SUMMARY
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a tick-borne virus with 12%−30% case mortality rates and is related to the Heartland virus (HRTV) identified in the United States. Together, SFTSV and HRTV are emerging segmented, negative-sense RNA viral (sNSV) pathogens with potential global health impact. Here, we characterize the amino-terminal cap-snatching endonuclease domain of SFTSV polymerase (L) and solve a 2.4-Å X-ray crystal structure. While the overall structure is similar to those of other cap-snatching sNSV endonucleases, differences near the C terminus of the SFTSV endonuclease suggest divergence in regulation. Influenza virus endonuclease inhibitors, including the US Food and Drug Administration (FDA) approved Baloxavir (BXA), inhibit the endonuclease activity in in vitro enzymatic assays and in cell-based studies. BXA displays potent activity with a half maximal inhibitory concentration (IC50) of ~100 nM in enzyme inhibition and an EC50 value of ~250 nM against SFTSV and HRTV in plaque assays. Together, our data support sNSV endonucleases as an antiviral target.
In Brief
Wang et al. solve the X-ray crystal structure of SFTSV L endonuclease domain and investigate the characteristics of SFTSV and HRTV endonuclease function. Resulting data support a mechanism for regulation. Baloxavir effectively inhibits the endonuclease activity of SFTSV and HRTV.
Graphical Abstract
INTRODUCTION
Severe fever with thrombocytopenia syndrome virus (SFTSV) is an emerging pathogen that was initially discovered in China in 2009 (Yu et al., 2011) and has spread throughout East Asia, including to Korea and Japan. SFTSV was isolated from patients who presented with fever, thrombocytopenia, leukocytopenia, and multiorgan failure (Chen et al., 2012; Yu et al., 2011; Zhang et al., 2012a, 2012b). Ticks are the potential vector responsible for the spread of SFTSV to humans, and among them, Haemaphysalis longicornis has been identified in the transmission of the virus (Luo et al., 2015). Heartland virus (HRTV), a related tick-borne virus, was found in the US in 2009 and infected individuals present with similar symptoms as SFTSV, highlighting the widespread impact of banyangviruses to global human health (McMullan et al., 2012). Currently, there are no US Food and Drug Administration (FDA)-approved therapies or vaccines available to counter bunyaviral infections. Most clinical treatments are limited to therapeutic plasma exchange (Oh et al., 2017) and supportive care. A standard nucleoside analog class of inhibitors, such as ribavirin and favipiravir, has been tested for antiviral activities against SFTSV and HRTV infections, as they have been tested experimentally for other bunyaviral infections, including Hantaan virus (HTNV), Crimean Congo hemorrhagic fever virus (CCHFV), and Rift Valley Fever virus (RVFV) (Beaucourt and Vignuzzi, 2014; Delang et al., 2018; Tani et al., 2018; Westover et al., 2017). However, these nucleoside analogs are clinically ineffective (Liu et al., 2013) and are associated with adverse side effects. Plus many nucleotide analog inhibitors are prone to the emergence of rapid resistant mutants. Ribavirin is not recommended for clinical use owing to its high toxicity (Lu et al., 2015; Russmann et al., 2006), and favipiravir possesses a risk of teratogenicity and embryotoxicity (Furuta et al., 2017). Thus, identification and characterization of antiviral targets within SFTSV and HRTV will facilitate the discovery of previously unrecognized opportunities to combat bunyaviral infections.
SFTSV is a segmented, negative-sense RNA virus (sNSV), which includes viruses from the Bunyavirales and Articulavirales orders. This virus is a member of the newly identified Banyangvirus genus in the Phenuiviridae family of the Bunyavirales order according to the Virus Taxonomy 2019, which is released by the International Committee on Taxonomy of Viruses (ICTV) (Abudurexiti et al., 2019). SFTSVs form spherical virions of approximately 80–100 nm in diameter with viral glycoprotein (Gn and Gc) at the membrane that facilitates entry (Lei et al., 2015; Yu et al., 2011). Inside the virion, the RNA genome is composed into three segments (S, small; M, medium; and L, large) that are encompassed by nucleoprotein and bound to L polymerase. These segments encode for nucleoprotein (N) and nonstructural protein (NSs), glycoprotein (Gn and Gc), and the RNA-dependent RNA polymerase (RdRp or L), respectively (Lei et al., 2015).
The bunyavirus L polymerase is required for viral replication and transcription and it does not encode a domain with capping activity. Instead, bunyaviruses employ the amino (N)-terminal endonuclease domain of the L polymerase as a cap-snatching mechanism that cannibalizes host cellular mRNA cap structures. These short, capped RNA fragments then serve as primers for viral mRNA transcription and allow for viral protein translation (Reguera et al., 2010, 2016). Although this process has been described extensively for influenza A virus (IAV) and to some degree for other bunyaviruses (Amroun et al., 2017; Dias et al., 2009; Duijsings et al., 2001; Garcin et al., 1995; Plotch et al., 1981), less is known structurally and biochemically about the SFTSV polymerase. Here, we report the identification of the N-terminal endonuclease domain of the L polymerase of SFTSV, which is crucial for cap-dependent viral mRNA transcription. We solved the X-ray crystal structure of the endonuclease domain at 2.4-Å resolution, which revealed features that are unique to SFTSV as well as those features that are shared among other cap-snatching viral endonuclease domains (Fernández-García et al., 2016; Jones et al., 2019; Reguera et al., 2010, 2016; Song et al., 2016). We also demonstrated the inhibition of SFTSV and HRTV endonuclease activities by using IAV polymerase acidic protein (PA) inhibitors, including Baloxavir acid (BXA) derived from a recently FDA-approved drug, Baloxavir marboxil (BXM), suggesting that the banyangvirus endonuclease domain is a major therapeutic target that can guide the development of broad-spectrum antivirals against sNSVs.
RESULTS
X-Ray Crystal Structure of the SFTSV L Endonuclease Domain Reveals a Mixed Alpha-Beta Fold
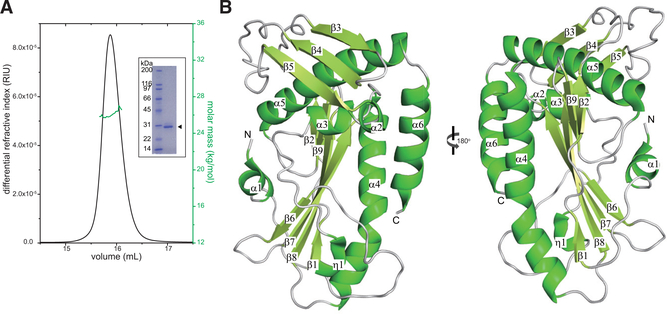
While the domain organization of SFTSV polymerase remains undefined, it is likely to be similar to other bunyaviruses (Fernández-García et al., 2016; Gogrefe et al., 2019; Jones et al., 2019; Reguera et al., 2016; Reguera et al., 2010), including Toscana virus (TOSV), Andes virus (ANDV), Lassa virus (LASV), La Crosse virus (LACV), and RVFV, which contain an N-terminal endonuclease domain, a central RdRp domain, and a C-terminal cap-binding domain (Figure S1A). SFTSV, HRTV, TOSV, ANDV, LASV, LACV, and RVFV are genetically distinct and belong to different bunyavirus families, while IAV is assigned to the Articulavirales order (Figure S1B). Sequence alignment of the N-terminal residues of several sNSVs suggests that SFTSV and HRTV polymerases also include an N-terminal endonuclease domain (Figure S1C). In order to biochemically characterize the SFTSV endonuclease domain, we recombinantly expressed and purified the N terminus (1–231 aa) of SFTSV. Size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) analysis determined a molecular mass of 2.71 ± 0.01 × 104 Da and the calculated protein molecular mass is 2.76 × 104 Da, suggesting that the endonuclease domain is a monomer in solution (Figure 1A).
Figure 1. The SFTSV L Endonuclease Domain Is a Monomer in Solution and Adopts a Mixed Alpha-Beta Fold.
(A) SEC-MALS analysis of SFTSV L 1–231 aa. The determined mass of SFTSV L 1–231 aa (green) is 2.71 ± 0.01 × 104 Da. The theoretical mass for the monomer is 2.76 × 104 Da. Independent experiments were carried out in triplicate and representative data are shown here. Inset: Coomassie-stained SDS-PAGE of purified, recombinant N-terminal His-tagged SFTSV L endonuclease domain 1–231 aa (arrowhead).
(B) X-ray crystal structure of the SFTSV L endonuclease domain. Secondary structural elements that form the α helices, the β sheets, and the loop regions are annotated based on the relative position from the N terminus.
We next initiated crystallization trials and obtained crystals of SFTSV L 1–231 aa in the presence of trypsin. Analysis of the mass of these crystals using liquid chromatography coupled to mass spectrometry (LC-MS) identified three major protein species: 1–226 aa (25143 Da), His-tagged (GSSHHHHHH SSGLVPRGSH) 1–226 with the first methionine cleaved (26893 Da), and Met-His-tagged (MGSSHHHHHHSSGLVPRGSH) 1–226 (27,071 Da) (Figure S2). However, we were unable to solve the structure of the SFTSV endonuclease domain by molecular replacement using other endonuclease domain structures (PDB: 5HSB, 5J1P, or 5DES) as search models. Therefore, we generated selenomethionine (SeMet)-labeled protein crystals, which grew in 0.1-M sodium citrate pH 5 and 10% polyethylene glycol (PEG) 20,000, and built a lower-resolution (2.5 Å) model using a single-wavelength anomalous dispersion (SAD) phasing method. This initial model was then used as the search model for molecular replacement on another SeMet dataset with better resolution and density to solve the X-ray crystal structure of the SFTSV endonuclease domain to 2.4-Å resolution (Figure 1B). The crystallographic asymmetric unit consists of three molecules; however, we did not observe any significant buried surface area that would indicate that the SFTSV endonuclease is an oligomer. Furthermore, we only observed electron density for residues 1–226, which corresponds to the 25,143-Da peak in our liquid chromatography-mass spectrometry (LC-MS) results (Figure S2). The overall structure features a central beta sheet composed of six mixed β strands (β1, β2, and β6-β9), forming a twisted plane that is surrounded by six α helices (α1-α6) and one 310-helix (η1). The two long parallel β7 and β8 strands are connected by a long helix α4, which runs parallel to the central β sheet and is flanked by α6 and η1. On the opposite side of the β sheet, helix α5 is covered by a surface-exposed antiparallel β sheet made of strands β3-β5. However, unlike other endonuclease domain structures, we did not observe any obvious density corresponding to a manganese ion in the active site even though 5 mM manganese chloride was supplied during crystallization. It is likely that the metal ions are chelated by the high concentration of sodium citrate in the crystallization buffer.
The Structural Fold of Endonuclease Domains from sNSVs Is Largely Conserved
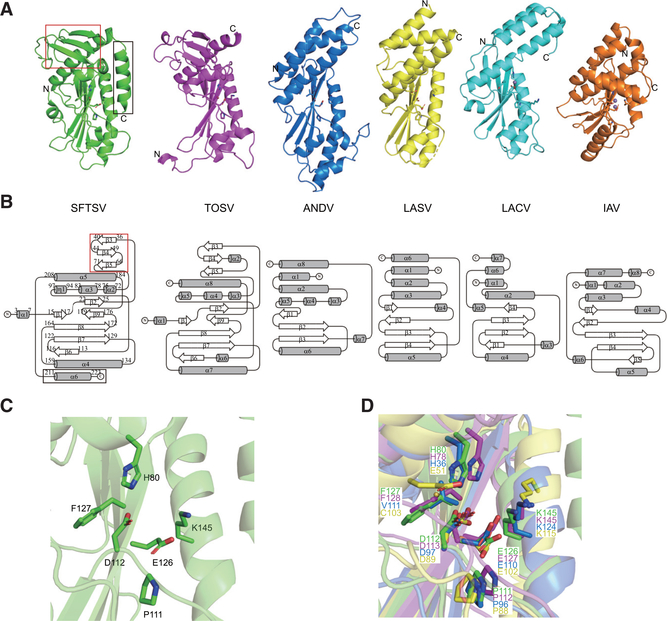
Analysis of the SFTSV endonuclease domain structure reveals significant similarities to the cap-snatching endonucleases of TOSV, ANDV, LASV, LACV, and IAV (Figures 2A and 2B). Structural alignments using the SuperPose tool (Krissinel and Henrick, 2004) show that all structures present root-mean-square deviations (RMSDs) below 3.5 Å for at least 94 residues (Table S1) despite having sequence identities below 20% (Figure S1C), suggesting that the core regions are conserved among these endonucleases. Furthermore, the (H/E)-PD-(E/D)X-K motif critical for the active site is conserved among these representative sNSVs (Figures 2A and S1C). In the SFTSV endonuclease structure, residues H80, D112, and E126, corresponding to the divalent-cation binding residues, and residue K145, corresponding to the catalytic lysine, are oriented with respect to each other (Figure 2C). These highly conserved residues are positioned similarly in an alignment with other endonuclease structures (Figure 2D). However, the SFTSV endonuclease contains an additional surface-exposed beta sheet (strands β3-β5) that is positioned on top of helix α5 (Figures 2A and 2B, red box outline). Only the recent TOSV endonuclease structure (the PDB of which was released while this manuscript was in review; PDB: 6QW0) (Jones et al., 2019) contains a similar feature. Other endonuclease structures lack this beta sheet entirely.
Figure 2. The Structural Fold of sNSV Endonuclease Domains Is Conserved among sNSVs.
(A) Comparisons of the endonuclease structures of SFTSV (green, PDB: 6NTV), TOSV (purple, PDB: 6QW0), ANDV (blue, PDB: 5HSB), LASV (yellow, PDB: 5J1P), LACV (cyan, PDB: 2XI5), and IAV (orange, PDB: 5DES). Manganese ions are shown as purple spheres in TOSV, ANDV, LASV, LACV, and IAV structures. Key catalytic residues are shown as sticks.
(B) Topological diagrams generated by TopDraw of endonuclease domains shown in (A) with the common core α helices represented as gray cylinders and the β strands as white arrows. The SFTSV L endonuclease domain contains a unique β sheet insertion at the N terminus (red box) and an extra C-terminal helix (black box).
(C) Close-up view of the active site of the SFTSV L endonuclease domain. Key catalytic residues are shown as sticks.
(D) Close-up view of superimposed endonuclease structures of SFTSV (green), TOSV (purple), ANDV (blue), and LASV (yellow). Key catalytic residues are shown as sticks.
The N-Terminal Domains of Banyangviruses Show Endonuclease Activity In Vitro
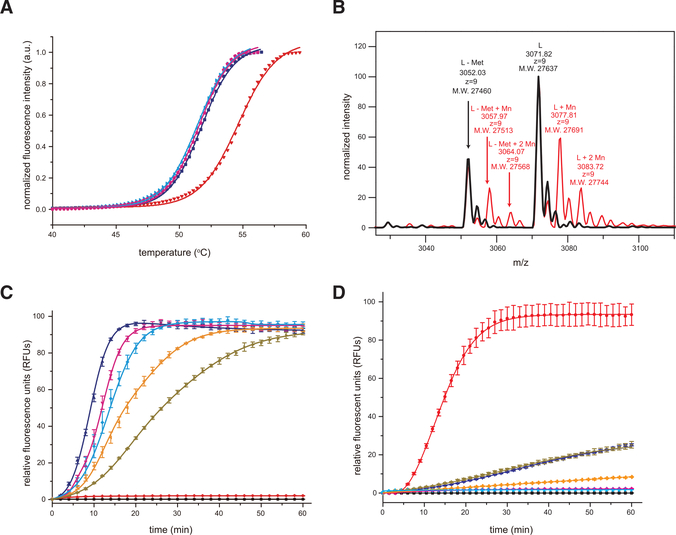
Several sNSV endonucleases require divalent metals for their enzymatic activity (Doan et al., 1999; Fernández-García et al., 2016; Reguera et al., 2016). To assess the metal requirements for SFTSV endonuclease activity, we first measured the stability of SFTSV endonuclease in the absence and presence of divalent cations (Mn2+, Mg2+, and Ca2+) using a fluorescence thermal-shift assay (TSA) (Figure 3A). We found that SFTSV endonuclease displayed a melting temperature (Tm) of 51.8°C in the absence of any metal ions. Addition of Mn2+ increased the thermal stability of SFTSV endonuclease to 54.2°C, whereas addition of Mg2+ or Ca2+ had no measurable difference in the thermal stability, suggesting that SFTSV endonuclease binds Mn2+ ions. Native MS supports this as two species were identified. One that bound one Mn2+ ion and a second that bound two Mn2+ ions (Figure 3B). To further assess the functional implications, we used a fluorescence resonance energy transfer (FRET)-based endonuclease assay to measure the dependence of Mn2+ binding on endonuclease activity. Mn2+ binding correlated with data from a FRET-based endonuclease assay using different metal ions (Figure S3). These results further support that SFTSV, and likely HRTV, endonuclease utilizes Mn2+ as a co-factor for its enzymatic activity. To determine the optimal concentration of endonuclease protein for FRET-based endonuclease assays, we tested different concentrations of protein ranging from 0.125 to 2 μM (Figure 3C), which identified 1 μM as the best concentration with the largest dynamic range. Next, we used the enzymatic assay to characterize the following SFTSV endonuclease mutants: H80A, D92A, D112A, or E126A. As these residues are important for either metal binding or for catalytic activity based on structural homology (Figure 2C), these mutants displayed a dramatic reduction in the enzymatic reaction rate (Figure 3D). Specifically, the D112A mutant showed near-complete loss of SFTSV endonuclease activity, whereas H80A, D92A, and E126A mutants displayed drastically reduced endonuclease activity (Figure 3D). These observations indicate that despite relatively low sequence similarities among the SFTSV L 1–231 aa, HRTV L 1–231 aa, and IAV PA 1–192 aa endonuclease domains, they contain the same key amino acid residues within their enzyme pockets and kinetic properties that suggest similar functions.
Figure 3. The SFTSV L Endonuclease Activity Is Sensitive to Divalent Cations.
(A) Thermal stability of SFTSV L 1–231 aa protein in the absence or presence of 2-μM divalent cations. No divalent cations, blue; manganese (Mn2+), red; magnesium (Mg2+), cyan; and calcium (Ca2+), magenta. The lines represent the non-linear fits of the normalized fluorescence curves to the Boltzmann equation.
(B) Intact mass measurement of SFTSV L 1–231 aa protein using LC-MS. Native mass spectra of SFTSV L 1–231 aa (black) and SFTSV L1–231 aa with MnCl2 (red) from charge state +9 are shown. 27,637, 27,691 and 27,744 Da are masses that represent SFTSV L residues 1–231 with first methionine modified (+46 Da) His6-tag, plus 0, 1, or 2 Mn2+. 27,460, 27,513 and 27,568 are masses that represent SFTSV L residues 1–231 with first methionine cleaved His6-tag, plus 0, 1, or 2 Mn2+.
(C) FRET-based endonuclease activity of SFTSV L 1–231 aa was measured by monitoring fluorescence intensity using λex/λem = 485 nm/535 nm. Increasing concentrations of SFTSV L 1–231 aa (2 μM, blue; 1 μM, magenta; 0.5 μM, cyan; 0.25 μM, orange; 0.125 μM, dark brown) were incubated with a 100-nM single-stranded RNA (ssRNA) probe. Buffer only (black) and substrate only (red) were included as blank and background controls, respectively. Each data point represents the average of three independent runs with error bars representing the standard deviation, which was then fit using a four-parameter log curve.
(D) Active site mutations severely impair SFTSV endonuclease activity. FRET-based endonuclease assays were performed in the presence of 1-μM SFTSV wild-type (red) or SFTSV mutants (H80A, blue; D92A, dark brown; D112A, magenta; E126A, orange). Buffer only (black) and substrate only (cyan) were included as blank and background controls, respectively. Experiments were carried out in triplicate and average data are shown with error bars representing the standard deviation.
Structural Elements Unique to SFTSV L Endonuclease Modulate Its Activity
In our X-ray crystal structure, we observed two structural elements that are unique to the SFTSV endonuclease domain, which are absent in the previously solved endonuclease domains. The first element is a beta sheet “cap” that sits on top of the endonuclease domain (Figure 2A, red box), which is an insertion that is absent in other endonucleases from this family. The TOSV endonuclease structure shares this feature, but its exact role in endonuclease activity has not been described thus far. Our attempts to evaluate the function of this domain led to the generation of several N-terminal truncation mutations. Not surprisingly, the N-terminal deletion mutations containing residues 22–231 aa or 75–231 aa resulted in a destabilized SFTSV L protein. Importantly, the β1 secondary structure element is part of the mixed alpha-beta structure. Given the structural importance of the beta “cap” to the structure, it is tempting to speculate that the cap may be important for protein-protein interactions, a notion supported by a prediction algorithm (Figure S4). These observations require future experimental validation.
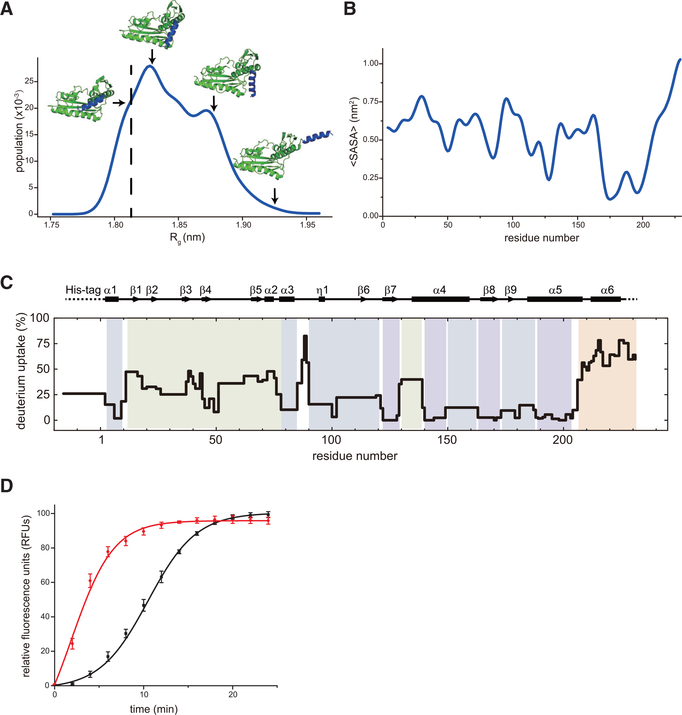
Next, we explored the significance of the additional structural element, helix α6, at the C terminus of the endonuclease domain (Figure 2A, black box). In the structure, it appears that the C-terminal α6 helix sits on the active site, likely to occlude access to substrates and that may undergo substantial dynamics to enable substrate to reach the active site. To test this hypothesis, we first performed a molecular dynamics simulation of the SFTSV endonuclease crystal structure. The Markov state models were built from the fluctuation amplification of specific traits (FAST) simulation data, and the solvent accessible surface area (SASA) of each residue was averaged with the preceding and subsequent residue to highlight concerted behaviors (Figures 4A and 4B). Our results predict that the C-terminal α6 helix is highly flexible. Specifically, we observed significant clusters of structures with varying radii of gyration (Figure 4A). Importantly, residues near the α6 helix displayed the largest change in SASA (Figure 4B). To complement the static X-ray structure and the computational simulation studies, we performed hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments to assess the dynamics of the protein. The results showed that the SFTSV endonuclease domain, based on exchange behavior, could be classified into four regions by its HDX kinetics (Figures 4C and S5). The first region (blue), composed of β7 and 8 of the center beta sheet and parts of α4 and α5, undergoes minimal deuterium exchange at 10 s and does not increase significantly with time. This first region makes up a rigid core that is highly structured and protected. An exception is that Leu87, Ser88, and Lys89 also show high levels of exchange across the HDX time range. These residues form a dynamic loop that stands out from the nearby core region. The second region (dark green), which includes residues from α1, α3, η1, and β6, as well as the loops associated with these elements, has slightly faster exchange from a low percentage of exchange at early times, indicating this region is also highly structured but slightly less rigid. Region 2 plus the previous rigid core surrounds and sandwiches the active site of SFTSV L protein. The third region (green) contains α2 and the first several residues of α4 and β1-β5. This group shows initial exchange rates between 10% and 20%. This region displays a more noticeable rapid increase than the previous two regions and displays exchange rates of 60%−80%. The third region also surrounds the C terminus of the endonuclease domain. The fourth region (orange) contains the last few residues of α5, α6, and a few C-terminal residues that lack electron density in our crystal structure. This is the most dynamic region, where deuterium exchange rates start between 40% and 50%. This observation is suggestive of rapid breathing motions, likely of the α6 helix at a time shorter than we can achieve in our HDX-MS experiment (i.e., exchange within the “dead” time, prior to detection, which is about 10 ms). In this fourth region, exchange rates rapidly increase to 80%−100% at 900 s. These results offer experimental confirmation that the fourth region, which contains α6, is highly dynamic and likely serves as a hinge and cap for the active site. Given the proximity of the third region to the hinge, its increased dynamics can be caused by opening and closing of the α6 helix. To further determine the role of the C-terminal α6 helix in the catalytic activity, we compared the endonuclease activities of wild-type (1–231 aa) and a C-terminal deletion mutant (1–208 aa) using a FRET-based assay (Figure 4D). The C-terminal deletion mutant (1–208 aa) showed higher endonuclease activity than did the wild-type (1–231 aa). Collectively, results from the FRET assay and HDX-MS studies closely mirror the molecular dynamics results, suggesting that our data are internally consistent and support a model where the C-terminal helix α6 structurally regulates endonuclease activity by controlling access to the active site.
Figure 4. Conformational Dynamics C-Terminal α6 Helix of SFTSV L Protein Modulates Its Endonuclease Activity.
(A) Dynamic simulation of SFTSV L 1–231 aa. The C-terminal α6 helix (colored in blue) is predicted to be very flexible.
(B) The average solvent accessible surface area (SASA) of each residue in SFTSV L 1–231 aa.
(C) Deuterium uptake in SFTSVL1–231 aa from HDX-MS. The deuterium uptake on SFTSV L1–231 aa is mapped onto its sequence and secondary structure with color for exchange time at 120 s of exposure in a D2O buffer (Figure S5). Groups of residues with similar differential deuterium exchange rates are colored according to a gradient for HDX-MS rates in Figure S5.
(D) FRET-based endonuclease assays of SFTSV L 1–231 aa (black) and SFTSV L 1–208 aa (red). Experiments were carried out in triplicate and average data are shown with error bars representing the standard deviation.
Cap-Snatching Endonucleases Are Antiviral Targets
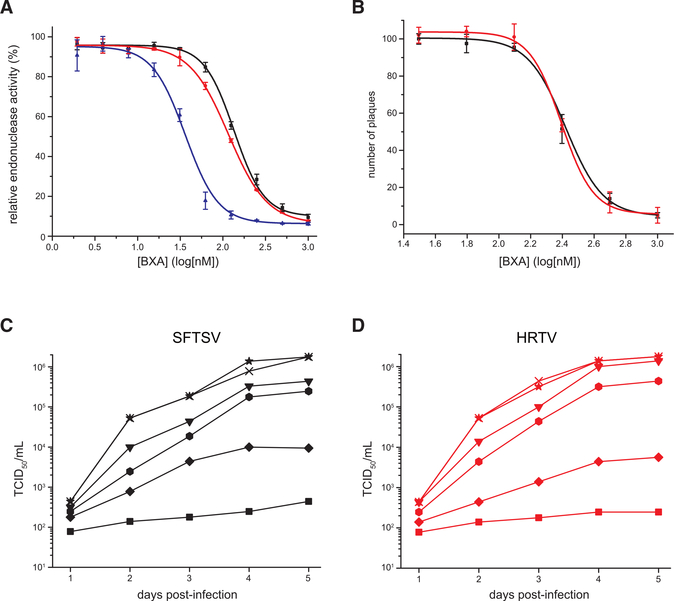
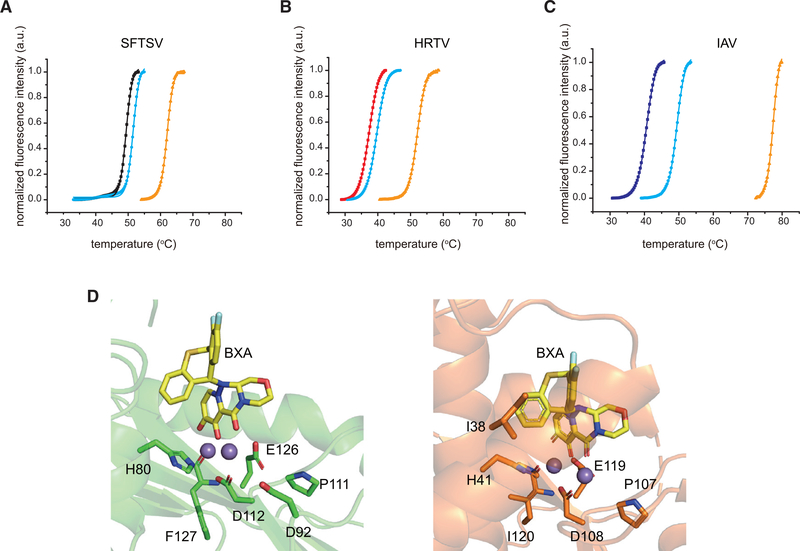
Drug repurposing can offer faster drug discovery and shorter development times with reduced risk than de novo drug discovery and development strategies (Ashburn and Thor, 2004). To test the potential use of SFTSV and HRTV endonuclease domains as antiviral targets, we tested the efficacy of several influenza endonuclease inhibitors on SFTSV and HRTV endonuclease domains. Our studies included the recently FDA-approved compound BXM (Omoto et al., 2018), which is a prodrug that is converted to its active acidic form BXA. Using an in vitro FRET-based assay, we found that BXA effectively inhibited the enzymatic activity of SFTSV L, HRTV L, and IAV PA endonucleases with half maximal inhibitory concentration (IC50) values of 135 ± 5, 116 ± 2, and 36 ± 3 nM, respectively (Table S2; Figure 5A). By comparison, L-742,001 (Song et al., 2016) inhibited with IC50 values of 2.1 ± 0.2, 3.3 ± 0.4, and 1.4 ± 0.1 μM, respectively (Table S2; Figure S6A). Corresponding virus growth inhibition assay revealed that BXA suppressed SFTSV and HRTV replication at a concentration above 250 nM, whereas L-742,001 only worked at high concentrations (>50 μM) (Figures 5C, 5D, S6B, and S6C). Similarly, BXA inhibited SFTSV and HRTV plaque formation with half maximal effective concentration (EC50) values of 263 ± 14 and 248 ± 5 nM, respectively (Figure 5B), whereas L-742,001 failed to inhibit plaque formation (data not shown). To further verify direct binding between the inhibitor and endonuclease domains, we conducted TSA experiments. These results revealed that the addition of 2 mM Mn2+ and 200 μM BXA increased the thermostability by 10.5°C,12.5°C, and 28.1°C for SFTSV, HRTV, and IAV endonucleases, respectively, compared to each protein with only Mn2+ added (Figures 6A–6C). These TSA results also suggest that BXA stabilizes the SFTSV and HRTV endonucleases in a Mn2+-dependent manner similar to IAV with approximately 2-fold lower stabilization of SFTSV and HRTV relative to IAV. While BXA has been shown to chelate the two metal ions in the active site of IAV and IBV endonucleases (Omoto et al., 2018), we currently lack direct structural evidence for SFTSV or HRTV endonuclease interactions with BXA or L-742,001. To further probe the binding mode between SFTSV and HRTV endonuclease and BXA and L-742,001 inhibitors, we used Surfelx-dock to dock the small molecules into the endonuclease active site (H80, D112, E126, F127, S128, K145, E219, and A223). Examination of the best poses, shown in Figure 6D (left) and Figure S6D, suggests that both compounds bind in a manner similar to that of influenza virus endonuclease (Figure 6D, right). Taken together, our data show that BXA and L-742,001 bind and stabilize SFTSV and HRTV endonuclease domains and that the binding results in inhibition of enzymatic activity. These studies also support the broad-spectrum efficacy of these endonuclease inhibitors.
Figure 5. Characterization of SFTSV L Endonuclease Inhibitors.
(A) FRET-based assays with SFTSV L 1–231 aa (black), HRTV L1–231 aa (red), or IAV PA 1–192 aa (blue) in the presence of 1 mM MnCl2 and increasing concentrations of BXA. The dose-response curves were fit to yield IC50 values using a nonlinear regression model. Experiments were carried out in triplicate and average data are shown with error bars representing the standard deviation.
(B) Plaque reduction assays for SFTSV (black) and HRTV (red). Plaques were counted by visual examination and the required concentration to reduce the 50% plaque number (EC50) was calculated as relative to the control without BXA. Experiments were carried out in duplicate and average data are shown with error bars representing the standard deviation.
(C and D) Virus growth inhibition assays of (C) SFTSV or (D) HRTV in the presence of BXA. BXA concentrations used are as follows: 1000 nM, square; 500 nM, diamond; 250 nM, hexagon; 125 nM, down triangle; 62.5 nM, cross; and 0 nM, star. The virus yield was visualized by immunostaining and expressed as TCID50/mL using the Reed-Muench method. Experiments were carried out in duplicate.
Figure 6. Inhibitor binding to SFTSV L endonuclease stabilizers its endonuclease domain.
Thermal stability was measured in the presence of 10 mM EGTA for: (A) SFTSV L 1–231 aa (black), (B) HRTV L 1–231 aa (red), or (C) IAV PA 1–192 aa (blue). Thermal stability for each viral endonuclease was also measured in the presence of 2 mM MnCl2 (cyan) and 2 mM MnCl2 with 0.2 mM BXA (orange). The lines represent the non-linear fits of the normalized fluorescence curves to Boltzmann equation. (D) Left, docking simulation of two manganese ions (purple) and BXA (yellow) with SFTSV L 1–231 aa (green). Key residues are shown in stick representation. Right, Structure of BXA bound to IAV PA (PDB 6FS6) is shown for comparison.
DISCUSSION
Bunyavirus L polymerases are composed of a single polypeptide protein containing multiple functions, including an N-terminal endonuclease that is important for cleaving host pre-mRNAs required for viral transcription to proceed (Gerlach et al., 2015). While much of what is known about the sNSV cap-snatching mechanism is largely derived from studies of the IAV polymerase complex (Hengrung et al., 2015; Pflug et al., 2014; Reich et al., 2014; Thierry et al., 2016; Walker and Fodor, 2019), less is known mechanistically regarding bunyavirus polymerases. Here, we present the 2.4-Å-resolution X-ray crystal structure and functional characterization of the SFTSV endonuclease. We find that the SFTSV endonuclease adopts a mixed alpha-beta fold and contains a (H/E)-PD-(E/D)X-K motif that is similar to other viral endonucleases, including TOSV L (Jones et al., 2019), IAV PA (Song et al., 2016), LACV L (Reguera et al., 2010), LASV L (Reguera et al., 2016), and ANDV L (Fernández-García et al., 2016). Closer examination of the structures reveals that the active site of the SFTSV endonuclease domain is more closely related to the ANDV and LASV endonuclease domains as it features a cluster of three acidic residues (D92, D112, and E126), a lysine residue (K145) neighboring E126, and a histidine residue (H80). Mutation of these residues dramatically reduced SFTSV endonuclease activity. Furthermore, mutation of D112 resulted in the largest reduction of endonuclease activity, likely due to coordination of the two manganese ions by D112, which are important for the stability of the endonuclease. We also proved the binding by native MS that SFTSV endonuclease could bind to one or two Mn2+. While in TOSV, without inhibitor only one Mn2+ is chelated rather than two dues to the flexibility of the loop, which D90 locates (Jones et al., 2019). Further experiments are needed to investigate if other sNSV endonucleases share similar properties.
Interestingly, the SFTSV endonuclease contains an additional surface-exposed, anti-parallel β sheet (β3-β5) that is orthogonal to the central β sheet and covers helix α5. This structural element is unique to the banyangviruses and phleboviruses, but absent in other sNSV endonuclease structures. Truncation of this element resulted in a destabilized protein (75–231 aa; data not shown), suggesting that the β sheet and the N-terminal region are important for the stability of the SFTSV endonuclease domain. Moreover, analysis using cons-PPISP, a 3D-structure-based protein-protein interaction predictor (Chen and Zhou, 2005) to predict the residues that will likely form the binding site for another protein, suggests that the neutral/acidic residues (T39, D41, T46, D48, G50, A51, V52, D54, S55, S62, I63, N64, A65, G66, L67, I68, T69, I70, Q71, and E74) located within the β sheet may be associated with protein-protein interaction (Figures S4A and S4B). While this manuscript was in preparation, Jones et al. (2019) published the endonuclease structure from TOSV, which also contains an anti-parallel β sheet capping helix α5. However, the TOSV endonuclease, as well as other viral endonuclease structures available to date, are lacking the C-terminal helix α6 that is unique to the SFTSV endonuclease structure. Our analysis of the functional significance revealed that helix α6 plays a critical role in modulating endonuclease activity. Specifically, deletion of helix α6 increased SFTSV endonuclease activity. Consistent with this observation, molecular dynamic simulations show a high degree of flexibility and conformational heterogeneity of helix α6. Our in vitro experimental attempts to test the dynamic nature of helix α6 and its potential role in the regulation of enzyme activity by HDX-MS further support the notion that the dynamics of helix α6 likely control substrate accessibility and endonuclease function in SFTSV and HRTV. Further studies are warranted to determine if similar regulatory elements exist at the C-termini of other viral endonucleases.
The metal binding properties of viral endonucleases provide considerable opportunities for the repurposing of FDA-approved drugs and candidates (Credille et al., 2018; Omoto et al., 2018). Previously, a number of inhibitors that target the IAV PA endonuclease domain have been characterized (Ju et al., 2017). Our initial studies reveal that SFTSV endonuclease is also inhibited by these inhibitors, albeit less effectively. In 2018, the FDA approved Baloxavir Marboxil, a prodrug that is represented by its active form BXA. Based upon available data, including structural studies, BXA binds the IAV endonuclease active site near two Mn2+ ions and this binding inhibits cleavage of a pre-mRNA substrate at the 3′ end of guanine (Omoto et al., 2018). This mechanism of inhibition is similar to dolutegravir, a BXA analog that inhibits human immunodeficiency virus (HIV) integrase (Johns et al., 2013). Our study reveals that BXA can effectively inhibit SFTSV and HRTV endonuclease activities, further supporting the broad utility of BXA as an antiviral agent against sNSV endonucleases. In addition to demonstrating the application of drug repurposing for endonucleases, our studies demonstrate the generality of metal chelating small molecules as specific inhibitors of microbial enzymes. While our data strongly support the potential use of BXM to treat SFTSV and SRTV infections, future studies are needed to assess the efficacy in in vivo models.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gaya K. Amarasinghe (gamarasinghe@wustl.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Cultures
For recombinant protein expression, we used the Escherichia coli strain BL21(DE3) (Novagen). Cells were grown at 37°C as described in the Method Details section.
Cells and Viruses
Vero E6 cells and 293-T cells were purchased from ATCC and maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, #11965118) containing 10% Fetal Bovine Serum (VWR Life Science Seradigm, #1500–500) and 1% penicillin-streptomycin (Thermo Fisher Scientific, #15140163). Transient transfections for virus rescue were performed with polyethylenimine (PEI) transfection reagent (Polysciences, #23966) according to the manufacturer’s instructions.
For SFTSV generation, 293-T cells were co-transfected with plasmid expressing T7 polymerase along with plasmids for SFTSV reverse genetics (pTM-ppL, pTM-N, pTVT-ppL, pTVT-ppM and pTVT-S) (Brennan et al., 2015) with PEI at 3:1 ratio. After 5 days post-transfection, the supernatant was collected and passed through a 0.2 μM polyethersulfone (PES) filter (Nalgene, #566–0020). The clarified supernatant was loaded into Vero cells and supernatant was collected after 9 days post-infection. After the supernatant was clarified, the virus suspension was aliquoted and stored at −80°C until use. For HRTV, virus was purchased from BEI Resources (https://www.beiresources.org/) with one passage in Vero E6 cells and the supernatant was aliquoted and stored at −80°C until use.
METHOD DETAILS
Compounds
All the compounds used in this study were purchased from a commercial vendor. Briefly, L-742,001 was purchased from Sigma-Aldrich (#SML1010–5MG) and Baloxavir acid (S-033447, BXA) was purchased from Probechem (#PC-35417–5MG).
Plasmids
The SFTSV (GenBank Accession AFK08658.1) 1–231 aa, HRTV L (GenBank Accession AIF75093.1) 1–231 aa and IAV PA (DuBois et al., 2012) 1–192 aa were synthesized (Genewiz) and subclones into a pET52b(+) expression vector between 5′ NcoI and 3′ NotI sites for expression in E. Coli. Mutant constructs were generated using GeneArt Site-Directed Mutagenesis System (Thermo Fisher Scientific, #A13282) according to the manufacturer’s instructions.
Protein Expression and Purification
Plasmids encoding SFTSV L 1–231 aa, HRTV L 1–231 aa, and IAV PA 1–192 aa were transformed in BL21(DE3) E. coli cells (Novagen), cultured in Luria Broth media or in M9 medium (for selenomethionine labeled proteins) at 37°C, induced with 0.5 mM IPTG, and grown for 12 hr at 15°C. Cells were harvested and resuspended in lysis buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, and were lysed using an EmulsiFlex-C5 homogenizer (Avestin). Lysates were clarified by centrifugation at 47,000 × g at 4°C for 40 min. Proteins were purified using a series of chromatographic columns and a size exclusion column as a final step. Protein purity was determined by Coomassie staining of SDS-PAGE.
SEC-MALS
SEC-MALS experiments were performed using a DAWN-HELEOS II detector (Wyatt Technologies) coupled to a Superdex 200 Increase 10/300 column (GE Healthcare) in 20 mM HEPES pH 7, 150 mM NaCl. 2 mg/ml protein was injected, and raw data were analyzed using ASTRA 6 software (Wyatt Technologies) to determine the weight averaged molecular mass (MW). Protein concentrations were determined using the refractive index measured by an Optilab T-rEX (Wyatt Technologies) and a dn/dc of 0.174 mL × g−d.
Protein Crystallization
The initial crystallization condition was obtained by sparse matrix screening with Hampton Research screens using the vapor diffusion method in sitting drop mode. In some conditions, 10 mg/ml of SFTSV L1–231 aa was mixed with trypsin in a 1:1,000 ratio. From the initial primary screens, we observed about 10 conditions in which protein crystals appeared within 7–12 days of setting up at 20 °C. Similar conditions were obtained for SeMet labeled SFTSV L protein construct 1–231 aa. The crystal from which the structure was solved came from the following growth condition: 0.1 M sodium citrate pH 5 and 10% PEG 20,000. Crystals appeared after 1 week at 20°C and were vitrified in a solution containing the mother liquor plus 25% PEG400.
Proteins in the crystals were analyzed by liquid chromatography-mass spectrometry (LC-MS) to identify their molecular weight before Se-Met incorporation. 10 protein crystals were picked up, washed with well solution, and subsequently dissolved in 10 μL water. 1 μL sample was then diluted in 50 μL 0.1% formic acid and injected into a two-valve LC device. The proteins were captured on a ZORBAX Eclipse XDB C8 column (2.1 × 15 mm, Agilent) for desalting (3 min) with 0.1% trifluoroacetic acid. The C8 column was then switched inline. A linear gradient (3% to 80% acetonitrile, 0.1% formic acid, 200 μL/min flow rate, over 5 min) was used to elute proteins subjected to electrospray ionization source equipped on a Maxis 4G Q-TOF mass spectrometer (Bruker). Mass spectra were deconvoluted with MagTran (Zhang and Marshall, 1998) to give zero-charge intact mass of proteins. The intact mass was used to match the SFTSV L sequence.
Data Collection and Structure Determination
Crystals were screened at Advanced Photon Source Beamline 19ID at the Argonne National Laboratory. Diffraction images were processed by HKL3000 (Minor et al., 2006). The initial model was determined by the single-wavelength anomalous diffraction phasing method from a single Se-Met substituted protein crystal to a maximum resolution of 2.50 Å. The SHELX C/D/E pipeline (Sheldrick, 2008) in CCP4i (Winn et al., 2011) was used to locate the selenium atoms and perform phasing. Then AutoSol (Terwilliger et al., 2009) in Phenix (Adams et al., 2010) was used to improve the experimental phasing and build initial model. Automated model-building and refinement was carried out using AutoBuild (Terwilliger et al., 2008) as implemented in Phenix. The initial model was used as the search model against another Se-Met dataset and solved to 2.40 Å resolution by molecular replacement using the Molrep program (Vagin and Teplyakov, 2010) as part of the CCP4i program suite. SFTSV L 1–226 residues were built by manual building in COOT (Emsley et al., 2010) and refined with Refmac5 (Murshudov et al., 1997) and Phenix.refine (Afonine et al., 2012). The structure quality was assessed by MolProbity (Chen et al., 2010). Data collection and refinement statistics are listed in Table S3. Structure figures were prepared using PyMOL (Schrodinger, 2018).
Sequences Alignment and Graphical Presentation
Alignment of the endonuclease domain of Severe fever with thrombocytopenia virus (SFTSV; GenBank Accession code AFK08658.1) with Heartland virus [Heartland virus (HRTV; GenBank Accession AIF75093.1), Toscana virus (TOSV; GenBank Accession ALS19628.1), Lassa mammarenavirus (LASV; GenBank Accession AAT48998.1), Andes orthohantavirus (ANDV; GenBank Accession NP_604473.1), Influenza A virus (A/H1N1; UniProtKB C3W5S0), Uukuniemi virus (UUKV; GenBank Accession NP_941973.1), Seoul orthohantavirus (SEOV; GenBank Accession NP_942558.1), Sin Nombre orthohantavirus (SNV; GenBank Accession NP_941976.1) and La Crosse virus (LACV; GenBank Accession ADH04736.1)]. The alignment was generated using Expresso software (Armougom et al., 2006; Di Tommaso et al., 2011), taking into account available structural information (PDB code 6NTV for SFTSV, 6QW5 for TOSV, 5J1P for LASV, 2XI5 for LACV, 5HSB for ANDV, 5IZE for SOEV, and 5W3I for IAV) and visualized using ESPript 3.0 (Robert and Gouet, 2014). Red triangles highlight the H/E-PD-E/DX-K motif in the active site.
Thermal Shift Assays (TSA)
The thermal stability of SFTSV L 1–231 aa, HRTV L 1–231 aa, and IAV PA 1–192 aa proteins were measured by TSA. The assay contained a final concentration of 5 μM protein (SFTSV 1–231 aa, HRTV L 1–231 aa, or IAV PA 1–192 aa), 4.5 × Sypro Orange, 20 mM HEPES pH 7.8, 150 mM NaCl, supplemented with either 2 mM MnCl2, 2 mM MgCl2, 2 mM CaCl2, 2 mM MnCl2 plus 200 μM BXA, 10 mM EGTA, or 10 mM EGTA plus 200 μM BXA as described previously (Ericsson et al., 2006). The plates were heated in StepOnePlus Real-Time PCR System (Thermo Fisher) from 20 to 90°C in increments of 0.3°C. The wavelengths for excitation and emission were 490 and 575 nm, respectively.
Endonuclease Assay
Enzymatic activity of purified N-terminal region of SFTSV and HRTV L was measured by using FRET-based endonuclease assay with a little modification of previously reported publication (Noble et al., 2012). Briefly, recombinant amino-terminal protein of L segment of either SFTSV or HRTV (1 μM) was mixed with a 100 nM single stranded RNA (ssRNA) probe (5’FAM-AGGAAGAUUAAUAAUUUUCCU-lowa Black® FQ3’) in reaction buffer containing 50 mM HEPES pH 7.8, 150 mM KCl and 1 mM MnCl2 (unless indicated otherwise). The reactions were measured by detecting fluorescence intensity (λex/λem = 485nm/535nm) using FilterMax F5 Multi-Mode Microplate Reader (Molecular Devices, LLC). To measure the enzyme kinetics, the reactions were measured every 5 min until reactions were saturated. In case of endonuclease inhibition assay, inhibitors were added at the indicated concentrations and an IC50 value was determined using a linear regression model. All assays performed at least in triplicate using at least two different protein preparations.
Hydrogen Deuterium Exchange Mass Spectrometry
SFTSV L 1–231 aa sample was buffer exchanged with 1 × PBS pH 7.4. HDX was initiated by diluting samples (25 μM, 2 μL) 10-fold with 1 × PBS prepared in D2O, or 1 × PBS H2O buffer for samples measured for no-deuterium control. At different time intervals (10, 30, 60, 120, 900, 3,600, and 14,400 s), the exchange reaction was quenched by rapidly decreasing the pH to 2.5 with 30 μL of quench buffer (3 M urea, 1% trifluoroacetic acid, H2O) at 4°C. The protein mixture was immediately injected into a custom-built HDX sample-handling device that enabled on-column digestion with resin-immobilized pepsin (2 × 20 mm) at a flow rate of 100 μL/min in 0.1% trifluoroacetic acid. The resulting peptic peptides were captured on a ZORBAX Eclipse XDB C8 column (2.1 × 15 mm, Agilent) for desalting (3 min). The C8 column was then switched inline, followed by a Hypersil Gold C18 column (2.1 mm × 50 mm, Thermo Fisher Scientific). A linear gradient (4%−40% acetonitrile, 0.1% formic acid, 50 μL/min flow rate, over 5 min) was used to separate the peptides and direct them to an electrospray ionization source equipped on an LTQ-FTICR mass spectrometer (Thermo Fisher Scientific, Santa Clara, CA). Valves, columns, and tubing for protein digestion and peptide separation were immersed in an ice-water bath to minimize back-exchange.
To map the peptic peptides, the digest, in the absence of HDX, was submitted to bottom-up proteomics analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with the LTQ-FTICR mass spectrometer. Digestion conditions (denaturant concentration in quench buffer) were optimized under “HDX assay” conditions with the absence of D2O. The peptic peptides were identified using Byonic and Byologic (Protein Metrics, Cupertino, CA). The peptide sets and raw HDX data were then submitted to HDExaminer (Sierra Analytics, Modesto, CA) for deuterium uptake calculation and data visualization in automated fashion with manual validation. Peptides for each run were assessed based on relative representation and statistical validation. The deuterium uptake at each time point was calculated by subtracting the centroid of the isotopic distribution of the non-deuterated peptide from that of the deuterated peptide. The relative deuterium uptake was plotted versus the labeling time to yield kinetic curves.
Native Mass Spectrometry
SFTSV L 1–231 aa was buffer exchanged to 150 mM ammonium acetate, pH = 6.8 with the Bio-Spin 6 Columns (Bio-Rad, Hercules, CA). Samples containing 2.5 μM SFTSV L or 2.5 μM SFTSV L plus 100 μM MnCl2 in the ammonium acetate buffer were loaded into the ES380 borosilicate nanoES spray emitters (Thermo Fisher Scientific, Santa Clara, CA). The emitters were mounted on the Exactive Plus EMR mass spectrometer (Thermo Fisher Scientific, Santa Clara, CA) to acquire mass spectra for 1 to 2 min. Some MS parameters are: 1.5 kV spray voltage, 20 eV in-source CID, 20 HCD cell energy, 100°C transfer capillary temperature. All assays performed at least in triplicate using at least two different protein preparations.
Molecular Dynamics Simulations
Molecular dynamics simulations were run with Gromacs 5.1.1 at 300 K using the AMBER03 force field with explicit TIP3P solvent (Abraham et al., 2015; Duan et al., 2003; Jorgensen et al., 1983). Simulations were prepared by placing the starting structure (PDB ID: 6NTV) in a dodecahedron box that extends 1.0 Å beyond the protein in any dimension. The system was then solvated (32770 atoms), and energy minimized with a steepest descents algorithm until the maximum force fell below 100 kJ/mol/nm using a step size of 0.01 nm and a cutoff distance of 1.2 nm for the neighbor list, Coulomb interactions, and van der Waals interactions. For production runs, all bonds were constrained with the LINCS algorithm and virtual sites were used to allow a 4 fs time step (Feenstra et al., 1999; Hess, 2008). Cutoffs of 1.0 nm were used for the neighbor list, Coulomb interactions, and van der Waals interactions. The Verlet cutoff scheme was used for the neighbor list. The stochastic velocity rescaling (v-rescale) (Bussi et al., 2007) thermostat was used to hold the temperature at 300 K. Conformations were stored every 20 ps.
The FAST algorithm (Zimmerman and Bowman, 2015, 2016) was used to enhance conformational sampling of the C-terminal helix (residues 209–226) when generating simulation data. FAST-distance simulations were run for 8 rounds, with 10 simulations per round, where each simulation was 40 ns in length (3.2 μs aggregate simulation). The FAST-distance ranking function favored restarting simulations from states that maximized pairwise distances between heavy-atoms on the C-terminal helix and their initial neighbors. Additionally, a similarity penalty was added to the ranking to promote conformational diversity in starting structures, as has been described previously (Zimmerman et al., 2017).
A Markov state model (MSM) was built from the FAST simulation data using enspara (https://github.com/bowman-lab/enspara). An MSM is a network representation of a free-energy landscape, where nodes are discrete conformational states and directed edges are conditional transition probabilities. The state space was defined using backbone heavy atoms (atoms C, Cα, Cβ, N, O), which was clustered with a k-centers algorithm based on RMSD between conformations until every cluster center had a radius less than 1.8 Å. Then 10 sweeps of a k-medoids update step was used to center the clusters on the densest regions of conformational space. Following clustering, an MSM was built by row-normalizing the observed transition counts, at a lag-time of 2 ns, with a small pseudo-count as a prior (Zimmerman et al., 2018). Radius of gyration and the solvent accessible surface areas for each state in the MSM were calculated using MDTraj and were weighted based on their determined populations (McGibbon et al., 2015). The solvent accessible surface area (SASA) of each residue was averaged with the preceding and subsequent residue to highlight concerted behaviors.
Surflex-Docking of BXA and L-742,001 to SFTSV L
The crystal structure of SFTSV L does not contain Mn2+ metal atoms, which are necessary for BXA and L-742,001 binding. Protein structures containing Mn2+ used for docking were generated by aligning the SFTSV L crystal structure (PDB 6NTV) to that of IAV H1N1 PA in complex with DPBA (PDB 4AWF) (Kowalinski et al., 2012) and using the resulting positions of the two Mn2+ metal atoms. The alignment was achieved using SFTSV L residues 112–130 to IAV H1N1 PA residues 108–123; this resulted in a good alignment of active site residues His80, Asp112, Glu126 and Phe127 that are critical for binding Mn2+.
MOL2 files for BXA and L-742,001 were generated using Open Babel (O’Boyle et al., 2011) and docked to the PDB files described above using Surflex-dock (Jain, 2007). Specifically, a threshold of 0.25 and bloat of 2.0 were used to generate the receptor protomols, and the default ‘-pgeom’ docking parameters were used to dock the compounds. To ensure that the 2,4-dioxobutanoic acid in BXA and L-742,001 were coordinated properly, we performed a fragmet-guided search using a 20 pKd/Å2 deviation penalty (Jain, 2007). The reference 2,4-dioxobutanoic acid fragment was created from the structure alignment to PDB 4AWF described above, with the phenyl ring removed. Active site residues important for binding were defined as His80, Asp112, Glu126, Phe127, Ser128, Lys145, Glu219, and Ala223.
Virus Growth Inhibition Assay
Confluent monolayers of Vero E6 cells in 24-well plates were washed once with DMEM and infected with either SFTSV or HRTV at 0.01 MOI. The plates were placed on a rocker in 37°C incubator for 1 hr for virus adsorption. The virus inoculum was removed and replaced by DMEM medium containing BXA or L-742,001 at various concentrations and incubated at 37°C in 5% CO2. Viruses were harvested every 24 hr until day 5 post-infection, and the virus yield was visualized by immunostaining and expressed as TCID50/ml using Reed-Muench method. Briefly, after infecting the virus samples in quadruplicate and incubated at 37°C in 5% CO2 for 3 days, Vero E6 cells in 96-well plate were fixed with 4% (w/v) paraformaldehyde (PFA) for 20 min at room temperature (RT). PFA-fixed cells were treated with 0.25% (v/v) Triton X-100 for 20 min at RT. Virus-infected cells were detected by probing with either Anti-NP (Sun et al., 2014) (pAB, for SFTSV) or 2AG8 (Calvert and Brault, 2015) (mAb, for HRTV) prepared in blocking buffer: DPBS containing 5% FBS for 1 hr at 37°C, followed by anti-rabbit Alexa Fluor 488 (for Anti-NP) or anti-mouse Alexa Fluor 488 (for 2AG8) (Thermo Fisher Scientific, USA) prepared in blocking buffer for 30 min at 37°C. Two washing cycles of DPBS was done after each step of the immunostaining. After the final washing, the cells were then visualized, and the virus infection were detected using Keyence BZ-X710 fluorescence microscope. As a control, the infected cells incubated in BXA-free medium were included throughout the experiment.
Plaque Reduction Assay
Confluent monolayers of Vero E6 cells in 6-well plates were washed once with DMEM and infected with either about 100 pfu of SFTSV or HRTV. The plates were placed on a rocker in 37°C incubator for 1 hr for virus adsorption. The virus inoculum was removed and replaced by overlay media (DMEM containing 1% low-melting agarose, without serum), containing BXA at various concentrations. After incubating cultures in 37°C incubator with 5% CO2, until plaques can be visualized under light, monolayers were fixed with 4% formaldehyde solution for 30 min and the agarose was then removed by flowing water and stained with 0.2% (w/v) crystal violet solution. The plaques were counted by visual examination and the required concentration to reduce the 50% plaque number (EC50), was calculated as relative to the control without BXA.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the exact value of n with the description of what n represents, the mean and the SEM are reported in Figures 1, 2, 3, 4, 5, and 6 and their legends. Statistical analyses were performed using Origin (OriginLab).
DATA AND CODE AVAILABILITY
The hydrogen deuterium exchange mass spectrometry plots are available at https://data.mendeley.com/datasets/f5t9tdvn5n/1. Protein Data Bank (PDB; https://www.rcsb.org/): 6NTV.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11001; RRID:AB_2534069 |
| Polyclonal serum to SFTSV NP (Anti-NP) | Wenhui Li (National Institute of Biological Sciences) | N/A |
| Monoclonal anti-HRTV NP (2AG8) | Amanda E. Calvert (U.S. Centers for Disease Control and Prevention) | N/A |
| Bacterial and Virus Strains | ||
| Severe Fever with Thrombocytopenia Syndrome virus (SFTSV) or Huaiyangshan banyangvirus | Brennan et al., 2015 | N/A |
| Heartland virus | BEI Resources | NR-49771 |
| Escherichia coli BL21(DE3) | Novagen | Cat#69450 |
| Biological Samples | ||
| Fetal Bovine serum (GIBCO) | Thermo Fisher Scientific | Cat#16140071 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-742,001 | Sigma-Aldrich | Cat#SML1010–5MG |
| Baloxavir acid | Probechem | Cat#PC-35417–5MG |
| Sypro Orange | Invitrogen | Cat#S6650 |
| TALON® Metal Affinity Resin | Takara | Cat#635504 |
| TransIT-LT1 | Mirus Bio | Cat#MIR2300 |
| Trypsin | Hampton Research | Cat#HR2–429-02 |
| Deposited Data | ||
| Structure of SFTSV L endonuclease domain | This paper | PDB: 6NTV |
| Experimental Models: Cell Lines | ||
| Vero E6 | ATCC | Cat#CRL-1586; RRID:CVCL_0574 |
| 293T | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| Oligonucleotides | ||
| 5’-FAM-AGGAAGAUUAAUAAUUUUCCU-Iowa Black® FQ-3’ |
This paper | N/A |
| Recombinant DNA | ||
| pRev (T7 RNA polymerase expressing plasmid) | Julie Pfeiffer (The University of Texas Southwestern Medical Center) | N/A |
| pTM-L | Brennan et al., 2015 | N/A |
| pTM-N | Brennan et al., 2015 | N/A |
| pTVT-L | Brennan et al., 2015 | N/A |
| pTVT-M | Brennan et al., 2015 | N/A |
| pTVT-S | Brennan et al., 2015 | N/A |
| SFTSV L 1–231aa | This paper | N/A |
| HRTV L 1–231aa | This paper | N/A |
| IAV L 1–192aa | DuBois et al., 2012 | N/A |
| Software and Algorithms | ||
| HKL3000 | Minor et al., 2006 | https://hkl-xray.com/hkl-3000 |
| CCP4i | Winn et al., 2011 | https://www.ccp4.ac.uk/ccp4i_main.php |
| Phenix | Adams et al., 2010 | https://www.phenix-online.org/ |
| WinCoot | Emsley et al., 2010 | http://bernhardcl.github.io/coot/ |
| Pymol 2.3 | Schrodinger | https://pymol.org/2/ |
| ASTRA 6 | Wyatt | https://www.wyatt.com/products/software/astra.html |
| Origin 7 | OriginLab | https://www.originlab.com/ |
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| MagTran | Zhang and Marshall, 1998 | N/A |
| Byonic and Byologic | Protein Metrics | https://www.proteinmetrics.com/ |
| HDExaminer | Sierra Analytics | http://massspec.com/hdexaminer/ |
| Gromacs 5.1.1 | Abraham et al., 2015 | http://manual.gromacs.org/5.1.1/index.html |
| FAST algorithm | Zimmerman and Bowman, 2015, 2016 | N/A |
| Surflex-dock | Jain, 2007 | https://omictools.com/surflex-dock-tool |
Highlights.
X-ray crystal structure of the endonuclease domain from SFTSV L protein is solved
SFTSV endonuclease domain active site is similar to those of other sNSVs
Alpha 6 helix in the endonuclease domain may control substrate access
FDA-approved drug Baloxavir inhibits SFTSV and HRTV virus endonuclease activity
ACKNOWLEDGMENTS
We thank Ms. S. Smith for general support and coordination. Research was supported by National Institutes of Health (NIH) grants R01AI107056 and R01AI140758 (D.W.L.); R01AI123926 (G.K.A.); U19AI109945 and U19AI109664 (G.K.A.); R01GM12400701 (G.R.B.); and CA200422, AI116585, AI129496, AI140705, and AI140718 (J.U.J.); as well as by the·Fletcher Jones Foundation (J.U.J.), National Science Foundation (NSF) CAREER Award MCB-1552471 (G.R.B.), and Department of Energy (DOE) Integrated Diffraction Analysis (IDAT) grant contract DE-AC02–05CH11231. The mass spectrometry was supported by the National Institute of General Medical Sciences (NIGMS) of the NIH (grant P41GM103422). Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC-CAT is operated by University of Chicago Argonne for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02–06CH11357. The content or interpretations do not necessarily reflect the position or policies of the federal government, and no official endorsement should be inferred.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.020.
REFERENCES
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, and Lindahl E (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. [Google Scholar]
- Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, et al. (2019). Taxonomy of the order Bunyavirales: update 2019. Arch. Virol 164, 1949–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amroun A, Priet S, and Querat G (2017). Toscana virus cap-snatching and initiation of transcription. J. Gen. Virol 98, 2676–2688. [DOI] [PubMed] [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, and Notredame C (2006). Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34, W604–W608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn TT, and Thor KB (2004). Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov 3, 673–683. [DOI] [PubMed] [Google Scholar]
- Beaucourt S, and Vignuzzi M (2014). Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr. Opin. Virol 8, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan B, Li P, Zhang S, Li A, Liang M, Li D, and Elliott RM (2015). Reverse genetics system for severe fever with thrombocytopenia syndrome virus. J. Virol 89, 3026–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, and Parrinello M (2007). Canonical sampling through velocity rescaling. J. Chem. Phys 126, 014101. [DOI] [PubMed] [Google Scholar]
- Calvert AE, and Brault AC (2015). Development and Characterization of Monoclonal Antibodies Directed Against the Nucleoprotein of Heartland Virus. Am. J. Trop. Med. Hyg 93, 1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, and Zhou HX (2005). Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins 61, 21–35. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Cong ML, Li MH, Kang YJ, Feng YM, Plyusnin A, Xu J, and Zhang YZ (2012). Infection and pathogenesis of Huaiyangshan virus (a novel tick-borne bunyavirus) in laboratory rodents. J. Gen. Virol 93, 1288–1293. [DOI] [PubMed] [Google Scholar]
- Credille CV, Dick BL, Morrison CN, Stokes RW, Adamek RN, Wu NC, Wilson IA, and Cohen SM (2018). Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease. J. Med. Chem 61, 10206–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delang L, Abdelnabi R, and Neyts J (2018). Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 153, 85–94. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, and Notredame C (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, and Ruigrok RW (2009). The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918. [DOI] [PubMed] [Google Scholar]
- Doan L, Handa B, Roberts NA, and Klumpp K (1999). Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry 38, 5612–5619. [DOI] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. (2003). A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem 24, 1999–2012. [DOI] [PubMed] [Google Scholar]
- DuBois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ, Webb TR, and White SW (2012). Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8, e1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijsings D, Kormelink R, and Goldbach R (2001). In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J. 20, 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson UB, Hallberg BM, Detitta GT, Dekker N, and Nordlund P (2006). Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem 357, 289–298. [DOI] [PubMed] [Google Scholar]
- Feenstra KA, Hess B, and Berendsen HJC (1999). Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem 20, 786–798. [DOI] [PubMed] [Google Scholar]
- Fernández-García Y, Reguera J, Busch C, Witte G, Sánchez-Ramos O, Betzel C, Cusack S, Günther S, and Reindl S (2016). Atomic Structure and Biochemical Characterization of an RNA Endonuclease in the N Terminus of Andes Virus L Protein. PLoS Pathog. 12, e1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Komeno T, and Nakamura T (2017). Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci 93, 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, and Kolakofsky D (1995). The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol 69, 5754–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach P, Malet H, Cusack S, and Reguera J (2015). Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter. Cell 161, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogrefe N, Reindl S, Günther S, and Rosenthal M (2019). Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PLoS Pathog. 15, e1007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart DI, Grimes JM, and Fodor E (2015). Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527, 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B (2008). P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput 4, 116–122. [DOI] [PubMed] [Google Scholar]
- Jain AN (2007). Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des 21, 281–306. [DOI] [PubMed] [Google Scholar]
- Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, et al. (2013). Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744). J. Med. Chem 56, 5901–5916. [DOI] [PubMed] [Google Scholar]
- Jones R, Lessoued S, Meier K, Devignot S, Barata-García S, Mate M, Bragagnolo G, Weber F, Rosenthal M, and Reguera J (2019). Structure and function of the Toscana virus cap-snatching endonuclease. Nucleic Acids Res. 47, 10914–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935. [Google Scholar]
- Ju H, Zhang J, Huang B, Kang D, Huang B, Liu X, and Zhan P (2017). Inhibitors of Influenza Virus Polymerase Acidic (PA) Endonuclease: Contemporary Developments and Perspectives. J. Med. Chem 60, 3533–3551. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OH, Ruigrok RW, and Cusack S (2012). Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8, e1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Lei XY, Liu MM, and Yu XJ (2015). Severe fever with thrombocytopenia syndrome and its pathogen SFTSV. Microbes Infect. 17, 149–154. [DOI] [PubMed] [Google Scholar]
- Liu W, Lu QB, Cui N, Li H, Wang LY, Liu K, Yang ZD, Wang BJ, Wang HY, Zhang YY, et al. (2013). Case-fatality ratio and effectiveness of ribavirin therapy among hospitalized patients in china who had severe fever with thrombocytopenia syndrome. Clin. Infect. Dis 57, 1292–1299. [DOI] [PubMed] [Google Scholar]
- Lu QB, Zhang SY, Cui N, Hu JG, Fan YD, Guo CT, Qin SL, Yang ZD, Wang LY, Wang HY, et al. (2015). Common adverse events associated with ribavirin therapy for Severe Fever with Thrombocytopenia Syndrome. Antiviral Res. 119, 19–22. [DOI] [PubMed] [Google Scholar]
- Luo LM, Zhao L, Wen HL, Zhang ZT, Liu JW, Fang LZ, Xue ZF, Ma DQ, Zhang XS, Ding SJ, et al. (2015). Haemaphysalis longicornis Ticks as Reservoir and Vector of Severe Fever with Thrombocytopenia Syndrome Virus in China. Emerg. Infect. Dis 21, 1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX, Schwantes CR, Wang LP, Lane TJ, and Pande VS (2015). MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J 109, 1528–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, et al. (2012). A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med 367, 834–841. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006). HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, and Dodson EJ (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Noble E, Cox A, Deval J, and Kim B (2012). Endonuclease substrate selectivity characterized with full-length PA of influenza A virus polymerase. Virology 433, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, and Hutchison GR (2011). Open Babel: An open chemical toolbox. J. Cheminform 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WS, Yoo JR, Kwon KT, Kim HI, Lee SJ, Jun JB, Ryu SY, Kim HA, Hur J, Wi YM, et al. (2017). Effect of Early Plasma Exchange on Survival in Patients with Severe Fever with Thrombocytopenia Syndrome: A Multicenter Study. Yonsei Med. J 58, 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, and Cusack S (2018). Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep 8, 9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug A, Guilligay D, Reich S, and Cusack S (2014). Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–360. [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, and Krug RM (1981). A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858. [DOI] [PubMed] [Google Scholar]
- Reguera J, Weber F, and Cusack S (2010). Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6, e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Gerlach P, Rosenthal M, Gaudon S, Coscia F, Günther S, and Cusack S (2016). Comparative Structural and Functional Analysis of Bunyavirus and Arenavirus Cap-Snatching Endonucleases. PLoS Pathog. 12, e1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crépin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, and Cusack S (2014). Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russmann S, Grattagliano I, Portincasa P, Palmieri VO, and Palasciano G (2006). Ribavirin-induced anemia: mechanisms, risk factors and related targets for future research. Curr. Med. Chem 13, 3351–3357. [DOI] [PubMed] [Google Scholar]
- Schrodinger (2018). The PyMOL Molecular Graphics System, Version 2.2.
- Sheldrick GM (2008). A short history of SHELX. Acta Crystallogr. A 64, 112–122. [DOI] [PubMed] [Google Scholar]
- Song MS, Kumar G, Shadrick WR, Zhou W, Jeevan T, Li Z, Slavish PJ, Fabrizio TP, Yoon SW, Webb TR, et al. (2016). Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor. Proc. Natl. Acad. Sci. USA 113, 3669–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Qi Y, Liu C, Gao W, Chen P, Fu L, Peng B, Wang H, Jing Z, Zhong G, and Li W (2014). Nonmuscle myosin heavy chain IIA is a critical factor contributing to the efficiency of early infection of severe fever with thrombocytopenia syndrome virus. J. Virol 88, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Komeno T, Fukuma A, Fukushi S, Taniguchi S, Shimojima M, Uda A, Morikawa S, Nakajima N, Furuta Y, and Saijo M (2018). Therapeutic effects of favipiravir against severe fever with thrombocytopenia syndrome virus infection in a lethal mouse model: Dose-efficacy studies upon oral administration. PLoS ONE 13, e0206416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, and Adams PD (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, and Hung LW (2009). Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, et al. (2016). Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol. Cell 61, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, and Teplyakov A (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr 66, 22–25. [DOI] [PubMed] [Google Scholar]
- Walker AP, and Fodor E (2019). Interplay between Influenza Virus and the Host RNA Polymerase II Transcriptional Machinery. Trends Microbiol. 27, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover JB, Rigas JD, Van Wettere AJ, Li R, Hickerson BT, Jung KH, Miao J, Reynolds ES, Conrad BL, Nielson S, et al. (2017). Heartland virus infection in hamsters deficient in type I interferon signaling: Protracted disease course ameliorated by favipiravir. Virology 511, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, et al. (2011). Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med 364, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, He YW, Dai YA, Xiong Y, Zheng H, Zhou DJ, Li J, Sun Q, Luo XL, Cheng YL, et al. (2012a). Hemorrhagic fever caused by a novel Bunyavirus in China: pathogenesis and correlates of fatal outcome. Clin. Infect. Dis 54, 527–533. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Zhou DJ, Qin XC, Tian JH, Xiong Y, Wang JB, Chen XP, Gao DY, He YW, Jin D, et al. (2012b). The ecology, genetic diversity, and phylogeny of Huaiyangshan virus in China. J. Virol 86, 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, and Marshall AG (1998). A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom 9, 225–233. [DOI] [PubMed] [Google Scholar]
- Zimmerman MI, and Bowman GR (2015). FAST Conformational Searches by Balancing Exploration/Exploitation Trade-Offs. J. Chem. Theory Comput 11, 5747–5757. [DOI] [PubMed] [Google Scholar]
- Zimmerman MI, and Bowman GR (2016). How to Run FAST Simulations. Methods Enzymol. 578, 213–225. [DOI] [PubMed] [Google Scholar]
- Zimmerman MI, Hart KM, Sibbald CA, Frederick TE, Jimah JR, Knoverek CR, Tolia NH, and Bowman GR (2017). Prediction of New Stabilizing Mutations Based on Mechanistic Insights from Markov State Models. ACS Cent. Sci 8, 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MI, Porter JR, Sun X, Silva RR, and Bowman GR (2018). Choice of Adaptive Sampling Strategy Impacts State Discovery, Transition Probabilities, and the Apparent Mechanism of Conformational Changes. J. Chem. Theory Comput 14, 5459–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The hydrogen deuterium exchange mass spectrometry plots are available at https://data.mendeley.com/datasets/f5t9tdvn5n/1. Protein Data Bank (PDB; https://www.rcsb.org/): 6NTV.