Abstract
Asian soybean rust (ASR), caused by the obligate fungal pathogen Phakopsora pachyrhizi, often leads to significant yield losses and can only be managed through fungicide applications currently. In the present study, eight urediniospore germination or appressorium formation induced P. pachyrhizi genes were investigated for their feasibility to suppress ASR through a bean pod mottle virus (BPMV)‐based host‐induced gene silencing (HIGS) strategy. Soybean plants expressing three of these modified BPMV vectors suppressed the expression of their corresponding target gene by 45%–80%, fungal biomass accumulation by 58%–80%, and significantly reduced ASR symptom development in soybean leaves after the plants were inoculated with P. pachyrhizi, demonstrating that HIGS can be used to manage ASR. In addition, when the in vitro synthesized double‐stranded RNAs (dsRNAs) for three of the genes encoding an acetyl‐CoA acyltransferase, a 40S ribosomal protein S16, and glycine cleavage system H protein were sprayed directly onto detached soybean leaves prior to P. pachyrhizi inoculation, they also resulted in an average of over 73% reduction of pustule numbers and 75% reduction in P. pachyrhizi biomass accumulation on the detached leaves compared to the controls. To the best of our knowledge, this is the first report of suppressing P. pachyrhizi infection in soybean through both HIGS and spray‐induced gene silencing. It was demonstrated that either HIGS constructs targeting P. pachyrhizi genes or direct dsRNA spray application could be an effective strategy for reducing ASR development on soybean.
Keywords: dsRNA, HIGS, Phakopsora pachyrhizi, SIGS, soybean rust
Double‐stranded RNA either produced inside soybean through a modified bean pod mottle virus or sprayed directly on soybean leaves reduced Phakopsora pachyrhizi infection and soybean rust disease.
1. INTRODUCTION
Phakopsora pachyrhizi is an aggressive obligate pathogen and the causal agent of Asian soybean rust (ASR). Unlike other highly specialized rust fungi, P. pachyrhizi has a wide host range and is able to infect more than 150 species of plants from more than 53 genera, including soybean, related Glycine species, and other hosts in the Fabaceae (Hershman et al., 2011). Soybean yield losses of up to 80% in experimental trials have been reported in Asia (Hartman et al., 1991), 63% in Brazil during 2003, 60% in Paraguay during 2001 (Yorinori et al., 2005), up to 100% in South Africa (Caldwell et al., 2004), and up to 55% in the USA (Mueller et al., 2009). Currently, all commercial soybean cultivars are susceptible to P. pachyrhizi, and the only available method to control ASR is multiple fungicide applications (Miles et al., 2007). The level of ASR control depends on disease pressure and the timing of fungicide applications (Mueller et al., 2009). However, continued use of fungicides has led to not only the development of fungicide resistance among P. pachyrhizi populations, but also increased operation cost. For example, soybean producers in Brazil spent close to $2 billion per year on fungicides to control ASR (Godoy et al., 2015). Therefore, there is an urgent need to develop soybean varieties that are resistant or tolerant to ASR to reduce its potential to cause severe yield losses.
Soybean lines with resistance to P. pachyrhizi infection have been reported (Bromfield, 1984) and so far seven resistance to P. pachyrhizi (Rpp) genes have been discovered in different soybean germplasm accessions (Childs et al., 2018), such as Rpp1 in PI 200,492 (McLean and Byth, 1980), Rpp2 in PI 230,970 (Walker et al., 2014), Rpp3 in PI 462,312 (Hartwig and Bromfield, 1983) and Rpp4 in PI 459025B (Hartwig, 1986). However, developing rust‐resistant soybean varieties through traditional plant breeding has been a slow, time‐consuming process (Saurabh et al., 2014) and single‐gene resistance can be overcome by the pathogen quickly (Grasso et al., 2006; Godoy, 2012; Schmitz et al., 2014).
Besides identifying ASR resistance‐related genes and incorporating them into soybean to enhance its resistance to infection by P. pachyrhizi, recent advances in RNA interference (RNAi) offer another possibility of managing fungal diseases via host‐induced gene silencing (HIGS). HIGS is an RNAi‐based approach in which small interfering RNAs (siRNAs) homologous to genes of fungal origin are produced in the host plant and subsequently silence their targets in the pathogen during its infection of the host plant (Nowara et al., 2010). In the past several years, HIGS has been shown to be an effective alternative to fungicide applications in managing various plant diseases, such as reducing powdery mildew on barley and wheat through targeting the Avra10 effector of Blumeria graminis f. sp. tritici (Nowara et al., 2010), suppressing wheat stripe rust through silencing a highly abundant Puccinia striiformis f. sp. tritici haustorial transcript (Yin et al., 2011), and reducing fusarium wilt disease in banana through expressing siRNAs targeting vital Fusarium oxysporum f. sp. cubense genes (Ghag et al., 2014), or aflatoxin contamination in maize by expressing a HIGS construct targeting the Aspergillus flavus aflC gene (Thakare et al., 2017; Sharma et al., 2018) or aflM gene (Raruang et al., 2020).
In addition, several studies have demonstrated that direct double‐stranded RNA (dsRNA) application can be used to manage plant fungal diseases. For example, foliar spray of in vitro synthesized CYP3 dsRNAs effectively reduced F. graminearum infection and resulted in much smaller head blight lesions on barley (Koch et al., 2016). External application of dsRNAs and small RNAs (sRNAs) targeting Dicer‐like protein DCL1 and DCL2 genes of Botrytis cinerea on vegetables, fruits, and flower petals significantly suppressed grey mould (Wang et al., 2016). McLoughlin et al. (2018) demonstrated exogenously applied dsRNA protected plants against Sclerotinia sclerotiorum and B. cinerea.
The obligate biotrophic nature of P. pachyrhizi makes it difficult to study in vitro. The fungal genome has not been completely sequenced despite repeated attempts (Loehrer et al., 2014). To date, only limited transcriptomic information is available on P. pachyrhizi, such as expressed sequence tag analysis (Posada‐Buitrago and Frederick, 2005), RNA‐Seq studies of infected soybean leaves at different stages of infection (Tremblay et al., 2010, 2012, 2013), transcriptome analysis during appressorium and haustorial formation (Stone et al., 2012; Link et al., 2014), and secretome analysis of P. pachyrhizi (de Carvalho et al., 2017).
A shortlist of potential P. pachyrhizi target genes that have been generated through bioinformatic analysis of data from the above ASR transcriptomic studies and that might be involved in infection and pathogenicity were selected for suppressing ASR on soybean via HIGS. A bean pod mottle virus (BPMV)‐based vector system developed by Zhang et al. (2010) was chosen to transiently express siRNA in soybean for this study. This vector system has been successfully used to test the candidate genes from soybean for their involvement in resistance to ASR (Meyer et al., 2009; Cooper et al., 2013; Liu and Whitham, 2013; Qi et al., 2016).
When soybean plants are infected by the BPMV‐based vector system, dsRNAs formed during virus replication will trigger siRNA production. BPMV constructs that contain sequences from P. pachyrhizi genes can then produce siRNAs against target genes during P. pachyrhizi infection. The main objectives of this study were to determine whether genes involved in urediniospore germination or appressorium formation can be used to manage ASR through HIGS, and to examine the effectiveness of the dsRNAs for three of the genes in ASR reduction through spray‐induced gene silencing (SIGS). In this study, eight genes from P. pachyrhizi were cloned into a BPMV‐based HIGS vector system and introduced into soybean through biolistic and rubbing inoculation. Among them, three HIGS constructs significantly reduced P. pachyrhizi infection and pustule formation compared to the controls after being introduced into soybean plants in both in vitro and in planta assays. Significant reduction of P. pachyrhizi infection was also demonstrated after directly spraying in vitro synthesized dsRNAs targeting P. pachyrhizi genes encoding an acetyl‐CoA acyltransferase (ATC), a 40S ribosomal protein S16 (RP_S16), and a glycine cleavage system H protein (GCS_H) onto detached soybean leaflets before P. pachyrhizi inoculation. To the best of our knowledge, this is the first report of suppressing P. pachyrhizi infection in soybean through HIGS and direct dsRNA application.
2. RESULTS
2.1. Construction of HIGS vectors and confirmation of successful HIGS construct expression
Eight P. pachyrhizi urediniospore germination or appressorium formation‐related genes (Table 1) were selected based on bioinformatic analysis of the available transcriptomic data of P. pachyrhizi (Stone et al., 2012; Link et al., 2014; de Carvalho et al., 2017) and a segment of 200–400 nucleotides (nt) in length for each of the genes was amplified and cloned into the BPMV vector (Zhang et al., 2013) (Figure 1a). Soybean seedlings developed mild mottling foliar symptoms 14 days after mechanical inoculation using primary inoculum prepared from plants bombarded with BPMV:EV (empty vector) whereas foliar photobleaching symptoms were observed on soybean seedlings inoculated with primary inoculum containing BPMV:PDS, which has a section of the sequence encoding soybean phytoene desaturase (PDS) in the modified BPMV, indicating that the expression of the target gene was specifically suppressed under the experimental conditions (Figure 1b). Mild mottling foliar symptoms were consistently observed on the second trifoliate leaves of soybean plants mechanically inoculated with eight primary inocula containing each of the eight HIGS constructs listed in Table 1 (Figure 1b). Furthermore, enzyme‐linked immunosorbent assay (ELISA) confirmed the presence of BPMV in the leaf tissues inoculated with the empty BPMV vector or any one of the eight HIGS constructs. Taken together, the mottling foliar symptoms, photobleached leaves of the positive control, and the ELISA results indicate that the BPMV‐HIGS constructs can be successfully delivered into soybean plants and expressed properly (Figure 1c).
TABLE 1.
List of candidate genes from Phakopsora pachyrhizi and primers used to construct HIGS vectors
| Candidate gene | Accession no. | Putative function | Primer sequence | Amplicon size (bp) | |
|---|---|---|---|---|---|
| ATC | EH237261.1 | Acetyl‐CoA acyltransferase | Forward | CGCGGATCCGGAAATGCGTCGCAAGTG | 351 |
| Reverse | CGCGGATCCCTATCTCCTCCTTTCTAAT | ||||
| UN_1 | EH249765.1 | Hypothetical protein | Forward | CGCGGATCCAGAGACGAAGTCTTCCGT | 202 |
| Reverse | CGCGGATCCATAGGAATGCCTAAGGGA | ||||
| UN_2 | EH249689.1 | Hypothetical protein | Forward | CGCGGATCCTTGTGGCTATCGGCTGTGT | 192 |
| Reverse | CGCGGATCCTTTGAGCTGAATTGCGACA | ||||
| UN_3 | EH236143.1 | Hypothetical protein | Forward | CGCGGATCCAGCTTGCTCAAGAGAGTG | 275 |
| Reverse | CGCGGATCCAGCCAGCTCTCCAGGCTT | ||||
| GCS_H | EH229082.1 | Glycine cleavage system H protein | Forward | CGCGGATCCGATCAGAAGTCCGGCAACGA | 304 |
| Reverse | CGCGGATCCTGATGCCGCTTTGACATCCT | ||||
| RP_S16 | EH249914.1 | 40S ribosomal protein S16 | Forward | CGCGGATCCTAGGTCCCGGTGTAGATGGG | 319 |
| Reverse | CGCGGATCCAGGATTGGAAAGGCCAGGAA | ||||
| CRP_6 | DN739834.1 | Conidiation‐related protein 6 | Forward | CGCGGATCCTTGCAAGGACGGAAAGGGTT | 257 |
| Reverse | CGCGGATCCCTTGGGTTCCATTCTCCGGG | ||||
| PHR | JK650639.1 | Putative 3‐hydroxy‐3‐methylglutaryl‐coenzyme A reductase | Forward | CGCGGATCCCTAGTCTCTTATGGCGGCGG | 326 |
| Reverse | CGCGGATCCGCGTTTGCTTGTCGTTCAGT | ||||
The restriction enzyme BamHI site was incorporated in the primer and is underlined.
FIGURE 1.
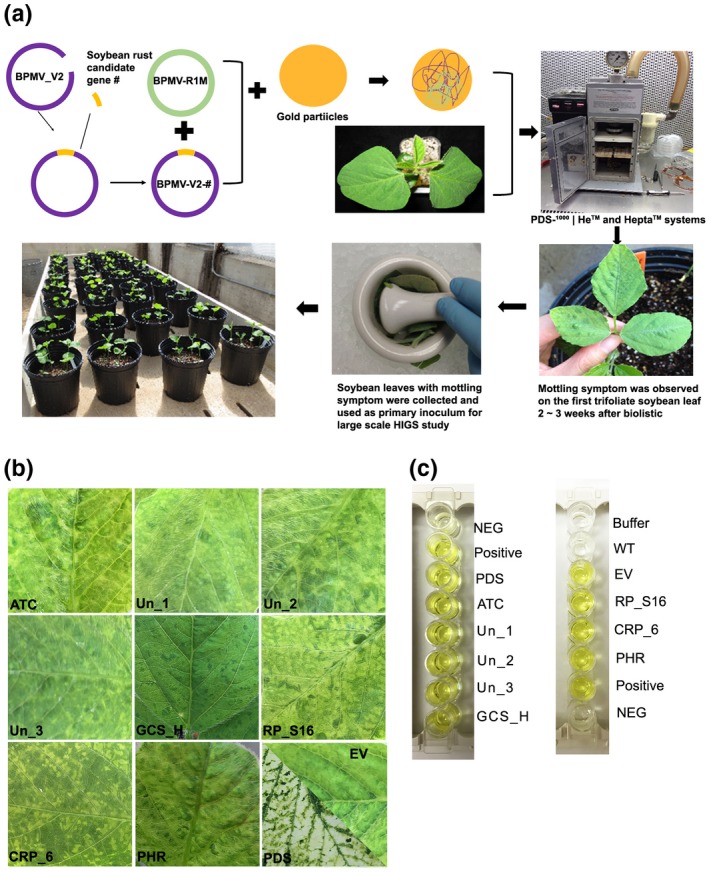
Suppression of Phakopsora pachyrhizi gene expression in soybean through a BPMV‐based transient host‐induced gene silencing (HIGS) approach. (a) Workflow of primary viral inoculum generation for various HIGS constructs. (b) Visual mottle symptoms on soybean leaves expressing various HIGS constructs. (c) Enzyme‐linked immunosorbent assay (ELISA) confirming the presence of bean pod mottle virus (BPMV) in leaf samples. The genes used in the HIGS constructs encode various proteins with putative functions, such as acetyl‐CoA acyltransferase (ATC), glycine cleavage system H protein (GCS_H), 40S ribosomal protein S16 (RP_S16), conidiation‐related protein 6 (CRP_6), putative 3‐hydroxy‐3‐methylglutaryl‐coenzyme A reductase (PHR) or unknown functions (Un_1, 2, and 3). The gene PDS (soybean phytoene desaturase) was used as a positive control to show the bleached leaf phenotype. The BPMV that was supplied with the ELISA kit was used as a positive control. Water (WT) and buffer were included as negative controls
2.2. Detection of gene‐specific sRNA in HIGS‐treated soybean leaf samples
RNA‐Seq was performed to examine whether the leaf samples expressing the HIGS constructs were associated with the production of gene‐specific sRNAs. Leaf samples containing HIGS constructs targeting the ATC, GCS_H, and RP_S16 genes were selected for sRNA library construction and sequencing, which yielded about 10 million reads per library. The sRNA sequencing data were analysed following the flowchart described in Figure 2. No detectable sRNA reads that matched the ATC, GCS_H or RP_S16 gene sequences were observed in libraries prepared from mock‐treated soybean leaves or BPMV:EV infected soybean leaves (Table 2). However, more than 1,000 sRNA reads in the soybean leaf samples expressing each of the HIGS constructs were aligned to their corresponding target gene sequences (Table 2), demonstrating that this transient HIGS approach was working as expected. These gene‐specific sRNAs were found to distribute unevenly across the target gene sequence, with most of the sRNAs appearing to be generated from a few hotspots in the target sequence for ATC (Figure 3a), GCS_H (Figure 3c) or RP_S16 (Figure 3e). Most of the sRNAs from leaf samples containing BPMV:ATC and BPMV:RP_S16 aligned to the antisense strand of the target sequence (Figure 3a,e) whereas the sRNAs from leaf samples containing BPMV:GCS_H appeared to align to both sense and antisense strands of the target sequence (Figure 3c). Further, the most abundant sRNA from these libraries was 21–22 nt in length (Figure 3b,d,f), which agrees with previous studies on sRNAs produced in tissues expressing HIGS constructs (Ghag et al., 2014; Jahan et al., 2015; Koch et al., 2016).
FIGURE 2.
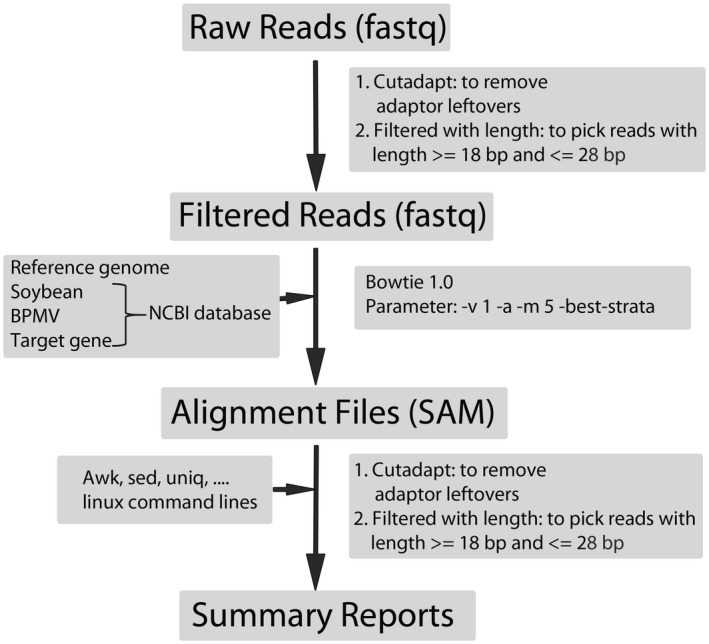
Analysis workflow used to identify gene‐specific small RNAs expressed in small RNA libraries prepared from HIGS‐ and empty vector (EV)‐treated soybean leaf samples. The bioinformatics pipeline used in the small RNA analysis was modified from Tian et al. (2017)
TABLE 2.
Number of small RNA reads matched to the intended target sequences in each of the libraries
| Libraries sequenced a | Number of reads aligned to reference sequences b | |||
|---|---|---|---|---|
| BPMV | ATC | GCS_H | RP_S16 | |
| WT | 552 | 2 | 4 | 1 |
| BPMV:EV | 388,893 | 2 | 2 | 1 |
| BPMV:ATC | 47,517 | 1,562 | 1 | 1 |
| BPMV:GCS_H | 44,721 | 5 | 3,146 | 1 |
| BPMV:RP_S16 | 84,249 | 1 | 2 | 1,889 |
Leaves from soybean plants (Williams 82) were collected 2 weeks after rubbing inoculation with buffer (WT), bean pod mottle virus (BPMV) empty vector (EV), BPMV:ATC, BPMV:GCS_H, or BPMV:RP_S16. ATC, acetyl‐CoA acyltransferase; GCS_H, glycine cleavage system H protein; RP_S16, 40S ribosomal protein S16.
Information on reference sequences is provided in Table 1.
FIGURE 3.
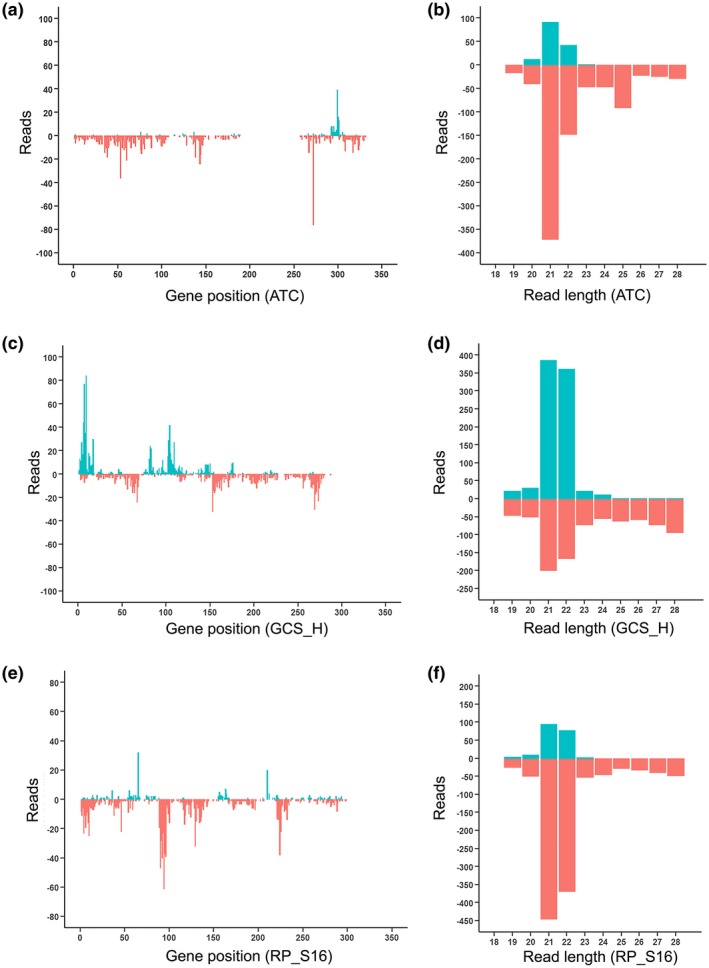
Small RNA (sRNA) profiling of Phakopsora pachyrhizi target genes in soybean leaf tissue that had been rub‐inoculated with modified BPMV containing either EV or HIGS constructs. Number of reads that mapped to various positions of the target genes (a), (c), and (e) and their corresponding read length distributions (b), (d), and (f) of gene‐specific sRNAs identified through RNA‐Seq analysis of the sRNA libraries prepared from leaf samples treated with BPMV:ATC (a) and (b), BPMV:GCS_H (c) and (d) or BPMV:RP_S16 (e) and (f). ATC, acetyl‐CoA acyltransferase; GCS_H, glycine cleavage system H protein; RP_S16, 40S ribosomal protein S16
2.3. HIGS constructs reduced P. pachyrhizi infection in greenhouse conditions
ASR development on the control and HIGS construct‐treated soybean plants was visually assessed 2 weeks after inoculation with P. pachyrhizi and the representative foliar symptoms of three repeated studies are presented (Figure 4a). Significant reduction (α = 0.01) in pustule number (by more than 41.6%–61.2%) was observed for soybean leaves treated with HIGS constructs targeting P. pachyrhizi ATC, unknown 3 (Un_3), GCS_H, and RP_S16 genes compared to the BPMV:EV treatment (Figure 4b). In addition, P. pachyrhizi biomass accumulation in soybean leaves containing HIGS constructs targeting P. pachyrhizi ATC, Un_3, GCS_H, and RP_S16 genes was reduced by 70.2%, 57.8%, 63.7%, and 63.6%, respectively, compared to the P. pachyrhizi biomass accumulation in soybean leaves containing the BPMV:EV without HIGS target genes (Figure 4c). The P. pachyrhizi biomass in soybean leaves inoculated with other HIGS constructs did not show significant differences compared to the control leaves inoculated with the BPMV:EV. The corresponding transcript was also found to be reduced by 49.6%, 75.2%, and 46.3% (α = 0.01), respectively, in the leaf samples containing HIGS construct targeting the P. pachyrhizi ATC, GCS_H, and RP_S16 genes, respectively, when compared to the control leaf sample containing BPMV:EV (Figure 4d). The transcript level for Un_3 in the leaf samples inoculated with HIGS construct BPMV:Un_3 did not change significantly compared to BPMV:EV control.
FIGURE 4.
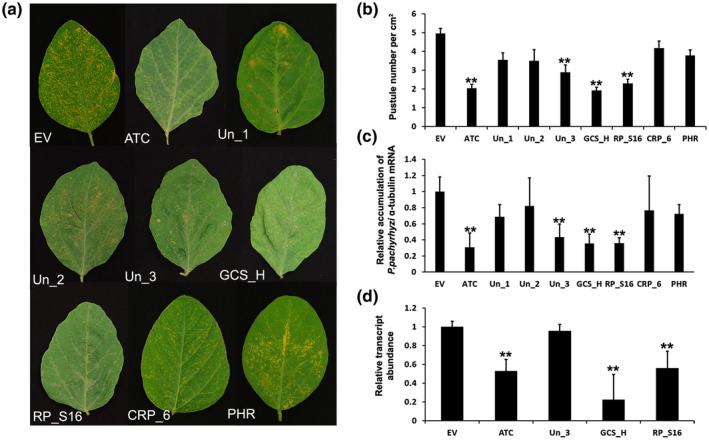
Responses of soybean plants (Williams 82) that had been rub‐inoculated with modified BPMV containing either EV or HIGS constructs prior to inoculation with Phakopsora pachyrhizi in a greenhouse. (a) Differences in soybean rust symptom development 2 weeks after P. pachyrhizi inoculation among soybean plants that had been inoculated with modified BPMV containing HIGS construct targeting various P. pachyrhizi genes encoding acetyl‐CoA acyltransferase (ATC), glycine cleavage system H protein (GCS_H), 40S ribosomal protein S16 (RP_S16), conidiation‐related protein 6 (CRP_6), putative 3‐hydroxy‐3‐methylglutaryl‐coenzyme A reductase (PHR), and other hypothetical proteins of unknown functions (Un_1, Un_2, Un_3) or with modified BPMV only containing an EV. (b) Pustule numbers per cm2 of soybean leaves that had been inoculated with modified BPMV containing HIGS construct targeting P. pachyrhizi genes or EV. (c) Relative expression of P. pachyrhizi α‐tubulin mRNA to soybean ubiquitin following BPMV and P. pachyrhizi inoculation. (d) Relative levels of target gene expression in soybean leaves 2 weeks post‐inoculation with P. pachyrhizi. Values are expressed relative to the endogenous P. pachyrhizi reference gene Cyt B, with the EV set at 1. Bars represent mean values ± SD of one representative experiment with five biological replicates. Asterisks indicate statistical significance: *p < .05, **p < .01 (Student's t test)
2.4. HIGS constructs reduced P. pachyrhizi infection in detached leaf assays
Similar suppression in ASR severity as determined by pustule number per cm2 of leaf was observed on detached soybean leaves expressing BPMV:ATC, BPMV:GCS_H, and BPMV:RP_S16, and was about 58.1%–70.8% of that of the BPMV:EV control (Figure 5a,b). Soybean leaves expressing BPMV:ATC, BPMV:GCS_H, and BPMV:RP_S16 also showed significant reduction (α = 0.01) in P. pachyrhizi biomass in inoculated soybean leaves, which was about 20.3%, 34.7%, and 25.4%, respectively, of that in the control leaves expressing BPMV:EV (Figure 5c). The expression of the corresponding P. pachyrhizi ATC, GCS_H, and RP_S16 genes was also reduced to 45.1%, 20.4%, and 54.6%, respectively, of that in the control leaves treated with BPMV:EV. However, the pustule number per cm2, P. pachyrhizi biomass, and target gene expression in soybean leaves expressing BPMV:Un_3 did not show significant differences compared to those in control leaves (Figure 5a,b), which is different from what was observed in the greenhouse study.
FIGURE 5.
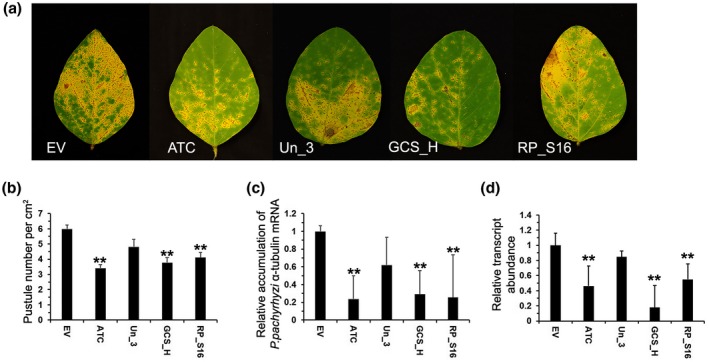
Responses of detached soybean (Williams 82) leaves that had been rub‐inoculated with modified BPMV containing either EV or HIGS constructs prior to inoculation with Phakopsora pachyrhizi urediniospores. (a) Differences in soybean rust symptom development 2 weeks after P. pachyrhizi inoculation among the detached soybean leaves that had been inoculated with modified BPMV containing EV or HIGS constructs targeting various P. pachyrhizi genes that encode an acetyl‐CoA acyltransferase (ATC), a glycine cleavage system H protein (GCS_H), a 40S ribosomal protein S16 (RP_S16), or a hypothetical protein of unknown function (Un_3). (b) Pustule numbers per cm2 of soybean leaves that had been inoculated with modified BPMV containing HIGS constructs targeting P. pachyrhizi genes or EV. (c) Relative expression of P. pachyrhizi α‐tubulin mRNA to soybean ubiquitin following BPMV and P. pachyrhizi inoculation. (d) Relative levels of target gene expression in soybean leaves inoculated with modified BPMV containing either EV or HIGS constructs 2 weeks after P. pachyrhizi inoculation. Values are expressed relative to the endogenous P. pachyrhizi reference gene Cyt B, with expression in the EV‐treated leaves set at 1. Bars represent mean values ± SD of one representative experiment with five biological replicates. Asterisks indicate statistical significance: *p < .05, **p < .01 (Student's t test)
2.5. Direct dsRNAs spray on detached soybean leaves reduced ASR development
Soybean leaves sprayed with dsRNA targeting P. pachyrhizi ATC, GCS_H, and RP_S16 genes before being inoculated with P. pachyrhizi were found to have a lower number of pustules per cm2 of leaf than the negative control 2 weeks after inoculation (Figure 6a). The control soybean leaves sprayed with water developed an average of 9.1 pustules per cm2, whereas the detached soybean leaves sprayed with dsRNA of ATC, GCS_H, and RP_S16 developed an average of 1.5, 1.6, and 1.6 pustules per cm2, respectively, which are significantly lower values than for the control (Figure 6b). Soybean leaves sprayed with dsRNA of ATC, GCS_H, and RP_S16 also showed significant reduction (α = 0.01) in P. pachyrhizi biomass, which was about 17.0%, 20.9%, and 25.1%, respectively, of that in the detached soybean leaves sprayed with water (Figure 6c). The expression of corresponding P. pachyrhizi genes for ATC, GCS_H, and RP_S16 was also reduced to 32.9%, 32.4%, and 28.2%, respectively, of that in detached soybean leaves treated with H2O based on quantitative reverse transcription PCR (RT‐qPCR) (Figure 6d).
FIGURE 6.
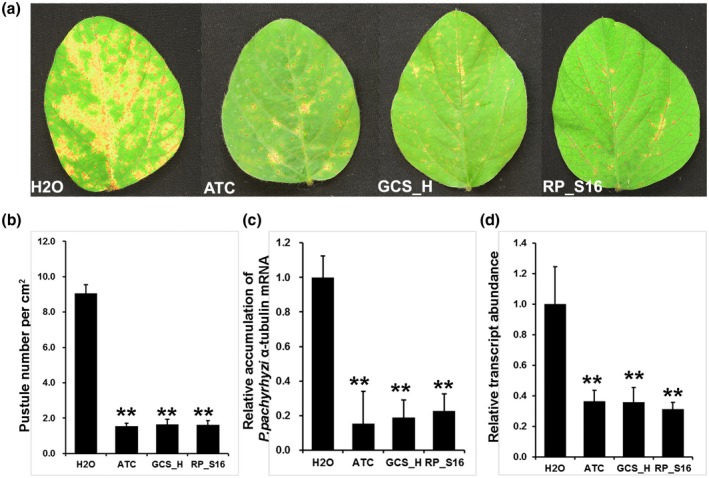
Responses of detached soybean (Williams 82) leaves that had been pretreated with or without dsRNAs of Phakopsora pachyrhizi genes prior to inoculation with P. pachyrhizi. (a) Differences in rust symptom development on soybean leaves 2 weeks after inoculation with H2O (negative control) or with dsRNAs targeting various P. pachyrhizi genes. ATC, acetyl‐CoA acyltransferase; GCS_H, glycine cleavage system H protein; RP_S16, 40S ribosomal protein S16. (b) Pustule numbers per cm2 of leaf 2 weeks post inoculation with P. pachyrhizi of soybean leaflets that had been pretreated with water or dsRNAs targeting various P. pachyrhizi genes. Bars represent mean values ± SD of one representative experiment with five biological replicates. Asterisks indicate statistical significance: *p < .05, **p < .01 (Student's t test)
3. DISCUSSION
Lack of resistance in commercial soybean varieties and the slow process in breeding lines with high level of resistance to ASR have created an urgent need to develop alternative approaches to fungicide applications to reduce environmental pollution, operating cost to farmers, and the chance of the pathogen becoming fungicide resistant (Childs et al., 2018). The emergence of RNAi and cross‐kingdom RNAi, which has been employed to manipulate gene expression to improve specific traits or to suppress pathogen infection and disease development in crops (Baulcombe, 2004; Saurabh et al., 2014), offers a new potential for enhancing soybean resistance to ASR. Enhanced crop resistance to various fungal and oomycete pathogens using HIGS through either stable or transient expression of genes from pathogens or even direct spray application of dsRNAs targeting pathogens has been reported (Nowara et al., 2010; Chen et al., 2016; Wang and Jin, 2017; Zhu et al., 2017; Song and Thomma, 2018). This study examined the feasibility of using HIGS and SIGS as an effective alternative approach to conventional soybean breeding and fungicide application for ASR disease management.
In the present study, eight genes from P. pachyrhizi that were up‐regulated either in urediniospore germination or appressorium formation were selected and screened for their potential in suppressing ASR development through the BPMV‐based HIGS vector system. Three of the HIGS constructs that target ATC, GCS_H, and RP_S16 reduced the endogenous P. pachyrhizi transcript abundance by at least 45.4% and fungal growth (biomass) by at least 57.8% in infected soybean leaves compared to control soybean leaves infected by the BPMV:EV in both greenhouse and detached leaf studies. However, when the construct BPMV:Un_3 was screened under more controlled laboratory detached leaf assay conditions, it did not significantly suppress ASR symptom development as in greenhouse conditions, indicating that the detached leaf assay, which allows a large number of candidate genes to be screened simultaneously, may be more reliable with fewer false positives.
The presence of gene‐specific siRNAs in soybean leaves treated with the HIGS constructs has also been demonstrated through analysis of sRNA‐Seq data. In addition, these gene‐specific siRNAs could only be found in soybean leaves inoculated with the corresponding HIGS constructs, indicating the successful expression of these constructs in soybean plants. This result further confirms that the reduction of ATC, GCS_H, and RP_S16 transcript abundance in respective HIGS construct inoculated leaves was due to the presence of gene‐specific siRNA expressed by those HIGS constructs.
Several recent studies have demonstrated that direct spraying of dsRNA can reduce F. graminearum infection on barley leaves (Koch et al., 2016) and S. sclerotiorum infection in canola roots (McLoughlin et al., 2018). In the present study, direct spraying of dsRNA resulted in a reduction in the number of pustules per cm2 of leaf, fungal biomass, and endogenous target gene expression by at least 68% compared to the control. The results from this study and previous reports demonstrate that this novel way of exploring RNAi is effective and has great potential for fungal disease control.
However, it is still unclear how the siRNA produced in the host cells or the dsRNA applied in vitro travels to the pathogen cells to suppress the expression of its target genes. Cai et al. (2018) reported that host plants secrete extracellular sRNA‐containing vesicles during pathogen infection, which have been observed in the extrahaustorial matrix (Micali et al., 2011). In addition, transport of RNA between plants and fungi appears to be bidirectional. sRNAs from B. cinerea have been shown to target host defence genes in Arabidopsis and tomato (Weiberg et al., 2013). Verticillium dahliae recovered from infected cotton plants contained 28 miRNAs from cotton, implying that host‐derived sRNAs were transmitted into the pathogen during infection (Liu et al., 2016). Furthermore, the transportation of endogenous sRNAs from host cells into fungal cells may not be a simple concentration‐dependent diffusion process but rather a more selective movement (Cai et al., 2018).
Several studies have revealed that the effects of HIGS are generally not observed until after the formation of haustoria, and silencing is more effective against genes that are highly expressed in haustoria than genes expressed in other cell types (Nowara et al., 2010; Yin et al., 2011; Panwar et al., 2013). Therefore, the transfer of siRNA into pathogens is thought to occur across haustoria or similar feeding structures and the plant extracellular vesicles are the most likely candidates for delivering RNAs into pathogens (Nowara et al., 2010; Micali et al., 2011; Panwar et al., 2013). In the soybean–P. pachyrhizi pathosystem, the HIGS signal molecules are likely to travel from host to the fungus via the haustorial interphase utilizing the mechanism of vesicle‐mediated (exosomes) transport (Qi et al., 2018). However, further experimental evidence is needed to support this hypothesis. Only three out of eight genes suppressed P. pachyrhizi infection in the present study, which is probably because P. pachyrhizi infection cannot be suppressed by silencing certain genes that are not critical during the initial infection process. It is also possible that sRNAs generated from the other five genes might not be able to be effectively transported into P. pachyrhizi. Further study is necessary to dissect the mechanism of siRNA or dsRNA transportation between soybean and P. pachyrhizi, which will help to select better candidate genes.
Once the candidate genes that are effective in reducing target gene expression, fungal biomass accumulation, and number of pustules per cm2 of leaf in the infected soybean leaves have been identified, one of the next natural steps is to develop stable transgenic soybean plants expressing a HIGS construct that targets multiple P. pachyrhizi genes simultaneously to obtain durable protection of soybean plants against P. pachyrhizi. However, managing soybean fungal diseases through a transgenic approach has its own limitations: acquiring regulatory approval of transgenic materials before going to market is a time‐consuming process and the negative public perceptions about genetically modified organisms in general are another concern. Therefore, the current study also explored the potential of SIGS in managing ASR following the successful studies on managing other plant fungal diseases using dsRNA as reported by Koch et al. (2016) and Wang et al. (2016). Spraying dsRNAs has been widely used previously as “oral insecticides” to control plant pests (San Miguel and Scott, 2016). SIGS can be designed to suppress multiple genes to achieve simultaneous control of a wide range of pests and pathogens without the need for a genetic engineering process (Koch et al., 2016; Wang and Jin, 2017). The present study also demonstrated that direct spraying of dsRNAs specific to pathogen genes, such as P. pachyrhizi ATC, GCS_H, and RP_S16, can be an effective means to manage soybean rust disease. Future studies should focus on improving the efficacy and duration of plant protection by the sprayed dsRNAs with the use of nontoxic, degradable, layered double hydroxide clay nanosheets (Mitter et al., 2017).
In summary, a BPMV‐based HIGS system was evaluated in the present study to explore the feasibility for ASR control. Among the eight selected P. pachyrhizi genes, three of them (ATC, GCS_H, and RP_S16) reduced the number of pustules per cm2 of leaf, fungal biomass accumulation, and target gene expression in the soybean leaves containing the corresponding HIGS constructs. This suppression was probably due to the presence of gene‐specific sRNAs produced by inoculated BPMV constructs. In addition, direct spraying of dsRNAs (ATC, RP_16S, and GCS_H) significantly reduced the number of pustules per cm2 of leaf, fungal biomass, and endogenous target gene transcript abundance in detached leaf assays. Our study demonstrated that HIGS or SIGS using genes from P. pachyrhizi can be a new and effective approach for managing ASR on soybean and possibly other fungal pathogens. To the best of our knowledge this is the first report of suppressing P. pachyrhizi infection in soybean through HIGS and direct dsRNA treatment. Future studies are needed to fine‐tune the procedures to improve the effectiveness of direct spray application of dsRNA for ARS management under field conditions.
4. EXPERIMENTAL PROCEDURES
4.1. Selection of fungal candidate genes and HIGS vector construction
The available P. pachyrhizi cDNA sequences that were up‐regulated in germinating urediniospores or during appressorium formation (Posada‐Buitrago and Frederick, 2005; Link et al., 2014) were searched through BLAST analysis to generate a shortlist of eight genes that might be involved in infection and pathogenicity (Table 1). Based on the cDNA sequences, a 200–400 bp fragment of each gene was selected and tested by the SI‐FI software tool (http://labtools.ipk-gatersleben.de/) to avoid off‐targets in the soybean transcriptome. Gene‐specific primers containing a BamHI restriction site at each end were designed for cloning into pBPMV‐IA‐V2, which was modified from pBPMV‐IA‐R2 with multiple cloning sites (Zhang et al., 2009).
Total RNA from P. pachyrhizi was isolated from germinated urediniospores (average viability 70%–80%) that were previously collected from a soybean field in 2013 at Ben Hur Research Station (Baton Rouge, USA) using the RNeasy Plant Mini Kit (Qiagen). First‐strand cDNA synthesis was performed with 2 µg of total RNA and a High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The primer pairs listed in Table 1 were used for PCR amplification with cDNA as templates, and the PCR products were cloned into pCR2.1‐TOPO (Invitrogen). Inserted sequences were confirmed by sequencing using M13 forward and reverse primers before being cloned into the pBPMV‐IA‐V2 vector.
4.2. BPMV virus inoculum production
To generate virus inoculum for the HIGS experiments, plasmid DNA of BPMV RNA1 (pBPMV‐IA‐R1M, which induces moderate symptoms on inoculation compared to wild‐type RNA1 clone pBPMV‐IA‐R1) and the recombinant RNA2 (pBPMV‐IA‐V2 containing P. pachyrhizi genes) were used to coat gold particles (cat. #165‐2263, Bio‐Rad) and bombarded into the primary leaves of Williams 82 soybean plants 14 days after sowing, as previously described (Zhang et al., 2009). BPMV‐infected leaf tissue was collected at 3–5 weeks after bombardment, lyophilized and stored at 4 °C before being ground in 20 ml of 50 mM potassium phosphate (pH 7.0) for 3 g of leaf tissue and used as primary BPMV inoculum.
4.3. RNA sequencing to confirm the presence of target gene‐specific sRNAs in HIGS‐treated soybean leaves
Fourteen days after P. pachyrhizi inoculation, leaves with ASR symptoms from greenhouse‐grown plants that had been previously rub‐inoculated with modified BPMV viruses were collected and frozen in liquid nitrogen before being stored at −80 °C until RNA extraction. Total RNAs extracted from five independent biological replicates of leaf samples containing EV only or BPMV with one of the three P. pachyrhizi genes (ATC, GCS_H or RP_S16) were checked for quality using NanoDrop and 1.5% agarose gel before being pooled together for sRNA library construction using a TruSeq Small RNA Library Preparation kit according to the manufacturer's instructions (Illumina). The single‐end 50 cycle sequencing was performed using an Illumina HiSeq 4,000 platform at Genome Sequencing Core at UC Davis (Davis, California, USA), which produces 50 single‐end reads. The bioinformatics pipeline used in the sRNA analysis was modified from Tian et al. (2017). After low‐quality and adapter sequences were removed by cutadapt (Martin, 2011), sRNA reads ranging from 18 to 28 nt were used for further analyses. Bowtie v. 1.2.0 (Langmead et al., 2009) was used to align sRNA reads, allowing non‐mismatch against sequences inserted into HIGS constructs. Linux command lines were used to extract sRNA specific to targeted P. pachyrhizi genes from alignment result files. R (R Development Core Team, 2013) was used to generate sRNA mapping figures.
4.4. HIGS study of target genes using whole plants in the greenhouse
To get even virus symptom and silencing efficiency, 20 primary leaves of 2‐week‐old Williams 82 seedlings grown in a growth chamber for each HIGS construct were dusted with carborundum and subsequently rub‐inoculated with primary inoculum corresponding to each HIGS construct. BPMV‐inoculated plants were maintained in the growth chamber at 20 °C with a 16‐hr photoperiod. Three days following BPMV infection, plants were transferred to the greenhouse. Three weeks after BPMV rubbing inoculation, the plants were inoculated with urediniospores (105/ml) of P. pachyrhizi resuspended in sterile water containing 0.01% Tween 20 and then placed in a dew chamber for 48 hr. Plants were then moved back to the greenhouse. In addition to the HIGS vector‐inoculated plants, three control treatments were included: the mock control (mock‐inoculation: the same experimental conditions as the HIGS‐treated plants but rub‐inoculated with inoculation buffer only), empty vector (EV) control (plants that were inoculated with a BPMV vector lacking an insert), and healthy control (plants that were not treated). All of these control plants were inoculated with P. pachyrhizi urediniospores as described for the experimental plants. For each construct/treatment, five plants (replicates) were usually used for data collection and final analysis.
4.5. HIGS constructs confirmation, target gene silencing efficiency, and fungal growth assessment
Changes in soybean resistance to P. pachyrhizi, such as pustule density, were evaluated using soybean leaves collected at 2 weeks post‐inoculation with P. pachyrhizi. After being photographed to allow counting of pustule density, soybean leaves were ground in liquid nitrogen, and total RNA was isolated and first‐strand cDNA was synthesized as described above. Fungal biomass was assessed by quantifying the constitutively expressed pathogen α‐tubulin gene (van de Mortel et al., 2007; Meyer et al., 2009). Expression data of fungal target genes were normalized to the soybean ubiquitin‐3 gene (GenBank accession number D28123.1), which showed stable expression following P. pachyrhizi infection (van de Mortel et al., 2007). The expression of P. pachyrhizi target genes was determined by RT‐qPCR with P. pachyrhizi in HIGS and EV‐treated plants at 14 days post‐inoculation (dpi) and was normalized to the P. pachyrhizi CytB gene (Table 3). Real‐time PCRs in a final volume of 20 µl containing 2 µl of cDNA and 0.3 µM of each primer were run on a CFX96 Real‐Time‐PCR System (Bio‐Rad) under standard conditions with melting curve analysis at the end of cycles. The ∆∆C t method was used to calculate the target gene relative expression level (Schmittgen and Livak, 2008). The data presented here are the mean values ± SD from five biological replicates.
TABLE 3.
List of primers used for real‐time quantitative reverse transcription PCR and in vitro double stranded RNA synthesis
| Gene product | Primer name and sequence (5ʹ–3ʹ) | Amplicon size (bp) | |
|---|---|---|---|
| Glycine max Ubiquitin‐3 | U3F | GTGTAATGTTGGATGTGTTCCC | 107 |
| U3R | ACACAATTGAGTTCAACACAAACCG | ||
| P. pachyrhizi α‐tubulin | TUBF | CCAAGGCTTCTTCGTGTTTCA | 67 |
| TUBR | CAAGAGAAGAGCGCCAAACC | ||
| TaqMan probe | 5ʹ FAM–3ʹ Blackhole1 | TCGTTTGGAGGCGGATCGGTTCA | |
| CytB_1_F | TCAAGACGCATCCAATTCTAGGTC | 89 | |
| CytB_1_R | GTGTTACACCCGTGATAATCTGAATGAT | ||
| PPATC_F | GAGGAGCTGCAAATGGGTGA | 115 | |
| PPATC_R | GAATGGGGATGGCAGCATCA | ||
| PP_Un_3_F | CGGATCTCAGGATCTCACACG | 91 | |
| PP_Un_3_R | GCCAGCTCTCCAGGCTTAAA | ||
| PP_GCS_H_F | AGCCGTCGAGAGTGTCAAAG | 110 | |
| PP_GCS_H_R | AGATTGGCCTGGTCGCTTAG | ||
| PP_RP_S16_F | AAATTTGGAGGAAAGGGAGCACG | 89 | |
| PP_RP_S16_R | ACAGCAGGAAATAAAAACCCAAACC | ||
| P. pachyrhizi ATC dsRNA | ATC_T7_F | GCGTAATACGATCCATCATAGGGAGAGTTTCCATCGATGATGGCATTC | 741 |
| ATC_T7_R | GCGTAATACGATCCATCATAGGGAGA CTAGCAAGCCAAACCATAAGC | ||
| P. pachyrhizi GCS_H dsRNA | GCS_H_T7_F | GCGTAATACGATCCATCATAGGGAGAAAACCATCGTAATCTGCCCG | 420 |
| GCS_H_T7_R | GCGTAATACGATCCATCATAGGGAGATCCTCTCCTTCACAATGAGCT | ||
| P. pachyrhizi RP_S16 dsRNA | RP_S16_T7_F | GCGTAATACGATCCATCATAGGGAGACCAGCCTCACAAAACGTTCA | 500 |
| RP_S16_T7_R | GCGTAATACGATCCATCATAGGGAGAAACCCAAACCTCCATCAACG | ||
4.6. HIGS study using detached soybean leaves
Five trifoliate leaves from greenhouse‐grown soybean plants that had been previously rub‐inoculated with various HIGS vectors were collected 2 weeks later for each construct and then inoculated with P. pachyrhizi urediniospores (105/ml) resuspended in sterile water containing 0.01% Tween 20. Following urediniospore inoculation, the leaves were placed in transparent square boxes (230 × 230 × 17 mm) with wetted paper towel and maintained under continuous light at 25 °C. Leaves were then evaluated for pustule number per cm2 2 weeks after inoculation with P. pachyrhizi. After evaluation, leaf tissues were ground in liquid nitrogen and total RNAs were isolated for quantification of target gene suppression and fungal biomass as described above.
4.7. Synthesis of dsRNA in vitro and spray application
Gene‐specific primer pairs with T7 promoter sequence at the 5′ end (Table 3) were used for in vitro dsRNA synthesis using MEGAscript High Yield Transcription Kit following the manufacturer’s instructions (Ambion). Synthesized dsRNA was quantified using NanoDrop and stored at −80 °C. For spray application, dsRNA was diluted to a final concentration of 20 µg/ml with diethyl pyrocarbonate‐treated water. Each box containing six detached individual leaflets was evenly sprayed with 1 ml of diluted dsRNA (20 μg dsRNA). After spraying, boxes were kept open until the surface of each individual leaflet was dry (approximately 1.5 hr). Leaves were then spray‐inoculated with 1 ml of 105 urediniospores/ml 2 hr after the dsRNA spray. Boxes were closed and incubated for 2 weeks at approximately 25 °C on the laboratory bench with continuous lights Two controls were included in this study: one was soybean leaflets that were sprayed with H2O and the other was sprayed with dsRNA targeting the soybean PDS gene. The latter was used to determine whether dsRNA can enter soybean leaves. No significant differences in pustule numbers per cm2, P. pachyrhizi tubulin level, and target gene expressions were observed between the water‐treated and PDS dsRNA‐treated soybean leaves, therefore only the data for the water‐treated control are presented here.
4.8. Statistical analysis
All experiments were repeated and yielded reproducible results. The most representative data are shown in this paper. A two‐tailed Student's t test was performed with data gained from greenhouse inoculation, detached leaf inoculation, dsRNA spray assays, and RT‐qPCR studies. Data are presented as means ± SD of the mean. p values <.05 were considered significant.
5. COMPETING INTERESTS
The authors declare no competing financial or non‐financial interests.
AUTHOR CONTRIBUTIONS
Z‐Y.C. and D.H. designed the study and wrote the manuscript. D.H. conducted the experiments and M.G. identified the ATC. C.Z. provided the BPMV constructs and advised the virus inoculation. D.H. performed the bioinformatics analysis of sRNA data. D.H. and Z‐Y.C. analysed all data and drafted the figures.
ACKNOWLEDGEMENTS
This study was supported by the Louisiana State Soybean and Small Grain Promotion Board from 2012 to 2019 and the Louisiana Board of Regents Grants LEQSF‐2008‐11‐RD‐A‐01 and LEQSF(2015‐16)‐ENH‐TR‐02. Published with the approval of the Director of the Louisiana State University Agricultural Center Agricultural Experiment Station as manuscript number 2019‐ 240‐34069.
Hu D, Chen Z‐Y, Zhang C, Ganiger M. Reduction of Phakopsora pachyrhizi infection on soybean through host‐ and spray‐induced gene silencing. Molecular Plant Pathology. 2020;21:794–807. 10.1111/mpp.12931
Funding information
This study was supported by the Louisiana State Soybean and Small Grain Promotion Board from 2012 to 2019 and the Louisiana Board of Regents Grants LEQSF‐2008‐11‐RD‐A‐01 and LEQSF(2015‐16)‐ENH‐TR‐02.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bromfield, K.R. (1984) Soybean Rust. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Cai, Q. , Qiao, L. , Wang, M. , He, B. , Lin, F.‐M. , Palmquist, J. et al (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science, 360, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, P. , McLaren, N. , Moscardi, F. , Hoffmann‐Campo, C. , Saraiva, O. , Galerani, P. et al (2004) Soybean rust research in South Africa In: Moscardi F., Hoffmann‐Campo C.B., Saraiva O.F., Galerani P.R., Krzyzanowski F.C., Carrão‐Panizzi M.C. (Eds.) Proceedings VII World Soybean Research Conference, IV International Soybean Processing and Utilization Conference, III Congresso Brasileiro de Soja (Brazilian Soybean Congress), 29 February‐5 March, 2004. Foz do Iguassu, PR, Brazil: Brazilian Agricultural Research Corporation, National Soybean Research Center, pp. 354–360. [Google Scholar]
- Chen, W.X. , Kastner, C. , Nowara, D. , Oliveira‐Garcia, E. , Rutten, T. , Zhao, Y.S. et al (2016) Host‐induced silencing of Fusarium culmorum genes protects wheat from infection. Journal of Experimental Botany, 67, 4979–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs, S.P. , Buck, J.W. and Li, Z. (2018) Breeding soybeans with resistance to soybean rust (Phakopsora pachyrhizi). Plant Breeding, 137, 250–261. [Google Scholar]
- Cooper, B. , Campbell, K.B. , McMahon, M.B. and Luster, D.G. (2013) Disruption of Rpp1‐mediated soybean rust immunity by virus‐induced gene silencing. Plant Signaling and Behavior, 8, e27543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho, M.C.D.C. , Costa Nascimento, L. , Darben, L.M. , Polizel‐Podanosqui, A.M. , Lopes‐Caitar, V.S. , Qi, M. et al (2017) Prediction of the in planta Phakopsora pachyrhizi secretome and potential effector families. Molecular Plant Pathology, 18, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghag, S.B. , Shekhawat, U.K.S. and Ganapathi, T.R. (2014) Host‐induced post‐transcriptional hairpin RNA‐mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnology Journal, 12, 541–553. [DOI] [PubMed] [Google Scholar]
- Godoy, C.V. (2012) Risk and management of fungicide resistance in the Asian soybean rust fungus Phakopsora pachyrhizi In: Thind T.S. (Ed.) Fungicide resistance in crop protection: risk and management. Wallingford, UK: CAB International, pp. 87–95. [Google Scholar]
- Godoy, C.V. , Bueno, A.D.F. and Gazziero, D.L.P. (2015) Brazilian soybean pest management and threats to its sustainability. Outlooks on Pest Management, 26, 113–117. [Google Scholar]
- Grasso, V. , Sierotzki, H. , Garibaldi, A. and Gisi, U. (2006) Relatedness among agronomically important rusts based on mitochondrial cytochrome b gene and ribosomal ITS sequences. Journal of Phytopathology, 154, 110–118. [Google Scholar]
- Hartman, G.L. , Wang, T.C. and Tschanz, A.T. (1991) Soybean rust development and the quantitative relationship between rust severity and soybean yield. Plant Disease, 75, 596–600. [Google Scholar]
- Hartwig, E.E. (1986) Identification of a 4th major gene conferring resistance to soybean rust. Crop Science, 26, 1135–1136. [Google Scholar]
- Hartwig, E.E. and Bromfield, K.R. (1983) Relationships among three genes conferring specific resistance to rust in soybeans. Crop Science, 23, 237–239. [Google Scholar]
- Hershman, D.E. , Sikora, E.J. and Giesler, L.J. (2011) Soybean rust PIPE: past, present, and future. Journal of Integrated Pest Management, 2, D1–D7. [Google Scholar]
- Jahan, S.N. , Åsman, A.K.M. , Corcoran, P. , Fogelqvist, J. , Vetukuri, R.R. and Dixelius, C. (2015) Plant‐mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans . Journal of Experimental Botany, 66(9), 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Biedenkopf, D. , Furch, A. , Weber, L. , Rossbach, O. , Abdellatef, E. et al (2016) An RNAi‐based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathogens, 12, e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biology, 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, T.I. , Lang, P. , Scheffler, B.E. , Duke, M.V. , Graham, M.A. , Cooper, B. et al (2014) The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Molecular Plant Pathology, 15, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Z. and Whitham, S.A. (2013) Overexpression of a soybean nuclear localized type–III DnaJ domain‐containing HSP40 reveals its roles in cell death and disease resistance. The Plant Journal, 74, 110–121. [DOI] [PubMed] [Google Scholar]
- Liu, S. , da Cunha, A.P. , Rezende, R.M. , Cialic, R. , Wei, Z. , Bry, L. et al (2016) The host shapes the gut microbiota via fecal microRNA. Cell Host and Microbe, 19, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehrer, M. , Vogel, A. , Huettel, B. , Reinhardt, R. , Benes, V. , Duplessis, S. et al (2014) On the current status of Phakopsora pachyrhizi genome sequencing. Frontiers in Plant Science, 5, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet. Journal, 17, 10–12. [Google Scholar]
- McLean, R.J. and Byth, D. (1980) Inheritance of resistance to rust (Phakopsora pachyrhizi) in soybean. Australian Journal of Agricultural Research, 31, 951–956. [Google Scholar]
- McLoughlin, A.G. , Wytinck, N. , Walker, P.L. , Girard, I.J. , Rashid, K.Y. , de Kievit, T. et al (2018) Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea . Scientific Reports, 8, 7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J.D.F. , Silva, D.C.G. , Yang, C. , Pedley, K.F. , Zhang, C. , van de Mortel, M. et al (2009) Identification and analyses of candidate genes for Rpp4‐mediated resistance to Asian soybean rust in soybean. Plant Physiology, 150, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, C.O. , Neumann, U. , Grunewald, D. , Panstruga, R. and O'connell, R. (2011) Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cellular Microbiology, 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Miles, M. , Levy, C. , Morel, W. , Mueller, T. , Steinlage, T. , Van Rij, N. et al (2007) International fungicide efficacy trials for the management of soybean rust. Plant Disease, 91, 1450–1458. [DOI] [PubMed] [Google Scholar]
- Mitter, N. , Worrall, E.A. , Robinson, K.E. , Li, P. , Jain, R.G. , Taochy, C. et al (2017) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nature Plants, 3, 16207. [DOI] [PubMed] [Google Scholar]
- van de Mortel, M. , Recknor, J.C. , Graham, M.A. , Nettleton, D. , Dittman, J.D. , Nelson, R.T. et al (2007) Distinct biphasic mRNA changes in response to Asian soybean rust infection. Molecular Plant‐Microbe Interactions, 20, 887–899. [DOI] [PubMed] [Google Scholar]
- Mueller, T.A. , Miles, M.R. , Morel, W. , Marois, J.J. , Wright, D.L. , Kemerait, R.C. et al (2009) Effect of fungicide and timing of application on soybean rust severity and yield. Plant Disease, 93, 243–248. [DOI] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , et al. (2010) HIGS: Host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . The Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar, V. , McCallum, B. and Bakkeren, G. (2013) Host‐induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the barley stripe mosaic virus. Plant Molecular Biology, 81, 595–608. [DOI] [PubMed] [Google Scholar]
- Posada‐Buitrago, M.L. and Frederick, R.D. (2005) Expressed sequence tag analysis of the soybean rust pathogen Phakopsora pachyrhizi . Fungal Genetics and Biology, 42, 949–962. [DOI] [PubMed] [Google Scholar]
- Qi, M. , Link, T.I. , Müller, M. , Hirschburger, D. , Pudake, R.N. , Pedley, K.F. et al (2016) A small cysteine‐rich protein from the Asian soybean rust fungus, Phakopsora pachyrhizi, suppresses plant immunity. PLoS Pathogens, 12, e1005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, T. , Zhu, X. , Tan, C. , Liu, P. , Guo, J. , Kang, Z. et al (2018) Host‐induced gene silencing of an important pathogenicity factor P s CPK 1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnology Journal, 16, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raruang, Y. , Omolehin, O. , Hu, D. , Wei, Q. , Han, Z.Q. , Rajasekaran, K. , et al. (2020) Host induced gene silencing targeting Aspergillus flavus aflM reduced aflatoxin contamination in transgenic maize under field conditions. Frontiers in Microbiology, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . (2013) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Available at: https://www.R-project.org/ [Accessed 11 March 2020]. [Google Scholar]
- San Miguel, K. and Scott, J.G. (2016) The next generation of insecticides: dsRNA is stable as a foliar‐applied insecticide. Pest Management Science, 72, 801–809. [DOI] [PubMed] [Google Scholar]
- Saurabh, S. , Vidyarthi, A.S. and Prasad, D. (2014) RNA interference: concept to reality in crop improvement. Planta, 239, 543–564. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative Ct method. Nature Protocols, 3, 1101. [DOI] [PubMed] [Google Scholar]
- Schmitz, H.K. , Medeiros, C.A. , Craig, I.R. and Stammler, G. (2014) Sensitivity of Phakopsora pachyrhizi towards quinone‐outside‐inhibitors and demethylation‐inhibitors, and corresponding resistance mechanisms. Pest Management Science, 70, 378–388. [DOI] [PubMed] [Google Scholar]
- Sharma, K.K. , Pothana, A. , Prasad, K. , Shah, D. , Kaur, J. , Bhatnagar, D. et al (2018) Peanuts that keep aflatoxin at bay: a threshold that matters. Plant Biotechnology Journal, 16, 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. and Thomma, B. (2018) Host‐induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Molecular Plant Pathology, 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, C.L. , McMahon, M.B. , Fortis, L.L. , Nunez, A. , Smythers, G.W. , Luster, D.G. et al (2012) Gene expression and proteomic analysis of the formation of Phakopsora pachyrhizi appressoria. BMC Genomics, 13, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare, D. , Zhang, J. , Wing, R.A. , Cotty, P.J. and Schmidt, M.A. (2017) Aflatoxin‐free transgenic maize using host‐induced gene silencing. Science Advances, 3, e1602382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B. , Wang, S. , Todd, T.C. , Johnson, C.D. , Tang, G. and Trick, H.N. (2017) Genome‐wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genomics, 18, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, A. , Hosseini, P. , Alkharouf, N.W. , Li, S. and Matthews, B.F. (2010) Transcriptome analysis of a compatible response by Glycine max to Phakopsora pachyrhizi infection. Plant Science, 179, 183–193. [Google Scholar]
- Tremblay, A. , Hosseini, P. , Li, S. , Alkharouf, N.W. and Matthews, B.F. (2012) Identification of genes expressed by Phakopsora pachyrhizi, the pathogen causing soybean rust, at a late stage of infection of susceptible soybean leaves. Plant Pathology, 61, 773–786. [Google Scholar]
- Tremblay, A. , Hosseini, P. , Li, S.X. , Alkharouf, N.W. and Matthews, B.F. (2013) Analysis of Phakopsora pachyrhizi transcript abundance in critical pathways at four time‐points during infection of a susceptible soybean cultivar using deep sequencing. BMC Genomics, 14, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.R. , Harris, D.K. , King, Z.R. , Li, Z. , Boerma, H.R. , Buckley, J.B. , et al. (2014) Evaluation of soybean germplasm accessions for resistance to Phakopsora pachyrhizi populations in the southeastern United States, 2009–2012. Crop Science, 54, 1673–1689. [Google Scholar]
- Wang, M. and Jin, H. (2017) Spray‐induced gene silencing: a powerful innovative strategy for crop protection. Trends in Microbiology, 25, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.‐M. , Thomma, B.P.H.J. , Huang, H.‐D. and Jin, H. (2016) Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature Plants, 2, 16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg, A. , Wang, M. , Lin, F.‐M. , Zhao, H. , Zhang, Z. , Kaloshian, I. et al (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science, 342, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, C. , Jurgenson, J.E. and Hulbert, S.H. (2011) Development of a host‐induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . Molecular Plant‐Microbe Interactions, 24, 554–561. [DOI] [PubMed] [Google Scholar]
- Yorinori, J.T. , Paiva, W.M. , Frederick, R.D. , Costamilan, L.M. , Bertagnolli, P.F. , Hartman, G.E. et al (2005) Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease, 89, 675–677. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Whitham, S.A. and Hill, J.H. (2013) Virus‐induced gene silencing in soybean and common bean In: Becker A. (Ed.) Virus‐Induced Gene Silencing: Methods and Protocols. Totowa, NJ: Humana Press, pp. 149–156. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Yang, C.L. , Whitham, S.A. and Hill, J.H. (2009) Development and use of an efficient DNA‐based viral gene silencing vector for soybean. Molecular Plant‐Microbe Interactions, 22, 123–131. [DOI] [PubMed] [Google Scholar]
- Zhang, C.Q. , Bradshaw, J.D. , Whitham, S.A. and Hill, J.H. (2010) The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiology, 153, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.G. , Qi, T. , Yang, Q. , He, F.X. , Tan, C.L. , Ma, W. et al (2017) Host‐induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiology, 175, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


