice were visually observed daily, and tumours were measured every second day using a digital calliper. Tumour dimensions were measured, and volume determined according to the following equation: Volume = L x W x H x π / 6. Mean (geometric) tumour volume (mm3) was plotted over time to monitor tumour growth. Additionally, the weight of the mice was measured for general health as well as potential side effects of the drug.
Bioinformatic analysis
Heat maps (Extended Data Figs. 1a and 4a) were generated using R (3.4.1) with previously published data of differential gene expression between WT and Tfam+/− MEFs4. For gene expression profiling of data from cancer cell lines and tumour samples were downloaded from CCLE (Cancer Cell Line Encyclopedia; https://portals.broadinstitute.org/ccle)23. Cancer cell lines were sorted according to the expression level of genes of interest (e.g. Tfam, mtDNA-metabolism-related genes, other mitochondrial genes) and the top and bottom quartiles were analysed for expression of ISGs, IFN genes and GAPDH (Extended Data Fig. 9). P values were calculated using Wilcoxon rank-sum test.
Statistical analysis
Figures were prepared using Prism8 or R (3.4.1). Unless otherwise indicated in the figure legends, statistical significance was calculated using an unpaired, two-tailed student’s t-test with 95% confident intervals. For multiple t tests, Holm-Sidak method was used to correct for multiple comparisons and adjusted P-values were used to determine the significance. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, n.s. not significant, (p > 0.05). Further statistic information of each individual figure including specific P-values, degree of freedom, adjustment for multiple comparisons, and boxplots information are provided as source data.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
CCLE RNA-seq data can be found through link https://portals.broadinstitute.org/ccle. TFAM microarray data are available in NCBI GEO database with accession number GSE63767. Additional qPCR biological replicate data can be found in the supplementary file. Other data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Code availability
Code used for data analysis was deposited in Github. http://github.com/kepbod/CCLE_analysis
Extended Data
Extended Data Fig. 1. Additional analysis of innate immune signalling in MEFs.
a, Heat map of normalized expression values of the indicated ISGs (red font), interferon genes (black font), and NF-κB target genes (blue font) from our previously published microarray analysis4 of WT and Tfam+/− MEFs (two of each). b-d, qRT-PCR analyses of the indicated ISGs (b), NF-κB target genes (c) and interferon genes (d) in WT MEFs transfected with control (Ctrl) or Tfam siRNA for 72 hours. e, Western blot probing the indicated NF-κB pathway proteins, TFAM and VDAC (loading control) in WT and Tfam+/− littermate MEFs. (n=3 independent experiments.) f, qRT-PCR analysis of interferon β (Ifnb) gene expression in WT MEFs transfected with 2 μg poly(I:C) or lipofectamine only (Mock) for 9 hours. g, qRT-PCR analysis of the indicated ISGs in WT MEFs cultured overnight in the presence of control media (plain), media conditioned by WT or Tfam+/− MEFs, or media conditioned by WT MEFs stimulated with poly(I:C) for 9 hours (PIC-stimulated). The data shown are from one of two (f and g) or three (b-d) biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other biological replicates are provided as Supplementary Figures. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, n.s. not significant (P > 0.05).
Extended Data Fig. 2. Additional analysis of the STAT1 null (Stat1−/−) condition in WT (Tfam+/+ Stat1+/+) and Tfam+/− MEFs.
a, Western blot of STAT1, TFAM and GAPDH (loading control) in littermate MEFs of the indicated Tfam and Stat1 genotypes (bottom) that were transfected with 2 μg of Poly(I:C) or lipofectamine only (Mock) for 12 hours. (n=3 independent experiments). MEFs described in a were analysed (all normalized to WT) for b, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers; c, mitochondrial mass using MitoTracker Green (MTG) and flow cytometry (mean fluorescence intensity, MFI, in arbitrary units, A.U., is plotted), and d, mitochondrial membrane potential using MitoTracker Deep Red (MTDR) and flow cytometry (MFI in A.U. is plotted). e, MEFs of the indicated genotypes were analysed by immunofluorescence for mitochondrial and mtDNA nucleoid morphology using antibodies against HSP60 (Mito., magenta) and DNA (DNA, cyan), respectively (n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 μm. f, qRT-PCR analysis of the ISGs Cxcl10 and Ifit1 in MEFs of the indicated genotypes. g, qRT-PCR analysis of the indicated ISGs in Tfam+/− MEFs transfected with control (Ctrl) or Irf3 siRNAs for 72 hours. h, qRT-PCR analysis of the indicated ISGs in Tfam+/− Stat1−/− MEFs transfected with control (Ctrl) or Irf3 siRNAs for 72 hours. i, Western blot showing siRNA knock-down of IRF9 or STAT2 in Tfam+/− MEFs compared to control (Ctrl) siRNA treated cells. GAPDH was probed as the loading control. (n=3 independent experiments). j, qRT-PCR analysis of the indicated ISGs in Tfam+/− Stat1−/− MEFs transfected with control (Ctrl) or Irf9 siRNAs for 72 hours. k, qRT-PCR analysis of the indicated ISGs in Tfam+/− Stat1−/− MEFs transfected with control (Ctrl) or Stat2 siRNAs for 72 hours. c-d, error bars indicate mean ± SD of n=3 biological replicates. For b the data shown are from one of two biological replicates with the error bars indicating the mean ± SD of three technical replicates. For g, h, j and k, the data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates and the FACS gating strategy are provided as Supplementary Figures. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001, n.s. not significant (P > 0.05).
Extended Data Fig. 3. Additional DNA damage and repair analysis in MEFs.
a, Pie charts showing the percentage of reported U-ISGF3-induced11 (green) and IRDS-related14 (blue) ISGs that are upregulated in Tfam+/− MEFs. The specific ISGs in these signatures are shown in the Table to the right, with the genes common to all three groups in bold. b-d, Stat1−/− and Tfam+/− Stat1−/− MEFs were treated with 1μM doxorubicin for 48 hours followed by (b) Apoptosis analysis by flow cytometry using Annexin V and Propidium Iodide (PI); (Contour plots represent n=3 biological replicates). (c) Quantification of flow cytometry analysis in b. for live cells (Annexin V and PI low), apoptotic cells (Annexin V high and PI low), and dead cells (Annexin V and PI high); (d) Cell viability analysis using the alamarBlue assay. c, d, Error bars indicate mean ± SD of (c) n=3 or (d) n=8 biological replicates. e and f, Analysis of nuclear DNA repair rate (i.e. the rate of γH2A.X and p-53BP1 foci resolution during recovery after 2 Gy IR). Nuclei are labelled with DAPI (blue), while γH2A.X (magenta), and p-53BP1 (green) were detected by immunofluorescence. Plotted to the right of the images is the average number of foci per nucleus at the indicated times. g and h, Analysis of the rate of induction of the nDNA damage response (i.e. the rate of γH2A.X and p-53BP1 foci formation) upon 2 Gy IR. Imaging was done as described in e and f. Plotted to the right is the percentage of positive cells (>20 γH2A.X foci per nucleus or >15 p-53BP1 foci per nucleus) at the indicated times. e-h, Scale bars represent 15 μm. e-h, Error bars indicate means ± SD of n=3 biological replicates, except for the 7.5 min time point in g and h, which are from n=2 biological replicates. In each replicate, 50 nuclei were quantified. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, n.s. not significant (P > 0.05).
Extended Data Fig. 4. Additional data supporting the role of PARP9 in mtDNA-stress-mediated enhancement of nDNA damage responses.
a, Heat map of PARP family gene expression from previously published microarray data4 in WT and Tfam+/− littermate MEFs (two of each). b, qRT-PCR analysis of Parp9 expression in WT (Tfam+/+ Stat1+/+), Tfam+/−, Stat1−/− and Tfam+/− Stat1−/− littermate MEFs. c, qRT-PCR analysis of Parp9 expression at the indicated times after transfection with two Parp9 siRNAs (#1 and #2). b and c, the data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates are provided as Supplementary Figures. d, Tfam+/− MEFs were transfected with control (Ctrl) or one of two Parp9 siRNAs (#1 and #2) for 48 hours and then assessed for cell viability using the alamarBlue assay after treatment with 1.0 or 1.5 μM doxorubicin (Dox) for 24 hours. Error bars indicate mean ± SD of n=4 biological replicates. e-h, Analysis of DNA repair rate (i.e. the rate of γH2A.X and p-53BP1 foci resolution during recovery after (e and f) 2 Gy IR or (g and h) 12 hours of 1μM doxorubicin-mediated damage. Nuclei are labelled with DAPI (blue), while γH2A.X (magenta), and p-53BP1 (green) were detected by immunofluorescence. (e-h) Plotted to the right of the images is the average number of foci per nucleus at the indicated times. Scale bars represent 15 μm. Error bars indicate means ± SD of n=3 biological replicates, with 50 nuclei quantified in each. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 5. Additional data supporting mtDNA stress-mediated ISG induction in MEFs.
a, Western blot probing STAT1, p-STAT1 (Y701), γH2A.X (DNA damage marker) and GAPDH (loading control) in WT MEFs treated with the indicated doses of doxorubicin (Dox) for 24 hours. (n=3 independent experiments). b, qRT-PCR analysis of the indicated ISGs in WT (Stat1+/+) and Stat1 null (Stat1−/−) littermate MEFs challenged with (+Dox) or without 500 nM Dox for 24 hours. c, WT MEFs treated with (Dox) or without (Ctrl) 1μM doxorubicin were analysed by immunofluorescence for mitochondrial and mtDNA nucleoid morphology by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan). (n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 μm. d and f, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in WT MEFs exposed to (d) 100 μM 2’−3’-dideoxycytidine (ddC) or (f) 450 ng/mL ethidium bromide (EtBr) for 10–12 days. e, g, qRT-PCR analysis of the ISGs Ifit1 and Ifit3 in (e) Control (Ctrl) and ddC-treated MEFs (described in d) challenged with (+Dox) or without (-Dox) 500nM for doxorubicin for 16 hours, and (g) Control (Ctrl) and EtBr-treated MEFs (described in f) transfected with 2 μg dsDNA90 or lipofectamine only (Mock) for 9 hours. h, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in LMTK- cells with (ρ+) or without (ρ˚) mtDNA. i, qRT-PCR analysis of the ISGs Ifit1 and Ifit3 in LMTK- ρ+ and ρ˚ cells (described in h) challenged with 500 nM Dox for 24 hours. j and k, MEFs were pre-treated with 100 μM chloramphenicol (Chlo) for 24 hours followed by 500 nM Doxorubicin (Dox) for 24 hours. j, Western blot using an OXPHOS complex cocktail (mtDNA-encoded subunit is in red, nucleus-encoded subunits in black), γH2A.X (DNA damage marker) or actin (loading control) (n=3 independent experiments). k, qRT-PCR analysis of the ISGs Ifit1 and Ifit3. For e and g the data shown are from one of two biological replicates with the error bars indicating the mean ± SD of three technical replicates. For b, d, f, h and i, the data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other biological replicates are provided as Supplementary Figures. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 6. Doxorubicin promotes mtDNA stress-mediated ISG induction in MC-38 mouse colon cancer cells.
MC-38 cells treated with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 hours and analysed by a, qRT-PCR analysis of the indicated ISGs and b, Western blot probing STAT1, p-STAT1 (Y701), γH2A.X (DNA damage marker) and GAPDH (loading control). (n=3 independent experiments) c, d, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in WT MEFs exposed to (c) 100μM 2’−3’-dideoxycytidine (ddC) or (d) 200ng/mL ethidium bromide (EtBr) for 48 hours. e, qRT-PCR analysis of the ISGs Ifit1 and Ifit3 in control (Ctrl) and ddC treated MC-38 cells (described in c) challenged with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 hours. f, qRT-PCR analysis of the ISGs Ifit1 and Ifit3 in control (Ctrl) and EtBr-treated MC-38 cells (described in d) challenged with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 hours. g, qRT-PCR analysis of the indicated ISGs in MC-38 cells treated with 3 μM mitochondria-targeted doxorubicin (mitoDox) or DMSO (Mock) for 48 hours. h, Western blot analysis in MC-38 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) gRNA control to determine the knockout efficiency. (n=1 only to validate) Vinculin was probed as the loading control. i-k, qRT-PCR analysis of the ISGs Ifit1 and Ifit3 in MC-38 cell pools transduced with the indicated gRNAs (described in h) that were transfected with (i) 2 μg dsDNA90 or (j) 2 μg Poly(I:C) for 8 hours or (k) challenged with 150 nM doxorubicin for 24 hours. For i and j, the data shown are from one of two biological replicates with the error bars indicating the mean ± SD of three technical replicates. For a, c-g and k, the data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other biological replicates are provided as Supplementary Figures. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 7. TFAM deficiency induces mtDNA stress and DNA-damage resistance in MC-38 mouse colon cancer cells.
a-e, WT and TFAM deficient (TFD) MC-38 cells (see Methods section) were analysed. a, qRT-PCR analysis of Tfam mRNA. b, western blot probing STAT1 (L.E., long exposure; S.E., short exposure), TFAM and GAPDH (loading control) (n=3 independent experiments). c, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers. d, mitochondrial and mtDNA nucleoid morphology analyses by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan) (n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 μm. e, qRT-PCR analysis of the indicated ISGs. f, Western blot analysis in WT and TFD MC-38 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) control gRNA to determine the knockout efficiency (n=1, only to validate). GAPDH was probed as the loading control. g, qRT-PCR analysis of the indicated ISGs in WT and TFD MC-38 cell pools transduced with the indicated gene-specific gRNAs (described in f). h, Cell survival analysis of WT and TFD MC-38 cells challenged with 500 nM doxorubicin for 48 hours using the alamarBlue assay (n=7 biological replicates). a, c, e and g, the data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates are provided as Supplementary Figures. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows:. ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 8. Additional analysis of mtDNA stress and DNA-damage resistance in YUMMER mouse melanoma cells.
a, Western blot of STAT1, TFAM and GAPDH (loading control) in WT and Tfam deficient (TFD) YUMMER cells (n=3 independent experiments). b, Colony formation analysis of WT and TFD YUMMER cells challenged with 250nM doxorubicin (Dox) for 4 hours. Plotted to the right is the quantification of relative cell survival (n=3 biological replicates). c, Western blot analysis in WT and TFD MC-38 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) control gRNA to determine the knockout efficiency (n=1, just to validate). GAPDH was probed as the loading control. d, Western blot of HA, TFAM and β-actin (Actin, loading control) in parental WT and TFD YUMMER cells, or WT and TFD YUMMER cells stably overexpressing empty vectors (EV) or HA-Flag-TFAM (OE) (n=3 independent experiments). e and f, YUMMER cells described in d were analysed as follows. e, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers. f, qRT-PCR analysis of the indicated ISGs. g, Mitochondrial and mtDNA nucleoid morphology analysis by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan)(n=3 independent experiments). Images are Z-stack projections and scale bar represents 10μm. The data shown are from one of three biological replicates with the error bars indicating the mean ± SD of three technical replicates. The other two biological replicates are provided as Supplementary Figures. ** P < 0.01, *** P < 0.001.
Extended Data Fig. 9. ISG expression inversely correlates with mRNA expression of Tfam and other mtDNA-metabolism-related genes.
a-c, RNA data from the CCLE23 (Cancer Cell Line Encyclopedia) was analysed. a, shown is the analysis of cells in the upper and lower quartiles of Tfam RNA expression, for expression the indicated ISGs, three interferon genes IFNA1, IFNB1 and IFNG (bold) and GAPDH (negative control). b, shown is the analysis of cells in the upper and lower quartiles of Lig3 (top row), Nthl1 (middle row), and Polg (bottom row) RNA expression, for expression of the indicated ISG. c, shown is the analysis of cells in the upper and lower quartiles of Sod2 (top row), SDHD (middle row) and VDAC1 (bottom row) RNA expression, for expression of the indicated ISGs. P values were calculated using Wilcoxon rank-sum test and displayed under each box plot.
Supplementary Material
Acknowledgements
The authors thank Dr. Akiko Iwasaki and Dr. Charles Dela-Cruz for reagents and advice, Albert Mennone Jr. and Dr. Tong Zhang for assistance with microscopy, Carolyn O’Connor for help with cell sorting and flow cytometry, Dr. Mathias Leblanc and Dr. Ya-Lan Chang for mouse tissue collection, Dr. Shana O. Kelley and Tanja Sack for providing mitochondria targeted doxorubicin, and Dr. Nissi Varki and the UCSD histopathology core for preparation and analysis of mouse tissues. This work was supported by NIH R01 AR069876 and the Audrey Geisel Chair in Biomedical Science to G.S.S., NIH R01 CA216101 to G.S.S and S.M.K., NIH R35 CA197574 to P.M.G, NIH R01 CA237586 to QY, NIH F31 AG062099 to A.G.S, and NIH P50 CA121974. A.P.W. was supported by grants RP170734 from the Cancer Prevention and Research Institute of Texas and W81XWH-17-1-0052 from the Office of the Assistant Secretary of Defence for Health Affairs, Peer Reviewed Medical Research Program, Z.W. was supported by the China Scholarship Counsel, K.C.M by the Salk Excellerators Postdoctoral Fellowship, and L.E.N. by the George E. Hewitt Foundation for Medical Research Postdoctoral Fellowship.
Footnotes
Competing Interests Statement. The authors have no competing interests to declare with regard to the data presented in the manuscript.
REFERENCES
- 1.Bogenhagen DF, Rousseau D & Burke S The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283, 3665–3675, 10.1074/jbc.M708444200 (2008). [DOI] [PubMed] [Google Scholar]
- 2.West AP, Shadel GS & Ghosh S Mitochondria in innate immune responses. Nat Rev Immunol 11, 389–402, 10.1038/nri2975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West AP & Shadel GS Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 17, 363–375, 10.1038/nri.2017.21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AP et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557, 10.1038/nature14156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong Z et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203, 10.1038/s41586-018-0372-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aarreberg LD et al. Interleukin-1beta Induces mtDNA Release to Activate Innate Immune Signaling via cGAS-STING. Mol Cell 74, 801–815 e806, 10.1016/j.molcel.2019.02.038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai J et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc Natl Acad Sci USA 114, 12196–12201, 10.1073/pnas.1708744114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider WM, Chevillotte MD & Rice CM Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32, 513–545, 10.1146/annurev-immunol-032713-120231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandvaux N et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol 76, 5532–5539, 10.1128/jvi.76.11.5532-5539.2002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda K & Taniguchi T IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6, 644–658, 10.1038/nri1900 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Cheon H et al. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J 32, 2751–2763, 10.1038/emboj.2013.203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheon H & Stark GR Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A 106, 9373–9378, 10.1073/pnas.0903487106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung PS et al. Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc Natl Acad Sci U S A 112, 10443–10448, 10.1073/pnas.1513341112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weichselbaum RR et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A 105, 18490–18495, 10.1073/pnas.0809242105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minn AJ Interferons and the Immunogenic Effects of Cancer Therapy. Trends Immunol 36, 725–737, 10.1016/j.it.2015.09.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang CS et al. Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol Cell 66, 503–516 e505, 10.1016/j.molcel.2017.04.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Q et al. BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol Cell Biol 33, 845–857, 10.1128/MCB.00990-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Q et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell 36, 110–120, 10.1016/j.molcel.2009.08.019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat Immunol 16, 1215–1227, 10.1038/ni.3279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe T & Barber GN Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88, 5328–5341, 10.1128/JVI.00037-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buondonno I et al. Mitochondria-Targeted Doxorubicin: A New Therapeutic Strategy against Doxorubicin-Resistant Osteosarcoma. Mol Cancer Ther 15, 2640–2652, 10.1158/1535-7163.MCT-16-0048 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Wang J et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 30, 428–435, 10.1111/pcmr.12591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607, 10.1038/nature11003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh G, Sharkey SM & Moorehead R Mitochondrial DNA damage by anticancer agents. Pharmacol Ther 54, 217–230 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Hämäläinen RH et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nature Metabolism 1, 958–965, 10.1038/s42255-019-0120-1 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Quiros PM, Mottis A & Auwerx J Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 17, 213–226, 10.1038/nrm.2016.23 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM 3rd & Bohr VA Protecting the mitochondrial powerhouse. Trends Cell Biol 25, 158–170, 10.1016/j.tcb.2014.11.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadel GS & Clayton DA Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66, 409–435, 10.1146/annurev.biochem.66.1.409 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Woo DK et al. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am J Pathol 180, 24–31, 10.1016/j.ajpath.2011.10.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durbin JE, Hackenmiller R, Simon MC & Levy DE Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450, 10.1016/s0092-8674(00)81289-1 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308, 10.1038/nprot.2013.143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefzadeh MJ et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet 10, e1004654, 10.1371/journal.pgen.1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oeck S et al. The Focinator v2–0 - Graphical Interface, Four Channels, Colocalization Analysis and Cell Phase Identification. Radiat Res 188, 114–120, 10.1667/RR14746.1 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Oeck S, Malewicz NM, Hurst S, Rudner J & Jendrossek V The Focinator - a new open-source tool for high-throughput foci evaluation of DNA damage. Radiat Oncol 10, 163, 10.1186/s13014-015-0453-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furda A, Santos JH, Meyer JN & Van Houten B Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol 1105, 419–437, 10.1007/978-1-62703-739-6_31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CCLE RNA-seq data can be found through link https://portals.broadinstitute.org/ccle. TFAM microarray data are available in NCBI GEO database with accession number GSE63767. Additional qPCR biological replicate data can be found in the supplementary file. Other data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.