Abstract
CASP13 has investigated the impact of sparse NMR data on the accuracy of protein structure prediction. NOESY and 15N-1H residual dipolar coupling data, typical of that obtained for 15N,13C-enriched, perdeuterated proteins up to about 40 kDa, were simulated for 11 CASP13 targets ranging in size from 80 to 326 residues. For several targets, two prediction groups generated models that are more accurate than those produced using baseline methods. Real NMR data collected for a de novo designed protein were also provided to predictors, including one data set in which only backbone resonance assignments were available. Some NMR-assisted prediction groups also did very well with these data. CASP13 also assessed whether incorporation of sparse NMR data improves the accuracy of protein structure prediction relative to non-assisted regular methods. In most cases, incorporation of sparse, noisy NMR data results in models with higher accuracy. The best NMR-assisted models were also compared with the best regular predictions of any CASP13 group for the same target. For 6 of 13 targets, the most accurate model provided by any NMR-assisted prediction group was more accurate than the most accurate model provided by any regular prediction group; however, for the remaining 7 targets, one or more regular prediction method provided a more accurate model than even the best NMR-assisted model. These results suggest a novel approach for protein structure determination, in which advanced prediction methods are first used to generate structural models, and sparse NMR data is then used to validate and/or refine these models.
Keywords: CASP, Structure Prediction, Sparse NMR Data, Simulated NMR Spectra, Residual Dipolar Coupling, Protein Modeling, Contact Prediction
INTRODUCTION
Since its inception, CASP has been a driving force in the field of contact prediction and contact-directed modeling (see for example references [1–5]). Conceptually, even a few accurate native contacts could reliably guide de novo fold predictions, or provide valuable information for selecting among alternate models. During the CASP10 International Meeting, it was suggested that rather than using predicted contacts, which at the time were not very reliable, it might be more productive to explore the impact of a few real experimental contacts, as can be obtained from NMR, cross-linking, fluorescence energy transfer, or other experimental methods. This concept developed in CASP11 into the first NMR-assisted contact prediction experiment [6–8]. In the meantime, the accuracy of contact prediction based on evolutionary sequence co-variance analysis and machine learning has increased dramatically [5, 9–14], making the original proposal of replacing predicted contacts with real, sparse experimental contacts moot. None the less, the concept of combining sparse experimental data with sophisticated modeling methods is an important and emerging area of integrative structural biology, and CASP provides an important venue for testing and developing such hybrid methods. In CASP12 and CASP13, this integrative approach was also explored using small angle X-ray scattering [15] and chemical cross-link data [15]. Such integrative data-driven protein prediction is evolving into an important approach for structural biology.
The NMR Community has also explored automated NMR structure analysis in the context of the Critical Assessment of Protein Structure Determination by NMR (CASD-NMR). In this series of studies [16–19], NOESY peak lists and NMR resonance assignments for 20 small proteins were distributed to several groups developing fully automated nuclear Overhauser effect spectroscopy (NOESY) assignment and structure determination methods. In the first phase, CASD-NMR 2010, NOESY peak lists for 10 proteins were preprocessed to be relatively free of noise peaks, and automated structure determination was carried out in a blind fashion, without knowledge of the manually refined reference structure. It was observed that with such data, several fully automated NOESY analysis program could consistently deliver structures with backbone rmsd’s < 2.5 Å from the manually-refined reference structure, demonstrating the feasibility of routine, fully automated protein structure determination by NMR. These results were extended in CASD-NMR 2013, a similar blinded study using uncurated, noisy NOESY peak lists. Across the entire set of more than 140 models submitted in this phase, 70% of all entries had a backbone accuracy relative to the reference NMR structure better than 1.5 Å backbone rmsd, with some methods having up to 100% of their submitted models within 1.5 Å rmsd. However, using these uncurated NOESY peak lists, some automated structure determination methods did not converge for some targets. These studies provide benchmark results demonstrating strengths and weaknesses of several programs for fully automated NOESY analysis and structure generation of small (< 15 kDa) proteins.
While these CASD-NMR studies were very successful with these relatively small proteins, determining larger-sized protein (20 – 70 kDa) structures by solution NMR is extremely challenging but highly feasible [20, 21]. For such larger proteins, perdeuteration becomes necessary to circumvent the efficient spin relaxation properties resulting from their slow rotational correlation times. Backbone and sidechain amide hydrogens (HN) can be exchanged back into the protein structure, allowing collection of HN-HN NOE data, and some methyl and/or aromatic groups can be protonated by biosynthetic methods [22, 23]. However, aside from such selectively protonated side-chain moieties, replacing most protons in the protein structure with deuterons also eliminates most long-range and sidechain NOESY information. The difficulty in determining accurate structures with no, or limited, side-chain information (i.e. sparse NMR data) is a major technological challenge to the modeling community that currently limits routine application of solution NMR to larger systems.
In CASP11, we explored this challenge together with the global CASP community [6–8], by providing interatomic contacts derived from sparse nuclear Overhauser effect (NOE) data simulated from the X-ray crystal structure coordinates of 19 CASP template-free modeling targets, assuming perdeuteration with selective protonation of backbone amide and certain methyl groups. These targets ranged in size from 108 to 462 residues. The results were compared with baseline modeling results using some of the more successful automated structure determination programs assessed in CASD-NMR, including the ASDP program [24, 25]. While most NMR-assisted CASP11 methods could not provide accurate models using these sparse experimental data, a few groups (e.g., Lee, Baker) submitted models for several targets that were more accurate than those generated using “conventional” baseline automated NOESY analysis methods. These results demonstrate the strong synergy between the computational NMR and protein prediction communities, as each has the potential to learn from one another.
In CASP13, we extended our NMR-assisted structure prediction study begun in CASP11. NMR data were simulated from CASP free modeling targets with realistic degrees of incompleteness and noise, typical to that observed in real NMR spectra of perdeuterated, selectively protonated proteins. These free modeling targets ranged from 80 to 326 residues. In addition to contacts based on simulated 3D NOESY data, simulated residual 15N-1H dipolar coupling (RDC) data, and dihedral restraints as derived from backbone chemical shift data, for a subset of residues were also provided. In some cases, contact predictions from evolutionary sequence co-variance analysis were also provided to predictors. Two real NMR data sets were also made available to the CASP13 prediction community. These results further drive the field of integrated protein structure modeling by exploring the impact of sparse experimental data in enhancing the power of protein structure prediction methods.
METHODS
Experimental NMR Structure Determination
NMR studies were performed using a uniformly 15N,13C-enriched sample of a de novo designed protein, named foldit3 [26], CASP13 target 1008. The synthetic codon optimized gene (Genscript, Inc), designed to exclude ACA nucleotide sequences [27, 28 ], was cloned into plasmid pET15TEV_NESG [29]. The resulting protein product includes a short N-terminal 6xHis purification tag, followed by a TEV protease cleavage site, which was removed prior to data collection. Details of the production and characterization of this sample have been described elsewhere [26], and are also provided in the Supplemental Material. Homogeneity (> 97%) was validated by SDS polyacrylamide gel electrophoresis. The purified protein was dialyzed against 20 mM potassium phosphate, pH 6.5, and the protein concentration was adjusted to between 0.3–0.4 mM for NMR studies.
All NMR spectra were recorded at 25 ºC using cryogenic NMR probes. NMR data were collected on a Bruker AVANCE III 600 MHz spectrometer, processed using the program NMRPipe [30], and analyzed using the programs SPARKY [31] and XEASY [32]. Spectra were referenced to external DSS. Sequence-specific resonance assignments were determined using AutoAssign software [33, 34] together with interactive manual analysis. NMR data collection included simultaneous 15N,13C-edited 3D NOESY and 15N-edited 3D NOESY, both recorded with mixing time τm = 120 ms. Backbone dihedral angle constraints were then derived from the assigned chemical shifts using the program TALOS_N [16–19] for residues located in well-defined secondary structure elements. The programs ASDP [24, 25] and CYANA [35, 36] were used to automatically assign NOEs and to generate 3D structures, respectively. NOESY peak lists used for NMR-assisted predictions in CASP13 were all based on fully-automated NOESY peak assignment with ASDP.
For structure refinement, RPF analysis [37, 38], comparing observed and predicted NOESY peak lists, was used to guide iterative cycles of noise/artifact peak removal, peak picking, and NOESY peak assignments. The 20 conformers with the lowest target CYANA function value were then refined by restrained molecular dynamics in explicit water [39] using the program CNS [40]. Structural statistics and global structure quality factors were assessed using the PSVS 1.5 [41] and PDBStat [42] software packages. The global goodness-of-fit of the final structure ensembles with the NOESY peak list data, the NMR DP score, was determined using the RPF program [37, 38].
Baseline Modeling with ASDP-Cyana
Baseline modeling was carried out using state-of-the-art “conventional” methods for modeling protein structures from NMR data. NMR structures were modeled using the ASDP program for NOESY peak assignment, together with Cyana for structure generation from the resulting restraints. This pipeline, described in detail elsewhere [25], was one of the top performing automated NOESY analysis methods in the CASD-NMR experiments [16–19]. ASDP uses expert system methods to assign NOESY cross peaks, and to generate distance restraints. These restraints are then input to a structure generation program. In this case, structures were generated from the restraints using the restrained molecular dynamics in torsion angle space module of the program Cyana. The resulting intermediate structure models are then used to iteratively rule in / rule out additional NOESY cross peak assignments [24]. The final structures generated in several cycles of NOESY peak assignment and model generation are then refined with these NMR restraints active using Rosetta, with loop remodeling and core repacking, as described elsewhere (Mao et al. 2013).
CASP Assessment Units and Assessment Metrics
Simulated sparse NMR data were provided to CASP13 predictors for 11 proteins and protein domains (Table 1, CASP Targets), ranging in size from 80 to 326 residues. In addition, real NMR data were provided for one target, protein T1008 (aka, foldit3) [26], of 80 residues. In this case, two different real NMR were made available, differing in the completeness of the assignment of the NMR frequencies. Submitted prediction models were assessed by standard CASP metrics [43–45]. Summed or averaged Z scores for each metric were computed by the CASP Prediction Center [46]. NMR DP scores [37], comparing the short 1H-1H distances in prediction models with the NOESY peak list, and 15N-1H RDC Q scores [47, 48] were provided for each model submitted to the CASP Prediction Center for statistical analysis.
Table 1.
CASP13 NMR-Assisted Prediction Targets, Assessment Units and NMR Data
| No. of Residues | CASP Assessment Unit (AU) | No. of RDCs | No. of Dihedral Restraints | No. of Possible Contacts | Average Ambiguity per Contact | Maximum Ambiguity per Contact | |
|---|---|---|---|---|---|---|---|
| Simulated NMR Data | |||||||
| N0957s1 | 163 | N0957s1-D1 | 95 | 202 | 5582 | 5 | 50 |
| N0957s1-D2 | |||||||
| N0968s1 | 123 | N0968s1 | 62 | 128 | 1506 | 2 | 16 |
| N0968s2 | 115 | N0968s2 | 59 | 118 | 2088 | 4 | 32 |
| N0980s1 | 105 | N0980s1 | 43 | 87 | 1489 | 3 | 18 |
| N0981-D1 | 86 | N0981-D1 | 32 | 66 | 538 | 2 | 10 |
| N0981-D2 | 80 | N0981-D2 | 26 | 54 | 504 | 2 | 8 |
| N0981-D3 | 203 | N0981-D3 | 64 | 130 | 4701 | 4 | 32 |
| N0981-D4 | 111 | N0981-D4 | 42 | 90 | 1093 | 2 | 10 |
| N0981-D5 | 127 | N0981-D5 | 58 | 122 | 1983 | 3 | 21 |
| N0989 | 246 | N0989-D1 | 100 | 194 | |||
| N0989-D2 | 7095 | 5 | 90 | ||||
| N1005 | 326 | N1005 | 154 | 320 | 49,887 | 11 | 92 |
| Real NMR Data | |||||||
| N1008 | 80 | N1008 | N/A | 148 | 2,273 | 5 | 54 |
| n1008 | 80 | a | N/A | 148 | 29,205 | 9 | 169 |
Target n1008 is a control real NMR data set with essentially complete backbone and sidechain resonance assignments, and was not included in the calculations of summed Z score metrics.
Accuracy of submitted models was evaluated at the domain level. Z scores were calculated for a total of 14 assessment units (domains) listed in the third column of Table 1. The Z score analysis excluded the combined domain constructs N0957s1-D1.D2 and N0989-D1.D2, and also the n1008 target for which full NMR assignments were available, although structure accuracy metrics for these are also available on the CASP13 Prediction Site. The relative performance of participants was established based on the combination of Z scores calculated from per-target distributions of evaluation scores.
Sidechain Rotamer Analysis
In order to assess the accuracy of predicted structures against a reference structure, a useful metric of structure quality is the accuracy of side-chain rotamer states for well-defined (i.e., converged), buried (i.e., not on the protein surface) side chains [49]. PDBStat [42] is a computer program originally developed as a universal coordinate and protein NMR restraint converter. Its primary function is to provide a user-friendly tool for interconverting between protein coordinate and NMR restraint data formats. It also provides an integrated set of computational methods for protein structure quality assessment. Here, the PDBStat program was extended for assessing the agreement of sidechain χ1 and χ2 rotamer states between predicted and reference protein structures. This automated sidechain analysis protocol of PDBStat was used to assess NMR-assisted protein structure predictions in CASP13.
The χ1 and χ2 rotamers for all residues in each reference structure were assigned to the nearest g+, t, or g− conformational state. Side chains with solvent accessible surface area (SASA) less than 40 Å2 in the reference structure (calculated using the program Molmol [50]) were considered as buried side chains. In considering NMR structure ensembles, side chains whose χ1 (or χ2) dihedral angle values had standard deviation of < 30 degrees were considered as ‘converged side chains’. For NMR-derived reference structures, the medoid conformer of the ensemble (i.e. the conformer most similar to all of the other conformers [42, 51]) was selected as the representative structure.
Organization of Simulated and Real Data for CASP Predictors
The NMR data packages distributed to CASP13 participants for each target are summarized in Supplementary Table S1. Data consisted of an Ambiguous Contact List (described below), a Table of Dihedral Angle Restraints, and a Table of RDC Values (for two alignments), where available. For several targets, residue-residue contact predictions, based on multiple sequence alignments (Evolutionary Contacts, ECs) from the Meta PSI COV server [14], were also provided. For bookkeeping, the participants also were provided a FASTA file with the protein sequence. All files were distributed as tab-separated text files, to facilitate data ingestion, compressed in a single archive. Simulated and experimental data were organized and distributed in essentially the same manner. All simulated and experimental data distributed to participants are available on the CASP13 web site (http://www.predictioncenter.org/casp13/index.cgi), as well as from the Zenodo web site (DOI: 10.5281/zenodo.3386805).
RESULTS
Simulation of Resonance Assignments
Sequence specific resonance assignments were simulated from the atomic coordinates of the X-ray crystal structures of 11 CASP-NMR targets (excluding targets N1008 and n1008 for which real NMR data were generated for this study). First, any selenomethione (MSE) residues in the original PDB coordinate file of the reference X-ray structures were changed to methionine (MET). Hydrogen atoms were then added to the coordinates of X-ray crystal structures with the program Reduce [52]. The resulting coordinates were then used to simulate 1H, 13C, and 15N chemical shift values using the program SHIFTX2 [53]. As one goal of the CASP13 experiment is to explore the impact of NMR data obtainable from NMR studies of larger proteins on the accuracy of structure prediction, resonance assignments were all simulated assuming a perdeuterated protein sample with typical selective reprotonation [22, 23]. Specifically, only backbone HN, N, Cα, and C’ atoms, sidechain Cβ atoms, and the C and H atoms of Ile(δ1), Leu, Val and Ala methyl groups were included in the simulated chemical shift table. This proton labeling pattern corresponds to that provided by the application of typical selective labeling strategies used for studies of proteins in the size range 20 to 70 kDa. It was assumed that individual stereospecific assignments of the isopropyl methyls of Leu and Val were not available, and no corrections were made to account for deuterium isotope effects on bound 13C chemical shift values.
Simulation of NOESY Peak Lists
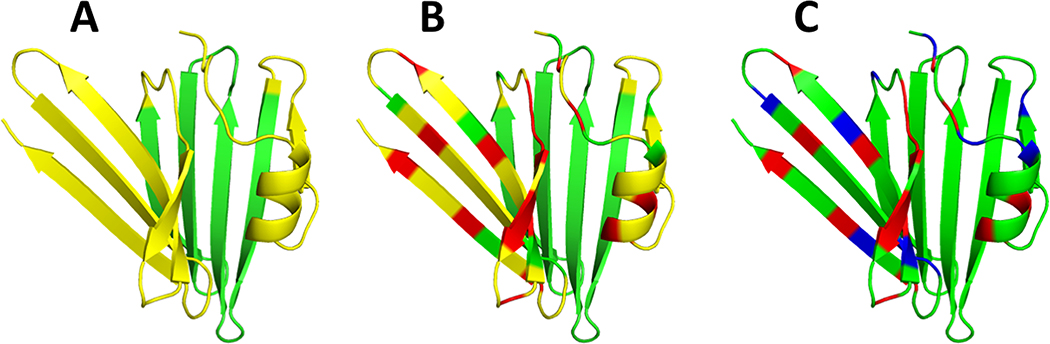
In order to create incomplete NOESY peak lists like those observed with real NMR data, a number of resonance assignments were deleted prior to simulating the NOESY spectra. In this process, illustrated for a representative target in Figure 1, our expertise in protein NMR studies was used to simulate the effects of line broadening due to conformational dynamics and/or weak spectra in causing “missing resonances”. First, we created a list containing selected regions for each CASP target proposed to exhibit missing resonances and/or NOESY cross peaks (e.g., yellow residues in Figure 1A). The choice of region to select was made so as to simulate the effects of local dynamics which could plausibly result in exchange broadening. This typically included surface loop residues and/or potentially dynamic secondary structures. Within each of these regions, we randomly selected 25% of the residues and deleted all chemical shift assignments for these residues (e.g., red residues in Figure 1B).
Fig. 1. Schematic Depiction of Process for Simulating Missing NOESY Peak and Resonance Assignment Data for CASP-NMR Target N0968s1.
(A) Ribbon representation of the backbone structure of T0968s1 showing the locations (in yellow) of candidate regions for NOESY peak and/or resonance assignment deletion. (B) Residues (red) for which all resonance assignments are deleted prior to generating the NOESY peak list. This results in the elimination of potential NOESY peaks from the simulated spectra due to, for example, conformational exchange broadening. (C) Residues (blue) for which, after the NOESY cross peaks are generated, all resonance assignments are deleted. These data simulate the case where NOESY peaks are observed, but one or more of the corresponding 1H resonances are not assigned. These NOESY cross peaks are present in the NOESY peak list, but cannot be correctly assigned, and often result in incorrect NOESY cross peak assignments due to accidental degeneracies with observable resonances.
These chemical shifts were then used, together with 1H-1H distances from the atomic coordinates, to simulate 3D 13C-edited and 3D 15N-edited NOESY peak lists (frequencies and intensities) which would be obtained for perdeuterated 13C,15Nenriched, backbone 1HN and ILVA 13C-1H3 methyl labeled proteins. For all potential 1H-1H NOEs, a summation distance [42] was calculated from the atomic coordinates of all degenerate proton resonances,
| Eqn (1) |
If the resulting rij was less than a cutoff distance Dcutoff, 3D 13C- or 15N-edited NOE cross peaks were simulated with intensity of 10,000 / r6ij. The maximum observable interproton distance was set to 5 Å. NOESY “cross peaks” (frequencies and intensities in a NOESY peak list), representing these short inter-proton distances, were then created between these resonances. Adjacent NOESY peaks created within tolerances of 0.02 ppm for the direct H dimension, 0.2 ppm for the C/N, and 0.03 ppm for the indirect H dimension were merged, and the corresponding resonance frequencies were averaged, to simulate overlapped regions in the NOESY spectra. Short 1H-1H distances between protons in residues for which resonance assignments were deleted did not generate any NOESY peak, corresponding to False Negative, FNs.
After simulating these NOESY peak lists (i.e., the resonance frequencies and intensities of observable NOESY cross peaks, excluding cross peaks involving resonances that are missing due to exchange broadening), an additional 15% of the residues in the resonance assignment list were randomly selected and their chemical shift values were removed from the chemical shift list, while preserving the corresponding NOESY peaks (e.g., blue residues in Figure 1C). This process simulates the situation where the NOESY cross peak is present, but one or both of the corresponding sequence-specific resonance assignments cannot be determined. These NOESY cross peaks cannot be correctly matched to the original 1H-1H pair, and have the potential to be incorrectly assigned. Finally, weak NOESY noise peaks were added to the NOESY peak lists, at frequency positions consistent with assigned resonances, but not corresponding to short 1H-1H distances in the reference structure. These are false positive NOESY peaks, FPs. The contacts indicated by these FP peaks are generally inconsistent with the native protein structure. This similar process of resonance assignment deletions (resulting in FNs and incorrect NOESY cross peak assignments) and random addition of weak NOESY peaks (resulting in FPs) was applied in simulating 13C- and 15N-edited 3D NOESY spectra peak lists from reference X-ray structures of 11 CASP-NMR targets (excluding targets N1008 and n1008 for which real NMR data were available).
NMR data were simulated assuming a monomer structure for all targets. No efforts were made to simulate interfacial X-filtered NOESY data; homodimers were simulated as the single protomer. Targets N0957s1 and N0989 are two-domain proteins, in which each domain is well-defined relative to the other. Hence, the complete two-domain coordinates were used to simulate NOE and RDC data assuming the same static orientation of domains as observed in the corresponding X-ray crystal structures. Target N0980 is a dimer of heterodimers (2:2 tetramer). Since one chain is a small polypeptide, we simply used the coordinates of the single protomer of the larger subunit as the target for simulating NMR data. Target N0981 is a five-domain structure, in which the domains are likely to be independent of one another. Hence, each domain was treated as an independent target, and NOESY and RDC data were simulated separately for each of them.
Statistics on Simulated and Real NOESY Peak Lists
Statistics on the NOESY peak list data for each target are summarized in Supplementary Table S1 and Supplementary Figure S1. The process of simulating NOESY peak lists outlined here provided data sets with properties similar to those generally obtained for uniformly 15N,13C-enriched, perdeuterated proteins with ILVA methyl 13C-1H3 labeling. Analysis of these NOESY peak lists against the reference atomic coordinates showed that 5 to 18% of short distances in the reference structures have no corresponding NOESY peak, and 5 to 10% of the NOESY peaks in these lists cannot be assigned to any true short 1H-1H distance. For these simulated NOESY peak lists, the data ranged from 3.5 to 9.3 NOESY peaks per residue. This compares to 8.4 NOESY peaks per residue for the sparse real data set N1008, and 43.4 NOESY peaks per residue for the generally complete experimental dataset of n1008. Generally speaking, the low restraint density (10 NOESY peaks / residue) of the simulated data sets, and of the real data set N1008, make these targets challenging for NMR-based structural determination using traditional methods.
These NOESY data sets (summarized in Supplementary Table S1) are not only sparse (incomplete), but they also include significant numbers of false peaks which cannot be satisfied by the correct structure. For the simulated datasets, 6.2 to 9.1% of the peaks in the NOESY spectra cannot be satisfied by the reference structure. For real data set N1008, the fraction of peaks with possible assignments for which none are consistent with the native structure is even higher, 19.7%. This is because this protein sample was fully protonated, but its NOESY peak list was analyzed using only the backbone resonance assignments, including HN and Hα resonances; cross peaks involving sidechain atoms with chemical shifts similar to backbone atoms were thus often incorrectly assigned (uniquely or ambiguously) as backbone-backbone NOEs, making this real data set particularly challenging. For the real data set n1008, which included essentially complete backbone and sidechain assignments, only about 0.2% of the NOESY peaks cannot be explained by the final, refined solution NMR structure, a hallmark of high quality NMR data and structures [54].
Generation of Ambiguous Contact Lists
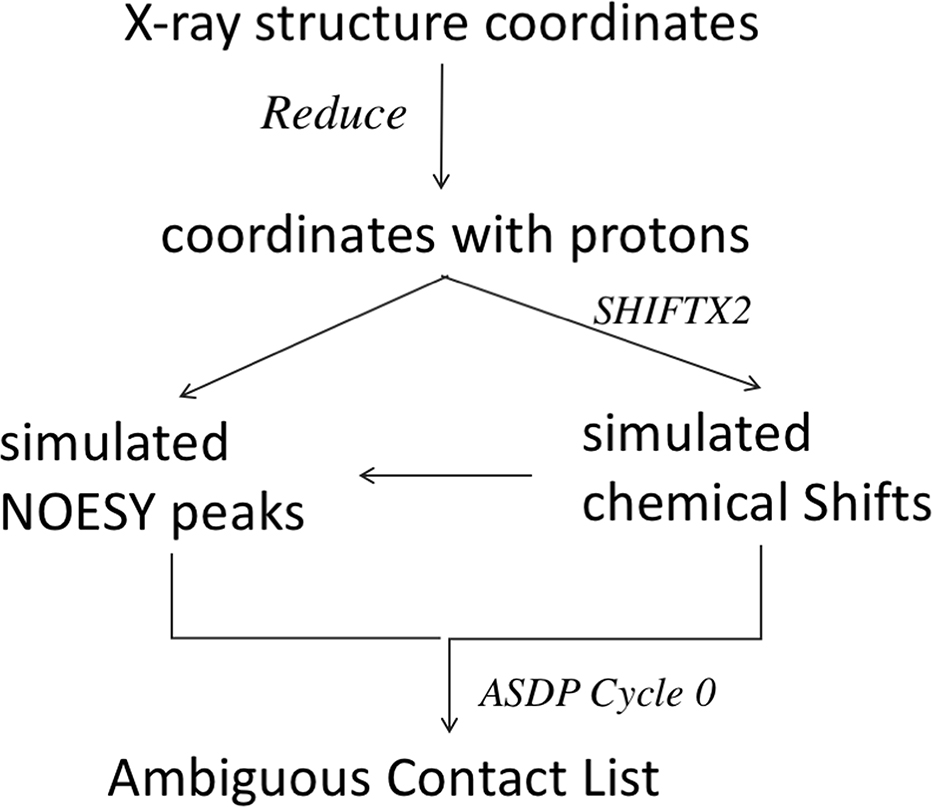
The ideal input for NMR-assisted prediction would be the unassigned NOESY peak lists together with sequence-specific NMR assignments and RDC data, as was done in the CASD-NMR project [16–19]. However, in order to reduce the extent of domain specific NMR spectroscopy knowledge required for participation in CASP13, the organizers instead provided these NOESY data as “Ambiguous Contact Lists” (Supplementary Figure S2). For each NOESY peak, the Ambiguous Contact List provides the set of 1H-1H pairs which, within a defined frequency tolerance of matching the NOESY peak frequencies to the chemical shift frequencies, are possible assignments for each NOESY peak.
Ambiguous Contact Lists were generated by analyzing simulated (or real) NOESY peak lists together with the corresponding resonance assignment lists, without knowledge of the target 3D structure. The resonance assignment list was first modified to simulate the small inconsistencies generally seen between peak frequencies measured in the NOESY spectrum and the corresponding frequencies in the resonance assignment list. Random noise shifts were added to the chemical shift values in each dimension, with standard deviation 0.01 ppm for direct 1H dimension, 0.20 ppm for indirect C/N, and 0.02 ppm for the indirect 1H dimension. 3D NOESY peak lists were then analyzed together with these resonance assignment lists using the Cycle 0 module of the program ASDP. This algorithm assigns NOESY cross peaks to one or more potential 1H-1H interactions based on chemical shift matching. These initial assignments use information, based on backbone chemical shift, on the locations of α-helices and β-strands, inter strand alignments, and other topological rules derived automatically from distances within standard secondary structures, to reduce the ambiguity of NOESY cross peak assignments, as described elsewhere [24]. The Cycle 0 ASDP analysis was executed with match tolerances of 0.03 ppm for H atoms and 0.30 ppm for C/N atoms, and with parameters Dcutoff = 5.0 Å and Dupper = 7.5 Å.
In this protocol, outlined in Figure 2, only one cycle of ASDP was executed. If peaks could be uniquely assigned by the algorithms of ASDP cycle 0, the unique assignments for these particular NOESY peaks, including any unique long-range HN-HN NOEs important for β-strand alignments, were included in the Ambiguous Contact Lists. Short-range intra-residue and sequential NOESY peak assignments (|i-j| ≤ 2) were excluded. For each remaining NOESY peak, the output of ASDP Cycle 0 provided all possible proton pair assignments within the defined resonance frequency match tolerances. In practice, each NOESY peak is assigned to a set of ambiguous 1H-1H pair assignments whose chemical shifts are compatible with the resonance frequencies associated with the 3D NOESY peak. In the absence of experimental errors, at least one of these 1H-1H pairs should correspond to a short-distance interproton interaction that is consistent with the native protein structure. However, NOESY peaks that arise from unassigned resonances, as well as random noise peaks, will provide a set of ambiguous contacts, or possibly even a unique assignment, none of which are consistent with the native structure. We calibrated the number of added noise peaks so that number of FP contacts due to these random noise peaks did not exceed 10% (except for real data set N1008) of the final ambiguous restraint list. The resulting Ambiguous Contact Lists (Supplementary Figure S2) were provided to CASP predictors.
Fig. 2.
Process Flow Chart for Generating Ambiguous Contact Lists for NMR-Assisted CASP13.
Backbone Dihedral Angle Restraints
For relatively static protein conformations, backbone chemical shift data can be used to make reliable predictions of backbone dihedral angle values based on statistical assessment against the database of protein chemical shifts and local structures [53]. In this work, we observed that dihedral angle restraints computed using the program Talos_N [55], from chemical shifts predicted from the atomic coordinates with the program SHIFTX2, were not always consistent with the X-ray crystal structure used to predict the chemical shifts. This probably reflects shortcomings in the accuracy of these chemical shift predictions. In order to provide the kind of restraint data based on backbone chemical shifts that would be available using real NMR data, the backbone dihedral angle restraints for residues with “observed” and “assigned” backbone chemical shifts (i.e. for residues that were not deleted from the chemical shift list) were also provided to CASP predictors. These dihedral restraints were provided as ranges, in which two random numbers between 5 and 30 degrees were added and subtracted from the dihedral angle value observed in the reference X-ray structure.
Simulation of 15N-1H Residual Dipolar Coupling (RDC) Data
RDCs arise from the interaction of two magnetically active nuclei in the presence of the external magnetic field of an NMR instrument. In solution NMR studies, this interaction is normally reduced to zero due to the isotropic tumbling of molecules in their aqueous environment. The introduction of partial order to the molecular alignment reintroduces dipolar interactions by minutely limiting isotropic tumbling. This partial order can be introduced in numerous ways, including inherent magnetic anisotropy susceptibility of molecules, incorporation of artificial tags (such as lanthanides) that exhibit magnetic anisotropy, or using a liquid crystal or otherwise partially-ordered aqueous solution.
The RDC interaction phenomenon has been formulated in different ways. To harness the computational synergy of RDC data, in this study we have utilized the matrix formulation of this interaction as shown in Eqn (2). The matrix S shown in Eq (2) and (3) represents the Saupe order tensor matrix (the ‘order tensor’) that can be described as a 3×3 symmetric and traceless matrix. Dmax in Eqn (2) is a nucleus-specific collection of constants, rij is the separation distance between the two interacting nuclei (in units of Å), and vij is the corresponding normalized internuclear vector
| Eqn (2) |
| Eqn (3) |
| Eqn (4) |
The software package REDCAT [56, 57] used this formalism to simulate 15N-1HN RDCs for the target proteins. REDCAT uses the protein structure and an order tensor S to calculate RDCs using Eqns. 2–4. For each of the target protein structures, two different order tensors were calculated using the software package PALES [58]. PALES utilizes a steric collision model to calculate order tensors in different simulated alignment media. In this work, two different simulated alignment media were utilized: bicelle (wall-like structures) and phage (rod-like structures). The concentration used for both simulations was 0.05 units. The resulting 15N-1H RDC for the “observed” and “assigned” residues of each target were provided to CASP13 predictors.
Summary of NMR Data
Table 1 also summarizes the numbers of RDCs, dihedral restraints, and ambiguous contacts (NOESY peaks) provided to predictors for each target. The number of all possible atom pair assignments for all NOESY peaks provide by ASDP Cycle 0 ranged from 504 (for target data set N0981-D2) to 49,887 (for target data set N1005). These NOE-based contacts, provided to CASP predictors. had an average of 2 to 11 possible atom pair assignments per contact, with maximum ambiguity of 92 possible assignments per contact.
Assessment of Baseline NMR Assisted Modeling with ASPD
Baseline modeling was carried out for each target using the ASDP software program for NOESY peak assignment and restraint generation. These “baseline structures” were modeled in “blinded fashion”, in which simulated NOESY and RDC data were provided to one of the authors (JH) without her knowledge of the reference structures, and structures were generated from these data using the conventional automated NOESY assignment and modeling algorithms of the ASDP software. ASDP takes as input the NOESY peak and resonance assignment lists, from which the Ambiguous Contact Lists were derived. For this reason, the ASDP baseline calculations used these NOESY peak lists, rather than the Ambiguous Contact Lists, as input. Peaks in these unassigned NOESY peak lists were labeled only by the corresponding resonance frequencies, and did not include any link to the table of chemical shift assignments. NOESY peaks were assigned and disambiguated using ASDP, these data were interpreted as calibrated distance restraints, and the resulting assigned distance restraints were used to generate structural models with the software CYANA [59]. The resulting structures were further refined using restrained Rosetta refinement [60], as outlined in the Methods section.
Baseline models were generated using three protocols. Baseline_Group 321 provided 5 models generated using the simulated (or real) sparse NOESY, dihedral, and RDC data, without EC contact predictions. Baseline_Group 459 provided 5 models generated using these same data, plus EC contact predictions from the Meta PSI COV server [14], which were also provided to all predictors. A third set of models was generated for each target using ECs from the EVFold contact prediction pipeline [61], run locally for this study. Alignments were generated using five jackhmmer [62] iterations against the Uniref100 sequence database (February 2018 release), with multiple normalized bitscore thresholds ranging from 0.1 to 0.9 (with T0981 subsequently run at 0.03). Alignments were chosen based on maximizing both the effective number of sequences and the non-gap coverage of each position. Pseudolikelihood maximization was then used to compute evolutionary couplings using the alignments, with the default settings found at the evcouplings.org webserver. These ECs were then combined with NMR data to generate EC-NMR structures, and the top 5 scoring models were selected. The resulting 15 models for each target (5 from NMR alone, 5 from EC-NMR using Meta PSI COV, and 5 from EC-NMR using EVFold ECs) were then assessed using the DP score “NMR R-factor” metric, which compares the contact map for the NMR-derived model against the NOESY peak list [37, 38]. The five models with highest DP score were then submitted as Baseline Group 313.
These baseline models were then assessed by the CASP Prediction Center. All three baseline groups (ASDP Baseline_Groups 313, 321, and 459) had similar overall accuracy performance based on GDT-TS, GDT_HA, GDT_ALL, GDC_SC, SphereGrinder, and RPF assessment metrics (these metrics are described in [45] and [44]). In general, modeling accuracy was highest for ASDP Baseline_Group 313 (best DP score), followed by ASDP Baseline_Group 459 (with Meta PSI COV ECs), and then ASDP Baseline_Group 321 (without ECs). Interestingly, using the knowledge-based MolProbity assessment score, the highest quality structures were those generated by protocol ASDP Baseline_Group 321 (without ECs), while ASDP Baseline_Group 459 (with PSI COV ECs) had significantly poorer MolProbity scores, suggesting that inclusion of contact predictions in these protocols can distort models from their best atomic packing conformations. For the sake of simplicity, in the remaining analysis we utilize only the ASDP Baseline_Group 321 (without ECs) and ASDP Baseline_Group 459 (with PSI COV ECs) as the baseline comparison results.
Initial Assessment of NMR Assisted Predictions
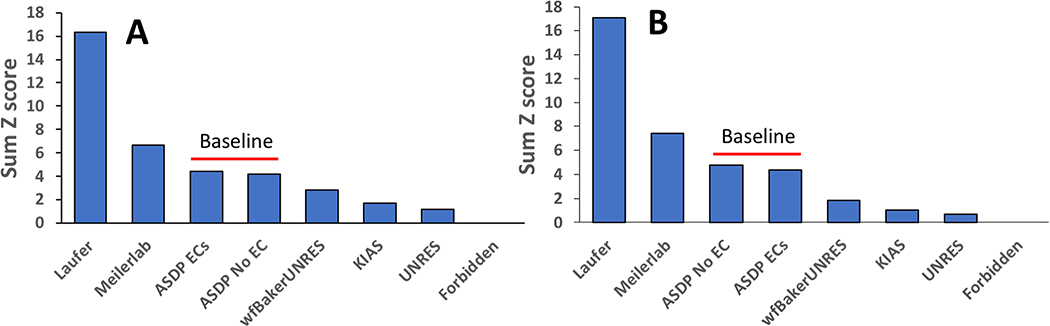
Six CASP13 predictor groups participated in this NMR-guided prediction experiment; Forbidden (122), KIAS-Gdansk (208), Meilerlab (250), UNRES (288), Laufer (431), and wf-Baker-UNRES (492). An initial ranking of the six NMR-guided prediction groups, along with two baseline groups, was done using summed GDT-TS Z scores for the first-ranked model submitted for each predictor group (Figure 3A), as described elsewhere [43, 63, 64]. For the calculation of the summed GDT-TS Z scores we used the common convention of setting the Z score = −2 for any model with Z score ≤ −2 [43, 63, 64]. This is done so as not to heavily penalize the worst models, and to encourage the exploration of new (perhaps less successful) methods in CASP.
Fig. 3. Summed Z Score Ranking of CASP13 NMR-Assisted Predictions.
(A) Summed GDT-TS Z scores. (B) Weighted summed Z scores for multiple metrics, as described in text.
Figure 3A demonstrates that two prediction groups (Laufer and Meilerlab) generally provided more accurate models than the baseline groups. The same conclusion was drawn by considering the ‘best of 5’ models from each group, and also for the individual assessment measures GDT_HA, GDT_All, GDC_SC, Sphere Grinder, and RPF. However, in ranking using only MolProbity scores, groups Laufer and Meilerlab are reversed in their relative order; apparently group Meilerlab, using the Rosetta force field and fragment libraries, does a better job of generating better packed and more physically plausible conformations, with better MolProbity scores.
Principal Component Analysis (PCA) on Assessment Metrics
A good CASP prediction should be both similar to its corresponding experimentally derived target structure and physically reasonable. Therefore, the ranking of predicted structures for a given CASP target should incorporate statistics, such as the GDT-TS [65], quantifying how accurately a structure models a target, as well as measures of biophysical structure quality such as the MolProbity score [66]. Incorporation of multiple measures of structure accuracy and quality into a single ranking involves either folding multiple statistics into a single composite score or using consensus methods to combine rankings based on multiple measures of structure quality and accuracy into a single composite ranking. As in previous template-based modeling CASP experiments [43, 63, 64], our final ranking of NMR data assisted predictions in CASP13 combines multiple structure evaluation statistics using a weighted sum. Thus, the question of how to rank predictions of a given target reduces to a question of finding appropriate weights to use in adding together a selection of measurements of structure quality and accuracy.
We chose to use for evaluation of NMR-data assisted structure predictions superposition-dependent global measures of structure accuracy (GDT-HA and GDT-SC), a local superimposition dependent local measure of structure accuracy (SphereGrinder), measurements of the accuracy of interatomic contact areas (CAD-AA) and contact distances (RPF), and a measurement of the physical reasonableness of the structure (MolProbity score).
A recent analysis [44] of protein structure evaluation scores indicates that most methods for structure evaluation are highly correlated. We observed this also for the set of metrics we used for assessing NMR-assisted structure predictions (Supplementary Table S2). The MolProbity score, however, is less correlated to the other scores, and provides complementary information. While the MolProbity score was distinct, none of the scores was inconsistent with each other according to Friedman’s test. In general, models with reasonable accuracy were (as judged by MolProbity score) physically reasonable structures, although some models with good Molprobity scores were not particularly accurate. Inaccurate models with good MolProbity scores have also been observed in assessments of incorrect homology models [67] and of inaccurate CASD-NMR experimental NMR structures [18, 19].
The high correlation between structure evaluation statistics suggests that Principal Component Analysis (PCA) may be a useful ad hoc method to calculate weights for summing multiple measurements of structure quality and accuracy. PCA identified that a composite score of 0.442*Z_GDT_HA + 0.449*Z_GDT_SC + 0.425*Z_RPF + 0.428*Z_SphGrdr + 0.433*Z_CAD_AA + 0.227*Z_MolProbity (where Z_[X] indicates the z-score calculated on a per target basis, using the first model provided by each predictor, from quality measure [X]) explains approximately 87% of the variance in structure evaluation scores (Supplementary Table S3). Setting all Z-scores below a certain threshold yields a similar composite statistic via PCA. We rounded the coefficients of the PCA score explaining the highest amount of variance down to the nearest tenths to create the following linear regression: 0.40 for GDT-HA, GDC-SC, RPF, SphereGrinder and CAD-AA and 0.20 for MolProbity. Note that while PCA is a useful tool for constructing a composite metric for assessing structure prediction quality, we do not necessarily expect PCA-derived weights will be similar from one CASP dataset to the next.
Overall Ranking Based on PCA-defined Combination of Scores
The final ranking of NMR-assisted predictions, using the combined weighted Z sores of GDT-HA (0.4), GDT-SC (0.4), RPF(0.4), SphereGrinder (0.4), CAD-AA (0.4) and MolProbity (0.2) is illustrated in Figure 3B for the predictor-designated “first” models. Again, in this analysis Z score is set to −2 for any model with Z score ≤ −2. Similar results were obtained by selecting the best-scoring model out of the 5 submitted. These rankings are essentially the same as those obtained using the GDT-TS (or other individual metrics) alone. Relative to the baseline groups, two prediction groups (Laufer and Meilerlab) generally provided more accurate models, while the remaining four groups had somewhat poorer accuracy performance. For the 14 assessment units (AUs, listed in column 3 of Table 1), the top prediction groups were Laufer 431 for 7 AUs, Meilerlab 250 for 3 AUs, ASDP Baseline_Group 321 (without ECs) for 2 AUs, and ASDP Baseline_Group 459 (with PSI COV ECs) for 2 AUs, respectively. Groups wf-Baker-UNRES and KIAS-Gdansk also outperformed the ASDP Baseline_Groups on 3 and 1 AU, respectively. These results demonstrate that for 10 of 14 AUs, two CASP13 predictor groups - Laufer and Meilerlab – submitted first-ranked models for many targets that are more accurate than those generated using our conventional automated ASDP modeling protocol.
Target N1008 – Real NMR Data with Backbone Assignments Only
Two real NMR data sets (N1008 and n1008) were provided for the data-guided prediction program of CASP13. Both data sets were for the CASP COMMONS target T1008 (foldit3), proposed by Brian Koepnick and David Baker as part of their project assessing de novo protein design by citizen scientists in the online protein-folding game Foldit (Koepnick et al, 2019). Foldit players were provided a set of general principles for protein design in the form of Foldit rules, and the resulting designs were assessed by Rosetta stability calculations. One hundred and fifty-six designs were encoded in synthetic genes, which were expressed, screened for stability, and (in 4 cases) experimental structures were determined (Koepnick et al, 2019). One of the protein designs, the 80-residue foldit3 protein, was produced for CASP13 with uniform 15N,13C-enrichment, and its structure was determined by conventional triple-resonance NMR in the context of this project. The structural statistics and global structure quality factors including Verify3D ([68]), ProsaII [69], PROCHECK ([70]), and MolProbity [66] raw and statistical Z-scores were computed using the Protein Structure Validation Suite Software PSVS 1.5 [67] and PDBStat [42] software packages. The global goodness-of-fit of the final structure ensembles with the NOESY peak list data, the NMR DP score, was determined using the RPF analysis program. The resulting reference 3D structure of foldit3, CASP target 1008, exhibits excellent convergence and structure quality statistics (Supplementary Table S4). This structure and the associated data have been deposited in the Protein Data Bank (PDB id 6msp) and chemical shifts have been deposited in the BioMagResDataBase (BMRB id 30527).
Ambiguous Contact Lists for target 1008 were provided to CASP13 predictors as two distinct NMR-assisted targets. In the first cycle, NMR-assisted target N1008, backbone resonance assignments (only) were combined with complete 15N- and 13C edited NOESY spectra and Talos_N backbone dihedral restraints (derived from backbone chemical shift data) as input to the program ASDP. Following one cycle of analysis with ASDP, the structure-independent NOESY peak assignments (most of which are assigned to multiple possible 1H-1H interactions) were used to generate the Ambiguous Contact List for target N1008. This CASP13 target explores a novel approach to NMR structure determination, in that 1H-1H NOE interactions due to backbone-sidechain and sidechain-sidechain contacts are present in the NOESY peak list, but cannot be correctly assigned as the sidechain resonances are not present in the chemical shift list. For the second target data set, NMR-assisted target n1008, the nearly complete backbone and sidechain resonance assignments were combined with complete 15N- and 13C-edited NOESY data and Talos_N backbone dihedral restraints, as input to ASDP. Following one cycle of analysis with ASDP, the structure-independent NOESY peak assignments (most of which, again, are assigned to multiple possible 1H-1H interactions) were used to generate Ambiguous Contact List n1008. In this case backbone-sidechain and sidechain-sidechain NOEs could generally be reliably assigned. Note that no RDC or EC data are available for target 1008. CASP13 predictor models provided using Ambiguous Contact Lists N1008 and n1008 were all assessed against the final manually-refined NMR structure.
Data set N1008 was designed to test the ability to combine backbone-only assignments with advanced structure prediction methods. Several studies have previously explored the combination of sparse NMR data obtained on perdeuterated protein samples with advanced molecular modeling methods [7, 8, 71–78]. For such data sets, the NOESY cross peak assignments are not complicated by the presence of unassigned sidechain resonances. However, in the case of N1008, the backbone resonances were assigned in a fully protonated 13C,15N-enriched protein sample; hence the NOEs may arise from backbone/backbone, backbone/sidechain, or sidechain/sidechain interactions. Since the sidechain resonances are not in the chemical shift list, many backbone/sidechain NOEs may be incorrectly assigned as unique backbone/backbone interactions; this is particularly problematic for NOEs involving resonances which are degenerate with assigned backbone HN and Hα protons. The resulting falsely assigned backbone – backbone contacts might be expected to corrupt the structure. The goal of this experiment is to assess if data-guided predictions could overcome such corruption and provide an accurate 3D structure without sidechain assignments. In practice, the complicating effects of NOEs involving sidechain protons can be overcome using protein samples with perdeuterated Hα and sidechain resonances. However, here we explored the potential of avoiding perdeuteration, completing backbone assignments, and using such noisy NOESY data for accurate structure determination.
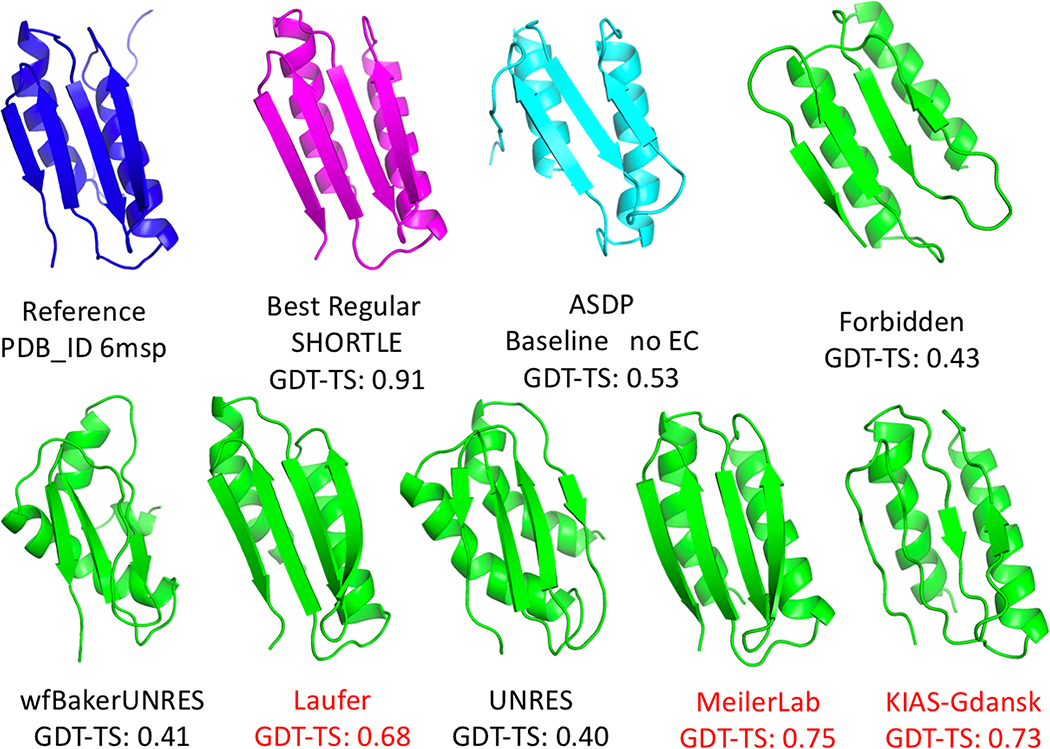
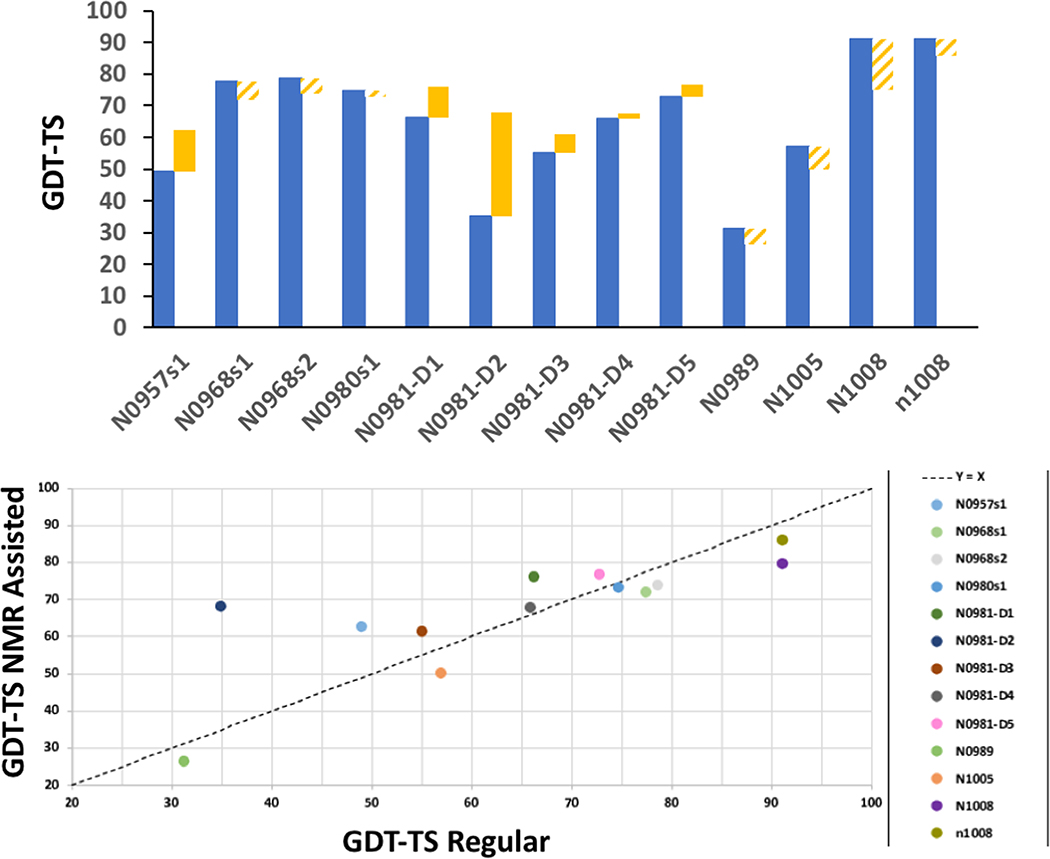
CASP13 results for target N1008 are illustrated in Figure 4. While these NOESY peak lists contain large numbers of NOE peaks which cannot be correctly assigned using the backbone chemical shift list, three prediction groups did very well with these data: Meilerlab 250 (GDT-TS 0.75), KIAS-Gdansk 208 (GDT-TS 0.73), and Laufer 431 (GDT-TS 0.68). The results of the ASDP baseline group 321 (GDT-TS 0.53) was significantly less accurate (note that Baseline group 459 did not contribute a distinct structure because no ECs are available for this de novo designed protein). For this N1008 data set, the first selected model of predictor groups were 15 to 22 GDT-TS points higher than the best models provided by the significant value of these modeling methods in obtaining accurate structures from sparse, noisy NOESY data.
Fig. 4. NMR-guided predictions for real NMR data target N1008.
Target N1008 was produced with uniform 15N-,13C-enrichment (without perdeuteration). The backbone chemical shift assignments were used together with 3D 15N-NOESY data to generate Ambiguous Contact List, which include many NOESY peaks involving sidechain resonances for which no assignment in list of possible assignments is correct. Blue – experimental solution NMR structure. Magenta – best non-assisted regular prediction. Lt blue – ASDP Baseline model. Green – NMR-assisted prediction results.
Although target N1008 was a NMR-data assisted CASP target, some of the “regular predictions”, which did not use any NMR data, were also quite good. In particular, SHORTLE 281 (GDT-TS 0.91) (Figure 4), A7D-DeepMind 043 (GDT-TS 0.81), and other regular predictor groups did remarkably well with this target, and significantly better than any group could do using these sparse NMR data. It should be noted that target 1008 is a de novo designed protein, and may be more amenable to accurate structure prediction compared to natural proteins. None the less, these results suggest hybrid methods in which models generated with regular methods are simply validated or refined against NMR data could be used for data sets like that provided for N1008.
Target n1008 – Real NMR Data with Extensive Backbone and Sidechain Assignments
Data set n1008 was provided as a control for performance with essentially complete NMR assignments. A reliable NMR-assisted prediction method should do well with these data. Because n1008 was among the very last data sets released for CASP13, only four predictor groups, plus Baseline_Group 321, ASDP without ECs, submitted models for n1008. For this data set, Baseline Group 321 provided the most accurate top ranked model, with GDT-TS 0.83. Laufer-431 also submitted a good first-ranked model (GDT-TS 0.57), followed by UNRES 288 (GDT-TS 0.41), Forbidden 122 (GDT-TS 0.40), and wf-Baker-UNRES 492 (GDT-TS 0.27). These results highlight the value of such control data in testing and developing NMR-assisted prediction methods, as the n1008 control data set is an important benchmark for testing various methods.
Overall Performance Per Target Per Group
Table 2 provides a summary of overall performance for the six predictor groups and two Baseline groups (321No EC or 459 Meta-PSI-COV EC), based on GDT-TS score of the top-ranked submission, per group, and per target. The table is color coded so that GDT-TS scores > 0.50 (correct fold) are colored in shades of green, and scores < 0.50 in shades of red. Certain targets (e.g. N0981-D4) appear to be relatively easy, as most groups submitted good models with GDT-TS > 0.50 while other targets (e.g. N0989, N0981-D2, N0981-D3, and N1005) were more difficult. Not surprisingly, the largest targets (> 200 residues) were all among the most difficult targets. These results suggest that more efforts are needed even by the best NMR-assisted prediction methods, for addressing larger perdeuterated proteins where only sparse NMR data can be collected.
Table 2.
GDT-TS Scores for First Ranked Models for Each Target and Each NMR-Assisted Predictor Group
| Target | Nres | Best Regular Prediction | 492 wfBaker UNRES | 431 Laufer | 288 UNRES | 250 Meilerlab | 208 KIAS-Gdansk | 122 Forbidden | ASDP Baseline No EC | ASDP Baseline With EC |
|---|---|---|---|---|---|---|---|---|---|---|
| N0957s1 | 162 | 45.2 A7D |
31.9 | 52.9 | 28.8 | 56.0 | NA | 20.5 | 32.2 | 30.2 |
| N0989 | 246 | 31.3 Zhang |
12.7 | 23.5 | 13.9 | 17.5 | NA | 10.7 | 15.4 | 16.6 |
| N0968s1 | 123 | 71.4 Elofsson |
64.6 | 59.5 | 45.3 | 69.0 | NA | 31.5 | 59.7 | 54.6 |
| N0968s2 | 115 | 78.7 A7D |
60.0 | 73.7 | 55.4 | 43.2 | NA | 30.4 | 33.7 | 49.5 |
| N0980s1 | 105 | 54.8 Multicom |
29.8 | 67.7 | 25.0 | 59.8 | NA | 28.6 | 62.0 | 72.1 |
| N0981-D1 | 86 | 66.2 slbio_server |
49.4 | 58.4 | 53.7 | 55.5 | 61.0 | NA | 70.3 | 69.7 |
| N0981-D2 | 80 | 34.0 Venclovas |
NA | 40.0 | 33.7 | 34.0 | 42.1 | NA | 64.3 | 67.5 |
| N0981-D3 | 203 | 55.1 A7D |
37.9 | 41.0 | 39.0 | 17.4 | 49.3 | NA | 55.7 | 55.1 |
| N0981-D4 | 111 | 65.9 Multicom |
50.6 | 65.7 | 47.7 | 61.7 | 59.6 | NA | 60.1 | 58.1 |
| N0981-D5 | 127 | 72.8 A7D |
53.1 | 76.5 | 39.7 | 59.8 | 40.9 | NA | 38.9 | 25.9 |
| N1005 | 326 | 56.3 A7D |
28.9 | 49.8 | 26.4 | 36.2 | 29.2 | NA | 33.9 | 29.4 |
| N1008 | 97 | 91.2 SHORTLE |
40.5 | 68.1 | 40.2 | 75.0 | 73.0 | 42.8 | 52.9 | NA |
| n1008 | 97 | 91.2 SHORTLE |
27.2 | 57.4 | 41.5 | NA | NA | 40.2 | 82.7 | NA |
Sidechain Rotamer Metrics
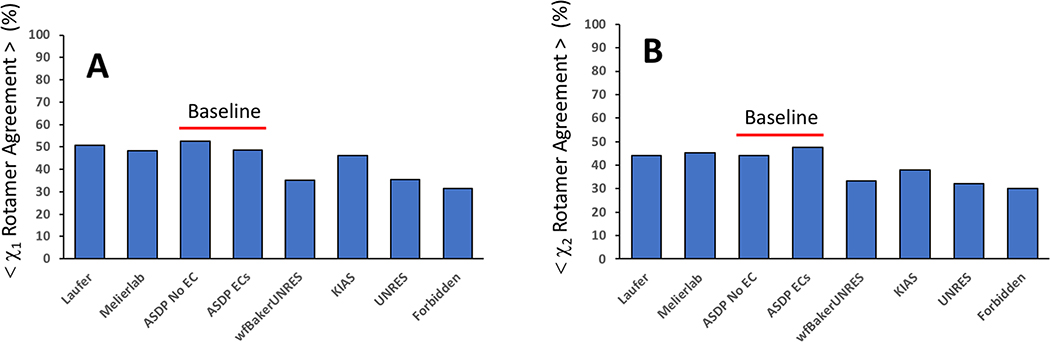
Another valuable structure quality metric involves comparing the sidechain conformations of buried residues in predictor models versus the reference structure [49]. Both χ1 and χ2 sidechain rotamer states for residues with buried side chains were compared between the first-ranked predicted models and the corresponding reference structure using the PDBStat program [42], as described elsewhere [49]. Groups Laufer 431, Meilerab 250, and KIAS-Gdansk 208, well as ASDP Baseline_Groups 321 and 459, provided models with significantly better than average χ1 and χ2 sidechain rotamer agreement with reference structures, compared with the other predictor groups (Figure 5). However, all of the predictor groups have average χ1 and χ2 rotamer agreement of only 30 to 60%, indicating that this is a valuable metric which should be focused on in future CASP experiments.
Fig. 5. Sidechain χ1 and χ2 Rotamer States.
Percent of (A) χ1 or (B) χ2 rotamer states for buried residues in each assessment unit that match the corresponding dihedral angle of the reference structure in the best prediction models submitted by each prediction group.
RDC Q-Scores for NMR-Assisted Prediction Models
The RDC Q-score is a measure of the agreement between RDC values calculated from the model, and the RDC data. The Q-score ranges from 1 to 0, with lower values indicating better agreement between calculated and observed RDCs. The average 15N-1H RDC Q-scores for each of the six predictor groups ranged from 0.49 to 0.83 (Supplementary Figure S3). These scores are significantly poorer than those of the baseline groups, which range from 0.19 to 0.21. Among the predictor groups, the best average RDC Q-scores were for models submitted by Meilerlab (average RDC Q-score 0.49) and Laufer (average RDC Q-score 0.63). The submitted models of the remaining four predictor groups have poor RDC Q-scores. These results demonstrate that all of the data-guided predictor groups could improve model accuracy by better consideration of RDC data in their prediction algorithms.
DP Scores for NMR-Assisted Prediction Models
The NMR DP score [37, 38] is a “NMR R factor”, comparing the short 1H-1H distances in a protein structure model with all possible assignments of peaks in the NOESY peak list, considering the available chemical shift data. The NMR DP score ranges from 0 to 1, and is correlated with structural accuracy. Correct structures generally have DP scores > 0.6 [37, 38, 54]. The DP scores for each of the six predictor groups ranged from 0.47 to 0.69 (Supplementary Figure S4). These scores are generally lower than those of the baseline groups, which range from 0.73 to 0.75. However, ASDP uses the DP score to guide the automated NOESY peak analysis process. Among the predictor groups, the best average DP scores were for NMR-assisted models submitted by Laufer (average DP score 0.69) and Meilerlab (average DP score 0.63). DP scores for each reference X-ray crystal and NMR structure, and for the best regular and NMR-assisted models submitted for each target are summarized in Supplementary Figure S5. These are generally consistent with the corresponding GDT-TS scores. Overall, the NMR-assisted predictor rankings based on DP scores are consistent with the rankings based on GDT-TS and other conventional CASP metrics, placing Laufer and Meilerlab as the best performing NMR-assisted prediction groups in CASP13.
DISCUSSION
Simulations of NOESY Data for CASP Targets
Simulated data provide an important tool for computational methods development. Although it is challenging to accurately simulate something as complex as a protein NOESY peak list, the powerful advantage of such simulated data is that the ground truth structure is known. Using real data has the advantage of including effects which are not captured in simulated data. For example, protein dynamics and signal overlap are primary causes of errors in the conversion of NMR observables into structural restraints, and may be difficult to account for in simulating NOESY data. However, with real data the “true” structural distribution from which these data arise is generally not known.
Normally, one cannot assign the frequencies of resonance of all nuclei in a protein. In practice, two scenarios may occur: either there are no observable peaks involving a given nucleus, because of local dynamics preventing their detection, or the resonance cannot be assigned to a unique atom with confidence, e.g. due to accidental degeneracies. In the latter case, peaks are observable but cannot be converted into the appropriate structural restraints. For this work, we introduced both types of problems in our simulations, by manually selecting for each target regions of protein sequence from which resonance assignment were deleted. To simulate missing NOESY data for assigned resonances, removed some resonance assignments before simulating the NOESY peak list. To simulate missing assignments of resonances which do provide NOESY peaks, we removed resonance assignments for some residues after simulating the NOESY spectrum, while retaining the NOESY peaks. This second situation is actually common in sparse NMR data sets, and can lead to restraints that are incorrect. For example, mis-assignments of NOESY peaks due to missing resonance assignments are particularly extensive for the real NMR data set N1008, in which only backbone resonance assignments were provided while the NOESY peak list includes NOEs with unassigned sidechain resonances. Importantly, some of the modeling methods used in this data assisted CASP13 experiment were able to overcome the challenges of these real data provided for target N1008, to provide accurate structures without the need for sidechain resonance assignments or perdeuteration of the protein sample. This represents a novel approach to small protein structure determination by NMR.
In CASP11, one successful strategy used by the Baker lab was to focus their initial NMR-guided predictions on the uniquely assigned NOE-based contacts provided in the Ambiguous Contact Lists (7). Supplementary Table S5 provides an analysis of the unique long-range, and unique HN-HN long-range, contacts in the Ambiguous Contact Lists provided to CASP13 predictors. These ranged from 0.8 to 1.6 uniquely-assigned long-range contacts per residue, and from 0.24 to 0.58 HN-HN long-range contacts per residue, similar to the distributions provided in the CASP11 Ambiguous Contact Lists, which were also based on simulated NOESY peak lists. For the real sparse NMR data set, target N1008, the Ambiguous Contact List has 0.54 and 0.28 unique long-range and unique HN-HN long-range contacts, respectively. These densities of long-range contacts in this real NMR data set are similar to, but at the lower-end, of the ranges provided in the simulated Ambiguous Contact Lists.
Impact of NMR Data in Improving Regular Predictions
An important question to be addressed in this NMR data guided prediction component of CASP13 is whether incorporation of sparse experimental data can improve the accuracy of prediction. To assess this, we compared the best “regular prediction” model with the best “NMR data assisted” model, where both the regular and assisted models were provided by the same predictor group; i,e, for Forbidden, KIAS-Gdansk, UNRES, Laufer, and wf-Baker-UNRES. Meilerlab did not provide a “regular prediction”, precluding this analysis. The CASP organizers recognize that predictor groups may have utilized different modeling methods for their regular and “NMR assisted” predictions, or even used models submitted by other predictor groups, which became available between the release of “regular” and “data assisted” targets, making these comparisons not as rigorous as we might like.
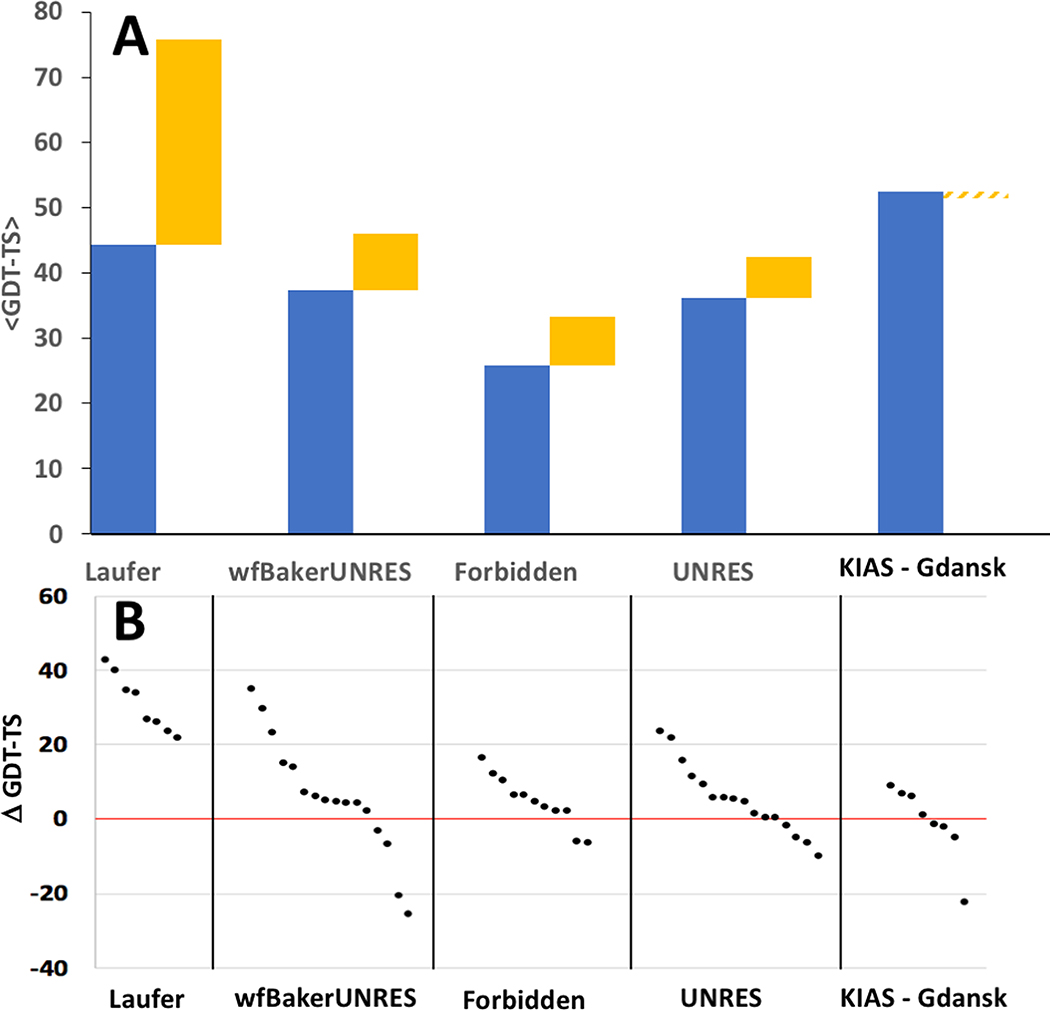
Our results show that, in most cases, incorporation of NMR data results in models with much higher accuracy predictions. NMR-assisted prediction models are, on average, more accurate than the corresponding regular prediction by the same group (Figure 6A). Modeling methods that used NMR data generally improved accuracy of prediction over modeling methods used by the same groups without NMR data. In some cases, the improvement for particular targets was as much as 40 GDT-TS points (Figure 6B). The improvement was particularly dramatic for groups Laufer (average improvement across all targets of 25 GDT-TS points; maximum improvement on a specific target of 42 GDT-TS points) and wf-Baker-UNRES (average improvement across all targets of 5 GDT-TS points; maximum improvement on a specific target of 39 GDT-TS points). Hence, we conclude that sparse, noisy NMR data can generally improve model prediction accuracy. In some cases, however, incorporating these simulated or real NMR data resulted in reduced model accuracy for some targets; e.g. most of the predictor groups submitted first-ranked “data assisted” targets which are less accurate than their corresponding “regular” predictions (Figure 6B). For group KIAS-Gdansk 208, this results in an average reduction in GDT-TS score across all targets of - 5 GDT-TS points (Figure 6A). These results suggest that more efforts are needed by predictor in implementing sparse NMR data in their data-guided prediction algorithms.
Fig. 6. Impact of Sparse NMR Data on Modeling Accuracy.
Where a NMR-assisted prediction group also submitted regular prediction models, the corresponding best regular and best NMR-assisted predictions were compared. (A) Stacked column plot showing the average GDT-TS scores for best models submitted for each assessment unit as a regular prediction (blue) or NMR-assisted prediction (yellow). (B) Δ GDT-TS = GDT-TSassisted – GDT-TSregular for the best NMR-assisted and best regular model submitted for each target by each group providing both regular and assisted predictions.
NMR-Assisted Predictions of Larger Proteins
None of the NMR-assisted groups did particularly well with the three larger (> 200 residue) targets (Table 2). For these targets, most NMR-assisted models have GDT-TS < 0.50. The most accurate predictions were those of the Laufer (2 of 3 targets) and ASDP baseline (1 of 3 targets) groups. This contrasts the results in CASP11, where two predictor groups, Lee and Baker, were particularly outstanding in modeling larger (>200 residues) proteins more accurately than baseline methods using sparse NMR data. Regrettably, neither these Lee or Baker groups of CASP11 participated in the NMR-assisted prediction component of CASP13.
The Best “Regular” Prediction for a Target was Often More Accurate than the Best “Data Assisted” Prediction
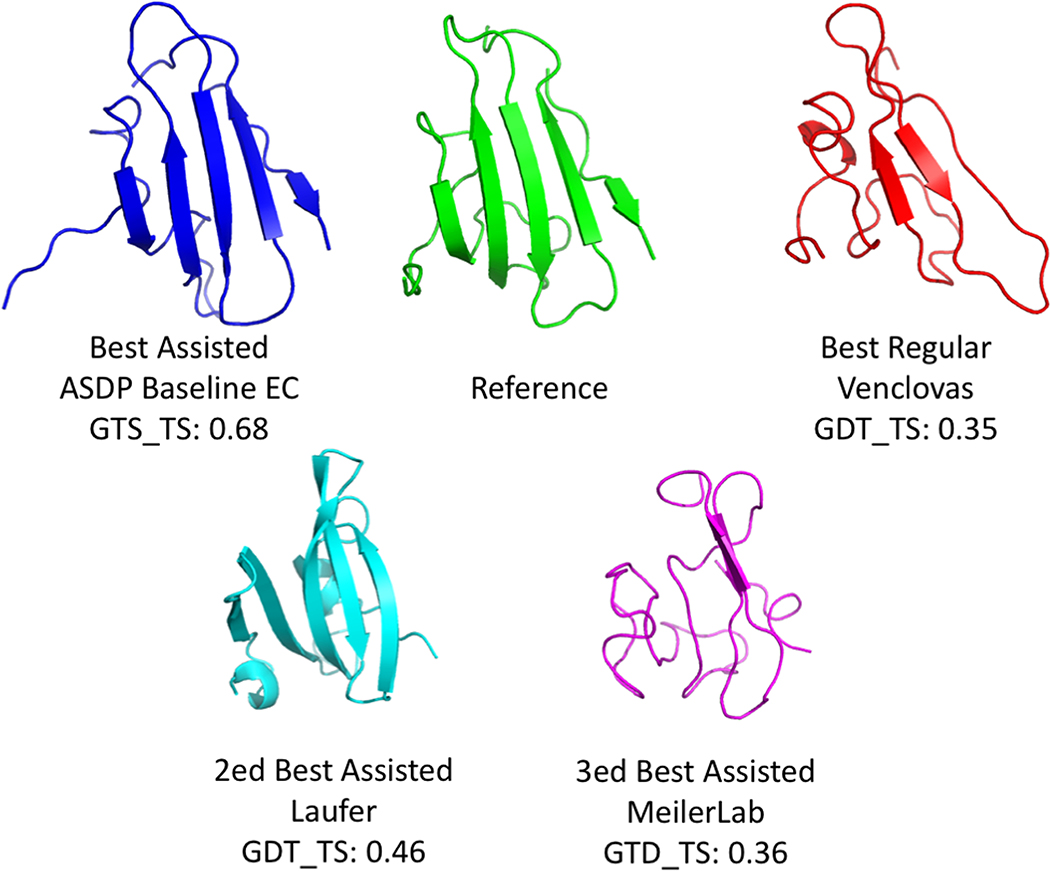
A second key question we wanted to address involves comparing the accuracy of all regular prediction methods with NMR data assisted predictions. For 6 of the 13 targets, the best NMR-assisted models were more accurate than any regular prediction (solid yellow histogram bars in Figure 7A). This improvement using the NMR data was particularly dramatic for target N0981-D2 (Figure 8). For this target, the best NMR-assisted model (Baseline_Group 313 ASDP No EC, GDT-TS 0.68) is significantly more accurate than the best unassisted regular model (Venclovas 366; GDT-TS 0.35). Interestingly, all 15 of the top-ranked assisted models for target N0981-D2 were from the Baseline groups 321, 459, and 313. This may reflect the nature of this fold, since the ASDP program uses algorithms designed to address the unique features of beta sheets [24]. The best NMR-assisted models submitted by predictor groups Laufer (GDT-TS 0.46) and Meilerlab (GDT-TS 0.36) were also more accurate than the best model from regular prediction groups. These results confirm the expectation that, generally speaking, inclusion of sparse NMR data improves the accuracy of predictions. Detailed descriptions of the methods used by Laufer [79] and Meilerlab [80] are presented in their own papers on NMR-assisted prediction in CASP13.
Fig. 7. Best NMR Assisted vs Best Regular Predictions.
(A) GDT-TS scores for the best model submitted by any NMR-assisted prediction group (blue bars) compared with the best model submitted by any regular prediction group (yellow bars). The solid yellow bars indicate targets for which the best model submitted by any regular prediction was more accurate than the best model submitted by any NMR-assisted prediction group. The hashed-yellow bars indicate targets for which the best model submitted by any regular prediction was less accurate than the best model submitted by any NMR-assisted prediction group. (B) Scatter plot comparing the GDT-TS scores for the best model submitted by any regular prediction versus the best model submitted by any NMR-assisted prediction.
Fig. 8. Comparison of Best Assisted and Best Regular Prediction Models Submitted for Target N0981-D2.
The best NMR-assisted models of the baseline ASDP (with EC) and Laufer groups are more accurate than the best models provided by any regular prediction group (Venclovas).
Although NMR-assisted modeling provide the best models generated by any methods for several targets, for 7 out of 13 targets used in the NMR-data assisted component of CASP13, the most accurate (best) model provided by a regular prediction group was actually more accurate than the most accurate model provided by any NMR-data assisted prediction group (hashed yellow histogram bars in Figure 7A). This is also evident by plotting the GDT-TS score for best model submitted by the NMR-assisted groups against the GDT-TS score for the best model of the corresponding target by any regular prediction group (Figure 7B); many of these comparisons fall below the diagonal indicating that at least one regular prediction group provided a more accurate model than the corresponding NMR-assisted model. Although in these cases, the improved accuracy of the non-assisted group is only marginal, they are non-the-less impressive because no sample-specific experimental data was used. The regular prediction groups providing these highly accurate “regular predictions” include groups A7D 043 (Deep Mind), Zhang 322, Venclovas 366, slbio_serve 266s, SHORTLE 281 and MULTICOM 083. Several of these regular prediction groups utilize novel machine learning methods to guide structural modeling. For the NMR-guided targets, group A7D 043 (Deep Mind) provided the most accurate models for 6 of 13 targets, and 10 of 16 assessment units; they also contributed 27 of 48 (i.e., 3 × 16 = 48) top three most accurate models. These remarkable results suggest a novel approach for structure determination using sparse NMR data, in which pure prediction methods, like the machine leaning methods being developed by Deep Mind, Zhang, MULTICOM, and other groups, are first used to generate structural models, and the sparse NMR data is then used validate and/or refine these models.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH NIGMS grants 1R01GM120574 (to G.T.M.), R01GM100482 (to K.F.), and P20 RR-016461 (to H.V.).
Footnotes
G. L. is an officer of Nexomics Biosciences, Inc. M. I. is a scientific advisor of Nexomics Biosciences, Inc. G. T. M. is a founder of Nexomics Biosciences, Inc.
REFERENCES
- 1.Lesk AM. CASP2: report on ab initio predictions. Proteins. 1997; Suppl 1:151–166. [DOI] [PubMed] [Google Scholar]
- 2.Monastyrskyy B, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact predictions in CASP9. Proteins. 2011; 79 Suppl 10:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins. 2014; 82 Suppl 2:138–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. New encouraging developments in contact prediction: Assessment of the CASP11 results. Proteins. 2016; 84 Suppl 1:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin A. Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age. Proteins. 2018; 86 Suppl 1:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin NV. Assessment of CASP11 contact-assisted predictions. Proteins. 2016; 84 Suppl 1:164–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ovchinnikov S, Park H, Kim DE, Liu Y, Wang RY, Baker D. Structure prediction using sparse simulated NOE restraints with Rosetta in CASP11. Proteins. 2016; 84 Suppl 1:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joo K, Joung I, Cheng Q, Lee SJ, Lee J. Contact-assisted protein structure modeling by global optimization in CASP11. Proteins. 2016; 84 Suppl 1:189–199. [DOI] [PubMed] [Google Scholar]
- 9.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011; 108:E1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011; 6:e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks DS, Hopf TA, Sander C. Protein structure prediction from sequence variation. Nat Biotechnol. 2012; 30:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ovchinnikov S, Kinch L, Park H, Liao Y, Pei J, Kim DE, Kamisetty H, Grishin NV, Baker D. Large-scale determination of previously unsolved protein structures using evolutionary information. Elife. 2015; 4:e09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovchinnikov S, Park H, Varghese N, Huang PS, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC, Baker D. Protein structure determination using metagenome sequence data. Science. 2017; 355:294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchan DWA, Jones DT. Improved protein contact predictions with the MetaPSICOV2 server in CASP12. Proteins. 2018; 86 Suppl 1:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogorzalek TL, Hura GL, Belsom A, Burnett KH, Kryshtafovych A, Tainer JA, Rappsilber J, Tsutakawa SE, Fidelis K. Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy. Proteins. 2018; 86 Suppl 1:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Guntert P, et al. Blind testing of routine, fully automated determination of protein structures from NMR data. Structure. 2012; 20:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Guntert P, Herrmann T, Huang YJ, Jonker HR, et al. CASD-NMR: critical assessment of automated structure determination by NMR. Nat Methods. 2009; 6:625–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosato A, Vranken W, Fogh RH, Ragan TJ, Tejero R, Pederson K, Lee HW, Prestegard JH, Yee A, Wu B, Lemak A, Houliston S, et al. The second round of Critical Assessment of Automated Structure Determination of Proteins by NMR: CASD-NMR-2013. Journal of biomolecular NMR. 2015; 62:413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragan TJ, Fogh RH, Tejero R, Vranken W, Montelione GT, Rosato A, Vuister GW. Analysis of the structural quality of the CASD-NMR 2013 entries. Journal of biomolecular NMR. 2015; 62:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A. 2012; 109:10873–10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi P, Monneau YR, Xia Y, Ishida Y, Kalodimos CG. Toolkit for NMR Studies of Methyl-Labeled Proteins. Methods Enzymol. 2019; 614:107–142. [DOI] [PubMed] [Google Scholar]
- 22.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. Journal of molecular biology. 1996; 263:627–636. [DOI] [PubMed] [Google Scholar]
- 23.Gardner KH, Rosen MK, Kay LE. Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry. 1997; 36:1389–1401. [DOI] [PubMed] [Google Scholar]
- 24.Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006; 62:587–603. [DOI] [PubMed] [Google Scholar]
- 25.Huang YJ, Mao B, Xu F, Montelione GT. Guiding automated NMR structure determination using a global optimization metric, the NMR DP score. Journal of biomolecular NMR. 2015; 62:439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koepnick B, Flatten J, Husain T, Ford A, Silva DA, Bick MJ, Bauer A, Liu G, Ishida Y, Boykov A, Estep RD, Kleinfelter S, et al. De novo protein design by citizen scientists. Nature. 2019; 570:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Molecular cell. 2005; 18:253–261. [DOI] [PubMed] [Google Scholar]
- 28.Schneider WM, Inouye M, Montelione GT, Roth MJ. Independently inducible system of gene expression for condensed single protein production (cSPP) suitable for high efficiency isotope enrichment. J Struct Funct Genomics. 2009; 10:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, Janjua H, Kornhaber G, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods in enzymology. 2011; 493:21–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 31.Goddard TD, Kneller DG. Sparky 3. San Francisco, CA: University of California; 2000. [Google Scholar]
- 32.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. Journal of biomolecular NMR. 1995; 6:1–10. [DOI] [PubMed] [Google Scholar]
- 33.Zimmerman DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997; 269:592–610. [DOI] [PubMed] [Google Scholar]
- 34.Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods in enzymology. 2001; 339:91–108. [DOI] [PubMed] [Google Scholar]
- 35.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997; 273:283–298. [DOI] [PubMed] [Google Scholar]
- 36.Guntert P Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004; 278:353–378. [DOI] [PubMed] [Google Scholar]
- 37.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005; 127:1665–1674. [DOI] [PubMed] [Google Scholar]
- 38.Huang YJ, Rosato A, Singh G, Montelione GT. RPF: a quality assessment tool for protein NMR structures. Nucleic Acids Res. 2012; 40:W542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003; 50:496–506. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007; 66:778–795. [DOI] [PubMed] [Google Scholar]
- 42.Tejero R, Snyder D, Mao B, Aramini JM, Montelione GT. PDBStat: a universal restraint converter and restraint analysis software package for protein NMR. Journal of biomolecular NMR. 2013; 56:337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009; 77 Suppl 9:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olechnovic K, Monastyrskyy B, Kryshtafovych A, Venclovas C. Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics. 2019; 35:937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kryshtafovych A, Monastyrskyy B, Fidelis K. CASP prediction center infrastructure and evaluation measures in CASP10 and CASP ROLL. Proteins. 2014; 82 Suppl 2:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kryshtafovych A, Monastyrskyy B, Fidelis K. CASP11 statistics and the prediction center evaluation system. Proteins. 2016; 84 Suppl 1:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clore GM, Schwieters CD. How much backbone motion in ubiquitin is required to account for dipolar coupling data measured in multiple alignment media as assessed by independent cross-validation? J Am Chem Soc. 2004; 126:2923–2938. [DOI] [PubMed] [Google Scholar]
- 48.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Magn Reson. 2004; 167:228–241. [DOI] [PubMed] [Google Scholar]
- 49.Tang Y, Huang YJ, Hopf TA, Sander C, Marks DS, Montelione GT. Protein structure determination by combining sparse NMR data with evolutionary couplings. Nat Methods. 2015; 12:751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996; 14:51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 51.Struyf Anja, Hubert Mia, Peter. Clustering in an object-oriented environment. J Stat Software. 1997; 1:1–30. [Google Scholar]
- 52.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007; 35:W375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. Journal of biomolecular NMR. 2011; 50:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosato A, Tejero R, Montelione GT. Quality assessment of protein NMR structures. Curr Opin Struct Biol. 2013; 23:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Y, Bax A. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol Biol. 2015; 1260:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. Journal of magnetic resonance (San Diego, Calif : 1997). 2004; 167:228–241. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt C, Irausquin SJ, Valafar H. Advances in the REDCAT software package. BMC bioinformatics. 2013; 14:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zweckstetter M NMR: prediction of molecular alignment from structure using the PALES software. Nat Protoc. 2008; 3:679–690. [DOI] [PubMed] [Google Scholar]
- 59.Güntert P Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004; 278:353–378. [DOI] [PubMed] [Google Scholar]
- 60.Mao B, Tejero R, Baker D, Montelione GT. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. J Amer Chem Soc. 2014; 136:1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheridan R, Fieldhouse RJ, Hayat S, Sun Y, Antipin Y, Yang L, Hopf T, Marks DS, Sander C. EVfold.org: Evolutionary couplings and protein 3D structure prediction. bioRxiv. 2015; bioRxiv 021022. [Google Scholar]
- 62.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011; 79 Suppl 10:37–58. [DOI] [PubMed] [Google Scholar]
- 64.Huang YJ, Mao B, Aramini JM, Montelione GT. Assessment of template-based protein structure predictions in CASP10. Proteins. 2014; 82 Suppl 2:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zemla A LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003; 31:3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya A, Wunderlich Z, Monleon D, Tejero R, Montelione GT. Assessing model accuracy using the homology modeling automatically software. Proteins. 2008; 70:105–118. [DOI] [PubMed] [Google Scholar]
- 68.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992; 356:83–85. [DOI] [PubMed] [Google Scholar]
- 69.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993; 17:355–362. [DOI] [PubMed] [Google Scholar]
- 70.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993; 26:283–291. [Google Scholar]
- 71.Zheng D, Huang YJ, Moseley HN, Xiao R, Aramini J, Swapna GV, Montelione GT. Automated protein fold determination using a minimal NMR constraint strategy. Protein Sci. 2003; 12:1232–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins. 2009; 75:147–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010; 327:1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109:10873–10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li W, Zhang Y, Kihara D, Huang YJ, Zheng D, Montelione GT, Kolinski A, Skolnick J. TOUCHSTONEX: protein structure prediction with sparse NMR data. Proteins. 2003; 53:290–306. [DOI] [PubMed] [Google Scholar]
- 76.Shealy P, Simin M, Park SH, Opella SJ, Valafar H. Simultaneous structure and dynamics of a membrane protein using REDCRAFT: membrane-bound form of Pf1 coat protein. Journal of magnetic resonance (San Diego, Calif : 1997). 2010; 207:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cole CA, Ishimaru D, Hennig M, Valafar H. An investigation of minimum data requirement for successful structure determination of Pf2048.1 with REDCRAFT. Proceedings of the International Conference on Bioinformatics & Computational Biology (BIOCOMP):: The Steering Committee of The World Congress in Computer Science, Computer Engineering and Applied Computing (WorldComp); 2015. p. 17–24. [Google Scholar]
- 78.Timko E, Shealy P, Bryson M, Valafar H. Minimum data requirements and supplemental angle constraints for protein structure prediction with REDCRAFT. BIOCOMP 2008. p. 738–744. [Google Scholar]
- 79.Robertson JC, Nassar R, Liu C, Brini E, Dill KA, Perez A. NMR-assisted protein structure prediction with MELDxMD. Proteins. 2019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuenze G, Meiler J. Protein structure prediction using sparse NOE and RDC restraints with Rosetta in CASP13. Proteins. 2019. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.