Abstract
Bat-borne zoonotic pathogens belonging to the family Paramxyoviridae, including Nipah and Hendra viruses, and the family Filoviridae, including Ebola and Marburg viruses, can cause severe disease and high mortality rates on spillover into human populations. Surveillance efforts for henipaviruses and filoviruses have been largely restricted to the Old World; however, recent studies suggest a potentially broader distribution for henipaviruses and filoviruses than previously recognized. In the current study, we screened for henipaviruses and filoviruses in New World bats collected across 4 locations in Trinidad near the coast of Venezuela. Bat tissue samples were screened using previously established reverse-transcription polymerase chain reaction assays. Serum were screened using a multiplex immunoassay to detect antibodies reactive with the envelope glycoprotein of viruses in the genus Henipavirus and the family Filoviridae. Serum samples were also screened by means of enzyme-linked immunosorbent assay for antibodies reactive with Nipah G and F glycoproteins. Of 84 serum samples, 28 were reactive with ≥1 henipavirus glycoprotein by ≥1 serological method, and 6 serum samples were reactive against ≥1 filovirus glycoproteins. These data provide evidence of potential circulation of viruses related to the henipaviruses and filoviruses in New World bats.
Keywords: Filovirus, Henipavirus, Trinidad, Bats, Screening, Serology, Luminex, RT-PCR
Since 1994, >350 human fatalities from Hendra (HeV) or Nipah virus (NiV) disease outbreaks have been reported [1–3]. Periodic outbreaks of Ebola and Marburg virus disease caused by members of the family Filoviridae have resulted in approximately 13 700 recorded human fatalities since 1976 [4, 5]. In addition to public health concerns, henipavirus and filovirus spillover events continue to have severe economic and ecological impacts [6–9]. Bats are natural reservoirs for some paramyxoviruses (NiV, Hendra virus, Cedar virus, Menangle virus, and Achimota virus 1 and 2 ) and some filoviruses (Marburg and Bombali viruses) and are the putative reservoirs for other paramyxovirus and filovirus species [10–21]. In the context of henipaviruses, the geographic distribution outside South and Southeast Asia, Africa, and Australia has yet to be determined [2, 22]. In the context of filoviruses, the broader ecology and circulation within their respective natural reservoirs and the extent of the geographic distribution of filoviruses are still largely unknown [23].
Henipaviruses have only been isolated from pteropid bats in Southeast Asia and Australia [13–15]. However, multiple studies have presented evidence for the presence of henipaviruses in Africa [16, 22, 24–30], with full genome sequences recovered for the bat-borne Ghana henipavirus in Ghana [18]. In addition, recent serological data suggest that African henipaviruses are capable of spillover into human and husbandry animal populations, although this data has not been associated with any recorded morbidity and mortality events [24, 28, 29]. A serological study by de Araujo et al found henipavirus-like antibodies in Brazilian bats. Given the distribution of bat species in Latin America that were serologically positive for the Brazilian henipa-like virus, it is possible that these viruses are circulating in Trinidad and Tobago.
The discovery of filoviruses outside Africa, including Reston virus (RESTV) in the Philippines, Lloviu virus (LLOV) in Spain, and Měnglà, Xīlǎng, and Huángjiāo viruses in China, demonstrates the broad geographic range of filoviruses [31–34]. Serological and polymerase chain reaction (PCR) evidence for filoviruses in China, Singapore, Bangladesh, and Hungary also suggest the possibility that uncharacterized filoviruses may circulate in bat populations beyond the currently described geographic range [35–39]. Han et al [40] used published filovirus surveillance data to predict bat species which may be potential filovirus reservoirs based on behavior, life history, and ecological traits; their study predicted that several New World bats, including several bat species with populations in Trinidad and Tobago, may be potential hosts of uncharacterized filoviruses.
In 2012, bats of 6 species were captured from 4 locations in Trinidad. Malmlov et al [41] screened these bat samples and found evidence of the circulation of Tacaribe virus. We describe here the results of surveillance efforts for evidence of henipa-like and filo-like viral infection in the same sample set, because the breadth of the host range and geographic distribution are still largely unknown for these virus families.
METHODS
Ethics Statement
All field work was performed under the approval of the Ethics Committee, Faculty of Medical Sciences, The University of the West Indies (UWI), St Augustine Campus, and under a special game license from the Wildlife Section, Forestry Division, Ministry of Agriculture, Land and Fisheries, Republic of Trinidad and Tobago. All work with infectious henipaviruses and filoviruses was performed under biosafety level 4 conditions at the Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, according to standard operating protocols approved by the Institutional Biosafety Committee.
Bat Capture
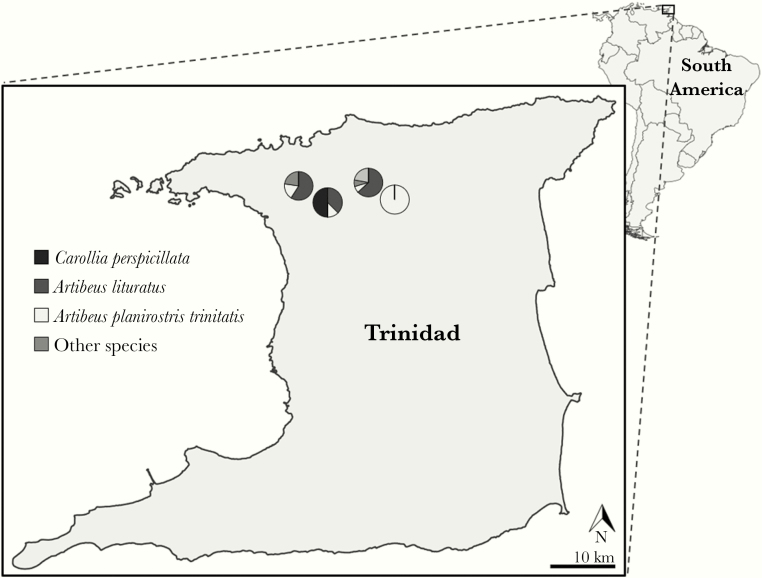
In February 2012, bats were captured with mist nets in Trinidad at 4 locations; Mount Hope (N 10.67120, W 061.28677), Lopinot (N 10.69792, W 061.32243), Santa Cruz (N 10.69596, W 061.44629), and Maracas Valley (N 10.70945, W 061.40177) (Figure 1). Cloth bags were used to individually confine and transport bats to laboratory facilities at the University of the West Indies, St Augustine for processing. Six bat species were obtained: 36 flat-faced fruit bats (Artibeus planirostris trinitatis), 31great fruit-eating bats (Artibeus lituratus), 3 Pallas’s long-tongued bat (Glossophaga soricina), 7 greater sac-winged bats (Sacropteryx bilineata), 3 little yellow-shouldered bats (Sturnira lilium), and 4 Seba’s short-tailed bats (Carollia perspicillata). Bats were euthanized through inhalation of isoflurane and exsanguination before necropsy. Tissue (lung, liver, kidney, spleen, brain, and blood) and serum samples were stored at −80° C before shipment on dry ice to Rocky Mountain Laboratories for further processing.
Figure 1.
Field sites of bat capture. Collections at Mt Hope were performed during the day at the University of West Indies. Collections at Santa Cruz, Maracas Valley, and Lopinot were performed over 3 nights.
Luminex Serology
The presence of immunoglobulins against henipavirus- and filovirus-soluble native-like oligomeric virus envelope glycoproteins was measured using a Luminex xMAP-based multiplex microsphere immunoassay (MIA) [37, 42]. Briefly, soluble tetrameric henipavirus receptor binding proteins (sGtet) (Yan et al in review) and soluble trimeric ectodomains of filovirus envelope glycoproteins were produced, as described elsewhere [37]. Purified sGtetand envelope glycoprotein antigens were coupled to Bio-Plex Pro magnetic COOH beads (Bio-Rad). Blood was collected into serum separating tubes by means of cardiac puncture with bats under deep anesthesia, and it was centrifuged at 1000g for 10 minutes before serum was collected and frozen at −80ºC. We performed the Luminex assay on serial dilutions of negative control serum samples from 14 captive-bred Rousettus aegyptiacus bats to determine an appropriate dilution for screening bat serum samples with the Luminex assay. All negative control serum samples were negative at a final dilution of 1:500. Field-collected bat serum samples were heat inactivated at 56oC for 30 minutes and diluted 1:500 before screening, and each sample was run in duplicate.
Enzyme-Linked Immunosorbent Assay
Nunc Maxisorp 96-well flat-bottom Immuno Plates (ThermoFisher) were coated with purified Nipah F and G glycoproteins (50 ng in 100 μL per well, diluted in phosphate-buffered saline [PBS]) overnight at 4°C. Plates were washed 3 times with PBS with 0.1% Tween 20 (PBS-T) and then blocked with 5% nonfat milk in PBS-T (100 μL per well) for 1 hour at room temperature. After being washed 3 times with PBS-T, diluted bat serum samples (1:100, 1:250, or 1:500 in 5% nonfat milk) were added to the wells in duplicate (100 μL) and incubated for 1 hour at room temperature. Plates were washed 5 times with PBS-T. Secondary antibody (goat anti-bat immunoglobulin G [IgG; heavy and light] horseradish peroxidase conjugate; Bethyl; 1:2500) was added to wells (100 μL) and incubated for 1 hour at room temperature. After 5 washes with PBS-T, 100 μL of a 1:1 ratio of 3,3’,5,5’-tetramethylbenzidine (TMB) solution and peroxide solution (Pierce TMB Substrate Kit; ThermoFisher) was added to wells. Plates were allowed to develop in the dark. After stopping the reaction with 100 μL of 2 mol/L sulfuric acid, plates were read at 450 nm.
In Vitro Transcription
Bombali virus and LLOV have not been isolated. Therefore, in vitro transcripts were generated as positive controls. RNA-dependent RNA polymerase coding sequence segments of Bombali virus and LLOV were synthesized into pUC57 cloning vectors (Biobasic). Plasmids were transformed into Stellar Competent Cells, following protocol PT5055-2 (Clontech). Plasmids were isolated using a PureLink HiPure Plasmid Midiprep kit (Invitrogen). Linear templates were generated by a single digestion with restriction enzyme EcoR1, according to the manufacturer’s protocol (New England Biolabs). Negative-sense RNA was transcribed using the MEGAscript T7 kit.
Nucleic Acid Extraction
RNA and DNA from Trinidad bat tissues were extracted using the Cador Pathogen 96 QIAcube HT Kit and QIAcube robot (Qiagen). The bat tissues were lysed in RLT buffer (Qiagen), followed by incubation in 95%–100% ethanol for 10 minutes before extraction. Extracted RNA from virus stocks of all currently isolated henipavirus and filovirus species were used for assay validation and positive controls. RNA was isolated using the QIAmp Viral RNA Kit (Qiagen) in a biosafety level 4 laboratory, with published modifications appropriate for virus inactivation in biosafety level 4 conditions [43]. Henipaviruses included were NiV, species Nipah henipavirus, isolate Malaysia; HeV, species Hendra henipavirus, isolate Hendra; and Cedar virus (CedV), species Cedar henipavirus, isolate Cedar. Filoviruses included were Ebola virus (EBOV), species Zaire ebolavirus, isolate Gabon; Sudan virus (SUDV), species Sudan ebolavirus, isolate Boniface; Taï Forest virus (TAFV), species Taï Forest ebolavirus, isolate Taï Forest; RESTV, species Reston ebolavirus, isolate Pennsylvania; Bundibugyo virus (BDBV), species Bundibugyo ebolavirus, isolate Bundibugyo; Marburg virus (MARV), species Marburg marburgvirus, isolate Angola; and Ravn virus (RAVV), species Marburg marburgvirus, isolate Ravn.
Henipavirus, Morbillivirus, and Respirovirus Assay
Complementary DNA (cDNA) was synthesized from 10 µL of RNA using the SuperScript III or IV First-Strand Synthesis System for reverse-transcription PCR (RT-PCR) (Invitrogen). RT-PCR was performed using TopTaq Master Mix Kit (Qiagen) 50-µL reactions, with 25 µL of TopTaq MasterMix, 5 µL of CoraLoad Dye, 1 µL of 10 µmol/L primers (final concentration 1.0 µmol/L), and 5 µL of cDNA template used for each reaction. Previously designed primers targeting a conserved region of the RNA-dependent RNA polymerase gene for henipaviruses, morbilliviruses, and respiroviruses [44] were used for PCR. Thermal cycling conditions were followed, according to the manufacturer’s protocol, with an annealing temperature of 50oC. PCR products were analyzed using a 1% agarose gel and SYBR Safe DNA Gel Stain (Fisher Scientific). The expected fragment size based on the position of the second primer set was approximately 600 base pairs.
Panfilovirus Assay
cDNA was synthesized as described above. Nested RT-PCR was performed using TopTaq Master Mix Kit (Qiagen) 50-µL reactions, including 25 µL of TopTaq MasterMix, 5 µL of CoraLoad Dye, 1 µL of 10 µmol/L primers (final concentration 0.2 µmol/L), and 5 µL of cDNA template for each reaction. Previously designed primers targeting a conserved region of the filovirus RNA-dependent RNA polymerase gene [19] was used for nested PCR, with the addition of a modified forward primer for the second reaction (5’-TYTCHVT/ideoxyI/CAAAA/ideoxyI/CAYTGGGG-3’). Thermal cycling conditions for both rounds were as follows: 94oC for 5 minutes; 15 cycles of 94oC, 60.9oC (−1oC/cycle), and 72oC for 1 minute each; 15 cycles of 94oC, 45.9 oC, and 72oC for 1 minute each; and a final extension at 72 oC for 7 minutes. PCR products were analyzed using a 1% agarose gel and GelRed Nucleic Acid Stain (Phenix Research Products) or SYBR Safe DNA Gel Stain (Fisher Scientific). The expected fragment size based on the position of the second primer set was approximately 680 base pairs.
RT-PCR Limit of Detection
The genome copy number from the respective henipavirus and filovirus controls was determined using a 1-step protocol for Droplet Digital PCR (ddPCR) and the Automated Droplet Generator (Bio-Rad), according to the manufacturer’s instructions. Eight representative filoviruses (Supplementary Figure 1) and 3 representative henipaviruses (NiV, HeV, and CedV) were used to determine the limit of detection (LOD) for the RT-PCR assay with ddPCR before bat screening. Primers and probes used are listed in Supplementary Table 1. The LOD was determined by means of serial 10-fold dilution of viral RNA–positive controls and further refined with serial 2-fold dilution. The LOD was determined based on the highest dilution from which an observable PCR product was obtained.
RESULTS
Serum samples from 84 Trinidad bats were screened with MIA for the presence of antibodies reactive to henipavirus or filovirus envelope glycoproteins. The median fluorescence intensity (MFI) cutoff value was set as 3 times the mean MFIs of a naive serum sample from a captive Egyptian fruit bat (R. aegyptiacus). The percentage of bat serum samples reactive against henipavirus- or filovirus-soluble glycoproteins was 3.57% (3 of 84) and 7.14% (6 of 84), respectively. Six serum samples from A. lituratus bats were reactive against the soluble glycoproteins of RAVV, SUDV, RESTVp (pig isolate), RESTVm (primate isolate), EBOV, NiV, GhV, or CedV (Table 1). Serum samples from 1 flat-faced fruit bat (A. planirostris trinitatis) and 1 greater sac-winged bat (S. bilineata) were reactive against RAVV-soluble glycoprotein (Table 1). The highest MFI value relative to negative control was from an A. lituratus bat (bat no. 41) against SUDV-soluble glycoprotein (Table 1). Serological reactivity was observed in sample 41 between SUDV, RESTVp, RESTVm, NiV, and GhV and in sample 64 between SUDV and EBOV (Table 1).
Table 1.
Samples Seropositive Against Henipavirus or Filovirus Glycoproteins With Enzyme-Linked Immunosorbent and Multiplex Luminex Serological Assaysa
| ELISA: NiV | Luminex Multiplex Assay | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bat No. | Sampling Site | Species | G | F | NiV (G) | HeV (G) | GhV (G) | CeV (G) | TAFV (GP) | SUDV (GP) | RAVV (GP) | EBOV (GP) | MARV (GP) | LLOV (GP) | RESTVp (GP) | RESTVm (GP) | BDBV (GP) |
| 12 | UWI Chapel | Artibeus planirostris trinitatis | … | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 14 | UWI Chapel | A. planirostris trinitatis | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 15 | UWI Chapel | A. planirostris trinitatis | … | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 17 | UWI Chapel | A. planirostris trinitatis | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 18 | UWI Chapel | A. planirostris trinitatis | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 20 | UWI Chapel | A. planirostris trinitatis | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 21 | UWI Chapel | A. planirostris trinitatis | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 25 | UWI Chapel | A. planirostris trinitatis | … | … | … | … | … | … | … | … | 10.6 | … | … | … | … | … | … |
| 30 | Lopinot | A. lituratus | 1:1000 | 1:500 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 33 | Lopinot | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 34 | Lopinot | A. lituratus | 1:100 | … | … | … | … | … | … | … | 538.1 | … | … | … | … | … | … |
| 35 | Lopinot | A. lituratus | 1:100 | … | … | … | … | 183.6 | … | … | … | … | … | … | … | … | … |
| 37 | Lopinot | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 38 | Lopinot | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 41 | Lopinot | A. lituratus | 1:100 | 1:100 | 57.3 | … | 319.5 | … | … | 2719.9 | … | … | … | … | 1305.6 | 175.0 | … |
| 42 | Lopinot | Glossophaga soricina | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 48 | Lopinot | G. soricina | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 53 | Lopinot | A. lituratus | 1:100 | … | 57.0 | … | … | … | … | … | … | … | … | … | … | ||
| 55 | Santa Cruz | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 56 | Santa Cruz | A. lituratus | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 58 | Santa Cruz | Sacropteryx bilineata | … | … | … | … | … | … | … | … | 33.8 | … | … | … | … | … | … |
| 59 | Santa Cruz | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 63 | Santa Cruz | A. planirostris trinitatis | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 64 | Santa Cruz | A. lituratus | 1:100 | 1:100 | … | … | … | … | … | 306.9 | … | 20.3 | … | … | … | … | … |
| 65 | Santa Cruz | A. lituratus | … | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 67 | Santa Cruz | S. bilineata | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 75 | Santa Cruz | A. lituratus | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 77 | Maracas Valley | Carollia perspicillata | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 78 | Maracas Valley | C. perspicillata | 1:100 | 1:100 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 81 | Maracas Valley | A. lituratus | 1:250 | 1:250 | … | … | … | … | … | … | … | 626.3 | … | … | … | … | … |
Only positive reactivity with the specific assay and glycoprotein is displayed.
Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; ELISA, enzyme-linked immunosorbent assay; F, fusion glycoprotein; G, attachment glycoprotein; GhV, Ghana henipavirus; G/GP, glycoprotein; HeV, Hendra virus; LLOV, Lloviu virus; MARV, Marburg virus; NiV, Nipah virus; RAVV, Ravn virus; RESTVm, primate isolate; RESTVp, pig isolate; SUDV, Sudan virus; TAFV, Taï Forest virus; UWI, University of West Indies.
aELISA results are reported as the highest dilution for which each sample was seropositive. We report the mean fluorescence intensity (MFI) of the multiplex Luminex assay after subtracting the value of 3 standard deviations plus the mean MFI of the naive serum sample for each antigen.
Serum samples were also screened by enzyme-linked immunosorbent assay (ELISA) for the presence of antibodies reactive to Nipah F and G glycoproteins. The MFI cutoff value was set as 3 times the standard deviation of the average MFI of naive bat serum from a captive Egyptian fruit bat. The proportions of bat serum samples reactive against Nipah G and F at 1:100 dilution were 29.76% (25 of 84) and 19.05% (16 of 84), respectively (Table 1). Only 2 samples were reactive against Nipah G and F at dilutions of 1:250 or greater. Twelve samples were reactive against Nipah G, but not Nipah F, and 3 were reactive against Nipah F but not Nipah G. All samples that showed reactivity with MIA were reactive to Nipah G at ELISA. However, only 1 sample (bat 41) was reactive to both Nipah G and F at ELISA and Nipah G at MIA.
Previously established panviral RT-PCR assays for high-throughput screening of biologically derived samples were used to detect respirovirus, morbillivirus, henipaivirus, and filovirus RNA [19]. The panfilovirus assay was modified by incorporating sequence information for recently identified filoviruses and validated for specificity and sensitivity. Eight representative filoviruses (Supplementary Figure 1) and 3 representative henipaviruses were used to determine the LOD for the assays by means of ddPCR before bat screening. The average LOD for the representative henipaviruses and filoviruses was 3.2 and 1.5 copies/µL, respectively (Supplementary Table 2). The L gene segment of LLOV generated product only at starting concentrations >1000 copies/μL and was considered an outlier for the LOD. Tissue samples from 78 Trinidad bats were screened for respiroviruses, morbilliviruses, henipaviruses, and filoviruses by means of RT-PCR. Tissues screened were lung, liver, kidney, spleen, and brain. No henipavirus or filovirus RNA was detected in this sample set.
Discussion
Worldwide virus discovery and surveillance efforts have led to the identification of a variety novel European, African and Chinese henipaviruses and filoviruses [16, 19, 21, 22, 24, 33, 36, 45]. In addition, they have identified potential henipavirus circulation in Latin America [46]. The zoonotic and cross-species spillover potential of these novel viruses is currently unknown. However, these discoveries highlight the importance of virus discovery and surveillance efforts for novel henipavirus and filovirus species given their potential public health, economic, and ecological impacts. Therefore, expanding surveillance efforts beyond the known geographic distributions of henipaviruses and filoviruses may shed further light on the ecology and evolutionary history of these important viruses.
In the current study, we screened phyllostomid and emballonurid bat serum and tissue samples from Trinidad for henipaviruses and filoviruses. Eight of 84 bat serum samples were positive at Luminex serology and reacted to ≥1 of the henipavirus or filovirus glycoproteins. Twenty-eight samples were positive for Nipah G, F, or both at ELISA. Of note, the 3 bat species (A. lituratus, A. planirostris trinitatis, and C. perspicillata) positive for a henipavirus-like antibodies at MIA or ELISA are 3 of the 6 species that were positive for henipavirus-like antibodies in a Brazilian study [46]. One bat species sampled in this study, A. lituratus, which was found to have antibodies reactive against filovirus-soluble glycoproteins, was among those predicted to be potential hosts of novel filoviruses based on a study by Han et al [40]. Several bats from this species showed reactivity to both filovirus and henipavirus antigens (including an individual with antibodies against both), a phenomenon also observed in pteropodid bats [24, 47, 48].
The serological IgG reactivity observed in our study is likely due to the circulation of viruses that have surface glycoproteins antigenically related to henipavirus and filovirus glycoproteins used in our assays. Similar serological cross-reactivity has been observed in a study of Rousettus bats experimentally challenged with filoviruses [49]. We found some discordance between the serological results of the ELISA against Nipah G and the multiplex Luminex assay that includes Nipah G, specifically that more samples were positive against Nipah G with the ELISA than with the Luminex (Table 1). Most of the samples showing some reactivity against Nipah G with ELISA but not the Luminex assay were not seropositive at dilutions above 1:100, with 2 exceptions in which A. lituratus bats (bats 30 and 81 [Table 1]) were seropositive by ELISA for both Nipah G and F at dilutions >1:100.
The multiplex nature of the Luminex assay complicates interpretation of the serological results, because we detected reactivity against the glycoproteins of unrelated viruses, including filovirus, SUDV, and GhV in serum collected from an A. lituratus bat (bat 41 [Table 1]). The specific history of viral exposure is inherently unknown in field-collected samples, and polyclonal serum samples are frequently cross-reactive,; therefore, the conclusions that we can draw from these data are limited. Further efforts to characterize the viral diversity circulating in South American bats are needed to refine these serological assays and allow for the development of specific target antigens.
Although we improved on the sensitivity and specificity of a previously established panfilovirus RT-PCR assay [50–53], we detected no henipavirus or filovirus viral RNA. This is not surprising, given our sample sizes and the comparatively low detection rate of virus shedding compared to that of IgG antibodies against these viruses observed in naturally infected bats in field studies and experimentally infected bats in laboratory studies [54–57]. The geographic distributions of several bat species sampled in our study extend as far as Brazil, where bat serum samples were found to be positive for exposure to henipa-like viruses, with ELISA and immunofluorescence assay [46], suggesting the possibility of widespread circulation of henipa-like viruses in Central and South America. Here we provide evidence for the potential circulation of henipa-like and filo-like viruses in Trinidad. No viral RNA was detected in this set of bat samples using RT-PCR. However, 35.7% of the samples were serologically positive. A primary limitation of our study is the low sample size; prior surveillance studies have found antibody-positive and PCR-positive prevalences for filoviruses as low as 1.7% and 1.9% respectively [11, 58]. Taken together, our findings provide evidence of more widespread geographic distribution of henipaviruses and filoviruses than previously appreciated.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank Friederike Feldmann for preparing the stocks of the respective filoviruses used in this study.
Financial support. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R15AI089419) and the Defense Threat Reduction Agency, Department of Defense (grant HDTRA1-17-10037).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed..
References
- 1. Centers for Disease Control and Prevention. Hendra virus disease (HeV). 28 June 2019. https://www.cdc.gov/vhf/hendra/index.html.
- 2. Centers for Disease Control and Prevention. Nipah virus (NiV). 28 June 2019. https://www.cdc.gov/vhf/nipah/index.html.
- 3. World Health Organization. Nipah virus outbreaks in the WHO South-East Asia Region. 28 June 2019. http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus_outbreaks_sear/en/.
- 4. Centers for Disease Control and Prevention. Ebola (Ebola virus disease). 14 January 2019. https://www.cdc.gov/vhf/ebola/index.html.
- 5. Centers for Disease Control and Prevention. Marburg hemorrhagic fever (Marburg HF). 14 January 2019. https://www.cdc.gov/vhf/marburg/index.html.
- 6. World Bank. 2014–2015 West Africa Ebola crisis: impact update. 14 January 2019. http://www.worldbank.org/en/topic/macroeconomics/publication/2014-2015-west-africa-ebola-crisis-impact-update.
- 7. Walsh PD, Abernethy KA, Bermejo M, et al. . Catastrophic ape decline in western equatorial Africa. Nature 2003; 422:611–4. [DOI] [PubMed] [Google Scholar]
- 8. Looi LM, Chua KB. Lessons from the Nipah virus outbreak in Malaysia. Malays J Pathol 2007; 29:63–7. [PubMed] [Google Scholar]
- 9. Degeling C, Gilbert GL, Annand E, et al. . Managing the risk of Hendra virus spillover in Australia using ecological approaches: a report on three community juries. PLoS One 2018; 13:e0209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Towner JS, Amman BR, Sealy TK, et al. . Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog 2009; 5:e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leroy EM, Kumulungui B, Pourrut X, et al. . Fruit bats as reservoirs of Ebola virus. Nature 2005; 438:575–6. [DOI] [PubMed] [Google Scholar]
- 12. Leendertz SA, Gogarten JF, Düx A, Calvignac-Spencer S, Leendertz FH. Assessing the evidence supporting fruit bats as the primary reservoirs for Ebola viruses. Ecohealth 2016; 13:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chua KB, Koh CL, Hooi PS, et al. . Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect 2002; 4:145–51. [DOI] [PubMed] [Google Scholar]
- 14. Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol 2000; 81:1927–32. [DOI] [PubMed] [Google Scholar]
- 15. Marsh GA, de Jong C, Barr JA, et al. . Cedar virus: a novel henipavirus isolated from Australian bats. PLoS Pathog 2012; 8:e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drexler JF, Corman VM, Müller MA, et al. . Bats host major mammalian paramyxoviruses. Nat Commun 2012; 3:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barr JA, Smith C, Marsh GA, Field H, Wang LF. Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. J Gen Virol 2012; 93:2590–4. [DOI] [PubMed] [Google Scholar]
- 18. Baker KS, Todd S, Marsh GA, et al. . Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum. J Virol 2013; 87:1348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldstein T, Anthony SJ, Gbakima A, et al. . The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 2018; 3:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forbes KM, Webala PW, Jaaskelainen AJ, et al. . Bombali virus in Mops condylurus bat, Kenya. Emerg Infect Dis 2019;25:955–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karan LS, Makenov MT, Korneev MG, et al. . Bombali virus in Mops condylurus bats, Guinea. Emerg Infect Dis 2019;25:1774–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drexler JF, Corman VM, Gloza-Rausch F, et al. . Henipavirus RNA in African bats. PLoS One 2009;4:e6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olival KJ, Hayman DT. Filoviruses in bats: current knowledge and future directions. Viruses 2014; 6:1759–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayman DT, Suu-Ire R, Breed AC, et al. . Evidence of henipavirus infection in West African fruit bats. PLoS One 2008;3:e2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weiss S, Nowak K, Fahr J, et al. . Henipavirus-related sequences in fruit bat bushmeat, Republic of Congo. Emerg Infect Dis 2012; 18:1536–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peel AJ, Baker KS, Crameri G, et al. . Henipavirus neutralising antibodies in an isolated island population of African fruit bats. PLoS One 2012;7:e30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muleya W, Sasaki M, Orba Y, et al. . Molecular epidemiology of paramyxoviruses in frugivorous Eidolon helvum bats in Zambia. J Vet Med Sci 2014;76:611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pernet O, Schneider BS, Beaty SM, et al. . Evidence for henipavirus spillover into human populations in Africa. Nat Commun 2014; 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atherstone C, Diederich S, Weingartl HM, et al. . Evidence of exposure to henipaviruses in domestic pigs in Uganda. Transbound Emerg Dis 2019;66:921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iehlé C, Razafitrimo G, Razainirina J, et al. . Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg Infect Dis 2007; 13:159–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jahrling PB, Geisbert TW, Dalgard DW, et al. . Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 1990; 335:502–5. [DOI] [PubMed] [Google Scholar]
- 32. Negredo A, Palacios G, Vázquez-Morón S, et al. . Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 2011; 7:e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang XL, Tan CW, Anderson DE, et al. . Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nature Microbiology 2019; 4:390–395. [DOI] [PubMed] [Google Scholar]
- 34. Shi M, Lin XD, Chen X, et al. . The evolutionary history of vertebrate RNA viruses. Nature 2018; 556:197–202. [DOI] [PubMed] [Google Scholar]
- 35. He B, Feng Y, Zhang H, et al. . Filovirus RNA in fruit bats, China. Emerg Infect Dis 2015; 21:1675–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang XL, Zhang YZ, Jiang RD, et al. . Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis 2017; 23:482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laing ED, Mendenhall IH, Linster M, et al. . Serologic evidence of fruit bat exposure to filoviruses, Singapore, 2011-2016. Emerg Infect Dis 2018; 24:114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olival KJ, Islam A, Yu M, et al. . Ebola virus antibodies in fruit bats, Bangladesh. Emerg Infect Dis 2013; 19:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kemenesi G, Kurucz K, Dallos B, et al. . Re-emergence of Lloviu virus in Miniopterus schreibersii bats, Hungary, 2016. Emerg Microbes Infect 2018; 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DT, Drake JM. Undiscovered bat hosts of filoviruses. PLoS Negl Trop Dis 2016; 10:e0004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malmlov A, Seetahal J, Carrington C, et al. . Serological evidence of arenavirus circulation among fruit bats in Trinidad. PLoS One 2017; 12:e0185308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan YP, Yan L, Feng YR, Broder CC. Preparation of recombinant viral glycoproteins for novel and therapeutic antibody discovery. Methods Mol Biol 2009; 525:31–58, xiii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haddock E, Feldmann F, Feldmann H. Effective chemical inactivation of Ebola virus. Emerg Infect Dis 2016; 22:1292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol 2008; 46:2652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu Z, Yang L, Yang F, et al. . Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis 2014; 20:1064–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Araujo J, Lo MK, Tamin A, et al. . Antibodies against henipa-like viruses in Brazilian bats. Vector Borne Zoonotic Dis 2017; 17:271–4. [DOI] [PubMed] [Google Scholar]
- 47. Brook CE, Ranaivoson HC, Broder CC, et al. . Disentangling serology to elucidate henipa- and filovirus transmission in Madagascar fruit bats. J Anim Ecol 2019; 88:1001–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayman DT, Yu M, Crameri G, et al. . Ebola virus antibodies in fruit bats, Ghana, West Africa. Emerg Infect Dis 2012; 18:1207–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schuh AJ, Amman BR, Sealy TS, et al. . Comparative analysis of serologic cross-reactivity using convalescent sera from filovirus-experimentally infected fruit bats. Sci Rep 2019; 9:6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panning M, Laue T, Olschlager S, et al. . Diagnostic reverse-transcription polymerase chain reaction kit for filoviruses based on the strain collections of all European biosafety level 4 laboratories. J Infect Dis 2007; 196 (suppl 2):S199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhai J, Palacios G, Towner JS, et al. . Rapid molecular strategy for filovirus detection and characterization. J Clin Microbiol 2007; 45:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pang Z, Li A, Li J, et al. . Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses. PLoS One 2014; 9:e95635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lu G, Zhang J, Zhang C, et al. . One-step reverse-transcription FRET-PCR for differential detection of five Ebolavirus species. PLoS One 2015; 10:e0126281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Epstein JH, Baker ML, Zambrana-Torrelio C, et al. . Duration of maternal antibodies against canine distemper virus and Hendra virus in pteropid bats. PLoS One 2013; 8:e67584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Epstein JH, Prakash V, Smith CS, et al. . Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg Infect Dis 2008; 14:1309–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schuh AJ, Amman BR, Sealy TK, Spengler JR, Nichol ST, Towner JS. Egyptian rousette bats maintain long-term protective immunity against Marburg virus infection despite diminished antibody levels. Sci Rep 2017; 7:8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Halpin K, Hyatt AD, Fogarty R, et al. ; Henipavirus Ecology Research Group Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 2011; 85:946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Nys HM, Kingebeni PM, Keita AK, et al. . Survey of Ebola viruses in frugivorous and insectivorous bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015-2017. Emerg Infect Dis 2018; 24:2228–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.