Abstract
Immune checkpoint inhibitor (ICI) therapy has shown a significant benefit in the treatment of a variety of cancer entities. However, immune-related adverse events (irAEs) occur frequently and can lead to ICI treatment termination. MicroRNA-146a (miR-146a) has regulatory functions in immune cells. We observed that mice lacking miR-146a developed markedly more severe irAEs compared with WT mice in several irAE target organs in 2 different murine models. miR-146a–/– mice exhibited increased T cell activation and effector function upon ICI treatment. Moreover, neutrophil numbers in the spleen and the inflamed intestine were highly increased in ICI-treated miR-146a–/– mice. Therapeutic administration of a miR-146a mimic reduced irAE severity. To validate our preclinical findings in patients, we analyzed the effect of a SNP in the MIR146A gene on irAE severity in 167 patients treated with ICIs. We found that the SNP rs2910164 leading to reduced miR-146a expression was associated with an increased risk of developing severe irAEs, reduced progression-free survival, and increased neutrophil counts both at baseline and during ICI therapy. In conclusion, we characterized miR-146a as a molecular target for preventing ICI-mediated autoimmune dysregulation. Furthermore, we identified the MIR146A SNP rs2910164 as a biomarker to predict severe irAE development in ICI-treated patients.
Keywords: Inflammation, Oncology
Keywords: Adaptive immunity, Cancer immunotherapy, Noncoding RNAs
Immune-related adverse events due to immune checkpoint inhibition is regulated in mice and patients by microRNA-146a, which may also serve as a biomarker predicting severe irAEs before treatment start.
Introduction
In the last decade, immune checkpoint inhibitors (ICIs) have dramatically changed the treatment algorithms of many solid tumors and hematological malignancies. These antibodies inhibit negative regulatory molecules of the immune response, including cytotoxic T lymphocyte–associated antigen 4 (CTLA-4) and programmed cell death protein-1 (PD-1) and its ligand (PD-L1), resulting in an enhanced T cell response against cancer cells. However, the increased immune activation can also induce immune-related adverse events (irAEs), which can affect any organ system, most commonly affecting the skin, gastrointestinal tract, endocrine glands, liver, and lung (1). The incidence of irAEs is high, with frequencies of up to 62% and 50% in patients treated with anti–CTLA-4 and anti–PD-1/PD-L1, respectively (2–4). Grades 3–4 irAEs, in particular, are a major concern, since patients oftentimes have to permanently discontinue ICI therapy, receive immunosuppressive therapy, including corticosteroids, and are at high risk of death. To date, it is still largely unknown why some patients develop severe irAEs while other patients do not (5). A better understanding of the pathogenesis of irAEs could help to identify robust biomarkers predicting ICI toxicity, which would be essential to tailor safety monitoring protocols and treatment decisions.
MicroRNAs (miRNAs) are small, double-stranded, noncoding RNA molecules, which regulate expression of their target genes at the posttranscriptional level. Due to their ability to modulate numerous different target mRNAs, therapeutic targeting of miRNAs has high potential, especially for diseases with a multifactorial origin, including inflammatory diseases and cancer. Several clinical trials on miRNA mimics and inhibitors are ongoing (6). MicroRNA-146a (miR-146a) exerts an immunoregulatory function in both myeloid cells and effector T cells. Previous studies have identified miR-146a as an important negative regulator of inflammation, myeloid cell proliferation, T cell receptor signalling, and cancer (7, 8). Based on our previous work showing a major role for miR-146a in the regulation of allogeneic immune responses (9, 10), we set out to understand if this miRNA is involved in the regulation of irAE pathogenesis in 2 murine models and in patients undergoing ICI therapy.
Results
miR-146a regulates irAE development.
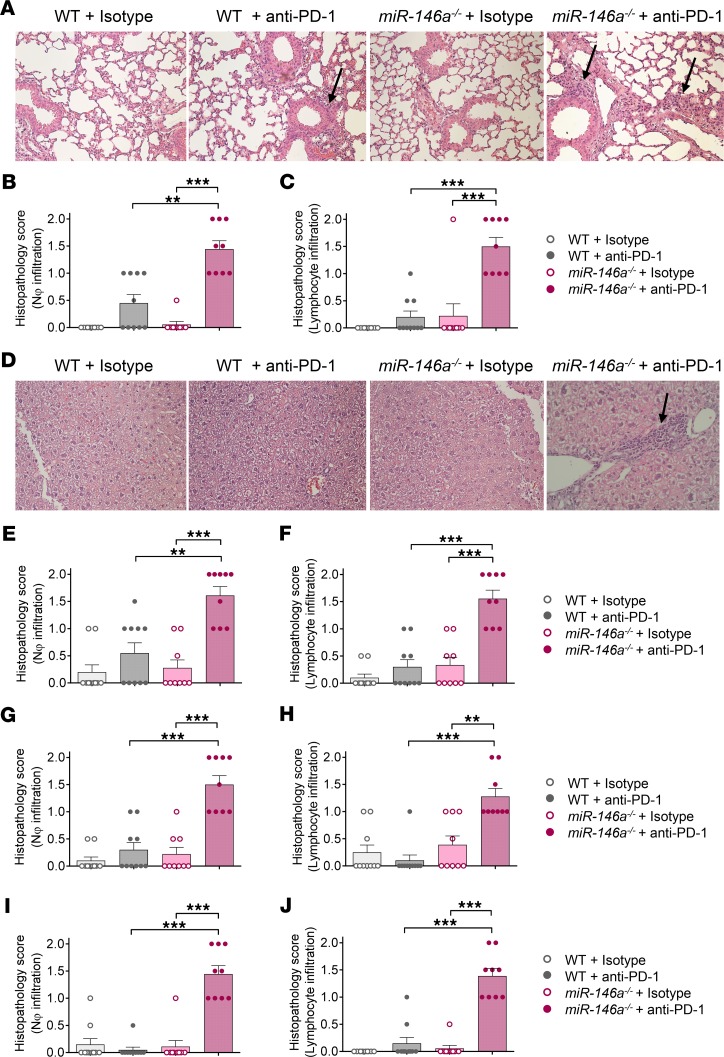
To assess the influence of miR-146a on irAE development, we used gene-targeted miR-146a–deficient mice and treated them with blocking anti–PD-1 antibody twice a week for 3 weeks and with low-dose LPS once a week for 3 weeks (for treatment schedule, see Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132334DS1). Low-dose LPS was given to mimic the human situation, in which the immune system is typically not as naive as in mice under specific pathogen–free conditions. irAE development was assessed by histopathology using a scoring system to quantify neutrophil and lymphocyte infiltration in different target organs that was established based on reports of the histopathological features of human irAEs (11, 12). Treatment of miR-146a–/– mice with anti–PD-1 led to significantly more severe irAEs compared with WT mice treated with anti–PD-1, indicated by increased neutrophil and lymphocyte infiltration in the major irAE target organs, comprising the lungs (Figure 1, A–C), liver (Figure 1, D–F), colon (Figure 1, G and H), and skin (Figure 1, I and J). To control for nonspecific effects of the antibody or low-dose LPS treatment, WT or miR-146a–/– mice treated with LPS and isotype control antibody were analyzed and did not develop signs of significant immune infiltration (Figure 1, A–J). In addition to the increased irAE severity, as assessed by histopathology, the inflammatory side effects caused reduced survival of the miR-146a–/– + anti–PD-1 group, compared with all other groups (Supplemental Figure 1B).
Figure 1. miR-146a deficiency increases irAE severity in anti–PD-1–treated mice.
WT or miR-146a–/– mice (n = 9–10 per group) were treated with LPS and anti–PD-1/isotype control antibody for 3 weeks as described. The lungs, liver, colon, and skin were isolated on day 22 after the first treatment for histopathological assessment. irAE grading was performed by an experienced pathologist blinded to the treatment groups. 0 = absent, 1 = mild, 2 = massive neutrophil/lymphocyte infiltration. Data were pooled from 2 independent experiments. Statistical significance was analyzed by Kruskal-Wallis test followed by 2-stage linear step-up procedure of Benjamini, Krieger and Yekutieli. Adjusted P value is depicted: **P < 0.01, ***P < 0.001. (A) Representative H&E staining of lung sections at original magnification ×200. Black arrows point towards inflammatory infiltrates. (B and C) Histopathology scores for neutrophil infiltration and lymphocyte infiltration into the lung. (D) Representative H&E staining of liver sections at original magnification ×200. Black arrows point towards inflammatory infiltrates. (E and F) Histopathology scores for neutrophil infiltration and lymphocyte infiltration into the liver. (G and H) Histopathology scores for neutrophil infiltration and lymphocyte infiltration into the colon. (I and J) Histopathology scores for neutrophil infiltration and lymphocyte infiltration into the skin. miR-146a, microRNA-146a, irAE, immune-related adverse event, PD-1, programmed cell death protein-1.
In a second, LPS-independent irAE model, we treated mice with the clinically relevant combination therapy of anti–PD-1 and anti–CTLA-4 antibody in order to induce irAEs. In line with our previous data, miR-146a deficiency in the hematopoietic system also increased checkpoint inhibitor–mediated autoimmunity in the major irAE target organs in mice treated with anti–PD-1 and anti–CTLA-4 combination therapy compared with WT mice (Supplemental Figure 2).
Taken together, our data indicate that miR-146a negatively regulates inflammatory side effects of ICI therapy while mice lacking miR-146a exhibit increased ICI-induced organ toxicity.
miR-146a–deficient mice treated with ICIs show increased CD4 T cell activation.
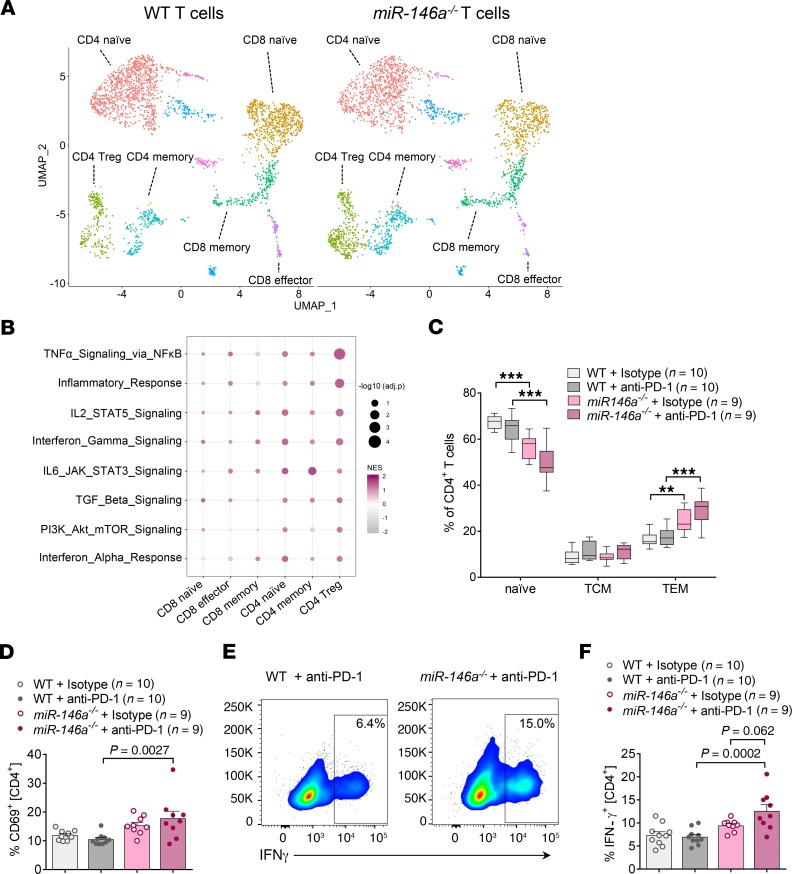
Since anti–PD-1 treatment is therapeutically used to enhance T cell activation, we next used an unbiased approach to assess whether miR-146a regulates the T cell response during ICI therapy. To analyze the T cell phenotype at the single cell level, we performed 10× Genomics’ single-cell RNA sequencing (scRNA-seq) from murine splenic T cells of WT or miR-146a–deficient mice treated with anti–PD-1 antibody. Following quality controls, 3083 single cells from 2 WT mice and 3235 single cells from 2 miR-146a–/– mice were used for further analysis. Unsupervised clustering showed that both WT and miR-146a–/– cells separated into clearly defined clusters expressing genes characteristic of distinct T cell subsets (Figure 2A and Supplemental Figure 3). Differential expression analysis followed by gene set enrichment analysis (GSEA) of the main T cell clusters using Hallmark immune gene sets revealed an enrichment of pathways involved in inflammation and immune activation in miR-146a–deficient T cells compared with WT T cells (Figure 2B). Strikingly, inflammatory gene sets were predominantly upregulated in the CD4+ T cell compartment of miR-146a–deficient mice.
Figure 2. Increased immune activation signature in miR-146a–deficient CD4 T cells of irAE mice.
(A and B) WT or miR-146a–/– mice (n = 2 per group) were treated with low-dose LPS and anti–PD-1/isotype control antibody for 3 weeks before capturing of MACS purified splenic T cells for scRNA-seq using 10× v3.1 Next GEM chemistry. Data were processed, visualized, and analyzed using the Seurat pipeline v3.0 (45, 46). (A) Uniform Manifold Approximation and Projection (UMAP) plots showing distinct T cell clusters in both miR-146a–/– and WT mice. (B) Gene set enrichment analysis of major T cell clusters. Bivariate heatmap depicts normalized enrichment score as color code and –log10 of the adjusted P value as dot size. Hallmark gene sets were derived from MSigDB. (C and D) WT or miR-146a–/– mice (n = 9–10 per group) were treated with LPS and anti–PD-1/isotype control antibody as indicated. Spleens were isolated on day 22 and CD4+ T cells analyzed by flow cytometry to differentiate naive T cells (CD44–CD62L+), central memory T cells (TCM, CD44+CD62L+, effector memory T cells (TEM, CD44+CD62L–), and activated T cells (CD69+). ***P < 0.001 by 1-way ANOVA followed by Tukey’s post hoc test. Box-and-whisker plot (C) depicts the 25th and the 75th percentiles as the bounds of the boxes, the median as the line within the box, and minimum to maximum as the whiskers. (D) Representative flow cytometry plots showing intracellular IFN-γ staining gated on CD4+ T cells. (E and F) WT or miR-146a–/– mice (n = 9–10 per group) were treated with LPS and anti–PD-1/isotype control antibody as indicated. Splenic CD4+ T cells were assessed by flow cytometry on day 22. (E) Pooled data from 2 independent in vivo experiments. Statistical significance was analyzed by 1-way ANOVA followed by Tukey’s post hoc test. miR-146a, microRNA-146a, irAE, immune-related adverse event, PD-1, programmed cell death protein-1, scRNA-seq, single cell RNA sequencing.
To confirm these data at the protein level, we next performed flow cytometric analysis of T cell activation markers. Analyzing CD44 and CD62L surface levels, we observed a significant expansion of CD4 effector memory T cells (CD62L–CD44+) in miR-146a–/– mice treated with anti–PD-1 antibody compared with WT mice treated with anti–PD-1 (Figure 2C). Correspondingly, the frequency of naive CD4 T cells (CD62L+CD44–) was significantly reduced in the miR-146a–deficient group (Figure 2C). Moreover, miR-146a–/– mice treated with anti–PD-1 displayed an increased proportion of CD69+ CD4 T cells when compared with anti–PD-1–treated WT mice, indicative of a shift toward a more activated T cell phenotype in miR-146a–/– mice treated with anti–PD-1 (Figure 2D).
Lack of miR-146a increases T cell effector function and neutrophil recruitment in mice treated with ICIs.
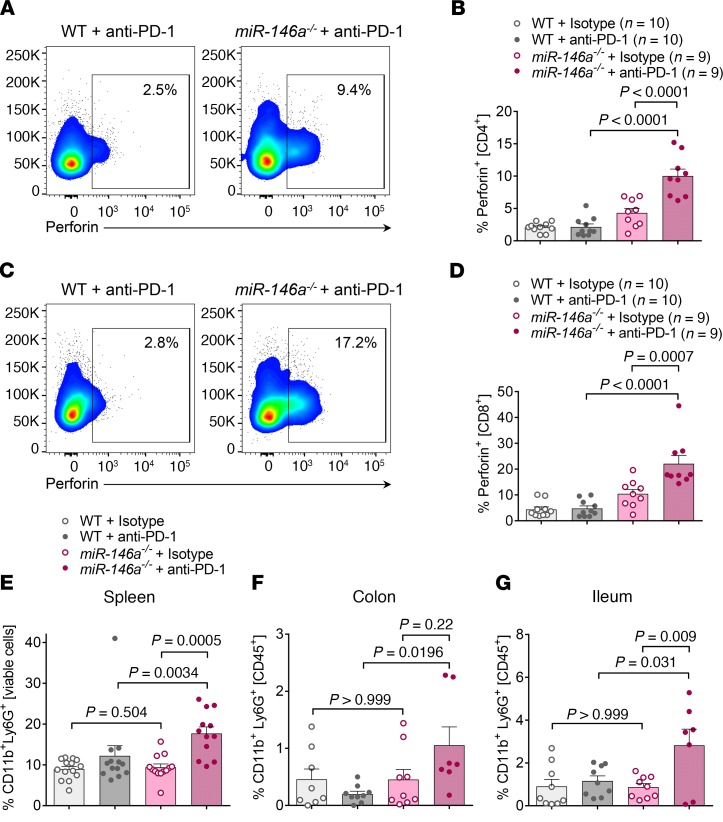
Based on the increased T cell inflammatory signature identified by scRNA-seq (Figure 2B), we further analyzed the splenic T cell effector function after ICI therapy to delineate the cellular mechanisms causing increased irAE severity in miR-146a–/– mice. Intracellular cytokine staining revealed increased numbers of IFN-γ+ CD4 T cells in the ICI treated miR-146a–deficient group compared with the ICI-treated WT group and the isotype control–treated miR-146a–deficient group (Figure 2, E and F). Strikingly, cytotoxic function of the T cells was also increased in anti–PD-1–treated miR-146a–/– mice, as demonstrated by strongly elevated frequencies of perforin+ CD4 T cells (Figure 3, A and B) and perforin+ CD8 T cells (Figure 3, C and D), indicating that lack of miR-146a increases T cell effector function during ICI therapy.
Figure 3. T cell effector function and neutrophil recruitment are regulated by miR-146a during irAE development.
(A–D) WT or miR-146a–/– mice (n = 9–10 per group) were treated with LPS and anti–PD-1/isotype control antibody as indicated and splenocytes assessed by flow cytometry on day 22. Statistical significance was analyzed by 1-way ANOVA followed by Tukey’s post hoc test. (A) Representative flow cytometry plots showing intracellular perforin staining gated on CD4+ T cells. (B) Pooled data from 2 independent in vivo experiments. (C) Representative flow cytometry plots showing intracellular perforin staining gated on CD8+ T cells. (D) Pooled data from 2 independent in vivo experiments. (E) Pooled spleen cell data from 3 independent experiments (n = 12–14 per group). Statistical significance was analyzed by Kruskal-Wallis test followed by 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. (F and G) WT or miR-146a–/– mice (n = 7–9 per group) were treated with LPS and anti–PD-1/isotype control antibody as indicated. Colon and ileum were isolated and digested on day 22. CD11b+Ly6G+ neutrophils were analyzed by flow cytometry. Data were from 2 independent experiments. Statistical significance was analyzed by 1-way ANOVA followed by Tukey’s post hoc test. miR-146a, microRNA-146a, irAE, immune-related adverse event, PD-1, programmed cell death protein-1.
In addition to T cells, neutrophils have been reported to be involved in the effector phase of irAE development (13). Moreover, high blood neutrophil counts and neutrophil-to-lymphocyte ratios have been shown to predict decreased survival and poor outcome to ICI therapy in patients with cancer (14–18). Therefore, we also analyzed levels of CD11b+Ly6G+ neutrophils in anti–PD-1–treated miR-146a–/– mice and found a strong increase compared with WT mice or isotype control–treated miR-146a–/– mice, in both the spleen (Figure 3E) and the colon (Figure 3F) and ileum (Figure 3G), which are major irAE target organs. In contrast, we did not detect any changes in monocyte and dendritic cell proportions (Supplemental Figure 4, A and B).
Our data show that both the T cell effector function and neutrophil recruitment are enhanced in mice lacking miR-146a in response to anti–PD-1 treatment.
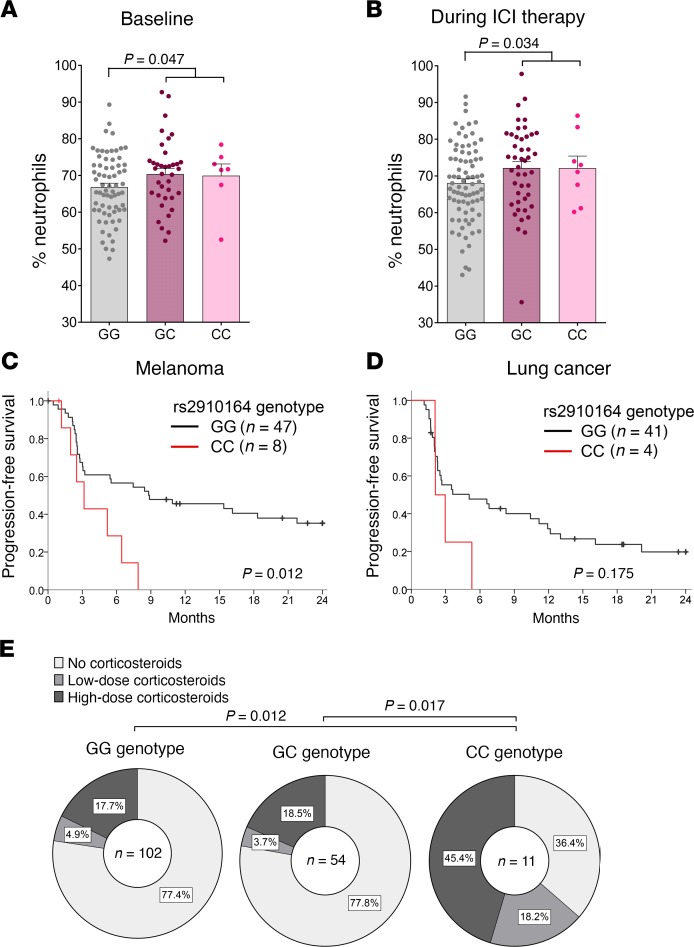
The MIR146A SNP rs2910164 is associated with increased irAE severity in patients undergoing ICI therapy.
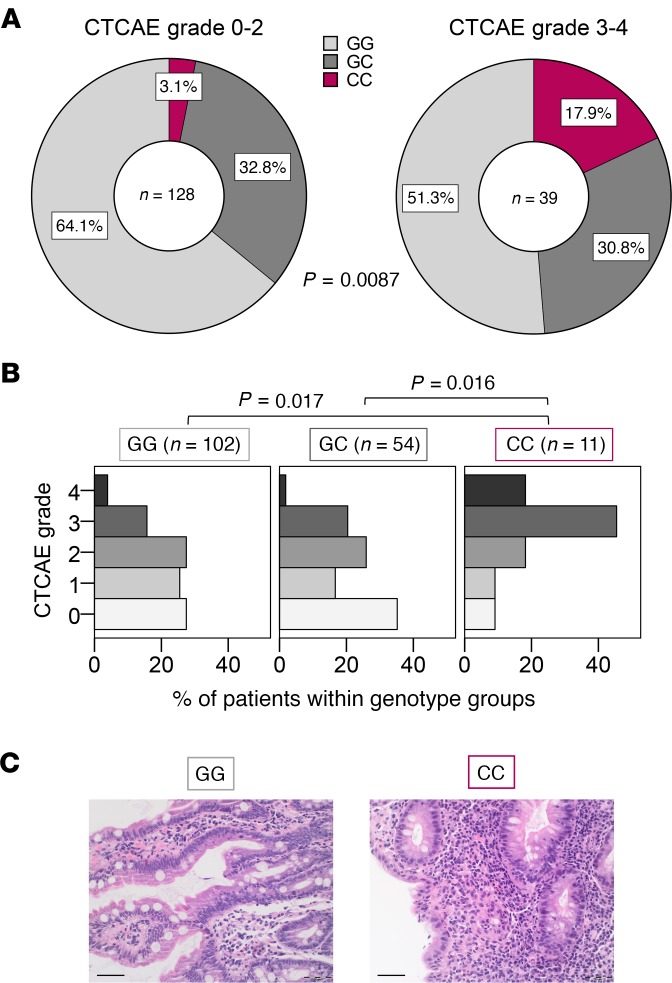
To assess the potential relevance of the findings from our murine model during human irAE development, we next performed a SNP analysis on genomic DNA from patients with solid tumors treated with checkpoint inhibitors. The SNP rs2910164 (C>G) within the MIR146A gene is known to affect processing of the miRNA, causing a reduced expression of the mature sequence (9, 19). In order to analyze a potential role of miR-146a during ICI therapy, we determined the MIR146A rs2910164 genotypes of 179 patients with cancer, who had been treated with an anti–PD-1 or anti–PD-L1 antibody. Of those, we selected 167 patients, who had received at least 3 cycles of therapy, for further analysis. The baseline patient characteristics were comparably distributed between the 3 genotypes (Supplemental Table 1). The genotype frequencies of the cohort were in accordance with the Hardy-Weinberg equilibrium (χ2 = 1.073; P = 0.773) and comparable to those reported in a large European multicenter study (20). We found that ICI-treated patients with cancer carrying the rs2910164 CC genotype, where both alleles carry the SNP, had a significantly higher risk of developing severe irAEs (Common Terminology Criteria for Adverse Events [CTCAE] grades 3–4) compared with those carrying the GC or GG genotype (P = 0.004, OR = 6.78, 95% CI = 1.87–24.60; Figure 4A; Supplemental Table 2). Similarly, the CTCAE grade was significantly higher in patients carrying the CC genotype compared with patients with a GC or GG genotype (Figure 4B). The main irAE target organs in our cohort were the skin, liver, intestine, lungs, and endocrine glands (Supplemental Table 3). The organ distribution of irAEs was comparable between the genotype groups, with a slight increase in gastrointestinal tract irAEs in the CC group (Supplemental Table 3). Histopathology of colon biopsies of patients affected by immune-related colitis showed a mixed infiltration of granular lymphocytic inflammatory cells with highly active inflammation, granulocytic lesion of the crypts, abscesses, and apoptosis, as well as a dense lymphoplasmocytic infiltration with intraepithelial lymphocytosis, particularly in patients with the CC genotype (Figure 4C).
Figure 4. SNP rs2910164 in the MIR146A gene is associated with increased irAE severity in patients.
Genotype of the SNP rs2910164 in the MIR146A gene was determined from 167 patients with cancer treated with ≥3 cycles anti–PD-1 or anti–PD-L1 therapy. irAEs were graded according to the Common Terminology Criteria for Adverse Events (CTCAE). (A) Genotype frequencies for the group of patients who developed no/mild irAEs (CTCAE grades 0–2) or severe irAEs (CTCAE grades 3–4), respectively, are shown. Fisher’s exact test was used to analyze the contingency table. (B) Percentage of patients with CTCAE grades 0–4 irAEs in the different genotype groups is shown. Data were statistically analyzed by Kruskal-Wallis test followed by post hoc pairwise comparison Dunn’s test and P values were adjusted by Bonferroni’s correction. (C) Exemplary colon histopathology pictures of a GG patient with grade 1 intestinal irAEs (left) and of a CC patient with grade 4 intestinal irAEs (right) are shown. Scale bar: 50 μm. irAE, immune-related adverse event, PD-1, programmed cell death protein-1.
The MIR146A SNP rs2910164 is associated with increased neutrophil numbers and mortality rate in patients undergoing ICI therapy.
Since we found increased neutrophil levels in miR-146a–deficient mice treated with ICIs, we next analyzed peripheral blood neutrophil levels of patients with solid tumors before and during ICI therapy. In line with our findings in the murine model and the histopathological representation, proportions of neutrophils were increased in patients with the SNP reducing miR-146a expression compared with patients without the SNP both at baseline (Figure 5A) and during ICI therapy (Figure 5B).
Figure 5. Patients with the MIR146A SNP rs2910164 display increased neutrophil numbers and reduced survival.
(A and B) Patient charts of patients treated with immune checkpoint inhibitors (ICIs) were retrospectively analyzed for neutrophil counts before ICI treatment (A) and during ICI treatment (B), where available. Percentage of neutrophils of total leukocyte count is depicted for patients with the rs2910164 GG, GC, and CC genotype, respectively. Data were analyzed by 2-tailed unpaired Student’s t test. (C and D) Progression-free survival of patients with melanoma (C) and patients with lung cancer (D) according to their rs2910164 genotype is shown. Data were analyzed by log-rank (Mantel-Cox) test. (E) Corticosteroid dose due to inflammatory side effects in the different rs2910164 genotype groups is depicted. Low-dose corticosteroids, <1 mg/kg/d prednisone or equivalent. High-dose corticosteroids, ≥ 1 mg/kg/d. Data were analyzed by Kruskal-Wallis test followed by Dunn-Bonferroni’s post hoc test.
Previous studies have suggested that patients developing irAEs have a better clinical response to ICI therapy, whereas others reported that treatment outcomes are not affected by irAE occurrence (21–23). Therefore, we next asked whether the miR-146a SNP not only affects irAE development but also influences therapy outcome. We analyzed progression-free survival (PFS) of patients with melanoma and lung cancer, respectively, starting from the first cycle of ICI therapy. Baseline and clinical patient characteristics were similar between the genotype groups, for both patients with malignant melanoma (Supplemental Table 4) and patients with lung cancer (Supplemental Table 5). Surprisingly, we found that melanoma patients with the CC genotype had a significantly reduced PFS after ICI therapy compared with patients without the SNP (Figure 5C). Although the number of patients with lung cancer was limited in our cohort, we also found a trend toward a shorter PFS for patients with the CC genotype in this population (Figure 5D). To assess whether the reduced PFS might be due to increased treatment with steroids in CC patients developing severe irAEs, we next analyzed corticosteroid dosage in our cohort. In line with the more frequent development of severe irAEs in patients with the CC genotype, this group of patients had to be treated with high-dose corticosteroids more often (Figure 5E). These data indicate that the patient population with the rs2910164 C>G SNP benefits less from ICI therapy, manifested by augmented irAE severity, increased need for steroid intervention, and shorter PFS.
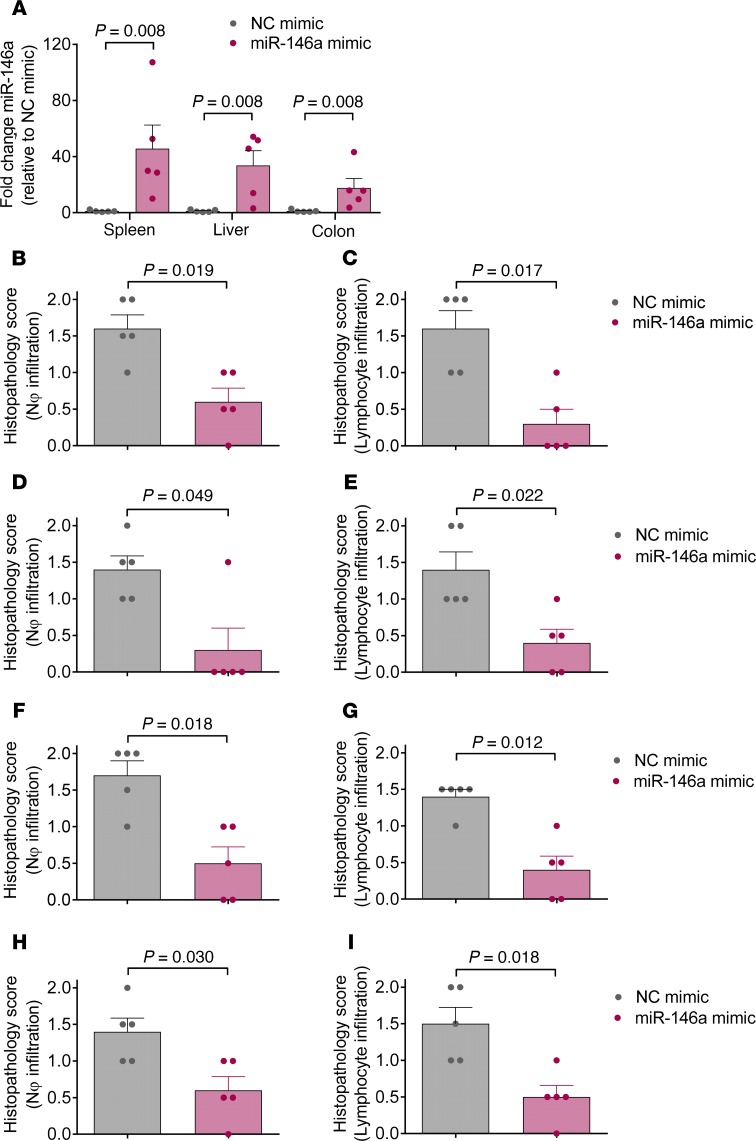
Exogenous increase of miR-146a using a miR-146a mimic ameliorates irAEs.
Having shown that miR-146a modulates irAE development in both mice and humans, we next investigated whether this finding could be used in a therapeutic setting to prevent irAEs. Therefore, we treated WT C57BL/6 mice developing irAEs with a specific miR-146a mimic or a negative control mimic on days 5, 9, 13, 17, and 21 after the first anti–PD-1 treatment. Treatment with the miR-146a mimic led to significantly increased miR-146a expression in the spleen, colon, and liver (Figure 6A). In line with our previous data, treatment with the miR-146a mimic significantly reduced irAE severity in the major irAE target organs, including the lung, liver, colon, and skin, as assessed by histopathological scoring (Figure 6, B–I). Taken together, we believe our results indicate that exogenous increase of miR-146a expression may be a potential novel strategy to prevent irAE development.
Figure 6. Treatment with a miR-146a specific mimic reduces irAE severity in anti–PD-1–treated mice.
WT C75BL/6 mice (n = 5 per group) were treated with LPS and 250 μg of anti–PD-1 antibody for 3 weeks as described. Additionally, mice were treated i.p. with negative control (NC) mimic or miR-146a mimic in in vivo-jetPEI on days 5, 9, 13, 17, and 21. (A) Expression of miR-146a in the spleen, liver, and colon was analyzed by qRT-PCR on day 22. (B–I) The lungs, liver, colon, and skin were isolated on day 22 for histopathological assessment and irAE grading by an experienced pathologist. Histopathology scores for neutrophil infiltration and lymphocyte infiltration into the lung (B and C), liver (D and E), colon (F and G), and skin (H and I) are shown. 0 = absent, 1 = mild, 2 = massive neutrophil/lymphocyte infiltration. Data were statistically analyzed by Mann Whitney U test. miR-146a, microRNA-146a, irAE, immune-related adverse event, PD-1, programmed cell death protein-1.
Discussion
The use of ICI therapy has revolutionized the therapy of immune-sensitive cancer entities. Today, one of the major objectives is to rationally combine different checkpoint inhibitors for even more efficient cancer treatment. However, by globally increasing the activity of the immune system, ICI therapy can also increase autoimmune responses and induce inflammatory side effects, known as irAEs. Grade 3–4 irAEs, in particular, are a major concern, as discontinuation of ICI therapy leads to tumor progression and death related to irAEs and cancer can occur (24, 25).
To date, no reliable clinical or molecular biomarkers to predict the occurrence of severe irAEs are available. It is still largely unknown why some patients develop severe irAEs while other patients do not (5) and the pathophysiology of irAEs has not been assessed in detail yet. In this study, we identified miR-146a as a crucial miRNA regulating irAE severity in 2 murine irAE models and in humans. This miRNA was chosen based on previous reports showing its role in autoimmunity, including Lupus erythematosus and rheumatoid arthritis (26–29) and GVHD (9, 10). One major finding was that mice lacking this miRNA exhibited stronger inflammation in the lungs, liver, colon, and skin, which are the organs most commonly affected by irAEs (1). To our knowledge, this is the first functional study showing that a single immune regulatory molecule can affect the risk of developing severe irAEs. Currently, there are published studies reporting a connection between patient-derived biomarkers, such as lymphocyte counts, polymorphisms and cytokine profiles, and the risk of developing irAEs after anti–CTLA-4 therapy (30) or B cell number changes predicting irAEs when patients were treated with anti–PD-1 and anti–CTLA-4 combination therapy (31). In contrast to these studies, we provide a mechanistic insight into the development of irAEs because we observed that miR-146a regulates IFN-γ and perforin production in T cells of mice treated with anti–PD-1 antibody, thereby ameliorating irAE severity. scRNA-seq revealed that CD4 T cells from miR-146–/–treated with ICIs showed a stronger immune activation signature compared with WT T cells. These findings are in line with the mode of action of ICIs and the concept that irAE development is thought to be mainly CD4 T cell driven (32, 33). Moreover, neutrophil numbers were significantly increased in miR-146a–/– mice developing irAEs, in both the spleen and the intestine, common irAE target organs. In line with our findings, gastrointestinal irAEs have been shown to be associated with increased levels of neutrophil activation markers CD177 and CEACAM1, which are correlated with neutrophilic inflammation (34).
In addition to the KO approach using miR-146a–/– mice, we showed that systemic delivery of a miR-146a mimic in cationic polyethylenimine nanoparticles can highly increase miR-146a expression in vivo, which is consistent with previous studies by our lab and others (10, 35). Our data show that treatment with the miR-146a mimic effectively reduced irAE severity, providing evidence that miR-146a replacement therapy might be effective in patients with severe irAEs. miRNA therapeutics such as miRNA mimics or antagomirs have already been shown to exert beneficial actions in preclinical models of inflammatory diseases and cancer (10, 36–38). Although the development of miRNA therapeutics has not yet translated into clinically approved therapies, multiple miRNA mimics or inhibitors are under investigation in clinical trials, underlining the potential of miRNA replacement therapy in the future (reviewed in ref. 39). One limitation of our study is that there is currently no miR-146a-specific mimic under clinical development and extensive, further studies will be needed before a miR-146a mimic could enter the clinical phase. These include the careful investigation of additional in vivo targets of miR-146a that might cause unfavorable side effects and the examination of a safe, efficient, and targeted in vivo delivery system in humans. However, downstream effects of miR-146a in the regulation of irAEs identified in our study might be more readily targetable in clinical practice. Pharmacological measures to interfere with both IFN-γ signaling and neutrophil activation and recruitment, such as JAK1/2 inhibitors, which potently reduced acute GVHD (40), could be a potentially novel approach to interfere with irAEs.
In addition to the functional studies in the irAE mouse model, we also provide strong evidence that a common allelic variant in the MIR146A gene leads to increased risk for irAE in patients. We found that patients with the MIR146A polymorphism rs2910164, which leads to an about 2-fold reduced miR-146a expression (19), had a significantly increased risk of developing severe irAEs during ICI therapy compared with patients without the SNP. In accordance with our data, this SNP has previously been shown to be associated with acute GVHD severity and autoimmune diseases, including multiple sclerosis and inflammatory bowel disease (9, 41, 42). In order to establish this SNP as a new biomarker for irAE development, further studies will be needed and our findings should be validated in a larger, independent cohort. Ideally, clinical validation would involve determination of the rs2910164 genotype in a large clinical trial of anti–PD-1 and –PD-L1 antibodies, where baseline patient characteristics, irAE grading, and efficiency outcomes are available in a controlled setting.
Our finding that patients with the rs2910164 CC genotype exhibited a reduced PFS rate may be related to the reduced number of administered ICI therapy cycles and the increased use of corticosteroids in patients with the CC genotype in our study cohort. Consistent with this hypothesis, it has been shown that steroid use reduces the efficacy of PD-1 and PD-L1 inhibitors (43, 44). Our data indicate that the patient population with the rs2910164 CC genotype benefits less from ICI therapy as revealed by increased irAE severity, greater need for steroid intervention, and shorter PFS. In summary, we believe we have identified a novel mechanism of irAE regulation by miR-146a in mice and patients and a potential new biomarker to predict severe irAE development before the start of ICI therapy.
Methods
Human subjects.
For this prospective clinical study, genomic DNA from peripheral blood of 179 patients with cancer treated with ICIs was isolated and the MIR146A rs2910164 genotype was determined (Clinicaltrials.gov NCT04038996). Histopathological analyses and scoring of human colon biopsies were performed by experienced pathologists from the Institute of Surgical Pathology of Freiburg. Medical history was analyzed retrospectively for irAEs, percentage of neutrophils, corticosteroid therapy, and disease progression. irAEs were graded according to CTCAE version 5.0.
Human SNP analysis.
Genomic DNA was extracted and purified from peripheral blood samples using the QIAamp DNA Micro Kit and a QIAcube System (QIAGEN). Quantitative real-time–PCR–based (qRT-PCR–based) TaqMan SNP Genotyping Assay (ASSAY ID C_15946974_10, SNP DB ID: rs2910164; Life Technologies) was used to identify the single nucleotide polymorphism of interest. Two primers for each sequence and 2 minor groove binder (MGB) probes for detecting the alleles were applied. MGB probes were labeled with different fluorescence dyes (VIC or 6FAM). Data were analyzed using LightCycler 480 Software, version 1.5.1.62.
Mice.
C57BL/6 WT mice were obtained from the local stock of the animal facility at Freiburg University Medical Center, and gene-targeted miR-146a–/– mice on a C57BL/6 background were originally purchased from Jackson Laboratories and bred in the same mouse facility as the WT mice. All mice were housed under specific pathogen–free conditions at the mouse facility of the University Medical Center Freiburg. Age- and gender-matched mice that were 7 to 14 weeks of age were used for all in vivo experiments.
Murine irAE model and histopathological analysis of irAE target organs.
C57BL/6 WT and miR-146a–/– mice were treated i.p. with 1 mg/kg LPS once a week and 200 μg anti–PD-1 (clone J43, provided by B. Blazar, University of Minnesota, Minneapolis, Minnesota, USA) or Armenian hamster isotype control antibody (polyclonal, Bio X Cell) twice a week for 3 weeks (Supplemental Figure 1). The mice were sacrificed for analysis on day 22 after the first treatment. Sections of skin, liver, colon, and lung collected on day 22 were stained with H&E and scored on the basis of histopathological characterization of human irAEs, including lymphocyte and neutrophil infiltration, crypt abscesses, and apoptotic cells (11, 12), by an experienced pathologist.
Treatment with miRNA mimics.
WT C57BL/6 mice were treated with LPS and anti–PD-1 according to the schedule described above. The dose was increased to 250 μg anti–PD-1 per injection in order to increase the irAE severity in WT mice. For in vivo delivery of miRNA mimics, in vivo-jetPEI (Polyplus Transfection) was used according to the manufacturer’s instructions. Recipient mice were treated i.p. with 2.4 mg/kg body weight of the mirVana miRNA mimic Negative Control #1 (Thermo Fisher) or the mirVana miRNA-146a mimic (Thermo Fisher) in in vivo-jetPEI/5% glucose (N/P ratio = 7) on days 5, 9, 13, 17, and 21 after the first anti–PD-1 treatment.
TaqMan qRT-PCR for miRNA expression.
For analysis of miR-146a expression, total RNA was isolated from whole splenocytes, liver, or colon on day 22 after the first anti–PD-1 treatment using the miRNeasy Mini Kit (QIAGEN). MultiScribe Reverse Transcriptase (Thermo Fisher) was used for reverse transcription and miR-146a expression was examined using the TaqMan MicroRNA Assay Kit (Thermo Fisher) and a LightCycler 480 (Roche). TaqMan primers for mmu-miR-146a-5p (assay ID 000468) were used to monitor miRNA expression. Gene expression was normalized to the reference gene snoRNA202 (assay ID 001232) and expressed relative to the negative control mimic group (ΔΔCT).
scRNA-seq and GSEA.
WT C57BL/6 mice and miR-146–KO mice (n = 2 per group) were treated with LPS and anti–PD-1 according to the treatment schedule described above. On day 22, total splenic CD4 and CD8 T cells were isolated using negative selection with the mouse Pan T Cell Isolation Kit II (Miltenyi Biotec) and then counted and washed in 0.04% BSA/PBS according to the 10x Genomics’ cell preparation protocol. Single cells were partitioned together with 10× barcoded Gel Beads Gel Bead-in-Emulsion and reverse transcriptase enzymatic reaction using the Chromium Controller Single-Cell Instrument (10× Genomics). scRNA-seq libraries were then prepared using Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 (10× Genomics) according to the manufacturer’s instructions (one sample per GEM well). Libraries were pooled and sequenced on a NovaSeq 6000 (Illumina) at 44658 mean reads per cell. Samples were demultiplexed using bcl2fastq version 2.20.0.422 (Illumina). Barcode processing, alignment, and single-cell 3′ gene counting were performed using Cell Ranger Software version 3.1.0 (10× Genomics). Further data processing, visualization, and analysis were done using the Seurat pipeline version 3.0 (45, 46). After log-normalization of raw gene counts, cells with unique gene counts <250 or >2500, as well as cells with >10% of mitochondrial genes, were removed from the analysis. Cells were clustered using the FindClusters function of the Seurat analysis package, which identifies clusters of cells by a shared nearest neighbor modularity optimization-based clustering algorithm. Clusters of cells expressing myeloid (Csf3r, Cxcr2, Ly6g, C1qa, C1qb), B cell (Ig genes, Mzb1), and NK cell (Klr genes, Ncr1) markers, and lacking T cell markers (Cd3, Cd4, and Cd8) represented 17.6% of the total population and were removed from the analysis. Pure T cells were clustered again using the Seurat FindClusters function. GSEA of major T cell clusters was performed using the fgsea R package (47). Hallmark gene sets used in these analyses were derived from the MSigDB and immune-related gene sets were selected (48). The scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus database (GEO GSE147327; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147327).
Murine LPS–independent combination therapy irAE model and histopathological analysis of irAE target organs.
To create BM chimeric mice, WT C57BL/6 recipients were irradiated with 12 Gy before i.v. injection of 5 × 106 WT C57BL/6 or miR-146–/– C57BL/6 BM cells. Starting 5 weeks after the syngeneic transplantation, C57BL/6 WT and miR-146a–/– BM chimera were treated i.p. with 250 μg anti–PD-1 (clone J43) and 150 μg anti–mouse CTLA-4 (CD152) antibody (clone 9D9, Bio X Cell) or the respective isotype control antibodies (polyclonal Armenian hamster IgG and IgG2b isotype control, clone MPC-11, Bio X Cell, respectively) diluted in PBS on days 0, 3, 6, 9, and 12. The mice were sacrificed for analysis on day 15 after the first treatment. Sections of skin, liver, colon, and lung collected on day 15 were stained with H&E and scored on the basis of histopathological characterization of human irAEs, including lymphocyte and neutrophil infiltration, crypt abscesses, and apoptotic cells (11, 12), by an experienced pathologist blinded to the experimental groups.
Intestinal leukocyte isolation.
Intestinal leukocytes were isolated as described before (49). In brief, 2-cm-long intestinal segments were isolated and Peyer’s patches removed. Segments were opened longitudinally and rinsed in PBS to remove feces. Epithelial cells were separated from the lamina propria using CD buffer (Hank’s Balanced Salt Solution [Gibco, Thermo Fisher] without Mg2+, Ca2+, 5 mm EDTA, 10 mm N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and the remaining tissue was digested with digestion buffer (Hank’s Balanced Salt Solution with Mg2+, Ca2+, collagenase D 0.5 mg/mL (Roche), DNAse (MilliporeSigma) 0.5 mg/mL, dispase 0.5 U/mL) to obtain single-cell suspensions that were further processed for flow cytometry.
Flow cytometry.
Spleens were isolated from WT and miR-146a–KO mice after treatment with anti–PD-1 and erythrocytes were lysed. Intestinal leukocytes were isolated as described above. Dead cells were stained using Zombie NIR fixable dye (BioLegend) and Fc receptors were blocked using anti–mouse CD16/CD32. Surface staining was performed using fluoro-conjugated antibodies for 20 minutes at 4°C. Intracellular staining was performed using the eBioscience Transcription Factor Staining Buffer Set (Thermo Fisher). All antibodies used for flow cytometry are listed in Supplemental Table 6. Data were acquired on a BD LSR Fortessa (BD Biosciences) and analysis was performed using FlowJo software version 10.
Statistics.
GraphPad Prism version 7.01 and IBM SPSS Statistics version 23 were used for statistical analysis. Unpaired 2-tailed Student’s t test was used for comparison of 2 groups of normally distributed data. The Mann-Whitney U test was used when the data did not conform to a normal distribution. Data are presented as mean ± SEM. Differences were considered statistically significant when P < 0.05.
For comparison of more than 2 groups, statistical significance was analyzed by 1-way ANOVA, followed by Tukey’s post hoc test for parametric data, and by Kruskal-Wallis test, followed by 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, if nonparametric testing was suggested. If multiple comparison tests were used, the adjusted P value is depicted.
The Hardy-Weinberg equilibrium for the polymorphism rs2910164 was analyzed using a χ2 test. Frequencies of genotypes were compared using Fisher’s exact test. Kruskal-Wallis test followed by Dunn-Bonferroni’s post hoc test was used to compare the CTCAE grade between the genotype groups. PFS was estimated using the Kaplan-Meier-method and compared by log-rank (Mantel-Cox) test.
Study approval.
We collected all human samples after approval by the ethics committee of ALU Freiburg (protocol 300/16) and after written informed consent in accordance with the Declaration of Helsinki. All animal experiments were performed in accordance to the respective national, federal, and institutional regulations. Animal protocols (G-19/36, X-15/10A) were approved by the Regierungspräsidium Freiburg, Freiburg, Germany (Federal Ministry for Nature, Environment and Consumers’ Protection of the state of Baden-Württemberg).
Author contributions
DM, MF, and KHM designed and performed the experiments, acquired and analyzed data. The authorship order of the 2 co–first authors was assigned based on the first key experiments that were performed and analyzed by DM. NRJ, EH, and S. Duquesne contributed to experimental design, conducted experiments, and analyzed the data. FS, S. Dicks, MK, MB, and RSN helped with scRNA-seq experimental design and data analysis. JR, KA, PM, DRS, and FM helped to collect and analyze patient data. ASG performed histopathological scoring; JD contributed to experimental design and data interpretation; and RZ developed the overall concept, analyzed the data, and helped to write the manuscript. NK designed the experiments, developed the overall concept, performed the experiments, analyzed the data, and wrote the manuscript.
Supplementary Material
Acknowledgments
PD-1 blocking antibody was provided by Bruce Blazar (Department of Pediatrics, Division of Blood and Marrow Transplantation, University of Minnesota, Minneapolis, Minnesota, USA). This study was supported by the Else Kröner-Fresenius-Stiftung (2019_A74 to NK); the DKMS Foundation for Giving Life (Mechtild Harf Research Grant 2019 to NK); the Deutsche Forschungsgemeinschaft, Germany, SFB850 (C6 and Z2 to RZ and Z1 to MB), TRR167 project B06 to RZ, SFB1160 TP B09 to RZ and Z02 to MB; European Research Council Consolidator grant (ERC-2015-COG 681012 GvHDCure to RZ); and the German Federal Ministry of Education and Research within the framework of the e:Med research and funding concept CoNfirm (FKZ 01ZX1708F to MB and S. Dicks).
Version 1. 03/03/2020
In-Press Preview
Version 2. 03/26/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(6):e132334.https://doi.org/10.1172/jci.insight.132334.
Contributor Information
Dominik Marschner, Email: do.marschner@klinikum-stuttgart.de.
Martina Falk, Email: Martina.falk.87@hotmail.de.
Nora Rebeka Javorniczky, Email: rebeka.javorniczky@uniklinik-freiburg.de.
Kathrin Hanke-Müller, Email: k.hanke-mueller@web.de.
Justyna Rawluk, Email: justyna.rawluk@uniklinik-freiburg.de.
Annette Schmitt-Graeff, Email: annette.schmitt-graeff@uniklinik-freiburg.de.
Federico Simonetta, Email: fedsimo@stanford.edu.
Eileen Haring, Email: eileen.haring@uniklinik-freiburg.de.
Severin Dicks, Email: severin.dicks@uniklinik-freiburg.de.
Manching Ku, Email: manching.ku@uniklinik-freiburg.de.
Sandra Duquesne, Email: sandra.duquesne@uniklinik-freiburg.de.
Konrad Aumann, Email: konrad.aumann@uniklinik-freiburg.de.
David Rafei-Shamsabadi, Email: david.rafei-shamsabadi@uniklinik-freiburg.de.
Frank Meiss, Email: frank.meiss@uniklinik-freiburg.de.
Patrick Marschner, Email: patrick.marschner@uniklinik-freiburg.de.
Melanie Boerries, Email: melanie.boerries@uniklinik-freiburg.de.
Robert S. Negrin, Email: negrs@stanford.edu.
Justus Duyster, Email: justus.duyster@uniklinik-freiburg.de.
Robert Zeiser, Email: robert.zeiser@uniklinik-freiburg.de.
Natalie Köhler, Email: natalie.koehler@uniklinik-freiburg.de.
References
- 1.Johnson DB, Chandra S, Sosman JA. Immune checkpoint inhibitor toxicity in 2018. JAMA. 2018;320(16):1702–1703. doi: 10.1001/jama.2018.13995. [DOI] [PubMed] [Google Scholar]
- 2.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378(2):158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 6.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209(9):1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stickel N, et al. MicroRNA-146a reduces MHC-II expression via targeting JAK/STAT signaling in dendritic cells after stem cell transplantation. Leukemia. 2017;31(12):2732–2741. doi: 10.1038/leu.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stickel N, et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood. 2014;124(16):2586–2595. doi: 10.1182/blood-2014-04-569046. [DOI] [PubMed] [Google Scholar]
- 11.Beck KE, et al. Enterocolitis in patients with cancer after antibody blockade of cytotoxic T-lymphocyte-associated antigen 4. J Clin Oncol. 2006;24(15):2283–2289. doi: 10.1200/JCO.2005.04.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johncilla M, et al. Ipilimumab-associated hepatitis: clinicopathologic characterization in a series of 11 cases. Am J Surg Pathol. 2015;39(8):1075–1084. doi: 10.1097/PAS.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 13.Hirschhorn D, et al. A delicate interplay between adaptive and innate immunity caused by immunotherapy triggers tumor immunity and aseptic inflammation. J Immunol. 2018;200(suppl_1):17842 [Google Scholar]
- 14.Templeton AJ, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106(6):dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 15.Shen M, Hu P, Donskov F, Wang G, Liu Q, Du J. Tumor-associated neutrophils as a new prognostic factor in cancer: a systematic review and meta-analysis. PLoS One. 2014;9(6):e98259. doi: 10.1371/journal.pone.0098259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrucci PF, et al. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. Br J Cancer. 2015;112(12):1904–1910. doi: 10.1038/bjc.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebhardt C, et al. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with ipilimumab. Clin Cancer Res. 2015;21(24):5453–5459. doi: 10.1158/1078-0432.CCR-15-0676. [DOI] [PubMed] [Google Scholar]
- 18.Kuzman JA, et al. Neutrophil-lymphocyte ratio as a predictive biomarker for response to high dose interleukin-2 in patients with renal cell carcinoma. BMC Urol. 2017;17(1):1. doi: 10.1186/s12894-016-0192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löfgren SE, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13(3):268–274. doi: 10.1038/gene.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attia P, et al. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J Clin Oncol. 2005;23(25):6043–6053. doi: 10.1200/JCO.2005.06.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downey SG, et al. Prognostic factors related to clinical response in patients with metastatic melanoma treated by CTL-associated antigen-4 blockade. Clin Cancer Res. 2007;13(22 pt 1):6681–6688. doi: 10.1158/1078-0432.CCR-07-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvat TZ, et al. Immune-related adverse events, need for systemic immunosuppression, and effects on survival and time to treatment failure in patients with melanoma treated with ipilimumab at Memorial Sloan Kettering Cancer Center. J Clin Oncol. 2015;33(28):3193–3198. doi: 10.1200/JCO.2015.60.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haanen JBAG, et al. Management of toxicities from immunotherapy: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(suppl_4):iv119–iv142. doi: 10.1093/annonc/mdx225. [DOI] [PubMed] [Google Scholar]
- 25.Brahmer JR, et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2018;36(17):1714–1768. doi: 10.1200/JCO.2017.77.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 27.Ji JD, Cha ES, Lee WJ. Association of miR-146a polymorphisms with systemic lupus erythematosus: a meta-analysis. Lupus. 2014;23(10):1023–1030. doi: 10.1177/0961203314534512. [DOI] [PubMed] [Google Scholar]
- 28.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63(6):1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, et al. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann Rheum Dis. 2015;74(6):1265–1274. doi: 10.1136/annrheumdis-2013-204377. [DOI] [PubMed] [Google Scholar]
- 30.Manson G, Norwood J, Marabelle A, Kohrt H, Houot R. Biomarkers associated with checkpoint inhibitors. Ann Oncol. 2016;27(7):1199–1206. doi: 10.1093/annonc/mdw181. [DOI] [PubMed] [Google Scholar]
- 31.Das R, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest. 2018;128(2):715–720. doi: 10.1172/JCI96798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaehler KC, Piel S, Livingstone E, Schilling B, Hauschild A, Schadendorf D. Update on immunologic therapy with anti-CTLA-4 antibodies in melanoma: identification of clinical and biological response patterns, immune-related adverse events, and their management. Semin Oncol. 2010;37(5):485–498. doi: 10.1053/j.seminoncol.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Anderson R, Rapoport BL. Immune dysregulation in cancer patients undergoing immune checkpoint inhibitor treatment and potential predictive strategies for future clinical practice. Front Oncol. 2018;8:80. doi: 10.3389/fonc.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahabi V, et al. Gene expression profiling of whole blood in ipilimumab-treated patients for identification of potential biomarkers of immune-related gastrointestinal adverse events. J Transl Med. 2013;11:75. doi: 10.1186/1479-5876-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morishita Y, et al. Delivery of microRNA-146a with polyethylenimine nanoparticles inhibits renal fibrosis in vivo. Int J Nanomedicine. 2015;10:3475–3488. doi: 10.2147/IJN.S82587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranganathan P, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119(20):4786–4797. doi: 10.1182/blood-2011-10-387522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima G, et al. In vivo delivery and therapeutic effects of a microRNA on colorectal liver metastases. Mol Ther. 2017;25(7):1588–1595. doi: 10.1016/j.ymthe.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114–127. [PMC free article] [PubMed] [Google Scholar]
- 40.Zeiser R, et al. Ruxolitinib in corticosteroid-refractory graft-versus-host disease after allogeneic stem cell transplantation: a multicenter survey. Leukemia. 2015;29(10):2062–2068. doi: 10.1038/leu.2015.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Common genetic variation within miR-146a predicts disease onset and relapse in multiple sclerosis. Neurol Sci. 2018;39(2):297–304. doi: 10.1007/s10072-017-3177-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhu M, Li D, Jin M, Li M. Association between microRNA polymorphisms and the risk of inflammatory bowel disease. Mol Med Rep. 2016;13(6):5297–5308. doi: 10.3892/mmr.2016.5157. [DOI] [PubMed] [Google Scholar]
- 43.Arbour KC, et al. Impact of baseline steroids on efficacy of programmed cell death-1 and programmed death-ligand 1 blockade in patients with non-small-cell lung cancer. J Clin Oncol. 2018;36(28):2872–2878. doi: 10.1200/JCO.2018.79.0006. [DOI] [PubMed] [Google Scholar]
- 44.Scott SC, Pennell NA. Early use of systemic corticosteroids in patients with advanced NSCLC treated with nivolumab. J Thorac Oncol. 2018;13(11):1771–1775. doi: 10.1016/j.jtho.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. doi: 10.1101/060012. Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016:060012. Posted . Posted June 20, 2019. Accessed February 25, 2020. [DOI]
- 48.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hulsdunker J, Zeiser R. In vivo myeloperoxidase imaging and flow cytometry analysis of intestinal myeloid cells. Methods Mol Biol. 2016;1422:161–167. doi: 10.1007/978-1-4939-3603-8_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.