Abstract
Grey mould is one of the most determinative factors of lily growth and plays a major role in limiting lily productivity. MicroRNA159 (miR159) is a highly conserved microRNA in plants, and participates in the regulation of plant development and stress responses. Our previous studies revealed that lre‐miR159a participates in the response of Lilium regale to Botrytis elliptica according to deep sequencing analyses; however, the response mechanism remains unknown. Here, lre‐miR159a and its target LrGAMYB gene were isolated from L. regale. Transgenic Arabidopsis overexpressing lre‐MIR159a exhibited larger leaves and smaller necrotic spots on inoculation with Botrytis than those of wild‐type and overexpressing LrGAMYB plants. The lre‐MIR159a overexpression also led to repressed expression of two targets of miR159, AtMYB33 and AtMYB65, and enhanced accumulation of hormone‐related genes, including AtPR1, AtPR2, AtNPR1, AtPDF1.2, and AtLOX for both the jasmonic acid and salicylic acid pathways. Moreover, lower levels of H2O2 and were observed in lre‐MIR159a transgenic Arabidopsis, which reduced the damage from reactive oxygen species accumulation. Taken together, these results indicate that lre‐miR159a positively regulates resistance to grey mould by repressing the expression of its target LrGAMYB gene and activating a defence response.
Keywords: Botrytis, GAMYB gene, Lilium, miR159, plant–pathogen interaction, transgenics
lre‐miR159a positively regulates resistance to grey mould by repressing the expression of its target LrGAMYB gene and activating a defence response in Lilium regale.

1. INTRODUCTION
Lily (Lilium spp.), as one of the most important ornamental plants in the word, can be used as both a cultivated flower crop and a potted plant. However, during both pre‐ and postharvest, especially in summer, high relative humidity provides an ideal environment for grey mould infection (Doss et al., 1988; Hsieh et al., 2001; van Baarlen et al., 2004). Lily grey mould is caused by the fungal pathogen Botrytis elliptica, which has a specialized interaction with lily (Hou and Chen, 2003; Furukawa et al., 2005). During the infection, spot lesions first occur on the leaves, and then the whole plant rapidly dies, causing large yield losses.
MicroRNAs (miRNAs) are small 20–24 nucleotides (nt), single‐stranded noncoding RNAs that regulate multiple biological pathways in complex organisms. Plant miRNAs mainly cleave target messenger RNAs (mRNAs) and thus regulate their expression to function in many plant biological and metabolic processes. Many plant miRNAs have been reported to respond to abiotic challenge, suggesting a broader involvement of miRNAs in defence (Jones‐Rhoades and Bartel, 2004; Phillips et al., 2007; Shanfa et al., 2010). For example, miR398 expression was down‐regulated by oxidative stresses, and the expression of its target genes Cu/Zn SUPEROXIDE DISMUTASES was increased to protect plants from oxidative damage during SO2 exposure (Li et al., 2017). In addition, the overexpression of miR393a causes plants to become tolerant to drought stress, salt stress, and heat stress (Zhao et al., 2018). Moreover, miRNAs have also been shown to be pivotal molecules in plant–pathogen interactions. For instance, miR396 was confirmed to target a set of transcription factors in the GROWTH‐REGULATING FACTOR (GRF) family in response to the necrotrophic fungi Plectosphaerella cucumerina and Botrytis cinerea, and the hemibiotrophic fungi Fusarium oxysporum f. sp. conglutinans and Colletotrichum higginsianum. The MIM396 plant, in which miR396 activity is reduced, was more resistant to these fungal infections, whereas the overexpression of MIR396 increased plant susceptibility to fungal infections (Soto‐Suárez et al., 2017). Moreover, slmiR482e‐3p, which targets FRG3 (a novel R gene) at the transcript level, is necessary for resistance to tomato wilt disease (Ji et al., 2018). Several miRNAs have been reported to respond to B. cinerea. In tomato, miR160 and miR171a were up‐regulated, and miR169 was down‐regulated after inoculation with B. cinerea (Jin et al., 2012). In addition, miR394 was verified as a negative regulator of Arabidopsis resistance to B. cinerea by targeting LEAF CURLING RESPONSIVENESS (LCR) (Tian et al., 2018). Furthermore, large numbers of miRNAs were identified in response to grey mould in Solanum lycopersicum, Paeonia lactiflora, and Lilium regale through high‐throughput sequencing (Jin and Wu, 2015; Zhao et al., 2015; Gao et al., 2017), which might be associated with resistance to Botrytis stress.
The miR159 family is a conserved miRNA that has been found in most land plants except bryophytes (Allen et al., 2007). miR159 has been found to negatively regulate GAMYB or GAMYB‐like transcription factors, which activate gibberellin (GA)‐responsive genes (Achard et al., 2004). In Arabidopsis, seven of the miR159 targets encode proteins similar to GAMYB, all of which share conserved putative miR159‐binding sites of analogous complementarity, such as MYB33, MYB65, and MYB101 (Millar, 2005; Zheng et al., 2017). The functional role of the miR159‐MYB pathway has been analysed during seed germination floral development (Tsuji et al., 2006), and primary root growth (Xue et al., 2017). Despite this, more research has focused on the function of miR159 in response to multiple environmental stresses. In sugarcane, a progressive increase in miR159 transcripts was observed under short‐term polyethylene glycol stress with a concomitant down‐regulation of target MYB genes (Patade and Suprasanna, 2010). Similarly, root endophytic fungi induced the accumulation of miR159, enhancing the tolerance of drought stress in rice (Ehsan et al., 2017). In wheat, miR159 was up‐regulated in leaves when challenged with Puccinia striiformis f. sp. tritici, resulting in a resistant phenotype through the regulation of TaMYB3 expression (Feng et al., 2013). The overexpression of miR159 caused increased resistance to powdery mildew in Arabidopsis (Sun, 2014). All of these indicate that miR159 is a potential positive regulator in plant response to a variety of biotic and abiotic stresses.
Previously, we systematically investigated the role of different miRNAs in the response of L. regale to B. elliptica and found that the miR159 family was significantly expressed after inoculation with B. elliptica (Gao et al., 2017). To further our knowledge about the functions of miR159, unravelling its role in response to Botrytis and the underlying molecular mechanisms, we carried out this study. The miR159a and target LrGAMYB genes were identified and characterized in L. regale, and transgenic Arabidopsis plants overexpressing lre‐pre‐MIR159a were generated. Particularly interesting effects were observed in transgenic Arabidopsis plants overexpressing miR159a, which had markedly increased resistance to grey mould in comparison with wild‐type controls. The target of lre‐miR159a, LrGAMYB, is repressed in the transgenic plants. Our results reveal that the miR159a acts as a positive regulator of grey mould tolerance in Lilium.
2. RESULTS
2.1. Identification of lre‐miR159a in L. regale
We took advantage of L. regale, which shows resistance to grey mould, to clone lre‐miR159a. The length of precursor lre‐MIR159a was 219 nt, containing a 21 nt mature miR159a sequence at the 3′ end. Sequence alignment of the MIR159a precursor revealed 52.97% identity between Arabidopsis thaliana and L. regale, as shown in Figure 1a. A characteristic stem‐loop structure of pre‐lre‐MIR159a was predicted; this structure folded into a typical secondary structure (Figure 1b), with −92.40 kal/mol negative minimal folding free energies. The mature miRNA of the precursor was located in the identical stem arm (Figure 1b). This implied that the pre‐lre‐MIR159a could be processed correctly to form mature lre‐miR159a.
Figure 1.
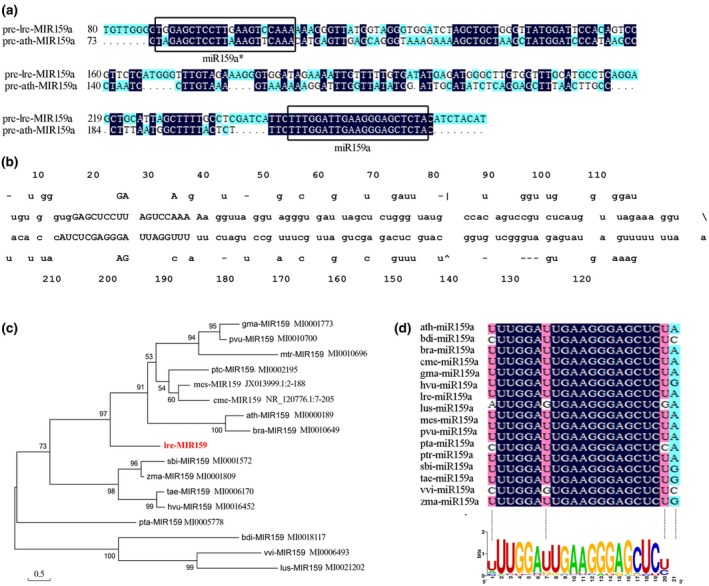
Mature and primary sequence analysis of miR159a in Lilium regale. (a) Sequence alignment of pre‐lre‐MIR159a and pre‐ath‐MIR159a. Boxes indicate the sequences of mature miR159a and miR159a*. (b) Proposed secondary structures for lre‐MIR159a precursors. (c) Phylogenetic tree of miR159a precursors in 17 species. (d) Sequence logo view of the mature miR159a sequence based on 106 miR159a sequences
Based on the stem‐loop sequences of the MIR159a precursors, a phylogenetic tree was conducted with 17 species, including L. regale and other species, as shown in Figure 1c. Two branches were observed in the phylogenetic tree: one branch consisted of bdi‐MIR159a, vvi‐MIR159a, and lus‐MIR159a, while the other branch consisted of the remaining members. MIR159a precursors were scattered among the different species in the phylogenetic tree without clustering of monocotyledonous plants, indicating the diversity of MIR159 sources. lre‐MIR159a formed an independent clade, suggesting a difference in MIR159a precursors from other species.
The mature miR159 sequence base conservation was analysed among 106 members, as shown in Figure 1d. The consensus mature miR159 sequence was 5′‐UUUGGAUUGAAGGGAGCUCUA‐3′ and shared high identity from the second to the 20th nucleotide. Mature lre‐miR159a shared the same sequence as the consensus sequence, indicating that lre‐miR159a might play a similar role as in other species.
2.2. lre‐miR159a response to B. elliptica infection
To elucidate the potential function of lre‐miR159a in the response to B. elliptica infection, the expression levels were detected by quantitative reverse transcription PCR (RT‐qPCR) among different B. elliptica infection times in the B. elliptica‐resistant L. regale and the B. elliptica‐susceptible Lilium Asiatic hybrid cultivar Tresor, the mock‐infected as control (Figure 2). The abundance of lre‐miR159a did not significantly change in the mock‐infected leaves at different time points. Compared with the expression level of lre‐miR159a in the control, the expression level of lre‐miR159a in resistant lines decreased before 24 hr post‐inoculation (hpi), and then a significantly up‐regulated expression pattern was exhibited following B. elliptica infection in resistant lilies, reaching a maximum of 2.7 times higher (Figure 2a). However, lre‐miR159a was slightly changed in susceptible Tresor during the infection process, except for a significant increase at 12 hpi. Furthermore, the levels of lre‐miR159a were significantly higher in resistant Lilium than in susceptible Lilium. The dynamic expression of lre‐miR159a in resistant Lilium during B. elliptica infection showed that lre‐miR159a was responding to B. elliptica, indicating that lre‐miR159a may have a role in resistance to grey mould.
Figure 2.
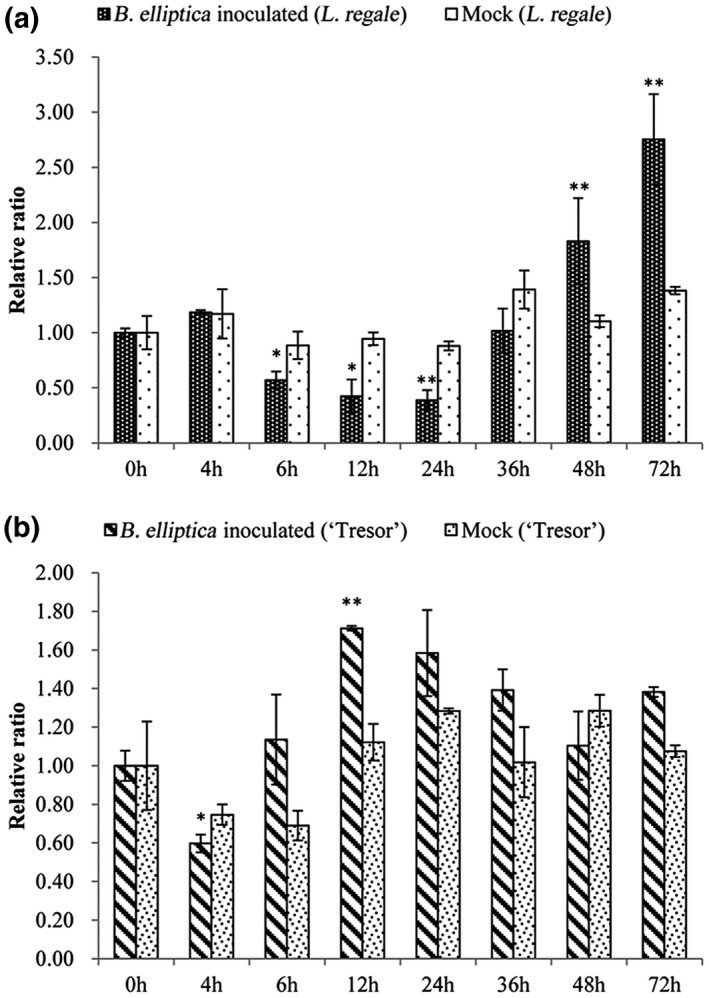
Quantitative reverse transcription PCR validations of Botrytis elliptica‐responsive miRNAs in the resistant Lilium regale and the susceptible Lilium cultivar Tresor. The level of expression was normalized to the level of 18S rRNA. The normalized miRNA levels at 0 hr were arbitrarily set to 1. Each bar shows the mean ± SE of triplicate assays. * or ** indicates a statistically significant difference as relative to the value at 0 hr for each miRNA at p < .05 or .01, respectively
2.3. LrGAMYB is targeted by miR159a through cleavage
It is well known that plant miRNAs function through cleaving target genes. Therefore, identifying target genes of mRNAs is important to understand their specific contributions. In plants, miR159 was predicted to target MYBs with a strongly conserved miR159‐binding site. To identify miR159 target genes in Lilium, psRNATarget was used to predict target genes based on the Lilium transcriptome, and one cDNA fragment, characterized as GAMYB, was selected for further analysis. Through rapid amplification of cDNA ends (RACE), the full‐length cDNA of the LrGAMYB gene was isolated, which contains a 1617 nucleotide (nt) open reading frame and encodes a 539 amino acid polypeptide. Sequence alignment of the LrGAMYB protein sequence with A. thaliana, Oryza sativa Japonica group and Zea mays sequences is shown in Figure 3a. Similar to other species, the LrGAMYB protein possesses an R2R3 DNA‐binding domain in the N‐terminus and three conserved motifs (Box1, Box2, and Box3). In addition, the LrGAMYB protein sequence shares high similarity with GAMYB in monocots, such as Elaeis guineensis (49%) and Phoenix dactylifera (48%).
Figure 3.
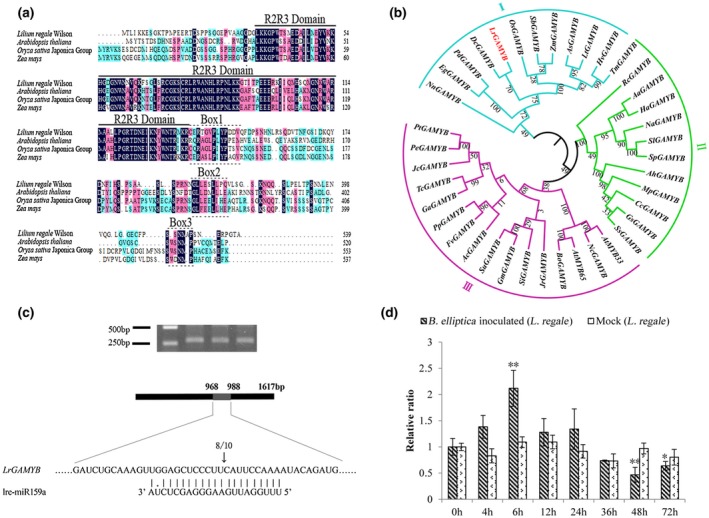
Identification and multiple sequence analysis of the miR159‐targeted LrGAMYB gene. (a) Amino acid alignment of the LrGAMYB protein with MYB homologues in Arabidopsis thaliana, Oryza sativa Japonica group, and Zea mays. (b) The phylogenetic tree of MYBs of Lilium regale and 37 species was constructed using the maximum‐likelihood method. (c) The RNA ligase‐mediated rapid amplification of cDNA ends (RLM‐RACE) products for the predicted LrGAMYB gene amplified and positions of the cleavage. Mapping of lre‐miR159a‐guided cleavage sites in LrGAMYB mRNA by RLM‐RACE. Vertical arrow indicates the 5′ position of the cleaved mRNA fragment, and the number indicates the number of independent clones analysed in the different tissues. (d) Quantitative reverse transcription PCR analysis of LrGAMYB in L. regale after Botrytis elliptica inoculation
A maximum‐likelihood phylogenetic tree was established with the LrGAMYB protein sequences and 37 homologues from several species (Figure 3b). The evolutionary tree of related genes was divided into three parts (I, II, and III). LrGAMYB formed a clade with DcGAMYB, which belongs to group I with members mainly from monocots (Figure 3b). Furthermore, the LrGAMYB gene exhibited base pairing with near‐perfect complementarity with lre‐miR159a, suggesting LrGAMYB was the putative target of lre‐miR159a.
To confirm whether LrGAMYB genes are direct targets of lre‐miR159a, a modified 5′ RNA ligase‐mediated rapid amplification of cDNA ends (RLM‐RACE) was conducted to examine the miR159‐directed cleavage sites of LrGAMYB transcripts. Generated cleavage products were amplified and cloned into a vector. The 5′ end sequencing of the amplified products was sequenced in independent clones, suggesting that cleavage sites are located in the middle of the lre‐miR159a complementary region. Cleavage occurred at an identical position, which corresponded to the 10th nucleotide position of the consensus mature miR159a sequence, in 8 of the 10 tested samples (Figure 3c). Furthermore, L. regale plants were inoculated with B. elliptica to investigate the expression pattern of LrGAMYB using RT‐qPCR analysis (Figure 3d). This revealed that LrGAMYB was up‐regulated in the initial B. elliptica infection and significantly reduced at later times. The expression level of LrGAMYB was negatively correlated with lre‐miR159a in L. regale. These results confirm that LrGAMYB is an authentic target of lre‐miR159a, and LrGAMYB is subject to miR159‐mediated down‐regulation.
2.4. lre‐miR159a positively regulates plant resistance to B. elliptica
To study the function of lre‐miR159a and LrGAMYB in plant pathogens, we constructed two plasmids overexpressing lre‐miR159a and LrGAMYB, respectively, and transformed these into the Arabidopsis Col‐0 plants, which was used as the wild type (WT). The stem‐loop precursor of lre‐MIR159a and LrGAMYB were under the control of the CaMV 35S promoter and linked to pCAMBIA1301 vector (35S:lre‐MIR159a and 35S:LrGAMYB). Multiple transgenic lines were identified through lre‐MIR159a and LrGAMYB amplification (Figure 4a). It seems that lre‐miR159a meditates plant development, resulting in increased plant size after 4 weeks of growth (Figure 4b). In addition, Arabidopsis overexpressing miR159a (OEmiR159a) was evaluated for stem elongation and early flowering. However, Arabidopsis plants overexpressing LrGAMYB (OELrGAMYB) exhibited short stature along with reduction in stem diameter and length of leaves.
Figure 4.

Phenotypic characteristics of wild‐type (WT), transgenic Arabidopsis plants overexpressing lre‐MIR159a (OEmiR159a), and overexpressing LrGAMYB (OELrGAMYB) under the control of the 35S promoter (35S:lre‐MIR159a and 35S:LrGAMYB). (a) Agarose gel analysis of lre‐miR159a and LrGAMYB genes in transgenic Arabidopsis. (b) Comparison of 4‐week‐old plants between transgenic lines and WT
To determine whether lre‐miR159a and LrGAMYB play a role in the response to Botrytis, both WT and overexpression plants were inoculated with discs of B. cinerea, and the disease progression was observed 2 days post‐inoculation (dpi). Minimal growth of Botrytis was observed on the leaf surface of the OEmiR159a, and water‐soaked symptoms appeared. However, infected lesions and necrosis were observed on WT and OELrGAMYB (Figure 5a). Necrosis reached nearly 70% on the leaves of OELrGAMYB Arabidopsis at 48 hpi (Figure 5c). Spores of B. cinerea were also used for the inoculation, and the results are shown in Figure S1, which were same for mycelial (disc) infection. Trypan blue staining further verified these results. The proliferation of the fungal mycelia was widespread in OELrGAMYB, accompanied by the development of spots and necrosis (Figure 5b). Fewer hyphae and lesions were observed in OEmiR159a, suggesting lre‐miR159a positively regulates resistance to grey mould.
Figure 5.
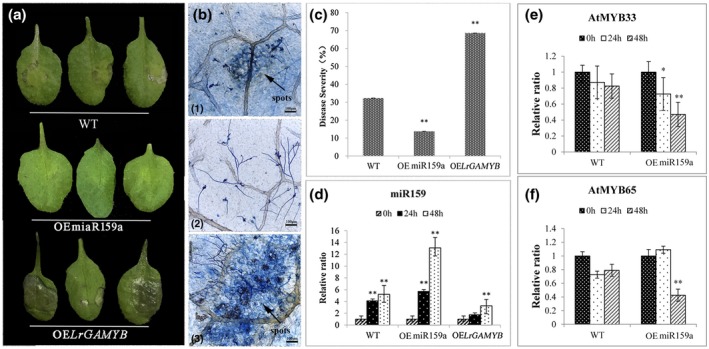
Plant disease assay following Botrytis cinerea inoculation. (a) Disease symptoms in Arabidopsis wild‐type (WT), overexpressing line OEmiR159a, and overexpressing line OELrGAMYB. Leaves were collected at 2 days after Botrytis inoculation. Similar results were observed in three to five replicate experiments. (b) Trypan blue staining of infected leaves after B. cinerea inoculation. (1) to (3) represent WT, OEmiR159a, and OELrGAMYB Arabidopsis, respectively. Scale bar = 100 μm. (c) Disease severity of WT, OEmiR159a, and OELrGAMYB Arabidopsis; (d–f) Relative expression levels of miR159a and miR159a‐targets, AtMYB33, and AtMYB65, in WT and OEmiR159a Arabidopsis after B. cinerea inoculation
Moreover, RT‐qPCR was used to verify the expression of lre‐miR159a in the WT and transgenic plants (OEmiR159a and OELrGAMYB Arabidopsis). miR159a was up‐regulated during the Botrytis infection process; however, the miR159a levels in the OEmiR159a lines were significantly higher than those in WT and OELrGAMYB Arabidopsis (Figure 5d). These results suggest that lre‐miR159a is successfully expressed in transgenic Arabidopsis and that overexpression could enhance the resistance to pathogen infection. Additionally, we further studied the expression of the miR159a‐targeted genes in Botrytis‐inoculated OEmiR159a Arabidopsis (Figure 5e,f). The miR159a‐targeted genes AtMYB33 and AtMYB65 were down‐regulated in OEmiR159a Arabidopsis during the pathogen infection process, and this expression pattern contrasted with miR159a, suggesting miR15a regulates the target GAMYB gene in response to Botrytis infection.
2.5. The expression of hormone‐related genes in lre‐miR159a overexpression Arabidopsis after infection with B. cinerea
Phytohormone signalling pathways, such as salicylic acid (SA) and jasmonic acid (JA), are involved in the control of the initiation of defence mechanisms against B. cinerea (Zhao et al., 2003; Blanco‐Ulate et al., 2013). To investigate the possible signalling pathways involved in miR159‐mediated resistance, the transcript levels of genes in the SA‐ and JA‐dependent pathways during fungal inoculation were detected. Both the SA‐responsive marker genes (AtNPR1, AtPR1, and AtPR2) and the JA‐responsive marker gene (AtPDF1.2) were significantly expressed higher in Botrytis‐inoculated OEmiR159a Arabidopsis (Figure 6) than in WT, which indicates that the OEmiR159a plants might mediate grey mould resistance by activating the SA and JA signalling pathways. The transcription of AtLOX in JA pathway was down‐regulated, suggesting that JA might play a role in later infection processes.
Figure 6.
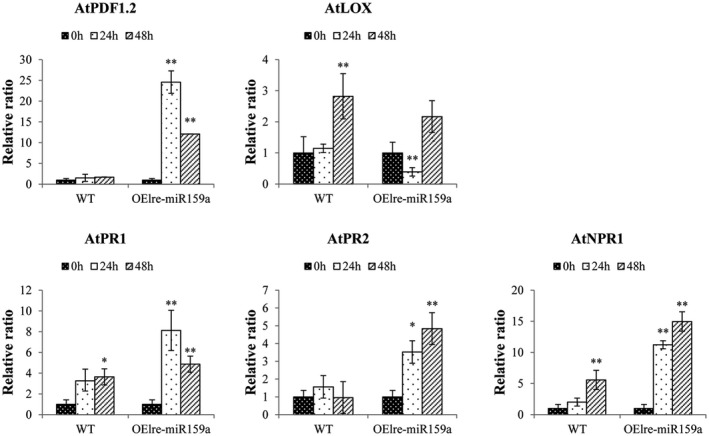
The expression of hormone‐related genes of wild‐type (WT) and Arabidopsis overexpressing miR159a (OElre‐miR159a) upon inoculation with Botrytis cinerea. The level of expression was normalized to the level of the AtActin gene. The normalized gene levels at 0 hr were arbitrarily set to 1. Each bar shows the mean ± SE of triplicate assays. * or ** indicates a statistically significant difference relative to the value at 0 hr for each gene at p < .05 or .01, respectively
2.6. miR159a overexpression balances ROS homeostasis and increased resistance to Botrytis
Because one of the earliest defence responses in B. cinerea is reactive oxygen species (ROS) production (Asselbergh et al., 2007; van Kan, 2006), we monitored H2O2 and level fluctuations when WT and transgenic Arabidopsis were inoculated with B. cinerea. After inoculation with B. cinerea for 2 days, the transgenic and WT Arabidopsis both had increased H2O2 levels, and the brown colour of OELrGAMYB leaves was deeper than that of WT leaves and OEmiR159a leaves (Figure 7a–c), suggesting that OELrGAMYB plants had higher ROS levels than WT and OEmiR159a Arabidopsis. The excess H2O2 generation was apparent in susceptible OELrGAMYB transgenic Arabidopsis, suggesting that infection accelerated H2O2 accumulation, resulting in cellular damage.
Figure 7.
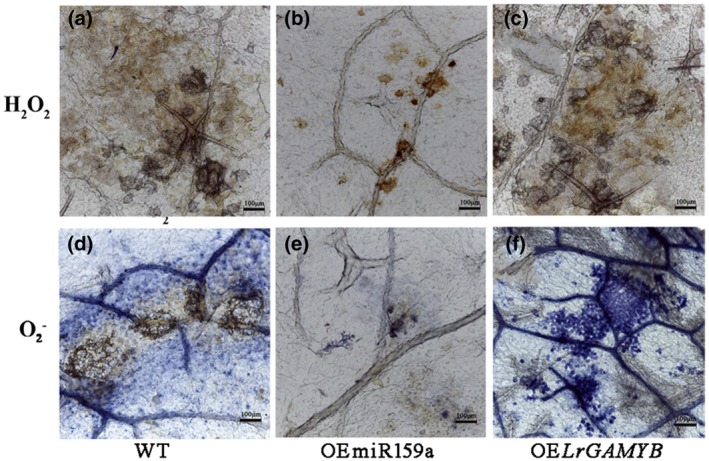
Temporal evolution of H2O2 and accumulation in the leaves of wild‐type (WT) and transgenic Arabidopsis overexpressing miR359a (OEmiR359a) or LrGAMYB (OELrGAMYB) following Botrytis cinerea inoculation for 48 hr. Scale bar = 100 μm. (a) to (c) are stained with 3,3′‐diaminobenzidine and show the accumulation of H2O2; (d) to (f) are stained with nitroblue tetrazolium and show the accumulation of
Similarly, the bluish violet staining was reduced in OEmiR159a transgenic Arabidopsis but widespread in the intercellular spaces and epidermal cells of WT and OELrGAMYB transgenic Arabidopsis leaves after inoculation (Figure 7d–f). accumulation expanded from the sites of fungal contact, where rot spots were apparent. These data indicate that altering miR159a expression enhanced H2O2 and homeostasis, which may ultimately increase resistance to Botrytis.
3. DISCUSSION
The miR159 family is one of the ancient miRNA families in not only in monocotyledons and dicotyledons but also gymnosperms and ferns, such as Picea abies (Xia et al., 2015), Pinus taeda (Lu et al., 2007), and Selaginella moellendorffii (Axtell et al., 2007). Furthermore, the miR159 family differs in the number of mature and precursor genes among species. In Arabidopsis, this family is encoded by three genes and forms three mature miR159 members (miR159a, miR159b, and miR159c) located in different regions of the genome (Allen et al., 2007). Further research has indicated that a mir159a mir159b double mutant has pleiotropic morphological defects, including altered growth habits, curled leaves, small siliques, and small seeds. Neither mir159a nor mir159b single mutants displayed any of these traits, indicating functional redundancy (Allen et al., 2007; Alonso‐Peral et al., 2010). In our previous research, five miR159 members were identified in L. regale through high‐throughput sequencing; however, lre‐miR159a was significantly more highly and differentially expressed than others members after inoculation with B. elliptica, suggesting that lre‐miR159a might respond to B. elliptica in L. regale (Gao et al., 2017). Therefore, we chose lre‐miR159a for further research in this study.
The results of sequence alignment analysis suggested that mature miR159a sequences are highly conserved in different species; however, the precursors have substantial differences. For instance, Z. mays and P. abies possess the most miR159 precursors with 11 genes, but 11 species hold only one. Furthermore, the phylogenetic tree showed that the evolutionary distance separating the branches of different species of miR159a precursors is long, and the sequence similarity is relatively low, suggesting varied sources of mature miR159a. pre‐lre‐MI159a formed an independent clade, suggesting it is different from other species. The highly conserved sequences of mature miR159 indicate that they may perform similar functions.
Studies have revealed that miR159 plays an important role in the response to stresses. For instance, miR159 responds to salinity stress (Kitazumi et al., 2015), drought stress (Mohsenifard et al., 2017), and heat stress (Li et al., 2016). Suppression of miR159 in plants resulted in short stature along with smaller stem, leaf, and grain size, which enhanced the hypersusceptibility to adverse environmental stress (Zhao et al., 2017). Furthermore, altered accumulation of miR159a levels was observed during tomato leaf curl virus infection in tomato, indicating that miR159 behaves as an active factor in pathogen resistance (Koundal et al., 2010). In our previous study, we also found by high‐throughput sequencing that lre‐miR159a was involved in the response to Botrytis. miR159 mainly contributes to the regulation of plant development and stress by targeting MYB transcription factors (Millar, 2005; Xue et al., 2017). The GAMYB or GAMYB‐like genes encode a highly conserved family of R2R3 MYB domain transcription factors implicated in GA signal transduction (Woodger et al., 2003). Studies have indicated that the GAMYB transcription factor superfamily is involved in plant development and metabolism (Alonso‐Peral et al., 2010; Sheldon et al., 2001) and response to pathogens and abiotic stress (Yang et al., 2014; Butt et al., 2017). Moreover, studies have revealed that MYB transcriptome factors participate in response to B. cinerea. For instance, the overexpression of AtMYB44 in Arabidopsis results in a stronger ROS burst, greater cell death, and severe necrosis symptoms, which enhances susceptibility to B. cinerea (Shi et al., 2011). Furthermore, MYB46, thought to regulate secondary cell wall biosynthesis in the vascular tissue of the stem, functions as a disease susceptibility modulator to B. cinerea. The MYB46‐mutant plants exhibited increased disease resistance to B. cinerea (Vicente et al., 2011). These results suggest that MYB transcription factors are negative regulators in response to B. cinerea. In this study, LrGAMYB transcription factors were verified as one target of lre‐miR159a in L. regale by RLM‐RACE. After inoculation with B. elliptica, lre‐miR159a was significantly up‐regulated in resistant Lilium, accompanied by decreased expression of LrGAMYB, indicating a negative relationship between lre‐miR159a and its target LrGAMYB. These results reveal that lre‐miR159a and its target LrGAMYB responded to Botrytis infection in lily.
To further study the role of lre‐miR159a and the target LrGAMYB gene in disease resistance, transgenic Arabidopsis of the lre‐miR159a and LrGAMYB genes were constructed. The OEmiR159a plants exhibited larger leaves and smaller necrotic spots than the WT and OELrGAMYB Arabidopsis plants after Botrytis inoculation (Figure 5). The results indicate that the molecular and physiological responses to Botrytis included ROS production and transcriptional responses. A stronger response of JA‐ and SA‐mediated defence genes was detected in the Botrytis‐infected OEmiR159a plants (Figure 6), implying that overexpression of miR159a enhances transgenic plant defence by activating the hypersensitive response. An SA‐dependent signalling pathway led to the expression of pathogenesis‐related (PR) proteins, including AtNPR1, AtPR1, and AtPR2, thus contributing to resistance. AtPDF1.2, which is dependent on the JA pathway, was also induced and was highly expressed. In agreement with previous reports, the SA and JA signalling pathways interact extensively and cooperatively in response to Botrytis infection (Glazebrook, 2005).
B. cinerea is a nonspecific necrotrophic fungal pathogen that triggers plants to generate large amounts of ROS and induces local cell death to facilitate infection (Su et al., 2011; Pietrowska et al., 2015). Previous studies have shown that the induction of H2O2 in plant cells, accompanied by generation, can promote programmed cell death in the host and the expansion of disease lesions to facilitate B. cinerea infection (Govrin and Levine, 2000; Patykowski, 2006; Asai and Yoshioka, 2009; Wan et al., 2015). During the early stages of Botrytis infection, the ROS burst can induce a defensive reaction. However, high H2O2 levels can disturb redox homeostasis, trigger initiation of cell death, and facilitate necrotrophic pathogen attack of host plants. In our study, H2O2 and levels were lower in OEmiR159a Arabidopsis than in WT and OELrGAMYB Arabidopsis, most probably due to the overexpression of lre‐miR159a. These results indicate that lre‐miR159a plays a positive role in resistance to Botrytis by suppressing the target LrGAMYB gene.
We also found an interesting phenomenon in which OEmiR159a Arabidopsis showed early flowering. Studies have indicated that miR159 is involved in floral organ development. For instance, miR159 expression regulates floral transition (Li et al., 2013), flower development (Wang et al., 2017), and timing of flowering (Guo et al., 2017). Furthermore, miR159 is required for fruit set (da Silva et al., 2017), and the accumulation of miR159 could affect seedling development. These results suggest multiple regulatory networks of miRNAs that participate in plant growth and development. Further research is needed to verify the functions of these miRNAs and their regulatory networks.
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials and B. elliptica inoculation
Fresh, uniform‐sized bulbs of L. regale were held in cold storage (approximately 1 °C) for a month before planting. After this vernalization period, bulbs were planted on culture medium under a 12 hr day/night cycle at 25/22 °C and maintained in a semi‐open house covered with a shading screen at Huairou, Beijing Province, China. The fungus, B. elliptica, isolated from diseased lily leaves, was maintained on potato dextrose agar at 22 °C under near‐UV light in a 100% relative humidity chamber. For the bioassay, 7‐day‐old cultures were used. Conidia were collected by gently vortexing in Tween 20 and adjusted to a concentration of 5 × 104 conidia/ml for inoculation.
Arabidopsis Columbia (Col‐0) was used for transformation by pre‐lre‐MIR159a and LrGAMYB plasmids. Seeds were sown on 1/2 × Murashige and Skoog (MS) medium, cold treated for 1 week at 4 °C, and transferred to a controlled environment incubator with a 16/8 hr light/dark photoperiod and a 25/18 °C day/night thermoperiod.
4.2. Sequence analysis of precursor miR159a from L. regale and prediction target genes
Total genomic DNA was extracted from leaves using the New Plant Genomic DNA Extraction Kit (Tiangen). The DNA sequence was amplified by PCR using primers designed based on the small‐RNA library previously constructed by our laboratory (Gao et al., 2017), and the primers are shown in Table S1. PCR amplification was performed with KOD‐Plus‐Neo polymerase (Totobo) according to the instruction manual, with a programme set as follows: 94 °C for 2 min; and 45 cycles of 98 °C for 10 s, 59 °C for 15 s, and 72 °C for 15 s. The PCR products were gel purified and cloned into the pLB vector (Tiangen) and then sequenced by Sangon Biotech (Shanghai).
Mfold web server (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) was used to examine the hairpin structure of the miR159a precursor. Precursors of miR159a from other plants were obtained from miRBase 21.0 and aligned with the miR159a precursor sequence by DNAMAN (Lynnon Biosoft) and ClustalW. The target genes of miR159a were predicted by psRNATarget web (http://plantgrn.noble.org/psRNATarget/) with default parameters (Dai and Zhao, 2011). The prediction of the lre‐miR159a targets was based on the transcriptome of L. regale, which was constructed by our laboratory. A phylogenetic tree was constructed with MEGA 6 using the neighbour‐joining method. The conserved domains of mature miR159a sequences were aligned using the Weblogo program with default parameters (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
4.3. Cloning the lre‐miR159a‐targeted GAMYB gene and sequence analysis
The lre‐miR159a‐targeted GAMYB partial sequence was deduced from the L. regale transcriptome. First, total RNA from L. regale leaves was extracted using EASYspin Plus RNA Kit (Aidlab). Full GAMYB cDNA fragments were then obtained with the SMARTer RACE 5′/3′ Kit (Takara) followed by the second round of nested PCR with specific outer and inner primers listed in Table S2. The full‐length of GAMYB gene sequence was amplified with designed primers (Table S1) and cloned into the pLB vector. These GAMYB protein sequences of other plants were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/). Multiple alignments of these protein sequences were aligned using DNAMAN.
4.4. RNA ligase‐mediated rapid amplification of cDNA ends
To detect the miRNA‐target cleavage site, RLM‐RACE was conducted using the 5′‐Full RACE Kit (Takara) with a 5′‐RACE adaptor (Lu et al., 2017). Total RNA for RLM‐RACE was obtained from L. regale leaves. The 5′‐RACE outer primer and gene‐specific outer primer (GSP1) were used for the first round of nested PCR, followed by the second round of nested PCR using the 5′‐RACE inner primer and gene‐specific inner primer (GSP2) (Table S2). Amplification products were gel purified, cloned into the pRACE vector, and sequenced.
4.5. Plasmid construction and Arabidopsis transformation
To overexpress miR159a and LrGAMYB, the sequence of the precursor lre‐miR159a and full cDNA fragments encoding LrGAMYB were amplified from the corresponding cloned vectors and then inserted downstream of the CaMV 35S promoter in pCAMBIA1301 (GenBank no. AF234297). The recombinant vectors were then introduced into Agrobacterium tumefaciens GV3101 and the Arabidopsis Col‐0 WT was used for transformation by the floral‐dip method. Transformants were selected with 50 mg/L hygromycin B and confirmed by reverse transcription PCR.
Three homozygous T3 lines were established and used for the treatments. For the bioassay, discs of B. cinerea mycelia and conidia solution were both used for the inoculation of detached leaves. Discs of mycelia were punched from the growing edge of colonies, and conidia were adjusted to a concentration of 5 × 105 conidia/ml for inoculation. Trypan blue staining for the presence of the fungal infection was performed as previously described (Gao et al., 2018). After the decolorization treatment, the mycelium in tissue was observed with a microscope (BME, Leica).
4.6. RNA insolation and RT‐qPCR
Total RNAs were extracted and reverse transcribed using the First Strand cDNA Synthesis Kit (Toyobo). RT‐qPCR was conducted in a total volume of 20 μl containing 10 μl SYBR Premix Ex Taq (Takara) using following programme: 5 min denaturation at 94 °C; followed by 30 cycles of 94 °C for 5 s, 60 °C for 20 s, and 72 °C for 20 s. The 18S rRNA and CLATHRIN genes were used as the reference genes for normalization in L. regale, and the AtActin gene was used for A. thaliana. Each sample was performed in triplicate and the mean value of technical replicates was recorded for each biological replicate. The primers used are listed in Table S3.
For RT‐qPCR analysis of lre‐miR159a and LrGAMYB to grey mould response in Lilium, leaves inoculated with B. elliptica were harvested at 0, 2, 6, 12, 24, 36, 48, and 72 hpi of the B. elliptica‐resistant L. regale and the B. elliptica‐susceptible cv. Tresor according to previous studies (Gao et al., 2018).
For RT‐qPCR analysis in Arabidopsis, inoculated leaves were harvested at 0, 24, and 48 hpi for RNA extraction.
4.7. 3,3′‐diaminobenzidine and nitroblue tetrazolium staining
H2O2 and were detected by previously described 3,3′‐diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining protocols, respectively, to compare the ROS responses of WT and transgenic Arabidopsis. To detect the production of H2O2, leaves were immersed in a DAB solution (1 mg/ml acidified with HCl to pH 3.8) under direct light for 2 hr after samples were inoculated with B. cinerea 48 hpi. The stained leaves were incubated in a solution of 70% ethanol and then photographed.
For observation of , leaves were collected directly into 0.1% NBT solution in 10 mM phosphate buffer (pH 7.8) prior to vacuum infiltration for 30 min and exposure to direct light for 20 min. The NBT‐stained samples were then observed.
4.8. Statistical analysis
All data are presented as the mean ± SE and were subjected to analysis of variance according to Student's t test (*p < .05, **p < .01). Each data set was independently compared with the control data set to determine significant differences.
Supporting information
ACKNOWLEDGEMENTS
This project was supported by the Fundamental Research Funds for the Central Universities (grant no. BLX201813), China Postdoctoral Science Foundation (grant no. 2019M650518), and National Key R&D Program of China (grant no. 2019YFD1000403).
Gao X, Zhang Q, Zhao Y‐Q, Yang J, Jia G‐X, He H‐B. The lre‐miR159a‐LrGAMYB pathway mediates resistance to grey mould infection in Lilium regale . Molecular Plant Pathology. 2020;21:749–760. 10.1111/mpp.12923
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Achard, P. , Herr, A. , Baulcombe, D.C. and Harberd, N.P. (2004) Modulation of floral development by a gibberellin‐regulated microRNA. Development, 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Allen, R.S. , Li, J. , Stahle, M.I. , Dubroué, A. , Gubler, F. and Millar, A.A. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proceedings of the National Academy of Sciences of the United States of America, 104, 16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Peral, M.M. , Li, J. , Li, Y. , Allen, R.S. , Schnippenkoetter, W. , Ohms, S. et al (2010) The microRNA159‐regulated GAMYB‐like genes inhibit growth and promote programmed cell death in Arabidopsis . Plant Physiology, 154, 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, S. and Yoshioka, H. (2009) Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 22, 619–629. [DOI] [PubMed] [Google Scholar]
- Asselbergh, B. , Curvers, K. , Franca, S.C. , Audenaert, K. , Vuylsteke, M. , Van Breusegem, F. et al (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid‐deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiology, 144, 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J. , Snyder, J.A. and Bartel, D.P. (2007) Common functions for diverse small RNAs of land plants. The Plant Cell, 19, 1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen, P. , Staats, M. and van Kan, J.A.L. (2004) Induction of programmed cell death in lily by the fungal pathogen Botrytis elliptica . Molecular Plant Pathology, 5, 559–574. [DOI] [PubMed] [Google Scholar]
- Blanco‐Ulate, B. , Vincenti, E. , Powell, A.L. and Cantu, T. (2013) Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea . Frontiers in Plant Science, 4, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H.I. , Yang, Z. , Gong, Q. , Chen, E. , Wang, X. , Zhao, G. et al (2017) GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biology, 17, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E. , Hon, G. , Chandonia, J.M. and Brenner, S.E. (2004) WebLogo: a sequence logo generator. Genome Research, 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. and Zhao, P.X. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research, 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss, R.P. , Christian, J.K. and Chastagner, G.A. (1988) Infection of easter lily leaves from conidia of Botrytis elliptica . Canadian Journal of Botany, 66, 1204–1208. [Google Scholar]
- Ehsan, M.F. , Mehdi, G. , Nastaran, M. and Behnam, B. (2017) Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. Plant Molecular Breeding, 5, 10–18. [Google Scholar]
- Feng, H. , Zhang, Q. , Li, H. , Wang, X. , Wang, X. , Duan, X. et al (2013) vsiRNAs derived from the miRNA‐generating sites of pri‐tae‐miR159a based on the BSMV system play positive roles in the wheat response to Puccinia striiformis f. sp. tritici through the regulation of taMYB3 expression. Plant Physiology and Biochemistry, 68, 90–95. [DOI] [PubMed] [Google Scholar]
- Furukawa, T. , Ushiyama, K. and Kishi, K. (2005) Botrytis blight of Taiwanese toad lily caused by Botrytis elliptica (Berkeley) Cooke. Journal of General Plant Pathology, 71, 95–97. [Google Scholar]
- Gao, X. , Cui, Q. , Cao, Q. , Zhao, Y.‐Q. , Liu, Q. , He, H.‐B. et al (2018) Evaluation of resistance to Botrytis elliptica in Lilium hybrid cultivars. Plant Physiology and Biochemistry, 128, 392–399. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Cui, Q. , Cao, Q. , Liu, Q. , He, H. , Zhang, D. et al (2017) Transcriptome‐wide analysis of Botrytis elliptica responsive microRNAs and their targets in Lilium regale Wilson by high‐throughput sequencing and degradome analysis. Frontiers in Plant Science, 8, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Current Biology, 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Guo, C. , Xu, Y. , Shi, M. , Lai, Y. , Wu, X. , Wang, H. et al (2017) Repression of miR156 by miR159 regulates the timing of the juvenile‐to‐adult transition in Arabidopsis . The Plant Cell, 29, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, P.F. and Chen, C.Y. (2003) Early stages of infection of lily leaves by Botrytis elliptica and B. cinerea . Plant Pathology Bulletin, 12, 103–108. [Google Scholar]
- Hsieh, T.F. , Haung, J.W. and Hsiang, T. (2001) Light and scanning electron microscopy studies on the infection of oriental lily leaves by Botrytis elliptica . European Journal of Plant Pathology, 106, 571–581. [Google Scholar]
- Ji, H.M. , Zhao, M. , Gao, Y. , Cao, X.X. , Mao, H.Y. , Zhou, Y. et al (2018) FRG3, a target of slmiR482e‐3p, provides resistance against the fungal pathogen Fusarium oxysporum in tomato. Frontiers in Plant Science, 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W. and Wu, F. (2015) Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biology, 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W. , Wu, F. , Xiao, L. , Liang, G. , Zhen, Y. , Guo, Z. et al (2012) Microarray‐based analysis of tomato miRNA regulated by Botrytis cinerea . Journal of Plant Growth Regulation, 31, 38–46. [Google Scholar]
- Jones‐Rhoades, M.W. and Bartel, D.P. (2004) Computational identification of plant microRNAs and their targets, including a stress‐induced miRNA. Molecular Cell, 14, 787–799. [DOI] [PubMed] [Google Scholar]
- van Kan, J.A.L. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends in Plant Science, 11, 247–253. [DOI] [PubMed] [Google Scholar]
- Kitazumi, A. , Kawahara, Y. and Onda, T.S. (2015) Implications of mir166 and mir159 induction to the basal response mechanisms of an andigena potato (Solanum tuberosum subsp. andigena) to salinity stress, predicted from network models in Arabidopsis . Genome, 58, 13–24. [DOI] [PubMed] [Google Scholar]
- Koundal, V. , Vinutha, T. , Haq, Q.M.R. and Praveen, S. (2010) Modulation of plant development and MYB down regulation: both during in planta expression of miR159a and in natural ToLCV infection. Journal of Plant Biochemistry and Biotechnology, 19, 171–175. [Google Scholar]
- Li, H. , Wang, Y. , Wang, Z. , Guo, X. , Wang, F. , Xia, X.J. et al (2016) Microarray and genetic analysis reveals that csa‐miR159b plays a critical role in abscisic acid‐mediated heat tolerance in grafted cucumber plants. Plant, Cell and Environment, 39, 1790–1804. [DOI] [PubMed] [Google Scholar]
- Li, L. , Yi, H. , Xue, M. and Yi, M. (2017) miR398 and miR395 are involved in response to SO2 stress in Arabidopsis thaliana . Ecotoxicology, 26, 1181–1187. [DOI] [PubMed] [Google Scholar]
- Li, X. , Bian, H. , Song, D. , Ma, S. , Han, N. , Wang, J. et al (2013) Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Annals of Botany, 111, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Sun, Y.H. , Amerson, H. and Chiang, V.L. (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. The Plant Journal, 51, 1077–1098. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Dun, H. , Lian, C. , Zhang, X. , Yin, W. and Xia, X. (2017) The role of peu‐miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica . Plant Physiology and Biochemistry, 115, 418–438. [DOI] [PubMed] [Google Scholar]
- Millar, A.A. (2005) The Arabidopsis GAMYB‐Like genes, MYB33 and MYB65, are microRNA‐regulated genes that redundantly facilitate anther development. The Plant Cell, 17, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsenifard, E. , Ghabooli, M. , Mehri, N. and Bakhshi, B.B. (2017) Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. Plant Molecular Breeding, 5, 10–18. [Google Scholar]
- Patade, V.Y. and Suprasanna, P. (2010) Short‐term salt and PEG stresses regulate expression of microRNA, miR159 in sugarcane leaves. Journal of Crop Science and Biotechnology, 13, 177–182. [Google Scholar]
- Patykowski, J. (2006) Role of hydrogen peroxide and apoplastic peroxidase in tomato–Botrytis cinerea interaction. Acta Physiologiae Plantarum, 28, 589–598. [Google Scholar]
- Phillips, J.R. , Dalmay, T. and Bartels, D. (2007) The role of small RNAs in abiotic stress. FEBS Letters, 581, 3592–3597. [DOI] [PubMed] [Google Scholar]
- Pietrowska, E. , Różalska, S. , Kaźmierczak, A. , Nawrocka, J. and Małolepsza, U. (2015) Reactive oxygen and nitrogen (ROS and RNS) species generation and cell death in tomato suspension cultures–Botrytis cinerea interaction. Protoplasma, 252, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanfa, L. , Ying‐Hsuan, S. and Chiang, V.L. (2010) Stress‐responsive microRNAs in Populus . The Plant Journal, 55, 131–151. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C. , Gubler, F. , Moritz, T. , Bagnall, D.J. , Parish, R.W. , Dennis, E.S. et al (2001) GAMYB‐like Genes, flowering, and gibberellin signaling in Arabidopsis . Plant Physiology, 127, 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Cui, R. , Hu, B. , Wang, X. , Zhang, S. , Liu, R. et al (2011) Overexpression of transcription factor AtMYB44 facilitates Botrytis infection in Arabidopsis . Physiological and Molecular Plant Pathology, 76, 90–95. [Google Scholar]
- da Silva, E.M. , Silva, G.F.F.E. , Bidoia, D.B. , da Silva Azevedo, M. , de Jesus, F.A. , Pino, L.E. et al (2017) microRNA159‐targeted SlGAMYB transcription factors are required for fruit set in tomato. The Plant Journal, 92, 95–109. [DOI] [PubMed] [Google Scholar]
- Soto‐Suárez, M. , Baldrich, P. , Weigel, D. , Rubio‐Somoza, I. and San, S.B. (2017) The Arabidopsis miR396 mediates pathogen‐associated molecular pattern‐triggered immune responses against fungal pathogens. Scientific Reports, 7, 44898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, J. , Tu, K. , Cheng, L. , Tu, S. , Wang, M. , Xu, H. et al (2011) Wound‐induced H2O2 and resistance to Botrytis cinerea decline with the ripening of apple fruit. Postharvest Biology and Technology, 62, 64–70. [Google Scholar]
- Sun, X. (2014) Analysis of Arabidposis miR159 and its Targets Genes in Disease Resistance to Powdery Mildew Fungus. Beijing: University of Chinese Academy of Sciences. [Google Scholar]
- Tian, X. , Song, L. , Wang, Y. , Jin, W. , Tong, F. and Wu, F. (2018) miR394 acts as a negative regulator of Arabidopsis resistance to B. cinerea infection by targeting LCR. Frontiers in Plant Science, 9, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H. , Aya, K. , Ueguchi‐Tanaka, M. , Shimada, Y. , Nakazono, M. et al (2006) GaMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. The Plant Journal, 47, 427–444. [DOI] [PubMed] [Google Scholar]
- Vicente, R. , Astrid, A. , Alberto, C. et al (2011) MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis . Plant Physiology, 155, 1920–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, R. , Hou, X. , Wang, X. , Qu, J. , Singer, S.D. , Wang, Y. et al (2015) Resistance evaluation of Chinese wild Vitis genotypes against Botrytis cinerea and different responses of resistant and susceptible hosts to the infection. Frontiers in Plant Science, 6, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Xie, Z. , Sun, X. and Li, X. (2017) Function analysis of miR159 and its target gene VvGAMYB in grape flower development. Acta Horticulturae Sinica, 44, 1061–1072. [Google Scholar]
- Woodger, F.J. , Millar, A. , Murray, F. , Jacobsen, J.V. and Gubler, F. (2003) The role of GAMYB transcription factors in GA‐regulated gene expression. Journal of Plant Growth Regulation, 22, 176–184. [Google Scholar]
- Xia, R. , Xu, J. , Arikit, S. and Meyers, B.C. (2015) Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Molecular Biology and Evolution, 32, 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, T. , Liu, Z. , Dai, X. and Xiang, F. (2017) Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101 . Plant Science, 262, 182–189. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Zhang, N. , Mi, X. , Wu, L. , Ma, R. , Zhu, X. et al (2014) Identification of miR159s and their target genes and expression analysis under drought stress in potato. Computational Biology and Chemistry, 53, 204–213. [DOI] [PubMed] [Google Scholar]
- Zhao, D. , Gong, S. , Hao, Z. and Tao, J. (2015) Identification of miRNAs responsive to Botrytis cinerea in herbaceous peony (Paeonia lactiflora Pall.) by high‐throughput sequencing. Genes, 6, 918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Yuan, S. , Man, Z. , Yuan, N. , Li, Z. , Hu, Q. et al (2018) Transgenic creeping bentgrass overexpressing osa‐mir393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnology Journal, 17, 233–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wen, H. , Teotia, S. , Du, Y. , Zhang, J. , Li, J. et al (2017) Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biology, 17, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Thilmony, R. , Bender, C.L. , Schaller, A. , He, S.Y. and Howe, G.A. (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. The Plant Journal, 36, 485–499. [DOI] [PubMed] [Google Scholar]
- Zheng, Z. , Reichel, M. , Deveson, I. , Wong, G. , Li, J. and Millar, A.A. (2017) Target RNA secondary structure is a major determinant of miR159 efficacy. Plant Physiology, 174, 1764–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


