SUMMARY
G protein-coupled receptors (GPCRs) mediate a wide range of human physiological functions by transducing extracellular ligand binding events into intracellular responses. GPCRs can activate parallel, independent signaling pathways mediated by G proteins or β-arrestins. Whereas “balanced” agonists activate both pathways equally, “biased” agonists dominantly activate one pathway, which is of interest for designing GPCR-targeting drugs because it may mitigate undesirable side effects. Previous studies demonstrated that β-arrestin activation is associated with trans-membrane helix VII (TM VII) of GPCRs. Here, single-molecule fluorescence spectroscopy with the beta 2 adrenergic receptor (β2AR) in the ligand-free state showed that TM VII spontaneously fluctuates between one inactive and one active-like conformation. The presence of the β-arrestin-biased agonist isoetharine prolongs the dwell time of TM VII in the active-like conformation compared with the balanced agonist formoterol, suggesting that ligands can induce signaling bias by modulating the kinetics of receptor conformational exchange.
Keywords: G protein-coupled receptor, beta 2 adrenergic receptor, biased signaling, single-molecule fluorescence spectroscopy, conformational dynamics
INTRODUCTION
G protein-coupled receptors (GPCRs) are integral membrane proteins that bind extracellular ligands and transmit signals across the cell membrane to intracellular effectors, such as G proteins and β-arrestins (Kobilka, 2011; Peterson and Luttrell, 2017; Shenoy and Lefkowitz, 2011; Thal et al., 2018). The GPCR superfamily contains more than 800 members in the human proteome and mediates a multitude of physiological functions. Accordingly, GPCRs are the targets for a myriad of drugs (Hopkins and Groom, 2002; Jacobson, 2015; Hauser et al., 2017; Sriram and Insel, 2018), which account for roughly 30% of all FDA-approved pharmaceutical agents. Many GPCRs signal through both G protein and β-arrestin signaling pathways (Rajagopal et al., 2010). Whereas some drug ligands stimulate both pathways equally (balanced agonists), other ligands selectively stimulate either the G protein or β-arrestin pathway, a phenomenon termed “functional selectivity” or “biased signaling” (Correll and McKittrick, 2014; Wisler et al., 2014; Smith et al., 2018). Selectively stimulating just one of the signaling pathways could mitigate undesirable side effects resulting from activation of the other pathway. As such, biased signaling is of keen interest for the next generation of GPCR-based therapeutics (Smith et al., 2018). For example, agonists of opioid receptors that selectively activate the G protein pathway could provide effective and safe pain relief while avoiding respiratory failure and dependency associated with activation of the β-arrestin pathway (Soergel et al., 2014; Schmid et al., 2017), which is of key importance in view of the opioid crisis in the United States (Bauman et al., 2018; Kurland, 2018). Further studies of the molecular basis of biased signaling are therefore timely and of keen interest.
At the structural level, functional selectivity may arise if different classes of ligands stabilize distinct receptor conformations that are differentially recognized by G proteins or β-arrestins. Consistent with this hypothesis, double electron-electron resonance spectroscopy has shown that Gq-biased ligands stabilize conformations of the angiotensin II type 1 receptor that are distinct from those stabilized by β-arrestin-biased ligands (Wingler et al., 2019). Moreover, fluorescence-based studies of the vasopressin type 2 receptor (V2R) have shown that Gsbiased ligands shift the position of TM VI relative to the receptor C-terminus, whereas β-arrestin-biased ligands alter the position of TM VII (Rahmeh et al., 2012). NMR spectroscopic studies of β2AR labeled with 19F probes at the intracellular tips of TM VI or TM VII (Liu et al., 2012) revealed that the β-arrestin-biased ligand isoetharine (Drake et al., 2008) preferentially impacts the conformational state of TM VII. These studies suggest that β-arrestin bias is mediated by the conformational state of TM VII. This hypothesis is corroborated by structural studies showing that β-arrestin engages GPCRs via TM VII. For example, the crystal structure of rhodopsin bound by arrestin reveals that a “finger-loop” of arrestin engages TM VII of rhodopsin (Kang et al., 2015). TM VII was also observed as a trigger for biased signaling in both kappa opioid receptor and serotonin receptor systems (Che et al., 2018; McCorvy et al., 2018).
The aforementioned 19F-NMR study of β2AR resolved inactive and active-like conformational states of TM VII that coexist in equilibrium, and compared the effects of balanced ligands (with respect to G-protein or β-arrestin pathways) and biased ligands (with respect to the β-arrestin pathway) on the conformational equilibrium (Liu et al., 2012). Notably, binding of the β-arrestin-biased agonist isoetharine shifted the conformational equilibrium almost completely to the active-like state of TM VII, whereas the balanced agonist formoterol induced a smaller population shift. Here we now used a single-molecule fluorescence assay (Lamichhane et al., 2015) to elucidate the kinetic basis of the observed conformational bias induced at TM VII by isoetharine. We chemically attached a Cy3 fluorophore to a cysteine residue on TM VII, and showed that this could be achieved without disrupting binding of prototypical β2AR ligands. The labeled receptor was reconstituted in phospholipid nanodiscs, attached to a quartz surface and monitored over time at the single-molecule level by total internal reflection fluorescence (TIRF) microscopy. We observed that TM VII spontaneously fluctuates between one inactive and one active-like conformation, both in the absence or presence of agonist ligands. From statistical analysis of a sizeable collection of receptor molecules, we determined the mean dwell times of inactive and active-like TM VII conformers in the apo state, with the balanced agonist formoterol bound and with the β-arrestin-biased agonist isoetharine bound. Notably, isoetharine prolonged the dwell time of the active conformation of TM VII to a greater extent than formoterol, explaining the large population shift towards the active state reported in the earlier NMR spectroscopic study (Liu et al. 2012). Since a prolonged dwell time in the active conformation would favor coupling of TM VII to a β-arrestin effector, our results suggest that ligand-dependent changes in the kinetics of receptor conformational exchange contribute to signaling bias.
RESULTS
β2AR constructs.
Three variants of β2AR were used, which are described in detail at the outset of the Methods section. “β2AR” is closely related to the wild-type protein, while in “β2AR [AA]” two surface-exposed cysteines were replaced by alanines, and in “β2AR [AA-Cy3]” the native cysteine at position 327, near the cytoplasmic tip of TM VII, was covalently labeled with the fluorophore Cy3.
Conformational modeling of Cy3-β2AR conjugate.
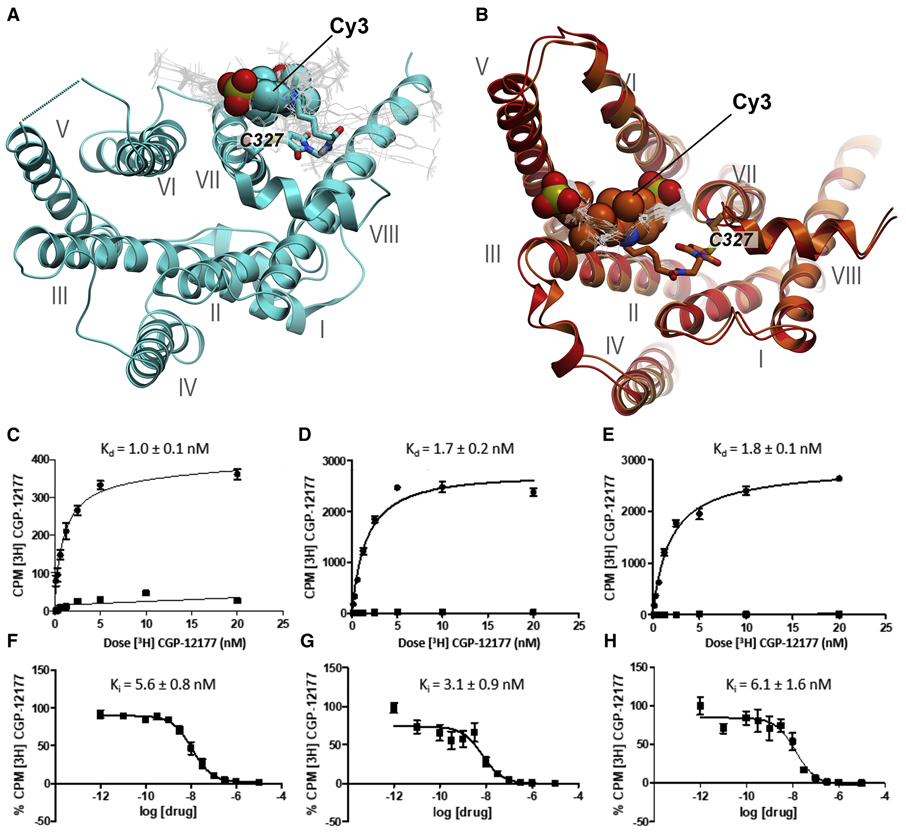
Since β2AR labeled with Cy3 at position 327 has to our knowledge not been prepared before, we performed molecular modeling to determine how the Cy3 moiety could be accommodated in the inactive and active receptor conformations, and to predict the corresponding local fluorophore environments. A model of the inactive receptor, based on the crystal structure of β2AR in complex with an antagonist (Cherzov et al., 2007), shows that the side chain of Cys327 is facing towards the receptor surface, such that the attached Cy3 moiety is fully solvent-exposed and can adopt a range of different positions of similar energy (Fig. 1A). In contrast, a model of the active receptor, based on a β2AR-agonist complex (Rasmussen et al. 2011), shows that Cys327 is oriented towards the interior of the TM helical bundle, such that the Cy3 moiety is sequestered within a narrow channel surrounded by TMs II, V and VI (Fig. 1B). Further, the modeling results suggest that the Cy3 probe can be accommodated within the active receptor conformation with only minor adjustments around the probe site (<2.0 Å for the Cys327-Cα atom), suggesting that the presence of the probe does not induce significant conformational strain in the receptor (Fig. 1B). These results suggested that the Cy3 moiety could be accommodated in either of the two receptor conformations.
Figure 1.
Conformational modeling of β2AR with Cy3 attached to position 327 and ligand binding activity of β2AR constructs reconstituted in lipid nanodiscs. A. Model of Cy3 fluorophore conjugated to Cys327 based on the crystal structure of a receptor-antagonist complex (PDB entry 2RH1, cyan ribbon). In the predicted lowest energy conformation, the Cy3 moiety (CPK representation) is located in an unrestricted solvent-exposed environment. Alternative Cy3 conformations lying within 4 kJ/mol of the lowest energy conformation are shown as thin grey lines. B. Model based on a receptor-agonist complex (PDB entry 4SN6, red ribbon). In the predicted lowest energy conformation, the Cy3 moiety fits into a shallow cavity formed by TMs II, V and VI. An energy-optimized conformation of the Cy3-conjugated receptor is also shown (orange ribbon). Comparison of the orange and red models suggests that, in spite of it’s location in the protein interior, the Cy3 probe can be accommodated within the active receptor conformation with only minor adjustments in the Cys327 region and does not lead to any substantial conformational strain in the receptor. C. Saturation binding of radioligand [3H]-CGP-12177 to β2AR in nanodiscs, as reported previously (Lamichhane et. al., 2015). Levels of total binding (circles) and non-specific binding (squares) are shown separately. The solid lines are best-fit curves, obtained as described in Methods. The corresponding equilibrium dissociation (Kd value) is indicated. D. Saturation binding of radioligand [3H]-CGP-12177 to β2AR [AA] in nanodisc. Same presentation as in C. E. Saturation binding of radioligand [3H]-CGP-12177 to β2AR [AA-Cy3] in nanodisc. Same presentation as in C. F. Competition binding of formoterol to β2AR in nanodiscs, in the presence of radioligand [3H]-CGP-12177. The solid line is a best-fit curve, obtained as described in Methods. The corresponding Ki value is indicated. G. Competition binding of formoterol to β2AR [AA] in nanodisc, in the presence of radioligand [3H]-CGP-12177. Same presentation as in F. H. Competition binding of formoterol to β2AR [AA-Cy3] in nanodisc, in the presence of radioligand [3H]-CGP-12177. Same presentation as in F.
Ligand binding properties.
Saturation binding assays employing the radiolabeled ligand [3H]-CGP-12177 and various β2AR constructs reconstituted in phospholipid nanodiscs showed that ligand binding is maintained in the receptor construct with attached Cy3 label (Figs. 1C to E, Table S1). Moreover, the Kd values are similar to the values reported for binding of this ligand to β2AR in membranes (Liu et al., 2012), indicating that the nanodisc environment preserves the ligand binding seen in the natural environment. Competition binding experiments confirmed that the affinity of the agonist formoterol for the receptor was also unaffected by the Cy3 label (Figs. 1F to H, Table S1). On the basis of these data, we conclude that the Cy3 fluorophore attached to the cysteine in position 327 does not impair the physiological ligand-binding ability of the receptor.
Single-molecule fluorescence measurements.
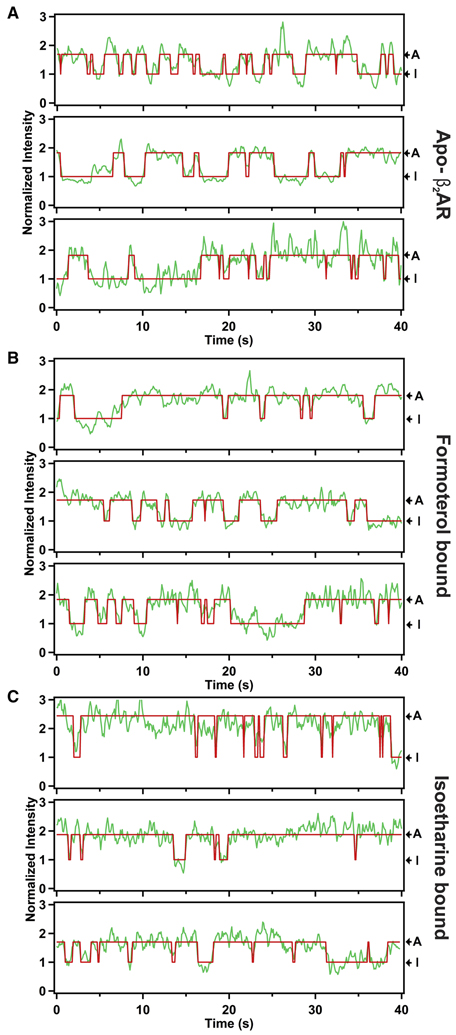
Individual Cy3-labeled receptor molecules reconstituted in phospholipid nanodiscs (Ritchie et al., 2009) were attached to a quartz microscope slide and monitored over time by single-molecule total internal reflection fluorescence (smTIRF) microscopy (Lamichhane et al., 2015). Traces of Cy3 emission intensity versus time of individual receptor molecules in the ligand-free apo-form show reversible transitions between two discrete intensity states (three representative examples are shown in Fig. 2A). Hidden Markov modeling showed that all time traces are adequately fitted with the assumption that there are two states (shown by the red lines in Fig. 2A). The higher-intensity state is assigned to an active-like conformation of TM VII (state A), based on the conformational modeling of the Cy3-β2AR conjugate, indicating that the Cy3 moiety is buried within the TM helical bundle in the conformation of the agonist complex (Fig. 1B). In such a restricted local environment, excited-state cis-trans isomerization of Cy3 will be inhibited, giving rise to an increased fluorescence quantum yield. This phenomenon is well known as protein-induced fluorescence enhancement (PIFE) (Stennett et al., 2015). The lower-intensity state is assigned to the receptor in an inactive conformation (state I), consistent with the modeling results showing that the Cy3 moiety is solvent-exposed and can adopt a range of different positions in the antagonist complex (Fig. 1A). No fluorescence enhancement is expected under these conditions. The observed effects of agonist ligands on the dwell times of the two intensity states (described below) are consistent with these assignments.
Figure 2.
Representative fluorescence intensity trajectories of individual molecules of β2AR [AA-Cy3] in nanodisc. A. In the absence of ligands (apo-form). Trajectories are in green and corresponding fits to a two-state Hidden Markov model are in red. The Cy3 emission intensity levels corresponding to inactive (I) and active-like (A) conformations of TM VII are indicated. The intensities at each time point are normalized to the mean intensity of state I. B. In the presence of the balanced agonist formoterol (1 mM concentration). Same presentation as in A. C. In the presence of the β-arrestin-biased agonist isoetharine (1 mM concentration). Same presentation as in A.
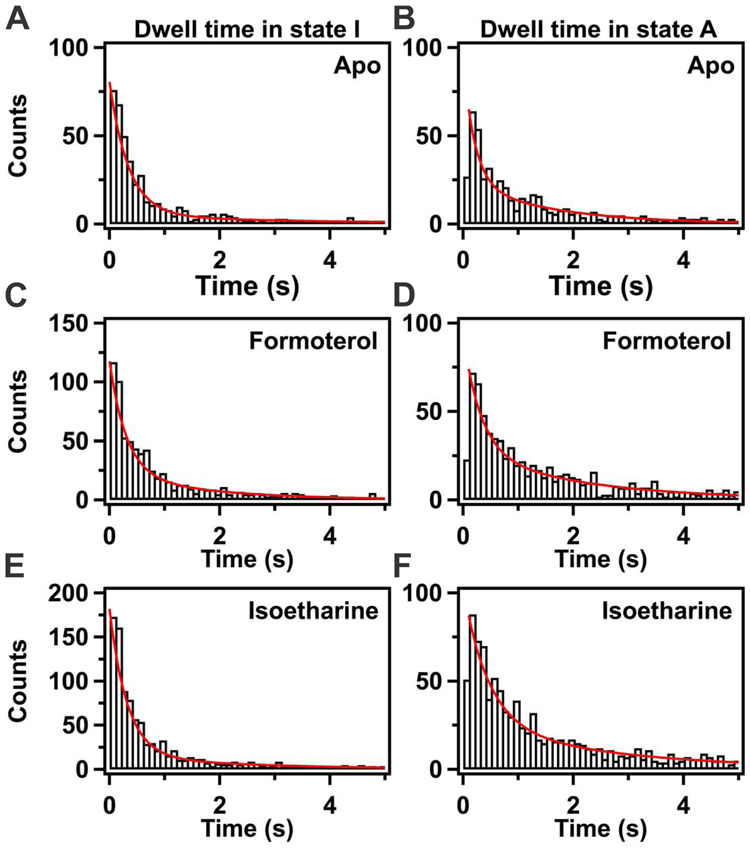
Information on the kinetics of switching between inactive and active-like states of TM VII was obtained from dwell time analysis. Dwell time histograms for the activation transition (state I to A) and deactivation transition (state A to I), compiled from multiple receptor molecules in the apo-form, are shown in Figs. 3A and B, respectively. Both histograms show a rapid initial decay, followed by a tail extending to longer dwell times; a precise fit for both histograms was obtained with a double exponential function (Eq. 1) (red lines in Figs. 3A and B). The slow kinetic phase accounts for 6 to 26% of the total amplitude and reflects a subpopulation of receptors that switch between states relatively slowly. Two populations were also observed for β2AR labeled with Cy3 in TM VI (Lamichhane et al., 2015). In that case, the slowly interconverting species was assigned to a non-functional population of β2AR molecules, since it has been reported that nanodisc reconstitution of β2AR results in the loss of ~30% of starting activity (Leitz et al., 2006). Because the minor kinetic component likely reflects a non-functional population of β2AR [AA-Cy3] molecules, hereafter we focus on the major population exhibiting rapid conformational switching. The mean dwell times of the inactive and active-like conformations of TM VII are presented graphically in Fig. 4.
Figure 3.
Kinetics analysis of conformational switching of TM VII in β2AR. A. Histogram of dwell times in state I for the apo-form. The red line shows the best fit to a double exponential function, according to Eq. 1. The best-fit parameters are τ1 = 345 ± 12 ms, τ2 = 3.2 ± 0.8 s, A1 = 94 ± 3 %, A2 = 6 ± 2 %. B. Histogram of dwell times in state A for the apo-form. The best-fit parameters are τ1 = 208 ± 35 ms, τ2 = 1.6 ± 0.2 s, A1 = 74 ± 6 %, A2 = 26 ± 4 %. C. State I in the presence of the balanced agonist formoterol (1 mM). The best-fit parameters are τ1 = 303 ± 37 ms, τ2 = 1.7 ± 0.6 s, A1 = 80 ± 5 %, A2 = 20 ± 5 %. D. State A in the presence of 1 mM formoterol. The best-fit parameters are τ1 = 323 ± 42 ms, τ2 = 2.1 ± 0.2 s, A1 = 70 ± 4 %, A2 = 30 ± 3 %. E. State I in the presence of 1 mM isoetharine. The best-fit parameters are τ1 = 323 ± 21 ms, τ2 = 2.4 ± 0.6 s, A1 = 91 ± 20 %, A2 = 9 ± 20 %. F. State A in the presence of the β-arrestin-biased ligand isoetharine (1 mM). The best-fit parameters are τ1 = 455 ± 62 ms, τ2 = 2.5 ± 0.3 s, A1 = 74 ± 6 %, A2 = 26 ± 4 %.
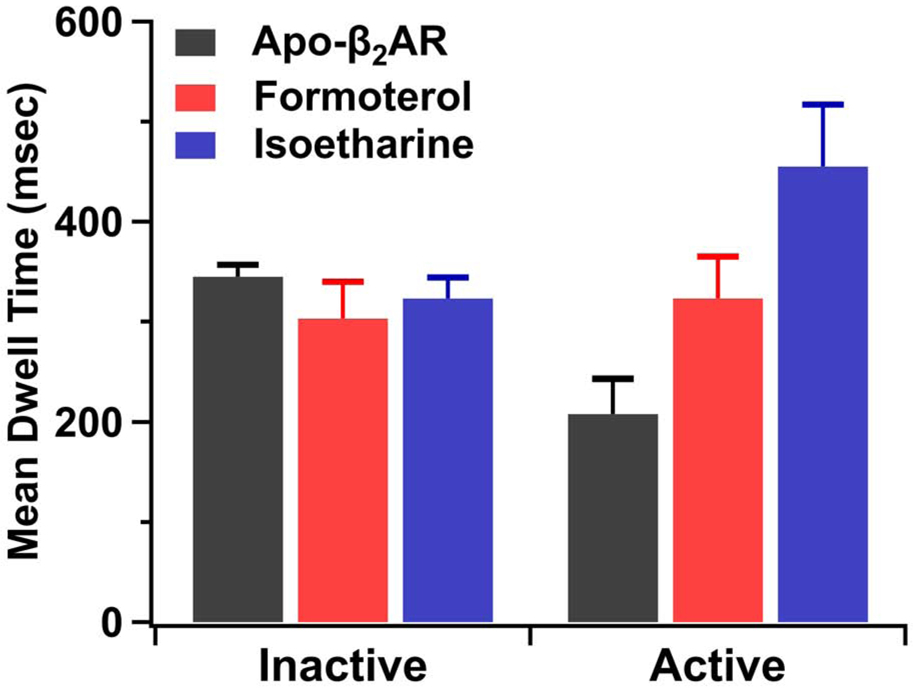
Figure 4.
Mean dwell times of inactive and active-like conformational states of TM VII in β2AR. Dwell times for the apo-form are shown as black bars, while the corresponding dwell times in the presence of balanced agonist formoterol (1 mM) or β-arrestin-biased agonist isoetharine (1 mM) are shown as red and blue bars, respectively. The error bars show the standard deviation in the best-fit values of the dwell time parameters, as reported by the Igor software.
Similar smTIRF measurements performed for complexes of β2AR with the balanced agonist formoterol or the β-arrestin-biased agonist isoetharine showed that there are also two intensity states, similar to those seen for the apo-form (Figs. 2B & C). The mean dwell times of the inactive state (Fig. 4) are unchanged relative to the apo receptor when either ligand is bound, indicating that these ligands have no effect on the rate of the activation transition. However, the mean dwell times of the active-like state are prolonged in the presence of either ligand (Fig. 4), showing that both ligands retard the deactivation transition. Since agonist ligands are known to stimulate the signaling activity of β2AR (Baker, 2010), these observations are consistent with the independently obtained assignments of the inactive and active-like states of TM VII. The key observation is that the β-arrestin-biased agonist isoetharine prolongs the dwell time of the active-like conformation of TM VII to a significantly greater extent than does the balanced agonist formoterol (Fig. 4).
DISCUSSION
Fluorescence spectroscopy has previously been used to monitor activation-related conformational changes of TM VI of β2AR at both ensemble (Ghanouni et al., 2001; Yao et al., 2009) and single-molecule (Lamichhane et al., 2015; Gregorio et al., 2017) levels. However, little was known about the conformational dynamics of TM VII. Here, we have shown that a Cy3 fluorophore can be covalently attached to Cys327, near the cytoplasmic end of TM VII, without disrupting the binding of prototypical β2AR ligands to the receptor (Fig. 1). These observations are consistent with the molecular modeling results, which suggest that the Cy3 moiety can be accommodated within the inactive or active receptor conformations, with only minor adjustments around the probe site in the case of the active conformation (Fig. 1B). The modeling results further suggest that the Cy3 moiety is solvent-exposed in the inactive receptor conformation (Fig. 1A) but is buried in a restricted environment in the active-like receptor conformation (Fig. 1B). The latter environment is expected to give rise to PIFE, resulting in a higher fluorescence intensity. We observed that the Cy3 intensity stochastically switches between two distinct intensity states in individual receptor molecules the absence of any ligands (Fig. 2A), indicating that TM VII samples both conformations. The ability of the receptor to sample an active-like conformation of TM VII in the absence of any ligands is consistent with the relatively high level of basal (ligand-independent) signaling activity of β2AR (Bond et al., 1995). The higher intensity state is favored in the presence of either formoterol or isoetharine agonists (Figs. 2B & C), confirming that this state reflects an active-like conformation of TM VII, consistent with the molecular modeling results.
We previously visualized the conformational dynamics of β2AR at the single-molecule level using a Cy3 reporter attached to the cytoplasmic end of TM VI (Lamichhane et al., 2015). In that study, TM VI also exhibited spontaneous transitions between inactive and active-like conformations in the absence of ligands. Combined with these previous observations on TM VI, the present data yield a consistent view of β2AR: the receptor is intrinsically dynamic and continually fluctuates between inactive and active-like conformations, even in the absence of ligands. Interestingly, the Cy3 reporter attached to TM VI exhibited lower fluorescence intensity in the active-like state compared with the inactive state (Lamichhane et al., 2015), whereas here we have observed the opposite behavior when Cy3 is attached to TM VII. This difference can be rationalized by the crystal structures of β2AR complexes with an antagonist (Cherezov et al., 2007) and an agonist (Rasmussen et al., 2011), which showed that the positions of TM VI and TM VII were the results of outward and inward movements, respectively, during activation.
Earlier studies with several different GPCRs suggest that TM VII mediates signaling bias and receptor coupling to β-arrestin (Rahmeh et al., 2012; Liu et al. 2012; Kang et al. 2015; Che et al. 2018; McCorvy et al., 2018). Hence, a major goal of the present study was to compare the influence of both balanced and β-arrestin-biased ligands on the conformational dynamics of TM VII. A previous NMR spectroscopic study of β2AR labeled with a 19F probe at C327 in TM VII (same site as used here) revealed two resonances with distinct chemical shifts, attributed to inactive and active-like conformations (Liu et al., 2012). Binding of the β-arrestin-biased agonist isoetharine produced a near-complete population shift towards the active-like state of TM VII, whereas the inactive state was still significantly populated (~35%) in the presence of the balanced agonist formoterol (Liu et al., 2012). This is in sharp contrast with the effects of these ligands on TM VI, where the populations of the active-like conformational state are ~50% regardless of which ligand is bound (Liu et al. 2012). Here, we have now elucidated the kinetics basis for the observed ligand bias at TM VII. In principle, agonist binding could destabilize the inactive state of TM VII, stabilize the active state or act through both mechanisms. The real-time kinetics data obtained in the present study now readily distinguish among these possibilities. The mean dwell times of the inactive state of TM VII are unchanged relative to apo-β2AR when either formoterol or isoetharine are present, indicating that the bound agonists neither stabilize nor destabilize this conformation (Fig. 4). However, the mean dwell times of the active-like conformation are prolonged when either agonist is bound (Fig. 4), indicating that the population shifts induced by agonist binding are specifically due to kinetic stabilization of the active-like receptor conformation. Notably, the β-arrestin biased agonist isoetharine prolongs the dwell time of the active-like state of TM VII to a greater extent than does the balanced agonist formoterol, explaining the large population shift towards this state observed in the 19F-NMR studies (Liu et al. 2012).
In the cellular context, signaling occurs in the presence of effector proteins, such as β-arrestin, which is absent in our in vitro experiments. Here we have shown that ligands can control the dwell times of specific receptor conformations in the absence of any effector proteins. By prolonging the dwell time of the active-like conformation of TM VII, a bound β-arrestin-biased ligand, such as isoetharine, would extend the time window available for β-arrestin to engage the receptor. Our results suggest that ligands are intrinsically capable of inducing signaling bias by modulating the conformational exchange kinetics of the receptor, which is an additional, new criterion to be considered in drug discovery projects. Future studies employing the present single-molecule fluorescence system with a range of synthetic and endogenous ligands, with or without β-arrestin present, will further explore the relationship between receptor conformational dynamics and biased signaling. In addition, by varying the lipid composition of the nanodiscs, the present system could also be used to investigate whether specific lipids can modulate the kinetic response of the receptor to balanced or biased ligands.
STAR METHODS
LEAD CONTACT AND MATERIAL AVAILABILITY
For further information, please direct enquiries to the lead contact, David Millar (millar@scripps.edu). This study did not generate unique new reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Not applicable.
METHOD DETAILS
Preparation of Cy3-Labeled β2AR.
The starting β2AR construct, “β2AR”, used in this study contains the thermostabilizing E122W mutation (Roth et al., 2008), is truncated at residue 348, a portion of the intracellular loop 3 (ICL3) not required for G protein binding (residues 245 to 249) is removed and it contains three reactive cysteine residues (Liu et al. 2012). To label only Cys327, located near the cytoplasmic end of helix VII, the other two reactive cysteine residues (Cys265 and Cys341) were removed, which yielded a C265A/C341A receptor mutant, “β2AR [AA]”. All proteins were expressed in Sf9 cells and extracted as described (Liu et al., 2012), before incubating with cobalt-based immobilized metal affinity chromatography beads (Talon) overnight. The receptor-loaded Talon beads were washed extensively with wash buffer 1 (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DDM, 0.2 mM CHS, 20 mM imidazole, 8 mM ATP) and wash buffer 2 (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DDM, 0.2 mM CHS, 20 mM imidazole) and then exchanged into 10 ml labeling buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DDM, 0.2 mM CHS). A 10-fold molar excess of Cy3 maleimide (G.E. Healthcare) (20 μl of a 5 mg/ml solution in DMSO) was then added and the reaction mixture incubated in the dark for one hour. After Cy3 labeling, β2AR was further purified using the standard protocol (Liu et al., 2012), yielding ‘β2AR [AA-Cy3]”
Modeling of the Cy3 Conjugated Receptors.
Energy-based conformational modeling of the β2AR conjugate was performed using the ICM molecular modeling suite (Molsoft, LLC). Modeling of the inactive state was based on the crystal structure of the β2AR complex with the inverse agonist carazolol (PDB entry 2RH1), where the T4 lysozyme fused to the ICL3 was removed. The active state model was based on the crystal structure of the ternary complex of β2AR complex with the agonist BI-167107 and a G-protein heterotrimer (PDB entry 3SN6). The Cy3 dye molecule was attached to the cysteine thiol moiety, with its covalent geometry optimized using the MMFF force field. The conformation of the conjugated dye was thoroughly sampled, using more than 106 steps of a Monte Carlo minimization procedure in internal coordinates, and assuming flexibility of all side chains at the receptor surface (intracellular side). For both receptor states, the lowest energy conformation and all non-redundant conformations within 4 kJ/mol from the lowest energy conformation were collected. In addition, a second active state model was created in which the receptor backbone was also treated as flexible, which resulted in minor conformational shifts in the Cys327 region.
Nanodisc Preparation.
Biobeads were added to a mixture of labeled β2AR, membrane scaffold protein 1 (MSP1) and phospholipids (1:10:700) in cholate buffer, following the reconstitution procedure described previously (Ritchie et al., 2009). The phospholipid mixture contained POPC, POPS and biotinyl CAP PE (67.5%:27.5%:5%) (Avanti Polar Lipids). The mixture of receptor, MSP1 and lipids was incubated overnight at 4° C, after which the biobeads were removed and the nanodisc-receptor complex was purified by size exclusion chromatography. The reconstituted receptor-nanodisc complexes were further separated from empty nanodiscs by capturing His-tag containing receptors in nanodiscs using Talon columns.
Radioligand Binding Assays.
The Kd value of the radioligand [3H]-CGP-12177 was determined immediately after nanodisc reconstitution by a saturation binding assay. This was carried out in a 96-well plate at a final volume of 125 mL per well. 25 μL of radioligand was added to each well (ranging from 0.16 nM – 20 nM), followed by the addition of either 25 μL binding buffer (total binding) or 25 μL of 10 μM alprenolol (to assess non-specific binding). Receptor-nanodisc complexes were added and incubated for 1 hour prior to vacuum filtration onto cold 0.3% polyethyleneimine (PEI) soaked glass fiber filter mats. Wax scintillation cocktail was melted on the filter mat and radioactivity was counted in a Microbeta2 counter (Perkin Elmer). Competition binding measurements were performed to determine the Ki value of the agonist formoterol. Nanodisc samples were incubated with 25 μL of 1 nM of [3H] CGP-12177 and 25 μL of 0 – 10 μM formoterol. All conditions were performed in triplicate at least three times. Kd values for saturation binding and Ki values for competition binding were calculated using the Prism GraphPad software according to established data analysis protocols (https://pdspdb.unc.edu/pdspWeb/content/PDSP%20Protocols%20II%202013-03-28.pdf).
Single-molecule Fluorescence Measurements.
Single-molecule data collection was performed using an inverted Axiovert 200 microscope (Zeiss) modified for prism-based TIRF imaging (TIRF Labs Inc., Cary, N.C.), as described (Berezhna et al., 2012; Lamichhane et al., 2013). Quartz slides were cleaned, passivated with polyethylene glycol and coated with streptavidin, as described (Lamichhane et al., 2010). Biotin conjugated receptor-nanodisc complexes in imaging buffer (50 mM HEPES pH 7.5, 150 mM NaCl, and 2 mM trolox) were introduced into the sample chamber and allowed to bind to the streptavidin-coated surface, after which unbound nanodiscs were washed away and the imaging buffer enriched with glucose oxidase and catalase oxygen scavenging system was introduced. Immobilized nanodisc-receptor complexes were excited using a green laser (532 nm) and the Cy3 emission intensity was recorded over time on an intensified CCD camera (Andor Technology, Belfast U.K.) with 100 ms integration time. Binary complexes were formed by incubating a saturating concentration (1 mM) of formoterol or isoetharine with the receptor on the slide surface for 1 hr prior to laser excitation and data recording. All measurements were performed at 298 K. A custom-written single-molecule data acquisition package (downloaded from https://physics.illinois.edu/cplc/software/) was used in combination with IDL software (ITT VIS, version 8.1) to record CCD camera data and generate fluorescence intensity time traces.
Single-molecule Data Analysis.
A small population of receptor-nanodisc aggregates was present on the surface, but these were readily identified by their unusually high fluorescence intensity and were excluded from further analysis. Trajectories displaying reversible intensity fluctuations prior to a single-step photobleaching event were selected for analysis. Each individual emission intensity trajectory was corrected for background and truncated prior to the photobleaching event. Trajectories were fitted with a Hidden Markov model assuming two distinct intensity states, using the program HaMMy (McKinney et al., 2006). The intensity levels observed before and after each transition were assigned to active or inactive states, based on a chosen threshold value. The dwell times spent in each state prior to transition to the other state were compiled in the form of histograms, using data from multiple receptor molecules. The resulting histograms were fitted with a double-exponential function (Eq. 1) by a non-linear regression algorithm using the Igor Pro software:
| (Eq. 1) |
where N(t) is the number of counts with dwell time t, A1 and τ1 are the amplitude and mean dwell time of component 1, respectively, and A2 and τ2 are the corresponding quantities for component 2. The Igor software reports the best-fit values of each parameter, as well as the associated standard deviation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Described in Method Details.
DATA AND CODE AVAILABILITY
This study did not generate any unique data sets or codes.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HA Epitope Tag Antibody, Alexa Fluor 488 conjugate (16B12) | Thermo Fisher Scientific | Cat#A-21287 RRID: AB_2535829 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| EDTA-free complete protease inhibitor cocktail tablets | Roche | Cat#5056489001 |
| Iodoacetamide | Sigma | Cat#I1149 |
| n-dodecyl-beta-D-maltopyranoside (DDM) | Anatrace | Cat#D310 |
| Cholesterol hemisucinate (CHS) | Sigma | Cat#C6512 |
| TALON IMAC resin | Clontech | Cat#635507 |
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti | Cat#850457 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) | Avanti | Cat#840034 |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) | Avanti | Cat#870273 |
| Bio-Beads™ SM-2 Resin | Bio-rad | Cat#1523920 |
| [3H]-CGP-12177 | Perkin Elmer | Cat# NET106250UC |
| MultiLex A | Perkin Elmer | Cat# 1450-441 |
| Filtermat A | Perkin Elmer | Cat# 1450-421 |
| Glucose oxidase | Sigma-Aldrich | Cat# G2133 |
| Catalase | Sigma-Aldrich | Cat# C3155 |
| Neutravidin | Thermo Fisher | Cat# 31000 |
| Cy3 maleimide | GE Healthcare | Cat# PA13131 |
| ß2AR | This work | N/A |
| ß2AR[AA] | This work | N/A |
| Membrane scaffold protein 1 (MSP1) | This work | Ritchie et al., 2009 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Spodoptera frugiperda (Sf9) insect cells | Gibco | |
| Bac-to-Bac™ Baculovirus Expression System | Thermofisher | Cat# 10359016 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Human ß2AR genes | genescript | N/A |
| Software and Algorithms | ||
| Prism GraphPad | https://www.graphpad.com | N/A |
| IDL | https://www.harrisgeospatial.com | version 8.1 |
| HaMMy | McKinney et al., 2006 | N/A |
| Single | https://physics.illinois.edu/cplc/software/) | N/A |
| Igor Pro | https://www.wavemetrics.com | Igor Pro 6.3 |
| Molsoft | Molsoft, LLC | |
| Other | ||
Acknowledgments.
This research was supported by NIH Road Map Initiative grant P50 GM073197.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests. The authors declare no competing interests.
REFERENCES
- Baker JG (2010) The selectivity of beta-adrenoceptor agonists at human beta1-, beta2- and beta3-adrenoceptors. Br. J. Pharmacol 160, 1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann MH, Kopajtic TA and Madras BK (2018) Pharmacological research as a key component in mitigating the opioid overdose crisis. Trends Pharmacol. Sci 39, 995–998. [DOI] [PubMed] [Google Scholar]
- Berezhna SY, Gill JP, Lamichhane R and Millar DP (2012) Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J. Am. Chem. Soc 134, 11261–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond RA, Leff P, Johnson TD, Milano CA, Rockman HA, McMinn TR, Apparsundaram S, Hyek MF, Kenakin TP, Allen LF et al. (1995) Physiological effects of inverse agonists in transgenic mice with myocardial overexpression of the beta 2-adrenoceptor. Nature 374, 272–276. [DOI] [PubMed] [Google Scholar]
- Che T, Majumdar S, Zaidi SA, Ondachi P, McCorvy JD, Wang S, Mosier PD, Uprety R, Vardy E, Krumm BE et al. (2018) Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 172, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK et al. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll CC and McKittrick BA (2014) Biased ligand modulation of seven transmembrane receptors (7TMRs): functional implications for drug discovery. J. Med. Chem. 57, 6887–6896. [DOI] [PubMed] [Google Scholar]
- Drake MT, Violin JD, Whalen EJ, Wisler JW, Shenoy SK and Lefkowitz RJ (2008) Beta-arrestin-biased agonism at the beta2-adrenergic receptor. J. Biol. Chem 283, 5669–5676. [DOI] [PubMed] [Google Scholar]
- Ghanouni P, Steenhuis JJ, Farrens DL and Kobilka BK (2001) Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A 98, 5997–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GG, Masureel M, Hilger D, Terry DS, Juette M, Zhao H, Zhou Z, Perez-Aguilar JM, Hauge M, Mathiasen S et al. (2017) Single-molecule analysis of ligand efficacy in beta2AR-G-protein activation. Nature 547, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB and Gloriam DE (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews. Drug Discovery 16, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL and Groom CR (2002) The druggable genome. Nature Reviews. Drug Discovery 1, 727–730. [DOI] [PubMed] [Google Scholar]
- Jacobson KA (2015) New paradigms in GPCR drug discovery. Biochem. Pharmacol 98, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW et al. (2015) Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka BK (2011) Structural insights into adrenergic receptor function and pharmacology. Trends Pharmacol. Sci 32, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland M (2018) The opioid epidemic: the crisis that hits home. Professional Case Management. 23, 280–281. [DOI] [PubMed] [Google Scholar]
- Lamichhane R, Solem A, Black W and Rueda D (2010) Single-molecule FRET of proteinnucleic acid and protein-protein complexes: surface passivation and immobilization. Methods 52, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Berezhna SY, Gill JP, Van der Schans E and Millar DP (2013) Dynamics of site switching in DNA polymerase. J. Am. Chem. Soc 135, 4735–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Liu JJ, Pljevaljcic G, White KL, van der Schans E, Katritch V, Stevens RC, Wuthrich K and Millar DP (2015) Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proc. Natl. Acad. Sci. U.S.A 112, 14254–14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitz AJ, Bayburt TH, Barnakov AN, Springer BA and Sligar SG (2006) Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques 40, 601–602. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC and Wüthrich K (2012) Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCorvy JD, Wacker D, Wang S, Agegnehu B, Liu J, Lansu K, Tribo AR, Olsen RHJ, Che T, Jin J et al. (2018) Structural determinants of 5-HT2B receptor activation and biased agonism. Nature Struct. & Mol. Biol 25, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C and Ha T (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J 91, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson YK and Luttrell LM (2017) The diverse roles of arrestin scaffolds in G protein-coupled receptor signaling. Pharmacol. Rev 69, 256–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh R, Damian M, Cottet M, Orcel H, Mendre C, Durroux T, Sharma KS, Durand G, Pucci B, Trinquet E et al. (2012) Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc. Natl. Acad. Sci. U.S.A 109, 6733–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S, Rajagopal K and Lefkowitz RJ (2010) Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nature Reviews. Drug Discovery 9, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D et al. (2011) Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM and Sligar SG (2009) Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth CB, Hanson MA and Stevens RC (2008) Stabilization of the human beta2-adrenergic receptor TM4-TM3-TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J. Mol. Biol 376, 1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CL, Kennedy NM, Ross NC, Lovell KM, You Z, Morgenweck J, Cameron MD, Bannister TD and Bohn LM (2017) Bias factor and therapeutic window correlate to predict safer opioid analgesics Cell 17, 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK and Lefkowitz RJ (2011) β-arrestin-mediated trafficking and signal transduction. Trends Pharmacol. Sci 32, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ and Rajagopal S (2018) Biased signaling: from simple switches to allosteric microprocessors. Nature Reviews. Drug Discovery 17, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soergel DG, Subach RA, Sadler B, Connell J, Marion AS, Cowan CL, Violin JD and Lark MW (2014) First clinical experience with TRV130: pharmacokinetics and pharmacodynamics in health vounteers. J. Clin. Pharmacol 54, 351–357. [DOI] [PubMed] [Google Scholar]
- Sriram K and Insel PA (2018) G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 93, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennett EM, Ciuba MA, Lin S and Levitus M (2015) Demystifying PIFE: The photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett 6, 1819–1823. [DOI] [PubMed] [Google Scholar]
- Thal DM, Glukhova A, Sexton PM and Christopoulos A (2018) Structural insights into Gprotein-coupled receptor allostery. Nature 559, 45–53. [DOI] [PubMed] [Google Scholar]
- Wingler LM, Elgeti M, Hilger D, Latorraca NR, Lerch MT, Staus DP, Dror RO, Kobilka BK, Hubbell WL and Lefkowitz RJ (2019) Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell 176, 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisler JW, Xiao K, Thomsen AR and Lefkowitz RJ (2014) Recent developments in biased agonism. Curr. Opin. Cell Biol 27, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XJ, Velez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, Sunahara RK and Kobilka B (2009) The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U.S.A 106, 9501–9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique data sets or codes.