Abstract
Acute myeloid leukemia (AML) is an aggressive hematologic neoplasm resulting from the malignant transformation of myeloid progenitors. Despite intensive chemotherapy leading to initial treatment responses, relapse caused by intrinsic or acquired drug resistance represents a major challenge. Here, we report that histone 3 lysine 27 demethylase KDM6A (UTX) is targeted by inactivating mutations and mutation-independent regulation in relapsed AML. Analyses of matched diagnosis and relapse specimens from individuals with KDM6A mutations showed an outgrowth of the KDM6A mutated tumor population at relapse. KDM6A expression is heterogeneously regulated and relapse-specific loss of KDM6A was observed in 45.7% of CN-AML patients. KDM6A-null myeloid leukemia cells were more resistant to treatment with the chemotherapeutic agents cytarabine (AraC) and daunorubicin. Inducible re-expression of KDM6A in KDM6A-null cell lines suppressed proliferation and sensitized cells again to AraC treatment. RNA expression analysis and functional studies revealed that resistance to AraC was conferred by downregulation of the nucleoside membrane transporter ENT1 (SLC29A1) by reduced H3K27 acetylation at the ENT1 locus. Our results show that loss of KDM6A provides cells with a selective advantage during chemotherapy, which ultimately leads to the observed outgrowth of clones with KDM6A mutations or reduced KDM6A expression at relapse.
Subject terms: Acute myeloid leukaemia, Cancer therapeutic resistance
Introduction
Acute myeloid leukemia (AML) is characterized by expansion of abnormal myeloid precursor cells in the bone marrow and blood. If not treated, AML can progress quickly and become fatal within a few months. Standard strategies for initial therapy, which have not changed substantially during the last 30 years [1], include cytarabine (AraC) in combination with anthracyclines like daunorubicin (DNR). Although the majority of AML patients achieve complete remission following induction chemotherapy, the disease reoccurs in 60–65% of younger adult patients (≤60 years) within 3 years after diagnosis [2]. Relapse represents the major cause for treatment failure and drug resistance is likely to play a role in its development. Recently, we have analyzed paired diagnosis and relapse samples of 50 cytogenetically normal (CN) AML patients and found KDM6A as a novel relapse-associated gene in AML [3]. KDM6A (or UTX) is a JmjC domain containing histone H3 lysine 27 (H3K27)-specific demethylase [4, 5] and belongs to the KDM6 family that include KDM6B and UTY [4, 6]. KDM6A can facilitate gene activation through the catalytic JmjC domain and is also a component of the COMPASS-like complex, which is important for chromatin enhancer activation [7–11]. KDM6A is frequently targeted by somatic loss-of-function mutations in cancer [12–15] including leukemia [16–18]. Dependent on the cancer type, KDM6A appears to possess distinct tumor-suppressive functions. In T-cell acute lymphoblastic leukemia (T-ALL), KDM6A mutations are located almost exclusively in the JmjC domain [16, 17] and inactivation of the single KDM6A copy in males is sufficient to contribute to T-ALL pathogenesis [17]. In contrast, hematopoietic-specific loss of Kdm6a induces leukemogenesis through demethylase-independent alterations in H3K27 acetylation, H3K4 monomethylation and chromatin accessibility [19].
Using diagnosis and relapse samples from AML patients, patient-derived xenografts (PDX), and leukemia cell lines, we investigated the status of KDM6A during disease progression and the impact of KDM6A loss on chemotherapy resistance. We found three AML patients with enrichment of KDM6A loss-of-function mutations at relapse and relapse-specific loss of KDM6A mRNA and protein expression in 45.7% of CN-AML patients and 44.4% of AML patients, respectively. Reduction or loss of KDM6A expression in myeloid cell lines leads to increased resistance towards AraC and DNR treatment. Whereas re-expression of KDM6A in KDM6A-null cell lines sensitizes cells to AraC treatment. AraC resistance is achieved by reduction of the drug influx transporter ENT1. Taken together, our findings highlight KDM6A as a novel mediator of drug resistance in AML.
Materials and methods
Cell culture
Cell lines (Supplementary Table 1) were obtained from DSMZ (Braunschweig, Germany) and cultured according to the supplier’s recommendation. PDX AML samples were recovered from mice and cultured as previously described [20]. Authentication of cell lines was performed using short tandem repeat typing. Testing for Mycoplasma contamination was done using the MycoAlert Mycoplasma detection kit (Lonza).
Patients
Our analysis was based on samples from AML patients from the AMLCG-99 trial (NCT00266136), AMLCG-2008 trial (NCT01382147), and the Department of Medicine III, University Hospital, LMU. Informed consent for scientific use of sample material was received from all study participants in accordance with the Declaration of Helsinki.
Proliferation assay
Cells were treated with cytarabine (Selleck Chemicals, Houston, TX, USA), DNR (in-house), 6-thioguanine (Sigma Aldrich), or NBMPR (Sigma Aldrich). After 72–96 h, viable cells were counted on Vi-Cell Cell Viability Analyzer (Beckman Coulter, Krefeld, Germany). For long-term proliferation, cells were treated three times (d0, d4, d8) and viable cells were counted every second day. Unpaired, two-tailed Student’s t-test and calculation of IC50 values were performed using GraphPad Prism version 6.07 (GraphPad Software, La Jolla, CA, USA). PiggyBac KDM6A cells were cultured +/− doxycycline (0.5 μg/mL) for 48 h followed by treatment with AraC +/− doxycycline for 72 h. For longer proliferation, cells were cultured +/− doxycycline (0.5 μg/mL) for 8 days. Every 2 days, cells were counted and cultured in fresh medium +/− doxycycline.
Additional methods are provided in supplementary methods.
Results
Gain of KDM6A mutations at relapse
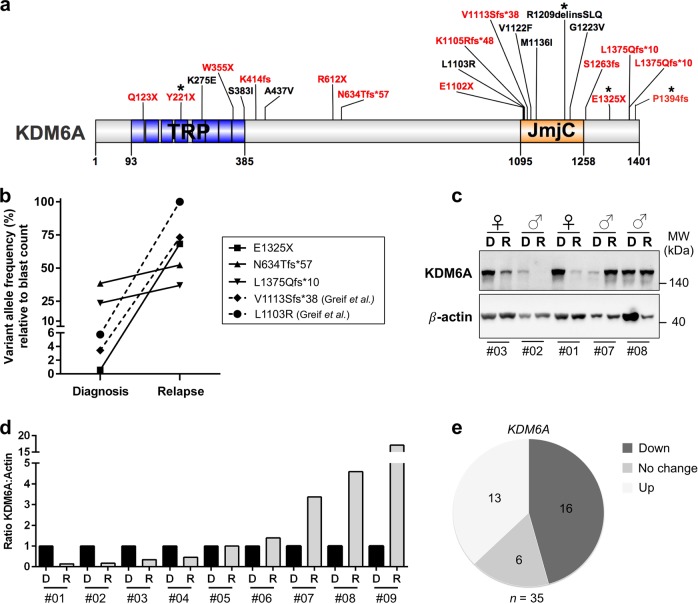
Despite their initial response to chemotherapy, the majority of AML patients will develop chemotherapy resistance and relapse. Acquired KDM6A mutations were reported at relapse [3] pointing towards a novel mechanism of resistance in AML. To get insight into the biological relevance of KDM6A mutations, we first analyzed their locations in 20 AML patients at diagnosis. Patients with KDM6A mutations were from the AMLCG-99 trial (n = 6), AMLCG-2008 trial (n = 9/664), a diagnosis-relapse cohort (n = 2/50) [3] and this work (n = 3). We found a broad location pattern with the majority of mutations either located at or within the proximity of the tetratricopeptide repeat (TRP) or the JmjC domain (Fig. 1a). 65% of patients harbor either frameshift insertions/deletions or nonsense mutations suggesting a loss-of-function phenotype. In the majority of patients (12/20), the mutation occurred only in a subpopulation of AML cells, with a variant allele frequency (VAF) below 15% (Supplementary Fig. 1a). In addition to two previously described CN-AML patients [3], we identified three patients with recurrent KDM6A mutations using matched diagnosis and relapse samples, which were available for 3/18 patients (Fig. 1b; Supplementary Fig. 1b–d). In all patients we observed an increase in VAF of KDM6A mutations at relapse (Fig. 1b). The mutant clone E1325X showed the most striking increase at relapse (68.2% VAF), as it was barely detectable at diagnosis (0.58% VAF). Transplantation of relapsed tumor cells from this patient into immunodeficient mice (PDX model [20]) resulted in stable regeneration of KDM6A E1325X mutant clone (PDX AML-393; Supplementary Fig. 1b), which was verified by Sanger sequencing (Supplementary Fig. 1e). A second KDM6A mutation, P1394fs, was present in the same diagnosed patient with a 12.8-fold greater VAF (8.1%) than E1325X, but was lost at relapse (Supplementary Fig. 1b).
Fig. 1.
Gain of recurrent KDM6A mutations at relapse and change in KDM6A RNA and protein expression at relapse. a Schematic overview of KDM6A protein structure (NP_066963.2) and mutations (red = truncating; black = missense) identified at diagnosis in 20 AML patients, illustrated using IBS software [40]. Location of mutations is displayed and amino-acid positions are indicated below the graph. Asterisk (*) signifies two patients harboring two mutations each. Presented KDM6A mutations are from AMLCG-99 trial (NCT00266136), AMLCG-2008 trial (NCT01382147), a CN-AML diagnosis-relapse cohort [3] and this work. TRP tetratricopeptide repeat, JmjC Jumonji C. b Comparison of variant allele frequency (VAF) between diagnosis and relapse in 5 AML patients with KDM6A mutations. Due to variations in blast count, VAF was calculated relative to the respective blast count. Raw data for mutation L1130R and V1113Sfs*38 originate from our previous study [3]. c, Immunoblotting for KDM6A expression in five AML patients at diagnosis (D) and relapse (R). Their respective gender is shown on top and the UPN is displayed below. MW, molecular weight; β-actin, loading control. d Comparison of KDM6A protein expression in nine AML patients without KDM6A mutations at diagnosis and relapse. The ratio of KDM6A to β-actin expression is displayed. Respective values at relapse were normalized to the corresponding diagnosis sample. e Pie chart illustrating the regulation of KDM6A mRNA expression in 35 CN-AML patients. The three groups, KDM6A-up, KDM6A-down and KDM6A-no change were defined as a change in expression between diagnosis and relapse of above or below 20%, respectively
Next, we compared KDM6A expression in matched diagnosis and relapse samples of 9 AML patients (listed in Supplementary Table 2) with no detectable KDM6A mutation (Fig. 1c, d; Supplementary Fig. 1f). A strong decrease in KDM6A protein expression at relapse was observed in four patients whereas three patients showed increased expression at relapse. Additional analysis of KDM6A mRNA regulation in 35 CN-AML patients revealed a downregulation of KDM6A in 45.7% of patients (n = 16/35) and an upregulation in 37.1% of patients (n = 13/35; Fig. 1e and Supplementary Fig. 2). Interestingly, DNA methylation levels of KDM6A varied between AML patients at diagnosis and AML patients with high DNA methylation levels of KDM6A (top 25%) showed significantly shorter overall survival in a publicly available dataset [21] (Supplementary Fig. 3).
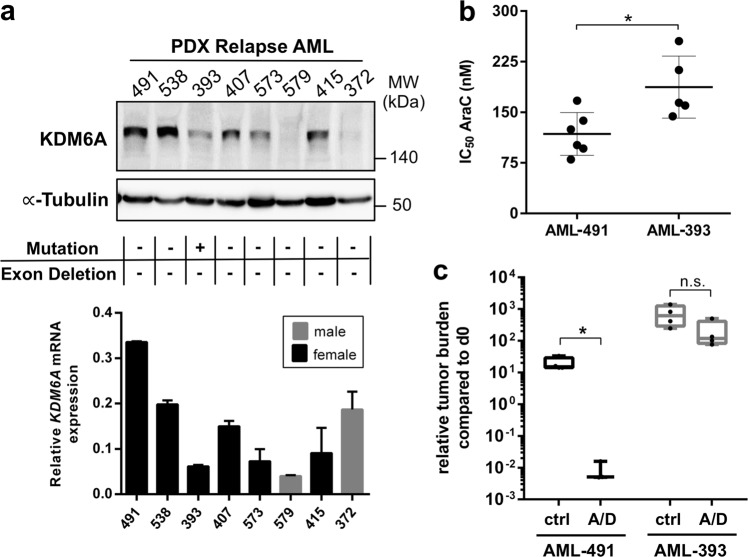
To further investigate the relevance of KDM6A at relapse, we analyzed KDM6A mRNA and protein expression in 8 PDX AML samples, established from primary patients′ cells at relapse. 50% of PDX AML samples showed low or undetectable protein expression, which, except for AML-372, correlated with mRNA expression (Fig. 2a). Mutational analysis by MLPA and targeted sequencing revealed only the above described KDM6A mutation (E1325X) in PDX AML-393 (Supplementary Fig. 1b). No additional KDM6A exon deletion mutations were detected (Supplementary Fig. 4). mRNA expression of the histone demethylase KDM6B and the histone methyltransferase EZH2 were slightly increased in AML-579 cells, whereas AML-538 showed low KDM6B and AML-491 low EZH2 mRNA expression (Supplementary Fig. 5a, b). Analysis of the mRNA expression of UTY in PDX AML samples showed normal UTY levels (Supplementary Fig. 6d). Since we were unable to detect a low molecular weight band corresponding to the premature stop mutation E1325X (estimated protein weight: 145 kDa) in the female PDX AML-393 cells, we treated these cells in vitro with the proteasomal inhibitor MG132. No increase in overall KDM6A expression, but also no appearance of an additional band corresponding to E1325X was observed (Supplementary Fig. 5c), which might point towards a nonsense-mediated mRNA decay. When we overexpressed E1325X, L1103R, and V1113Sfs*38 KDM6A mutants in HEK293T cells, proteasomal inhibition resulted in a significantly elevated expression of these mutants whereas wildtype (WT) and the demethylase-dead mutant H1146A did not change (Supplementary Fig. 5d, e). These results suggest that reduction of KDM6A expression might be either facilitated by nonsense-mediated mRNA decay or by proteasomal degradation.
Fig. 2.
Heterogeneous KDM6A expression in relapsed PDX AML cells and decreased drug sensitivity in KDM6A mutant PDX AML-393 cells. a KDM6A mRNA and protein expression was analyzed in eight PDX samples from relapsed AML patients. For Western Blot, α-Tubulin was used as a loading control. KDM6A mRNA expression is shown relative to GAPDH expression. Male samples are highlighted as gray bar plots. The occurrence of KDM6A mutations or exon deletions is indicated with the “+” symbol. b Comparison of AraC IC50 values between KDM6A WT PDX AML-491 and KDM6A mutant PDX AML-393 in vitro. Mean ± s.d. are given for at least five independent experiments performed in duplicates. Unpaired, two-tailed Student’s t-test; *P = 0.016. c Comparison of tumor load reduction under in vivo chemotherapy in AML-491 and AML-393 bearing animals. Tumor burden was quantified by bioluminescence before (d0) and after (d28) two cycles of treatment with AraC (days 1–4, 14–17) and DNX (days 1, 4, 14, and 17) (A/D) or control treated animals (ctrl). Relative tumor burden at day 28 compared to d0 was calculated. Unpaired, two-tailed Student’s t-test; *P = 0.0157; n.s., not significant
Next, we asked whether drug resistance might have resulted in the outgrowth of the KDM6A mutated population at relapse. Since AraC was a component of the induction therapy, we investigated whether KDM6A mutant cells are less sensitive to AraC treatment. Therefore, we used PDX AML models of the same adverse ELN classification [20] with (i) KDM6A WT (AML-491) and (ii) KDM6A mutant (AML-393) and compared the half-inhibitory concentration (IC50) of AraC. In vitro treatment for 72 h revealed a 0.63-fold decreased sensitivity towards AraC treatment for KDM6A mutant AML-393 compared to KDM6A WT AML-491 cells (187.3 nM vs. 117.9 nM; Fig. 2b). We observed the same effect in vivo after treating mice bearing AML-491 or AML-393 with two cycles of AraC and DaunoXome (DNX; liposomal DNR; Fig. 2c). Treatment dramatically decreased the tumor burden in KDM6A WT AML-491 bearing mice compared to control (P = 0.0157; Fig. 2c), whereas only a modest drop in tumor burden was observed in treated AML-393 bearing mice. Overall, these results indicate that a reduced KDM6A expression is associated with a decreased AraC sensitivity.
Decreased AraC sensitivity in KDM6A mutant cells
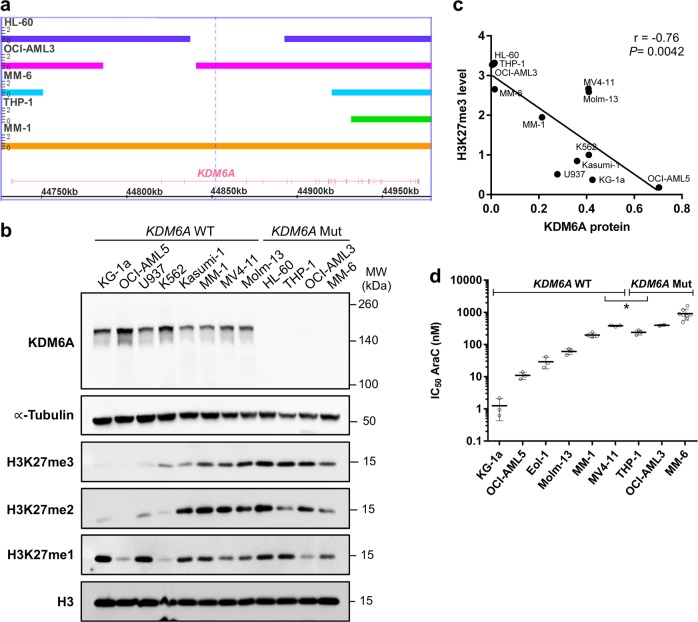
KDM6A exon deletion mutations have been observed in AML cell lines, MONO-MAC-6 (MM-6) and THP-1 [12]. To identify the frequency of KDM6A deletions in leukemia, we performed MLPA analysis for the KDM6A gene in 40 myeloid leukemia cell lines (Supplementary Table 1). We identified two additional AML cell lines, OCI-AML3 and HL-60, with in-frame deletions in exon 3–4 and 5–6, respectively (Supplementary Fig. 4). These deletions were confirmed by independent CytoScan HD Array hybridization analysis (Fig. 3a). Both in-frame deletions lead to a truncated protein of approximately 147 kDa (WT: 154 kDa). Although low to intermediate mRNA expression was detectable in the mutant cell lines (Supplementary Fig. 6a), KDM6A protein expression was completely absent (Fig. 3b). KDM6A mutant cells showed increased H3K27 tri-methylation, whereas H3K27 di- and mono-methylation levels were similar between WT and mutant cells (Fig. 3b). Global H3K27me3 was inversely correlated with KDM6A levels (r = −0.76; P = 0.0042; Fig. 3c) indicating a KDM6A dependent epigenetically altered phenotype. Additionally, we analyzed expression of KDM6B and EZH2 via qPCR as these genes might compensate for KDM6A loss. For both genes, mRNA expression was similar between mutant and WT cells (Supplementary Fig. 6b, c). Analysis of UTY mRNA expression showed loss of UTY mRNA expression in two KDM6A mutant and three KDM6A WT cell lines (Supplementary Fig. 6d).
Fig. 3.
AML cell lines with KDM6A loss show high H3K27 tri-methylation and increased AraC resistance. a Identification of KDM6A aberrations in AML cell lines HL60, OCI-AML3, MM-6, and THP-1. MM-1 serves as a WT control. Bar thickness ranging from 0 to 2 indicates the copy number (CN) status. Haploidy (CN = 1) in cell lines from male patients is due to the X-chromosomal localization of KDM6A. HL-60, the only cell line derived from a woman has lost one of its X chromosomes. b Western blot for KDM6A expression and global H3K27 mono-, di- and tri-methylation levels in KDM6A WT and mutant human leukemia cell lines. α-Tubulin and total H3 were used as loading controls. Blots are representative of three independent experiments. MW, molecular weight. c Negative correlation between KDM6A protein expression and global H3K27me3 level in KDM6A WT and mutant human leukemia cell lines (Pearson’s correlation; r = −0.76, *P = 0.0042). Mean values of three independent analyzes are shown. d Comparison of AraC IC50 values between KDM6A WT and mutant male AML cell lines. To determine their respective IC50 values, AML cell lines were treated with increasing concentrations of AraC for 72 h. Mean of IC50 values from at least three independent experiments ± standard deviation (s.d.) are shown. Unpaired, two-tailed Student’s t-test; *P = 0.044
Next, we investigated whether KDM6A loss leads to increased AraC resistance. KDM6A mutant cells showed a trend towards higher AraC IC50 values compared to WT (Supplementary Fig. 7a). To eliminate gender-specific effects of KDM6A WT cells (higher expression in females as KDM6A escapes X inactivation [3, 17]), we compared the IC50 values of male AML cell lines. Male KDM6A mutant AML cell lines had significantly increased IC50 values compared to WT cells (P = 0.0441; Fig. 3d). We demonstrated previously [3] that MM-1 cells (KDM6A WT) are more sensitive to AraC treatment than the KDM6A mutant sister cells MM-6. To investigate if MM-6 also has a competitive growth advantage compared to MM-1 under AraC therapy, we performed a competitive assay mixing MM-1 with MM-6 cells in a 9:1 ratio. Native conditions as well as treatment with AraC significantly increased the number of MM-6 cells to 32.3% (native conditions) or 52.6% (270 nM AraC) after 9 days (Supplementary Fig. 7b). Treatment of MM-1 cells with AraC, DNR or 6-TG for 72 h, applying concentrations in the range of their respective IC50 values, induced an upregulation of KDM6A protein expression (Supplementary Fig. 8).
Knockdown of KDM6A confers decreased AraC and DNR sensitivity in K562 cells
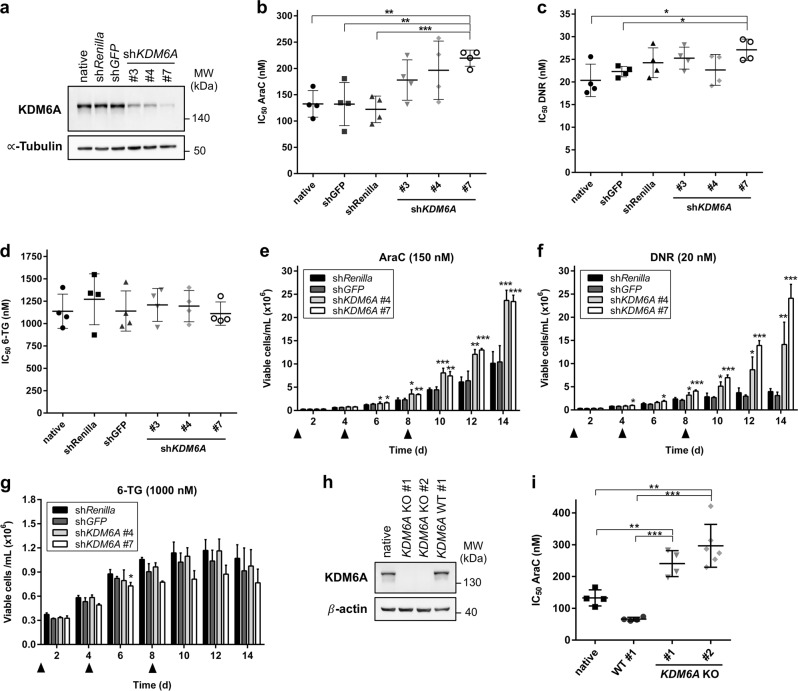
Although AraC was part of the induction regimen in the five investigated patients with KDM6A mutations, the composition of induction therapy varied between patients and in certain cases also included the drugs DNR and/or 6-thioguanine (6-TG). To investigate if reduced expression of KDM6A leads to increased resistance towards multiple drugs, we performed lentiviral shRNA-mediated knockdown (KD) of KDM6A in the myeloid cell line K562. Of several tested shRNAs, shKDM6A #3, #4, and #7 decreased KDM6A expression by 70% (#3, #4) or 90% (#7; Fig. 4a). Next, KDM6A KD or control cells were treated for 72 h with AraC, DNR, or 6-TG. KDM6A KD cells displayed decreased sensitivity towards AraC treatment (Fig. 4b) applying doses within the range of reported AraC plasma concentrations in patients [22] (Supplementary Fig. 9a). Only KD with the most potent shKDM6A #7 resulted in a significantly increased resistance to AraC (Fig. 4b). However, the effect of KDM6A KD on response towards DNR or 6-thioguanine (6-TG) was not as prominent or even absent: only KD cells shKDM6A #7 were slightly more resistant to DNR treatment (Fig. 4c), and no change in IC50 values was observed after 6-TG treatment (Fig. 4d). The applied DNR concentrations (5–75 nM) are within the lower range of the reported concentrations in AML patients (403.8 ± 349 nM) [23]. Since induction therapy typically involves continuous treatment for seven (“7 + 3”) or 10 days (TAD regime), we next applied a prolonged time course with multiple treatments. Prolonged treatment with 6-TG showed no difference in the amount of viable cells between control and KD after 14 days (Fig. 4g). Differences in growth under AraC treatment started at day 4, and resulted in a significant proliferative advantage for KDM6A KD cells compared to control (Fig. 4e). Growth of both control groups was completely arrested under DNR treatment after day 8, whereas KDM6A KD cells were strongly proliferating (Fig. 4f). KDM6A KD efficiency and proliferative advantage under DNR were positively correlated. To compare the impact of KDM6A KD vs. knockout (KO), we applied CRISPR/Cas9 genome editing to ablate KDM6A expression in K562 cells. We established single cell KDM6A KO clones (Supplementary Fig. 9c), leading to complete loss of KDM6A expression (Fig. 4h). After 72 h AraC treatment, IC50 values were significantly increased for both KDM6A KO clones compared to controls (Fig. 4i). We observed a trend towards higher IC50 values or no difference between KO and control cells after DNR or 6-TG treatment, respectively (Supplementary Fig. 9d, e). These data indicate that reduction or loss of KDM6A expression in K562 cells increases resistance to AraC and DNR but not 6-TG.
Fig. 4.
Reduction or loss of KDM6A expression confers decreased AraC and DNR sensitivity in K562 cells. a Immunoblot showing knockdown (KD) of KDM6A expression in K562 cells lentiviral transduced with three different shRNAs against KDM6A. shRenilla and shGFP serve as controls. Blot is representative of three independent experiments. MW molecular weight, α-Tubulin loading control. b–d Comparison of IC50 values for AraC (b), DNR (c), and 6-TG (d) between control and KDM6A KD in K562 cells. Cells were treated for 72 h with the respective drug. Mean of IC50 values from four independent experiments ± s.d. are shown. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001. e–g Long-term proliferation assay measuring the amount of viable K562 cells, shControl and shKDM6A, every 2 days for 14 days in total. Cells were treated with 150 nM AraC (e), 20 nM DNR (f), or 1000 nM 6-TG (g) on Day 0, 4, and 8 as indicated with the triangle. Mean ± s.d. are given for three independent experiments. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001. h, Immunoblot showing loss of KDM6A protein in KDM6A knockout (KO) K562 cells. Results of one representative experiment are shown (n = 3 experiments). MW molecular weight, β-actin loading control. i Increase in AraC IC50 values in both KDM6A KO K562 clones compared to WT #1 clone or native cells. Mean of IC50 values ± s.d. from at least four independent experiments are shown. Unpaired, two-tailed Student’s t-test; **P < 0.01; ***P < 0.001
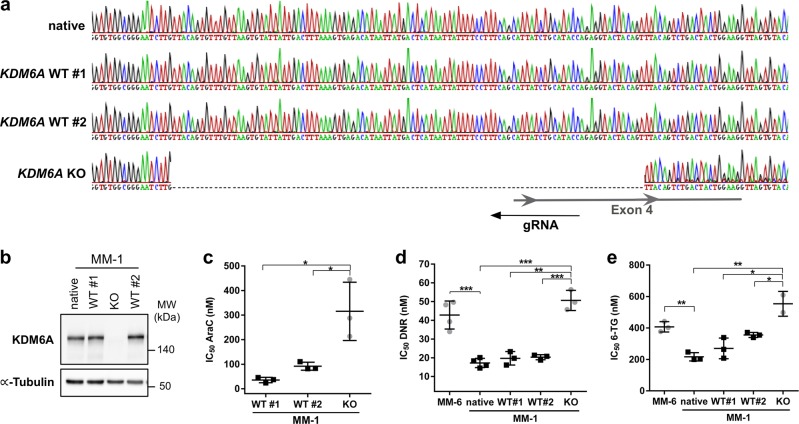
Loss of KDM6A in MM-1 recapitulates the drug phenotype of the KDM6A mutant sister cells MM-6
The sister cell lines MM-1 and MM-6 have originally been established in culture from the same male AML patient at relapse [24]. Whereas MM-1 cells express KDM6A, MM-6 cells harbor a KDM6A exon deletion, rendering them a good model to examine the implications of KDM6A loss within a similar genetic background. To investigate whether KDM6A KO in MM-1 cells results in the same drug resistance phenotype observed in MM-6 cells (Fig. 5d, e), we applied CRISPR/Cas9 targeting the KDM6A locus in MM-1 cells (Fig. 5a). Compared to MM-1 parental and WT single cell clones, KDM6A expression was lost in one clone (Fig. 5b). In agreement with our observation that MM-6 cells are 4.3-fold more resistant to AraC after 72 h than MM-1 (Supplementary Fig. 7a), KDM6A KO in MM-1 increased the AraC IC50 compared to both WT clones (3.4 to 8.8-fold increase after 96 h treatment; Fig. 5c). Comparison of the IC50 values after DNR (Figs. 5d) and 6-TG (Fig. 5e) treatment indicated that KDM6A KO MM-1 are significantly less sensitive to DNR and 6-TG than KDM6A WT MM-1 cells. Strikingly, KO of KDM6A in MM-1 conferred a similar resistance to DNR (MM-1: KO vs. WT#1/#2: 2.5 to 2.6-fold; native MM-1 vs. MM-6: 2.5-fold increase in IC50) and 6-TG (MM-1: KO vs. WT#1/#2: 1.6 to 2.1-fold; native MM-1 vs. MM-6: 1.9-fold) as in MM-6 cells. Together these data suggest that loss of KDM6A in MM-1 is responsible for a decreased sensitivity towards AraC, DNR, and 6-TG.
Fig. 5.
KDM6A loss in MM-1 recapitulates the drug phenotype of the KDM6A mutant sister cell line MM-6. a DNA sequencing chromatogram showing KDM6A frameshift mutation A112Vfs*3 of KDM6A knockout (KO) MM-1 clone which is absent in native MM-1 cells and two KDM6A WT clones. Last 74 bp of Intron 3 and 29 bp of exon 4 are deleted. WT #1 and #2 clones were tested negative for KDM6A KO after CRISPR/Cas9 targeting. b Immunoblotting for KDM6A expression in KDM6A WT and KO cells. Blot is representative of three independent experiments. MW molecular weight, α-Tubulin loading control. c–e Comparison of IC50 values for AraC (c), DNR (d), and 6-TG (e) between MM-6 and MM-1 cells including MM-1 native, two KDM6A WT and one KDM6A KO clone. Cells were treated for 96 h with the respective drug. Mean ± s.d are given for three independent experiments. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001
Re-expression of KDM6A sensitizes cells to AraC treatment
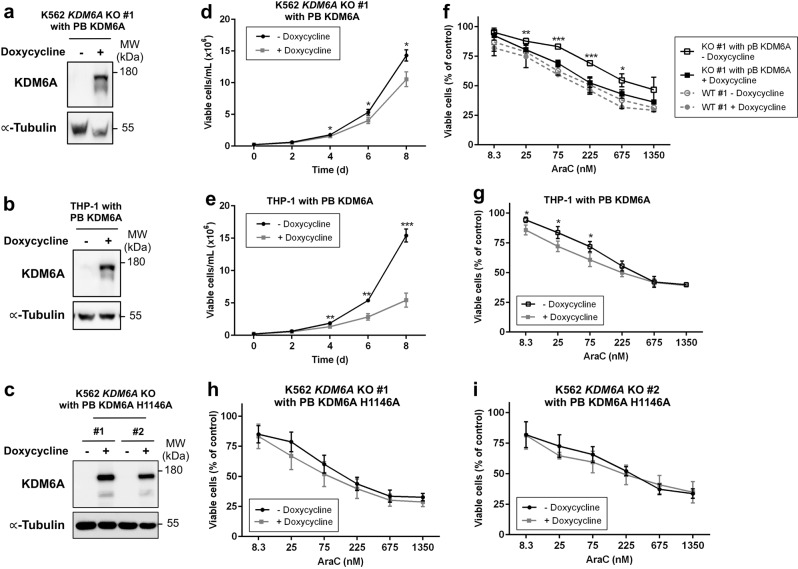
To investigate if re-introduction of KDM6A in KDM6A-null cell lines sensitizes them to AraC treatment, we generated stable cell lines with a doxycycline inducible PiggyBac (PB) KDM6A expression system (Fig. 6a–c). Re-expression of KDM6A significantly suppressed proliferation in THP-1 and K562 KDM6A KO cells (Fig. 6d, e). Furthermore, re-expression of KDM6A significantly decreased the amount of viable cells after treatment with AraC in both cell lines (Fig. 6f, g). KDM6A KO K562 cells, which were more resistant to AraC than KDM6A WT cells, were sensitized again to AraC treatment by re-expressing KDM6A (Fig. 6f). To investigate if the demethylase activity of KDM6A is essential for re-sensitizing cells to AraC treatment, we expressed a catalytically dead KDM6A mutant, H1146A [25], in the K562 KDM6A KO #1 and #2 cells. Expression of KDM6A H1146A showed only a trend in decreasing the amount of viable cells after treatment with AraC (Fig. 6h, i).
Fig. 6.
Re-expression of KDM6A suppresses cell growth and sensitizes cells to AraC therapy. a–c Immunoblot showing doxycycline inducible re-expression of KDM6A in K562 KDM6A KO #1 PB KDM6A (a), KDM6A mutant THP-1 PB KDM6A (b) and K562 KDM6A KO #1 and #2 PB KDM6A H1146A cells (c) after treatment with 0.5 μg/mL doxycycline for 48 h. Blot is representative of two independent experiments. MW molecular weight, α-Tubulin loading control. d, e Proliferation assay measuring the amount of viable K562 KDM6A KO #1 PB KDM6A (d) and KDM6A mutant THP-1 PB KDM6A cells (e) in the absence or presence of doxycycline (0.5 μg/mL) every 2 days for 8 days. Mean ± s.d. are given for three independent experiments. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001. f–i AraC dose-response analysis after treatment for 72 h in K562 KDM6A KO #1 PB KDM6A and K562 KDM6A WT #1 (f), KDM6A mutant THP-1 PB KDM6A (g) and K562 KDM6A KO PB KDM6A H1146A cells (h,i) in the absence or presence of doxycycline (0.5 μg/mL). Mean ± s.d. are given for at least three independent experiments. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001
Decreased ENT1 expression by KDM6A loss mediates increased AraC resistance
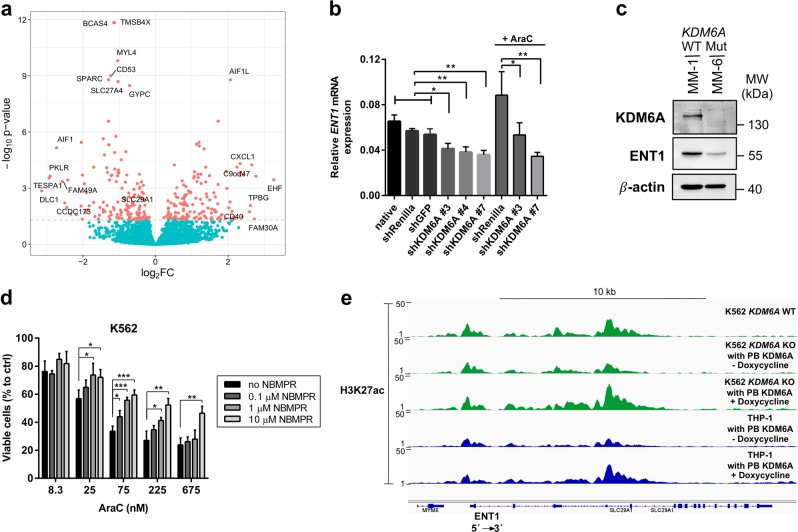
To identify genes involved in KDM6A-mediated drug resistance, we performed RNA-Seq analysis in K562 cells treated with siRNA (Supplementary Fig. 10a, b) or shRNA against KDM6A (Fig. 4a). Transient KDM6A KD with siRNA, which showed a trend towards higher IC50 for AraC compared to control (Supplementary Fig. 10c), resulted in transcriptional downregulation of 39 genes and upregulation of 7 genes (Supplementary Fig. 10d). For the most potent shKDM6A #7 we detected transcriptional deregulation of 295 genes compared to 7 or 54 deregulated genes during shKDM6A #3 or #4 mediated KD, respectively (Supplementary Fig. 10e). Whereas the majority of differentially expressed genes (39/46) was downregulated in the siRNA-mediated KD (Supplementary Fig. 10d), shKDM6A #7 KD resulted in similar transcriptional down- (150, 50.8%) and upregulation (145, 49.2%; Fig. 7a). Treatment with AraC (150 nM) during shRNA-mediated KD led to increased transcriptional deregulation (shKDM6A #7: 2,357; shRenilla: 2,272) in comparison to the native state with 40.3% of genes exclusively being deregulated in shKDM6A #7 (Supplementary Fig. 10f). Next, we compared KDM6A regulated genes with known candidate genes in drug metabolic pathways and found that ENT1 was consistently downregulated in KDM6A KD cells in both RNA-Seq screenings (Fig. 7a, Supplementary Fig. 10d). ENT1, or SLC29A1, is a membrane transporter important for the cellular uptake of nucleosides and its analogues [26]. ShKDM6A K562 cells showed significantly reduced ENT1 mRNA compared to controls (Fig. 7b). AraC treatment slightly increased ENT1 mRNA expression in shRenilla cells. In shKDM6A #7 cells ENT1 expression was reduced even after AraC administration (Fig. 7b). Additionally, decreased ENT1 expression was detected in KDM6A KO K562 (Supplementary Fig. 11a) and MM-6 cells (Fig. 7c). Treatment of K562, MM-1, and MM-6 cells with a selective ENT1 inhibitor, NBMPR, in combination with AraC resulted in increased cell survival compared to AraC alone (Fig. 7d, Supplementary Fig. 11b, c). By contrast, no change was observed when we combined ENT1 inhibition with DNR or 6-TG in K562 cells (Supplementary Fig. 11d, e). To further elucidate the mechanism of ENT1 regulation by KDM6A, we performed ChIP-seq analysis for H3K27me3 and H3K27ac in MM-1 and MM-6 cells as recent studies have reported that the tumor suppressor effect is largely demethylase independent [19, 27]. ChIP-seq for H3K27me3 showed no enrichment on the ENT1 locus, however, we detected differential H3K27ac peaks in the promoter and a putative enhancer region of ENT1 in MM-1 compared to MM-6 (Supplementary Fig. 12). Additionally, K562 and THP-1 cells with loss of KDM6A showed low H3K27ac peaks on the ENT1 locus, which were increased upon doxycycline inducible re-expression of KDM6A in both cell lines (Fig. 7e). In summary, our data demonstrate that strong reduction or complete loss of KDM6A decreases ENT1 expression, probably through direct or indirect effects of KDM6A on enhancer regions, which then promotes increased AraC resistance.
Fig. 7.
Downregulation of ENT1 by KDM6A loss is regulated by H3K27 acetylation and increases AraC resistance. a Volcano plot showing log2 fold change on the x-axis and adjusted P value on the y-axis for the differential gene expression between shRNA-mediated KD of KDM6A (shKDM6A #7) and control (shGFP) in K562 cells (n = 6). Genes with adjusted P value < 0.05 are highlighted in red and those with a log2 FC >2.5 or <−2.5 are labeled with the gene name. In addition, genes with adjusted P value < 1e-8 and the gene SLC29A1 are labeled. b qRT-PCR for ENT1 in K562 cells: native, shRenilla, shGFP and three different shKDM6A KDs. ENT1 mRNA for shRenilla, shKDM6A #3 and shKDM6A #7 K562 cells is also shown after treatment with 150 nM AraC for 72 h. The mean ± s.d. for ENT1 mRNA relative to GAPDH for three independent experiments is shown. c Immunoblotting showing strong reduction of ENT1 protein in KDM6A mutant MM-6 cells compared to the KDM6A WT sister cell line MM-1. Immunoblots are representative of three independent experiments. MW, molecular weight; β-actin, loading control. d Inhibition of ENT1 by NBMPR increases the amount of viable cells during AraC treatment. K562 cells were treated with different AraC concentrations in combination with 0, 0.1, 1 and 10 μM of NBMPR for 72 h. Mean ± s.d are given for three independent experiments. Unpaired, two-tailed Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001. e Genomic snapshot of H3K27ac ChIP-seq in K562 KDM6A WT, K562 KDM6A KO PB KDM6A and KDM6A mutant THP-1 cells in the absence or presence of doxycycline at the ENT1 locus. Cells were treated with media +/− doxycycline (0.5 μg/mL) every 24 h for 72 h
Discussion
In cancer, except for bladder cancer [28], the frequency of mutation in the KDM6A gene is rather low [29, 30]. In AML, frequency ranges from 0.7% [31] to 4.0% [3, 32, 33] and the majority of mutations are missense mutations. In our study, KDM6A mutations were exclusively missense and truncating mutations. One of these mutations, E1325X, has been previously described in an AML patient at diagnosis and was present in a subclone only [31]. Reduced expression or mutations/deletions of KDM6A correlate with poor overall survival in patients with CN-AML [3] or myeloma [29], respectively. We identified three AML patients harboring KDM6A mutations at diagnosis and observed an outgrowth of the KDM6A mutated population at relapse. These results are in agreement with our previous study [3] and suggest that KDM6A loss may contribute to increased chemo-resistance in AML.
As KDM6A is not X-inactivated [34], females with T-ALL benefit from two functional copies [17], and compared to female patients shortened relapse-free survival is seen in male CN-AML patients [3]. Dependent on expression dosage, KDM6A deficiency was found to accelerate and promote cancer progression in a mouse lymphoma model [35]. We observed that KDM6A mRNA and protein expression is reduced in 45.7% and 44.4% of AML patients at relapse, respectively. As most of these samples showed no KDM6A mutation, another mechanism contributing to KDM6A regulation might be DNA methylation. We found that AML patients with high DNA methylation levels of KDM6A have a significantly shorter overall survival. These results are in line with our previous study [3] showing a correlation of low KDM6A expression and poor overall survival in CN-AML patients.
In agreement with our data showing higher AraC resistance in KDM6A mutant AML cell lines, we observed that a KDM6A mutant PDX sample is less sensitive towards in vitro AraC treatment compared to a KDM6A WT PDX sample. Prominent differences in treatment response during in vivo therapy indicate that even small differences in drug sensitivity observed in vitro can significantly impact long-term in vivo therapy.
We have recently demonstrated that KDM6A mutant MM-6 are less sensitive to AraC than MM-1 cells [3]. Under competitive growth conditions, we observed a selective growth advantage of MM-6 during AraC treatment. Furthermore, inducible re-expression of KDM6A in KDM6A-null cells sensitized to AraC treatment. Very recently, Gozdecka et al. [19] could show that lentiviral expression of KDM6A in MM-6 cells suppressed spontaneous cell proliferation. The data presented here extend these findings and show that KDM6A acts as a tumor suppressor and mediates drug resistance in AML.
UTY, a catalytically inactive KDM6A homolog that is encoded on the Y chromosome, was recently shown to suppress myeloid leukemogenesis in KDM6A-deficient male mice [19]. It was also reported to be lost or reduced in conjunction with KDM6A mutations in pancreatic cancers with squamous differentiation in male patients [27]. We found that UTY mRNA expression was lost or barely detectable in MM-6 and MM-1 cells suggesting that the drug resistant phenotype of MM-6 is not affected by UTY.
Various mechanisms of drug resistance in AML have been proposed in the last decades. AraC for instance can only exert its cytotoxic effect after cellular uptake and conversion into its active form. Among the key candidate genes in AraC metabolism, we consistently found differential expression of the drug influx transporter gene ENT1. We demonstrate that KD/KO of KDM6A leads to decreased expression of ENT1 linking decreased KDM6A levels to increased AraC resistance. Previous studies showed that KD or inhibition of ENT1 in AML cell lines confers AraC resistance [36, 37]. In AraC based therapy, AML patients with low ENT1 levels were reported to have shorter disease-free or overall survival [38]. In agreement with previous results, inhibition of ENT1 by NBMPR increased AraC resistance. Furthermore, we demonstrate that transport of DNR and 6-TG across the cell membrane is ENT1 independent. Recently, KDM6A was reported to regulate gene expression during myeloid leukemogenesis mainly by modifying levels of H3K27 acetylation, H3K4 monomethylation and chromatin accessibility [19]. Our ChIP-seq analysis suggests that ENT1 expression is regulated by H3K27 acetylation whereas H3K27 demethylase activity is dispensable for ENT1 expression. As re-expression of a catalytically dead mutant in K562 KDM6A KO cells had not the same effect as KDM6A WT in re-sensitizing cells to AraC, it remains to be determined if disruption of the catalytic domain impacts the regulation of ENT1 expression.
Prolonging the treatment time for DNR, resulted in a significant increase in DNR resistance in MM-6 cells compared to a shorter treatment as previously described [3]. Additionally, we demonstrate that deletion of KDM6A in MM-1 recapitulates the same drug resistant phenotype observed in MM-6. The mechanisms leading to DNR resistance need further investigation, but loss of KDM6A-mediated upregulation of the DNR metabolizing enzymes AKR1C1 and AKR1C2 (data not shown) might be involved in DNR resistance. A study demonstrated that upregulation of AKR1C1/3 facilitated reduction of DNR efficacy in leukemic U937 cells [39].
Taken together, our results show that KDM6A inactivation either by loss-of-function mutations or protein downregulation mediates drug resistance in AML.
Supplementary information
Acknowledgements
The authors thank all study participants. We thank M. Fritschle for animal handling. KHM, PAG, GS, HL, IJ, and KS were supported by the German Research Council (DFG) within the SFB 1243 “Cancer Evolution”. SMS is a fellow of the Life Science Munich (LSM) graduate school. MDB is a fellow of IMPRS-LS. SMS, RM, ALH, SW, HDM and JK are members of the IRTG-1243 within the SFB 1243.
Author contributions
SMS designed and performed the experiments, collected, analyzed and interpreted the data, and wrote the manuscript; ALH, RM, MDB, BT, JK, and HDM performed the experiments, collected, and analyzed the data. SW performed and analyzed the RNA-seq experiments. HDM performed and analyzed the numerical aberration analysis. MC and WHL designed and established the knockdown system. BV and IJ established and provided serially transplantable PDX AML samples, and performed and analyzed in vivo therapy trial. MRT, KM, SV, and PAG were involved in the analysis of patient data. HL supervised the study. KS designed, interpreted the data and supervised the study. All authors discussed the results and commented on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41375-019-0497-6) contains supplementary material, which is available to authorized users.
References
- 1.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 3.Greif PA, Hartmann L, Vosberg S, Stief SM, Mattes R, Hellmann I, et al. Evolution of cytogenetically normal acute myeloid leukemia during therapy and relapse: an exome sequencing study of 50 patients. Clin Cancer Res. 2018;24:1716–26. doi: 10.1158/1078-0432.CCR-17-2344. [DOI] [PubMed] [Google Scholar]
- 4.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 6.Min GL, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 7.Hu D, Gao X, Morgan MA, Herz H-M, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–54. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar SS, Zhao D, Lin T, Gu B, Pal K, Wu SJ, et al. MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol Cell. 2018;70:825–41.e6. doi: 10.1016/j.molcel.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337–42. doi: 10.1016/j.gene.2017.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–69. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross JS, Badve S, Wang K, Sheehan CE, Boguniewicz AB, Otto GA, et al. Genomic profiling of advanced-stage, metaplastic breast carcinoma by next-generation sequencing reveals frequent, targetable genomic abnormalities and potential new treatment options. Arch Pathol Lab Med. 2015;139:642–9. doi: 10.5858/arpa.2014-0200-OA. [DOI] [PubMed] [Google Scholar]
- 14.Nickerson ML, Dancik GM, Im KM, Edwards MG, Turan S, Brown J, et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin Cancer Res. 2014;20:4935–48. doi: 10.1158/1078-0432.CCR-14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:1–7. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–22. doi: 10.1182/blood-2014-05-577270.H.-G.W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolli N, Manes N, McKerrell T, Chi J, Park N, Gundem G, et al. Characterization of gene mutations and copy number changes in acute myeloid leukemia using a rapid target enrichment protocol. Haematologica. 2015;100:214–22. doi: 10.3324/haematol.2014.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gozdecka Malgorzata, Meduri Eshwar, Mazan Milena, Tzelepis Konstantinos, Dudek Monika, Knights Andrew J., Pardo Mercedes, Yu Lu, Choudhary Jyoti S., Metzakopian Emmanouil, Iyer Vivek, Yun Haiyang, Park Naomi, Varela Ignacio, Bautista Ruben, Collord Grace, Dovey Oliver, Garyfallos Dimitrios A., De Braekeleer Etienne, Kondo Saki, Cooper Jonathan, Göttgens Berthold, Bullinger Lars, Northcott Paul A., Adams David, Vassiliou George S., Huntly Brian J. P. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nature Genetics. 2018;50(6):883–894. doi: 10.1038/s41588-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vick B, Rothenberg M, Sandhöfer N, Carlet M, Finkenzeller C, Krupka C, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients’ cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS ONE. 2015;10:e0120925. doi: 10.1371/journal.pone.0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Early AP, Preisler HD, Slocum H, Rustum YM. A pilot study of high-dose 1-beta-D-arabinofuranosylcytosine for acute leukemia and refractory lymphoma: clinical response and pharmacology. Cancer Res. 1982;42:1587–94. [PubMed] [Google Scholar]
- 23.Bogason A, Quartino AL, Lafolie P, Masquelier M, Karlsson MO, Paul C, et al. Inverse relationship between leukaemic cell burden and plasma concentrations of daunorubicin in patients with acute myeloid leukaemia. Br J Clin Pharmacol. 2011;71:514–21. doi: 10.1111/j.1365-2125.2010.03894.x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLeod RA, Voges M, Drexler HG. Mono Mac 6: a mature monoblastic leukemia cell line with t(9;11)(p21; q23) Blood. 1993;82:3221–2. doi: 10.1182/blood.V82.10.3221.3221. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Lee J-E, Cho Y-W, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109:15324–9. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M Pastor-Anglada, S Perez-Torras, Nucleoside transporter proteins as biomarkers of drug responsiveness and drug targets, Front. Pharmacol. 2015; 6. 10.3389/fphar.2015.00013. [DOI] [PMC free article] [PubMed]
- 27.Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–26.e8. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, Siew EY, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med. 2017;9:1–14. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 29.Pawlyn C, Kaiser MF, Heuck C, Melchor L, Wardell CP, Murison A, et al. The spectrum and clinical impact of epigenetic modifier mutations in Myeloma. Clin Cancer Res. 2016;22:5783–94. doi: 10.1158/1078-0432.CCR-15-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J-F Spinella, P Cassart, C Richer, V Saillour, M Ouimet, S Langlois, et al. Genomic characterization of pediatric T-cell acute lymphoblastic leukemia reveals novel recurrent driver mutations, Oncotarget. 2016;7. 10.18632/oncotarget.11796. [DOI] [PMC free article] [PubMed]
- 31.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Dennis G, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–99. doi: 10.1182/blood-2016-01-693879.Presented. [DOI] [PubMed] [Google Scholar]
- 33.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7:737–42. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat Commun. 2018;9:2720. doi: 10.1038/s41467-018-05084-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Lee C, Cheong HS, Koh Y, Ahn KS, Kim HL, et al. SLC29A1 (ENT1) polymorphisms and outcome of complete remission in acute myeloid leukemia. Cancer Chemother Pharmacol. 2016;78:533–40. doi: 10.1007/s00280-016-3103-x. [DOI] [PubMed] [Google Scholar]
- 37.Macanas-Pirard P, Broekhuizen R, González A, Oyanadel C, Ernst D, García P, et al. Resistance of leukemia cells to cytarabine chemotherapy is mediated by bone marrow stroma, involves cell-surface equilibrative nucleoside transporter-1 removal and correlates with patient outcome. Oncotarget. 2017;8:23073–86. doi: 10.18632/oncotarget.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan H, Zhu J, Chen F, Xiao F, Huang H, Han X, et al. SLC29A1 single nucleotide polymorphisms as independent prognostic predictors for survival of patients with acute myeloid leukemia: an in vitro study. J Exp Clin Cancer Res. 2014;33:90. doi: 10.1186/s13046-014-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsunaga T, Yamaguchi A, Morikawa Y, Kezuka C, Takazawa H, Endo S, et al. Induction of aldo-keto reductases (AKR1C1 and AKR1C3) abolishes the efficacy of daunorubicin chemotherapy for leukemic U937 cells. Anticancer Drugs. 2014;25:868–77. doi: 10.1097/CAD.0000000000000112. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–61. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.