Abstract
PURPOSE
For the advances of pediatric oncology next-generation sequencing (NGS) research to equitably benefit all children, a diverse and representative sample of participants is needed. However, little is known about demographic and clinical characteristics that differentiate families who decline enrollment in pediatric oncology NGS research.
METHODS
Demographic and clinical data were retrospectively extracted for 363 pediatric patients (0-21 years) with cancer approached for enrollment in Genomes for Kids (G4K), a study examining the feasibility of comprehensive clinical genomic analysis of tumors and paired normal samples. Demographic and clinical factors that significantly differentiated which families declined were subsequently compared, for 348 families, with enrollment in Clinical Implementation of Pharmacogenetics (PG4KDS), a pharmacogenomics study with more explicit therapeutic benefit examining genes affecting drug responses and metabolism.
RESULTS
Fifty-three families (14.6%) declined enrollment in G4K. Race/ethnicity was the only variable that significantly differentiated study refusal according to multivariable logistic regression, with families of black children more likely to decline enrollment compared with families of non-Hispanic or Hispanic white children. Reasons for declining G4K were generally consistent with other pediatric genomics research: feeling overwhelmed and insurance discrimination fears were most frequently cited. Families of black children were also more likely to decline enrollment in PG4KDS. Thirteen (3.7%) of the 348 families approached for both studies declined PG4KDS.
CONCLUSION
Race/ethnicity differentiated study declination across two different pediatric oncology genomics studies, suggesting enrollment disparities in the context of pediatric oncology genomics research. Genomics research participant samples that do not fully represent racial and ethnic minorities risk further exacerbating health disparities. Additional work is needed to understand the nuances of parental decision making in genomic research and facilitate enrollment of diverse patient populations.
INTRODUCTION
Precision medicine holds enormous promise for improving pediatric cancer cure rates. With implementation of next-generation sequencing (NGS) approaches, it is currently estimated that 5%-15% of children with cancer harbor an underlying predisposition.1-3 However, the benefits of genomic research may not be distributed equitably.4,5 To the extent that certain populations are underrepresented in genomic research (eg, racial minorities, patients with specific cancer types), findings may not generalize.6-8 To increase the likelihood that all children with cancer have equal opportunity to benefit from genomic advances, genomics research must include a representative population.
Prior studies have found that parents are interested in genomic testing in the context of pediatric cancer.9-11 Nonetheless, refusal rates in pediatric NGS studies range from 12%-30%.12 Research examining predictors of participation in genomic research has focused predominantly on adults.13-16 However, different reasons for declining participation have emerged in pediatric versus adult NGS studies, including more frequently cited concerns about privacy/discrimination and psychological impact.12 Moreover, NGS presents unique considerations that differ from single-gene testing, including possibly revealing predisposition for adult-onset cancers in pediatric patients, incidental and secondary findings, and more variants of uncertain significance.17 Identifying demographic and clinical factors that predict refusal to participate in pediatric oncology NGS research is critical to elucidating whether subsets of the population may be underrepresented. This can then inform interventions to decrease enrollment barriers and improve representativeness so that all patients can capitalize on the potential benefits of precision medicine.
This study examined demographic and clinical factors in relation to enrollment in Genomes for Kids (G4K), a research protocol in which children with cancer were offered comprehensive genomic analysis of tumor and germline tissues (ClinicalTrials.gov identifier: NCT02530658). We aimed to identify which demographic and clinical factors significantly differentiated families who declined from those who enrolled. As a second aim, the identified differentiating factors were subsequently examined in Clinical Implementation of Pharmacogenetics (PG4KDS), a separate clinical pharmacogenomics research study, to determine whether findings were reproducible across genomics studies with varying clinical utility. To better understand possible barriers to participation in pediatric genomics research, an exploratory aim was to qualitatively identify families’ reasons for declining.
CONTEXT
Key Objective
For children to benefit from next-generation sequencing (NGS) equitably, pediatric NGS research must be representative of the larger population. This retrospective chart review sought to identify factors related to declining to participate in a prospective study of tumor/germline NGS of pediatric patients with cancer while controlling for other potentially relevant demographic and clinical variables.
Knowledge Generated
Only patient race/ethnicity significantly related to enrollment, with families of black patients significantly more likely to decline participation. Patient race/ethnicity also significantly differentiated enrollment in a second pharmacogenomics study with more explicit therapeutic benefit for patients, suggesting that enrollment discrepancies may extend throughout pediatric genomics research.
Relevance
Findings from pediatric NGS research may not generalize to black children given underrepresentation in genomics investigations. Explicit steps should be taken to recruit a more racially diverse sample in pediatric NGS research, with efforts to facilitate enrollment of black patients in particular.
METHODS
Participants and Procedures of G4K
Data were collected retrospectively for 363 children approached for enrollment in G4K at St Jude Children’s Research Hospital (SJCRH) from August 2015-April 2017. G4K is an institutional review board–approved study incorporating NGS technologies (whole-genome sequencing, whole-exome sequencing, and RNA sequencing of tumors; whole-genome sequencing and whole-exome sequencing of paired normal samples) into the clinical care of children.18 Variant analysis was limited to genes previously associated with cancer. The study generated clinical results for tumor mutations (approximately 1,000 cancer-related genes) and/or germline reports for cancer-predisposing variants, which were placed in the medical record (unless families opted out of learning the sequencing results). Families consented to germline sequencing and disclosure of results for 63 cancer predisposition genes and to sequencing of additional genes on a research basis. Families also consented to be recontacted with updated results on the basis of new research findings, with 93 genes added during the course of the study, yielding a total of 156 cancer predisposition genes.
Eligible participants were 0-21 years with a diagnosis of liquid, non-CNS solid, or CNS solid tumor. Except for patients with retinoblastoma, craniopharyngioma, optic pathway glioma, or diffuse intrinsic pontine glioma (for which biopsy of the tumor was not performed because of concerns about morbidity), patients needed to have frozen tumor tissue available for analysis. Exclusion criteria included history of allogeneic hematopoietic stem-cell transplantation (or other condition resulting in blood DNA failing to match host tissue DNA), insufficient tumor or germline tissue, or attending physician’s objection to enrollment.
Consent conversations were conducted by a trained clinical research nurse using a 2-step consenting process. The first visit introduced the study, followed by a second visit to obtain informed consent.18 Study declination was defined as verbally declining or passively refusing to enroll in the study. Families that explicitly declined were asked their reason for declining, and the reason was documented in their consent note or study introduction note. Passive refusal was inferred from several missed or no-show appointments and/or no response from phone contact attempts (3 unsuccessful phone call attempts with voicemails without a return call).
Demographic and clinical data were obtained by medical record review. Demographic variables included patient sex, race, ethnicity, and region of the United States where the family resided, as documented in the medical record. Mother and father age at diagnosis, education level, marital status, sibling status (yes/no), and religion (Christian/other) were obtained from social work notes. Need for an interpreter referred to using an interpreter for the consent discussion. Clinical variables included patient age at diagnosis, tumor type (liquid/non-CNS solid/CNS solid tumor), new versus relapsed/refractory/secondary cancer as reported by physician(s), and time since diagnosis when approached. Reason for declination was extracted from chart review of consent notes and study introduction notes.
Participants and Procedures of PG4KDS
Enrollment in G4K was compared with enrollment in PG4KDS, a separate clinical pharmacogenomics trial at SJCRH. PG4KDS was designed to genotype patient samples and report pharmacogenetic test results in the medical record to guide pharmacotherapy as part of clinical care.19 Specifically, genomic DNA was obtained from a blood sample and genotyped on the DMET Plus array (ThermoFisher Scientific, Waltham, MA) supplemented with a CYP2D6 copy number assay. Genotyping was performed for 230 pharmacogenes.19 Patients were informed that the researchers would decide which of these gene test results would be placed in the medical record and that these gene test results would be placed in the medical record only as evidence supported their clinical use. Patients were also told that test results would be placed in the medical record across an extended period of time (as long as the participant stayed enrolled and the study was open); patients were given the option of receiving notifications each time a new test result was placed in their medical record. As of June 2019, results of 9 pharmacogenes coupled to 35 drugs have been implemented in medical records. SJCRH institutional review board approval was received in 2011.
Eligibility criteria for PG4KDS included receiving active therapy at SJCRH. Exclusion criteria included the following: previous allogeneic stem-cell transplantation (or other condition resulting in blood DNA failing to match host tissue DNA), history of liver transplantation, or attending physician’s objection to enrollment. Comparative analyses between PG4KDS and G4K include only the 348 families approached for both studies to control for different study enrollment time periods and differing study populations. Fifteen families approached for G4K were not approached for PG4KDS.
Consent conversations were conducted in person by trained clinical research nurses. Study declination was defined as verbally declining or passively refusing to enroll in the study. The study team did not solicit a reason for declination; however, if a declination reason was provided, it was documented. Passive refusal was inferred from 3 missed or no-show appointments and/or if the caregivers had not reached a decision for participation after 3 meetings.
Statistical Analysis
Descriptive statistics for patients who declined versus enrolled in G4K were compared using 2-sample t tests or Wilcoxon rank sum tests for continuous variables on the basis of the normality of the data and Fisher’s exact test for categoric variables as appropriate. G4K refusal was examined in relation to demographic (patient sex, patient race/ethnicity, caregiver age, caregiver education, caregiver marital status, siblings, religion, need for interpreter, region of country) and clinical (patient age at diagnosis, tumor type, new versus relapsed/refractory/secondary cancer diagnosis, time since diagnosis when approached) factors using univariable logistic regression models (refusal = 1, enrollment = 0). Variables for which the overall test statistic significantly predicted G4K refusal at P < .10 were assessed in a multivariate logistic regression using backward model selection and Bayesian information criteria.20
Qualitative reasons for declining G4K were categorized by trained research assistants. Categories were first created using consensus conversations between 2 trained research assistants and the first author. Reasons for declining were then coded into categories by both research assistants together using consensus conversation as needed.
Participation proportions (refusal v enrollment) were compared between G4K and PG4KDS using the McNemar test. Refusal rates were compared across studies for patients who were approached for both G4K and PG4KDS. Demographic and/or clinical variables found to significantly differentiate refusal for G4K were subsequently examined in relation to study refusal for PG4KDS using the Fisher exact test. Statistical analyses were performed using SAS, version 9.4 (SAS Institute, Cary, NC) and R, version 3.6.1 (R Core Team, Vienna, Austria) with 2-sided P values (significant if P < .05).
RESULTS
Preliminary Analyses for G4K
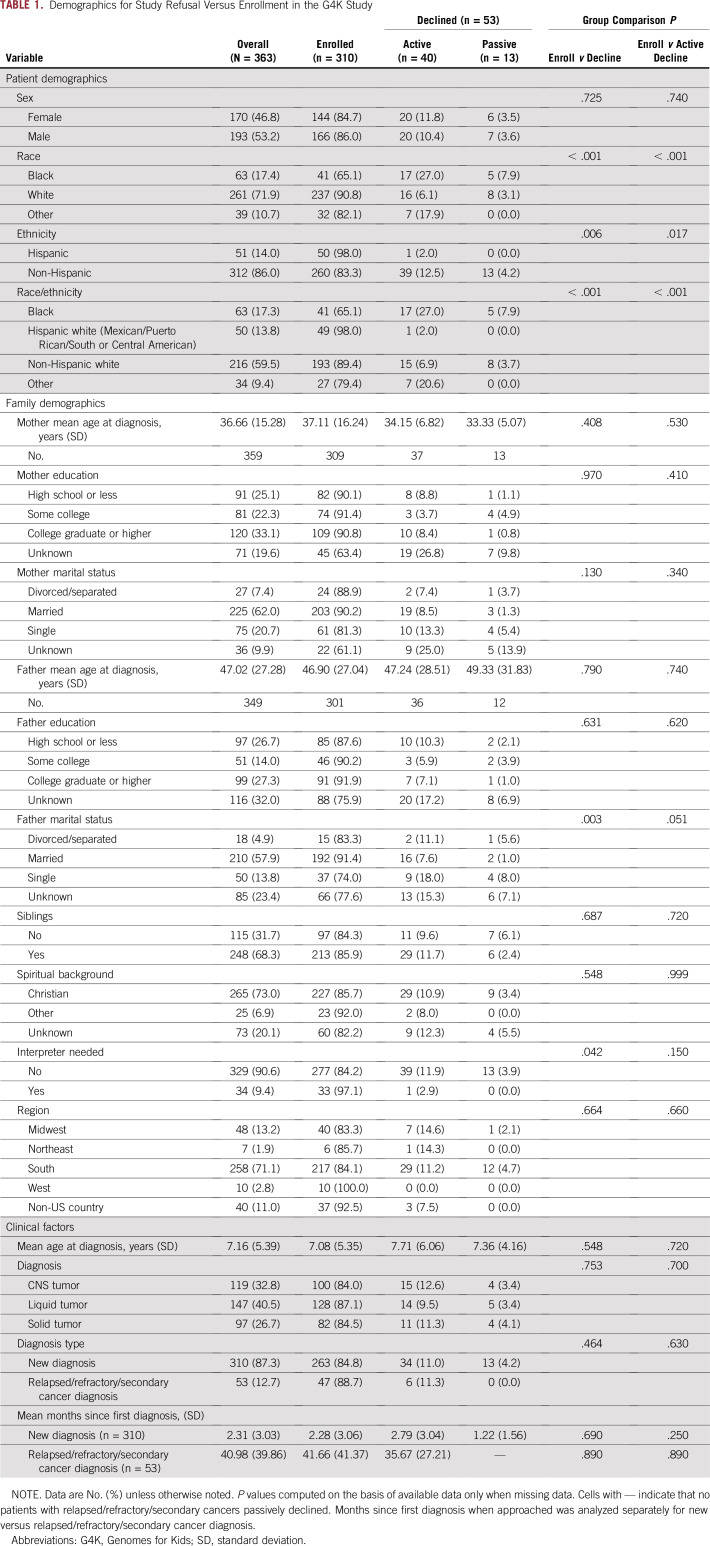
Of 363 families approached for participation in G4K, 53 (14.6%) declined enrollment (13 passively declined). Demographics are listed in Table 1 for patients who enrolled in G4K versus those who actively or passively declined. Of note, comparisons between active declination versus enrollment and overall declination versus enrollment were comparable, suggesting that passive declination was not responsible for group differences.
TABLE 1.
Demographics for Study Refusal Versus Enrollment in the G4K Study
Demographic and Clinical Variable Comparisons for G4K
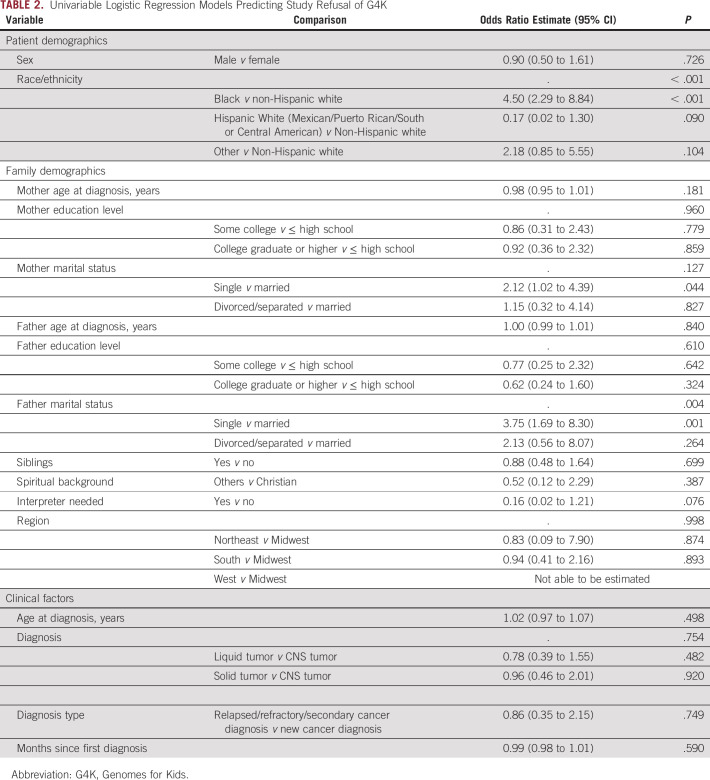
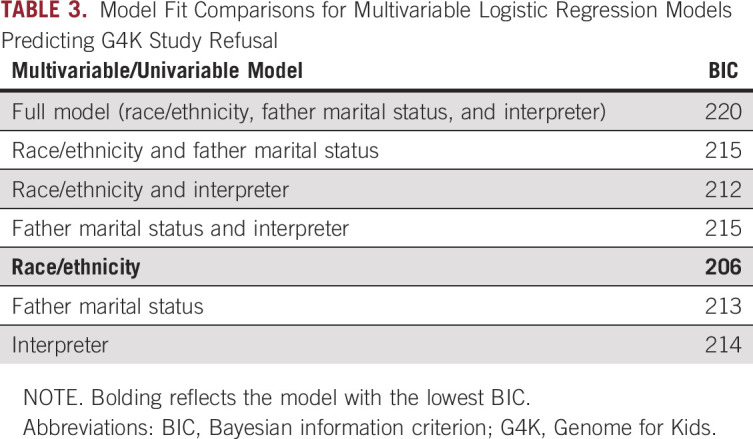
Demographic and clinical factors were compared for families who refused versus enrolled using separate logistic regression models (Table 2). Families of black children were significantly more likely to refuse enrollment compared with families of non-Hispanic white children (odds ratio [OR], 4.50; 95% CI, 2.29 to 8.84; P < .001). Although non-Hispanic white children were the reference group, it is noteworthy that families of black children were also significantly more likely to refuse compared with families of Hispanic white children (OR, 26.29; 95% CI, 3.40 to 203.52; P = .002). The father’s marital status was also associated with refusal. Families with a single father were more likely to refuse sequencing compared with families with married fathers (OR, 3.75; 95% CI, 1.69 to 8.30; P = .001); however, no significant differences emerged between divorced/separated versus married fathers (P = .264). Need for an interpreter was marginally significant, with families needing an interpreter less likely to refuse (OR, 0.16; 95% CI, 0.02 to 1.21; P = .076). Multivariate logistic regression analyses (Table 3) revealed that the best fitting model (ie, lowest Bayesian information criteria) was a model predicting refusal from only race/ethnicity. Thus, race/ethnicity was the only significant predictor of study refusal, with families of black children more likely to decline.
TABLE 2.
Univariable Logistic Regression Models Predicting Study Refusal of G4K
TABLE 3.
Model Fit Comparisons for Multivariable Logistic Regression Models Predicting G4K Study Refusal
Reasons for G4K Declination
Of the 40 families who actively declined participation in G4K, 35 (87.5%) provided a reason. The most common reason for refusal was feeling overwhelmed (n = 8; 22.9%; Table 4). Some families opted for clinical testing targeted to their child’s cancer and family history as opposed to testing a broad panel of genes through G4K (n = 7; 20.0%). Other families cited concerns about insurance discrimination (n = 6; 17.1%), perceiving that participating would not directly benefit them or their child (n = 3; 8.6%), the child or young adult patient not wanting to participate (n = 6; 17.1%), not wanting to learn sequencing results (despite being informed that they could participate without sequencing results being disclosed or in the patient’s medical record; n = 4; 11.4%), and opposition to the skin biopsy required for germline sequencing for some patients (n = 1; 2.9%).
TABLE 4.
Reasons for G4K Study Declination

Comparison With PG4KDS
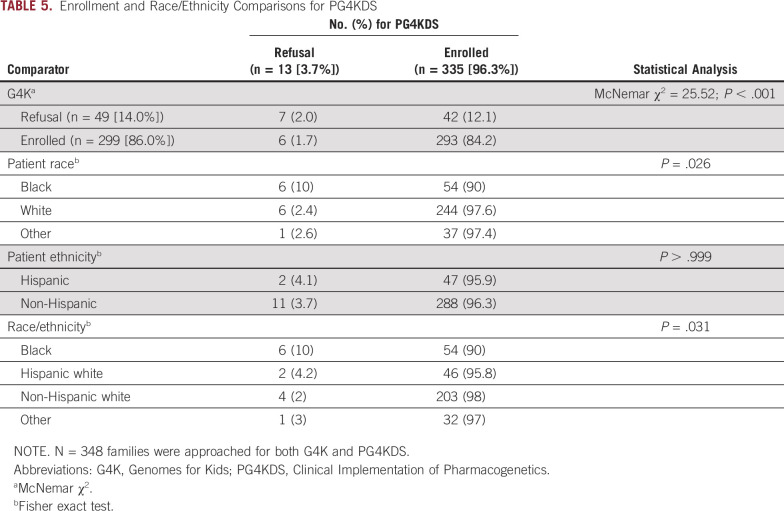
Of 348 families also approached for PG4KDS, 13 (3.7%) declined enrollment in PG4KDS. Refusal rates were compared across studies for patients approached for both G4K and PG4KDS. More families enrolled in PG4KDS but refused G4K (n = 42; 12.1%) compared with families who enrolled in G4K but refused PG4KDS (n = 6; 1.7%; χ2 = 25.52; P < .001; Table 5). Refusal for participation in PG4KDS in the cohort approached for both studies was significantly more common for families with a black child (P = .026; Table 5).
TABLE 5.
Enrollment and Race/Ethnicity Comparisons for PG4KDS
DISCUSSION
This retrospective study examined demographic and clinical factors in relation to declination for a pediatric oncology NGS study offering genetic sequencing for cancer predisposition (G4K) and compared the results with those of a separate pharmacogenomics study (PG4KDS). Rate of G4K refusal was comparable to prior pediatric NGS studies.12 Only patient race/ethnicity significantly differentiated G4K refusal, with families of black children more likely to decline than families of non-Hispanic or Hispanic white children. Among 348 families approached for both studies, families of black children were also less likely to enroll in PG4KDS. Although G4K examined genes that may be important in understanding individual and family risk for future malignancies, the relevance of some of these genes for therapeutic intervention is less clear. In contrast, PG4KDS has direct clinical utility, including modifying medication dosage or selecting alternative pharmacotherapy. Findings of race/ethnicity differences for study refusal in a genomics study with more direct clinical implications further bolsters these findings and highlights the importance of considering race/ethnicity in recruitment to pediatric genomics research regardless of the type of genomics study.
Racial differences in study refusal are consistent, with lower participation rates in genetics research among black patients.13,16,21-23 Underrepresentation of black patients has also been observed more broadly in clinical trials24,25 and has persisted in the context of National Institutes of Health efforts and techniques to increase minority representation.16,21,26 Race has remained a robust predictor of refusal even when controlling for relevant socioeconomic variables (eg, education, income).16,21 This study expands these findings in pediatric genomic research by demonstrating that race/ethnicity remains the only significant predictor of study declination even when controlling for other demographic and clinical factors, highlighting the robustness of race/ethnicity disparities. Although the disparity is well documented among adult populations,13,21,23 this is the first study, to our knowledge, to document race/ethnicity enrollment disparities in pediatric oncology genomics research. However, these findings are in line with reports that black parents express less interest than white parents in their child participating in NGS research.22
Among minorities, concerns have been raised that genetic data will be used in a harmful and/or discriminatory manner.27 Experiences of discrimination in medical settings and infamous research practices (ie, Tuskegee Syphilis Study, Henrietta Lacks) have, understandably, fostered mistrust in medical research specifically and medical relationships more broadly.27 Although underrepresentation of black patients in genomic research generally has been attributed to mistrust,24,28-30 black patients are also less likely to participate in clinical genetic testing and counseling (ie, when there is clear potential benefit to health).31 In the clinical context, adult patients have cited cost, among other barriers, as reasons not to pursue genetic testing28,29; however, this disparity remains even when logistic barriers (ie, cost and access) are reduced,30 suggesting that personal beliefs also play a role. Indeed, race differences have emerged in attitudes about genetic testing, perceptions of clinical utility of genetic screening, and responses to genetic sequencing results.22,32 Black parents have expressed more concerns about possible psychological impacts of mutation disclosure, access to care (ie, whether they would be capable of addressing health concerns), and possible misuse of genetic information by government and law enforcement; conversely, nonblack parents were more likely to express concerns about future insurance discrimination, privacy violations, and long-term care for future health risks.22
Focus groups have yielded recommendations for increasing diversity in genomics research, such as clarifying how data will be used and allowing participants to opt out of future research studies or decline having data shared with certain groups.27 Minority participants also expressed a desire for more explicit information about how privacy would be ensured, recommending institutional oversight to ensure accountability.27 Thus, participants want transparency and more extensive, ongoing informed consent collaborations. This literature calls for an emphasis on building trust between medical institutions and the communities they serve, including community-based participatory research, inclusion of community and religious leaders in protocol development to integrate local values, recruiting research assistants from local minority groups, and remodeling institutional culture to build patient-physician trust.27 Of note, neither study intentionally used research assistants from local minority groups; however, it should also be noted that SJCRH patients are from across the United States and other countries. Additional research using alternative methodologies (eg, discrete choice experiments) will also elucidate circumstances of genomic research that might influence minority participation.
Common reasons for study refusal were generally consistent with prior pediatric genetic research: feeling overwhelmed, possible psychological impact, privacy/discrimination concerns, disinterest in research, and study logistics (eg, time constraints).12 The relatively high number of families who cited feeling overwhelmed underscores the importance of considering when families are approached about participation. Time since diagnosis did not significantly predict refusal in this study; however, considerable heterogeneity existed in the timing of approach and illness/treatment characteristics. Future research should examine whether illness and treatment factors predict when during the cancer trajectory families are most receptive to genomics research. Although potential psychological impact of disclosing positive germline results has been cited as a concern for pediatric sequencing research,17,33 this was not a cited reason for refusal in G4K.
This study is limited by retrospective chart review design. Reliance on social work notes resulted in missing demographic data; thus, several demographic variables were less powered to detect a significant effect compared with race and ethnicity. However, effect sizes (odd ratios) also did not suggest an effect for demographic variables that had more missing data. In addition, other potentially relevant demographic variables (eg, income) were not available in the medical record. Examining demographic and clinical factors prospectively in future research would allow for more nuance in assessing sociodemographic variables, such as parent income, education, and religion. Race/ethnicity also may have differed if based on patient report or genomically derived, with the collection of race/ethnicity by chart review a limitation to this study. Of note, only patient race/ethnicity, and not caregiver race/ethnicity, was used, and caregivers did not always share the same race/ethnicity. The single-site nature of the design also may limit the generalizability of findings. However, it is noteworthy that SJCRH is unique in reducing financial and access to care barriers for patients, highlighting that underrepresentation of black patients persists beyond these barriers. Less is known about why passive decliners did not participate, with this subset perhaps differing from other patients in unique ways (eg, transportation challenges, elevated psychosocial stressors). Also, qualitatively examining reasons for study refusal was exploratory, and patients were not extensively queried. Future research should use alternative methodologies (eg, discrete choice experiments) to better understand families’ preferences for participating in pediatric oncology NGS research.
To our knowledge, this study is unique in documenting race/ethnicity disparities in enrollment in clinical genomic trials among pediatric oncology families, demonstrating a need to better understand and address barriers to minority participation in genomics research. As cancer treatment and surveillance decisions are increasingly affected by precision medicine, it is critical that genomics research be representative of patient populations, lest findings not generalize to minority populations and further widen health disparity gaps.
ACKNOWLEDGMENT
We thank Manish Kubal for his work as database analyst for this study, Sylwia Feibelman for assistance with table formatting, and members of the PG4KDS team. We also thank the oncologists, pathologists, and computational biologists. Last, we thank the patients and families who participated in this study.
Footnotes
PG4KDS is supported by NIGMS (5 P50GM115279-05) and The American Lebanese Syrian Associated Charities.
AUTHOR CONTRIBUTIONS
Conception and design: Niki Jurbergs, Annastasia Ouma, Kayla Hamilton, Regina Nuccio, Emily Quinn, Jami S. Gattuso, Michelle Pritchard, Belinda Mandrell, Liza M. Johnson, Kim E. Nichols
Collection and assembly of data: Annastasia Ouma, Lynn Harrison, Elsie Gerhardt, Leslie Taylor, Kayla Hamilton, Regina Nuccio, Emily Quinn, Stacy Hines-Dowell, Jami S. Gattuso, Michelle Pritchard, Belinda Mandrell, Cyrine E. Haidar, Liza M. Johnson
Data analysis and interpretation: Katianne M. Howard Sharp, Kayla Hamilton, Rose B. McGee, Regina Nuccio, Chimene Kesserwan, Anusha Sunkara, Jami S. Gattuso, Belinda Mandrell, Mary V. Relling, Cyrine E. Haidar, Guolian Kang, Liza M. Johnson
Provision of study material or patients: Elsie Gerhardt, Stacy Hines-Dowell
Administrative support: Elsie Gerhardt, Stacy Hines-Dowell
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Mary V. Relling
Research Funding: Servier Pharmaceutical
Kim E. Nichols
Research Funding: Incyte, Novartis, Alpine Immune Sciences
No other potential conflicts of interest were reported.
REFERENCES
- 1.Mody RJ, Wu Y-M, Lonigro RJ, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314:913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2:616–624. doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts JS, Dolinoy D, Tarini B. Emerging issues in public health genomics. Annu Rev Genomics Hum Genet. 2014;15:461–480. doi: 10.1146/annurev-genom-090413-025514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabatello M, Callier S, Garrison NA, et al. Trust, precision medicine research, and equitable participation of underserved populations. Am J Bioeth. 2018;18:34–36. doi: 10.1080/15265161.2018.1431328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korngiebel DM, Thummel KE, Burke W. Implementing precision medicine: The ethical challenges. Trends Pharmacol Sci. 2017;38:8–14. doi: 10.1016/j.tips.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcus PM, Pashayan N, Church TR, et al. Population-based precision cancer screening: A symposium on evidence, epidemiology, and next steps. Cancer Epidemiol Biomarkers Prev. 2016;25:1449–1455. doi: 10.1158/1055-9965.EPI-16-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt CL, Hussain A, Wachbroit R, et al. Precision medicine across the cancer continuum: Implementation and implications for cancer disparities. JCO Precis Oncol. 2017;1:1–5. doi: 10.1200/PO.17.00102. [DOI] [PubMed] [Google Scholar]
- 9.Malek J, Slashinski MJ, Robinson JO, et al. Parental perspectives on whole-exome sequencing in pediatric cancer: A typology of perceived utility. JCO Precis Oncol. 2017;1:1–10. doi: 10.1200/PO.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alderfer MA, Zelley K, Lindell RB, et al. Parent decision-making around the genetic testing of children for germline TP53 mutations. Cancer. 2015;121:286–293. doi: 10.1002/cncr.29027. [DOI] [PubMed] [Google Scholar]
- 11.Marron JM, DuBois SG, Glade Bender J, et al. Patient/parent perspectives on genomic tumor profiling of pediatric solid tumors: The Individualized Cancer Therapy (iCat) experience. Pediatr Blood Cancer. 2016;63:1974–1982. doi: 10.1002/pbc.26137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amendola LM, Robinson JO, Hart R, et al. Why patients decline genomic sequencing studies: Experiences from the CSER consortium. J Genet Couns. 2018;27:1220–1227. doi: 10.1007/s10897-018-0243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McQuillan GM, Porter KS, Agelli M, et al. Consent for genetic research in a general population: The NHANES experience. Genet Med. 2003;5:35–42. doi: 10.1097/00125817-200301000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Jones T, McCarthy AM, Kim Y, et al. Predictors of BRCA1/2 genetic testing among black women with breast cancer: A population-based study. Cancer Med. 2017;6:1787–1798. doi: 10.1002/cam4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McQuillan GM, Pan Q, Porter KS. Consent for genetic research in a general population: An update on the National Health and Nutrition Examination Survey experience. Genet Med. 2006;8:354–360. doi: 10.1097/01.gim.0000223552.70393.08. [DOI] [PubMed] [Google Scholar]
- 16.Hensley Alford S, McBride CM, Reid RJ, et al. Participation in genetic testing research varies by social group. Public Health Genomics. 2011;14:85–93. doi: 10.1159/000294277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilfond BS, Fernandez CV, Green RC. Disclosing Secondary Findings From Pediatric Sequencing to Families: Considering the “Benefit to Families.”. Los Angeles, CA: SAGE Publications; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson LM, Sykes AD, Lu Z, et al. Speaking genomics to parents offered germline testing for cancer predisposition: Use of a 2-visit consent model. Cancer. 2019;125:2455–2464. doi: 10.1002/cncr.32071. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman JM, Haidar CE, Wilkinson MR, et al. doi: 10.1002/ajmg.c.31391. PG4KDS: A model for the clinical implementation of pre‐emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 0(1):45-55, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 21.Moorman PG, Skinner CS, Evans JP, et al. Racial differences in enrolment in a cancer genetics registry. Cancer Epidemiol Biomarkers Prev. 2004;13:1349–1354. [PubMed] [Google Scholar]
- 22.Yu JH, Crouch J, Jamal SM, et al. Attitudes of non–African American focus group participants toward return of results from exome and whole-genome sequencing. Am J Med Genet A. 2014;164A:2153–2160. doi: 10.1002/ajmg.a.36610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bussey-Jones J, Garrett J, Henderson G, et al. The role of race and trust in tissue/blood donation for genetic research. Genet Med. 2010;12:116–121. doi: 10.1097/GIM.0b013e3181cd6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivers D, August EM, Sehovic I, et al. A systematic review of the factors influencing African Americans’ participation in cancer clinical trials. Contemp Clin Trials. 2013;35:13–32. doi: 10.1016/j.cct.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Heller C, Balls-Berry JE, Nery JD, et al. Strategies addressing barriers to clinical trial enrollment of underrepresented populations: A systematic review. Contemp Clin Trials. 2014;39:169–182. doi: 10.1016/j.cct.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson GM, Ward AJ. Recruiting minorities into clinical trials: Toward a participant-friendly system. J Natl Cancer Inst. 1995;87:1747–1759. doi: 10.1093/jnci/87.23.1747. [DOI] [PubMed] [Google Scholar]
- 27.Kraft SA, Cho MK, Gillespie K, et al. Beyond consent: Building trusting relationships with diverse populations in precision medicine research. Am J Bioeth. 2018;18:3–20. doi: 10.1080/15265161.2018.1431322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong K, Micco E, Carney A, et al. Racial differences in the use of BRCA1/2 testing among women with a family history of breast or ovarian cancer. JAMA. 2005;293:1729–1736. doi: 10.1001/jama.293.14.1729. [DOI] [PubMed] [Google Scholar]
- 29.Hayden S, Mange S, Duquette D, et al. Large, prospective analysis of the reasons patients do not pursue BRCA genetic testing following genetic counseling. J Genet Couns. 2017;26:859–865. doi: 10.1007/s10897-016-0064-5. [DOI] [PubMed] [Google Scholar]
- 30.Susswein LR, Skrzynia C, Lange LA, et al. Increased uptake of BRCA1/2 genetic testing among African American women with a recent diagnosis of breast cancer. J Clin Oncol. 2008;26:32–36. doi: 10.1200/JCO.2007.10.6377. [DOI] [PubMed] [Google Scholar]
- 31.Levy DE, Byfield SD, Comstock CB, et al. Underutilization of BRCA1/2 testing to guide breast cancer treatment: Black and Hispanic women particularly at risk. Genet Med. 2011;13:349–355. doi: 10.1097/GIM.0b013e3182091ba4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaphingst KA, Stafford JD, McGowan LDA, et al. Effects of racial and ethnic group and health literacy on responses to genomic risk information in a medically underserved population. Health Psychol. 2015;34:101–110. doi: 10.1037/hea0000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakefield CE, Hanlon LV, Tucker KM, et al. The psychological impact of genetic information on children: A systematic review. Genet Med. 2016;18:755–762. doi: 10.1038/gim.2015.181. [DOI] [PubMed] [Google Scholar]