Abstract
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumour, which has posed a heavy health and financial burden worldwide. Due to limited symptoms at the early stage and the limitation in current biomarkers, HCC patients are usually diagnosed at the advanced stage with a pessimistic overall survival rate. Circular RNAs (circRNAs) are a subclass of single‐stranded RNAs characterized by a covalently closed loop structure without 3’‐ or 5’‐end. With advances in high‐throughput sequencing technology and bioinformatics, accumulating studies have demonstrated the promotor or suppressor roles of circRNAs in the carcinogenesis, progression, and metastasis of HCC. Moreover, circRNAs are characteristic of higher abundance, stability and conservation compared with linear RNAs. Therefore, circRNAs have emerged as one of the most promising diagnostic and prognostic biomarkers for HCC with reliable accuracy, sensitivity and specificity. In this review, we briefly introduce the characteristics of circRNAs and summarize the roles of circRNAs in the biological procedures of HCC. Furthermore, we provide an overview on the potential diagnostic and prognostic value of circRNAs as biomarkers for patients with HCC. Finally, we discuss future perspectives of circRNAs in cancer research.
Keywords: circular RNAs, diagnostic biomarker, hepatocellular carcinoma, prognostic biomarker
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumour with an increasing incidence. 1 , 2 As a major subtype of primary liver cancer, HCC accounts for approximately 90% in general and is now the fourth leading cause of cancer‐related death worldwide, imposing severe health and financial burden. 2 , 3 Current treatment strategies for HCC include hepatic resection, percutaneous thermal ablation, radiotherapy, systemic treatment and immunotherapy. 4 However, since HCC patients show limited symptoms at the early stage, more than 75% of HCC patients are diagnosed at the advanced stage with tumour cell metastasis or diffusion. 5 Also, the treatment response of patients in the advanced stage is usually poor, and they commonly suffer from a low survival rate. Hence, it is essential to diagnose HCC at the early stage, and there is an urgent need to develop novel diagnostic or prognostic biomarkers for HCC.
Circular RNAs (circRNAs) are a subclass of single‐stranded RNAs characterized by a covalently closed loop structure. Base on whether they can be translated, circRNAs can be divided into non‐coding circRNAs and coding circRNAs. 6 With the advances in high‐throughput sequencing technology and bioinformatics, a variety of circRNAs have been demonstrated to play essential roles in regulating gene expression at transcriptional or post‐transcriptional levels 7 , 8 , 9 , 10 and participate in multiple biological processes of HCC. 11 Moreover, owing to the single‐stranded closed circular structure, circRNAs show higher abundance, stability and conservation compared with linear RNAs. These advantages make circRNAs one of the most promising biomarkers in the diagnosis and prognosis of HCC. 12 , 13
In this review, we briefly introduce the characteristics of circRNAs and summarize their roles in the biological processes of HCC. Furthermore, we provide an overview on the potential diagnostic and prognostic value of circRNAs as biomarkers. Finally, we give an insight into future perspectives of circRNAs in cancer research.
2. CHARACTERISTICS OF CIRCRNAS
CircRNAs were initially discovered as aberrant by‐products or abnormally spliced transcripts in human cells, while their specific roles remained vague due to the limitation of traditional RNA sequencing technologies. 14 , 15 Owing to the improvement of specialized computational pipelines in the last decade, accumulating information has been gathered in this area, and the roles of circRNAs in various diseases are becoming increasingly evident. 16 , 17 , 18 , 19
CircRNAs are single‐stranded transcripts arisen from exons (ecircRNAs), 20 introns (ciRNAs) 21 or both (EIciRNAs) 18 , 22 (Figure 1). CircRNAs are extensively expressed in eukaryotic cells and characterized by high abundance, stability, conservation, as well as a tissue‐ or developmental‐specific expression pattern. 11 , 23 , 24 , 25 The covalently closed circular structure makes circRNAs more stable and resistant to exonuclease‐induced degradation compared with linear RNAs. 26 Moreover, circRNAs display evolutionary conservation across multiple species and the expression profiles are tissue‐ or developmental stage‐specific, suggesting the broad participation in various physiological and pathophysiological processes. 23 , 27 , 28 , 29
Figure 1.
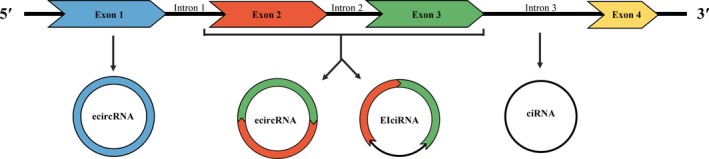
The biogenesis of circRNAs. CircRNAs are single‐stranded transcripts arising from exons (ecircRNAs), introns (ciRNAs), or both exons and introns (EIciRNAs)
Accumulating evidence has revealed the function of circRNAs in regulating gene transcription and expression, and the miRNA sponge activity is the hallmark function of circRNAs. 30 CircRNAs have a significant number of binding sites for miRNAs and can act as sponges of miRNAs, which would subsequently influence the stability and translation of target RNAs. 31 Apart from miRNA sponge activity, circRNAs can also interact with RNA binding proteins, 32 function as transcriptional or translational regulators, 22 influence splicing of pre‐mRNAs 33 and participate in protein translation. 34 , 35
3. ROLES OF CIRCRNAS IN HCC
Recently, the knowledge of circRNAs is accumulating at an increasing pace, and plenty of studies have revealed the association between circRNAs and various biological processes in cancers, including cell proliferation, apoptosis, invasion, as well as metastasis. In HCC, numerous circRNAs are dysregulated in tumour tissues and may play oncogenic or suppressor roles in cancer development and progression. 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 Generally, most of the up‐regulated circRNAs are positively associated with HCC progression, whereas down‐regulated circRNAs usually act as suppressors preventing the development of HCC. Table 1 summarizes recent studies on the regulating effect of circRNAs in HCC. 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54
Table 1.
Recent studies on the regulating effect of circRNAs in HCC
| circBase ID (Alias) | Alteration | Target | Function | Ref. |
|---|---|---|---|---|
| hsa_circ_0001946 (Cdr1as/ciRS‐7) | ↑ | miR‐7 | Oncogene | 55, 56, 58 |
| hsa_circRNA_101368 | ↑ | HMGB1/RAGE | Modulate the migration of HCC | 42 |
| Circ‐CDYL | ↑ | miR‐892a and miR‐328‐3p | Oncogene | 9 |
| hsa_circRNA‐104718 | ↑ | miR‐218‐5p/TXNDC5 | Promote HCC progression | 54 |
| hsa_circ_0067934 | ↑ | miR‐1324/FZD5/Wnt/β‐catenin | Promote tumour growth and metastasis | 60 |
| hsa_circ_0015756 | ↑ | miR‐7 | Promote proliferation, invasion and migration | 61 |
| hsa_circ_0020007 (circ‐ADD3) | ↓ | EZH2 | Inhibit HCC metastasis | 63 |
| hsa_circ_0001445 (cSMARCA5) | ↓ | miR‐17‐3p and miR‐181b‐5p | Inhibit growth and metastasis in HCC | 10 |
| hsa_circ_0000284 (circHIPK3) | ↑ | miR‐124 | Regulate cell proliferation and migration | 44 |
| hsa_circ_0000567 (circSETD3) | ↓ | miR‐421 | Inhibit the proliferation of HCC cells | 8 |
| hsa_circ_0000847 (circSMAD2) | ↓ | miR‐629 | Inhibit the migration and epithelial‐mesenchymal transition of HCC cells. | 45 |
| hsa_circ_0016788 | ↑ | miR‐486/CDK4 | Oncogene | 59 |
| hsa_circ_0128298 | ↑ | ‐ | Promote proliferation and metastasis | 72 |
| hsa_circ_0001727 (circZKSCAN1) | ↓ | Multiple cancer‐related signalling pathways | Inhibit HCC cell growth, migration and invasion | 48 |
| hsa_circ_0007144 (circPTPRM) | ↑ | ‐ | Promote proliferation and migration | 46 |
| hsa_circRNA8662‐12 (circTRIM33‐12) | ↓ | miR‐191 | Inhibit HCC proliferation, metastasis and immune evasion | 64 |
| hsa_circ_0002768 (circRNA‐MYLK) | ↑ | miR‐362‐3p | Promote the proliferation, invasion and migration | 47 |
| hsa_circRNA_102034 (circRHOT1) | ↑ | NR2F6 | Promote HCC growth and metastasis | 49 |
| hsa_circRNA_103809 | ↑ | miR‐377‐3p | Promote the proliferation, cycle progression and migration of HCC cells | 36 |
| hsa_circ_0001649 | ↓ | miR‐127‐5p, miR‐612 and miR‐4688 | Inhibit proliferation and migration of HCC | 50 |
| hsa_circ_001013 (circHIAT1) | ↓ | miR‐3171 | Inhibit the growth of HCC cells | 51 |
| Circβ‐catenin | ↑ | Wnt pathway | Promote HCC development | 52 |
| hsa_circ_0000267 | ↑ | miR‐646 | Facilitate cell growth, migration and invasion | 75 |
| hsa_circ_0008450 | ↑ | miR‐548p | Promote cell viability, migration and invasion | 37 |
| CircADAMTS13 | ↓ | miR‐484 | Suppress cell proliferation | 38 |
| hsa_circRNA_104075 | ↑ | miR‐582‐3p | Promote HCC development | 39 |
| hsa_circ_0018665 (circADAMTS14) | ↓ | miR‐572 | Inhibit tumour growth | 40 |
| hsa_circ_101280 | ↑ | miR‐375 | Promote HCC | 41 |
| hsa_circ_0016788 | ↑ | miR‐486 | Promote the proliferation, invasion and inhibit the apoptosis | 59 |
| Circ‐FOXP1 | ↑ | miR‐875‐3p and miR‐421 | Promote HCC progression | 53 |
| hsa‐circ‐0046600 | ↑ | miR‐640/HIF‐1α | Promote HCC progression | 43 |
Abbreviations: ‐, not provided; ↑, Up‐regulated; ↓, Down‐regulated; HCC, hepatocellular carcinoma.
Cdr1as (hsa_circ_0001946), a sponge of miR‐7, is one of the classic circRNAs acting as a promotor in the progression of HCC. Cdr1as was reported to be significantly up‐regulated in HCC cell lines and HCC tissues compared with the non‐tumour ones, and the expression level of Cdr1as was positively associated with hepatic microvascular invasion as well as deterioration. 55 , 56 As a target of Cdr1as, miR‐7 targets several oncogenes directly, and the up‐regulation of miR‐7 inhibits the proliferation and invasion of HCC cells. 57 Moreover, knockdown of Cdr1as would suppress the HCC cell proliferation and invasion, 56 while overexpression of Cdr1as could lead to the opposite effect. 58 Importantly, exosomes acquired from HCC cells overexpressing Cdr1as could also accelerate the proliferation and migration of surrounding normal cells. 58
Guan et al 59 conducted circRNA microarray analysis in three pairs of HCC and adjacent healthy tissues. Hsa_circ_0016788 was significantly up‐regulated both in HCC tissue and cell lines. The silence of hsa_circ_0016788 could retard tumour growth and inhibit the proliferation and invasion of tumour cells. Also, the bioinformatics analysis showed that hsa_circ_0016788 accelerated HCC progression via miR‐486/CDK4. These results indicated the oncogenic role of hsa_circ_0016788 in HCC.
Moreover, Wei et al 9 revealed a circRNA‐centric non‐coding regulatory RNA network activated in the early stage of HCC based on RNA expression profiles. Circ‐CDYL was reported to be highly expressed in the early stage of HCC, which promoted the properties of epithelial cell adhesion molecule‐positive liver tumour‐initiating cells. Additionally, circ‐CDYL could promote the expression of several proto‐oncogenes via PI3K‐AKT‐mTORC1/β‐catenin and NOTCH2 pathways in HCC cells. Similarly, various circRNAs were also reported as promotors for carcinogenesis and cancer progression, such as hsa_circ_0067934, 60 circHIPK3 (hsa_circ_0000284), 44 hsa_circ_0015756, 61 hsa_circ_0001955 62 and so forth.
Apart from oncogenic roles, circRNAs could also act as tumour suppressors preventing the occurrence and development of HCC. For example, Sun et al 63 showed a significant decrease of circ‐ADD3 (hsa_circ_0020007) in HCC by circRNA microarray expression profile in matched HCC and para‐cancerous tissues. Increased expression of circ‐ADD3 could effectively weaken the vascular invasion, intrahepatic metastasis and distant metastasis of HCC via regulating EZH2 stability. Moreover, Zhang et al 64 demonstrated that circTRIM33‐12(has_circRNA8662‐12), a sponge of miR‐191, was down‐regulated in HCC tissues and cell lines, and the reduced expression of circTRIM33‐12 could significantly promote tumour proliferation, migration, invasion as well as immune evasion abilities of HCC cells.
4. DIAGNOSTIC VALUE OF CIRCRNAS IN HCC
Due to limited symptoms at the early stage and the limitation in current biomarkers, HCC patients are usually diagnosed at the advanced stage with a pessimistic overall survival rate. 65 Moreover, current biomarkers, such as α‐fetoprotein (AFP) and AFP‐L3, show modest diagnostic performance for HCC. 65 Since circRNAs are closely associated with multiple biological processes in HCC and characteristic with high stability and abundance in HCC tissue as well as in body fluid, circRNAs have been proposed as diagnostic biomarkers for HCC. For example, Yao et al 66 revealed the close association between hsa_circ_0068669 expression level and microvascular invasion, and they demonstrated hsa_circ_0068669 as a potential biomarker for HCC metastasis. Table 2 summarizes recent studies on circRNAs as diagnostic biomarkers for HCC. 67 , 68 , 69
Table 2.
Recent studies on circRNAs as diagnostic biomarkers of HCC
| circBase ID (Alias) | Gene symbol | Genomic position | AUC | Sensitivity (%) | Specificity (%) | Ref. |
|---|---|---|---|---|---|---|
| Circ‐CDYL, plus HDGF and HIF1AN | ‐ | ‐ | 0.73 | 75.4 | 66.67 | 9 |
| hsa_circ_001565 | B4GALT2 | chr1:44446997‐44447136 | ‐ | 73.5 | 82.29 | 71 |
| hsa_circ_000224 | C17orf107 | chr17:4803230‐4803902 | ‐ | 95.6 | 92.7 | 71 |
| hsa_circ_000520 | VIM | chr10:17271723‐17271867 | ‐ | 97.1 | 89.6 | 71 |
| hsa_circ_0016788 | TRIM11 | chr1:228581376‐228594517 | 0.851 | ‐ | ‐ | 59 |
| hsa_circ_0128298 | SPINK1 | chr5:147210311‐147211162 | 0.668 | 71.6 | 81.5 | 72 |
| hsa_circ_0001727 (CirZKSCAN1) | ZKSCAN1 | chr7:99621041‐99621930 | 0.834 | 82.2 | 72.4 | 48 |
| hsa_circ_0000976, plus hsa_circ_0007750 and hsa_circ_0139897 | HPCAL1, RABGGTA, and MTM1, respectively | chr2:10559859‐10560261, chr14:24735635‐24737825, and chrX:149761066‐149787612, respectively | 0.843 | 87.5 | 81.2 | 73 |
| hsa_circRNA_104075 | NUP153 | chr6:17669524‐17669777 | 0.973 | 96.0 | 98.3 | 39 |
| hsa_circ_0005075 | EIF4G3 | chr1:21377358‐21415706 | 0.94 | 83.3 | 90.0 | 67 |
| hsa_circ_0028502 | SLC24A6 | chr12:113753139‐113754806 | 0.675 | 72.1 | 58.0 | 70 |
| hsa_circ_0076251 | ZFAND3 | chr6:38050167‐38084515 | 0.738 | 71.3 | 64.0 | 70 |
| hsa_circ_0027089 | PTGES3 | chr12:57059987‐57064148 | 0.784 | 57.8 | 84.8 | 68 |
| hsa_circ_0003998 | ARFGEF2 | chr20:47570092‐47580435 | 0.894 | 84.0 | 80.0 | 69 |
Abbreviations: ‐, not provided; AUC, area under the curve.
Circ‐CDYL is a promotor for HCC, and its up‐regulation could increase the expression of several proto‐oncogenes. Recently, Wei et al 9 showed the diagnostic performance of circ‐CDYL in the early stage of HCC with an area under the curve (AUC) of 0.64 (95% CI = 0.55‐0.72). When comprehensively analysing the expression levels of circ‐CDYL plus HDGF and HIF1A, the results showed an improved diagnostic performance with an AUC of 0.73 (95% CI = 0.65‐0.80), a sensitivity of 75.36% and specificity of 66.67%. Compared with circ‐CDYL plus HDGF and HIF1A, AFP only showed an AUC of 0.59 (95% CI = 0.49‐0.70), a sensitivity of 50.72% and a specificity of 83.78%. This study indicated that circ‐CDYL plus HDGF and HIF1AN might be more reliable diagnostic biomarkers than AFP.
Jiang et al 70 analysed the expression levels of hsa_circ_0028502 and hsa_circ_0076251 in cancer and adjacent para‐cancerous tissues. The results showed that both the hsa_circ_0028502 and hsa_circ_0076251 levels were significantly lower in HCC tissues (P < .001). Importantly, hsa_circ_0028502 level was related to tumour node metastasis stage (P = .015), while hsa_circ_0076251 expression was related to Barcelona Clinic Liver Cancer stage (P = .038). When distinguishing HCC tissues from the liver cirrhosis tissues and chronic hepatitis tissues, the AUCs of hsa_circ_0028502 and hsa_circ_0076251 were 0.675 and 0.738, respectively. Additionally, Matboli et al 71 assessed the diagnostic performance of hsa_circ_001565, hsa_circ_000224 and hsa_circ_000520 for HCC, and the results showed a higher sensitivity and specificity compared with AFP. When combined with these three biomarkers, the diagnostic performance was further improved to an accuracy of 80.49%, a sensitivity of 100%, a specificity of 83.3%. Moreover, the receiver operating characteristic (ROC) curve analysis of hsa_circ_0016788 was performed based on 40 HCC patients and 40 healthy controls, and the AUC of 0.851 was acquired. 59 Similarly, hsa_circ_0128298, which was significantly up‐regulated in HCC tissues, showed an AUC of 0.668, a sensitivity of 0.716 and specificity of 0.815. 72
Furthermore, Yao et al 48 analysed the expression of circZKSCAN1 (hsa_circ_0001727) in a cohort of 102 patients with HCC, and its expression was significantly lower in tumour tissues than matched adjacent non‐tumorous samples (P < .05). Importantly, circZKSCAN1 level was associated with various clinical characteristics, including tumour numbers (P < .01), cirrhosis (P = .031), vascular invasion (P = .002), microvascular invasion (P = .002) and tumour grade (P < .001). Also, as a diagnostic biomarker, circZKSCAN1 showed reliable performance with an AUC of 0.834, a sensitivity of 82.2% and specificity of 72.4%.
Apart from HCC tissue, the alteration of circRNAs in plasma could also be used as diagnostic biomarkers for HCC. In a large‐scale multicenter study, Yu et al 73 designed and evaluated a plasma circRNA panel (circPanel) consisting of hsa_circ_0000976, hsa_circ_0007750 and hsa_circ_0139897 to diagnose HBV‐related HCC. The newly created circPanel showed a better diagnostic performance (AUC = 0.843, 95% CI = 0.796‐0.890) than AFP (AUC = 0.747, 95% CI = 0.691‐0.804) in the validation set containing 306 individuals. Moreover, circPanel also showed a reliable performance in diagnosing small‐HCC (solitary ≤ 3 cm) and AFP‐negative HCC with AUCs of 0.838 (95% CI = 0.776‐0.900) and 0.857 (95% CI = 0.793‐0.921), respectively.
5. PROGNOSTIC VALUE OF CIRCRNAS IN HCC
Since circRNAs are involved in multiple biological processes in HCC, the prognostic value of circRNAs also attracts wide attention. Table 3 summarizes recent studies on circRNAs as prognostic biomarkers of HCC.
Table 3.
Recent studies on circRNAs as prognostic biomarkers of HCC
| circBase ID (Alias) | Gene symbol | Genomic position | Prognosis | Univariate analysis | Multivariate analysis | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |||||
| hsa_circ_0001445 (cSMARCA5) | SMARCA5 | chr4:144464661‐144465125 | OS | ‐ | ‐ | <.001 | 2.47 | 1.46‐4.18 | .001 | 10 |
| RFS | ‐ | ‐ | .001 | 1.67 | 1.08‐2.59 | .021 | 10 | |||
| hsa_circ_0001946 (Cdr1as/ciRS‐7) | CDR1 | chrX:139865339‐139866824 | MVI | 2.65 | 1.06‐6.63 | .037 | 4.08 | 1.06‐15.74 | .041 | 55 |
| hsa_circ_0128298 | SPINK1 | chr5:147210311‐147211162 | OS | 1.98 | 1.34‐3.02 | .009 | 6.66 | 2.66‐8.42 | .014 | 72 |
| hsa_circRNA8662‐12 (circTRIM33‐12) | ‐ | ‐ | OS | ‐ | ‐ | .001 | 0.50 | 0.93‐1.94 | .007 | 64 |
| hsa_circ_0001727 (circZKSCAN1) | ZKSCAN1 | chr7:99621041‐99621930 | OS | 0.09 | 10.04‐0.19 | <.001 | 0.10 | 0.05‐0.23 | <.001 | 32 |
| RFS | 0.22 | 0.12‐0.41 | <.001 | 0.22 | 0.11‐0.42 | <.001 | 32 | |||
| hsa_circ_0000267 | FAM53B | chr10:126370175‐126370948 | OS | 2.64 | 1.43‐4.87 | .002 | 2.11 | 1.09‐4.06 | .025 | 75 |
| Hsa_circ_0076251 | ZFAND3 | chr6:38050167‐38084515 | OS | 0.46 | 0.22‐0.98 | <.05 | ‐ | ‐ | ‐ | 70 |
| Hsa_circ_0003998 | ARFGEF2 | chr20:47570092‐47580435 | OS | 0.60 | 0.42‐0.86 | .006 | 0.58 | 0.41‐0.84 | .003 | 69 |
Abbreviations: ‐, not provided; CI, concordance index; HR, hazard ratios; MVI, microvascular invasion; OS, overall survival; RFS, relapse‐free survival.
A study on 112 patients with HCC showed that circZKSCAN1 expression was closely associated with various HCC characteristics. In the Kaplan‐Meier survival analysis, the expression level of circZKSCAN1 in cancer tissues was positively correlated with HCC prognosis (overall survival, P < .001). The further univariate and multivariate analysis showed that circZKSCAN1 expression was an independent factor for overall survival rate (HR = 0.104; 95% CI: 0.046‐0.234, P < .001) as well as RFS rate (HR = 0.219; 95% CI: 0.114‐0.420, P < .001) for HCC patients. 32
Moreover, Matboli et al 71 demonstrated that HCC patients with negative hsa_circ_001565, hsa_circ_000224 or hsa_circ_000520 had relatively more prolonged relapse‐free survival (RFS) after a median follow‐up of 26 months. Moreover, Kaplan‐Meier analysis suggested a significant decrease in RFS and an increase in cumulative hazards among hsa_circ_000520 in patients with HCC. And the Cox multivariate analysis indicated the expression level of hsa_circ_000520 as an independent prognostic factor of RFS. 71 Similarly, low expression of circTRIM33‐12 was closely correlated with poor prognosis and was demonstrated as an independent predictor for OS (P = .001 in univariate analysis; P = .007 in multivariate analysis) as well as post‐operative recurrence (P = .001 in univariate analysis; P = .005 in multivariate analysis). 64
In addition, circASAP1, a competing endogenous RNA for miR‐326 and miR‐532‐5p, was closely associated with pulmonary metastasis of HCC after curative resection. 74 In vitro, the overexpressed circASAP1 could promote cell proliferation, migration and invasion, as well as enhancing tumour growth and pulmonary metastasis in vivo. The analysis of clinical tumour samples showed a positive association between up‐regulated circASAP1 and tumour‐associated macrophages (eg CSF‐1 and MAPK1).
Besides, Yu et al 10 demonstrated that cSMARCA5 (hsa_circ_0001445) could inhibit the proliferation and migration of HCC cells, and down‐regulated cSMARCA5 expression was significantly correlated with aggressive clinicopathological characteristics. The Kaplan‐Meier survival analysis suggested that HCC patients with low cSMARCA5 level had a significantly reduced overall survival (P = .0004) and RFS (P = .0008). Further univariate and multivariate analysis also indicated that the cSMARCA5 level was an independent risk factor for overall survival and RFS after hepatectomy. 10 Similarly, the prognostic ability of hsa_circ_0128298 was evaluated by Cox regression analysis, and the results showed that the expression level of hsa_circ_0128298 could be used as a prognostic factor to predict poor OS in HCC patients (P = .009 in univariate analysis; P = .014 in multivariate analysis). 72 The Kaplan‐Meier survival analysis also showed a statistically better overall survival in patients with low expression of hsa_circ_0128298 compared to those with high expression (P = .003). 72
CircRHOT1 (hsa_circRNA_102034), which was prominently up‐regulated in HCC tissues, was demonstrated to promote tumour growth and metastasis significantly, 49 and patients with high‐level expression of circRHOT1 were associated with a more reduced overall survival rate (P = .02) and recurrence‐free survival rate (P = .02). 49 Moreover, hsa_circ_0000267 was up‐regulated in HCC tissue and cell line, and it was considered as a critical oncogene to facilitate the initiation and progression of HCC. In the multivariable analysis performed by Pan et al 75 the results suggested hsa_circ_0000267 could be used as an independent prognostic indicator for overall survival (HR = 2.107; 95% CI: 1.093‐4.064, P = .025).
6. CONCLUSIONS AND FUTURE PERSPECTIVES
HCC is the sixth most common malignant tumour with a pessimistic survival, 76 which accounts for approximately 90% of primary liver cancers. Due to limited symptoms at the early stage and the lack of satisfying biomarkers, more than 75% of HCC patients are diagnosed at the advanced stage with a reduced overall survival rate. 65 Despite the advances in treatment strategies against HCC, such limitation makes HCC remain one of the most lethal cancers, which accounts for at least 700,000 deaths worldwide annually. 77 , 78 Numerous circRNAs have been found to be dysregulated in tumour tissues, and accumulating studies have demonstrated the oncogenic or suppressor roles of circRNAs in the carcinogenesis and progression of HCC. 79 Moreover, circRNAs are characteristic of higher abundance, stability and conservation compared with linear RNAs. Therefore, circRNAs have recently emerged as one of the most promising diagnostic and prognostic biomarkers for HCC.
However, the research of circRNAs in HCC remains in its infancy stage, the mechanisms underlying the contribution of circRNAs to HCC generation and progression remain vague, and their overall function has not yet been fully understood. Compared with miRNAs or lncRNAs, 80 only a small amount of functional circRNAs have been discovered in HCC. So far, multiple theories have been proposed, and circRNAs may serve their functions in carcinogenesis, progression and metastasis by various mechanisms. The improved understanding of the molecular mechanisms and associated signalling pathways of these functional circRNAs will positively facilitate the identification of biomarkers and even therapeutic targets for HCC. In the next step, it is essential to study the function of circRNAs in HCC thoroughly and uncover the exact mechanisms of how circRNAs promote or suppress the progression of HCC, which will accelerate the clinical application of circRNAs in the diagnosis, prognosis and treatment of HCC.
Moreover, the number of HCC patients recruited in the diagnosing test is relatively small, and therefore, the results may be over‐interpreted. More studies on a large cohort are needed to validate the actual diagnostic or prognostic effect of circRNAs.
This review primarily provides an overview of circRNAs with a focus on the diagnostic or prognostic value, while the application of circRNAs as therapeutic targets has also attracted wide attention. For example, the knockdown of Cdr1as 56 or circ‐TCF4.85 81 was demonstrated to suppress the proliferation and invasion of HCC cells. Further, the subcutaneous injection of si‐circ‐TCF4.85‐transfected HCC cells could inhibit xenograft tumour formation in nude mice, which suggested the potential roles of circRNAs in cancer treatment. With the accumulating findings, circRNAs might be further applied in HCC as therapeutic targets in the future.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTION
Jin‐Yu Sun, Xiao‐Yu Zhang, Xiao Zhou and Xiao‐Xin Mu involved in conceptualization. Jin‐Yu Sun and Yi‐Zhi Cao involved in writing‐original draft. Jian Gu and Xiao‐Xin Mu involved in writing‐review and editing.
ACKNOWLEDGEMENTS
None.
Sun J‐Y, Zhang X‐Y, Cao Y‐Z, Zhou X, Gu J, Mu X‐X. Diagnostic and prognostic value of circular RNAs in hepatocellular carcinoma. J Cell Mol Med. 2020;24:5438–5445. 10.1111/jcmm.15258
Jin‐Yu Sun, Xiao‐Yu Zhang, and Yi‐Zhi Cao are Co‐first authors.
Contributor Information
Jian Gu, Email: Gujian@njmu.edu.cn.
Xiao‐Xin Mu, Email: mux@njmu.edu.cn.
REFERENCES
- 1. Global Burden of Disease Liver Cancer C , Akinyemiju T, Abera S, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 1990;2017(3):1683‐1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Kulik L, El‐Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477‐491.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun JY, Yin T, Zhang XY, Lu XJ. Therapeutic advances for patients with intermediate hepatocellular carcinoma. J Cell Physiol. 2019;234:12116‐12121. [DOI] [PubMed] [Google Scholar]
- 5. Costentin C. Hepatocellular carcinoma surveillance. Presse Med. 2017;46:381‐385. [DOI] [PubMed] [Google Scholar]
- 6. Li Z, Ruan Y, Zhang H, et al. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151‐1164. [DOI] [PubMed] [Google Scholar]
- 8. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA‐421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ‐CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71(1):130‐147. [DOI] [PubMed] [Google Scholar]
- 10. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214‐1227. [DOI] [PubMed] [Google Scholar]
- 11. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 12. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125‐1134. [DOI] [PubMed] [Google Scholar]
- 13. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167‐1176. [PMC free article] [PubMed] [Google Scholar]
- 14. Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64:607‐613. [DOI] [PubMed] [Google Scholar]
- 15. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis‐splicing yields circular RNA molecules. FASEB J. 1993;7:155‐160. [DOI] [PubMed] [Google Scholar]
- 16. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elia L, Quintavalle M, Condorelli G. Circular RNAs and heart failure: new players for an old disease. Cardiovasc Res. 2017;113(3):254‐255. [DOI] [PubMed] [Google Scholar]
- 18. Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu L, Wu S, Yao T, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018;32:e22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414‐2417. [DOI] [PubMed] [Google Scholar]
- 21. Monat C, Quiroga C, Laroche‐Johnston F, Cousineau B. The Ll.LtrB intron from Lactococcus lactis excises as circles in vivo: insights into the group II intron circularization pathway. RNA. 2015;21:1286‐1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 23. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li HM, Ma XL, Li HG. Intriguing circles: conflicts and controversies in circular RNA research. Wiley Interdiscip Rev RNA. 2019;10:e1538. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki H, Tsukahara T. A view of pre‐mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331‐9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salzman J, Chen RE, Olsen MN, et al. Cell‐type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westholm JO, Miura P, Olson S, et al. Genome‐wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep. 2014;9:1966‐1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR‐130b/miR‐494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 32. Zhu YJ, Zheng B, Luo GJ, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526‐3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56:55‐66. [DOI] [PubMed] [Google Scholar]
- 34. Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non‐coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919‐3931. [DOI] [PubMed] [Google Scholar]
- 35. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805‐1814. [DOI] [PubMed] [Google Scholar]
- 36. Zhan W, Liao X, Chen Z, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR‐377‐3p/FGFR1/ERK axis. J Cell Physiol. 2020;235:1733‐1745. [DOI] [PubMed] [Google Scholar]
- 37. Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR‐548p. J Cell Biochem. 2019;120:9487‐9494. [DOI] [PubMed] [Google Scholar]
- 38. Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR‐484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13:441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP‐dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song C, Li D, Liu H, et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA‐572/regulator of calcineurin 1. J Cell Physiol. 2019;234:2460‐2470. [DOI] [PubMed] [Google Scholar]
- 41. Cao S, Wang G, Wang J, et al. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR‐375/JAK2. Immunol Cell Biol. 2019;97:218‐228. [DOI] [PubMed] [Google Scholar]
- 42. Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR‐200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17:2349‐2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhai Z, Fu Q, Liu C, et al. Emerging roles of hsa‐circ‐0046600 targeting the miR‐640/HIF‐1alpha signalling pathway in the progression of HCC. Onco Targets Ther. 2019;12:9291‐9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR‐124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X, Luo P, Jing W, et al. circSMAD2 inhibits the epithelial‐mesenchymal transition by targeting miR‐629 in hepatocellular carcinoma. Onco Targets Ther. 2018;11:2853‐2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo Z, Mao X, Cui W. Circular RNA expression and circPTPRM promotes proliferation and migration in hepatocellular carcinoma. Med Oncol. 2019;36:86. [DOI] [PubMed] [Google Scholar]
- 47. Li Z, Hu Y, Zeng Q, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR‐362‐3p. Cancer Cell Int. 2019;19:211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su Y, Xu C, Liu Y, et al. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY). 2019;11:3362‐3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z, Zhao Y, Wang Y, Jin C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR‐3171/PTEN axis. Biomed Pharmacother. 2019;116:108932. [DOI] [PubMed] [Google Scholar]
- 52. Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circbeta‐catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Li Y, Li X, et al. Circular RNA circ‐FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR‐875‐3p and miR‐421. Biomed Pharmacother. 2020;121:109517. [DOI] [PubMed] [Google Scholar]
- 54. Yu J, Yang M, Zhou B, et al. CircRNA‐104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA‐218‐5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133:1487‐1503. [DOI] [PubMed] [Google Scholar]
- 55. Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS‐7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17‐27. [DOI] [PubMed] [Google Scholar]
- 56. Yu L, Gong X, Sun L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR‐7 expression. PLoS ONE. 2016;11:e0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR‐7 in cancer. Cancer Res. 2013;73:5609‐5612. [DOI] [PubMed] [Google Scholar]
- 58. Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8182‐8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR‐486/CDK4 pathway. J Cell Physiol. 2018;234:500‐508. [DOI] [PubMed] [Google Scholar]
- 60. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR‐1324/FZD5/Wnt/beta‐catenin axis. Biochem Biophys Res Commun. 2018;497:626‐632. [DOI] [PubMed] [Google Scholar]
- 61. Liu L, Yang X, Li NF, et al. Circ_0015756 promotes proliferation, invasion and migration by microRNA‐7‐dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18:2939‐2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR‐516a‐5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun S, Wang W, Luo X, et al. Circular RNA circ‐ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1‐mediated ubiquitination. Am J Cancer Res. 2019;9:1695‐1707. [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang PF, Wei CY, Huang XY, et al. Circular RNA circTRIM33‐12 acts as the sponge of MicroRNA‐191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Juarez‐Hernandez E, Motola‐Kuba D, Chavez‐Tapia NC, et al. Biomarkers in hepatocellular carcinoma: an overview. Expert Rev Gastroenterol Hepatol. 2017;11:549‐558. [DOI] [PubMed] [Google Scholar]
- 66. Yao T, Chen Q, Shao Z, et al. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;32:e22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shang X, Li G, Liu H, et al. Comprehensive circular RNA Profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95:e3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu K, Zhan H, Peng Y, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus‐related hepatocellular carcinoma. Carcinogenesis. 2019. 10.1093/carcin/bgz154. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qiao GL, Chen L, Jiang WH, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. 2019;12:5849‐5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jiang Z, Shen L, Wang S, et al. Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med. 2019;8:7278‐7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Matboli M, Shafei AE, Ali MA, et al. circRNAs (hsa_circ_00156, hsa_circ _000224, and hsa_circ _000520) are novel potential biomarkers in hepatocellular carcinoma. J Cell Biochem. 2018. 10.1002/jcb.28045. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 72. Chen D, Zhang C, Lin J, et al. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1275‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yu J, Ding WB, Wang MC, et al. Plasma circular RNA panel to diagnose hepatitis B virus‐related hepatocellular carcinoma: a large‐scale, multicenter study. Int J Cancer. 2020;146(6):1754‐1763. [DOI] [PubMed] [Google Scholar]
- 74. Hu ZQ, Zhou SL, Li J, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2019. 10.1002/hep.31068. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 75. Pan H, Tang L, Jiang H, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR‐646. J Cell Biochem. 2019;120(7):11350‐11357. [DOI] [PubMed] [Google Scholar]
- 76. Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: a comprehensive review. World J Hepatol. 2015;7:2648‐2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. El‐Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118‐1127. [DOI] [PubMed] [Google Scholar]
- 78. Sun JY, Yin TL, Zhou J, et al. Gut microbiome and cancer immunotherapy. J Cell Physiol. 2020;235:4082‐4088. [DOI] [PubMed] [Google Scholar]
- 79. Huang XY, Huang ZL, Huang J, et al. Exosomal circRNA‐100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang J, Sun J, Yang F. The role of long non‐coding RNA H19 in breast cancer. Oncol Lett. 2020;19:7‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gao J, Dai C, Yu X, et al. circ‐TCF4.85 silencing inhibits cancer progression through microRNA‐486‐5p‐targeted inhibition of ABCF2 in hepatocellular carcinoma. Mol Oncol. 2019;14:447‐461. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


