Serological assessment of IgG4 responses to a combination of biomarkers for onchocerciasis enables enhanced case detection (at individual and community level), a current and urgent need towards elimination of the disease as a global health problem.
Keywords: OV-16, OVOC3261, onchocerciasis, diagnostics, biomarkers
Abstract
Background
Serological assessments for human onchocerciasis are based on IgG4 reactivity against the OV-16 antigen, with sensitivities of 60-80%. We have previously identified 7 novel proteins that could improve serodiagnosis.
Methods
IgG4 responses to these 7 proteins were assessed by luciferase immunoprecipitation (LIPS) and enzyme-linked immunosorbent (ELISA) immunoassays.
Results
OVOC10469 and OVOC3261 were identified as the most promising candidates by IgG4-based immunoassays with sensitivities of 53% for rOVOC10469 and 78% for rOVOC3261 while specificity for each was >99%. These 2 antigens in combination with OV-16 increased the sensitivity for patent infections to 94%. The kinetics of appearance of these IgG4 responses based on experimentally infected non-human primates indicated that they were microfilarial- driven. Further, the IgG4 responses to both OVOC10469 and OVOC3261 (as well as to OV-16) drop significantly (p<0.05) following successful treatment for onchocerciasis. A prototype lateral flow rapid diagnostic test to detect IgG4 to both Ov-16 and OVOC3261 was developed and tested demonstrating an overall 94% sensitivity.
Conclusion
The combined use of rOVOC3261 with OV-16 improved serologic assessment of O. volvulus infection, a current unmet need toward the goal of elimination of transmission of O. volvulus.
(See the Major Article by Bartsch et al., on pages 1782–94.)
Onchocerciasis or river blindness, caused by the filarial parasite Onchocerca volvulus, is one of the 13 neglected tropical diseases selected by the World Health Organization (WHO) for elimination or control as a public health problem. Annual community-based distribution of ivermectin, a drug that targets primarily the microfilariae, is a strategy that benefits both the community (for transmission interruption) and the individual (reduction in ocular and skin pathology). However, despite mass drug administration programs with ivermectin, it remains a major problem in sub-Saharan Africa where an estimated 205 million people are at risk of infection [1]. Disease control/elimination efforts across the world have had variable success, with elimination in the Americas (Ecuador, Guatemala, Mexico, and Colombia) [2] and anticipated elimination in Sudan, Senegal, Mali, and parts of Uganda [2–7]. However, onchocerciasis still remains endemic in many countries (mostly in Africa).
The diagnosis of active O. volvulus infection is based on direct microfilariae detection in skin snips by microscopy, a technique that is now actively discouraged, primarily for its invasive nature and lack of sensitivity when microfilarial densities are low, as in O. volvulus-hypoendemic areas. Although polymerase chain reaction (PCR) or real-time PCR from DNA extracted from the skin snips can significantly increase sensitivity [8], these approaches are not suited for community-based surveillance or point-of-contact (POC) testing.
The monitoring of elimination efforts for onchocerciasis and subsequent country-wide verification (at least in the Americas) relies on 2 important pillars: (1) absence of IgG4 antibodies to (OV-16) [9], and (2) absence of O. volvulus DNA in pools of Simulium spp., the insect vectors responsible for O. volvulus transmission. To verify elimination, countries must demonstrate that all transmission zones have successively passed through high-coverage mass drug administration regimens and at least 3 years of posttreatment surveillance [10].
The IgG4 anti-OV-16 antibody threshold established by the WHO guidelines [10] for verification of elimination requires that antibody prevalence be less than 0.1% among children younger than 10 years of age, with a current recommended sample size of ≥ 3000 [11]. These threshold calculations relied on an enzyme-linked immunosorbent assay (ELISA) developed in the mid-1990s [9] and used quite successfully in Guatemala, Mexico, Columbia, Ecuador, and Uganda [12–16]. This assay has a high specificity (>99%) but a very low sensitivity (<45%) for microfilariae-positive individuals. Immunoassays currently in use, including a more sensitive ELISA [17], the commercial version (Standard Diagnostics) or the available rapid diagnostic tests (RDTs), have specificities >99% and improved sensitivities (75%–85%). More recently, modelling studies indicated that with these newer more sensitive assays, if <2% of children under 10 years of age are positive for anti-OV-16 IgG4, there is a high probability of elimination [18].
Because a number of O. volvulus-infected individuals have some degree of genetic restriction that prevents them from eliciting any immune response to OV-16 [19], it is assumed that any OV-16–based serology will fail to identify approximately 20% of the microfilariae-positive infected individuals [20]. Thus, there is a consensus that immunoassays that retain >99% specificity but improve upon the 80% sensitivity of IgG4 anti-OV–16 assays are needed, particularly as elimination targets are being selected.
To identify alternative (non-OV-16–based) biomarkers, we previously used stage-specific transcriptomic, proteomic analyses to find multiple candidates [21]. Based on IgG4 responses to proteins spotted on an O. volvulus protein array containing 360 O. volvulus proteins, 7 candidates (OVOC10469, OVOC11950, OVOC10602, OVOC3261, OVOC5127, OVOC8491, and OVOC9988) were then identified as the most promising antigens for detecting O. volvulus infection in humans [21]. The objective of this work was to screen and define which novel biomarkers would improve the sensitivity of current OV-16 assays. We show that of the 7 previously described novel biomarkers, OVOC10469 and OVOC3261, in conjunction with OV-16 increased the sensitivity for detection of O. volvulus infection to 94%. Further, as proof of concept, we demonstrate the utility of combining OVOC3261 with OV-16 in a prototype lateral-flow strip test. Our data suggest that combining OVOC3261 with OV-16 can help not only in elimination mapping [22] but also to better define the serological thresholds needed to ensure the successful elimination of onchocerciasis [20].
MATERIALS AND METHODS
Serum Samples
All human serum samples were obtained using protocols approved by the Institutional Review Board of National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) for filaria-infected patients (NCT00001230) and healthy donors (NCT00090662). Well-characterized and archived serum samples were obtained from previous studies that had been approved by the institutional review boards at NIAID/NIH (protocol numbers 92-I-0155; 88-I-83), New York Blood Center (protocol number 0321), Tropical Research Station, Kumba, Cameroon (protocol number 001), or the Centers for Disease Control and Prevention (CDC; protocol number 6196). Samples used for validation of the biomarkers consisted of individuals with onchocerciasis (n = 457), loiasis (n = 34), lymphatic filariasis (n = 45), strongyloidiasis (n = 34), mansonellosis (n = 10), or cysticercosis (n = 5); endemic normals (n = 36); and healthy US blood bank volunteers (n = 29). Uninfected individuals residing in O. volvulus-endemic areas (Ecuador and Guatemala) were used as endemic normals. Filariae-infected samples were all microfilariae-positive; strongyloidiasis and cysticercosis samples were positive by PCR and/or ELISA. A more detailed exposition of these samples can be found in Table 1.
Table 1.
Panel of Serum Samples Tested
| Infecting Agent | No. of Samples | Origin | Testing Site |
|---|---|---|---|
| Onchocerca volvulus (n =457) | 89 | DRC | CDC |
| 35 | Ethiopia | CDC | |
| 25 | Tanzania | CDC | |
| 10 | Togo | CDC | |
| 53 | Uganda | CDC | |
| 40 | Guatemala | NIH | |
| 69 | Ecuador | NIH | |
| 60 | Ghana | NIH | |
| 47 | Cameroon | NIH | |
| 29 | Expatriatesa | NIH | |
| Wuchereria bancrofti (n = 45) | 15 | Cook Islands | NIH |
| 15 | India | NIH | |
| 10 | Haiti | CDC | |
| 5 | Expatriatesb | NIH | |
| Loa loa | 34 | Expatriatesc | NIH |
| Strongyloides stercoralis (n = 34) | 8 | Expatriatesd | NIH |
| 16 | Southeast Asia | NIH | |
| 10 | Argentina | CDC | |
| Mansonella sp. | 10 | Peru | CDC |
| Taenia solium | 5 | Peru | CDC |
| Endemic normal (n = 36) | 19 | Ecuador (Quito) | NIH |
| 17 | Guatemala (Guatemala City) | NIH | |
| Blood bank normals | 29 | North America | NIH/CDC |
Expatriate samples are from individuals originally from endemic countries living in the United States.
Abbreviations: CDC, Centers for Disease Control and Prevention; NIH, National Institutes of Health.
aSierra Leone, Guatemala, and Cameroon.
bIndia, Comoros Island, and Guyana.
cCameroon, Gabon, Central African Republic, and Benin.
dSoutheast Asia.
To evaluate the kinetics of the antibody responses to the new biomarker antigens, we used archived serum samples of experimentally infected nonhuman primates (Pan troglodytes, NHP) from studies [23, 24] conducted between 1987 and 1995 at the Yerkes Regional Primate Research Center in Atlanta, Georgia (Yerkes protocol numbers Y90/3/05 and Y91/2/06). We used monthly samples from NHP-5, NHP-6, NHP-7, and NHP-8 that developed patent infections, defined by the detection of microfilariae in skin snips. The samples were evaluated for antibody reactivity over time to selected O. volvulus antigens. These antibody responses were compared to those against OV-16.
Luciferase Immunoprecipitation System Assay
Fusion proteins for OVOC10469, OVOC3261, OVOC5127, OVOC11950, OVOC10602, and OV-16 were made by cloning the full-length genes coding for the proteins into a FLAG epitope-tagged mammalian Renilla reniformis luciferase (Ruc)-containing expression vector pREN2 [25]. Lysates containing the fusion proteins were prepared by transfecting 293F cells (Thermo Fisher Scientific, Waltham, MA) as per the manufacturer’s instructions. Briefly, 30 µg of plasmid was used to transfect 293F cells at a final concentration of 1 µg of plasmid for 1 × 106 cells in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) and cultured for 48 hours at 37°C, 8% CO2, and shaking at 125 rpm. The cells were centrifuged, the pellet was lysed as described previously [25], and the lysate frozen until used.
A standard luciferase immunoprecipitation system (LIPS) antibody assay [26, 27] was used for the evaluation of IgG and IgG4 responses to each of the antigens. The output was measured as relative light units using a Berthold LB 960 Centro microplate luminometer and coelenterazine substrate mixture (Promega, Madison, WI).
Production and Purification of Recombinant Proteins
Selected proteins (OVOC10469 [A0A044QSR3] and OVOC3261 [A0A2K6VY22]) were expressed recombinantly as C-terminal His-tagged proteins (Genscript, Piscataway, NJ) by cloning the genes encoding the proteins into pET30a expression vectors. BL21 Star (DE3) strains of Escherichia coli were transformed with the recombinant plasmids. The proteins were obtained from the lysates of 1L cultures of E. coli transformed with the individual plasmids and induced with isopropyl β-d-1-thiogalactopyranoside at 15°C for 16 hours. The cells were harvested by centrifugation and resuspended in lysis buffer followed by sonication. Supernatants from centrifuged cultures containing the recombinant proteins were purified over a nickel metal affinity column (Supplementary Figure 1), dialyzed, filter sterilized (0.2 µm), and stored in aliquots at −80°C in phosphate-buffered saline (PBS) with 10% glycerol until use. For clarity, only the E. coli expressed proteins have been termed with the recombinant prefix r; the fusion proteins (lysates) intended for LIPS assay are denoted by their original protein name.
IgG4 ELISA to O. volvulus Peptides and Recombinant Proteins
Potentially immunogenic peptides were synthesized based on predictions using PROTEAN (Lasergene Suite, DNASTAR). Immulon 4 HBX flat-bottom 96-well microplates (Thermo Fisher Scientific) were coated with 100 μL of 1 μg/mL of the immunogenic peptides or the recombinant proteins in PBS and incubated overnight at 4°C. All the wash steps were carried out 6 times in PBS with 0.05% Tween 20 (PBST). The plates were washed and blocked with 5% bovine serum albumin in PBST for 1 hour at 37°C. The plates were washed and 100 μL of sera (1:100 dilution) from O. volvulus-infected and uninfected controls were then added in duplicates and incubated for 1 hour at 37°C. The plates were washed and incubated with 1:1000 of biotinylated anti-human IgG4 (Southern Biotech, Birmingham, AL) for 1 hour at 37°C. The plates were washed and incubated with 100 μL of (1:50 000) of streptavidin conjugated to horse radish peroxidase (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 1 hour at 37°C. The microplates were washed and 50 μL of 1-Step Ultra TMB-ELISA substrate solution (Thermo Fisher Scientific) was added. The reaction was stopped after 10 minutes with 50 μL of 2N H2SO4 and the absorbance of the reaction was measured at 450 nm.
OV-16/OVOC3261 Lateral-Flow Strip Prototype Immunoassay
Based on preliminary testing of both rOVOC10469 and rOVOC3261 on lateral-flow strip prototypes, only rOVOC3261 could be immobilized effectively on test strips. Prototype lateral-flow test strips were assembled as described previously [28]. A BioDot XYZ reagent dispenser was used to apply OV-16, rOVOC3261, and control antibody (goat anti-mouse IgG, Jackson ImmunoResearch) onto the nitrocellulose membrane, and to spray anti-human IgG4 antibody (Hybridoma Reagent Laboratory) conjugated to colloidal gold onto the conjugate pad. For detection of IgG4 to OV-16 and/or rOVOC3261, 3 μL of patient serum/plasma was added to each strip near the conjugate pad. The conjugate ends of the strips were placed in wells of a flat-bottomed 96-well plate with 100 μL of lateral-flow test buffer, and the strips were read at 20 minutes and when dry (24 hours). The strips were read by 2 or more readers and scored based on a color intensity-level chart. The scores of the readers were averaged.
Statistical Analyses
Unless otherwise stated, geometric means were used as a measure of central tendency. The cutoffs for LIPS and ELISA were determined based on the receiver operating characteristic (ROC) analyses performed with “other helminth” group of samples. The sensitivity values are based on 100% specificity as determined by the ROC curves. All analyses were performed using GraphPad Prism 7.
RESULTS
Screening and Validation of Potential Biomarkers
Of all the IgG4 reactive promising candidates identified [21], we focused on OVOC10469, OVOC11950, OVOC10602, OVOC3261, and OVOC5127, because neither OVOC8491 (fatty acid retinol binding protein 2) nor OVOC9988 (Ov17; SXP) had the specificity needed for further development. These 5 predicted hypothetical proteins were next screened with sera from O. volvulus-infected and -uninfected individuals using LIPS assays to measure total IgG and IgG4 antibody responses. Although each of the 5 proteins had significant IgG reactivity in O. volvulus-infected individuals (P < .0001) compared to normal healthy blood bank control sera (Supplementary Figure 2A), preliminary screening using IgG4 (an antibody isotype known to provide antigenic specificity) indicated significant cross-reactivity of OVOC11950 and OVCO10602 with sera from subjects with Wuchereria bancrofti and/or Loa loa infections (Supplementary Figure 2B). Based on these results, further analysis was conducted only for OVOC10469, OVOC3261, and OVOC5127.
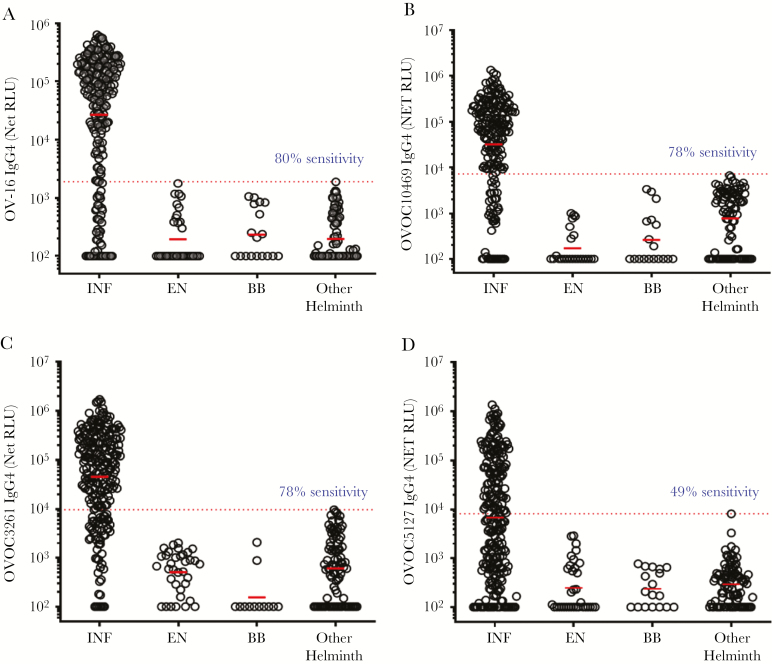
IgG4 responses to OVOC10469, OVOC3261, and OVOC5127, and also OV-16, were analyzed in sera of O. volvulus-infected, endemic normals, healthy US blood bank volunteers, and other-helminth–infected individuals (W. bancrofti, L. loa, and Strongyloides stercoralis) using the LIPS format. Similar to the sensitivities of 80% observed for OV-16 in ELISA or RDT formats at >99% specificity, we observed 80% sensitivity in IgG4 responses in O. volvulus-infected individuals (Figure 1A) compared to sera from other-helminth–infected individuals. Likewise, the sensitivity of assays using OVOC10469 and OVOC3261 for detection of O. volvulus-infected individuals was 78% (Figure 1B and 1C). In contrast, the sensitivity of the assay with OVOC5127 in identifying O. volvulus-infected individuals was 49% (Figure 1D).
Figure 1.
Screening of OVOC biomarkers by luciferase immunoprecipitation system (LIPS). The dot plots show the IgG4 net reactivity (value + 100) of the individual sera from Onchocerca volvulus-infected and microfilariae-positive (INF), endemic normal (EN), healthy US blood bank volunteers (BB), and Wuchereria bancrofti/ Loa loa/ Strongyloides stercoralis infections (other helminth) in a LIPS assay to OV-16 (A), OVOC10469 (B), OVOC3261 (C), and OVOC5127 (D). Dotted red lines denote the receiver operating characteristic (ROC) cutoff values (based on 100% specificity using the other helminth group as control). Sensitivity values are based on ROC cutoff with 100% specificity. The horizontal red bars denote geometric means. Negative values were assigned zero prior to adding 100 for all values. The values denote the net relative light units (RLU).
Combination of OVOC10469 and OVOC3261 Improves Sensitivity
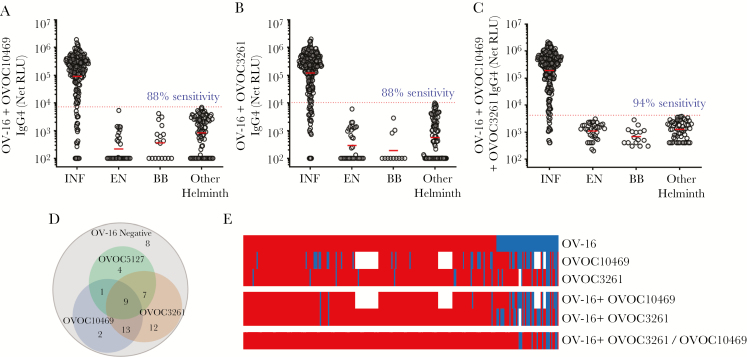
As OV-16 is the current serologic standard, we evaluated the additive values of OVOC10469, OVOC3261, and/or OVOC5127 to OV-16 positivity in detecting O. volvulus-infected individuals by testing combinations of OV-16 with OVOC10469, OVOC3261, and OVOC5127 in LIPS assays. As shown in Figure 2, compared to the observed sensitivity of 80% with OV-16 alone (Figure 1A) the combination of OV-16 with either OVOC10469 (Figure 2A) or OVOC3261 (Figure 2B) improved the sensitivity to 88%. The use of both OVOC10469 and OVOC3261 along with OV-16 resulted in 94% sensitivity (Figure 2C). Of the 56 OV-16–negative samples, 44 (78.5%) were positive for either OVOC3261 and/or OVOC10469. The addition of OVOC5127 was able to pick up 4 additional OV-16–negative samples as positive, while 8 of 245 O. volvulus-infected samples were negative for all proteins tested (Figure 2D and 2E).
Figure 2.
Validation of combinations of biomarkers by luciferase immunoprecipitation system (LIPS). The dot plots show the IgG4 net reactivity (value + 100) of the individual sera from Onchocerca volvulus-infected and microfilariae-positive (INF), endemic normal (EN), healthy US blood bank volunteers (BB), and Wuchereria bancrofti/ Loa loa/ Strongyloides stercoralis infections (other helminth) in a LIPS assay to OV-16 in combination with OVOC10469 (A), OVOC3261 (B), and OVOC10469 and OVOC3261 (C). The dotted red lines denote the receiver operating characteristic (ROC) cutoff values (based on 100% specificity using the other helminth group as control). Sensitivity values are based on ROC cutoff with 100% specificity. The horizontal red bars denote the geometric means. Negative values were assigned zero prior to adding 100 for all values. The values denote the net relative light units (RLU). D, The Venn diagram shows the differential reactivity of the OV-16–negative individuals that were positive to OVOC10469, OVOC3261, and/or OVOC5127. E, The heatmap depicts the samples classified as positive (red) or negative (blue) for OV-16, OVOC10469, OVOC3261, or combinations. Blank spaces denote that the sample was not tested.
Development of ELISA
Because the LIPS assay is an efficient system for high-throughput screening but not well-suited for immunoassays that could be developed for point of contact assays, we evaluated the use of peptides derived from OVOC10469, OVOC3261, and OVOC5127, or recombinant proteins (rOVOC10469 and rOVOC3261) in ELISA-based assays. Preliminary screening with pooled sera indicated that among the tested peptides, OVOC10469-Pep2, OVOC3261-Pep1, OVOC3261-Pep3, and OVOC5127-Pep5 had the best performance characteristics (Supplementary Figure 3). However, even in combination with OV-16, these peptides yielded sensitivities that were not better than OV-16 alone.
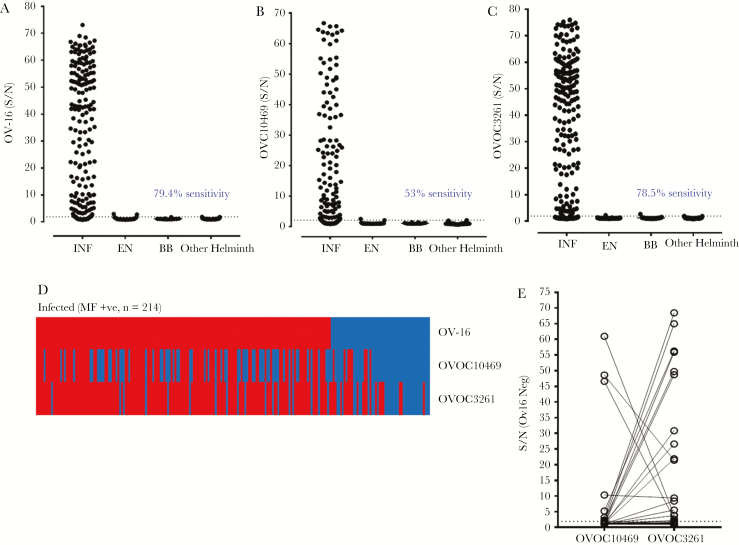
Using purified His-tagged rOVOC10469 and rOVOC3261 produced in E. coli (Supplementary Figure 1), we developed IgG4-based ELISAs (Figure 3A and 3B). Although generally good correlations were observed between the IgG4 reactivity in LIPS and the IgG4 reactivity in the ELISA format (for rOVOC3261 r = 0.67, P < .0001; and for rOVOC10469 r = 0.86, P < .0001; Supplementary Figure 4), the sensitivity of the assay with OVOC10469 for identifying O. volvulus-infected individuals dropped from 78% (as seen in LIPS) to 53% in the ELISA. Assays using the recombinant protein rOVOC3261, in contrast, retained high sensitivity when compared to that seen using the LIPS platform (Figure 1C). Based on the individual ROC curves (using the “other helminth” group as the control), of the 46 samples that were OV-16 negative (Figure 3C), 19 could be classified as true positive depending on their reactivity to rOVOC3261 and/or rOVOC10469 (Figure 3D). In the ELISA format, while rOVOC10469 does not appear to qualitatively contribute in identifying additional OV-16–negative samples (compared with positivity with rOVOC3261) (Figure 3D), the IgG4 signal to OVOC10469 was several-fold higher in some of the OV-16–negative samples (Figure 3E).
Figure 3.
Comparison of IgG4 responses to OV-16, rOVOC10469, and rOVOC3261 by enzyme-linked immunosorbent assay (ELISA). The scatter plots depict the signal to noise values (S/N) of OV-16 (A), rOVOC10469 (B), and rOVOC3261 (C) reactivity in sera of Onchocerca volvulus-infected (INF), endemic normal (EN), healthy US blood bank volunteers (BB), and Wuchereria bancrofti, Loa loa, Strongyloides stercoralis (other helminth) infections by ELISA. The dotted lines denote the receiver operating characteristic (ROC) cutoff values using the other helminth group as control. D, Heat map depicts the samples classified as positive (red) or negative (blue) for OV-16, rOVOC10469, and rOVOC3261 based on ROC. E, Signal to noise ratio of OV-16–negative sera (blue segment from heat map shown in Figure 3D) showing the differential reactivity to rOVOC10469 and rOVOC3261.
Kinetics: Seroreactivity and Seroconversion are Patency Dependent
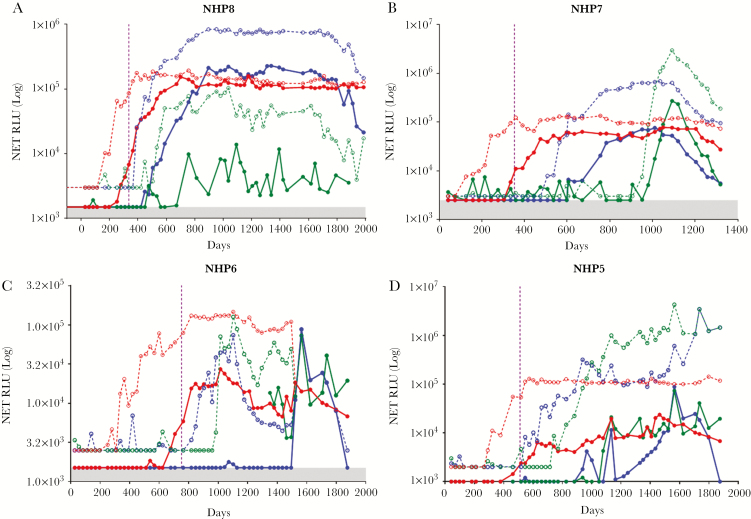
We used the archived NHP samples to evaluate the kinetics of the antibody responses to the antigens OV-16, OVOC10469, and OVOC3261 in O. volvulus infections. As shown in Figure 4, all the NHPs became microfilariae-positive (in skin snips) between 336 and 763 days following inoculation of L3 larvae; these NHPs remained microfilariae-positive almost to the end of the study (1323–1987 days). While the total IgG responses to OV-16 could be detected early in infection, IgG4 seroconversion occurred 50–100 days before the onset of patency. In contrast, IgG responses to both OVOC10469 and OVOC3261 were detectable only after the onset of patency, with the seroconversion to IgG4 following 100–150 days later. These results suggest that responses to OVOC10469 and OVOC3261 are microfilariae-driven, a finding that parallels the mRNA expression pattern of the corresponding 2 genes [21].
Figure 4.
Kinetics of development of antibody responses to OV-16, OVOC3261, and OVOC10469 in nonhuman primates (NHP). The kinetics of IgG (dotted) and IgG4 (solid) responses to OV-16 (red), OVOC3261 (blue), and OVOC10469 (green) in serum of 4 experimentally infected NHPs (A–D) by luciferase immunoprecipitation system. The values on the y-axis denote the net relative light units (RLU). The vertical violet dotted line denotes the time of appearance of skin microfilariae.
Significant Reduction in rOVOC10469-, rOVOC3261-, and OV-16–Specific IgG4 Posttreatment
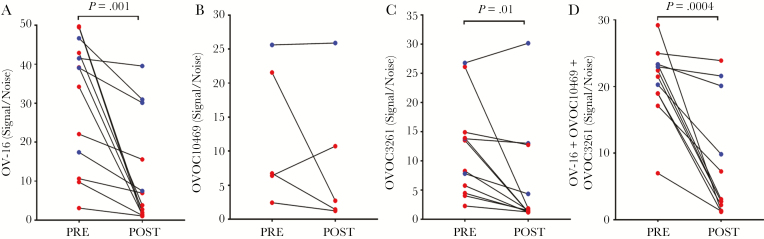
To evaluate further the utility of using the IgG4 responses to rOVOC10469, rOVOC3261, and OV-16 following presumed cure of onchocerciasis, we tested sera obtained from a limited number of O. volvulus-infected individuals that were diagnosed and treated at the NIH and followed longitudinally for 5 years or more [29]. To be able to compare the pre- and posttreatment data, only samples that had IgG4 signals more than twice the background were analyzed for responses to any given protein. The levels of antigen-specific IgG4 to OV-16 dropped significantly (P = .001) in the posttreatment samples. This was more profound in the individuals with >10 years posttreatment (Figure 5A, red). Although fewer samples were reactive to rOVC10469 (Figure 5B), the levels of IgG4 to rOVOC10649 (Figure 5B) and rOVOC3261 (Figure 5C) also decreased posttreatment, and the decrease was significant for rOVOC3261 (P = .01). When a cocktail of all 3 antigens was tested (Figure 5D) the IgG4 responses decreased significantly posttreatment (P < .0004). These data suggest that such a cocktail of proteins could be used for postcontrol surveillance as well as for mapping of endemic regions and individual diagnosis.
Figure 5.
IgG4 responses pre- and posttreatment. The paired IgG4 responses by enzyme-linked immunosorbent assay to OV-16 (A), rOVCO10469 (B), rOVOC3261 (C), and combination of OV-16, rOVOC10469, and rOVOC3261 (D) in patients that were treated and followed-up longitudinally for over 10 years (red) or between 5 and 7 years (blue). Signal to noise values are plotted on the y-axis.
Development and Screening of Prototype Lateral-Flow Test Using rOVOC3261 and OV-16
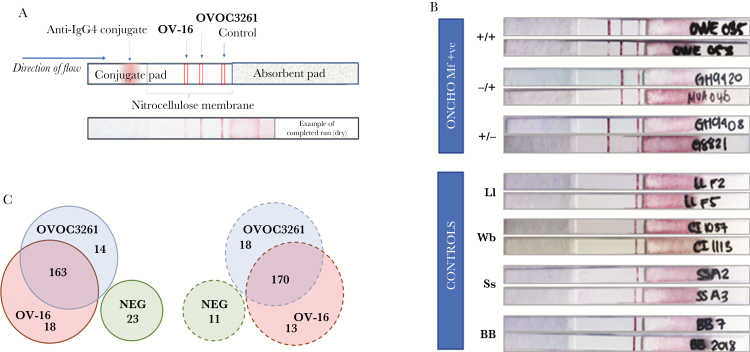
To evaluate the functional utility of complementing the existing OV-16 RDT, the better performing antigen rOVOC3261 was striped on the lateral-flow test strips along with OV-16 (Figure 6A). The lateral-flow test strips were initially tested at the NIH and later validated with a different panel at the CDC. The controls showed no reactivity to either OV-16 or rOVOC3261, while the O. volvulus-infected samples were positive for either OV-16, rOVOC3261, or both (Figure 6B). Of the 218 O. volvulus-infected samples tested at the NIH with the test strips, 83% were positive for OV-16, while 81% were positive for rOVOC3261. Of the samples that were negative for OV-16 by the test strip, 14 (38%) were positive for rOVOC3261. The sensitivity for detecting an infected sample increased to 89% when considering positivity for either OV-16 or rOVOC3261 (Figure 6C). Similarly, of the 212 O. volvulus-infected samples tested independently at CDC, 85% were positive for OV-16, while 87% were positive for rOVOC3261. Eighteen (58%) of the OV-16–negative samples from the CDC were positive for rOVOC3261, resulting in 94% of the infected individuals being positive for either OV-16 and/or rOVOC3261 (Figure 6C).
Figure 6.
Testing of prototype rapid diagnostic test. Prototype rapid diagnostic test strips with OV-16 and rOVOC3261 were developed (A) and tested with infected and uninfected or control sera. B, The top panel shows representative images of strips from Onchocerca volvulus microfilariae (mf)-positive sera that were positive for both OV-16 and rOVOC3261 (+/+), or positive only for OV-16 (+/−), or positive only for rOVOC3261 (−/+). The bottom panel shows representative images from reactivity against serum from healthy blood bank volunteers (BB) or people infected with Loa loa (Ll), Wuchereria bancrofti (Wb), or Strongyloides stercoralis (Ss). C, The Venn diagrams shows the number of samples positive for OV-16 and/or rOVOC3261 in the test strips tested at National Institutes of Health (solid lines) and at Centers for Disease Control and Prevention (dotted lines).
DISCUSSION
Although the existing methods and targets available to test for infection with O. volvulus have been useful for verifying suppression and interruption of transmission in the Americas and in several foci in Africa, more sensitive tools could greatly accelerate program elimination activities targeting onchocerciasis [30]. Because the sensitivity of the current POC OV-16 RDT using whole blood in low prevalence areas is poor, negative results currently require confirmatory ELISA tests [11]. A more sensitive POC test could obviate the need for this second step.
The major drawback of antibody-based biomarkers is that they do not clearly distinguish active from past infections, although they can be used for monitoring for infection in children born after transmission is believed to have been interrupted. Although the combination of OVOC9988 (Ov17; SXP), OVOC9984 (Ov33), and OVOC8491 (O. volvulus-FAR2) with OV-16 as multiplexed assays in laboratory settings was described as a sensitive platform for surveillance [31, 32], we excluded these well-characterized immunogenic proteins due to their cross-reactivity. We favored the newly identified hypothetical proteins OVOC10469, OVOC3261, OVOC10602, OVOC11950, and OVOC5127 because they were predicted to have few to no known homologues in other filariae. Based on their sequences, these newly identified proteins, in addition to having a signal peptide, were likely to be secreted [21]. The stage-specific enriched expression of OVOC10469 and OVOC3261 in microfilariae [21] is corroborated by the detectability of OVOC3261- and OVOC10469-specific antibodies only upon the onset of patency (as seen in experimentally infected nonhuman primates). Assays that detect antibodies to these markers not only overcome the sensitivity barrier but also indicate the presence of fertile females and possibly active transmission. Moreover, for at least OVOC10469 and OVOC3261, there were no nonsynonymous single-nucleotide polymorphisms in their coding sequences [33, 34] and hence it is likely that the responses to these proteins would be invariant across all geographic regions.
The existing OV-16 RDT is an IgG4-based immunoassay. Therefore, for an appropriate comparison, we focused on IgG4 responses to the selected hypothetical proteins OVOC10469 and OVOC3261, both of which were expressed in bacteria for recombinant antigen production. When we examined the additional benefit of either OVOC3261 or OVOC10469 for detecting patent infection in previously tested OV-16–negative O. volvulus-infected samples, an overwhelming number of the tested samples were positive for both OVOC3261 and OVOC10469. However, rOVOC10469 did not perform as well as rOVOC3261 in some solid-phase immunoassay formats.
The new immunoassay using OV-16 and rOVOC3261 is likely to provide a better serological tool to enhance the O. volvulus elimination efforts. First, it could be used in areas that need to undergo onchocerciasis mapping (including elimination mapping) given that the increased sensitivity will more accurately detect evidence of transmission in areas previously considered free of disease or hypoendemic. Secondly, the improved sensitivity of immunoassays using 2 O. volvulus-specific antigens may lead to a reduction in the number of children needing to be tested to meet elimination thresholds. Thirdly, because the positivity of IgG4 to both OVOC3261 and OVOC10469 are driven by microfilariae, these new tests may be able to drive better models for the decision to stop treatment. Further, if the limited longitudinal data on patients treated successfully (>10 years) who demonstrated loss of reactivity (Figure 5) can be repeated in a prospective study of large cohorts of samples from multiple rounds of mass drug administration, these new immunoassays may actually also be useful biomarkers of cure.
While these new antibody-based biomarkers drive the detection sensitivity of truly infected individuals to 94%, it remains to be seen if other antigen combinations can further increase the sensitivity to the desired level of 99%. More than likely, highly sensitive O. volvulus antigen detection assays will be needed, particularly as communities move toward their elimination targets. Nevertheless, the new IgG4-based combination immunoassays represent a major step forward toward the effort of onchocerciasis elimination.
SUPPLEMENTARY DATA
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health; and the Bill and Melinda Gates Foundation (grant number OPP1083910).
Potential conflicts of interest. A patent (WO2017173369A1) has been filed for the usage of the antigens described in the development of the lateral-flow assay. S.B., S.L. and T.B.N. are the named individuals in the patent.
All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: American Society of Tropical Medicine and Hygiene Meeting, 28 October–1 November 2018, New Orleans, Louisiana, abstract No. 717.
References
- 1. World Health Organization. Progress report on the elimination of human onchocerciasis, 2017–2018. Wkly Epidemiol Rec 2018; 47:633–48. [Google Scholar]
- 2. World Health Organization. Progress towards eliminating onchocerciasis in the WHO Region of the Americas: elimination of transmission in the north-east focus of the Bolivarian Republic of Venezuela. Wkly Epidemiol Rec 2017; 92:617–23. [PubMed] [Google Scholar]
- 3. World Health Organization. Progress towards eliminating onchocerciasis in the WHO Region of the Americas: verification of elimination of transmission in Guatemala. Wkly Epidemiol Rec 2016; 91:501–5. [PubMed] [Google Scholar]
- 4. Tekle AH, Zouré HG, Noma M, et al. . Progress towards onchocerciasis elimination in the participating countries of the African Programme for Onchocerciasis Control: epidemiological evaluation results. Infect Dis Poverty 2016; 5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ndyomugyenyi R, Lakwo T, Habomugisha P, Male B. Progress towards the elimination of onchocerciasis as a public-health problem in Uganda: opportunities, challenges and the way forward. Ann Trop Med Parasitol 2007; 101:323–33. [DOI] [PubMed] [Google Scholar]
- 6. Walker M, Stolk WA, Dixon MA, et al. . Modelling the elimination of river blindness using long-term epidemiological and programmatic data from Mali and Senegal. Epidemics 2017; 18:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Traore MO, Sarr MD, Badji A, et al. . Proof-of-principle of onchocerciasis elimination with ivermectin treatment in endemic foci in Africa: final results of a study in Mali and Senegal. PLoS Negl Trop Dis 2012; 6:e1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fink DL, Kamgno J, Nutman TB. Rapid molecular assays for specific detection and quantitation of Loa loa microfilaremia. PLoS Negl Trop Dis 2011; 5:e1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lobos E, Weiss N, Karam M, Taylor HR, Ottesen EA, Nutman TB. An immunogenic Onchocerca volvulus antigen: a specific and early marker of infection. Science 1991; 251:1603–5. [DOI] [PubMed] [Google Scholar]
- 10. World Health Organization. Guidelines for stopping mass drug administration and verifying elimination of human onchocerciasis: criteria and procedures. Geneva: World Health Organization, 2016. [PubMed] [Google Scholar]
- 11. World Health Organization. Report of the second meeting of the WHO Onchocerciasis Technical Advisory Subgroup. Geneva: World Health Organization, 2018. [Google Scholar]
- 12. Lindblade KA, Arana B, Zea-Flores G, et al. . Elimination of Onchocercia volvulus transmission in the Santa Rosa focus of Guatemala. Am J Trop Med Hyg 2007; 77:334–41. [PubMed] [Google Scholar]
- 13. Gonzalez RJ, Cruz-Ortiz N, Rizzo N, et al. . Successful interruption of transmission of Onchocerca volvulus in the Escuintla-Guatemala focus, Guatemala. PLoS Negl Trop Dis 2009; 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oguttu D, Byamukama E, Katholi CR, et al. . Serosurveillance to monitor onchocerciasis elimination: the Ugandan experience. Am J Trop Med Hyg 2014; 90:339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodríguez-Pérez MA, Fernández-Santos NA, Orozco-Algarra ME, et al. . Elimination of onchocerciasis from Mexico. PLoS Negl Trop Dis 2015; 9:e0003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nicholls RS, Duque S, Olaya LA, et al. . Elimination of onchocerciasis from Colombia: first proof of concept of river blindness elimination in the world. Parasit Vectors 2018; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cama VA, McDonald C, Arcury-Quandt A, et al. . Evaluation of an OV-16 IgG4 enzyme-linked immunosorbent assay in humans and its application to determine the dynamics of antibody responses in a non-human primate model of Onchocerca volvulus infection. Am J Trop Med Hyg 2018; 99:1041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lont YL, Coffeng LE, de Vlas SJ, et al. . Modelling anti-Ov16 IgG4 antibody prevalence as an indicator for evaluation and decision making in onchocerciasis elimination programmes. PLoS Negl Trop Dis 2017; 11:e0005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gbakima AA, Nutman TB, Bradley JE, et al. . Immunoglobulin G subclass responses of children during infection with Onchocerca volvulus. Clin Diagn Lab Immunol 1996; 3:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gass KM. Rethinking the serological threshold for onchocerciasis elimination. PLoS Negl Trop Dis 2018; 12:e0006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bennuru S, Cotton JA, Ribeiro JM, et al. . Stage-specific transcriptome and proteome analyses of the filarial parasite Onchocerca volvulus and its Wolbachia endosymbiont. MBio 2016; 7:pii:e02028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rebollo MP, Zoure H, Ogoussan K, Sodahlon Y, Ottesen EA, Cantey PT. Onchocerciasis: shifting the target from control to elimination requires a new first-step-elimination mapping. Int Health 2018; 10:i14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eberhard ML, Dickerson JW, Boyer AE, et al. . Experimental Onchocerca volvulus infections in mangabey monkeys (Cercocebus atys) compared to infections in humans and chimpanzees (Pan troglodytes). Am J Trop Med Hyg 1991; 44:151–60. [DOI] [PubMed] [Google Scholar]
- 24. Eberhard ML, Dickerson JW, Tsang VC, et al. . Onchocerca volvulus: parasitologic and serologic responses in experimentally infected chimpanzees and mangabey monkeys. Exp Parasitol 1995; 80:454–62. [DOI] [PubMed] [Google Scholar]
- 25. Burbelo PD, Goldman R, Mattson TL. A simplified immunoprecipitation method for quantitatively measuring antibody responses in clinical sera samples by using mammalian-produced Renilla luciferase-antigen fusion proteins. BMC Biotechnol 2005; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burbelo PD, Ramanathan R, Klion AD, Iadarola MJ, Nutman TB. Rapid, novel, specific, high-throughput assay for diagnosis of Loa loa infection. J Clin Microbiol 2008; 46:2298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramanathan R, Burbelo PD, Groot S, Iadarola MJ, Neva FA, Nutman TB. A luciferase immunoprecipitation systems assay enhances the sensitivity and specificity of diagnosis of Strongyloides stercoralis infection. J Infect Dis 2008; 198:444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steel C, Golden A, Kubofcik J, et al. . Rapid Wuchereria bancrofti-specific antigen Wb123-based IgG4 immunoassays as tools for surveillance following mass drug administration programs on lymphatic filariasis. Clin Vaccine Immunol 2013; 20:1155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry NL, Law M, Nutman TB, Klion AD. Onchocerciasis in a nonendemic population: clinical and immunologic assessment before treatment and at the time of presumed cure. J Infect Dis 2001; 183:512–6. [DOI] [PubMed] [Google Scholar]
- 30. Unnasch TR, Golden A, Cama V, Cantey PT. Diagnostics for onchocerciasis in the era of elimination. Int Health 2018; 10:i20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feeser KR, Cama V, Priest JW, et al. . Characterizing reactivity to Onchocerca volvulus antigens in multiplex bead assays. Am J Trop Med Hyg 2017; 97:666–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burbelo PD, Leahy HP, Iadarola MJ, Nutman TB. A four-antigen mixture for rapid assessment of Onchocerca volvulus infection. PLoS Negl Trop Dis 2009; 3:e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Norice-Tra CT, Ribeiro J, Bennuru S, et al. . Insights into Onchocerca volvulus population biology through multilocus immunophenotyping. J Infect Dis 2017; 216:736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi YJ, Tyagi R, McNulty SN, et al. . Genomic diversity in Onchocerca volvulus and its Wolbachia endosymbiont. Nat Microbiol 2016; 2:16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.