Abstract
It is of uttermost importance that the global health community develops the surveillance capability to effectively monitor emerging zoonotic pathogens that constitute a major and evolving threat for human health. In this study, we propose a comprehensive framework to measure changes in (1) spillover risk, (2) interhuman transmission, and (3) morbidity/mortality associated with infections based on 6 epidemiological key indicators derived from routine surveillance. We demonstrate the indicators’ value for the retrospective or real-time assessment of changes in transmission and epidemiological characteristics using data collected through a long-standing, systematic, hospital-based surveillance system for Nipah virus in Bangladesh. We show that although interhuman transmission and morbidity/mortality indicators were stable, the number and geographic extent of spillovers varied significantly over time. This combination of systematic surveillance and active tracking of transmission and epidemiological indicators should be applied to other high-risk emerging pathogens to prevent public health emergencies.
Keywords: emerging pathogens, monitoring, Nipah virus, surveillance
Emerging zoonotic pathogens represent an important and growing risk to humans. As cross-species interactions increase at the human-animal interface, more opportunities arise for pathogens to spillover. Large-scale epidemics of human immunodeficiency virus, Ebola, or Middle East respiratory syndrome coronavirus can trace their roots back to spillovers from a zoonotic reservoir [1]. The timely deployment of targeted interventions to prevent public health emergencies would clearly benefit from the real-time evaluation of temporal trends suggestive of altered spillover risk or evolutionary changes linked to increased transmission or disease. Given that many emerging pathogens spill over infrequently, changes in pathogen characteristics may go undetected unless there are systematic efforts to track these changes. Therefore, enhanced monitoring approaches that go beyond tracking case numbers to identify changes in disease risk and underlying mechanisms would help to improve global preparedness and response capacities. The International Health Regulations and Global Health Security Agenda both call for stronger surveillance and trend monitoring [2, 3]. However, it has rarely been possible to develop a comprehensive monitoring framework for emerging zoonotic pathogens due to the lack of stable surveillance collecting detailed case information.
Nipah virus (NiV) is an emerging zoonotic pathogen found in fruit bats throughout South and Southeast Asia. NiV is considered by the World Health Organization as an important health threat to humans due to the severity of disease it causes, the absence of treatments or vaccines, and its ability to be transmitted between people [4, 5]. Bangladesh is the only country reporting regular spillovers of NiV, which are almost exclusively detected through a systematic, hospital-based surveillance system, implemented in 2007 after the ad hoc identification of ~100 cases during 2001–2006 [6]. The surveillance system is based on 3 sentinel hospitals where all meningoencephalitis cases are routinely tested for NiV. Identification of an NiV case triggers detailed investigations in affected communities including the identification of transmission networks and the tracing of contacts. The hospital-based surveillance system leverages clinical services and is therefore less expensive and easier to maintain than other population-based surveillance systems.
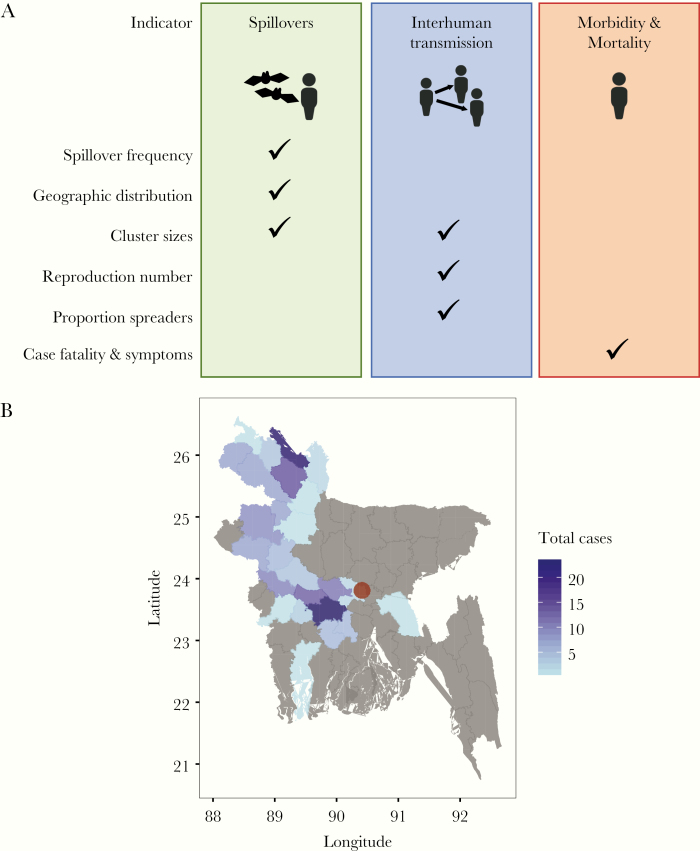
In this study, we propose 6 epidemiological indicators of emerging zoonotic pathogens and their transmission that can be derived from such routine surveillance data to measure changes in (1) spillover risk, (2) interhuman transmission, and (3) morbidity/mortality associated with infections (Figure 1A). Although many of these measures are commonly used in epidemiological studies, their role in the routine monitoring of emerging infectious diseases is not yet defined. The NiV surveillance system in Bangladesh provides a unique opportunity to collect comparable data on cases and their characteristics over time. The objective of this study was to demonstrate how these indicators can be obtained from routine surveillance data and used to monitor changes in disease transmission and epidemiology to inform future public health policy and practice.
Figure 1.
(A) Indicators for changes in spillover risk, interhuman transmission, and morbidity/mortality of NiV and other zoonotic pathogens. (B) Total number of Nipah cases ever reported by districts in Bangladesh (2007 to 2018).
METHODS
Establishing Baseline Measures and Assessing Historical Changes
We used data from the systematic NiV surveillance system collected during 2007–2018 to establish baseline measures of the 6 indicators and assess historical changes over time. Details of the surveillance methods have been previously published [7, 8], but, in short, at 3 tertiary care hospitals, all patients admitted with signs and symptoms consistent with febrile neurological illness during December–March had clinical and epidemiological information collected and serum samples obtained for testing for immunoglobulin M antibodies against NiV. We further assessed all 6 indicators using data collected during 2001–2006, a time period before routine hospital-based surveillance was implemented, and cases were not systematically detected (see Supplementary Material). During this time period, cases were detected through community investigations after the notification of case clusters, and NiV cases not associated with case clusters were likely to be missed.
Spillover Risk
In Bangladesh, humans usually acquire NiV through drinking date palm sap contaminated with the virus by fruit bats [4]. It is important to monitor spillover frequency because any increase may result in higher disease burden, or it may indicate emergence of new transmission pathways. In addition, each spillover event represents an opportunity for more transmissible or virulent strains to emerge. A spillover event is defined as a single case infected from the reservoir or a cluster of cases that can be traced back to a single spillover source (i.e., 1 or several individuals infected through a contaminated palm sap pot plus subsequent cases infected by interhuman transmission), and the annual spillover rate is defined as the number of spillover events in a given transmission season (December of the previous year until May of the given year). We assumed that spillover events follow a Poisson distribution, and we estimated the spillover rate observed in a given year with Poisson exact 95% confidence intervals (CIs) and the average annual spillover rate during a given time period (i.e., for the duration of stable surveillance or time periods between change points) with approximate Poisson 95% CIs. We tested the statistical significance of changes in spillover frequency before and after a specific time point using exact Poisson test, in which we moved the time point by 1-year increments over the duration of stable surveillance. If several time points resulted in significant differences, we selected the time point resulting in the lowest P value. We further assessed linear changes in annual spillover rates using Poisson regression: we evaluated statistical significance of linear changes using a likelihood ratio test and compared this model to step-change models using the Akaike Information Criterion.
Geographic Extent
The geographic extent of case occurrence informs the size and location of the population at risk. Moreover, pathogen introductions into new ecological areas or populations (e.g., higher population density, mobility) can affect transmission dynamics and potentially boost the spread and impact of the pathogen [9]. Using surveillance data on the geographic location of cases, we can monitor the geographic extent of NiV risk. The geographic extent of case reports is defined as the number of districts from which cases are reported. We quantified the geographic extent using 3-year sliding windows over the duration of the surveillance dataset and assessed spatial patterns based on administrative boundaries obtained from the Database of Global Administrative Areas (www.gadm.org).
Cluster Size
The NiV surveillance data can also be used to monitor indicators for changes in spillover mechanisms. The number of individuals who are infected from the reservoir during each spillover event may vary depending on the spillover source. For example, clusters associated with drinking palm sap may differ in size from clusters associated with pig exposure, which was the main spillover route in the Malaysian NiV outbreak [10]. Cluster sizes are also affected by interhuman transmission through the number of secondary cases associated with a spillover event (see section on Proportion of Cases Who Transmit Infection). We defined a cluster of cases as individuals who acquired infection through a single spillover event (either through 1 or more bat-to-human transmission events in one time and place or through subsequent interhuman transmission). We estimated the median and interquartile range (IQR) of cluster sizes for a given year or a given time period. We used Wilcoxon rank-sum test to assess changes in outbreak sizes by comparing cluster size distributions before and after a specific time point that we moved along the time of stable surveillance.
Reproduction Number
The reproduction number R (i.e., the average number of individuals infected by a case) is the standard measure of the interhuman transmission potential of a pathogen [11]. Whereas sustained transmission of a pathogen in a population can occur only if R ≥ 1, even increases in R to values <1 can lead to larger cluster sizes with transmission ceasing only after a substantial number of transmission generations. Therefore, tracking this parameter can provide an early warning sign of a change in transmission [12–14]. For NiV in Bangladesh, this essential parameter can be directly estimated from the transmission trees that are reconstructed during outbreak investigations. We estimated R and 95% CIs based on the observed number of secondary cases caused by a case, assuming that the number of secondary cases follows a negative binomial distribution with mean R and a dispersion parameter k [14]. We used a likelihood ratio test to assess changes in R before and after a specific time point that we moved along the duration of stable surveillance. We further assessed linear changes in R by year using negative binomial regression.
Proportion of Persons Who Transmit Infection
Interhuman transmission of emerging zoonotic pathogens is often highly heterogeneous, meaning that a large part of secondary cases originates from only a few superspreading events as previously noted for NiV [8, 15]. The mechanisms leading to superspreading events can be of social (e.g., a higher number of contacts resulting in more transmission opportunities) or biological nature (e.g., higher levels of virus shedding or stronger symptoms facilitating transmission) [14]. The extent of transmission heterogeneity has important implications for the optimization of control strategies [14]. For instances in which detailed information on transmission trees is available, we can quantify transmission heterogeneity as the percentage of cases who transmit the pathogen [14]. For a given reproduction number, a smaller percentage of cases who transmit will indicate a stronger transmission heterogeneity [14]. We estimated the proportion of spreaders among NiV cases and exact binomial 95% CIs for a given year or a given time period. We used an exact binomial test to assess changes in the proportion of spreaders before and after a specific time point that we moved along the time of stable surveillance. We further assessed linear changes in proportion of spreaders by year using logistic regression. We also assessed the probability of observing multiple superspreading events (i.e., an NiV patient infecting ≥5 individuals) during outbreaks of various sizes (Supplementary Material).
Morbidity and Mortality
Changes in the case fatality ratio (CFR) and the prevalence of specific symptoms such as difficulty breathing (a symptom previously found associated with interhuman transmission [6]), can indicate an adaptation of a pathogen to the human host. We estimated the CFR (ie, the proportion of cases who died among cases) and exact binomial 95% CIs for a given year or a given time period separately for cases infected by the reservoir (primary cases) and cases infected through interhuman transmission (secondary cases). We used an exact binomial test to assess changes in the case-fatality rate before and after a specific time point that we moved along the time of stable surveillance. We further assessed linear changes in the case-fatality rate by year using logistic regression. We applied the same methods to assess changes in the proportion of cases with difficulty breathing.
Real-Time Monitoring of Indicators
Changes in the transmission and epidemiology of NiV can also be assessed in real-time using the proposed framework by plotting new observations against baseline distributions of indicators established above. To facilitate identification of epidemiologic changes prospectively within this system, we identified thresholds for “highly unlikely” events by quantifying the 2.5th and 97.5th percentiles of these baseline distributions.
RESULTS
Monitoring Spillover Risk
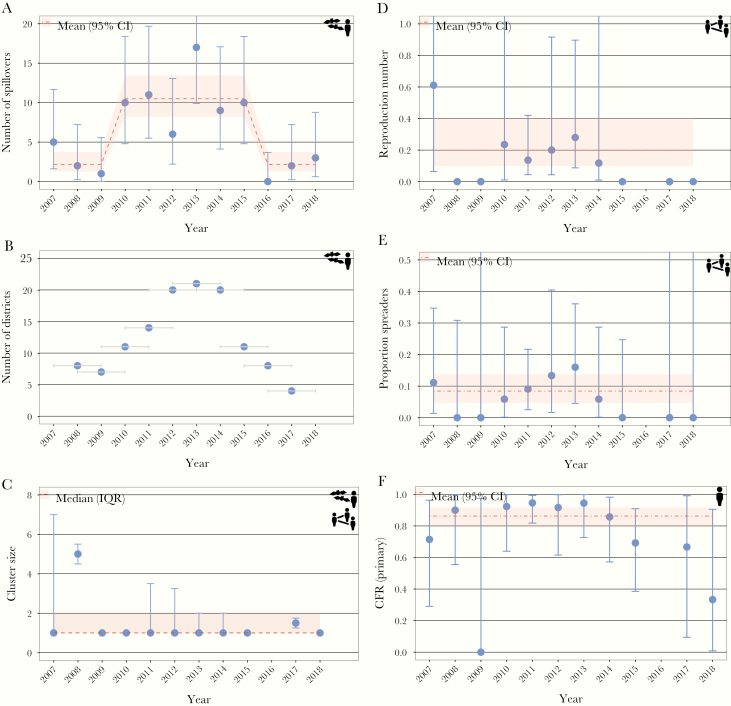
During 2007–2018, 76 NiV spillover events were detected in Bangladesh resulting in 166 NiV cases (Figure 1B). We observed that the average annual number of spillovers detected in Bangladesh varied significantly over time; it increased from 2.2 (95% CI, 1.3–3.7) in 2007–2009 to 10.5 (95% CI, 8.2–13.4) in 2010–2015, and it returned to the lower level in 2016–2018 (Figure 2A).
Figure 2.
Baseline estimates and historical changes of key indicators to monitor NiV epidemiology and transmission. Average number of spillovers (Poisson exact 95% confidence interval [CI]) (A), number of districts reporting NiV cases within 3-year window (time windows are indicated as gray bars) (B), median cluster size (interquartile range) (C), reproduction number (negative binomial 95% CI) (D), proportion spreaders (binomial exact 95% CI) (E), and case fatality ratio among primary cases, i.e., cases infected by the reservoir (binomial exact 95% CI) (F). The year represents a transmission season, which lasts from December of the previous year until May of the indicated year.
Coinciding with the temporary increase in spillover frequency, we found that the number of districts reporting cases was more than 2 times greater in 2011–2015 compared with the other time windows (Figure 2B). However, this increase does not seem to reflect a gradual spread of NiV into previously unaffected areas; instead, there were more regular spillovers in geographic regions where NiV had previously been observed (Supplementary Figure S1). Two NiV cases who acquired infections in rural areas were reported in Dhaka, the capital city of Bangladesh.
We observed a constant median cluster size of 1 case (IQR, 1–2) over time (Figure 2C). The median cluster size was higher in 2008 (with 2 clusters of 6 and 4 spillover cases, respectively) than in other years.
Monitoring Interhuman Transmission
Transmission potential of NiV did not vary significantly in Bangladesh during 2007–2018, with an average reproduction number of 0.20 (95% CI, 0.10–0.40) (Figure 2D). No interhuman transmission events were observed in 2008–2009 and 2015–2018, which is consistent with the estimated average reproduction number and may be explained by a smaller number of NiV cases during these years (Supplementary Figure S2).
Eight percent (95% CI, 5–14) of cases transmitted NiV to another person, which was stable over time (Supplementary Figure 2E). During 2007–2018, 2 outbreaks were driven by a superspreading event. The occurrence of more than 1 superspreading event in a single outbreak has never been observed and is unlikely given the current reproduction number (Supplementary Figure S3).
Monitoring Morbidity/Mortality Associated With Infections
We observed a stable CFR over time among cases infected by the reservoir (86%; 95% CI, 79–92) (Figure 2F) and those infected through interhuman transmission (46%; 95% CI, 29–63) (Supplementary Figure S4A). The percentage of NiV patients who developed breathing difficulties also remained constant over time with an average of 57% (95% CI, 49–64) (Supplementary Figure S4B). No significant difference in the proportion with breathing difficulties were detected between patients infected by the reservoir and those infected through interhuman transmission (χ 2P = .37). The CFR was higher among patients with breathing difficulty (90%) than those without (61%; χ 2P < .001).
Based on data collected during 2001–2006, a period before routine hospital-based surveillance was implemented and cases were not systematically detected, we observed a much higher variability in some of these indicators (in particular for spillover frequency, outbreak size, and reproduction number) (Supplementary Figure S5).
Real-Time Monitoring of Indicators
The baseline measures for these 6 indicators that we established here can be used by public health officials to detect future changes in NiV characteristics (Supplementary Figure S6). Such changes can be evaluated in real time by comparing expected values (e.g., percentiles of estimated distributions) to new observations, either for a single outbreak or for a transmission season. For example, based on current estimates of spillover frequency, observing more than 17 spillovers in a season is very unlikely (based on the 97.5th percentile of the high spillover frequency period), whereas half of the time, more than 10 spillovers are expected to occur (Supplementary Figure S6A).
DISCUSSION
Systematic NiV surveillance in Bangladesh has been an important step forward in the response to the emerging threat of this virus. These routinely collected data enable the establishment of baseline measures that comprehensively describe current NiV spillover risk, interhuman transmission, and morbidity/mortality, which can now be used by public health authorities to guide rapid and reliable decisions to respond to NiV outbreaks. Using these baseline measures in real-time assessments allows the detection of deviations from these at early stages of a transmission season or an ongoing outbreak.
Although interhuman transmission and morbidity/mortality of NiV has been stable since the start of systematic surveillance, frequency and geographic extent of spillovers temporarily increased during that time period before returning to initial levels. The temporary increase may reflect NiV transmission and shedding dynamics in bat populations [16], changes in bat behavior (e.g., moving to new areas to look for food), or changes in human behavior (eg, sap drinking, access to hospitals). A study of weather associations with spillovers between 2007 and 2013 identified a significant correlation between increasing spillovers and colder winter temperatures, suggesting that climatic factors could be driving some of these patterns over longer time scales [17]. The monitoring framework has thereby provided insights that can (1) form the basis of studies to test different hypotheses of what drives these patterns and (2) measure changes in patterns that could result from primary or secondary prevention programs. Continuing surveillance efforts to monitor time trends or cyclical dynamics in this disease system are critical and should be prioritized for funding.
Observed cluster sizes were stable over time, suggesting that no changes in spillover mechanisms had occurred during the time period of surveillance. This finding is consistent with other epidemiological evidence that identified palm sap as the main spillover route over time, including during spillovers in 2008 that were larger in size [18, 19]. No increase in the interhuman transmission potential has been suggested by any of the indicators, and the reproduction number currently is lower than what is required for a large outbreak. Based on the established baseline measures, the occurrence of multiple superspreading events during a single outbreak is highly unlikely and may therefore represent an early warning sign for the emergence of a more transmissible strain. For monitoring situations in which details on transmission networks are unavailable, R could also be derived from the distribution of cluster sizes or the proportion of cases infected by the reservoir [13, 20]. The detection of 2 NiV cases in Dhaka, the capital of Bangladesh, demonstrates the risk of virus introductions into densely populated areas where large-scale outbreaks may be more likely to occur. The observed heterogeneity in some indicators during 2001–2006, a time period before hospital-based surveillance started in Bangladesh, highlights that systematic surveillance for NiV, as introduced in Bangladesh in 2007, is key for establishing reliable baseline estimates of transmission characteristics and will remain crucial for the detection of departures from these trends in the future. However, based on established estimates of R, it would be highly unlikely to observe superspreading events as reported before 2007 (with 11 and 22 secondary cases generated by an NiV case) (Supplementary Figure S6C).
CONCLUSIONS
The analysis of the collected epidemiological data also provided other insights that can be investigated in future studies, such as the difference in the CFR between cases infected through the reservoir and those infected through interhuman transmission. This difference may be due to a higher dose of virus received through date palm sap than through contact with a patient. Developing capacities to efficiently detect and respond to unusual public health events is key for improved global epidemic preparedness [2, 3]. Therefore, in 2015, the World Health Organization advocated for prioritizing several emerging zoonotic diseases for urgent research and development that would allow for improved disease control. In addition to NiV, these diseases included Crimean Congo hemorrhagic fever, Ebola, Marburg, Lassa fever, Middle East respiratory syndrome and other coronaviruses, and Rift Valley fever [5]. We believe that systematic surveillance for these diseases, as exemplified by NiV surveillance in Bangladesh, should be implemented to enable the collection of comparable and reliable data over time. The framework proposed here would also allow tracking outbreaks of these diseases and targeting disease control measures. The global community would be much better prepared for these threats through investments in systematic surveillance coupled with active tracking and reporting of transmission and epidemiological indicators.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. icddr,b acknowledges with gratitude the commitment of the National Science Foundation, the National Institutes of Health (NIH).
Disclaimer. The opinions expressed in this article are the authors’ own and do not reflect the view of the Centers for Disease Control and Prevention, the Department of Health and Human Services, or the United States Government. The donors had no role in preparation or review of this manuscript.
Financial support. This work was funded by an National Institutes of Health grant for Ecology and Evolution of Infectious Diseases (Grant Numer 2R01-TW005869) and the US Centers for Disease Control and Prevention to its research efforts. icddr,b is thankful to the Governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support. B. N., H. S., and S. C. acknowledge the support from the Laboratory of Excellence Integrative Biology of Emerging Infectious Diseases (Grant ANR-10-LABX-62-IBEID), the National Institute of General Medical Sciences Models of Infectious Disease Agent Study Initiative, and the AXA Research Fund and the INCEPTION project (PIA/ANR-16-CONV-0005). This research was supported by the Defense Advanced Research Projects Agency DARPA PREEMPT # D18AC00031.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature 2007; 447:279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. International Health Regulations (IHR). Available at: http://www.who.int/topics/international_health_regulations/en/. Accessed 15 December 2019. [Google Scholar]
- 3. About | Global Health Security Agenda Available at: https://www.ghsagenda.org/about. Accessed 15 December 2019.
- 4. Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis 2009; 49:1743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. List of Blueprint priority diseases. Available at: http://www.who.int/blueprint/priority-diseases/en/. Accessed 20 December 2019. [Google Scholar]
- 6. Luby SP, Hossain MJ, Gurley ES, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerging Infect Dis 2009; 15:1229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naser AM, Hossain MJ, Sazzad HM, et al. Integrated cluster- and case-based surveillance for detecting stage III zoonotic pathogens: an example of Nipah virus surveillance in Bangladesh. Epidemiol Infect 2015; 143:1922–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nikolay B, Salje H, Hossain MJ, et al. Transmission of Nipah virus - 14 years of investigations in Bangladesh. N Engl J Med 2019; 380:1804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alexander KA, Sanderson CE, Marathe M, et al. What factors might have led to the emergence of Ebola in West Africa? PLoS Negl Trop Dis 2015; 9:e0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parashar UD, Sunn LM, Ong F, et al. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998–1999 outbreak of severe encephalitis in Malaysia. J Infect Dis 2000; 181:1755–9. [DOI] [PubMed] [Google Scholar]
- 11. Anderson RM, May RM.. Infectious Diseases of Humans: Dynamics and Control. Oxford, UK:Oxford University Press, 1991. [Google Scholar]
- 12. Ferguson NM, Fraser C, Donnelly CA, Ghani AC, Anderson RM. Public health. Public health risk from the avian H5N1 influenza epidemic. Science 2004; 304:968–9. [DOI] [PubMed] [Google Scholar]
- 13. Blumberg S, Lloyd-Smith JO. Inference of R(0) and transmission heterogeneity from the size distribution of stuttering chains. PLoS Comput Biol 2013; 9:e1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature 2005; 438:355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arunkumar G, Chandni R, Mourya DT, et al. Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J Infect Dis 2019; 219:1867–78. [DOI] [PubMed] [Google Scholar]
- 16. Plowright RK, Parrish CR, McCallum H, et al. Pathways to zoonotic spillover. Nat Rev Microbiol 2017; 15:502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortes MC, Cauchemez S, Lefrancq N, et al. Characterization of the spatial and temporal distribution of nipah virus spillover events in Bangladesh, 2007–2013. J Infect Dis 2018; 217:1390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hegde ST, Sazzad HMS, Hossain MJ, et al. Investigating rare risk factors for Nipah virus in Bangladesh: 2001–2012. Ecohealth 2016; 13:720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rahman MA, Hossain MJ, Sultana S, et al. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis 2012; 12:65–72. [DOI] [PubMed] [Google Scholar]
- 20. Cauchemez S, Epperson S, Biggerstaff M, Swerdlow D, Finelli L, Ferguson NM. Using routine surveillance data to estimate the epidemic potential of emerging zoonoses: application to the emergence of US swine origin influenza A H3N2v virus. PLoS Med 2013; 10:e1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.