Supplemental Digital Content is available in the text.
Background.
Primary graft dysfunction, infections, and acute rejection (AR) worsen lung transplantation (LTx) outcome and patient survival. Despite significant efforts, reliable biomarkers of acute lung allograft dysfunction are lacking. To address this issue, we profiled the bronchoalveolar lavage (BAL) miRNome in LTx patients.
Methods.
BAL-microRNAs (miRNAs) from 16 patients were collected 7 days (T0), 15 days (T1), and 3 months (T2) after bilateral LTx and profiled on low-density array. Unsupervised and supervised analyses were used to identify miRNAs associated with clinical features, pneumonia, or AR. Prognostic markers were identified using the Cox model. Targeted signaling pathways were predicted in silico. A second series of 11 patients were used to validate AR-associated miRNAs.
Results.
Variation in BAL-miRNAs was associated with acute lung allograft dysfunction. Increased levels of miR-23b-3p at T2 were detected in patients with pneumonia, whereas let-7f-5p, miR-146b-3p, miR-22-5p, miR-29c-5p, miR-362-5p, and miR-452-5p were upregulated at T2 in patients with AR. miR-148b-5p and miR-744-3p distinguished LTx patients with AR in both cohorts. Low miR-148b-5p and high miR-744-3p expression levels were significantly associated with a shorter time to AR either within the first year after LTx or during follow-up. Combination of the 2 miRNAs identified LTx patients with higher AR risk independently of clinical variables.
Conclusions.
Our data provide new insights into the roles of BAL-miRNAs in regulating the pulmonary environment after transplantation and suggest that these miRNAs could serve as biomarkers of early- or mid-stage events. If validated, these findings could pave the way to a personalized clinical approach in LTx patients.
Lung transplantation (LTx) is the gold standard treatment for chronic end-stage pulmonary diseases. Survival of LTx patients has improved recently due to advances in surgical techniques and pharmacologic management of complications. However, the mortality rate in the first year after transplant remains high, mostly due to acute lung allograft dysfunction (ALAD) events, such as acute cellular rejection or infections.1 The pathogenetic mechanisms underlying this phenomenon remain puzzled, and the correct clinical approach has yet to be firmly established. The influence of ALAD in this early period on mid- and late-term outcomes is a topic of intense interest.
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression, usually by silencing their target mRNAs. These molecules are powerful regulators of various diseases, with potential critical effects on disease initiation, progression, and treatment. A single miRNA can alter the expression of many target genes, thereby influencing entire signaling pathways rather than a single gene. This unique function makes miRNAs attractive targets for translational medicine. miRNAs are ideal circulating diagnostic, prognostic, and predictive biomarkers of disease status and are also present in bronchoalveolar lavage (BAL).2,3 Various studies have provided evidence that miRNAs play a fundamental role in respiratory disorders and several other physiologic and pathologic mechanisms, including immune cell development and function, inflammation, and oncogenesis. Preliminary work has shown that miRNAs are dysregulated following LTx.4
Hence, we sought to preliminary characterize differences in BAL-miRNA profiles, or their variation over time, according to features related to the recipient, donor, surgical procedure, or posttransplantation complications and to identify predictive markers of the onset of acute rejection (AR) in the first year after transplantation.
MATERIALS AND METHODS
Patients
We conducted a single-institution prospective study at Fondazione IRCCS Ca’ Granda—Ospedale Maggiore Policlinico of Milan. The local Ethical Committee approved the study (protocol No. 640/2017bis) and informed written consent was obtained from all subjects. For the study, we enrolled all patients who underwent bilateral LTx at our institution between May 2017 and July 2018. A first set of 16 patients was used for miRNA screening, whereas additional 11 patients were included in data validation analyses.
Exclusion criteria were as follows: patients who declined to participate, age < 18 years, previous solid organ transplantation, intensive care unit (ICU) stay > 15 days, and death within 1 week after surgery. Patients underwent bronchoscopic surveillance with BAL fluid withdrawal at 7 (T0) and 15 days (T1) after surgery. Pulmonary trans-bronchial biopsies with BAL sampling were scheduled at 3 (T2), 6, and 12 months after transplantation, as well as in cases of lung function decline or another clinical suspicion of ALAD. Our protocols for donor and recipient selection, graft procurement, and transplantation surgical procedures are described in the online supplement http://links.lww.com/TXD/A248.
For each LTx recipient, the following clinical data were collected: data regarding the primary lung disease, age, sex, lung allocation score, microbial colonization (in patients with cystic fibrosis [CF]), and cytomegalovirus serology. Donor data obtained during organ procurement included donor age and sex, smoking history, type of donation (donation after brain death or donation after circulatory death, respectively), Oto score, cytomegalovirus serology, donor/recipient mismatch, and use of static or portable machine perfusion (MP). We also recorded data on intraoperative extracorporeal membrane oxygenation, mechanical ventilation (MV), and warm ischemic time. For postoperative variables, we recorded primary graft dysfunction (PGD) score, best forced expiratory volume in the first second (FEV1), pneumonia, histological (grade A and B), and clinical AR (c-AR) in the first year after transplantation. c-AR occurrence during the complete follow-up time (ended in July 2019) was also recorded. Definition of clinical variables is provided in the Supplementary Methods.
Sample Collection
T0, T1, and T2 BALs were centrifuged at 2500 revolutions per minute at 4°C for 15 minutes to remove cell debris; the supernatant was aliquoted into RNase-free tubes and stocked at –80°C until RNA purification. miRNA profiling procedures were described previously5,6 and are also provided in the online supplement http://links.lww.com/TXD/A248.
Data Collection and Statistical Analysis
miRNA data were analyzed using both unsupervised and supervised approaches. Calculations were carried out in the R statistical computing environment using the libraries Biobase, perm, nparLD, Hmisc, survival, factoextra, FactoMineR, NbClust, and ComplexHeatmap.1,7 For unsupervised analysis, normalized miRNA quantities were imported in Rstudio, and the ComplexHeatmap package in Bioconductor was used for hierarchical clustering analysis. Principal component analysis plots were generated in R (https://cran.r-project.org/) to visualize LTx patients’ distribution according to the miRNome at individual times, as described.8 The K-means clustering algorithm was used to visualize sets of co-regulated miRNAs at individual time points.
Differences in miRNA expression according to clinical features (all treated as categorical variables) were performed either separately at individual time points (T0, T1, and T2) or as variations of each miRNA during follow-up (ie, the following 3 combinations: T1–T0, T2–T0, and T2–T1). For the first analysis, differentially expressed miRNAs according to clinical or outcome variables were identified by computing the fold change (log2Ratio) between the 2 classes and applying the Kolmogorov-Smirnov test. Time-dependent variations of miRNA levels during T0, T1, and T2 were examined in relation to clinical features using nonparametric repeated-measures ANOVA. Specifically, a model with an interaction of time and clinical feature, such as pneumonia or c-AR, was considered for each miRNA. Pneumonia or c-AR were treated as a binary variables. We evaluated if levels of miRNAs at T0, T1, and T2 schedules varied differently in patients with c-AR compared with patients who did not experienced acute graft rejection. The accuracy of the data was assessed by a permutation-based estimate of the false discovery rate (FDR). Accordingly, all P values were corrected for multiple comparisons using the FDR approach, with the FDR threshold set to 10%. The associations between pairs of variables were investigated using Fisher exact test (MedCalc software).
The impact of clinical or molecular variables on the incidence of c-AR was analyzed by univariate or multivariate analysis using the Cox proportional-hazards regression model. The miRNA level cutoffs used to sort LTx patients into low and high expression groups were generated using receiver operating characteristic (ROC) curves and Youden’s J statistic, as described.8 Kaplan-Meier survival curves were compared using log-rank and Gehan-Breslow-Wilcoxon tests. Logistic regression model was used to calculate odds ratio associated to c-AR (dependent variable). When a multivariate model was performed, the stepwise method was used. All of these analyses were performed using MedCalc software.
Lists of target genes were created using the miRTargetLink Human tool (https://ccb-web.cs.uni-saarland.de/mirtargetlink/), which provides a list of experimentally validated miRNA targets or with the multiMiR R package and database in Bioconductor when validated targets were unavailable. Gene lists were then either analyzed using the Over Representation Analysis within the WEB-based GEne SeT AnaLysis Toolkit using the human protein-coding genome as reference set (www.webgestalt.com) and enrichment of significant Gene Ontology category Biological Process was performed.
Data are presented as mean ± SEM or median ± interquartile range, as specified in the legend.
RESULTS
Clinical Data
From May 2017 to July 2018, 37 patients underwent bilateral LTx at our institution. One patient was excluded for previous liver transplantation, 2 patients were either < 18 or > 65 years old at the time of transplantation, 1 patient received a single LTx, 1 patient did not consent to participate in the study, and 2 patients had a prolonged ICU stay. Moreover, for 3 patients, the BAL at T0 was not suitable for RNA purification because of hemolysis. Therefore, 27 patients were ultimately enrolled in the study, of whom 16 were included in the discovery analysis and 11 were available to data confirmation (Table 1 and Table S1, SDC, http://links.lww.com/TXD/A248). Patients’ median age was 34 years (range, 21–64 y), and 19 were men. Nineteen subjects suffered from CF, whereas 8 had interstitial lung disease. Mean follow-up time was 18 months (range, 12–25 mo), during which 4 patients died. Patient characteristics are presented in Table 1 and Table S1 (SDC, http://links.lww.com/TXD/A248). For 1 and 4 patients, respectively, T1-BAL or T2-BAL was not suitable for molecular analysis due to hemolysis. A Consort flow chart is provided in the online supplement (Figure S1, SDC, http://links.lww.com/TXD/A248).
TABLE 1.
Clinical features of patients enrolled in this study (additional information is available in Table S1, SDC, http://links.lww.com/TXD/A248)
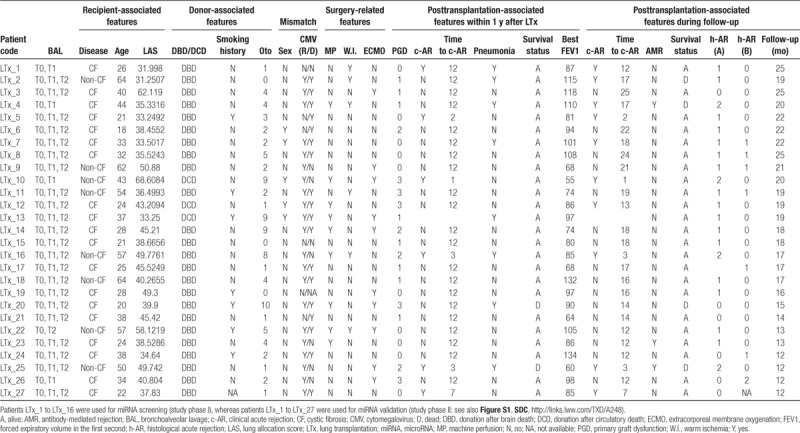
miRNA Expression in BAL Undergoes a Global Change 3 Months After Transplantation
Initially, we examined the BAL-miRNome in an unsupervised manner to determine whether any phenotypes were shared between classes of patients or timing of BAL withdrawal. To this end, the first series of patients was used (n = 16; discovery set). In total, 317 miRNAs were available for analysis. We observed a global change in miRNA levels 3 months after transplantation (T2-BAL) relative to T0 and T1 (Figure 1A). To determine which signals were most affected at earlier (T0 and T1) versus later times (T2), we performed k-means clustering analysis of miRNAs, which identified 3 main groups of miRNAs (Figure 1A). BAL-miRNAs belonging to clusters 1 and 2 were more likely to be represented at earlier times, whereas miRNAs belonging to cluster 3 were more likely to be expressed at T2. When we analyzed potentially targeted signaling pathways, we found that regulation of responses to stress, immunity, wounding, and vasculature development were predicted as targets of T0- and T1-miRNAs (Figure 1B; Table S2, SDC, http://links.lww.com/TXD/A248), whereas genes related to cell death and gene silencing were identified as potential targets of T2-miRNAs (Figure 1C; Table S3, SDC, http://links.lww.com/TXD/A248). These data, although preliminary, shed light on the dynamic biology in the lung microenvironment after LTx.
FIGURE 1.
Exploratory analysis of the bronchoalveolar lavage (BAL)-miRNome in lung transplantation (LTx) patients. A, Unsupervised hierarchical clustering of all BAL-microRNAs (miRNAs) (n = 317) detected in lung-transplanted patients (study phase I, n = 16). BAL samples were collected 7 d (T0), 15 d (T1), or 3 mo (T2) after transplantation. The heatmap shows miRNA levels in all patients at the indicated times. Red and blue indicate high and low expression, respectively. To identify groups of miRNAs with similar behavior, k-means clustering analysis was performed, yielding 3 miRNA clusters (color bar to the right of the heatmap). B and C, Biological processes potentially affected by the top miRNAs in k-means clusters 1 and 2 (B) or cluster 3 (C) target prediction was performed for top miRNAs using the miRTargetLink Human tool, and gene lists were annotated using WebGestalt and GeneOntology as functional database. Significant terms networks are presented as directed acyclic graph (DAG). D–F, Principal component analysis was performed for BAL-miRNAs at T0 (D), T1 (E), and T2 (F) to evaluate the sample distribution according to the indicated clinical variable. The variance explained by the first 3 components was as follows: T0 (PC1: 29%, PC2: 19%, PC3: 11%); T1 (PC1: 33%, PC2: 17%, PC3: 13%); and T2 (PC1: 47%, PC2: 10%, PC3: 9%). c-AR, clinical acute rejection; CF, cystic fibrosis; CMV, cytomegalovirus; ECMO, extracorporeal membrane oxygenation; h-AR, histological acute rejection, grade A; IPF, idiopathic pulmonary fibrosis; N, no; PC, principal component; PGD, primary graft dysfunction, Y, yes.
The BAL-miRNome Can Distinguish Patients With Distinct Clinical Phenotypes
Next, we analyzed the BAL-miRNome at individual time points by unsupervised principal component analysis to determine whether miRNAs could discriminate among LTx patients according to their clinical features. miRNA profiles obtained at T0 discriminated according to disease, Oto score, and PGD (Figure 1D; variance explained by the first 3 components = 59%), whereas T1-miRNAs discriminated according to PGD (Figure 1E; variance explained by the first 3 components = 63%), and T2-miRNAs discriminated according to c-AR (Figure 1F; variance explained by the first 3 components = 66%).
Variation in BAL-miRNAs Over 3 Months, Rather Than Evaluation of miRNAs at Individual Time Points, Provides Preclinical Biomarkers of Lung Allograft Outcome
Next, we sought to identify BAL-miRNA signatures associated with recipient-, donor-, and surgery-related clinical features (Tables S4–S6, SDC, http://links.lww.com/TXD/A248) or posttransplantation variables including PGD grade 2 and 3, the best FEV1, c-AR, or infections (Table S7, SDC, http://links.lww.com/TXD/A248). For this purpose, we considered either BAL-miRNAs obtained at individual time points (Tables S3–S7, SDC, http://links.lww.com/TXD/A248) or variation in miRNA levels across the 3 periods (nonparametric repeated-measures ANOVA; Table S8, SDC, http://links.lww.com/TXD/A248). According to the FDR criterion, 55 T0-miRNAs were associated with MP (Table S6, SDC, http://links.lww.com/TXD/A248), and 10 T2-miRNAs were associated with prolonged warm ischemia, but no miRNAs were significantly associated with either pneumonia or c-AR (Table S7, SDC, http://links.lww.com/TXD/A248). Conversely, repeated-measures ANOVA showed significant time-dependent interactions of miRNAs levels according to pneumonia or c-AR. Indeed the variation in miRNA levels across different time points provided signatures of 1 and 9 BAL-miRNAs associated with pneumonia and c-AR (treated as binary variables), respectively (Table S8, SDC, http://links.lww.com/TXD/A248). In general, the increase in expression of the identified BAL-miRNAs at T2 versus T1 was associated with a clinical event. Specifically, miR-23b-3p expression was elevated at T2 in patients with pneumonia (Figure 2A), whereas let-7f-5p, miR-146b-3p, miR-22-5p, miR-29c-5p, miR-362-3p, and miR-452-5p levels were higher at T2 than at T1 in patients with AR (Figure 2B). Interestingly, miR-29c-5p levels in BAL-T1 were increased in cases with PGD grade 2 and 3 (Table S7, SDC, http://links.lww.com/TXD/A248) and in BAL-T2 of patients with c-AR (Figure 2B).
FIGURE 2.
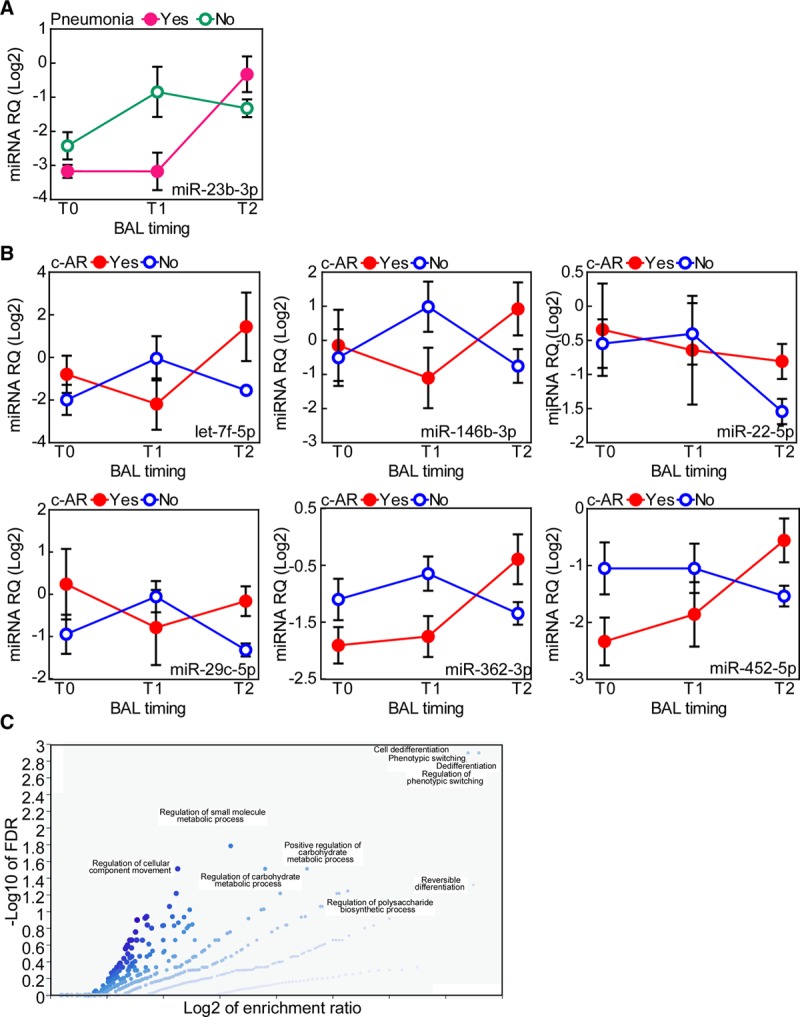
Changes in bronchoalveolar lavage (BAL)-microRNA (miRNA) levels are associated with pneumonia or clinical acute rejection (c-AR). The time-dependent interaction of BAL-miRNAs with presence of pneumonia (A) or c-AR (B) was analyzed by nonparametric repeated-measures ANOVA with correction for multiple comparisons (see also Table S8, SDC, http://links.lww.com/TXD/A248). This analysis was performed using the first series of patients (n = 16; see also Figure S1, SDC, http://links.lww.com/TXD/A248). miRNAs whose levels were significantly elevated at T2 are shown. C, Vulcano plot showing the biological processes potentially affected by c-AR-miRNAs. Target prediction was performed using the miRTargetLink Human tool, and gene lists were annotated using WebGestalt and GeneOntology as functional database. FDR, false discovery rate; RQ, relative quantity.
Based on our in silico analysis of potential predicted targets (Table S9, SDC, http://links.lww.com/TXD/A248), mRNAs involved in cell dedifferentiation and metabolism were targeted by c-AR-associated BAL-miRNAs (Figure 2C; Table S10, SDC, http://links.lww.com/TXD/A248).
Clinical Variables Are Not Able to Discriminate Patients With Clinical AR
Next, we explored the possible associations between pairs of features related to recipient, donor, or surgical procedure, or their associations with patient outcome. Based on Fisher exact test, we found that a lung allocation score above 40 was significantly associated with extracorporeal membrane oxygenation, whereas MP procedure was associated with donation after circulatory death and with an Oto score > 5 (Table S11, SDC, http://links.lww.com/TXD/A248). In regard to patient outcome, an Oto score > 5 or prolonged MV was associated with PGD. Prolonged warm ischemia was associated with pneumonia and pneumonia with AR (P = 0.04; Figure S2A, SDC, http://links.lww.com/TXD/A248). Finally, we had concordance between c-AR and histological AR (grade A) evaluated on trans-bronchial biopsies during patients’ surveillance (Figure S2B, SDC, http://links.lww.com/TXD/A248).
miR-148b-5p and miR-774-3p Are Novel Potential Predictive Biomarkers of Clinical AR
Lastly, we sought to determine whether clinical or molecular variables could predict the occurrence of c-AR during the follow-up period using the Cox model. In univariate analyses, no clinical features were significantly associated with patient outcome (Table 2). Using the first set of patients, we found that 4 T0-BAL miRNAs, miR-27a-5p, miR-148b-5p, miR-222-5p and miR-744-3p, and the T1-BAL miRNA miR-1285-3p (Table S12, SDC, http://links.lww.com/TXD/A248), were associated with c-AR risk within the first year after LTx. However, these molecular markers did not meet the criterion for significance following the FDR correction for multiple testing (Table S12, SDC, http://links.lww.com/TXD/A248). ROC analysis showed that T0-BAL miR-148b-5p and miR-744-3p could classify LTx patients according to c-AR with an area under the curve > 75% (Figure 3A and B), whereas miR-27a-5p, miR-222-5p, and miR-1285-3p did not (Figure S3, SDC, http://links.lww.com/TXD/A248). To support this preliminary observation, we extended miR-148b-5p and miR-744-3p evaluation including additional 11 LTx patients (total cohort n = 26 because the patient LTx_16 was excluded from c-AR analysis due to concurrent pneumonia infection; see also Table 1) for whom at least 1-year of follow-up was available (range, 12–17 mo; Table 1). Firstly, the ROC identified cutoffs for the 2 miRNAs (Youden’s J statistic; Figure 3A and B) significantly categorized patients according to AR occurrence (Figure 3C and D). Moreover, the ROC-generated cutoffs for both miRNAs could sort patients into 2 groups with significantly different risks of c-AR occurrence either in the first year from LTx (Figure 3E and F) or during the entire follow-up (range 12–25 mo; Figure 3G and H). Specifically, patients with higher levels of miR-148-5p in T0-BAL (miR-148b-5pHIGH; Figure 3E–G) had a lower risk of c-AR (P = 0.009 or P = 0.004, hazard ratio = 0.1 or 0.13 by log-rank test) than miR-148b-5pLOW subjects. By contrast, lung-transplanted patients with high miR-744-3p levels measured in BAL at T0 were characterized by a higher risk of c-AR (P = 0.03 and P = 0.04, hazard ratio = 5.6 or 3.8; Figure 3F–H).
TABLE 2.
Association of clinical variables with clinical AR during the first y of follow-up in LTx patients (n = 26), as determined by Cox regression model
FIGURE 3.
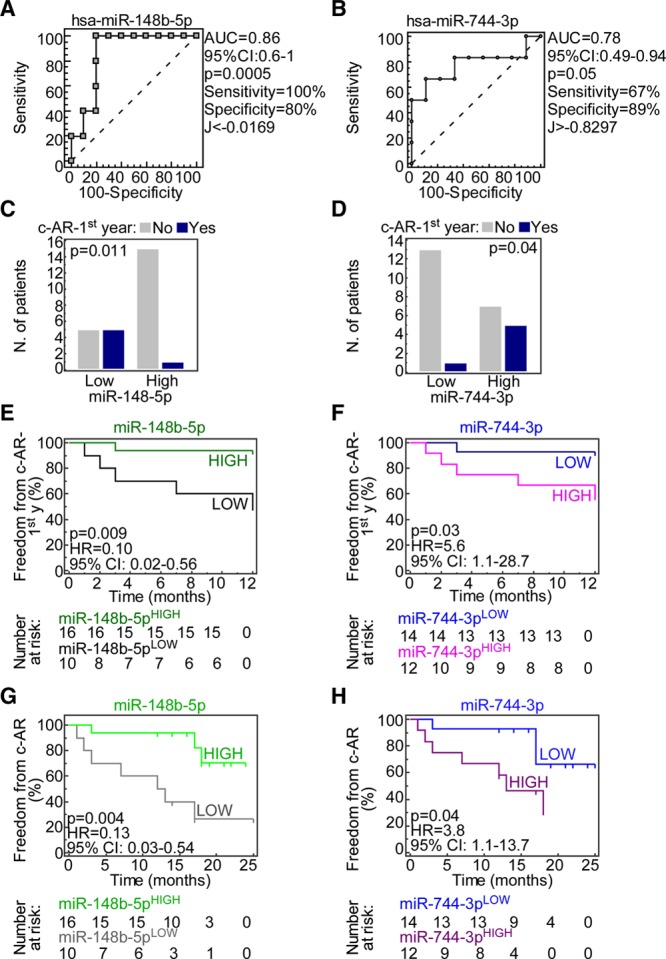
miR-148b-5p and miR-744-3p are novel potential predictive biomarkers of acute rejection in lung transplantation (LTx) patients. A and B, ROC curves were generated to test the accuracy of the indicated microRNAs (miRNAs) in classifying patients (n = 26) according to presence of clinical acute rejection (c-AR) within the first y after LTx. The cutoff for miRNA expression was calculated using Youden’s J statistic. C and D, The ROC-generated cutoffs were used to sort LTx patients into LOW or HIGH expression groups for both miR-148b-5p and miR-744-3p and their ability to correctly classify LTx patients according to c-AR was analyzed using the enlarged cohort (n = 27). P values are from Fisher exact test. E–H, Freedom from c-AR (probability) was estimated using Kaplan-Meier curves in LTx patients within 1 y from transplantation (E and F) or during follow-up (G and H). Patients were sorted according to the receiver operating characteristic (ROC)-generated cutoff into high- and low-miRNA groups. The number of patients at risk is reported below the curves for all the groups. Indicated P values were computed by the log-rank test. AUC, area under the curve; CI, confidence interval; HR, hazard ratio.
The ability of these 2 miRNAs to identify patients at higher risk of AR was further strengthened when we combined them, categorizing patients as miR-148b-5pHIGH and miR-744-3pLOW (both associated with better outcome; n = 10), miR-148b-5pLOW and miR-744-3pHIGH (both associated with poorer outcome; n = 7), or discordant (either miR-148b-5pLOW and miR-744-3pLOW or miR-148b-5pHIGH and miR-744-3pHIGH; n = 9). LTx patients with combinations of “good,” “discordant,” or “bad” miRNA classes had respectively smaller (18%), intermediate (28%), and larger (85%) incidence of c-AR (P = 0.01 by chi-square test; Figure 4A). Furthermore, using a logistic regression model we determined that the “bad” miRNA class had the strongest predictive value for c-AR within the first year after LTx (odds ratio, 25; 95% confidence interval, 1.8-346.7; P = 0.01; Figure 4B) also when a multivariate analysis was performed (Table S13, SDC, http://links.lww.com/TXD/A248). These observations, although preliminary, identify miR-148b-5p and miR-744-3p as novel players in lung allograft dynamics and open the possibility of using these miRNAs as early biomarkers of c-AR.
FIGURE 4.
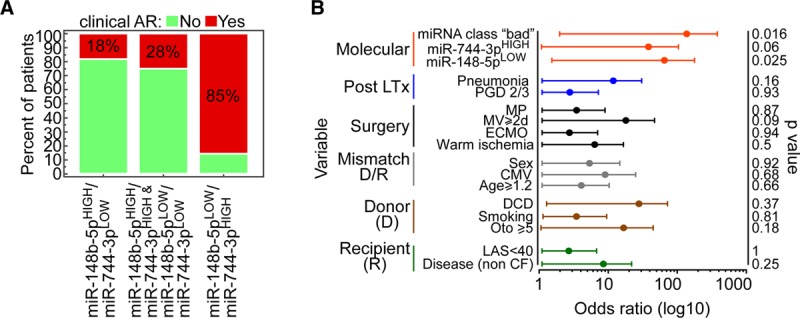
Combination of miR-148b-5p and miR-744-3p classes identifies LTx patients with higher risk of c-AR. A, Combination of the 2 microRNAs (miRNAs) categories stratifies lung transplantation (LTx) patients (n = 26) in 3 groups with different acute rejection (AR) incidence (indicated in the bars). P = 0.01 by chi-square test. B, Forest plot shows the odds ratio of the indicated variables. Bars, 95% confidence interval. P values are from logistic regression analysis. CF, cystic fibrosis; CMV, cytomegalovirus; DCD, donation after circulatory death; ECMO, extracorporeal membrane oxygenation; LAS, lung allocation score; MP, machine perfusion; MV, mechanical ventilation; PGD, primary graft dysfunction.
DISCUSSION
For the first time, our study provides an overview of miRNA expression in BAL at different times after bilateral LTx. Although preliminary, our data show that the BAL-miRNome is significantly modulated 3 months after transplantation and that changes in miRNA expression levels in each patient, rather than detection at individual schedule, provide clues about primary outcome events such as pneumonia and AR. In addition, we identified 2 T0-BAL miRNAs, miR-148b-5p and miR-744-3p, associated with c-AR risk irrespective of the presence of previous infections or other clinical variables.
miRNAs are implicated in the onset, progression, and prognosis of almost all human diseases, and thus represent potential novel drug targets. Several lines of evidence have revealed an association between miRNA deregulation and inflammation, and preliminary studies have analyzed miRNA content in LTx patients.9 Nevertheless, a deeper understanding of miRNA dynamics in LTx patients is required to identify the mechanism of disease and potential noninvasive biomarkers of graft rejection. Indeed, although LTx is the only therapy for patients with end-stage pulmonary disorders, the rate of chronic rejection is still high, and the 5-year survival rate is only 54% (according to International Socienty of Heart and Lung Transplantation [ISHLT] registry). The ISHLT registry has also reported that 1-year survival between 2000 and 2009 was significantly higher than in 2 preceding eras (1988–1994 and 1995–1999). However, the improvement in 5-year survival over this period was smaller, suggesting that management strategies have been more effective at addressing early complications than later ones, possibly via refinements in surgical techniques and postoperative care. By contrast, survival beyond the first year is primarily determined by chronic rejection and infection, and the incidences of these complications have not changed substantially since 1988. Therefore, the clinical management of LTx patients could benefit from the adoption of molecular biomarkers of graft rejection and infections. From this standpoint, we preliminarily investigated these issues by identifying several potential miRNA markers of early LTx failure, and also predicted the signaling pathways that these miRNAs target.
Pneumonia-associated miR-23b was previously described as a key factor in autoimmune diseases10 and sepsis.11 The targets of miR-23b include inflammatory cytokines such as tumor necrosis factor-α, interleukin-1β, interleukin-17, and vascular adhesion molecules intercellular adhesion molecule-1, E-selectin, and vascular cell adhesion molecule-1. Targeting of these molecules by miR-23b protects against autoimmunity but is detrimental in systemic inflammatory states such as sepsis, where it contributes to disease progression.11
Among the top pathways predicted to be targeted by AR-associated miRNAs, we identified dedifferentiation, phenotypic switch, and metabolic changes. These pathways might be related to tissue healing and remodeling, processes known to be important for organ engrafting and tissue regeneration. Remarkably, when tissue regeneration fails fibrosis might prevail stimulating aberrant lung remodeling and organ dysfunction.12
In keeping with this, we found that miR-29c-5p is significantly higher in T1-BAL of patients who experienced PGD and it is increased in patients who developed AR. miR-29c has been described as an effector of the transforming growth factor-β pathway and it is implicated in liver and kidney allografts fibrosis, a major hallmark of progressive organ dysfunction.13,14
Finally, we identified 2 possible markers predictive of c-AR, the BAL-miRNAs miR-148b-4p and miR-744-3p. The levels of these 2 molecules 7 days after surgery (T0) could stratify patients by AR risk. Grouping of LTx patients based on the levels of both miRNAs revealed that the 2 markers can be used simultaneously to improve identification of patients likely to have poorer graft outcomes.
Both of these miRNAs have been previously associated with inflammatory states. Specifically, miR-744 is upregulated in keratinocytes of psoriatic patients15 and the glomeruli of diabetic mice,16 and has been shown to boost type I interferon signaling in renal mesangial cells.17 Furthermore, miR-744 regulates transforming growth factor-β–mediated signal transduction.18 On the other hand, deregulation of miR-148b has been documented in lung diseases such as CF, where it contributes to inflammation and alveolar injury.19 Indeed, downregulation of miR-148b promotes endothelial-to-mesenchymal transition in endothelial cells, resulting in fibrosis and impaired wound closure.20 Consistent with these previous reports, we found that high levels of miR-744-3p and low levels of miR-148b-5p were significantly associated with shorter time to AR in LTx patients.
One limitation of our study is the small number of enrolled patients and the lack of recipients with chronic obstructive pulmonary disease as a pretransplantation lung dysfunction. Further, opposite to previous literature,21 we could not determine a correlation between PGD and the development of acute graft rejection, although prolonged MV and a poor donor Oto score were associated to PGD severity. This could be because we excluded from the study subjects that had a prolonged ICU stay or that die within the first week after LTx.
Nevertheless, we could confirm our initial data obtained with 16 patients including in the cohort 11 additional LTx. Altogether, our data provide a preliminary overview of molecular alterations in BAL of LTx patients over a 3-month period and suggest novel noninvasive potential markers for patient stratification and prognostication. Indeed, the miRNAs-based classification could support the definition of a patient-tailored immunosuppressive regimen in the first year after LTx.
As demonstrated for other clinical settings, such as kidney inflammatory diseases,17 deregulated expression of miRNAs could significantly impact inflammatory signal cascades in the pulmonary environment. Although our results need to be validated in a larger series, they open new opportunities for personalization of lung allograft management and understanding the biological basis of ALAD.
Supplementary Material
Footnotes
Published online 9 April, 2020.
A.P. and G.G. contributed equally to this study.
A.P., G.G., V.M., M.N., and V.V. participated in research design. A.P. and V.V. participated in the writing of the article. G.G., V.M., S.F., L.R., P.T., L.C.M., S.F., and M.N. participated in the performance of the research. A.P., V.E., A.T., F.A., and V.V. participated in data analysis.
The authors declare no conflicts of interest.
This work was supported by the Italian Ministry of Health “Ricerca Corrente 2018/19” program to S.F. and M.N.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Verleden GM, Raghu G, Meyer KC, et al. A new classification system for chronic lung allograft dysfunction. J Heart Lung Transplant. 2014; 33:127–133 [DOI] [PubMed] [Google Scholar]
- 2.Bibaki E, Tsitoura E, Vasarmidi E, et al. miR-185 and miR-29a are similarly expressed in the bronchoalveolar lavage cells in IPF and lung cancer but common targets DNMT1 and COL1A1 show disease specific patterns. Mol Med Rep. 2018; 17:7105–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z, Nayak D, Yang W, et al. Dysregulated microRNA expression and chronic lung allograft rejection in recipients with antibodies to donor HLA. Am J Transplant. 2015; 15:1933–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kishore A, Navratilova Z, Kolek V, et al. Expression analysis of extracellular microRNA in bronchoalveolar lavage fluid from patients with pulmonary sarcoidosis. Respirology. 2018; 23:1166–1172 [DOI] [PubMed] [Google Scholar]
- 5.Russo MV, Faversani A, Gatti S, et al. A new mouse avatar model of non-small cell lung cancer. Front Oncol. 2015; 5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augello C, Colombo F, Terrasi A, et al. Expression of C19MC miRNAs in HCC associates with stem-cell features and the cancer-testis genes signature. Dig Liver Dis. 2018; 50:583–593 [DOI] [PubMed] [Google Scholar]
- 7.Suwara MI, Vanaudenaerde BM, Verleden SE, et al. Mechanistic differences between phenotypes of chronic lung allograft dysfunction after lung transplantation. Transpl Int. 2014; 27:857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terrasi A, Bertolini I, Martelli C, et al. Specific V-ATPase expression sub-classifies IDHwt lower-grade gliomas and impacts glioma growth in vivo. Ebiomedicine. 2019; 41:214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Ramachandran S, Gunasekaran M, et al. MicroRNA-144 dysregulates the transforming growth factor-β signaling cascade and contributes to the development of bronchiolitis obliterans syndrome after human lung transplantation. J Heart Lung Transplant. 2015; 34:1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R, O’Connell RM. MiR-23b is a safeguard against autoimmunity. Nat Med. 2012; 18:1009–1010 [DOI] [PubMed] [Google Scholar]
- 11.Wu M, Gu JT, Yi B, et al. microRNA-23b regulates the expression of inflammatory factors in vascular endothelial cells during sepsis. Exp Ther Med. 2015; 9:1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beers MF, Morrisey EE. The three R’s of lung health and disease: repair, remodeling, and regeneration. J Clin Invest. 2011; 121:2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Kwan BC, Lai FM, et al. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol. 2012; 36:412–418 [DOI] [PubMed] [Google Scholar]
- 14.Ladak SS, Ward C, Ali S. The potential role of microRNAs in lung allograft rejection. J Heart Lung Transplant. 2016; 35:550–559 [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Zong J, Li Y, et al. MiR-744-3p regulates keratinocyte proliferation and differentiation via targeting KLLN in psoriasis. Exp Dermatol. 2019; 28:283–291 [DOI] [PubMed] [Google Scholar]
- 16.Chen YQ, Wang XX, Yao XM, et al. Abated microRNA-195 expression protected mesangial cells from apoptosis in early diabetic renal injury in mice. J Nephrol. 2012; 25:566–576 [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Han X, Tang Y, et al. miR-744 enhances type I interferon signaling pathway by targeting PTP1B in primary human renal mesangial cells. Sci Rep. 2015; 5:12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin J, Jenkins RH, Bennagi R, et al. Post-transcriptional regulation of transforming growth factor beta-1 by microRNA-744. PLoS One. 2011; 6:e25044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal R, Altamura S, Stanke F, et al. 138 Dysregulation of epithelial miR-148b contributes to goblet cell metaplasia, inflammation and alveolar damage in cystic fibrosis lung disease. J Cyst Fibros. 2015; 14:S93 [Google Scholar]
- 20.Miscianinov V, Martello A, Rose L, et al. MicroRNA-148b targets the TGF-β pathway to regulate angiogenesis and endothelial-to-mesenchymal transition during skin wound healing. Mol Ther. 2018; 26:1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitson BA, Prekker ME, Herrington CS, et al. Primary graft dysfunction and long-term pulmonary function after lung transplantation. J Heart Lung Transplant. 2007; 26:1004–1011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


