Abstract
SETDB1, a histone H3K9 methyltransferase, has been reported to be upregulated in a variety of tumors and promotes cancer development. However, the exact pathogenesis of SETDB1 in human colorectal cancer (CRC) is hitherto unknown. Here, we showed that SETDB1 expression was highly amplified in CRC. Functionally, SETDB1 downregulation in SW480 and HCT116 cells reduced cell proliferation, migration, invasion, and increased CRC cells apoptosis. In contrast, SETDB1 overexpression promoted CRC cells proliferation, migration, and invasion. High expression of SETDB1 was associated with a more aggressive phenotype in vitro. Flow cytometry showed that cell cycle was arrested in G1 phase after SETDB1 silencing. Furthermore, depletion of SETDB1 in vivo suppressed CRC cells proliferation. Mechanistically, p21 was identified as the target of SETDB1. After transfected with siSETDB1, expression of p21 was distinctly increased. In contrast, expression of p21 was significantly decreased after overexpression SETDB1. We also showed that SETDB1 could be involved in the regulation of epithelial–mesenchymal transition (EMT) in HCT116 cells. Moreover, we confirmed that SETDB1 could regulate the activity of p21 promoter by dual-luciferase repoter assay, and proved that SETDB1 could bind to the promoter of p21 and regulate its H3K9me3 enrichment level by ChIP-PCR experiment. Finally, we verified that silencing of SETDB1 inhibited CRC tumorigenesis in vivo. In conclusion, our results indicate that SETDB1 is a major driver of CRC development and might provide a new therapeutic target for the clinical treatment of CRC.
Subject terms: Cell proliferation, Cancer genetics
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer death in men and the second leading cause of cancer death in women, according to the latest global cancer statistic1. It is well known that CRC carcinogenesis is complicated and still has not been defined, involving multiple genomic variations and biological processes. Therefore, the molecular mechanism must be further studied to deepen the understanding of CRC.
Epigenetics plays a central role in the pathogenesis of cancer2. Epigenetics changes are independent of DNA sequence, mainly including DNA methylation and histone modification, which are reversible features3,4. Remarkably, histone can be modified at many sites5. Recent evidence indicates that histone methyltransferases (HMTs) and histone acetyltransferase such as EZH2, JMJD3, UTX, SMYD3, and KAT2A are dysregulated in human cancers6–8. Previous study showed that p300, a histone acetylase, cooperates with CREPT in CRC activating Wnt/β-catenin signaling pathway, thus promoting the development of CRC9. Increasing the level of H3K27me3 can improve drug sensitivity of CRC patients10. In addition, histone deacetylase (HDAC) inhibitors against CRC are already available in clinical11,12. Moreover, a recent study in vivo showed that the offsprings of KDM6A conditional knockout male mice exhibited increased incidence of tumors13. These studies suggest that histone modifications play important roles during the occurrence of human cancer. Cancer genome sequencing evidenced that heterochromatin-associated histone modification H3K9me3 accounts for more than 40% of mutation-rate variation14, and more recently study exhibited the increasing expression of H3K9me3 is associated with treatment-resistant phenotypes15. These suggest that H3K9me3 and its enzymes may be potential therapeutic targets.
SET domain bifurcated 1 (SETDB1) is also known as ESET/KMT1E and located on human chromosome 1q2116. SETDB1 is characterized by histone H3K9 methyltransferase activity17, which can catalyze the trimethylation of H3K918,19. H3K9me3 represents a specific tag for epigenetic repression by recruiting heterochromatin protein1 to methylated histones20. Dysregulation of SETDB1 can affect the normal development of embryos and participate in the pathogenesis of tumors21,22. Recent years, increasing evidence showed that abnormal expression of SETDB1 is closely related to tumorigenesis, including melanoma, lung cancer, breast cancer, and liver cancer23–26, suggesting that SETDB1 is involved in the pathogenesis of a variety of human cancers. Moreover, recent literatures have reported that SETDB1 expression is associated with poor clinical outcomes in CRC and is thought to be a dysregulated epigenetic regulator27–29. However, the role of SETDB1 in CRC is still controversial and has not been defined, the exact pathogenesis needs further investigations.
p21, also known as CDKN1A and a member of the Cip/Kip family, is a key regulator of cell cycle and cell senescence30,31. A recent study reveals that mesenchymal stem cells help CRC cells resist senescence through p53/p21 pathway32. In addition, histone modification has an important role in regulating p21 expression. EZH2 promotes tumor progression in melanoma cells through epigenetically silencing p21 expression33. 4-Nonylphenol could interact with RNF2 and EZH2 resulting in decreased p21 expression via increasing H3K27me3 repressive marks at p21 promoter and promoting MCF-7 cells proliferation34. In multiple myeloma (MM), hydroxamicacid-based small-molecule N-hydroxy-4-(2-[(2-hydroxyethyl) (phenyl) amino]-2-oxoethyl) benzamide (HPOB), a novel HDAC inhibitor, could induce MM cells death via transcriptional activation of p2135. Furthermore, lncRNA CRNDE binding with EZH2 promotes the proliferation of CRC cells by epigenetic silencing of DUSP5/p21 expression36, and another lncRNA HOXA-AS2 can inhibit the expression of p21 and KLF2 in CRC cells by binding with EZH2 and LSD1, which both regulate histone methylation37.
Given the great importance of epigenetic regulators in CRC, in the current study, we investigated the oncogenicity of SETDB1 in CRC and shed light on the importance of SETDB1 in the proliferation, cell cycle, apoptosis, migration, and invasion of CRC cells. We defined SETDB1 as an oncogene via elucidating the epigenetic regulation of the p21 expression. In addition, alteration of epithelial–mesenchymal transition (EMT) markers was observed after inhibition of SETDB1 in HCT116 cells. These results suggest that SETDB1 has the potential to serve as a novel biomarker and therapeutic target in CRC.
Results
SETDB1 is upregulated in CRC tissues
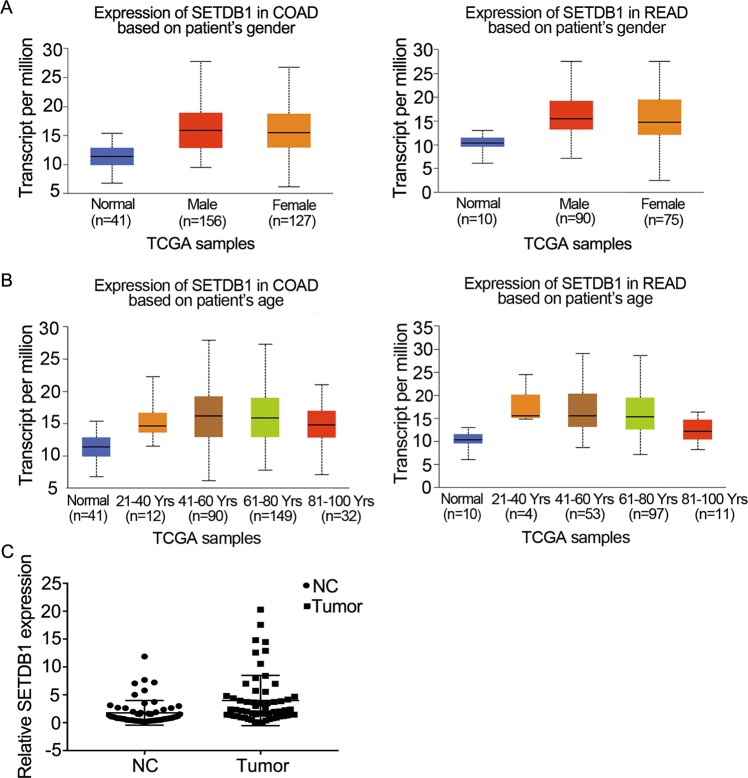
We applied UALCAN database38 to analyze the expression of SETDB1 in different genders and ages of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ), and the result showed that SETDB1 was upregulated in both colon and rectal cancer regardless of genders and ages (Fig. 1a, b). To further confirm the expression of SETDB1 in CRC, we examined the expression of SETDB1 in tumor and adjacent normal tissues of 60 CRC patients, the result of qRT-PCR showed that SETDB1 was highly expressed in cancer tissues compared to the controls (Fig. 1c). However, the expression of SETDB1 was not related to the clinical characteristic of CRC, including age, gender, location, pathological differentiation, depth of tumor, lymph node metastasis, distant metastasis, and tumor stage (Table 1). The results indicate that SETDB1 expression is associated with the development of CRC.
Fig. 1. SETDB1 expression in CRC tissues.
a, b Higher SETDB1 expression in COAD and READ tumor tissues of different genders and ages according to UALCAN analysis. c Relative expression of SETDB1 in 60 pairs CRC tissues. P = 0.0090, P < 0.05.
Table 1.
Correlation between SETDB1 expression and clinicopathologic variables in patients with CRC (n = 60).
| Characteristics | Number | Percent | SETDB1 | Chi-square test | |
|---|---|---|---|---|---|
| (n = 60) | High | Low | P value | ||
| Ages (years) | 0.481 | ||||
| <60 | 16 | 27% | 14 | 2 | |
| ≥60 | 44 | 73% | 33 | 11 | |
| Gender | 0.609 | ||||
| Male | 36 | 60% | 29 | 7 | |
| Female | 24 | 40% | 18 | 6 | |
| Tumor location | 0.561 | ||||
| Right colon | 13 | 21.7% | 9 | 4 | |
| Left colon | 22 | 36.7% | 17 | 5 | |
| Rectum | 25 | 41.6% | 21 | 4 | |
| Pathological differentiation | 0.578 | ||||
| Poor | 13 | 21.7% | 10 | 3 | |
| Well and moderate | 47 | 78.3% | 37 | 10 | |
| Depth of tumor | 0.673 | ||||
| T1 and T2 | 8 | 13% | 7 | 1 | |
| T3 and T4 | 52 | 87% | 40 | 12 | |
| Lymph node metastasis | 0.330 | ||||
| N0 | 35 | 58.4% | 26 | 9 | |
| N1 | 17 | 28.3% | 13 | 4 | |
| N2 | 8 | 13.3% | 8 | 0 | |
| Distant metastasis | 0.783 | ||||
| M0 | 59 | 98.3% | 46 | 13 | |
| M1 | 1 | 1.7% | 1 | 0 | |
| Tumor stage | 0.368 | ||||
| I and II | 35 | 58% | 26 | 9 | |
| III and IV | 25 | 42% | 21 | 4 | |
SETDB1 promotes CRC cell proliferation in vitro
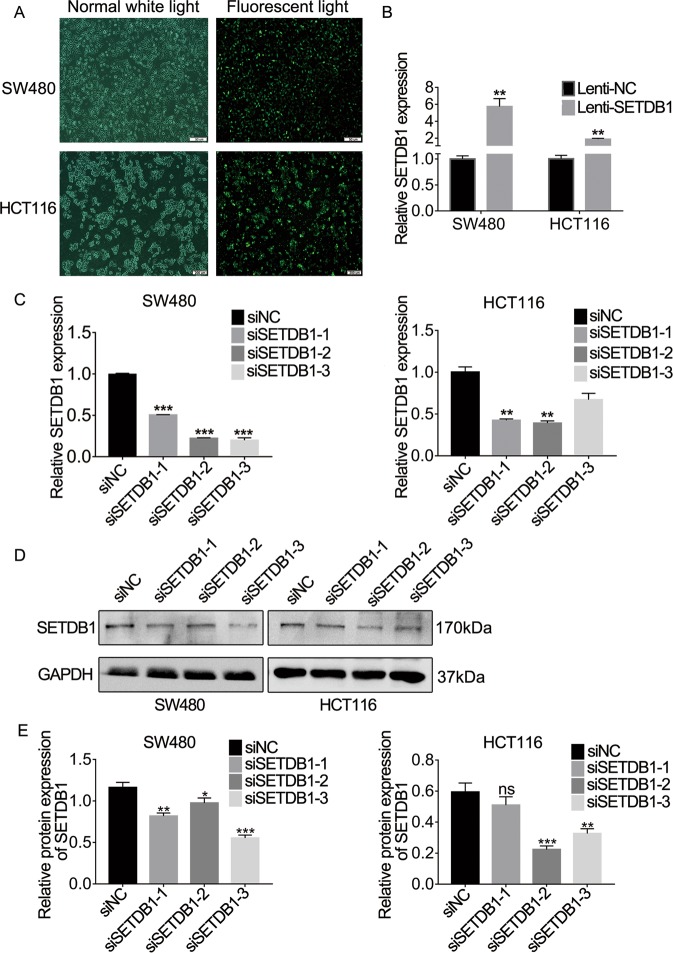
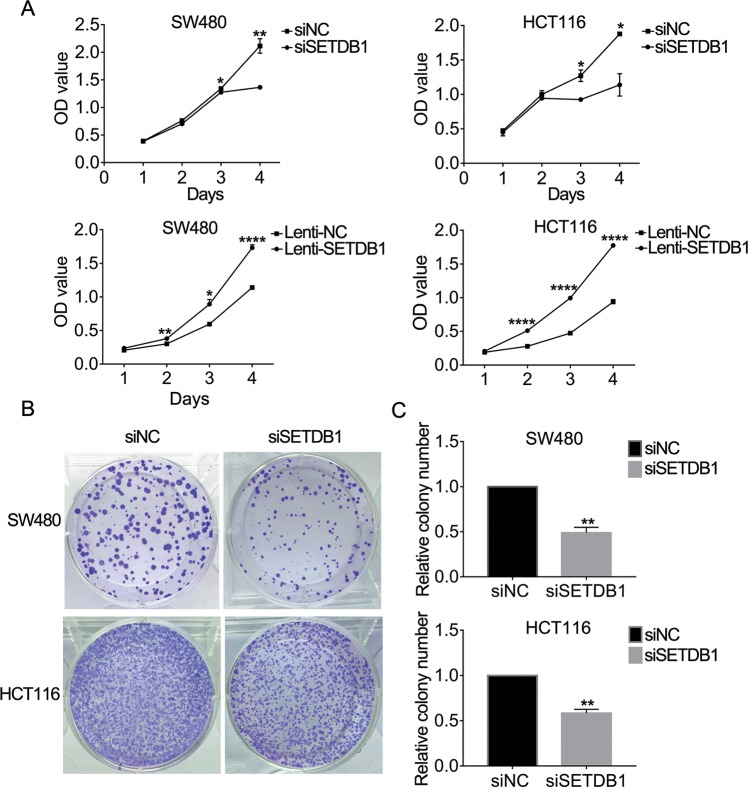
Since the expression of SETDB1 was increased in CRC, we investigated the biological function of SETDB1. We silenced or overexpressed the expression of SETDB1 by siRNA or lentivirus. We preliminarily evaluated the siRNA transfection efficiency in CRC cells through fluorescence image 24 h after transfection (Fig. 2a), qRT-PCR and WB were performed to verify the silencing or overexpression efficiency of SETDB1 (Fig. 2b–d). According to the results of qRT-PCR and protein gray analysis (Fig. 2c, e), we selected the most effective siRNA for further study, which is siSETDB1-3 and siSETDB1-2 in SW480 and HCT116 cells, respectively. In addition, we recorded them as siSETDB1 in the following experiments. To detect whether SETDB1 regulates the proliferation of CRC cells, we performed CCK8 and colony formation assay. The results showed that growth capacity and colony number of SW480 and HCT116 cells dramatically decreased following silence of SETDB1 (Fig. 3–c). Overexpressed SETDB1 promotes CRC cells proliferation (Fig. 3a). As a result, SETDB1 was associated with CRC cell proliferation.
Fig. 2. siRNA was selected for silencing SETDB1 or lentivirus overexpressing SETDB1 in CRC cells.
a At 24 h post transfection, fluorescence photographs of CRC cells transfected with FAM-labeled siRNA using lipo2000. SW480: Scale bar, 50 µm; HCT116: Scale bar, 200 µm. b qRT-PCR analysis of SETDB1 expression following the treatment of SW480 and HCT116 cells with lenti-SETDB1 or the lenti-NC. c, d SETDB1 mRNA and protein expression in SW480 and HCT116 after transfected with siSETDB1-1, 2, 3 or the negative control. e Gray analysis of WB results by image J software. Error bars indicate mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ns means no significance.
Fig. 3. SETDB1 promotes CRC cells proliferation in vitro.
a CCK8 assay was performed to detect the cell viability of siSETDB1 or Lenti-SETDB1 transfected CRC cells. b Colony assay was performed to analyze the proliferation of SW480 and HCT116 cells following treatment with siSETDB1 or siNC. c The number of cell clones was counted and analyzed. All the data come from three independent experiments. Error bars indicate mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Silence of SETDB1 promotes G1-phase arrest and causes apoptosis of CRC cells
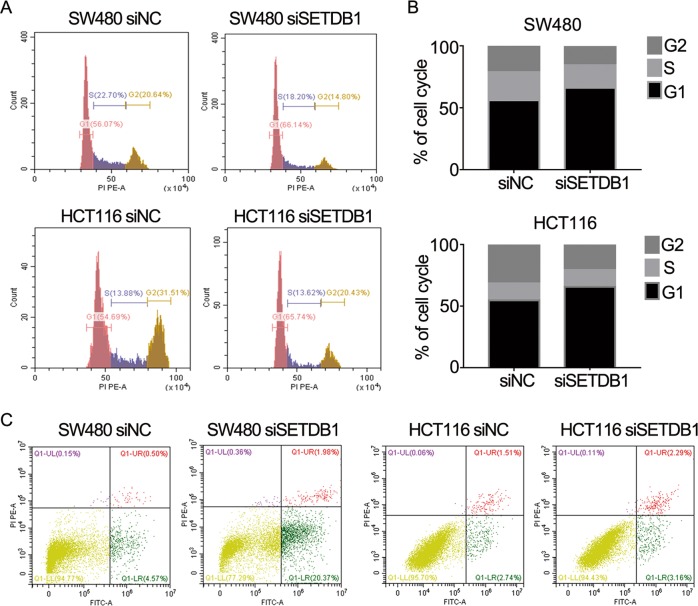
To assess whether the effect of SETDB1 on CRC cells proliferation was mediated by cell cycle progression change, flow cytometry was performed to measure the cell cycle of CRC cells. As shown in (Fig. 4a, b), downregulation of SETDB1 significantly increased the G1-phase ratio and reduced the S-phase ratio. In addition, the apoptosis level of CRC cells was assayed by flow cytometry, which also reflected the growth activity of CRC cells. The results exhibited that knockdown of SETDB1 notably increased the apoptosis level of CRC cells (Fig. 4c). These studies implicate that SETDB1 has an important effect on CRC cells through cell cycle and apoptosis.
Fig. 4. SETDB1 silencing promotes G1-phase arrest and induces apoptosis of CRC cells.
a Changes of CRC cells cycle after transfection with siSETDB1 or siNC. b Cell cycle distribution of CRC cells transfected with siSETDB1 was represented in a histogram. c Flow cytometry was used to measure the apoptosis rate of SW480 and HCT116 cells transfected with siSETDB1.
SETDB1 promotes the migration and invasion of CRC cells
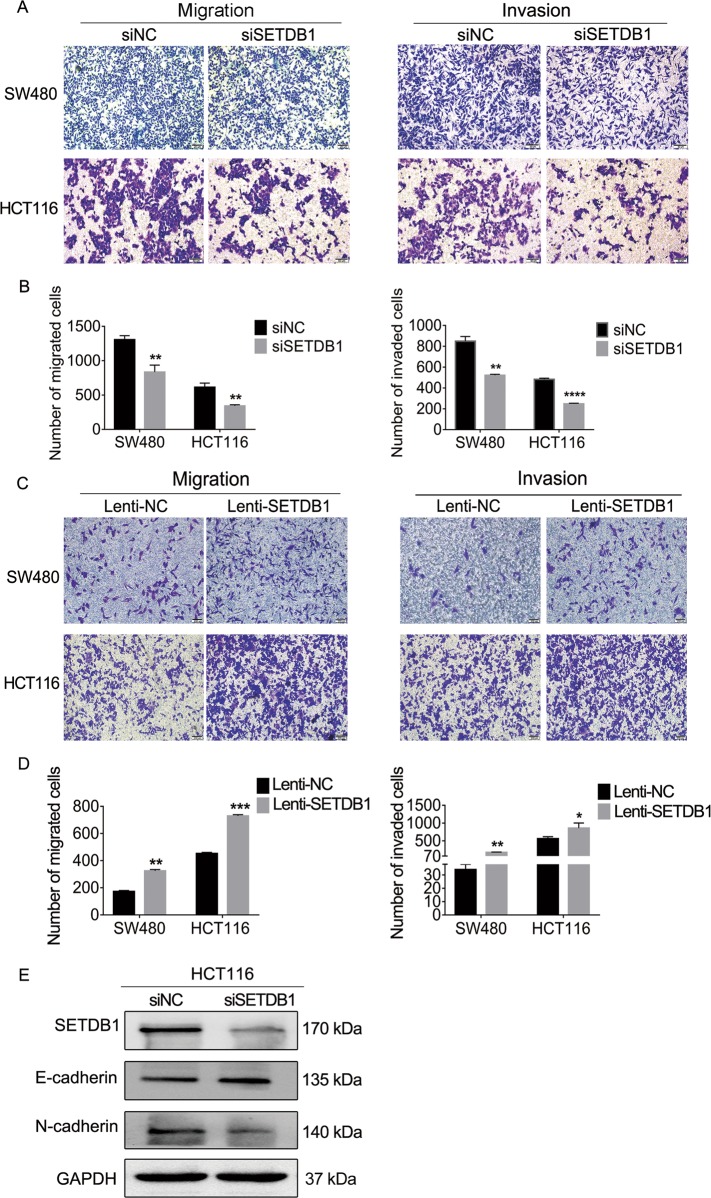
SETDB1 has been reported to be involved in the regulation of EMT in breast cancer26,39,40. However, there is no report about the relationship between SETDB1 and EMT in CRC. In our experiment, we proved that SETDB1 did influence the EMT process in CRC cells. Cell migration and invasion assay indicated that the numbers of migrated and invaded cells were reduced in siSETDB1 group compared to the control, respectively (Fig. 5a, b). Ectopic expression of SETDB1 promoted CRC cells migration and invasion (Fig. 5c, d). In addition, the epithelial characteristic protein E-cadherin was increased, while the expression of mesenchymal characteristic protein N-cadherin was decreased after silencing SETDB1 in HCT116 (Fig. 5e). Therefore, our results indicate that SETDB1 may play an important role in the process of EMT in CRC cell lines.
Fig. 5. SETDB1 regulates migration/invasion and EMT progression in CRC cells.
a Migration and invasion assays were performed after SETDB1 knockdown. SW480 migration: Scale bar, 20 µm. HCT116 migration and invasion: Scale bar, 50 µm. b The numbers of migrated/invaded cells were counted in three random fields by microscope. c Cell migration and invasion assays after transfected with Lenti-SETDB1 in SW480 and HCT116 cells. Scale bar, 200 µm. d Migrated and invaded cells of overexpressing SETDB1 were counted. e Protein of EMT markers in siSETDB1-knockdown HCT116 cells. The data are means from three independent experiments ±SD. *P < 0.05, **P < 0.01, ***P < 0.001.
SETDB1 epigenetically silences p21 transcription
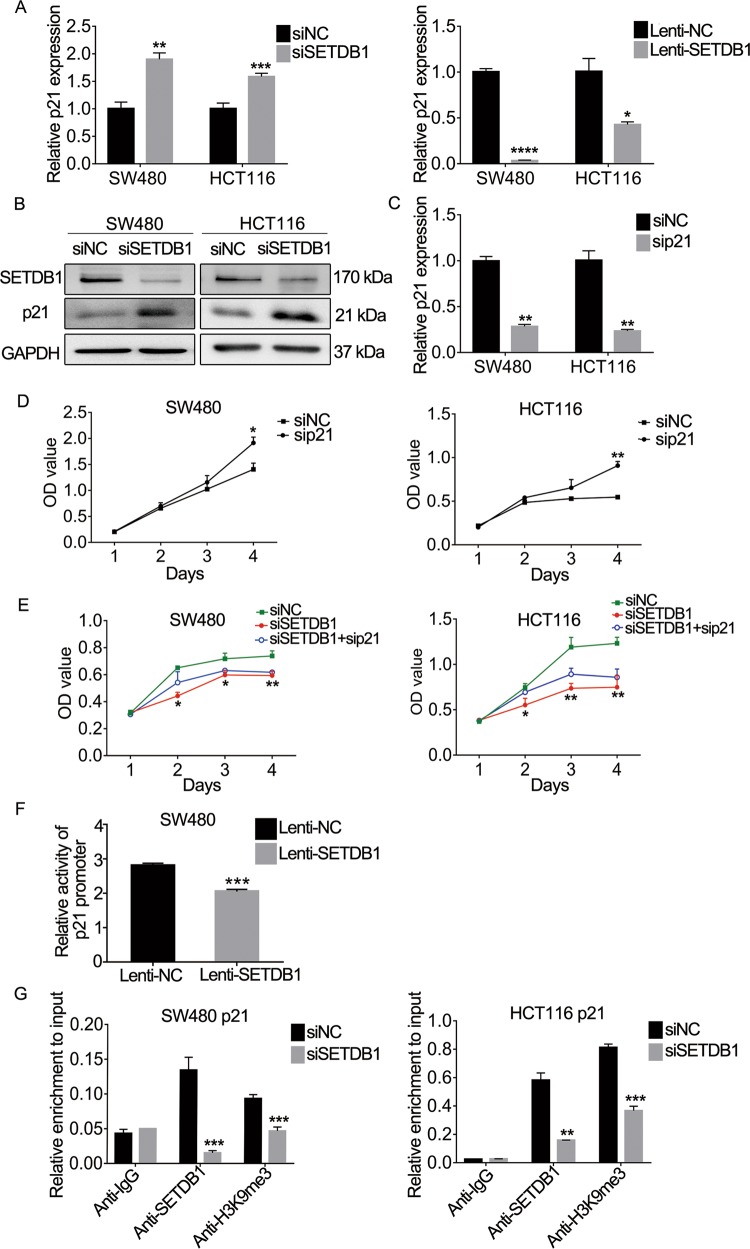
Although SETDB1 has been reported to play a critical role in CRC, the underlying mechanism remain unclear. To further explore the molecular mechanism, we used the UNIHI online database to predict the protein interactions and defined the target gene regulated by SETDB141,42. We identified a tumor suppressive gene p21, which involves in cell cycle progression. According to the previous report and our data, we hypothesized that SETDB1 plays a role via regulating the expression of p21. To verify this hypothesis, we performed qRT-PCR and WB in CRC cells transfected with siSETDB1 or Lenti-SETDB1. The results showed that the expression of p21 was significantly upregulated after transfecting with siSETDB1 in SW480 and HCT116 cells but markedly decreased following SETDB1 overexpression (Fig. 6a, b). The efficiency of silencing p21 expression was verified by qRT-PCR (Fig. 6c). CCK8 assay indicated that downregulation of p21 promoted CRC cell proliferation and cells viability co-transfected with siSETDB1 and sip21 were higher than those transfected with siSETDB1 or sip21 alone (Fig. 6d, e). These results suggested that p21 could inhibit CRC cell proliferation and may be a downstream target gene of SETDB1. In order to clarify how SETDB1 regulates the expression of p21, we performed the dual-luciferase reporter assay in SW480 cells, as the result showed that overexpression of SETDB1 could inhibit the activity of p21 promoter in SW480 (Fig. 6f). Furthermore, we carried out the ChIP experiment using the specific antibodies of SETDB1 and H3K9me3 to enrich the DNA fragments combined with them and designing the specific primer of p21 promoter to detect the enrichment of p21, we found that the p21 promoter sequences enriched by the two antibodies were significantly reduced in the siSETDB1 group compared with the siNC group (Fig. 6g). Combined with the results of dual-luciferase reporter assay, we speculated that SETDB1 could bind to p21 promoter and regulate its activity, and silencing SETDB1 could reduce the enrichment of H3K9me3 on p21 promoter. In conclusion, SETDB1 contributes CRC development at least partly by epigenetically silencing p21 expression.
Fig. 6. SETDB1 epigenetically silences p21 expression.
a mRNA expression of p21 following the treatment with siSETDB1 or LV-SETDB1 in SW480 and HCT116 cells. b Protein expression levels of SETDB1 in SW480 and HCT116 cells after transfected with siSETDB1. c p21 expression in SW480 and HCT116 after treated with sip21. d Cell proliferation was analyzed by CCK8 in SW480 and HCT116 cells transfected with sip21 or siNC. e CCK8 assay was performed to monitor the proliferation of siSETDB1 and sip21 co-transfected CRC cells. f Detection of p21 promoter activity after overexpression SETDB1 in SW480 cells by dual-luciferase reporter assay. g ChIP-PCR was performed to validate the occupancy of SETDB1 and H3K9me3 modification at the p21 promoter in SW480 and HCT116 cells transfected with siSETDB1 or siNC, IgG as a negative control. the results represent the mean ± SD. All experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
Silencing of SETDB1 inhibits CRC tumorigenesis in vivo
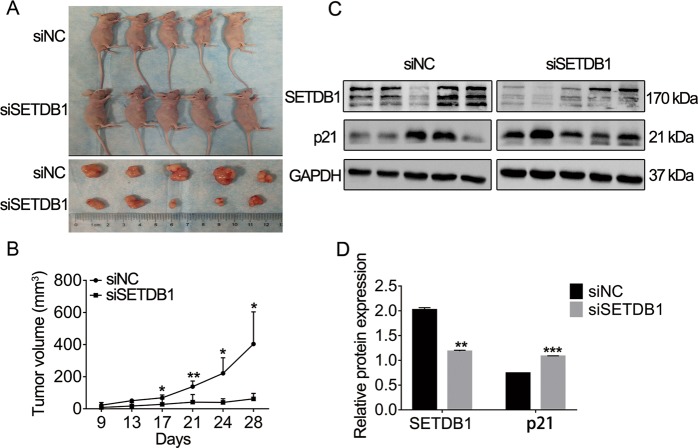
Then, using a xenograft model in nude mice, we investigate the function of SETDB1 on the CRC in vivo. BALB/c nude mice were injected subcutaneously with HCT116 cells. When subcutaneous tumors were observed 9 days later, siSETDB1 or siNC were injected intratumorally every three days and tumors volume were measured. All mice were sacrificed 28 days after HCT116 cells injection. The results exhibited that tumors size in the siSETDB1 group were much smaller than that in the siNC group (Fig. 7a). Similarly, the tumor growth rate of siSETDB1 was slower compared with control group (Fig. 7b). WB experiment results showed that the expression of SETDB1 was reduced in siSETDB1 group, and p21 expression was upregulated in the siSETDB1 group compared with control (Fig. 7c, d). Taken together, our results suggested that silencing of SETDB1 in vivo could inhibit the proliferation of CRC cells and upregulate the expression of the tumor suppressive gene p21.
Fig. 7. Silencing SETDB1 inhibits the CRC cells proliferation in vivo.
a Tumor images of SETDB1-knockdown and control nude mice (n = 5/group). b Tumor volumes of the SETDB1-knockdown and control groups. The tumors were measured every 3 days. c SETDB1 and p21 protein expression in xenograft tumors transfected with siSETDB1 or siNC. d Grayscale analysis of WB results by image J software. The results show as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Recent years, epigenetics attracted more attention and its effects in the diseases have been studied intensively, particularly in carcinogenesis. Some epigenetic markers have been included in medical guidelines43. Histone methylation modification is mediated by HMTs and histone demethylases and plays critical role in gene expression regulation. In general, H3K27me3 and H3K9me3 lead to gene repression, whereas H3K4me3, H3K79me3, and H3K36me3 are associated with active transcription44. SETDB1 is a lysine 9-specific methyltransferase, specifically trimethylating H3K918,45. SETDB1 can promote the development of tumors by inducing H3K9me3 modification at promoters of some genes and has been demonstrated abnormally expressed in various human cancer conditions46. In hepatocellular carcinoma, SETDB1 is amplified in tumor tissues and regulates the proliferation of liver cancer cells through methylation of p5347. In lung cancer, the expression of SETDB1 is upregulated in cancer tissues and cooperates with SMAD2/3 to inhibit lung cancer metastasis48. In addition, recent studies have found that SETDB1 can trigger AKT K64 methylation to activate AKT kinase activity, leading to cancer49,50. In addition, a previous study showed that SETDB1 promotes CRC progression via binding to the promoter of TP5327. More recently, Yu et al. reported that SETDB1 promotes CRC proliferation through the STAT1-CCND1/CDK6 axis28. Using UALCAN, an online cancer database and specimens of CRC patients, we found the expression of SETDB1 was upregulated in CRC. In addition, our data suggested that SETDB1 significantly influenced CRC cells proliferation, apoptosis, migration, and invasion in vitro. These results are consistent to previous studies and suggest that SETDB1 contributes to the pathogenesis of various cancers and plays an oncogenic role in CRC.
In current study, we also discovered that the classic tumor suppressor genes p21 is a target gene of SETDB1. p21 is involved in cell cycle regulation, which has been reported as a critical molecule for inhibiting cell proliferation in CRC cells51–54. Previous reports have showed that increased DNA methyltransferase 1 (DNMT1) in Ppara∆IE mice could promote carcinogenesis of CRC via repressing the transcription of p21 through DNA methylation55. Long noncoding RNA CRNDE promotes the proliferation of CRC cells partly through epigenetically silencing of DUSP5 and p21 expression by binding with EZH2 and induce H3K27me3 modification at the promoter of these genes36. LncRNA HOXA-AS2 promotes the progression of CRC via repressing the expression of p21 and KLF2 by binding with EZH2 and LSD1, which both regulate the histone methylation37. Therefore, p21 acts as a tumor suppressor gene in CRC, and histone methylation plays an important role in regulating its expression. In our study, we verified that SETDB1 could regulate the promoter activity of p21 in CRC cells through the dual-luciferase reporter assay. Moreover, the ChIP experiment further proved that SETDB1 could regulate the enrichment of H3K9me3 on p21 promoter. Bao et al. reported that lncRNA-p21 prevents cell reprogramming via associating with SETDB1 and DNMT to maintain the heterochromatin state of the gene56, which implicated that the epigenetic regulation of p21 is one of the important mechanisms to contribute to the physiological and pathological function. Here, we reveal a new regulatory mechanism of SETDB1 and p21 in CRC.
Furthermore, we demonstrated that downregulation of SETDB1 could inhibit HCT116 cells proliferation in vivo. It suggests that SETDB1 may be a potential therapeutic target. Dysregulation of epigenetic markers has been extensively reported in tumors and inhibitors targeting these epigenetic enzymes have been developed57. For example, some HDAC inhibitors vorinostat, romidepsin, and belinostat have been approved for some T-cell lymphoma and panobinostat for MM. Other HDAC inhibitors such as resminostat, practinostat are in phase I and II clinical trials for the treatment of solid malignancies including hepatocellular and prostate cancer58. Sulforaphane is one of the most potent HDAC inhibitors that can inhibit proliferation and induce apoptosis of CRC cells59. In addition, an early report showed that more than 20 HMT enzymes have been reported to be associated with the occurrence of CRC and drugs targeting histone methylation have been demonstrated promising therapeutic effects in preclinical CRC treatment60. For example, EZH2 and SUV39H1 inhibitors have shown efficacy for CRC treatment in preclinical. EZH2 inhibitor GSK346 and chaetocin, a fungal metabolite inhibiting SUV39H1 which regulates the methylation of H3K9, could reduce migration of CRC cells61,62. In conclusion, the treatment targeting epigenetic modification is a promising clinical therapeutic strategy.
In summary, our study illustrated that SETDB1 was upregulated in CRC tissues and serve as an oncogene in CRC. The function role of SETDB1 on CRC cell proliferation and tumorigenesis is partly through epigenetic silencing of p21 expression. SETDB1 might become a promising target for the diagnosis and therapeutic strategy of CRC.
Materials and methods
Specimen collection
A total of 60 pairs primary tumor tissues and corresponding normal tissues were collected from CRC patients who received surgical treatment at Zhongnan Hospital of Wuhan University (Wuhan, China) between August 2017 and February 2018. All patients were diagnosed by original histopathological detection and none of them received preoperative adjuvant chemotherapy or radiotherapy. The patients with non‐curative resection, cancer recurrence, severe injury of vital organs, or a history of autoimmune diseases were excluded. Samples of the collected tissues were preserved in liquid nitrogen. Clinical case data of patients were also collected. The collection of clinical specimens was approved by the clinical research institution review committee and ethics review committee of the Zhongnan Hospital, and every patient was informed of their consent.
Cell culture and transfection
Cell lines SW480 and HCT116 were purchased from the China Center for Type Culture Collection (Wuhan, China), and cells had been authenticated for STR profiling and tested for mycoplasma by the vendor. All the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, USA) containing 10% fetal bovine serum (FBS) (HyClone, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (Genom, China), at 37 °C with 5% CO2. Purinicin-inducible GFP-tagged lentiviral SETDB1 (Lenti-SETDB1) were designed and synthesized by Shanghai Genechem (Shanghai, China). Cells infected with lentivirus were selected by 2 μg/ml of puromycin (Sigma, USA) to obtain stably infected cell lines. siSETDB1 and FAM-labeled siNC were purchased from Guangzhou RiboBio (Guangzhou, China), and transfected into cancer cell lines using Lipofectamine 2000 (lipo2000) (Invitrogen, USA). Transfection efficiency of siRNA with lipo2000 was preliminarily assessed by fluorescence microscope (Olympus U-RFL-T, Japan) 24 h after transfection and the silencing effect of siSETDB1 was further verified by qRT-PCR and WB experiments.
RNA extraction and qRT-PCR
The total RNA in tissues and cells were extracted with Trizol reagent (Invitrogen, USA), and the reverse transcription was performed using TOYOBO ReverTra Ace kit (TOYOBO, Japan). mRNA expression was quantified using quantitative reverse transcription PCR (qRT-PCR) on Biorad CFX (Biorad, USA), and GAPDH were selected as a housekeeping gene. The primers were designed and synthesized by TSINGKE Biological Technology (Wuhan, China), primer sequences were as follows: GAPDHF-5′GGAGCGAGATCCCTCCAAAAT3′, R-5′GGCTGTTGTCATACTTCTCATGG3′, SETDB1F-5′GGTGGTCGCCAAATACAAA3′, R-5′ GGAAGCATAGCCATCATCAA3′, P21F-5′ TGCCGAAGTCAGTTCCTTGT3′, R5′CATTAGCGCATCACAGTCGC3′. The expression levels of mRNA were calculated using the comparative CT (2-∆∆CT), and all experiments were performed with three biological replicates.
Protein extraction and western blotting
Total protein in CRC lines and subcutaneous tumors was extracted using RIPA lysis buffer (Boyotime, China) according to the reagent instructions. Western blotting was performed with the specific antibody, SETDB1(1:1000, Proteintech, 11231-1-AP), p21(1:1000, proteintech, 10355-1-AP), GAPDH (1:1000, Proteintech, 60004-1-Ig), E-cadherin (1:1000, Cell Signaling Technology, 3195 T), N-cadherin (1:1000, Cell Signaling Technology, 13116-T).
CCK8 assay
Cell viability was detected using CCK8 kit (Dojindo Molecular Technologies, Japan). After transfection of HCT116 and SW480 cells with siSETDB1 and Lenti-SETDB1 for 48 h, 1000 cells per well were seeded in 96-wells plates with five replicate wells. CCK8 was added into the wells mixing with cell culture medium and incubate for 3 h. The absorbance at 450 nm was measured with a microplate reader (ELX-800; BioTek, USA), and the growth curve was drawn according to OD value.
Migration and invasion assay
Migration and invasion assay were performed using the transwell chambers (Corning, USA). One hundred microliters matrigel matrix (Corning, USA) was added into upper chamber and incubated at 37 °C for 1 h for the invasion assay. Two hundred microliters cell suspension about 1 × 105 were seeded in upper chambers with serum-free DMEM medium while the lower chamber was filled with 10% FBS. Cells were stained with crystal violet (sigma-Aldrich, USA) and photographed by microscope (Olympus, Japan).
Apoptosis and cell cycle assay
Cell apoptosis assay was performed using Annexin V-FITC/PI apoptosis detection kit (BestBio, China), and cell cycle assay was performed using cell cycle and apoptosis detection kit (Boyotime, China). Cells were collected after 50 h transfecting siRNA, and all operations were performed according to the reagent instructions.
Colony formation assay
After 24 h of siRNA transfection, 500 cells were seeded into a six-well culture plate and incubated at 37 °C with 5% CO2 and we performed the transfection again in these cells 4 days later, visible colonies could be observed at 9 days. The colonies were fixed with 4% paraformaldehyde and stained with crystal violet. Counting the number of colonies that made up of at least 50 cells.
Tumor xenograft in nude mice
siRNAs were synthesized and purchased from Guangzhou RiboBio and chemically modified in the form of 2 ‘Ome + 5’ Chol (Guangzhou, China). Chemically modified siRNAs are very stable which could be used in vivo assays63,64. The sample size of animal experiment was determined as five nude mice in siSETDB1 group and siNC group and it was carried out randomly. We were not blinded for animal experiment. Two hundred microliters cell suspension (about 5 × 106 HCT116 cells in PBS) were injected into female BALB/C nude mice at 4 weeks old, five mice per group. siRNA was injected into tumors when the subcutaneous tumor is visible at 9 days and we injected the siRNA into the tumors every 3 days to maintain its function. The tumor size was measured every 3 days using calipers. Mice were sacrificed at 28 days and the subcutaneous tumor was excised and preserved at −80°C. Animal experiments were approved by the Animal Ethics committee of Wuhan University.
ChIP-PCR assay
CRC cells were fixed with 1% formaldehyde and incubated at room temperature for 10 min to make DNA-protein cross-links. Then glycine was added to stop the crosslinking and incubated at room temperature for 5 min. One milliliter cell lysis containing protease inhibitors (MCE, USA) was added to suspend cells and then cell lysates were sonicated using EPISONIC (USA) to get 200–300 bp of chromatin fragments. Immunoprecipitation was performed with SETDB1 (1:100, Proteintech, 11231-1-AP), H3K9me3 (1:50, CST, D4W1U) specific antibodies and IgG (1:100, CST, 2729S). The chromatin DNA was extracted using DNA purification kit (TIANGEN, China) and the specific primers of p21 promoter were used for PCR. The primer sequences were as following: p21F 5′-TGCATTGGGTAAATCCTTGCC-3′, R5′-AATGAGTTGGCACTCTCCAGG-3′.
Dual-luciferase reporter assay
The high expression cell line of SETDB1 was constructed by infecting SW480 cells with lentivirus, and then the recombinant plasmid pGL4-basic-p21 was co-transfected with the control plasmid pRL-TK, 24 h after transfection, promoter activity was analyzed using a dual-luciferase assay kit (Promega, USA) according to the manufacturer’s instructions.
Statistics analysis
In this study, all the experiments were performed in three times and data were exhibited as mean ± SD. The sample sizes for relevant experiment were determined by power analysis. When the variance between the two groups was similar, student’s t test was used to analysis data difference between two groups, if not the same, welch’s t-test was used, and chi-squared test was used to analysis clinical data. Statistics analysis was performed using SPSS 17.0 software (IBM, USA) and GraphPad prism 7.0 software (GraphPad software, USA). p < 0.05 was considered to be statistically significant.
Acknowledgements
This project was supported by grant from the National Natural Science Foundation of China (No.81270468, No.81870391), Medical Science Advancement Program (Basic Medical Science) of Wuhan University (Grant No. TFJC2018004) and Science and technology innovation cultivation fund of Zhongnan hospital of Wuhan University (cxpy2017031).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Stephanou
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Brien GL, Valerio DG, Armstrong SA. Exploiting the epigenome to control cancer-promoting gene-expression programs. Cancer Cell. 2016;29:464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitaker JW, Chen Z, Wang W. Predicting the human epigenome from DNA motifs. Nat. Methods. 2015;12:265–272. doi: 10.1038/nmeth.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Pediconi N, et al. EZH2, JMJD3, and UTX epigenetically regulate hepatic plasticity inducing retro-differentiation and proliferation of liver cells. Cell Death Dis. 2019;10:518. doi: 10.1038/s41419-019-1755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Amplification of SMYD3 promotes tumorigenicity and intrahepatic metastasis of hepatocellular carcinoma via upregulation of CDK2 and MMP2. Oncogene. 2019;38:4948–4961. doi: 10.1038/s41388-019-0766-x. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. CREPT facilitates colorectal cancer growth through inducing Wnt/beta-catenin pathway by enhancing p300-mediated beta-catenin acetylation. Oncogene. 2018;37:3485–3500. doi: 10.1038/s41388-018-0161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, Q. et al. Elevating H3K27me3 level sensitizes colorectal cancer to oxaliplatin. J Mol Cell Biol. 10.1093/jmcb/mjz032 (2019). [DOI] [PMC free article] [PubMed]
- 11.Raynal NJ, et al. Repositioning FDA-approved drugs in combination with epigenetic drugs to reprogram colon cancer epigenome. Mol. Cancer Ther. 2017;16:397–407. doi: 10.1158/1535-7163.MCT-16-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patnaik S, Anupriya Drugs. Targeting epigenetic modifications and plausible therapeutic strategies against colorectal cancer. Front Pharm. 2019;10:588. doi: 10.3389/fphar.2019.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesch, B. J. et al. Intergenerational epigenetic inheritance of cancer susceptibility in mammals. Elife8, 10.7554/eLife.39380 (2019). [DOI] [PMC free article] [PubMed]
- 14.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 15.Torrano J, Al Emran A, Hammerlindl H, Schaider H. Emerging roles of H3K9me3, SETDB1 and SETDB2 in therapy-induced cellular reprogramming. Clin. Epigenetics. 2019;11:43. doi: 10.1186/s13148-019-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harte PJ, Wu W, Carrasquillo MM, Matera AG. Assignment of a novel bifurcated SET domain gene, SETDB1, to human chromosome band 1q21 by in situ hybridization and radiation hybrids. Cytogenet Cell Genet. 1999;84:83–86. doi: 10.1159/000015220. [DOI] [PubMed] [Google Scholar]
- 17.Loyola A, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- 20.Becker JS, Nicetto D, Zaret KS. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann F, et al. KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells. 2010;28:201–212. doi: 10.1002/stem.278. [DOI] [PubMed] [Google Scholar]
- 22.Rivera C, et al. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 2015;43:9097–9106. doi: 10.1093/nar/gkv929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orouji, E. et al. Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int J Cancer. 10.1002/ijc.32432 (2019). [DOI] [PubMed]
- 24.Wong CM, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Paredes M, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33:2807–2813. doi: 10.1038/onc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryu TY, et al. SETDB1 regulates SMAD7 expression for breast cancer metastasis. BMB Rep. 2019;52:139–144. doi: 10.5483/BMBRep.2019.52.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K, et al. Histone methyltransferase SETDB1 promotes the progression of colorectal cancer by inhibiting the expression of TP53. J. Cancer. 2017;8:3318–3330. doi: 10.7150/jca.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Y. et al. Histone methyltransferase SETDB1 promotes colorectal cancer proliferation through the STAT1-CCND1/CDK6 Axis. Carcinogenesis. 10.1093/carcin/bgz131 (2019). [DOI] [PubMed]
- 29.Ho YJ, et al. Significance of histone methyltransferase SETDB1 expression in colon adenocarcinoma. APMIS. 2017;125:985–995. doi: 10.1111/apm.12745. [DOI] [PubMed] [Google Scholar]
- 30.Wu HL, et al. Demystifying the mechanistic and functional aspects of p21 gene activation with double-stranded RNAs in human cancer cells. J. Exp. Clin. Cancer Res. 2016;35:145. doi: 10.1186/s13046-016-0423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Li, G. et al. Human colorectal cancer derived-MSCs promote tumor cells escape from senescence via P53/P21 pathway. Clin Transl Oncol. 10.1007/s12094-019-02152-5 (2019). [DOI] [PubMed]
- 33.Fan T, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol. Cancer Res. 2011;9:418–429. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh K, Chatterjee B, Maheswari U, Athifa M, Kanade SR. 4-Nonylphenol-enhanced EZH2 and RNF2 expression, H3K27me3 and H2AK119ub1 marks resulting in silencing of p21(CDKN1A) in vitro. Epigenomics. 2019;11:899–916. doi: 10.2217/epi-2018-0175. [DOI] [PubMed] [Google Scholar]
- 35.Liu, L. et al. Anti-Proliferative activity of HPOB against multiple myeloma cells via p21 transcriptional activation. Molecules23, 10.3390/molecules23051044 (2018). [DOI] [PMC free article] [PubMed]
- 36.Ding J, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding J, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrashekar DS, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W, et al. SETDB1 induces epithelialmesenchymal transition in breast carcinoma by directly binding with Snail promoter. Oncol. Rep. 2019;41:1284–1292. doi: 10.3892/or.2018.6871. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 41.Kalathur RK, et al. UniHI 7: an enhanced database for retrieval and interactive analysis of human molecular interaction networks. Nucleic Acids Res. 2014;42:D408–D414. doi: 10.1093/nar/gkt1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaurasia G, et al. UniHI: an entry gate to the human protein interactome. Nucleic Acids Res. 2007;35:D590–D594. doi: 10.1093/nar/gkl817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat. Rev. Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications—writers that read. EMBO Rep. 2015;16:1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, J. H. et al. Cockayne syndrome group B deficiency reduces H3K9me3 chromatin remodeler SETDB1 and exacerbates cellular aging. Nucleic Acids Res. 10.1093/nar/gkz568 (2019). [DOI] [PMC free article] [PubMed]
- 46.Karanth AV, et al. Emerging role of SETDB1 as a therapeutic target. Expert Opin. Ther. Targets. 2017;21:319–331. doi: 10.1080/14728222.2017.1279604. [DOI] [PubMed] [Google Scholar]
- 47.Fei Q, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat. Commun. 2015;6:8651. doi: 10.1038/ncomms9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu PC, et al. H3K9 histone methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3 pathway to suppress lung cancer metastasis. Cancer Res. 2014;74:7333–7343. doi: 10.1158/0008-5472.CAN-13-3572. [DOI] [PubMed] [Google Scholar]
- 49.Guo J, et al. AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat. Cell Biol. 2019;21:226–237. doi: 10.1038/s41556-018-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G, et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat. Cell Biol. 2019;21:214–225. doi: 10.1038/s41556-018-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou Z, et al. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol. Cancer. 2018;17:172. doi: 10.1186/s12943-018-0922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang MR, et al. miR-6734 up-regulates p21 gene expression and induces cell cycle arrest and apoptosis in colon cancer cells. PLoS One. 2016;11:e0160961. doi: 10.1371/journal.pone.0160961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang R, et al. Long noncoding RNA MLK7AS1 promotes proliferation in human colorectal cancer via downregulation of p21 expression. Mol. Med Rep. 2019;19:1210–1221. doi: 10.3892/mmr.2018.9702. [DOI] [PubMed] [Google Scholar]
- 54.Ji H, et al. Long noncoding RNA MAPKAPK5-AS1 promotes colorectal cancer proliferation by partly silencing p21 expression. Cancer Sci. 2019;110:72–85. doi: 10.1111/cas.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo Y, et al. Intestinal PPARalpha protects against colon carcinogenesis via regulation of methyltransferases DNMT1 and PRMT6. Gastroenterology. 2019;157:744–759. doi: 10.1053/j.gastro.2019.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao X, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett RL, Licht JD. Targeting epigenetics in cancer. Annu Rev. Pharm. Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eckschlager, T., Plch, J., Stiborova, M. & Hrabeta, J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci18, 10.3390/ijms18071414 (2017). [DOI] [PMC free article] [PubMed]
- 59.Martin SL, Kala R, Tollefsbol TO. Mechanisms for the inhibition of colon cancer cells by sulforaphane through epigenetic modulation of microRNA-21 and human telomerase reverse transcriptase (hTERT) down-regulation. Curr. Cancer Drug Targets. 2018;18:97–106. doi: 10.2174/1568009617666170206104032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang T, et al. Targeting histone methylation for colorectal cancer. Ther. Adv. Gastroenterol. 2017;10:114–131. doi: 10.1177/1756283X16671287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferraro A, Boni T, Pintzas A. EZH2 regulates cofilin activity and colon cancer cell migration by targeting ITGA2 gene. PLoS One. 2014;9:e115276. doi: 10.1371/journal.pone.0115276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yokoyama Y, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013;104:889–895. doi: 10.1111/cas.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ku SH, Jo SD, Lee YK, Kim K, Kim SH. Chemical and structural modifications of RNAi therapeutics. Adv. Drug Deliv. Rev. 2016;104:16–28. doi: 10.1016/j.addr.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 64.Zhao JF, et al. The ASH1-miR-375-YWHAZ signaling axis regulates tumor properties in hepatocellular carcinoma. Mol. Ther. Nucleic Acids. 2018;11:538–553. doi: 10.1016/j.omtn.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]