Abstract
Hypoxia (i.e., low oxygen (O2) levels) is a common environmental challenge for several aquatic species, including fish and invertebrates. To survive or escape these conditions, these animals have developed novel biological mechanisms, some regulated by neuropeptides. By utilizing mass spectrometry (MS), this study aims to provide a global perspective of neuropeptides in the blue crab, Callinectes sapidus, and their changes over time (0, 1, 4, and 8 hours) due to acute, severe hypoxia (~10% O2 water saturation) stress using a 4-plex reductive dimethylation strategy to increase throughput. Using both electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) MS, this study provides complementary coverage, allowing 88 neuropeptides to be identified. Interesting trends include (1) an overall decrease in neuropeptide expression due to hypoxia exposure, (2) a return to basal levels after 4 or 8 hours of exposure following an initial response, (3) changes only after 4+ hours exposure, and (4) an oscillating pattern. Overall, this study boosts the power of multiplexed quantitation to understand the large-scale changes due to severe hypoxia stress over time.
Keywords: Callinectes sapidus, Quantitative peptidomics, Neuropeptide, Hypoxia, Reductive dimethylation, isotopic labeling
Graphical abstract

Introduction
Estuaries and coastal ecosystems are increasingly threatened by climate change, poorly managed wastewater, and agricultural and industrial runoff.1 These factors often lead to eutrophication of coastal waters, causing large algal blooms and subsequent hypoxic (i.e., low oxygen (O2)) episodes that can last for hours to days.2 Aquatic hypoxia also occurs naturally from a multitude of hydrodynamic and meteorological effects.3 During hypoxic episodes, the dissolved O2 in the water greatly decreases, causing massive dead zones and a reduction in biodiversity as organisms are deprived of oxygen. Environmental hypoxia occurs most frequently in the spring and summer and can last for months.3 As commercially-fished species rapidly perish during these times, the repercussions of hypoxia become economic as well as environmental.4
Although many aquatic organisms are affected by hypoxia, the blue crab, Callinectes sapidus, is of particular interest. The blue crab possesses both environmental and economic relevance as it is frequently fished from estuaries plagued by eutrophication and hypoxia.2 In the literature, hypoxia has been shown to cause decreased rates of reproduction, growth, and feeding, and increased mortality rates in aquatic species.5 Due to the adverse effects of hypoxia, the blue crab has developed interesting ways of surviving the low levels of dissolved O2. Prior studies have observed hypoxia-initiated defensive behaviors, including inactivity, self-burying, and migration towards shallower, more O2-rich waters.5 Additionally, the composition of hemocyanin (i.e., O2 transport protein analogous to hemoglobin) has been shown to change in response to hypoxia,6, 7 demonstrating physiological defensive mechanisms as well.
The variable behavioral and physiological changes in C. sapidus suggest the presence of complex signaling pathways involved in survival. Neuropeptides are short amino acid chains that act as signaling molecules within the nervous and neuroendocrine system. Previously, neuropeptides have been implicated in a range of environmental stress responses, including temperature8 and salinity fluctuations.9 They can have highly diverse effects within the body while also maintaining low in vivo concentrations.10 Prior research has shown that the crustacean hyperglycemic hormone (CHH) is a neuropeptide involved in regulating the response to hypoxia in the blue crab,11 while global dynamic changes of neuropeptides during acute hypoxia have not been systematically evaluated. By examining the neuropeptide expression changes in the blue crab, their role in survival can be better understood.
Unfortunately, the high chemical diversity, low in vivo concentrations, and rapid degradation of neuropeptides makes their study challenging. Mass spectrometry (MS)–using both matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI)–has proven to be an effective method of analyzing neuropeptides as it offers high sensitivity, high specificity, and can provide both quantitative and sequence information. Additionally, because it requires no prior knowledge of the analyte, MS is ideal for discovering novel neuropeptides involved in response to hypoxia. Relative quantitation of neuropeptides by MS is typically achieved by employing either MS1-based labeling strategies (e.g., reductive dimethylation (also known as dimethyl labeling), iDiLeu, and mTRAQ),12–14 or tandem MS (MS/MS or MS2-based) labels (e.g., iTRAQ, TMT, and DiLeu).15–17 MS/MS reporters require the neuropeptide be selected for fragmentation to be quantified. The low abundance of many neuropeptides, however, makes their selection for MS/MS less likely. For this reason, MS1-based labeling strategies are often selected for neuropeptide quantitative analyses.
Previously, experiments have utilized duplex reductive dimethylation to analyze neuropeptidomic changes in crustaceans.8, 9 Stable isotopes, supplied by isotopic formaldehyde, are added to the N-termini and lysine side chains of peptides by reductive dimethylation, adding two methyl groups, which add either 28.03130 or 32.05641 Daltons (Da) to each primary amine, depending on the stable isotopes being incorporated. The heavy-and light-labeled samples are analyzed simultaneously to provide relative quantitation information between experimental and control conditions. Though effective, duplex labeling requires an individual control sample for each experimental sample. Expanding the multiplexing capabilities of reductive dimethylation strategies greatly reduces the number of samples needed as multiple samples can be compared to a single control. Simultaneous analysis of the differentially labeled samples also reduces the instrument time required and the run-to-run variability. A 4-plex reductive dimethylation method is achieved by selecting formaldehyde with different combinations of 12C/13C and H/D, providing four distinct mass additions (+28.03130, +30.03801, +32.05641, and +34.06312 Da) that can be incorporated at the N-termini and lysine residues via reductive dimethylation.18–20 This is a cost-effective approach to increase both throughput and quantitative abilities.
In this study, 4-plex reductive dimethylation was used to quantify the relative changes in expression of neuropeptides in Callinectes sapidus after 1, 4, and 8 hours of hypoxia exposure. These exposure durations are reflective of hypoxia exposure before blue crabs manage to escape hypoxic episodes and have been studied previously.5 The multiplexed samples were analyzed by both MALDI-and ESI-MS to provide enhanced, complementary coverage of the crustacean neuropeptidome.21 In fact, 88 identified neuropeptides were found using both ESI-and MALDI-MS analyses. Several trends were revealed in this time course study, including an oscillating expression pattern. A qualitative approach was also taken to investigate neuropeptides that only expressed themselves after hypoxia stress.
Materials and Methods
Methanol (MeOH), acetonitrile (ACN), glacial acetic acid (GAA), ammonium bicarbonate, and all crab saline components (see below) were purchased from Fisher Scientific (Pittsburgh, PA). Formaldehyde (CH2O), 13C-formaldehyde (13CH2O), D2-formaldehyde (CD2O), 13CD2-formaldehyde (13CD2O), and borane pyridine complex (~8M BH3) were acquired from Sigma-Aldrich (St. Louis, MO). 2,5-dihydroxybenzoic acid (DHB) was obtained from Acros Organics (Morris, New Jersey), and formic acid (FA) was purchased from Fluka (Mexico City, Mexico). All water (H2O) used in this study was either HPLC grade or doubly distilled on a Millipore filtration system (Burlington, MA), and C18 Ziptips were purchased from Millipore (Burlington, MA). All LC solvents were Fisher Optima Grade.
Animals and Stress Experiment
All female blue crabs, Callinectes sapidus, were purchased from LA Crawfish Company (Natchitoches, LA). After transport, crabs were allowed to recover in artificial seawater made to be 35 parts per thousand (ppt) salinity, 17–18 °C, and 8–10 parts per million (ppm) (~80–100%) O2 for several days (>5) prior to being exposed. To mimic severe hypoxia (1 ppm, ~10% O2), a tank was sparged with N2 gas for 30–40 minutes to reduce the dissolved O2 to 1 ppm as measured by a Pinpoint II Oxygen Monitor. A plastic tarp was placed on top of the water’s surface to minimize water-air oxygen exchange during sparging. A crab was then placed in the tank for the desired amount of time (i.e., 1 hour, 4 hours, or 8 hours), anesthetized on ice for 20 minutes, and sacrificed for its organs of interest as previously described.22 All dissections were performed in chilled (~ 10 °C) physiological saline (composition: 440 mM NaCl; 11 mM KCl; 13 mM CaCl2; 26 mM MgCl2; 10 mM Trizma acid; pH 7.4 (adjusted with NaOH)).
Sample Preparation
For each bioreplicate, one set of each tissue of interest (i.e., sinus gland (SG) (2), brain, pericardial organ (PO) (2), commissarial ganglion (CoG) (2), and thoracic ganglion (TG)) was extracted with a Fisherbrand Model 120 probe sonicator/sonic dismembrator with chilled acidified MeOH (90:9:1 MeOH:H2O:GAA; volume (v):v:v). Each sample was sonicated three times for 8 seconds at 50% amplitude with a 15 second break in between each sonication. After centrifugation at 20,000 rpm for 20 minutes at 4 °C, the supernatant was collected and dried down in a Savant SCV100 Speedvac. All crude extracts were purified using C18 ZipTips following the manufacturer’s protocol. All samples were centrifuged at high speed (>10,000 rpm) briefly prior to purification. Control and hypoxia-exposed samples were differentially labeled via reductive dimethylation using a previously published protocol18–20 with slight modifications. In short, the labeling uses acidic conditions to preferentially label the N-terminus. The samples were all differentially labeled as follows: (a) control (i.e, 0 hours) with formaldehyde (CH2O, +28.03130 Da), (b) 1 hr exposure with 13C-formaldehyde (13CH2O, +30.04391 Da), (c) 4 hr exposure with D2-formaldehyde (CD2O, +32.05641 Da), and (d) 8 hr exposure with 13C, D2-formaldehyde (13CD2O, +34.06902 Da). Borane pyridine was the reducing agent. All samples were mixed 1:1:1:1 after being quenched with ammonium bicarbonate. The multiplexed samples were then processed two different ways: (a) spotted with 150 mg/mL DHB (in 50:50 MeOH:H2O with 0.1% FA) on a stainless-steel plate to be analyzed by a Thermo MALDI-LTQ-Orbitrap XL or (b) purified again with C18 ZipTips and analyzed by a Thermo Q Exactive (QE) coupled to a Waters nanoAquity system.
MS Data Collection
MALDI samples were spotted in triplicate and analyzed in the mass-to-charge ratio (m/z) 500–2000 range at a resolution of 60,000 on the MALDI-LTQ-Orbitrap XL. ESI samples were injected in triplicate onto a homemade C18 column (14–16 cm), from which the analytes were eluted using a 90-minute gradient (10% B to 35% B) with H2O (0.1% FA) (A) and ACN (0.1% FA) (B) and analyzed by the QE in a mass range of m/z 200–2000 with a top 15 data-dependent acquisition method with high-energy collision dissociation. MS1 and MS/MS spectra were collected at a 70,000 and 17,500 mass resolution, respectively. All data collection parameters for MALDI-and ESI-MS are included in Tables S1 and S2, respectively.
Data Analysis
Data collected by MALDI-MS was analyzed by exporting all the m/z values from Xcalibur and processed using a custom program written in Java by accurate mass matching (±5 ppm) with an intensity threshold of 100. Neuropeptides were identified by matching their masses to an in-house database, accounting for the addition of +28.03130, +30.03801, +32.05641, and +34.06312 Da on the N-terminus from isotopic reductive dimethylation. Isotopic correction was performed manually post-extraction of the m/z intensities using previously published correction factors.19 ESI-MS raw data were imported into PEAKS 8.5 software for de novo sequencing and database matching. Database search results initially were filtered using a 1% false discovery rate. Isotopic corrections were performed automatically within the software prior to the database search. Peak areas were then extracted for corresponding tandem MS-identified neuropeptide peak sets if they were detected in at least 1 technical replicate (n=3), 3 biological replicates (n=8 for brain, CoG, PO, and TG; n=7 for SG), were unique to that neuropeptide, and eluted within ± 2 minutes from each other. Dimethylation of both the N-terminus and lysine ε-amino groups were considered for ESI data which has MS/MS data for validation; conversely, only labeling of one location (i.e., N-terminus) was considered for MALDI data (e.g., mass increase of one dimethyl group) due to the acidic conditions used during labeling. Peptides known to be amidated (e.g., RFamide, RYamide, allatostatin) were only considered in this data if they were identified in their amidated form. All fragments of a neuropeptide identified by ESI-MS were equally weighted for the calculation ratios. For both MALDI-and ESI-MS analyses, all channels were normalized by taking individual intensity or peak areas divided by the total intensity or peak area. Ratios were then calculated by dividing the normalized intensity of either the +30.03801, +32.05641, or +34.06312 channel by the +28.03130 channel’s normalized intensity. Statistical significance between experimental and control samples was determined by a Dunnett’s test, which is utilized for comparing multiple experimental conditions to a single control.23, 24 If only the control and one other time point were detected, a t-test was used to determine significance. All parameters used for data processing are included for MALDI-and ESI-MS in Table S1 and S3, respectively.
Results and Discussion
Method Development
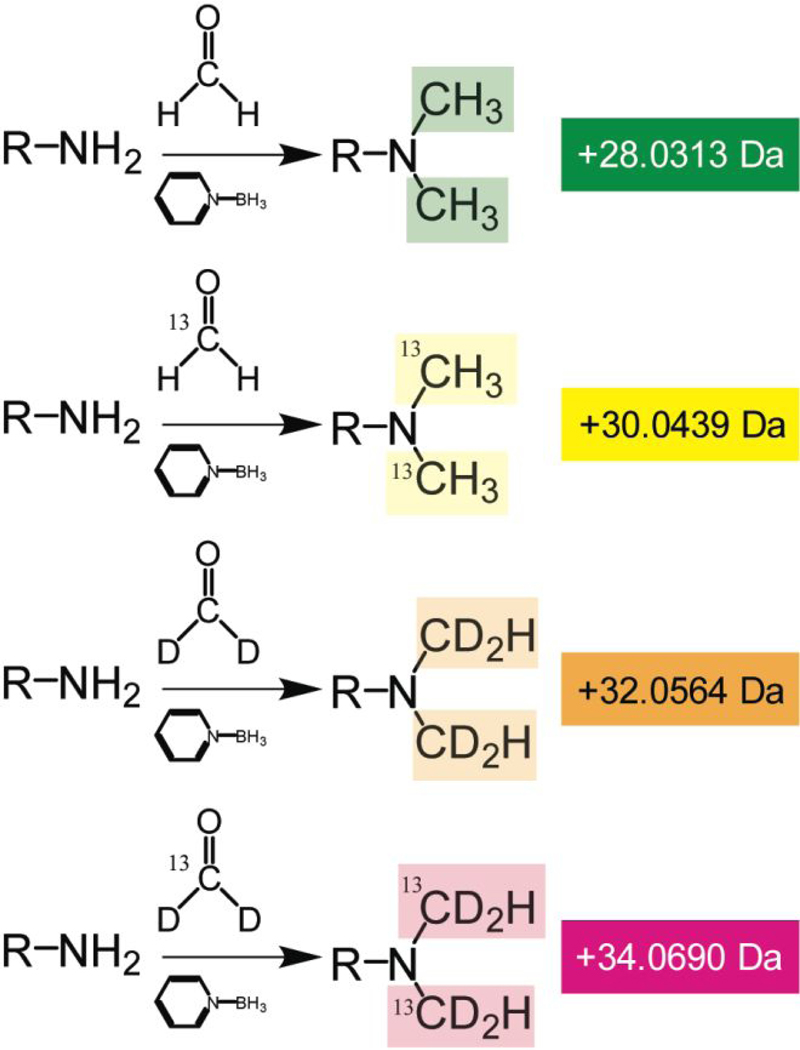
Hypoxia is rampant in coastal estuaries, and profiling the molecules (e.g., neuropeptides) that are implicated in the stress response, especially in crustaceans that tend to reside in these areas, is a priority.4, 25 In particular, a temporal component is important to consider more than just the immediate response to a stress. Short-term changes could be due to hyperarousal, and long-term exposure could reveal an alternative, possibly novel mechanism for surviving these stressful conditions until the hypoxic episode ends, which could be a few hours to days.26 In order to examine four time points (i.e., 0, 1, 4, and 8 hours), a multiplexing strategy was implemented using reductive dimethylation. In the literature, this technique has been utilized in a 2-plex, 3-plex, and 5-plex form, but a 4-plex version has not been investigated further.9, 12, 18, 27–30 To utilize a 4-plex version of reductive dimethylation, deuterium is not required to be present in the reducing agent, unlike the 3-and 5-plex. Thus, a borane pyridine complex is used as a reducing agent, which has already been proven to be successful for the 2-plex reductive dimethylation model (Figure 1).8, 9, 20, 30 One concern for multiplexing beyond 2-plex is isotopic overlap, and formulas have been derived to handle this issue specifically for 5-plex reductive methylation.19 These published corrections values were used for the processing of the MALDI data but was unnecessary for ESI data as the pre-processing performed in PEAKS automatically includes correcting the raw data for isotopic interferences.
Figure 1.
Reaction scheme for each channel of the 4-plex reductive dimethylation utilized in this study. 13C and D were the only isotopes utilized.
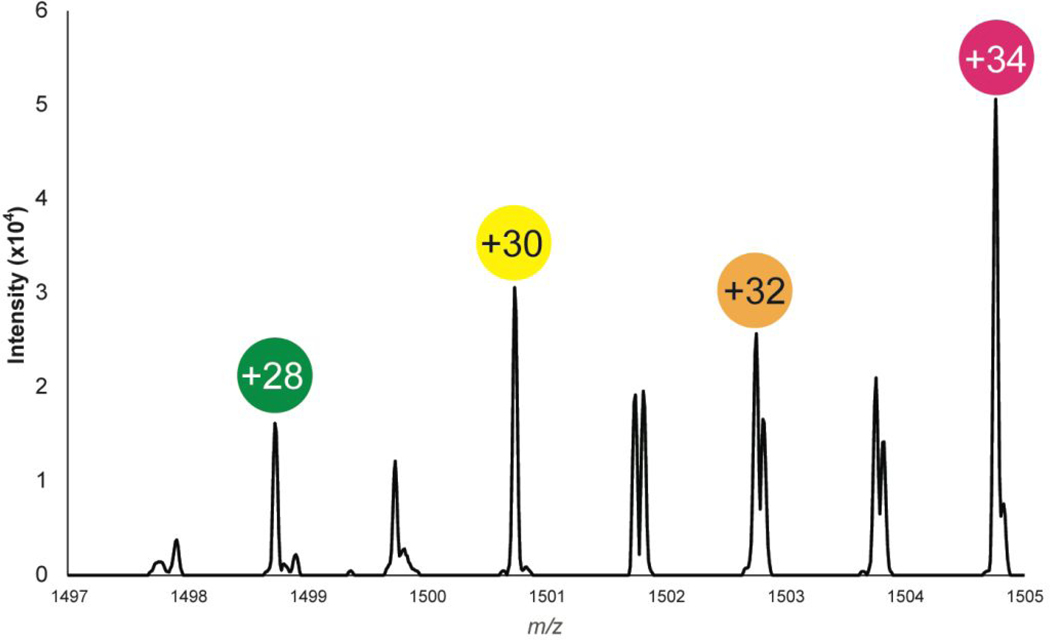
Figure 2a shows an example spectrum of allatostatin B-type VPNDWAHRFGSWamide (m/z 1470.703) found in the PO from the 4-plex labeled experimental data set. As expected, four distinct peaks in the spectra that were separated by ~2 Da are seen. In this spectrum, a dynamic, overall increase in neuropeptide expression due to increased time exposure of hypoxia stress is also observed. To test the quantitative accuracy of this system a 1:1:1:1 labeling experiment was performed, and the average abundance ratios were all within 15% of the expected ratio (+30/+28: 0.89; +32/+28: 0.93; +34/+28: 0.99). Compared to 2-plex reductive dimethylation, which traditionally has been used for crustacean neuropeptidomic studies,8, 9, 30 one particular challenge is identifying all channels due to spectral complexity, as seen in Tables S4–7. It is important to note that detecting the control channel is imperative for quantitative analysis, but there is value in also investigating those neuropeptides that are not expressed in the control channel (see Qualitative Analysis) (Table S8).
Figure 2.
Sample spectrum of 4-plex reductive dimethylation. Each label is spaced by ~2 (2.0126 Da to be exact). The representative neuropeptide is allatostatin B-type VPNDWAHRFGSWamide (m/z 1470.703) found in the PO. The accurate mass increases are described in Figure 1 and as follows: +28=+28.0313 Da, +30=+30.0439 Da; +32 =+32.0564 Da; +34=34.0690 Da.
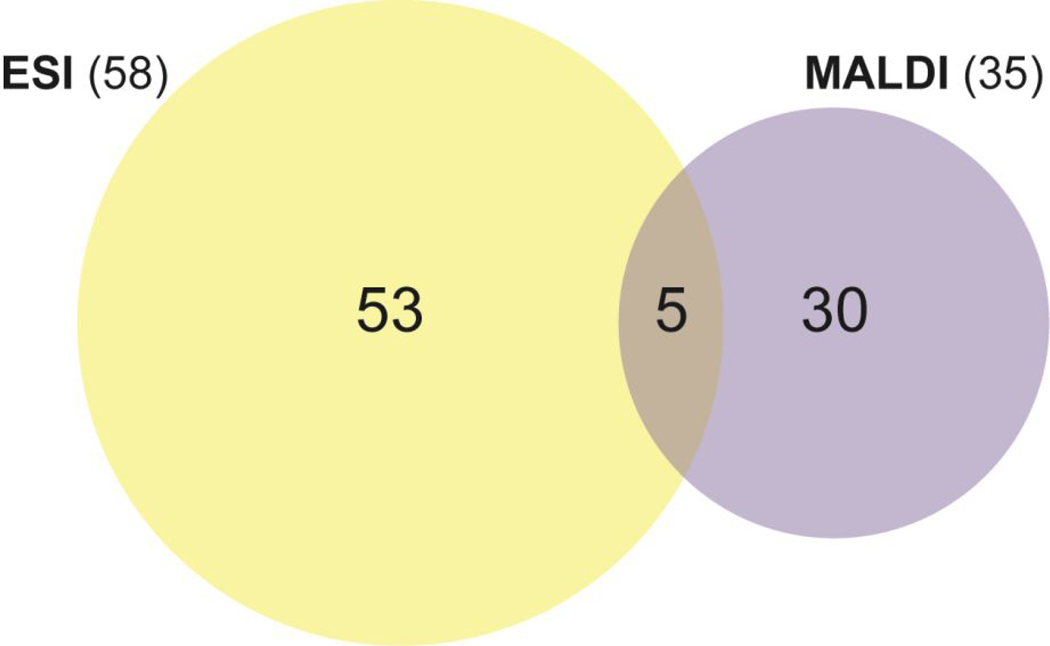
Overall, in this study, 88 neuropeptides across five different tissues (i.e., SG, brain, PO, CoG, and TG) were identified using both MALDI-and ESI-MS, with a majority of the identifications coming from ESI-MS only (~60%) (Figure 3). This small overlap is likely due to the following factors: (1) Different neuropeptide masses that can be identified between ESI-and MALDI-MS. MALDI-MS is known to primarily produce singly-charged ions, which limits the instrument’s ability to analyze neuropeptides with m/z 2000 or lower (i.e., due to our mass range being m/z 500–2000). Compared to MALDI-MS, ESI-MS has the inherent advantage of producing multiply charged ions, allowing for the identification of larger neuropeptides. (2) MALDI-and ESI-MS have different identification strategies. No MS/MS is performed during MALDI-MS analysis; only accurate mass matching (±5 ppm) is done to identify neuropeptides. On the other hand, ESI-MS relies upon MS/MS fragmentation to de novo sequence the MS/MS spectra and then match to a provided database. This translates to point (3) and (4). Accurate mass matching limits the identifications to only full-length neuropeptides and only those with single dimethylation sites to reduce false discovery rates. Since ESI-MS/MS relies upon de novo sequencing, truncated fragments of the neuropeptides can be identified as well as peptides with multiple dimethylation sites can be identified accurately. It should be noted that our acidic labeling conditions cause dimethylation to preferentially occur at the N-terminus. While labeling of lysine residues in the MALDI-MS data was not considered, we only have 4 of the 35 neuropeptides identified contained internal lysine residues, and most crustacean neuropeptides <2000 Da do not contain lysine residues. Finally, (5) MALDI-and ESI-MS have different ionization mechanisms, leading to largely different ionization efficiencies for different peptides between these two techniques and producing complementary identifications. Taking all of these factors into consideration, it is not surprising that only 5 neuropeptides were found in common. The five neuropeptides that overlapped included (1) RFamide SMPTLRLRFamide (m/z 1119.646), (2) orcomyotropin FDAFTTGRGHS (m/z 1186.516), (3) orcokinin NFDEIDRSGFA (m/z 1198.549), (4) orcokinin NFDEIDRSGFA (m/z 1270.570), and (5) allatostatin B-type VPNDWAHFRGSWamide (m/z 1470.703). Although they may overlap, in some cases they were identified in different tissues (i.e., orcomyotropin FDAFTTGRGHS (m/z 1186.516)) or were identified in different channels (i.e., RFamide SMPTLRLRFamide (m/z 1119.646)), boasting the power of using complementary methodology to improve the understanding of the temporal effects of hypoxia stress.
Figure 3.
A Venn diagram depicting the neuropeptide overlap (regardless of expression changes) between ESI-and MALDI-MS. Only neuropeptides that were found in at least 3 biological replicates in both the control and at least one other channel in the SG, brain, PO, CoG, or TG were included. The five neuropeptides that overlapped included (1) RFamide SMPTLRLRFamide (m/z 1119.646), (2) orcomyotropin FDAFTTGRGHS (m/z 1186.516), (3) orcokinin NFDEIDRSGFA (m/z 1198.549), (4) orcokinin NFDEIDRSGFA (m/z 1270.570), and (5) allatostatin B-type VPNDWAHFRGSWamide (m/z 1470.703).
Quantitative Analysis
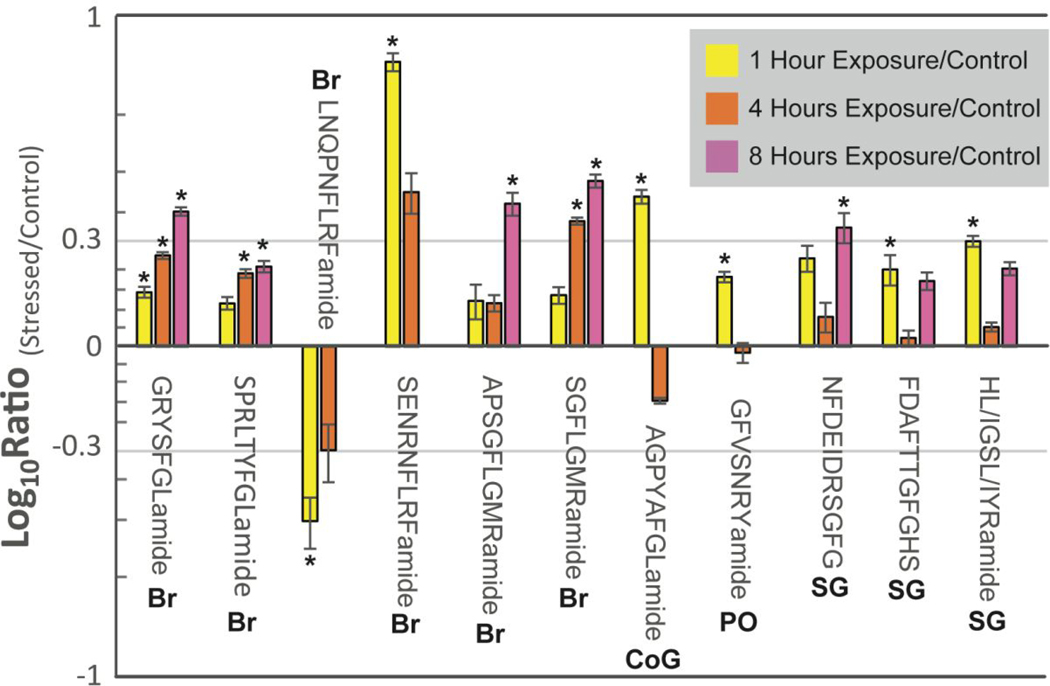
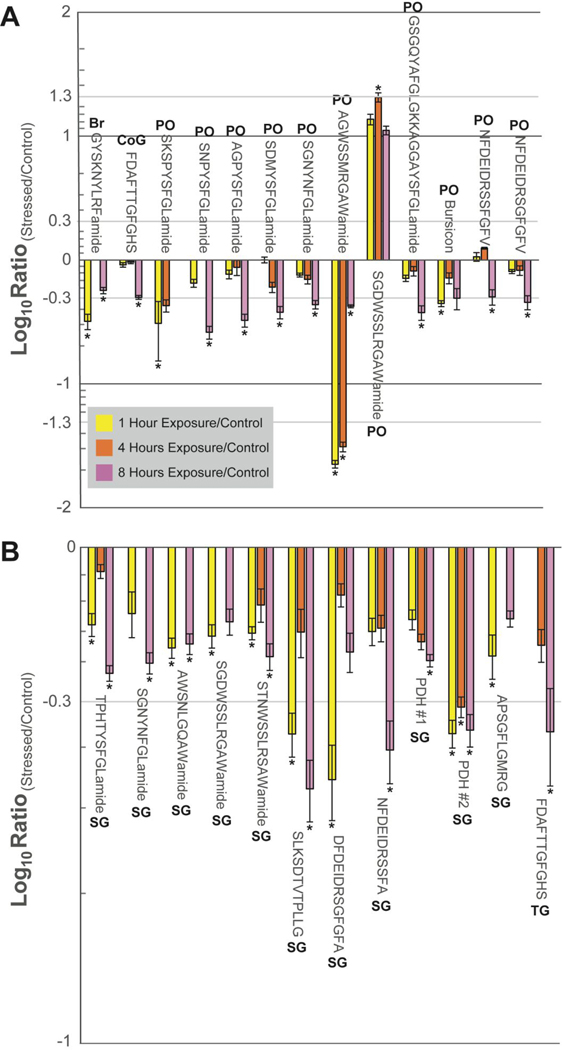
Neuropeptides identified in the ESI-and MALDI-MS datasets in at least three biological replicates are shown in Tables S4–S7. All neuropeptides that had a statistically significant change at in multiple time points during the hypoxia exposure time course are highlighted in Figures 4 and 5. From these results, several trends were revealed: (1) Of the neuropeptides quantified, the bulk majority are decreased in expression in response to hypoxia. This could be interpreted as a decrease in overall activity to conserve energy to try to outlive the hypoxic episode. This is substantiated by prior publications demonstrating decreased gene expression in response to hypoxia.31 (2) Although an initial response was observed at 1 hour of exposure, after 4 or 8 hours of exposure the neuropeptide expression was no longer significantly different than the control (i.e., basal levels). This suggests a hyperarousal response and is seen in the allatostatin A-type SKSPYSFGLamide, RFamide SENRNFLRFamide, and the RYamide GFVSNRYamide. The dysregulation of these neuropeptides could provide an initial survival mechanism for short bouts of hypoxia. For example, in crustaceans, allatostatin A-type neuropeptides are well documented for being inhibitory neuro/myomodulators,32 so their expression changes could serve to protect the crab from hypoxia by decreasing heart rate. (3) Some neuropeptides were only increased or decreased after a certain duration of hypoxia exposure (i.e., after 4 or 8 hours). This trend was observed across different tissue types but most notably in the PO. Four allatostatin A-type neuropeptides (i.e., SNPYSFGLamide, AGPYSFGLamide, SDMYSFGLamide, and SGNYNFGLamide) and two orcokinins (i.e., NFDEIDRSSFGFV and NFDEIDRSGFGFV) showed significant decreases after 8 hours of hypoxia exposure. The down regulation of these neuropeptides could suggest a survival mechanism better suited to longer hypoxia exposure (as opposed to those in trend 2). Alternatively, the hypoxia-exposed crab could be approaching death after 8 hours of severe hypoxia, so the downregulation of these neuropeptides could be a result of that process. (4) An oscillating expression pattern was observed. After 1 hour of exposure, there was a significant change in the neuropeptide content, followed by a return to the basal level at the 4-hour time point. Finally, after eight hours of exposure, the expression levels are similar to what was observed at the 1-hour time point. This trend is most prevalent in the SG, with both up and downregulation observed. Moreover, the change is not limited to specific neuropeptide families and was seen in the allatostatin A-type TPHTYSFGLamide, allatostatin B-type neuropeptides AWSNLGQAWamide and STNWSSLRSAWamide, CHH-precursor related peptide SLKSDTVTPLLG, orcokinin NFDEIDRSGFG, and orcomyotropin FDAFTTGFGHS. The variety of neuropeptides and families suggests that, in general, the crab may be going in and out of either escape or coma-like activities in order to survive the harsh, hypoxic environment.
Figure 4.
A bar graph of the ratios of all neuropeptides found to have significant differences between a control and at least two of the three time points (i.e., 1 hour (yellow), 4 hour (orange), and 8 hour (pink)) for severe (i.e., 1 ppm O2) hypoxia exposure identified by MALDI-MS. Neuropeptides were found in the brain (Br), CoG, PO, and SG. The x-axis shows the neuropeptides’ sequences, and the y-axis represents the log10 ratio of hypoxia-stressed to a control. The asterisk (*) represents statistical significance determined by a Dunnett’s test. The error bars reflect the standard error of the mean (SEM).
Figure 5.
Bar graphs of the ratios of all neuropeptides found to have significant differences between a control and at least two of the three time points (i.e., 1 hour (yellow), 4 hour (orange), and 8 hour (pink)) for severe (i.e., 1 ppm O2) hypoxia exposure identified by ESI-MS. (A) Neuropeptides found in the brain (Br), CoG, and PO. (B) Neuropeptides found in the SG and TG. The x-axis shows the neuropeptides’ sequences, while the y-axis represents the log (to the power of 10) ratio of hypoxia-stressed to a control. The asterisk (*) represents statistical significance determined by a Dunnett’s test. The error bars represent standard error of the mean (SEM). Bursicon (PO) refers to the sequence DECSLRPVIHILSYPGCTSKPIPSFACQGRCTSYVQVSGSKLWQTERSMCCQESGEREAAI TLNCPKPRPGEPKEKKVLTRAPIDCMCRPCTDVEEGTVLAQKIANFIQDSMPDSVPFLK. PDH #1 (SG) and PDH # 2 (SG) refer to neuropeptides QELKYQEREMVAELAQQIYRVAQAPWAAAVGPHKRNSELINSILGLPKVMNDAamide and QEDLKYFEREVVSELAAQILRVAQGPSAFVAGPHKRNSELINSLLGIPKVMNDAamide found in the pigment dispersing hormone family.
Of interest are also neuropeptides that do not have statistically significant changes, especially those that do not change across the entire time course. When looking through both the ESI-and MALDI-MS data, the bulk of neuropeptides identified did not have any significant changes at any point in the time course (20/35 and 31/58 neuropeptides for MALDI-and ESI-MS data, respectively). This overall trend is not unique to any tissue, but many neuropeptide families, including calcitonin-like hormone, cryptocyanin, proctolin, pyrokinin, SIFamide, and RYamides, showed no significant changes throughout the entire study, indicating that these families may contain neuropeptides whose expression levels were resistant to hypoxia stress-induced biochemical changes. These neuropeptide families have diverse or unknown functions; for example, proctolin has widespread modulatory function throughout the entire nervous system, while functional roles of pyrokinins, SIFamides, and RYamides have not been studied in the same detail.32 Recent studies have shown that pyrokinins influence the cardiac neuromuscular system.41 Conversely, there is evidence that SIFamide neuropeptides directly affect the feeding-related gastric mill and pyloric circuits in the crab C. borealis.42 RYamides have been found to be a member of the bilaterian family of LUQIN/RYamide-type neuropeptides. Although their direct functionally in crustaceans is yet to be explored thoroughly, this family of neuropeptides has also been found to be important in feeding.43–44 Overall, since changes in these neuropeptide families would directly impact viability and thus indicate the crustacean’s life declining, the significantly quantified changes in neuropeptides are likely due to the direct effects of hypoxia exposure.
Qualitative Analysis
For relative quantitation of a neuropeptide, the neuropeptide must be identified in at least the control and one other channel. Unfortunately, this does not allow quantification of neuropeptides that were expressed only after hypoxia exposure or ceased to be expressed after hypoxia exposure. Table S8 provides a representative sample of those neuropeptides that were unable to be quantified. All tissues except the TG had neuropeptides that were only measurable in the control condition, such as allatostatin A-type GPYSFGLamide (m/z 739.377) in the brain, CoG, and PO. This could indicate these neuropeptides were released from the tissue into the hemolymph or degraded after hypoxia exposure. Similar to trend 3 (see Qualitative Analysis), there were several examples of neuropeptides only appearing after hypoxia exposure. Some neuropeptides were detected over the entire time course (e.g., allatostatin A-type TPHTYSFGLamide (m/z 1021.510) in the PO and crypotocyanin KIFEPLRDKN (m/z 1259.711) in the SG), while others may only appeared at specific time points. For example, allatostatin A-type AGPYAFGLamide (m/z 794.420) in the PO and RFamide LAPQRNFLRFamide (m/z 1260.732) in the SG were only detected after 1 hour of severe hypoxia exposure but were not detected in the other channels. Overall, due to the variable neuropeptide families, their functions, and various tissues, it is difficult to discern the true physiological implications of each of these trends. It is clear, however, that they all play distinct roles in how the crustacean survives both short-and long-term hypoxia stress.32
Of particular interest to hypoxia stress are RFamides, RYamides, and tachykinins, because their mammalian homologs, neuropeptide Y (NPY) and substance P (SP), have been implicated in hypoxia stress.33–39 All three families are known for their dynamic functional roles in the nervous system, and it is expected that they will have a diverse response due to stress, including hypoxia stress.32 In the brain, PO, and SG, several RFamide and RYamide isoforms consistently appeared only after hypoxia stress. Many of these were highlighted in the trends above, including RFamide AYNRSFLRFamide (m/z 1172.632) in the brain, RFamide DPSFLRFamide (m/z 853.409) in the PO, RFamide SGRNFLRFamide (m/z 995.553) in the SG, RYamide LGRVSNRYamide (m/z 954.516) in the PO, and RYamide LSSRFVGGSRYamide (m/z 1227.659) in the PO. The range of changes observed in RFamides and RYamides, which are homologs to NPY,37, 38 emphasizes the importance of analyzing isoforms due to their possible different functions within the body, especially in understanding stress. It should also be noted that two tachykinin family neuropeptides (i.e., APSGFLGMRG (m/z 992.498) in the brain and SGFLGMRamide (m/z 766.403) in the CoG), which are homologous to SP,40 were identified only after exposure to severe hypoxia. Either way, validation studies with hemolymph analysis are of interest to truly characterize if these neuropeptides were being released after hypoxia exposure (in cases where the neuropeptides were only detected in the control condition) or were already present in the hemolymph to target these tissues after exposure to hypoxia stress (in cases where they appear only after hypoxia stress).
Conclusions and Future Directions
Neuropeptidomic studies offer new possibilities in untangling complex signaling pathways involved in a wide range of biological processes, such as environmental stress response. Characterization of neuropeptides, however, presents many challenges as these signaling molecules are highly diverse and the analysis is often sample-limited. By utilizing isotopic reductive dimethylation, the efficacy of using multiplex labeling to quantify neuropeptidomic changes in the blue crab, Callinectes sapidus after exposure to different durations of severe hypoxia is demonstrated. Several statistically significant changes were observed in both the MALDI-and ESI-MS datasets. Complementing the data presented here and offering further validation of the results, analyzing the spatial distribution of neuropeptides using MS imaging would allow observation of changes that would otherwise be missed by analyzing only abundance changes. Furthermore, analysis of the crustacean circulating fluid (i.e., hemolymph) could demonstrate the secretion and transport of specific signaling molecules.
Supplementary Material
Table S1. MALDI-MS instrument acquisition and data analysis settings.
Table S2. Chromatography and ESI-MS instrument acquisition.
Table S3. ESI-MS data analysis settings.
Table S4. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates in the MALDI-MS results.
Table S5. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the brain, CoG, and TG in the ESI-MS results.
Table S6. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the PO in the ESI-MS results.
Table S7. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the SG in the ESI-MS results.
Table S8. Select neuropeptides that were unquantifiable due to no detection in either the control tissue or the hypoxia-stressed tissue.
Acknowledgements
This work was supported by a National Science Foundation grant (CHE-1710140) and the National Institutes of Health (NIH) through grant 1R01DK071801. A.R.B. would like to thank NIH for a General Medical Sciences National Ruth L. Kirschstein Research Service Award (NRSA) Fellowship 1F31GM119365. C.S.S. and N.Q.V. would like to thank the National Institute of General Medical Sciences of the NIH under Award Number T32GM008349 for the Biotechnology Training Program Predoctoral Traineeship support. K.D. acknowledges a predoctoral fellowship supported by the NIH under NRSA T32 HL 007936 from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center and the National Institutes of Health-General Medical Sciences F31 National Research Service Award (1F31GM126870-01A1) for funding. The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting Information
The following supporting information is available free of charge at ACS website http://pubs.acs.org.
Data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD014688.
Conflicts of Interest
The authors declare no competing financial interest.
References
- 1.Barbier EB; Hacker SD; Kennedy C; Koch EW; Stier AC; Silliman BR, The value of estuarine and coastal ecosystem services. Ecological Monographs 2011, 81 (2), 169–193. [Google Scholar]
- 2.Rabalais NN; Diaz RJ; Levin LA; Turner RE; Gilbert D; Zhang J, Dynamics and distribution of natural and human-caused hypoxia. Biogeosciences 2010, 7 (2), 585–619. [Google Scholar]
- 3.Diaz RJ; Rosenberg R, Marine Benthic hypoxia: a review of its ecological effects and the behavioural responses of benthic macrofauna. Oceanography and Marine Biology -an Annual Review, Vol 33 1995, 33, 245–303. [Google Scholar]
- 4.Santana R; Lessa GC; Haskins J; Wasson K, Continuous Monitoring Reveals Drivers of D Oxygen Variability in a Small California Estuary. Estuaries and Coasts 2018, 41 (1), 99–113. [Google Scholar]
- 5.Stover KK; Burnett KG; McElroy EJ; Burnett LE, Locomotory fatigue and size in the Atlantic blue crab, Callinectes sapidus. Biological Bulletin 2013, 224 (2), 68–78. [DOI] [PubMed] [Google Scholar]
- 6.Bell GW; Eggleston DB; Noga EJ, Environmental and physiological controls of blue crab avoidance behavior during exposure to hypoxia. Biological Bulletin 2009, 217 (2), 161–172. [DOI] [PubMed] [Google Scholar]
- 7.Bell GW; Eggleston DB; Noga EJ, Molecular keys unlock the mysteries of variable survival responses of blue crabs to hypoxia. Oecologia 2010, 163 (1), 57–68. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y; Buchberger A; Muthuvel G; Li L, Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics 2015. 15 (23–24), 3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen RB; Xiao MM; Buchberger A; Li LJ, Quantitative neuropeptidomics study of the effects of temperature change in the crab cancer borealis. Journal of Proteome Research 2014, 13 (12), 5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li LJ; Sweedler JV, Peptides in the Brain: Mass Spectrometry-Based Measurement Approaches and Challenges In Annual Review of Analytical Chemistry, Annual Reviews: Palo Alto, 2008; Vol. 1, pp 451–483. [DOI] [PubMed] [Google Scholar]
- 11.Chung JS; Zmora N, Functional studies of crustacean hyperglycemic hormones (CHHs) of the blue crab, Callinectes sapidus -The expression and release of CHH in eyestalk and pericardial organ in response to environmental stress. Febs Journal 2008, 275 (4), 693–704. [DOI] [PubMed] [Google Scholar]
- 12.Kovanich D; Cappadona S; Raijmakers R; Mohammed S; Scholten A; Heck AJR, Applications of stable isotope dimethyl labeling in quantitative proteomics. Analytical and Bioanalytical Chemistry 2012, 404 (4), 991–1009. [DOI] [PubMed] [Google Scholar]
- 13.Greer T; Lietz CB; Xiang F; Li LJ, Enable Absolute Quantification of Peptides and Proteins Using a Standard Curve Approach. Journal of the American Society for Mass Spectrometry 2015, 26 (1), 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang CY; Kim K; Choi JY; Bahn YJ; Lee SM; Kim YK; Lee C; Kwon KS, Quantitative proteome analysis of age-related changes in mouse gastrocnemius muscle using mTRAQ. Proteomics 2014, 14 (1), 121–132. [DOI] [PubMed] [Google Scholar]
- 15.Wiese S; Reidegeld KA; Meyer HE; Warscheid B, Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7 (3), 340–350. [DOI] [PubMed] [Google Scholar]
- 16.Werner T; Becher I; Sweetman G; Doce C; Savitski MM; Bantscheff M, High-resolution enabled TMT 8-plexing. Analytical Chemistry 2012, 84 (16), 7188–7194. [DOI] [PubMed] [Google Scholar]
- 17.Xiang F; Ye H; Chen RB; Fu Q; Li LJ, N,N-Dimethyl leucines as novel Isobaric tandem mass tags for quantitative proteomics and peptidomics. Analytical Chemistry 2010, 82 (7), 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y; Wang FJ; Liu ZY; Qin HQ; Song CX; Huang JF; Bian YY; Wei XL; Dong J; Zou HF, Five-plex isotope dimethyl labeling for quantitative proteomics. Chemical Communications 2014, 50 (14), 1708–1710. [DOI] [PubMed] [Google Scholar]
- 19.Tashima AK; Fricker LD, Quantitative Peptidomics with Five-plex Reductive Methylation labels. J Am Soc Mass Spectrom 2018, 29 (5), 866–878. [DOI] [PubMed] [Google Scholar]
- 20.DeLaney K; Buchberger A; Li L, Identification, Quantitation, and Imaging of the Crustacean Peptidome. Methods Mol Biol 2018, 1719, 247–269. [DOI] [PubMed] [Google Scholar]
- 21.Stapels MD; Barofsky DF, Complementary use of MALDI and ESI for the HPLC-MS/MS analysis of DNA-binding proteins. Analytical Chemistry 2004, 76 (18), 5423–5430. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez GJ; Grashow RG, Cancer borealis stomatogastric nervous system dissection. J Vis Exp 2009, (25), e1207, doi: 10.3791/1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHugh ML, Multiple comparison analysis testing in ANOVA. Biochemia Medica 2011, 21 (3), 203–209. [DOI] [PubMed] [Google Scholar]
- 24.Jaki T; Hothorn LA, Statistical evaluation of toxicological assays: Dunnett or Williams test -Take both. Archives of Toxicology 2013, 87 (11), 1901–1910. [DOI] [PubMed] [Google Scholar]
- 25.Gray JS; Wu RSS; Or YY, Effects of hypoxia and organic enrichment on the coastal marine environment. Marine Ecology Progress Series 2002, 238, 249–279. [Google Scholar]
- 26.Wasson K; Lyon BE, Flight or fight: Flexible antipredatory strategies in porcelain crabs. Behavioral Ecology 2005, 16 (6), 1037–1041. [Google Scholar]
- 27.Hsu JL; Huang SY; Chen SH, Dimethyl multiplexed labeling combined with microcolumn separation and MS analysis for time course study in proteomics. Electrophoresis 2006, 27 (18), 3652–3660. [DOI] [PubMed] [Google Scholar]
- 28.Boersema PJ; Raijmakers R; Lemeer S; Mohammed S; Heck AJR, Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nature Protocols 2009, 4 (4), 484–494. [DOI] [PubMed] [Google Scholar]
- 29.Boersema PJ; Aye TT; van Veen TAB; Heck AJR; Mohammed S, Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 2008, 8 (22), 4624–4632. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y; DeLaney K; Hui L; Wang J; Sturm RM; Li L, A Multifaceted Mass Spectrometric Method to Probe Feeding Related Neuropeptide Changes in Callinectes sapidus and Carcinus maenas. J Am Soc Mass Spectrom 2018, 29 (5), 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown-Peterson NJ; Larkin P; Denslow N; King C; Manning S; Brouwer M, Molecular Indicators of Hypoxia in the Blue Crab Callinectes sapidus. Marine Ecology-Progress Series, 2005; Vol. 286, pp 203–215. [Google Scholar]
- 32.Christie AE; Stemmler EA; Dickinson PS, Crustacean neuropeptides. Cell Mol Life Sci 2010, 67 (24), 4135–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poncet L; Denoroy L; Dalmaz Y; Pequignot JM; Jouvet M, Alteration in central and peripheral substance P-and neuropeptide Y-like immunoreactivity after chronic hypoxia in the rat. Brain Research 1996, 733 (1), 64–72. [DOI] [PubMed] [Google Scholar]
- 34.Wang ZZ; Dinger B; Fidone SJ; Stensaas LJ, Changes in tyrosine hydroxylase and substance P immunoreactivity in the cat carotid body following chronic hypoxia and denervation. Neuroscience 1998, 83 (4), 1273–1281. [DOI] [PubMed] [Google Scholar]
- 35.Lee EW; Michalkiewicz M; Kitlinska J; Kalezic I; Switalska H; Yoo P; Sangkharat A; Ji H; Li LJ; Michalkiewicz T; Ljubisavljevic M; Johansson H; Grant DS; Zukowska Z, Neuropeptide Y induces ischemic angiogenesis and restores function of ischemic skeletal muscles. Journal of Clinical Investigation 2003, 111 (12), 1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moss IR; Laferriere A, Central neuropeptide systems and respiratory control during development. Respiratory Physiology & Neurobiology 2002, 131 (1–2), 15–27. [DOI] [PubMed] [Google Scholar]
- 37.Husson SJ; Mertens I; Janssen T; Lindemans M; Schoofs L, Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog Neurobiol 2007, 82 (1), 33–55. [DOI] [PubMed] [Google Scholar]
- 38.Dockray GJ, The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol 2004, 89 (3), 229–35. [DOI] [PubMed] [Google Scholar]
- 39.Coast GM; Schooley DA, Toward a consensus nomenclature for insect neuropeptides and peptide hormones. Peptides 2011, 32 (3), 620–31. [DOI] [PubMed] [Google Scholar]
- 40.Satake H; Kawada T; Nomoto K; Minakata H, Insight into Tachykinin-Related Peptides, Their Receptors, and Invertebrate Tachykinins: A review. Zoolog Sci 2003, 20 (5), 533–49. [DOI] [PubMed] [Google Scholar]
- 41.Dickinson PS; Sreekrishnan A; Kwiatkowski MA; Christie AE, Distinct or shared actions of peptide family isoforms: I. Peptide-specific actions of pyrokinins in the lobster cardiac neuromuscular system. JEB 2015, 218, 2892–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blitz DM; Christie AE; Cook AP; Dickinson PS; Nusbaum MP, Similarities and differences in circuit responses to applied Gly1-SIFamide and peptidergic (Gly1-SIFamide) neuron stimulation. J Neurophysiol 2019, 121, 950–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mekata T; Kono T; Satoh J; Yoshida M; Mori K; Sato T; Miyazato M; Ida T, Purification and characterization of bioactive peptides RYamide and CCHamide in the kuruma shrimp Marsupenaeus japonicus. Gen Comp Endocrinol 2017, 246, 321–330. [DOI] [PubMed] [Google Scholar]
- 44.Yañez-Guerra LA; Delroisse J; Barreiro-Iglesias A;Slade SE; Scrivens JH; Elphick MR, Discovery and functional characterisation of a luqin-type neuropeptide signalling system in a deuterostome. Scientific Reports 2018, 8 (7220). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. MALDI-MS instrument acquisition and data analysis settings.
Table S2. Chromatography and ESI-MS instrument acquisition.
Table S3. ESI-MS data analysis settings.
Table S4. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates in the MALDI-MS results.
Table S5. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the brain, CoG, and TG in the ESI-MS results.
Table S6. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the PO in the ESI-MS results.
Table S7. Ratios (stressed/control) of neuropeptides that were detected in at least three biological replicates for the SG in the ESI-MS results.
Table S8. Select neuropeptides that were unquantifiable due to no detection in either the control tissue or the hypoxia-stressed tissue.