Abstract
Background
Noroviruses are a leading cause of acute gastroenteritis. Genogroup 2 type 4 (GII.4) has been the dominant norovirus genotype worldwide since its emergence in the mid-1990s. Individuals with a functional fucosyltransferase-2 gene, known as secretors, have increased susceptibility to GII.4 noroviruses. We hypothesized that this individual-level trait may drive GII.4 norovirus predominance at the human population level.
Methods
We conducted a systematic review for studies reporting norovirus outbreak or sporadic case genotypes and merged this with data on proportions of human secretor status in various countries from a separate systematic review. We used inverse variance-weighted linear regression to estimate magnitude of the population secretor-GII.4 proportion association.
Results
Two hundred nineteen genotype and 112 secretor studies with data from 38 countries were included in the analysis. Study-level GII.4 proportion among all noroviruses ranged from 0% to 100%. Country secretor proportion ranged from 43.8% to 93.9%. We observed a 0.69% (95% confidence interval, 0.19–1.18) increase in GII.4 proportion for each percentage increase in human secretor proportion, controlling for Human Development Index.
Conclusions
Norovirus evolution and diversity may be driven by local population human host genetics. Our results may have vaccine development implications including whether specific antigenic formulations would be required for different populations.
Keywords: genotype, GII.4, norovirus, population, secretor
Using systematic literature review data, a positive population-level relationship was identified between the proportion of human secretor status in a population and predominance of the GII.4 norovirus genotype.
Noroviruses are a leading cause of acute gastroenteritis worldwide [1]. They are nonenveloped, positive sense, single-stranded ribonucleic acid viruses in the Caliciviridae family [2, 3]. Noroviruses are highly genetically diverse and grouped into 10 genogroups (GI–GX) based on polymerase and capsid gene sequences with strains from genogroups GI, GII, and GIV, GVIII, and GIX infecting humans [4]. Genogroup 2 is associated with approximately 90% of cases and outbreaks globally [5]. Within genogroups, human noroviruses are categorized into 9 GI, 27 GII, and 2 GIV capsid genotypes [4]. Genogroup 2 type 4 (GII.4) is the most prevalent genotype, associated with 50% to 80% of all norovirus outbreaks depending on the year [5].
Noroviruses bind to histoblood group antigens (HBGAs) on surfaces of gastroduodenal epithelial cells [6]. The HBGAs are oligosaccharides present in gut, urinary tract, and respiratory cells, as well as other bodily fluids including milk, blood, and saliva [7]. Two enzymes encoded by the fucosyltransferase (FUT)1 and FUT2 genes are responsible for the synthesis of certain HBGAs in humans. Individuals without a functional FUT2 gene are termed nonsecretors, because they lack these specific HBGAs on certain cell surfaces [8]. Although not all norovirus genotypes appear to be “secretor-dependent,” secretor individuals who have functional FUT2 genes are at markedly heightened risk for GII.4 infections as well as some less common types (eg, GI.1) [6, 9].
Among noroviruses, GII.4 viruses uniquely evolve rapidly and are able to bind to a variety of HBGAs [10]. Human population immunity against GII.4 appears to drive antigenic shifts, which in some cases may allow these viruses to expand their binding repertoire [11]. Novel GII.4 variants may spread rapidly and globally, with novel GII.4 strains replacing previously dominant strains in the course of 1 season [11]. The global dissemination (ie, pandemics) of novel GII.4 variants may result from antigenic escape, coupled with short-term natural immunity to norovirus [12, 13].
Several studies have demonstrated an association between secretor status and genotype-specific susceptibility, suggesting an interaction between human and viral genetic diversity [14, 15]. In both human challenge studies and observational outbreak studies, secretors have increased susceptibility to GII.4 noroviruses, whereas nonsecretors have strong innate resistance to GII.4 norovirus infection and disease [16, 17]. In our previous systematic review and meta-analysis on the association between secretor status and norovirus infection, we found that secretors had 4.2 times the odds of any norovirus infection (95% confidence interval [CI], 2.3–7.9) and 9.9 times the odds of GII.4 infection (95% CI, 3.9–24.8) compared with nonsecretors [9]. However, nonsecretors may not be completely resistant to GII.4 noroviruses [18].
We aimed to investigate the association between the frequency of secretor status in human populations with the preponderance of GII.4 norovirus among norovirus cases/outbreaks in that population. We developed a dataset by conducting a systematic literature review on studies reporting genotype distributions and synthesized it with estimates of country-level secretor prevalence. We hypothesized that a higher human population-level proportion of secretors was associated with greater GII.4 predominance.
METHODS
Data Sources
We searched PubMed for norovirus genotype studies published in English from 1994 until June 2017 using the search terms “norovirus” coupled with any of “surveillance,” “genotype,” or “strain.” All identified references were initially screened by title; single outbreak reports, studies of environmental samples that did not include human specimens, studies in animals, and other irrelevant studies were excluded. Included titles were then screened by abstract, and abstracts without genogroup or genotype data were excluded. We subsequently reviewed all screened abstracts by full text and extracted data from all included studies. Studies that reported only selected genotypes for genetic or epidemiologic analyses without reporting a genotype distribution were also excluded. Studies that did not report norovirus genotypes and studies from which the proportion of GII.4 among all genotypes could not be determined were excluded. Finally, genotyping studies from countries without corresponding secretor status data were also excluded from analysis (see below).
We used Evidence Partners DistillerSR (Ottawa, Canada) systematic review software to automate metadata collection and then manually entered data from the papers into a database. We extracted genogroup and genotype counts or percentages from each included study. For studies in which data of interest were only contained in figures, we requested data from corresponding authors by email. Other variables collected from the studies included sampling unit (ie, whether the study included outbreaks, sporadic cases, or both), setting, age distribution, study period, detection method, and typing region. In cases in which data were stratified by time period, typing region, or setting, multiple data entries were made from the same study.
Duplicate data were identified by reviewing full text articles of any observations with common within-country locations and overlapping time periods. Multiple papers that presented different analyses using data that came from the same study were considered duplicate. In general, when duplicate data were identified, larger studies were retained unless there was a reason to keep smaller ones. Smaller studies were included over larger ones if they were more representative of the country’s population or were published in multiple installments over a larger period of time than the larger study.
Secretor status data were extracted from a systematic literature review of studies that described the proportion of secretors among a specified population. Search terms used in this review included “secretor” or “secretor status.” Studies that reported results of secondary data analysis, nonhuman data, and those that were not published in English were excluded. Data were abstracted on the proportion of secretors among the population, as well as the country in which the study was conducted, the size of the study population, and, where specified, the ethnic group from which the population was drawn. For studies that examined the association between HBGA and a specific condition (eg, Helicobacter pylori, rotavirus), data from only the control group (ie, individuals without that condition) were abstracted. Studies that presented only the proportion of secretors among a group of individuals with such conditions and did not have a comparison group were excluded, because these populations may have inflated or deflated secretor prevalence compared with the general population. Furthermore, any studies in which participants were preferentially selected or screened out based on HBGA status were also excluded.
Data Management
Country-level secretor status proportion estimates were calculated by summing the number of secretors across all included studies in a country and dividing by the summed study populations across all included studies within that country. This aggregate secretor dataset was then merged with the genotype dataset assembled from this current systematic literature review.
All studies that reported genotypes in proportion were converted to counts by multiplying by the study sample size. The GII.4, GII.non-4, and GI proportions were calculated as the count of outbreaks or sporadic cases genotyped as that group divided by the total number of genotyped outbreaks or sporadic cases. Because GI and GII.non-4 could not be differentiated in some studies, GII.non-4 and GI proportions were only calculated for studies in which the GI and GII.non-4 were presented separately (n = 349).
Covariates assessed in the analysis were study type, age group, pandemic variant period, World Health Organization (WHO) region, Human Development Index (HDI), detection method, and genotyping region. The study type variable combined sampling unit (outbreaks vs sporadic cases) and setting into 1 classification scheme that included outbreak studies separately from different settings of sporadic case studies. Study population ages were categorized into ≤5 years, >5 years, and mixed. Because of the patient ages included in most studies, we were unable to stratify into smaller age groups. Observations were classified according to the dominant pandemic GII.4 variant of that time period, such that each time period was mutually exclusive. Time periods used were 1994–2001 (Lordsdale and US 95/96 variants), 2002–2003 (Farmington Hills 2002), 2004–2005 (Hunter 2004), 2006–2008 (Yerseke 2006a and Den Haag 2006b), 2009–2011 (New Orleans 2009), 2012–2017 (Sydney 2012), and observations whose study period spanned multiple pandemic periods [2]. The HDI data were obtained from the United Nations Development Program website (http://hdr.undp.org/en/data), and both the HDI categories and numeric indices were merged with the secretor and genotype data by country [19]. Likewise, WHO region classifications were obtained from the WHO website (http://www.who.int/about/regions/en/) for each country in the dataset and merged with the full dataset by country [20]. Typing region categories were classified as open reading frame (ORF)1, ORF2, ORF1 and ORF2, ORF1/ORF2 overlap only, and unknown. Studies that used both ORF1 and the ORF1/ORF2 overlap or ORF2 and the ORF1/ORF2 overlap were categorized as ORF1 and ORF2, respectively.
Genotype and secretor data aggregated by country were used for mapping and data visualization. Maps were created in Esri ArcGIS 10.5.1 (Redlands, CA) and projected using the World Geodetic System 1984 World Mercator projection.
Data Analysis
All statistical analyses were performed using SAS 9.4 (Cary, NC). Medians and interquartile ranges (IQRs) were calculated for GII.4, GII.non-4, and GI proportions across all observations and for each predictor variable category or quartile.
We used weighted linear regression to quantify the association between country-level secretor proportion and observation GII.4 proportion. All models were weighted by estimated inverse variance of GII.4 proportion with weights calculated using the inverse of the binomial proportion variance formula:
where σ2 is the estimated variance, p is the observation GII.4 proportion, and n is the observation sample size. For observations where GII.4 proportion was 1 or 0, proportions were adjusted by adding or subtracting 0.05 to calculate a defined, positive inverse variance. To assess the sensitivity of our results to inflated weights at the bounds of the binomial variance equation, we refit our final model omitting observations where GII.4 proportion was 1 or 0.
Simple regression models were fit for secretor proportion and each covariate individually to estimate associations with GII.4 proportion. Our multivariable models aimed to independently estimate the association between population secretor proportion and GII.4 proportion while controlling for potential confounding factors. Both WHO region and HDI were hypothesized a priori to be confounders of the secretor-GII.4 association; higher HDI countries and wealthier WHO regions tend to have higher secretor proportions and likely have more robust outbreak surveillance where GII.4 is more common. However, we suspected collinearity of WHO region and HDI, which indeed were highly associated (analysis of variance F = 120.1, P < .01). Because we had country-specific HDI values, and HDI was determined to be a more important predictor of GII.4 proportion than WHO region due to a higher R2 in simple regression (Table 2), HDI was included in the multivariable regression models. To assess the sensitivity of our final model to country-level nonindependence, we fit a generalized estimating equation (GEE) model to estimate the association between secretor and GII.4 proportions while controlling for HDI and accounting for country-level clustering.
Table 2.
Inverse Variance Weighted Simple Linear Regression Models of Each Predictor on GII.4 Proportion
| Predictor | R2 | β | 95% CI |
|---|---|---|---|
| Secretor Proportion | 0.05 | 1.17 | 0.69–1.64 |
| Study Type | 0.02 | ||
| Outbreaks | 0.00 | Reference | |
| Inpatient Cases | 0.03 | −0.06 to 0.11 | |
| Outpatient Cases | 0.00 | −0.10 to 0.11 | |
| Inpatient and Outpatient Cases | −0.03 | −0.12 to 0.06 | |
| Community Cases | −0.10 | −0.23 to 0.04 | |
| Mixed Setting | 0.06 | −0.03 to 0.14 | |
| Not Specified | 0.12 | −0.10 to 0.34 | |
| Age Category | 0.02 | ||
| ≤5 Years | 0.00 | Reference | |
| >5 Years | 0.05 | −0.11 to 0.20 | |
| Mixed | 0.11 | 0.03–0.19 | |
| Pandemic Variant Period | 0.24 | ||
| 2012–2017 | 0.00 | Reference | |
| 1994–2001 | −0.12 | −0.22 to −0.02 | |
| 2002–2003 | 0.15 | 0.03–0.27 | |
| 2004–2005 | 0.29 | 0.17 to 0.42 | |
| 2006–2008 | 0.32 | 0.26–0.39 | |
| 2009–2011 | 0.12 | 0.01–0.23 | |
| Multiple Periods | 0.13 | 0.06–0.20 | |
| WHO Region | 0.05 | ||
| Western Pacific | 0.00 | Reference | |
| Africa | −0.18 | −0.46 to 0.09 | |
| Americas | 0.04 | −0.03 to 0.11 | |
| Southeast Asia | −0.11 | −0.36 to 0.14 | |
| Europe | 0.11 | 0.04 to 0.18 | |
| Eastern Mediterranean | −0.36 | −0.61 to −0.10 | |
| HDI | 0.10 | 1.01 | 0.71–1.31 |
| Detection Method | 0.00 | ||
| RT-PCR | 0.00 | Reference | |
| Other | 0.00 | −0.14 to 0.15 | |
| Unknown | 0.06 | −0.05 to 0.17 | |
| Typing Region | 0.04 | ||
| ORF1 | 0.00 | Reference | |
| ORF2 | −0.11 | −0.18 to −0.04 | |
| ORF1 and ORF2 | −0.03 | −0.13 to 0.06 | |
| Overlap Only | −0.01 | −0.12 to 0.10 | |
| Unknown | 0.03 | −0.07 to 0.14 |
Abbreviations: CI, confidence interval; GII.4, genogroup 2 type 4; HDI, Human Development Index; ORF, open reading frame; RT-PCR, reverse transcription polymerase chain reaction; WHO, World Health Organization.
RESULTS
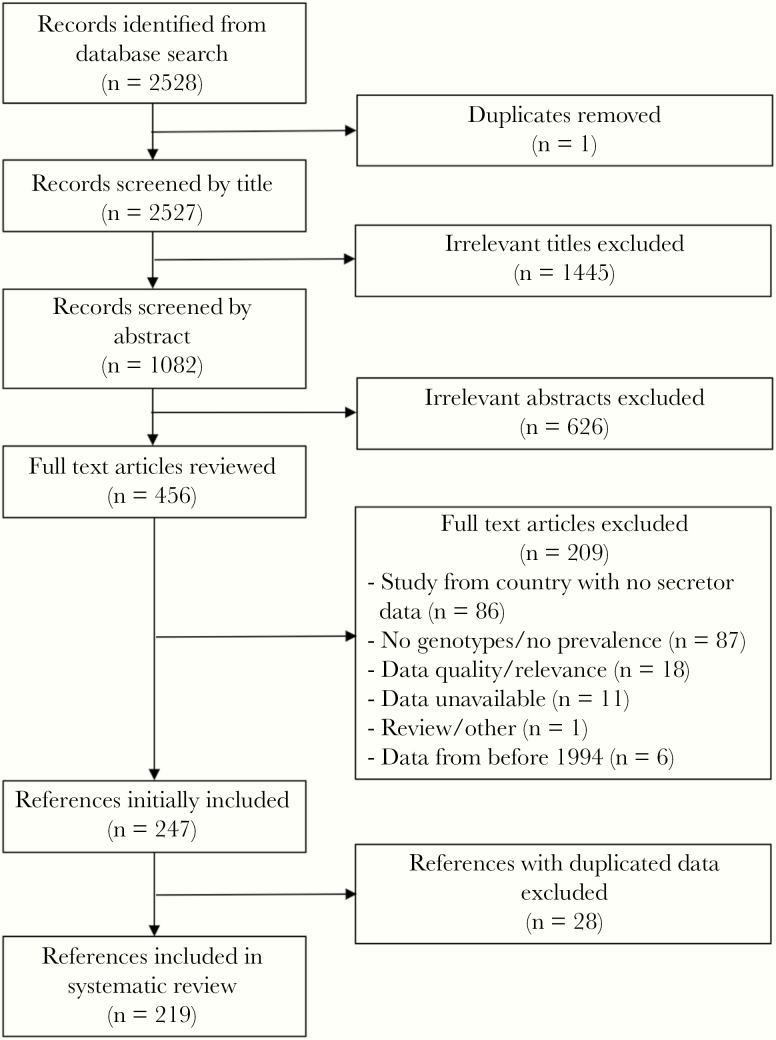
Our literature search for genotype data identified a total of 2528 references, 1 of which was duplicated. After screening titles and abstracts, 456 full text articles with potentially relevant information were reviewed. Data from 248 articles were initially included. After screening for duplicate data, 219 articles representing 38 countries were included (Figure 1). Scotland was included separately from the United Kingdom because we had a secretor proportion estimate and genotype data specifically for that Scotland. Secretor status data from 112 studies from the same 38 countries were also included.
Figure 1.
Flow diagram of references included in systematic literature review of norovirus genotype studies.
After extracting genotype data stratified by time period, typing region, or setting, 411 observations were included in the final dataset (Table 1). Most studies used reverse-transcription polymerase chain reaction detection (384, 93%), and approximately half used ORF2 to characterize noroviruses (203, 49%). Observation sample sizes (number of sporadic cases or outbreaks) ranged from 1 to 3960 with a median of 45. Almost half of all observations came from Western Pacific countries with the majority of those coming from China (n = 78), Japan (n = 56), South Korea (n = 36), and Australia (n = 20). A quarter of the observations’ study periods (103 of 411) coincided with global dominance of the Sydney 2012 pandemic GII.4 variant. One hundred eighteen observations were from outbreak studies (29%), 243 (59%) were from sporadic case studies, and the rest were mixed or unspecified (12%). Most included data came from countries with a very high HDI according to the United Nations Development Programme (292 of 411, 71%).
Table 1.
Characteristics of Studies Included in Final Analysis and Unweighted Median Proportions of Norovirus Genotype Among All Noroviruses by Characteristics
| Characteristics | N | Median GII.4 Proportion (IQR) | Median GII.non-4 Proportion (IQR)a | Median GI Proportion (IQR)a |
|---|---|---|---|---|
| Total | 411 | 0.64 (0.38–0.80) | 0.25 (0.11–0.48) | 0.03 (0.00–0.11) |
| Secretor Proportion Quartiles | ||||
| Quartile 1 (0.49–0.72) | 119 | 0.58 (0.36–0.79) | 0.30 (0.14–0.51) | 0.01 (0.00–0.11) |
| Quartile 2 (0.72–0.77) | 96 | 0.67 (0.48–0.78) | 0.23 (0.11–0.35) | 0.04 (0.02–0.15) |
| Quartile 3 (0.77–0.80) | 105 | 0.67 (0.40–0.81) | 0.26 (0.12–0.48) | 0.02 (0.00–0.10) |
| Quartile 4 (0.80–0.94) | 91 | 0.67 (0.31–0.88) | 0.25 (0.10–0.56) | 0.02 (0.00–0.12) |
| Study Type | ||||
| Outbreaks | 118 | 0.66 (0.33–0.84) | 0.21 (0.08–0.44) | 0.06 (0.00–0.16) |
| Inpatient Cases | 76 | 0.66 (0.45–0.81) | 0.26 (0.14–0.54) | 0.00 (0.00–0.05) |
| Outpatient Cases | 48 | 0.70 (0.51–0.85) | 0.27 (0.12–0.47) | 0.00 (0.00–0.09) |
| Inpatient and Outpatient Cases | 80 | 0.64 (0.43–0.75) | 0.27 (0.14–0.46) | 0.04 (0.00–0.11) |
| Community Cases | 39 | 0.57 (0.12–0.80) | 0.32 (0.15–0.51) | 0.01 (0.00–0.13) |
| Mixed Setting | 39 | 0.56 (0.33–0.73) | 0.28 (0.14–0.57) | 0.06 (0.00–0.15) |
| Not Specified | 11 | 0.77 (0.55–0.85) | 0.21 (0.19–0.32) | 0.03 (0.00–0.07) |
| Age Category | ||||
| ≤5 Years | 105 | 0.57 (0.37–0.75) | 0.28 (0.18–0.54) | 0.00 (0.00–0.11) |
| >5 Years | 15 | 0.79 (0.50–0.87) | 0.10 (0.05–0.26) | 0.08 (0.03–0.19) |
| Mixed | 291 | 0.67 (0.38–0.83) | 0.25 (0.10–0.48) | 0.03 (0.00–0.11) |
| Pandemic Variant Period | ||||
| 1994–2001 (Lordsdale and US95/96) | 52 | 0.49 (0.14–0.67) | 0.35 (0.20–0.69) | 0.08 (0.00–0.25) |
| 2002–2003 (Farmington Hills) | 20 | 0.85 (0.67–0.90) | 0.10 (0.00–0.19) | 0.03 (0.02–0.25) |
| 2004–2005 (Hunter) | 28 | 0.72 (0.52–0.88) | 0.16 (0.06–0.26) | 0.01 (0.00–0.13) |
| 2006–2008 (Yerseke and Den Haag) | 69 | 0.78 (0.59–0.89) | 0.14 (0.05–0.28) | 0.02 (0.00–0.06) |
| 2009–2011 (New Orleans) | 56 | 0.67 (0.49–0.82) | 0.29 (0.13–0.44) | 0.01 (0.00–0.06) |
| 2012–2017 (Sydney) | 103 | 0.59 (0.25–0.73) | 0.32 (0.18–0.58) | 0.03 (0.00–0.11) |
| Multiple Periodsb | 83 | 0.59 (0.35–0.73) | 0.30 (0.18–0.50) | 0.04 (0.00–0.13) |
| WHO Region | ||||
| Africa | 20 | 0.53 (0.46–0.65) | 0.28 (0.00–0.45) | 0.12 (0.00–0.33) |
| Americas | 79 | 0.67 (0.31–0.84) | 0.24 (0.08–0.50) | 0.02 (0.00–0.12) |
| Southeast Asia | 19 | 0.44 (0.25–0.75) | 0.43 (0.25–0.62) | 0.00 (0.00–0.13) |
| Europe | 83 | 0.72 (0.47–0.85) | 0.16 (0.10–0.33) | 0.04 (0.00–0.11) |
| Eastern Mediterranean | 14 | 0.33 (0.15–0.70) | 0.53 (0.17–0.72) | 0.11 (0.00–0.17) |
| Western Pacific | 196 | 0.64 (0.44–0.78) | 0.28 (0.17–0.48) | 0.02 (0.00–0.10) |
| UNDP Human Development Index Category | ||||
| Very High Development | 292 | 0.68 (0.48–0.82) | 0.23 (0.11–0.40) | 0.03 (0.00–0.11) |
| High Development | 73 | 0.42 (0.12–0.78) | 0.41 (0.11–0.87) | 0.00 (0.00–0.11) |
| Medium Development | 32 | 0.49 (0.24–0.67) | 0.43 (0.21–0.60) | 0.07 (0.00–0.15) |
| Low Development | 14 | 0.50 (0.29–0.67) | 0.25 (0.00–0.34) | 0.19 (0.00–0.38) |
| Detection Method | ||||
| RT-PCR | 384 | 0.64 (0.38–0.80) | 0.26 (0.11–0.48) | 0.03 (0.00–0.11) |
| Other | 25 | 0.67 (0.36–0.80) | 0.21 (0.06–0.49) | 0.00 (0.00–0.12) |
| Unknown | 2 | 0.76 (0.72–0.80) | 0.13 (0.10–0.11) | 0.10 (0.10–0.11) |
| Typing Region | ||||
| ORF 1 | 90 | 0.70 (0.38–0.87) | 0.19 (0.08–0.38) | 0.03 (0.00–0.13) |
| ORF 2 | 203 | 0.63 (0.41–0.79) | 0.28 (0.13–0.49) | 0.03 (0.00–0.11) |
| ORF 1 and ORF 2 | 89 | 0.57 (0.33–0.75) | 0.33 (0.16–0.56) | 0.02 (0.00–0.12) |
| ORF 1/ORF 2 Overlap Only | 16 | 0.68 (0.49–0.83) | 0.23 (0.12–0.48) | 0.00 (0.00–0.12) |
| Unknown | 13 | 0.60 (0.51–0.89) | 0.15 (0.07–0.28) | 0.04 (0.00–0.11) |
Abbreviations: GII.4, genogroup 2 type 4; IQR, interquartile range; ORF, open reading frame; RT-PCR, reverse-transcription polymerase chain reaction; UNDP, United Nations Development Programme; WHO, World Health Organization.
aPrevalence of GI and GII.non-4 were only calculated for 349 observations.
bObservations that span multiple pandemic variant periods.
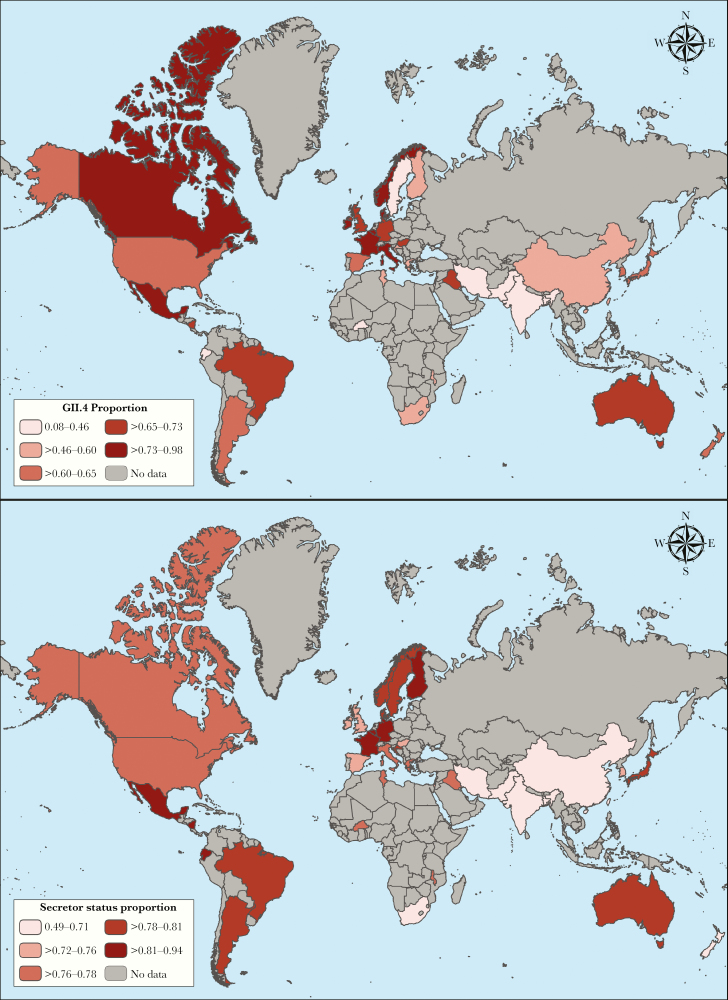
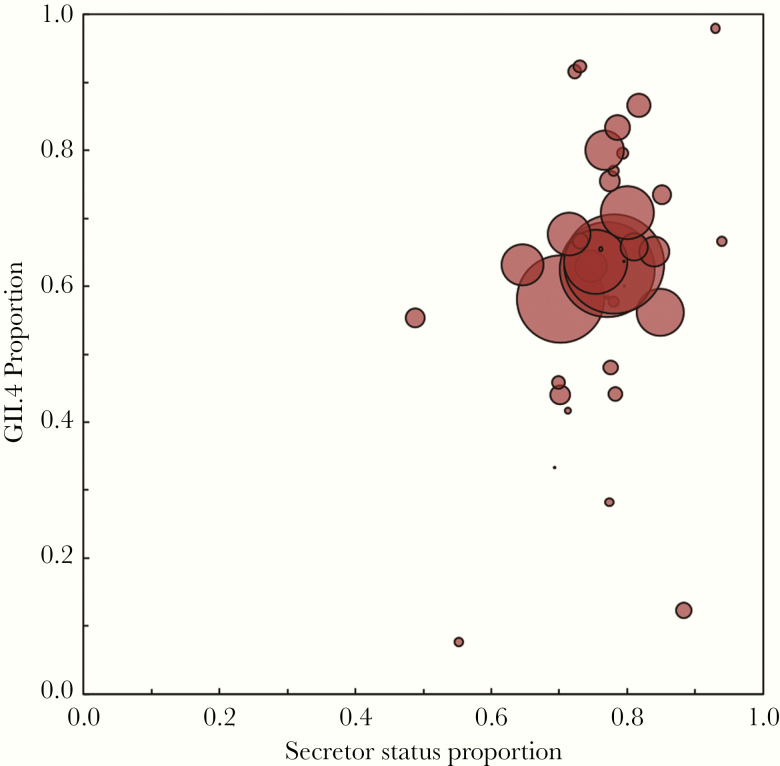
Overall median secretor prevalence was 77% (IQR, 72%–80%). Unweighted median GII.4 proportions were highest in outpatient sporadic case studies (70%; IQR, 51%–85%), studies with subjects over 5 years of age (79%; IQR, 50%–87%), and studies that coincided with the 2002–2003 period (85%; IQR, 67%–90%). The WHO European region and countries with a very high HDI had the highest unweighted median GII.4 prevalence (Table 1 and Figure 3A). Secretor proportion was highest in the Americas and parts of Europe, whereas lower secretor proportions were more common in Southeast Asia (Figure 3B). Country-level aggregated GII.4 proportions tended to be higher among countries with higher secretor proportions with some visual outliers that tended to occur in countries with less data (Figure 3 and Figure 4). Unweighted median GII.4 proportions were the same for countries with secretor proportions in quartiles 2, 3, and 4 (Table 1); however, these unweighted proportions do not account for study size.
Figure 3.
Distribution of aggregated genogroup 2 type 4 (GII.4) norovirus proportion among all noroviruses (A) and aggregated secretor proportion (B) for all countries included in the analysis. Darker colors represent higher aggregated GII.4 proportions (A) and higher aggregated secretor proportions (B), respectively.
Figure 4.
Scatterplot of aggregated country-level genogroup 2 type 4 (GII.4) proportion among all noroviruses and country-level secretor proportion. Aggregate secretor proportion tended to increase as GII.4 proportion increased. Marker size is proportional to total country genotype sample size with countries with more data represented by larger markers.
Linear Regression
In simple regression, country-level GII.4 proportion was associated with a 1.17% increase for every 1% increase in secretor prevalence (β = 1.17; 95% CI, 0.69–1.64) (Table 2 and Figure 3). The HDI was also positively associated with GII.4 proportion (β = 1.01; 95% CI, 0.71–1.31). Categorical variables that varied significantly with GII.4 proportion were WHO region, age category, typing region, and pandemic variant period, with the latter variable able to explain the most variance (R2 = 0.24).
In multivariable regression, for every percentage increase in population secretor prevalence, there was a 0.69% increase in GII.4 proportion (95% CI, 0.19%–1.18%), controlling for HDI (Table 3). We observed similar model results when excluding 100% or 0% GII.4 proportion values (β = 0.65; 95% CI, 0.16–1.13). Using a GEE model controlling for HDI and accounting for country-level clustering, we observed a stronger, but somewhat less precise, estimate of the association between secretor and GII.4 (β = 0.84; 95% CI, −0.04 to 1.74).
Table 3.
Inverse Variance Weighted MLR Model Estimating the Association Between Secretor and GII.4 Proportion, Controlling for HDI (Model 1), and GEE Accounting for Country-Level Clustering and Controlling for HDI (Model 2)
| Variable | Model 1: MLR | Model 2: GEE | ||
|---|---|---|---|---|
| β | 95% CI | β | 95% CI | |
| Intercept | −0.60 | −0.98 to −0.21 | −0.82 | −1.43 to −0.22 |
| Secretor proportion | 0.69 | 0.19 to 1.18 | 0.84 | −0.04 to 1.71 |
| HDI | 0.85 | 0.52 to 1.17 | 1.00 | 0.44 to 1.55 |
Abbreviations: CI, confidence interval; GEE, generalized estimating equation; GII.4, genogroup 2 type 4; HDI, Human Development Index; MLR, multiple linear regression.
Discussion
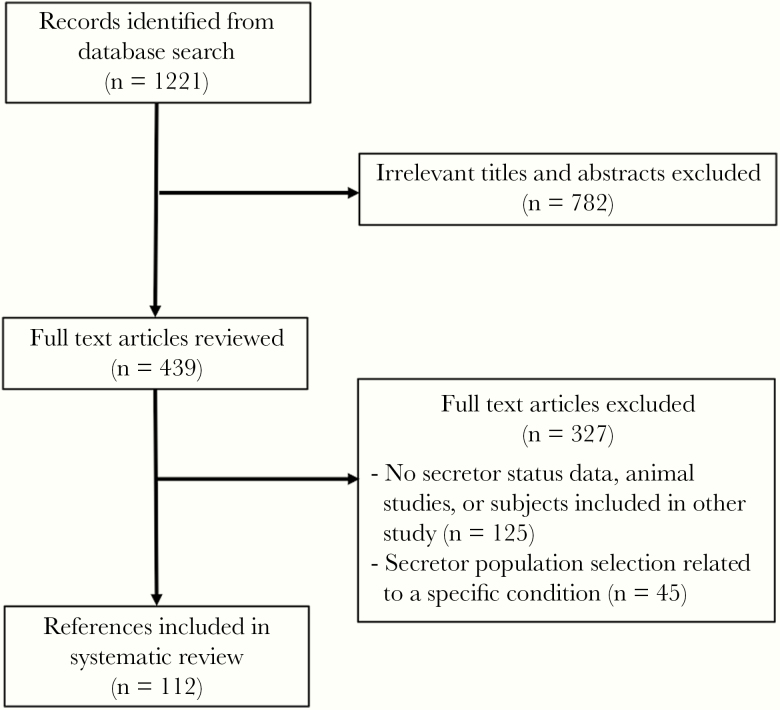
We identified a positive relationship between country-level secretor status prevalence and the proportion of norovirus cases and outbreaks caused by GII.4. This finding is consistent with our stated hypothesis that a higher proportion of secretors in a human population will result in higher proportion of GII.4 norovirus. Our study made use of a large body of literature and included all available published genotype data from countries with secretor data published at the time of the review. This dataset spanned a long period of GII.4 dominance since its global emergence during the mid-1990s [21]. The 38 included countries were geographically diverse, with every continent represented. Multivariable methods allowed us to control for HDI that may have confounded the relationship between secretor prevalence and GII.4 proportion. We were concerned that higher income countries tended to have populations with more secretor individuals (Figure 2B) and are also more likely to have more robust outbreak surveillance. Our modeling approach aimed to independently estimate the relationship between secretor status and GII.4 proportion.
Figure 2.
Flow diagram of references included in systematic literature review of secretor studies.
Genetic resistance to GII.4 among nonsecretors at the individual level is well established [16, 22–25], but studies have not examined the relationship at the population level. Our findings illustrate that this relationship also operates at the population level and suggest that human genetic diversity drives viral genotype selection. Similar population-level relationships have been observed in several other infectious diseases including cholera and malaria for which there are host genetic determinants of disease susceptibility [26]. Individuals with blood group O are at increased risk of severe cholera compared with those with A and B blood groups. Cholera-endemic regions are associated with low blood group O prevalence due to interaction between population genetics and cholera [27]. Similarly, sickle cell heterozygosity confers resistance to malaria, and higher prevalence of sickle cell trait has been observed in areas of higher malaria prevalence [28]. We cannot be certain of the direction of the selective pressure. Norovirus exposure may select for nonsecretor phenotype through increased mortality of secretors, or the human host populations genetics may give certain norovirus genotypes a fitness advantage in a given population. Given that norovirus infection is less fatal than the examples of cholera and malaria, we suspect the latter may be more likely, but we are not aware of instances in which the viral population is determined by the human host population. Moreover, because norovirus does not confer sterilizing immunity and there is a limited degree of cross-protection, the host-virus population genetic dynamic is likely to be complex.
This analysis is subject to at least 4 limitations. First, as a secondary analysis our findings are limited by the design of the original studies. Our analysis is subject to any selection bias that generated the secretor status or genotype data, which is likely more an issue for the former. Careful selection of secretor studies likely limited selection bias among included references. Second, only 38 countries were included in the analysis due to availability of both secretor and GII.4 data. The Western Pacific region is overrepresented, therefore raising some concerns about generalizability. More data from other regions could make this analysis more representative of human and viral diversity. Third, country-level secretor proportion does not account for any within-country heterogeneity. Thus, individual studies on secretor proportion may not be representative of the entire country, especially for countries with little data or diverse populations. Finally, this analysis does not consider spatial effects. Human and viral diversity do not conform to country boundaries, but for practicality, countries were chosen as the spatial unit. These last 2 limitations would essentially result in misclassification of secretor status and would therefore bias the relationship between secretor status and GII.4 toward the null, resulting in an underestimate of the true relationship.
Conclusions
Norovirus vaccines are moving through the development pipeline [29, 30], and our findings of an association between secretor proportion and GII.4 proportion may have implications for vaccination. Nonsecretors may be less likely to respond to vaccination, as has been observed for monovalent rotavirus vaccine [31, 32]. However, just as for norovirus, nonsecretors are less likely to be infected by rotavirus, which highlights a complex relationship between host genetics, vaccination, and natural infection. Nonsector individuals may be less likely to acquire immunity from vaccination, but they are also naturally resistant to infection, at least to a degree. Finally, a remaining question is whether populations with a higher proportion of secretors have higher overall incidence of norovirus gastroenteritis. Although it is clear that nonsecretor status confers strong resistance to GII.4 infections, some studies suggest that nonsecretor individuals are more frequently infected with other genotypes, such that they experience similar overall disease incidence [16, 33]. Accordingly, GII.4 vaccines may be less effective at lowering overall norovirus disease burden in high nonsecretor populations compared with high secretor populations because they may prevent less disease in nonsecretor individuals than in secretors. In summary, we found a positive population-level relationship between secretor proportion and GII.4 norovirus predominance. Future studies should identify the public health consequences of this coevolution and implications for vaccine development.
Notes
Acknowledgments. We thank Janet Mans, Leesa Bruggink, Miao Jin, Shobha Chitambar, Yasumasa Iwatani, Aksara Thongprachum, and Ushijima Hiroshi for providing genotype data contained in figures in their published papers.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was funded by the National Institutes of Health/National Institute of General Medical Sciences (R01GM124280; to B. L.) and the Centers for Disease Control and Prevention (IPA 48195; to B. L.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Kirk MD, Pires SM, Black RE, et al. . World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015; 53:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xi JN, Graham DY, Wang KN, Estes MK. Norwalk virus genome cloning and characterization. Science 1990; 250:1580–3. [DOI] [PubMed] [Google Scholar]
- 4. Chhabra P, de Graaf M, Parra GI, et al. . Updated classification of norovirus genogroups and genotypes. J Gen Virol 2019; 100:1393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Beek J, de Graaf M, Al-Hello H, et al. . Molecular surveillance of norovirus, 2005–16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 2018; 18:545–53. [DOI] [PubMed] [Google Scholar]
- 6. Marionneau S, Ruvoën N, Le Moullac-Vaidye B, et al. . Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002; 122:1967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan M, Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol 2005; 13:285–93. [DOI] [PubMed] [Google Scholar]
- 8. Henry S, Oriol R, Samuelsson B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang 1995; 69:166–82. [DOI] [PubMed] [Google Scholar]
- 9. Kambhampati A, Payne DC, Costantini V, Lopman BA. Host genetic susceptibility to enteric viruses: a systematic review and metaanalysis. Clin Infect Dis 2016; 62:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parra GI, Bok K, Taylor R, et al. . Immunogenicity and specificity of norovirus consensus GII.4 virus-like particles in monovalent and bivalent vaccine formulations. Vaccine 2012; 30:3580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bull RA, White PA. Mechanisms of GII.4 norovirus evolution. Trends Microbiol 2011; 19:233–40. [DOI] [PubMed] [Google Scholar]
- 12. Lopman B, Zambon M, Brown DW. The evolution of norovirus, the “gastric flu”. PLoS Med 2008; 5:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindesmith LC, Donaldson EF, Lobue AD, et al. . Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med 2008; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordgren J, Sharma S, Kambhampati A, Lopman B, Svensson L. Innate resistance and susceptibility to norovirus infection. PLoS Pathog 2016; 12:e1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thorven M, Grahn A, Hedlund KO, et al. . A homozygous nonsense mutation (428G–>A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J Virol 2005; 79:15351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lopman BA, Trivedi T, Vicuña Y, et al. . Norovirus infection and disease in an Ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis 2015; 211:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rydell GE, Kindberg E, Larson G, Svensson L. Susceptibility to winter vomiting disease: a sweet matter. Rev Med Virol 2011; 21:370–82. [DOI] [PubMed] [Google Scholar]
- 18. Carlsson B, Kindberg E, Buesa J, et al. . The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 Norovirus infection. PLoS One 2009; 4:e5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. UNDP. Human development data (1990–2015) Available at: http://hdr.undp.org/en/data. Accessed 1 April 2018.
- 20. World Health Organization. WHO Regional Offices Available at: http://www.who.int/about/regions/en/. Accessed 1 April 2018.
- 21. Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J Infect Dis 1999; 179:1334–44. [DOI] [PubMed] [Google Scholar]
- 22. Currier RL, Payne DC, Staat MA, et al. . Innate susceptibility to norovirus infections influenced by FUT2 genotype in a United States pediatric population. Clin Infect Dis 2015; 60:1631–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One 2013; 8:e69557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu LT, Liu FP, Huang YC, et al. . Genetic susceptibility to norovirus GII.4 sydney strain infections in Taiwanese children. Pediatr Infect Dis J 2017; 36:353–7. [DOI] [PubMed] [Google Scholar]
- 25. Van Trang N, Vu HT, Le NT, Huang P, Jiang X, Anh DD. Association between norovirus and rotavirus infection and histo-blood group antigen types in Vietnamese children. J Clin Microbiol 2014; 52:1366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nature Reviews Genetics 2012; 13:175–88. [DOI] [PubMed] [Google Scholar]
- 27. Glass RI, Holmgren J, Haley CE, et al. . Predisposition for cholera of individuals with O blood group. Possible evolutionary significance. Am J Epidemiol 1985; 121:791–6. [DOI] [PubMed] [Google Scholar]
- 28. Elguero E, Délicat-Loembet LM, Rougeron V, et al. . Malaria continues to select for sickle cell trait in Central Africa. Proc Natl Acad Sci U S A 2015; 112:7051–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leroux-Roels G, Cramer JP, Mendelman PM, et al. . Safety and immunogenicity of different formulations of norovirus vaccine candidate in healthy adults: a randomized, controlled, double-blind clinical trial. J Infect Dis 2018; 217:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riddle MS, Walker RI. Status of vaccine research and development for norovirus. Vaccine 2016; 34:2895–9. [DOI] [PubMed] [Google Scholar]
- 31. Kazi AM, Cortese MM, Yu Y, et al. . Secretor and salivary ABO blood group antigen status predict rotavirus vaccine take in infants. J Infect Dis 2017; 215:786–9. [DOI] [PubMed] [Google Scholar]
- 32. Lee B, Dickson DM, deCamp AC, et al. . Histo-blood group antigen phenotype determines susceptibility to genotype-specific rotavirus infections and impacts measures of rotavirus vaccine efficacy. J Infect Dis 2018; 217:1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Menon VK, George S, Sarkar R, et al. . Norovirus gastroenteritis in a birth cohort in Southern India. PLoS One 2016; 11:e0157007. [DOI] [PMC free article] [PubMed] [Google Scholar]