To the Editor:
Monoclonal Gammopathy of Undetermined Significance (MGUS) and Smoldering Myeloma (SMM) consistently precede symptomatic Multiple Myeloma (MM) requiring systemic therapy [1]. Several factors have been identified to predict higher risk of progression in asymptomatic patients [2]. High-risk chromosomal aberrations (CA) are associated with an increased risk of progression into symptomatic myeloma [2–4]. The impact of subclones and clonal evolution during progression of asymptomatic myeloma has not been extensively studied. So far only two published studies analyzed paired samples acquired at the time-point of MGUS/SMM and after progression into MM [5, 6]. In the current study, we investigated the presence of subclonal CA in patients with SMM and correlated findings with established baseline risk factors for disease progression as well as outcome. Second, we analyzed paired samples from SMM patients with and without progression into MM to characterize clonal evolution. We identified 191 eligible SMM patients for our analyses. Longitudinal samples were available in 67 SMM patients, in 43 patients after progression into MM and in 24 patients still at the stage of SMM without progression. We also included 139 patients with iFISH analyses at primary diagnosis and relapse after autologous stem cell transplantation (ASCT) who were previously reported [7] to illustrate the impact of selection pressure imposed by systemic therapy. Analyses were approved by the local ethics committee and performed in accordance with the Declaration of Helsinki.
iFISH was performed using the following probes: 1q21, 5p15, 5q35, 8p21, 9q34, 11q23, 13q14, 15q22, 17p13, and 19q13, the translocation probes t(4;14), t(11;14), and t(14;16) as well as a probe for IgH rearrangements. Threshold for all aberrations was 10%. As described previously [8, 9], subclones were defined by a clone size smaller than two thirds of the largest clone and an absolute difference of at least 30%. Hyperdiploidy (HD) was defined by gains of at least two odd-numbered chromosomes, the presence of gain 1q21, del 17p13, t(4;14) and t(14;16) defined high-risk CA. Detailed statistical analyses can be found in the supplemental material.
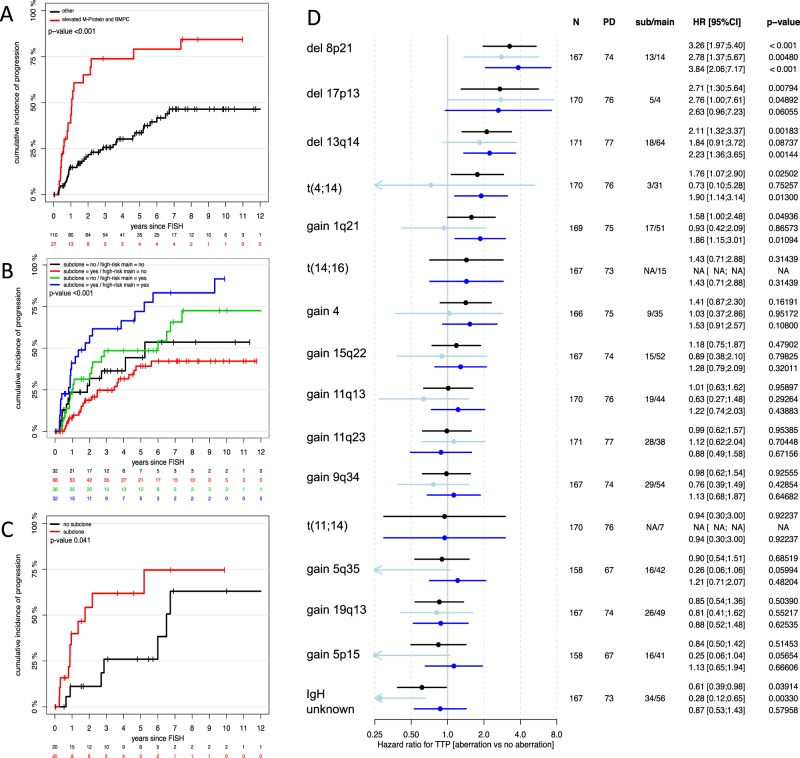
Of the 191 patients, 114 harbored subclones. Analyses of baseline characteristics revealed no significant differences between SMM patients with or without subclones at primary diagnosis (Supplementary Table 1). Follow-up data were available in 171 SMM patients. During a median follow-up of 61 months, 76 events occurred. Rate of progression was 29.5% at two years for the entire cohort. Patients with an elevated M-Protein and BMPC had a two-year progression rate of 65.1% (Fig. 1a).
Fig. 1.
a–d Summaries of univariate analysis. a–c Kaplan–Meier plots showing cumulative incidence of progression in a patients with elevated M-Protein (≥30 g/l) and bone marrow plasma cells (BMPC ≥ 10%; red line) and patients without the respective factors (black line); b high-risk patients with (blue line) and without subclones (green line) as well as standard-risk patients with (red line) and without subclones (black line); c high-risk patients with (red line) and without (black line) subclones and no elevated M-Protein and BMPC. d Forrest plot of single aberrations according to type of clone (any = black bars; light blue = subclone; dark blue = main clone); N number of patients tested for the respective aberration, PD progressive disease, sub subclone, main main clone, HR hazard ratio, CI confidence interval, TTP time to progression
We confirmed that the presence of t(4;14), del 17p13, gain 1q21, del 8p21, and del 13q14 were linked to higher risk of progression. The effects were more pronounced if the respective CA were present as main clone instead of subclone. Figure 1d summarizes results of univariate analyses.
The presence of any subclone was not prognostic in the overall cohort (HR 0.91 [0.58;1.44], p = 0.7), but the prognostic subclone effect was different for patients with HR-CA in main clone (interaction p = 0.04). Patients harboring high-risk CA were at even higher risk, when additional subclones were found (1.85 [1.01, 3.4]; p = 0.048, Fig. 1b). This effect was observed regardless whether subclones were confined to additional high- or standard-risk chromosomes and was also found in patients without elevated M-Protein and BMPC according to the MAYO clinic model (Fig. 1c). Multivariate analysis accounting for elevated M-Protein, BMPC, and an HD karyotype confirmed subclones to be an independent risk factor for progression in patients with high-risk main clone (2.34 [1.16, 4.71]; p = 0.02, Supplementary Table 2).
In 89 patients we found IgH translocations with unknown partners (main clone: n = 56; subclone: n = 33). Patients with the respective CA exhibited a lower risk of progression into symptomatic disease (0.62 [0.39; 0.98]; p = 0.041), especially if present as subclone (0.28 [0.12, 0.66]; p = 0.004; Fig. 1d).
We investigated changes in CA after progression from SMM into MM in 43 patients with longitudinal iFISH analyses. No changes between both time points were found in 22 patients. De novo CA after progression were detected in seven patients. In eleven patients a subclone present at initial diagnosis of SMM grew out to become the main clone at time of progression into symptomatic MM and three patients showed both, a de novo CA as well as an evolving subclone.
Analyses of single CA showed especially in chromosomes associated with HD evolution of initially present subclones to main clone in progressive SMM (Supplementary Fig. 1). Extensive changes in both directions were observed in paired samples from primary diagnosis and relapse after ASCT (Supplementary Fig. 1). Patients with progressive disease showed evolving subclones and de novo lesions for del 17p13 and gain 1q21. This pattern was more pronounced in patients with relapse after ASCT than progression from SMM to MM (Supplementary Fig. 2).
Defined IgH translocations remained stable in progressive SMM and MM. However, de novo and evolving IgH translocations with unknown partners occurred in subsets of patients with progressive SMM and relapsed MM (Supplementary Fig. 3).
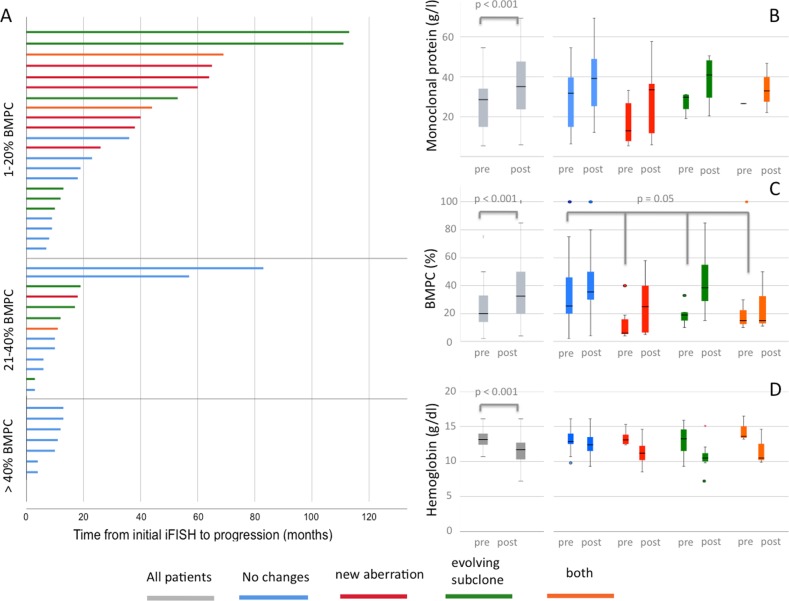
When analyzing median time from first iFISH to progression into symptomatic disease, we found significant differences based on the occurrence and evolution of CA. Patients without any changes or evolving subclones had the shortest TTP (11.2 months and 16.2 months, respectively) while patients with de novo CA or both de novo CA and evolving subclones had longer median TTP (39.8 months and 43.5 months, respectively; p = 0.02). In patients with symptomatic MM and relapse after ASCT, analysis of median time from initial iFISH to progression showed no significant differences when comparing patients with and without de novo CA (31.6 months and 27.6 months; p = 0.1). Figure 2a shows individual times to progression according to clonal evolution and initial BMPC.
Fig. 2.
Analysis of progressive SMM patients with longitudinal samples. a Swimmer’s plots showing individual time from initial iFISH analysis to progression according to pattern of cytogenetic evolution and bone marrow plasma cell (BMPC) percentage. Longitudinal changes of serum parameters at primary diagnosis (pre) and after progression (post) for b monoclonal protein, c bone marrow plasma cells (BMPC), and d hemoglobin
After progression from SMM to MM, patient exhibited higher BMPC and M-Protein as well as lower hemoglobin levels compared with baseline (Fig. 2b). Progressive patients without new CA had significantly higher BMPC at baseline compared with patients with de novo CA (21.0% versus 11.5%, respectively; p = 0.02, Fig. 2).
Two recent studies that investigated clonal evolution in a small number of patients with progressive asymptomatic plasma cell disorders identified two different types of progression [5, 6]. Both studies described clonal stability, i.e., no significant changes in clonal architecture, as one major mode of progression. In accordance with our findings, clonal stability was associated with a shorter time to progression, while in case of spontaneous evolution median TTP was longer [5]. Authors hypothesized that patients with the respective evolution pattern needed a longer time to accumulate enough malignant plasma cells [5]. This is supported by the fact that we found lower numbers of BMPC at baseline in patients with evolving CA, while no significant differences for BMPC and M-Protein were found after progression.
When looking at longitudinal changes of single CA over time, we observed evolution of subclones into main clones especially in chromosomes associated with HD and to a lesser extend in chromosomes linked to hypodiploidy and high-risk disease. This underlines the hypothesis that SMM patients with high-risk CA or an HD karyotype experience accelerated disease progression with a shorter time to surpass the threshold of disease burden needed to cause secondary end-organ damage (or fulfillment of Slim-CRAB criteria) [3].
In accordance with a recent subgroup analysis of the Myeloma XI trial, we did not observe differences in time from initial iFISH analyses to relapse after ASCT according to the pattern of cytogenetic evolution [10]. In contrast to progressive SMM, this underlines the importance of selection pressure imposed by chemotherapy that alters the natural history of cytogenetic evolution.
IgH translocations with unknown partners occur especially in patients with relapse after multiple lines of therapy [11]. Correspondingly, de novo and evolving clones with IgH translocations with unknown partners were observed in progressive SMM and relapsed MM. cMYC is one of the potential translocation partners and associated with adverse prognosis and refractory disease [11, 12].
In contrast to the reported adverse prognosis in symptomatic disease, SMM patients with subclonal IgH translocations with unknown partners were the only patients with a lower risk of progression. A possible explanation for this finding might be that the respective translocation occurred before legitimate class switch recombination creating nonmalignant long-lived B- or plasma cells [13]. However, further sequencing studies are warranted to proof this hypothesis and to identify translocation partners.
Taken together, we show that subclonal CA are of prognostic significance, especially in cytogenetically defined high-risk SMM. Based on our findings, the time needed to accumulate a certain tumor load is partially defined by baseline disease burden and CA. However, risk of progression is not constant but might actually change during the course of the disease due to clonal evolution. This is supported by a recent study that investigated serum immune markers in MGUS patients over time [14]. Authors found for the first time a conversion from low- to high-risk MGUS based on changes in monoclonal proteins in up to 70% of patients with high-risk disease before progression [14]. These findings and our current study support repeated risk assessments in patients with asymptomatic plasma cell diseases.
Supplementary information
Acknowledgements
The authors thank Maria Dörner, Ewelina Nickel, Hendrike Seidt, and Marie-Louise Brygider for technical assistance in the enrichment of CD138-positive plasma cells and Michaela Brough, Michelle Ebentheuer, Stephanie Pschowski-Zuck, Marie-Christin Meffert, and Annekathrin Borowski for performing interphase FISH analysis. This work was supported by grants from the German Federal Ministry of Education (BMBF) “CLIOMMICS” (01ZX1309) and “CAMPSIMM” (01ES1103), the Deutsche Forschungsgemeinschaft (SFB/TRR79), the Dietmar Hopp Stiftung “Heidelberger Konzept zur Optimierung der Diagnostik und Therapie des Multiplen Myeloms”, the 7th EU-framework program “OverMyR” and by Takeda Pharma Vertrieb GmbH & Co. KG, Germany (IISR2016-101654).
Author contributions
MM, AJ, and HG designed the study and interpreted data. TH performed statistical analyses. MM, DS, EKM, MSR, JH, AS, DH, and HG collected patient samples and data. AS and DH performed plasma purification. MG and AJ performed iFISH analyses. All authors wrote and approved the manuscript
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41375-019-0634-2) contains supplementary material, which is available to authorized users.
References
- 1.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113:5412–7. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakshman A, Rajkumar SV, Buadi FK, Binder M, Gertz MA, Lacy MQ, et al. Risk stratification of smoldering multiple myeloma incorporating revised IMWG diagnostic criteria. Blood Cancer J. 2018;8:59. doi: 10.1038/s41408-018-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neben K, Jauch A, Hielscher T, Hillengass J, Lehners N, Seckinger A, et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J Clin Oncol J Am Soc Clin Oncol. 2013;31:4325–32. doi: 10.1200/JCO.2012.48.4923. [DOI] [PubMed] [Google Scholar]
- 4.Rajkumar S, Gupta V, Fonseca R, Dispenzieri A, Gonsalves W, Larson D, et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia. 2013;27:1738–44. doi: 10.1038/leu.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolli N, Maura F, Minvielle S, Gloznik D, Szalat R, Fullam A, et al. Genomic patterns of progression in smoldering multiple myeloma. Nat Commun. 2018;9:3363. doi: 10.1038/s41467-018-05058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta AK, Fink JL, Grady JP, Morgan GJ, Mullighan CG, To LB, et al. Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia. 2019;33:457–68. doi: 10.1038/s41375-018-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merz M, Hielscher T, Seckinger A, Hose D, Bertsch U, Neben K, et al. Longitudinal fluorescence in situ hybridization at primary diagnosis and relapse reveals clonal evolution after autologous stem cell transplantation in multiple myeloma. Blood. 2016;128:4415–4415. doi: 10.1182/blood.V128.22.4415.4415. [DOI] [Google Scholar]
- 8.Bochtler T, Merz M, Hielscher T, Granzow M, Hoffmann K, Krämer A, et al. Cytogenetic intraclonal heterogeneity of plasma cell dyscrasia in AL amyloidosis as compared with multiple myeloma. Blood Adv. 2018;2:2607–18. doi: 10.1182/bloodadvances.2018023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merz M, Jauch A, Hielscher T, Bochtler T, Schönland SO, Seckinger A, et al. Prognostic significance of cytogenetic heterogeneity in patients with newly diagnosed multiple myeloma. Blood Adv. 2018;2:1–9. doi: 10.1182/bloodadvances.2017013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JR, Weinhold N, Ashby C, Walker BA, Wardell C, Pawlyn C, et al. Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica. 2019;2018:202200. doi: 10.3324/haematol.2018.202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2016. 10.1038/nrclinonc.2016.122. [DOI] [PubMed]
- 12.Weinhold N, Kirn D, Seckinger A, Hielscher T, Granzow M, Bertsch U, et al. Concomitant gain of 1q21 and MYC translocation define a poor prognostic subgroup of hyperdiploid multiple myeloma. Haematologica. 2016;101:e116–119. doi: 10.3324/haematol.2015.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González D, Burg M, van der, García-Sanz R, Fenton JA, Langerak AW, González M, et al. Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood. 2007;110:3112–21. doi: 10.1182/blood-2007-02-069625. [DOI] [PubMed] [Google Scholar]
- 14.Landgren O, Hofmann JN, McShane CM, Santo L, Hultcrantz M, Korde N et al. Association of immune marker changes with progression of monoclonal gammopathy of undetermined significance to multiple myeloma. JAMA Oncol. 2019. 10.1001/jamaoncol.2019.1568. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.