Abstract
It has been commonly accepted that soybean domestication originated in East Asia. Although East Asia has the historical merit in soybean production, the USA has become the top soybean producer in the world since 1950s. Following that, Brazil and Argentina have been the major soybean producers since 1970s and 1990s, respectively. China has once been the exporter of soybean to Japan before 1990s, yet she became a net soybean importer as Japan and the Republic of Korea do. Furthermore, the soybean yield per unit area in East Asia has stagnated during the past decade. To improve soybean production and enhance food security in these East Asian countries, much investment has been made, especially in the breeding of better performing soybean germplasms. As a result, China, Japan, and the Republic of Korea have become three important centers for soybean genomic research. With new technologies, the rate and precision of the identification of important genomic loci associated with desired traits from germplasm collections or mutants have increased significantly. Genome editing on soybean is also becoming more established. The year 2019 marked a new era for crop genome editing in the commercialization of the first genome-edited plant product, which is a high-oleic-acid soybean oil. In this review, we have summarized the latest developments in soybean breeding technologies and the remarkable progress in soybean breeding-related research in China, Japan, and the Republic of Korea.
Introduction
Cultivated soybean (Glycine max [L.] Merr.) is believed to be domesticated from annual wild soybean (Glycine soja) thousands of years ago (Carter et al. 2004; Hymowitz 1970). The question of whether cultivated soybean originated from a single (Jeong et al. 2019; Zhou et al. 2015) or multiple domestication events (Lee et al. 2011a; Xu et al. 2002) remains unanswered. However, it is generally agreed that ancient China, Korea, and Japan were the three pioneering societies to domesticate and cultivate soybean (Hymowitz 1970; Jeong et al. 2019; Lee et al. 2011a; Sedivy et al. 2017; Xu et al. 2002).
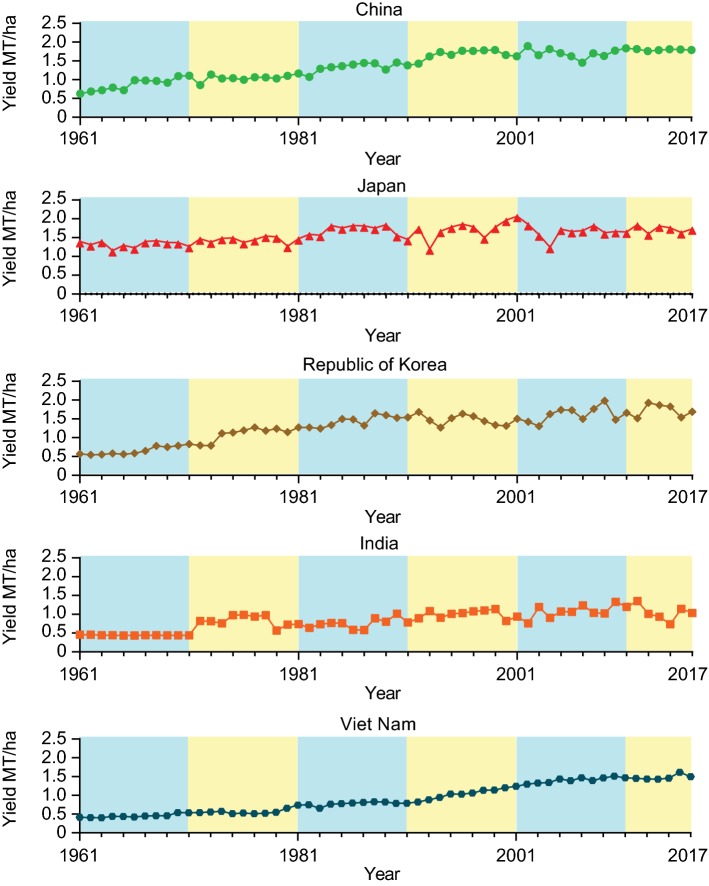
As a major economic crop of high plant oil and protein contents, there has been an escalating demand for soybean. Consequently, increasing soybean yield has always been the main objective of crop improvement. With the advents of modern farming practices and genetic improvements, the yield of soybean has steadily increased in Asia since 1961 (Fig. 1). The soybean yield in China increased from 0.6 metric tons per hectare (MT/ha) in 1961 to a record high of 1.89 MT/ha in 2002 and then plateaued at around 1.8 MT/ha for 15 years after (Fig. 1). Similarly, the soybean yield in Korea steadily increased from 0.6 MT/ha in 1961 to a peak of 1.98 MT/ha in 2009 and has stabilized at an average of 1.7 MT/ha in subsequent years (Fig. 1). The yield of soybean in Japan was 1.3 MT/ha in 1961, doubling those of the nearby countries during the same period. Nevertheless, the increase in soybean yield in Japan has occurred at a slower pace and the yield is now at a similar level to those in China and Korea (Fig. 1). Seemingly, the soybean yield in China, Japan, and Korea and nearby countries such as Vietnam and India has reached a plateau in recent years as well (Fig. 1). The stagnation of soybean yield growth in these countries could be due to multiple factors such as the relocation of soybean production from optimum (like Middle Northeast China) to harsh environments (like high-latitude cold regions in China) and the limitations in soil fertility and water resources. To further boost the soybean yield, improved soybean cultivars with higher yield potential and stronger tolerance to environmental stresses may provide a partial solution. However, the genetic bottleneck has hampered the progress in soybean breeding over the last century (Hyten et al. 2006). The recent rapid development in genome sequencing technology, the release of high-quality genome sequences, and a better understanding of the genomic regions associated with agronomically important traits provide important tools for accelerating the progress of marker-assisted and molecular breeding programs in soybean.
Fig. 1.
Per-hectare yield of soybean in selected Asian countries. Data were obtained from FAOstat (retrieved on May 20, 2019, from http://www.fao.org/faostat/en/#home)
Milestones of soybean genomic research in the three original soybean-domesticating countries
The first reference genome of the cultivated soybean is based on the cultivar, Williams 82 (Wm82) (Schmutz et al. 2010), which has been extensively used in various soybean genomic studies in the past decade (Chung et al. 2014; Kim et al. 2010; Lam et al. 2010; Li et al. 2014; Qi et al. 2014; Qiu et al. 2014; Zhou et al. 2015). However, Wm82 is a cultivated soybean bred in North America (Schmutz et al. 2010) and is distinct from those in South America and Asia (Vaughn and Li 2016; Wolfgang and An 2017; Zhou et al. 2015). Therefore, using Wm82 as the only reference for population genomic analyses may not be sufficient to capture all the genetic variations from diverse origins. As a result, genome assemblies of soybean accessions from different regional niches or ones bred for unique purposes were built.
In order to characterize the genomic structure of Japanese soybean, the genome of a leading Japanese cultivar Enrei was sequenced (Shimomura et al. 2015). Enrei is a breeding variety released in 1971 with the aim to boost the soybean production in central Japan (Shurtleff and Aoyagi 2014). It quickly became a leading cultivar with a high seed quality for food processing (Shurtleff and Aoyagi 2014). Enrei is therefore the major soybean used in soybean research in Japan. Approximately two million single-nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs) were identified between Enrei and Wm82 (Shimomura et al. 2015). The sequence information of Enrei is available at DAIZUbase (Katayose et al. 2011). Information from the Enrei genome has been used for gene discovery and, later, for breeding purposes. A chromosomal segment substitution lines (CSSLs) collection was produced by crossing Enrei with the Chinese landrace, Peking (Watanabe et al. 2018). Peking is a founder line for many disease-resistant cultivars. The low-complexity CSSL collection, with Peking genome segments in the Enrei background, could facilitate the identification of useful genetic resources from Peking and reduce the undesired attributes of the Peking background in later breeding manipulations. Furthermore, an ethyl methanesulfonate (EMS) mutant library of Enrei has been made (Tsuda et al. 2015). After two rounds of mutagenesis, 1437 M2 plants were able to set viable seeds with an average mutation density of 1 mutation/74 kb (Tsuda et al. 2015). This mutation rate should be enough for identifying mutants for forward genetic studies. Other than Enrei-based researches, the resequencing of Japanese and world soybean mini-core collections (Kaga et al. 2011) is also ongoing in Japan. Meanwhile, resequencing using AmpliSeq technology has identified novel variants of flowering time-related genes from the Japanese mini-core consisting of 96 soybean accessions, of which 65 are of Japanese origin (Ogiso-Tanaka et al. 2019).
Korea was among the pioneers to carry out deep sequencing of undomesticated soybean accessions using next-generation sequencing (Kim et al. 2010). A Korean wild soybean IT182932 was sequenced at a high depth of ~43-fold sequence coverage. By mapping the resequencing reads to the Wm82 reference genome and assembling the unmapped reads de novo, it was found that the genomes of the wild soybean and the cultivated one differ by around 3.76% (Kim et al. 2010). The wild-specific contigs of IT182932 were used in subsequent population genomic analyses in the absence of a wild soybean reference genome (Lee et al. 2013; Zhou et al. 2015). The INDEL information from IT182932 deep sequencing has been used to build a high-density linkage map of an F15 population derived from G. max Hwangkeum and G. soja IT182932 (Lee et al. 2013), providing supporting evidence that introgression is not uncommon in soybean. The resequencing approach enabled the marker interval to be reduced to under 6 cM, and consequently, these authors were able to identify seven previously misplaced markers, two absent ones, and six INDEL markers placed on the scaffolds in the Wm82 assembly owing to the increased resolution (Lee et al. 2013). Later, a resequencing study on nine Korean G. max accessions and five Korean G. soja accessions was also able to assemble unmapped reads to contigs for gene identification (Chung et al. 2014). In total, 1326 putative genes were identified in 31 Mb of de novo contigs and at least 343 of them showed a high degree of homology with known plant proteins (Chung et al. 2014), providing further support that some genes are conserved in wild germplasms.
A few hallmark resequencing studies have been done in China (Lam et al. 2010; Li et al. 2013; Zhou et al. 2015). Efforts have also been made to assemble draft genomes of wild soybeans and semi-wild soybeans (Qi et al. 2014; Qiu et al. 2014). Although the causal gene for salt tolerance was obtained through one of these draft genomes (Qi et al. 2014), the contiguity of these genomes makes them ineffective for genome comparisons. In a hope to better represent the diversity of wild soybeans, a pan-genome of seven geographically distinct G. soja accessions was also built (Li et al. 2014). The pan-genome has a size of 986.3 Mb, consisting of 59,080 annotated gene families. More than 20% of the pan-genome is dispensable, presumably related to environmental adaptations. The dispensable portion of the genome consists of a collection of genes for crop improvement. Yet with all this new information, the pan-genome is still far from being usable in breeding applications.
Recently, there are two breakthroughs in soybean genomic research in China: the assembly of a high-quality reference genome of Zhonghuang 13 (ZH13), a popular Chinese cultivated soybean and that of a high-quality reference genome of a wild soybean W05 (Shen et al. 2018; Xie et al. 2019). ZH13 is a bred released in Anhui Province, China, in 2001 as a multi-resistant soybean cultivar and became the most popular soybean cultivar in China since 2007 (Chinese Academy of Agricultural Sciences 2016). The cumulative cultivated area under ZH13 has reached 6.7 million hectares in 2018, demonstrating its importance in Chinese agriculture (Wei 2018). By using a bundle of state-of-the-art technologies, including PacBio single-molecule real-time sequencing, Hi-C sequencing, and optical mapping, the genome of ZH13 was assembled into 21 superscaffolds corresponding to the 20 chromosomal and the chloroplast genomes (Shen et al. 2018). The total assembled size of the genome is 1025 Mb with the contig N50 of 3.46 Mb. Assembled genome of ZH13 shows 84% synteny with Wm82. There are 1404 translocation events and 12,170 present–absent variations (PAVs) between the American and Chinese soybean reference genomes. The discrepancies may be due to the advancement in technologies between the two generations of genome assembly. Regardless, the variations between Wm82 and ZH13 also show that a single reference genome is not sufficient to capture all the genomic variations across soybeans of different geographical origins.
The other breakthrough in soybean genomic research is the release of the first reference-grade genome of the wild soybean, W05 (Xie et al. 2019). Wild soybean has been shown to be the reservoir for rich genetic resources (Chung et al. 2014; Lam et al. 2010; Li et al. 2013, 2014; Qiu et al. 2014; Zhou et al. 2015). Since the release of the Wm82 genome, there have been a few attempts to assemble the genomes of wild soybeans (Li et al. 2014; Qi et al. 2014; Qiu et al. 2014). Yet these assemblies are too fragmented for effective analyses. The genome of W05 was the first complete reference genome for wild soybean. W05 exhibits typical wild soybean morphologies (Lam et al. 2010; Qi et al. 2014). Thus, the reference genome of W05 could possibly be used to study the genomic differences between wild and cultivated soybeans. In fact, the genomic variation that caused the transition in seed coat color as a result of domestication has been identified using the W05 genome (Xie et al. 2019).
The existing genome assemblies represent soybeans from different geographical regions and backgrounds. In summary, Wm82 was released in North America in 1981 (Registration of Crop Cultivars 1988). It is a relatively old variety and has been used as the immediate parent for over 100 newer cultivars in the Americas (www.soybase.org). Thus, it is a good reference for marker identification and genetic mapping involving accessions with related pedigrees. However, breeding and research are catching up in East Asian countries in the past decade. Although in comparison with Wm82, the genome assembly of Enrei is not of reference grade, it is nevertheless a historical Japanese cultivar bred specifically for local conditions and therefore useful as a reference genome for further soybean research in that country. On the other hand, ZH13 is a widely planted cultivar in China after the year 2000, and the release of the ZH13 genome has provided a very important genetic resource for breeding high-quality soybean cultivars. W05 was an undomesticated soybean. Although the diversity of wild soybean is much higher than that of cultivated soybean, W05 possesses many typical wild traits and therefore can be used as a representative wild soybean genome to study the processes of domestication and to identify useful alleles from wild accessions.
Recent development in soybean breeding technologies
For crop breeding in general, there are multiple ways of using genetic resources to achieve improvement. So far, crossing cultivars with known desired traits is the most common practice as it is relatively easy to obtain lines with desirable traits while maintaining crop productivity. However, if cultivars show low genetic diversity, their existing genes/alleles may not be able to cope with new environmental stresses. New genes/alleles may then be identified by screening the wild relatives or created by random mutagenesis. Yet, introducing a large portion of the genome of wild relatives into cultivars or generating random mutants could be detrimental to crop production, and therefore, a long breeding cycle is needed to purify the genetic background while maintaining the introduced traits. Marker-assisted breeding (MAB) for plant improvement can help speed up the process. The universal application of MAB is marker-assisted selection (MAS) and genomic selection (GS) and the principle of MAS and GS has been well documented (Collard et al. 2005; Crossa et al. 2017). As modern breeding approaches, MAS exploits the markers associated with simple traits, while GS using genome-wide markers has the potential to address complex traits and domestication (Akond et al. 2018; Zhang et al. 2018). Transgenesis is normally a quick way to introduce a new trait into the crop, but controversies surrounding this method hamper public acceptance, especially in East Asia and Europe. Currently, genome editing has been a quick, precise, and promising way for crop improvement with better consumer acceptance. With high-quality genome information, the precision in genome editing is also improved, and off-target editing can also be predicted and avoided. Nonetheless, even with new technologies, soybean breeding is still full of challenges.
The growth period of soybean is significantly governed by more than ten quantitative trait loci (QTLs). With different combination of alleles in these loci, photosensitivity, flowering time, maturation time, and flower number could vary a lot. Furthermore, the relatively long growing period of soybean allows for only 1–3 breeding cycles per year. Thus, crossing of distinct varieties would require careful planning to match the flowering time of the two parental lines and to maximize the number of generations per year. To solve these problems, a soybean breeding protocol using CO2-supplemented growth chambers has greatly shortened the time of each breeding cycle from over 100 days to around 70 days (Nagatoshi and Fujita 2019). The protocol shortens the flowering time by controlling the light/dark cycle and temperature and improves flower number and quality to increase crossing efficiency by supplementing the plant with CO2 and by reducing the seed-to-seed period by growing immature seeds (Nagatoshi and Fujita 2019).
Genome editing has been a better received molecular breeding method than transgenesis by society in general. The first soybean product produced by genome editing was announced in early 2019 and will be available on the market soon. By using the transcription activator-like effector nuclease (TALEN) technique, a new high-oleic-acid soybean cultivar has been made without introducing foreign DNA into the soybean genome (Calyxt Inc. 2019; Splitter 2019). Vegetable oil produced with this new soybean cultivar contains 80% oleic acid and has 20% less saturated fat, which makes the oil healthier for human consumption, and the shelf life of the oil is also extended (Calyxt Inc. 2019). It is expected that the high-oleic-acid soybean oil will be the first gene-edited plant product to be commercialized (Calyxt Inc. 2019; Splitter 2019).
Gene editing in somatic cells or in the hairy roots system has received a lot of attention in soybean research (Cai et al. 2015; Du et al. 2016; Jacobs et al. 2015; Michno et al. 2015; Sun et al. 2015). A DNA-free genome editing protocol using Cfp1 (Cas12a) has been developed for soybean, but it is limited to protoplasts (Kim et al. 2017). A protocol for whole plant transformation has been well documented (Liu et al. 2019). Successful stable lines have been generated to disable the GmFT2a family, the GmSPL9 family, and two copies of PEAPOD genes to alter flowering time, root nodule number, and seed pod morphology, respectively (Bao et al. 2019; Cai et al. 2018a, 2019; Kanazashi et al. 2018). Nonetheless, the generation of stably edited lines largely relies on the soybean transformation efficiency, editing efficiency, and inheritability. So far, much effort has been made to screen the soybean endogenous U6 promoters to improve the expression of single-guide RNAs (sgRNAs) (Sun et al. 2015) and to optimize the codon usage of the cas9 gene in soybean to improve the translation efficiency of the enzyme (Michno et al. 2015). Constructs harboring multiple sgRNAs for editing multiple targets in the soybean genome are also available (Liu et al. 2019). There is a successful case of creating large deletions (1–4.5 kb) in the soybean genome using multiple sgRNAs targeting both GmFT2a and GmFT5a, genes that regulate flowering time and maturity (Cai et al. 2018b).
In addition, strategies used in other plants may also be adapted for soybean study. Transformation through pollen magnetofection (Zhao et al. 2017), biolistic bombardment (Rech et al. 2008), or the haploid induction system (Kelliher et al. 2019) can possibly help deliver the gene editing construct into germline cells and requires no explant regeneration. Furthermore, efforts have also been made to improve Cas9 activity. The original Cas9 gene was isolated from Streptococcus pyogenes. Research has demonstrated that heat shocking the Arabidopsis plant at 37 °C at the reproductive stage could increase the editing efficiency in pollen from 4.3–87% to over 95%, probably owing to the activation of the Cas9 enzyme at its optimal temperature (LeBlanc et al. 2018). Genome editing through the double-strand breakage of DNA by CRISPR/Cas9 is somewhat uncontrolled, which results in many alleles after editing. To correct this problem, base editing is achieved by fusing a base editing enzyme to Cas9 with no nuclease activity (Kang et al. 2018). Currently, base editing has been carried out successfully in rice and wheat (Li et al. 2018a; Yan et al. 2018).
Germplasm collections in China, Japan, and the Republic of Korea
Germplasm collections in China
The National Genebank (China) was established in 1986 to conserve the genetic resources of crop plants in China. It has preserved the genetic resources of 400,000 accessions of more than 200 crop species. There are currently over 40,000 soybean accessions in the Genebank, making up a significant portion of its collection (Table 1).
Table 1.
Resource centers for soybean genetic resources available in specific countries/regions
| Resource center | Country/region | Number of accessions | Website/contact method | |
|---|---|---|---|---|
| Description | Number | |||
| National Genebank | China | Glycine max | 31,432 |
Chinese Crop Germplasm Information System |
| Glycine soja | 9684 | |||
| NARO Genebank | Japan/Ibaraki Prefecture | Glycine soja | 2127 | http://www.gene.affrc.go.jp/databases-plant_search_en.php |
| Glycine max | 7683 | |||
| G. tabacina | 8 | |||
| G. tomentella | 1 | |||
| G. wightii | 2 | |||
| LegumeBase | Japan/Miyazaki Prefecture | Glycine soja | 715 | https://www.legumebase.brc.miyazaki-u.ac.jp/top.jsp |
| Glycine max | 265 | |||
| RILs | 263 | |||
| Mutant | 28 | |||
| National Agrobiodiversity Center | Republic of Korea | Glycine max | 19,338 | |
| Glycine soja | 3229 | |||
| Chung’s Wild Legume Germplasm Collection | Republic of Korea | Glycine soja | > 3000 | Email: chung@chonnam.ac.kr |
Germplasm collections in Japan
Approximately 13,400 soybean accessions (accessed on May 22, 2019) including local landraces, improved cultivars, and breeding lines developed by regional Japanese agricultural research institutes or introduced from agricultural research institutes overseas are maintained at the NARO Genebank (Table 1), operated by the Ministry of Agriculture, Forestry, and Fisheries (MAFF) of Japan.
LegumeBase, maintained by the National BioResource Project (NBRP), distributes approximately one thousand wild and cultivated soybean accessions via an online system. NBRP is directed by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This project also catalogs and distributes other experimental genetic resources such as EMS mutant lines and recombinant inbred lines (RILs) with the associated genotype information.
Germplasm collections in the Republic of Korea
The National Agrobiodiversity Center of Korea had its beginnings in 1987 when a seed management office of Rural Development Administration (RDA) was established (Table 1). Four years later, in 1991, the seed management office was upgraded to the Genetic Resource Department of the National Institute. In 2008, the Genetic Resource Department was transformed into the National Agrobiodiversity Center under the National Academy of Agricultural Sciences, part of the RDA of the Republic of Korea. In addition to the National Agrobiodiversity Center, a well-maintained and highly diverse legume germplasm collection is being held at Chonnam National University under the Chung’s Wild Legume Germplasm Collection (Table 1).
Molecular breeding of soybean in East Asia to meet region-specific demands
Progress on soybean molecular breeding in China
Like many other regions under soybean cultivation, the Chinese soybean yields are largely hampered by the soilborne pathogen Phytophthora sojae. Multiple races of P. sojae have been identified in China since 1989. At the same time, certain single dominant resistance genes (Rps) have been found to confer complete immunity toward certain races of P. sojae. So far though, there is no one Rps gene that confers absolute immunity against all P. sojae races, and thus, finding a broad-spectrum Rps gene/allele has become the Holy Grail for soybean breeding. A Chinese cultivar, Zaoshu 18, is able to resist ten out of 14 tested P. sojae isolates (Zhong et al. 2018b). A P. sojae resistance locus RpsZS18 was mapped to a 71.3 kb region on chromosome 2, which can be traced with two newly developed SSR markers (Table 2) (Zhong et al. 2018b). Another Chinese cultivar, Huachun 18, confers resistance toward at least 12 P. sojae isolates (Zhong et al. 2018a). After QTL mapping and fine mapping, a P. sojae resistance locus RpsHC18 was narrowed down to a 157-kb region on chromosome 3 that contains seven exonic non-synonymous SNPs. Six of these SNPs are located on two NBS-LRR genes, making them the best candidates of the causal gene in RpsHC18. Consequently, two SNP markers (Table 2) were developed for marker-assisted breeding for P. sojae resistance (Zhong et al. 2018a).
Table 2.
Potential universal markers for marker-assisted breeding
| Trait | Allele | Allele phenotype | Donor accessionsa | Potential universal markersb | Details for markers | References |
|---|---|---|---|---|---|---|
| Biological nitrogen fixation | ||||||
| Super-nodulation | SS2-2 | 2–3 times more nodules compared with normal soybean cultivars | SS2-2 (mutant of Sinpaldalkong 2) |
A-specific allele Fw: CAACCTCACCGGCGTACTTCCGA Rv: CCTCAGCGTCTTCAACTTCGACAAACTC T-specific allele Fw: GAACAACCTCACCGGCGTACTTCCTT Rv: CCTCAGCGTCTTCAACTTCGACAAACTC |
The ‘A-allele’ encodes a full-length GmNARK protein. The ‘T-allele’ encodes a truncated GmNARK protein which is the mutant allele from SS2-2 |
Kim et al. (2005) |
| Abiotic stress tolerance | ||||||
| Salt tolerance | GmCHX1 | Salt-tolerant | W05 (Qi et al. 2014), Tiefeng 8 (Guan et al. 2014) |
Sequence marker: Conserved coding sequence of GmCHX1 Fw: ATGACGTTCAACGCGAGCACC Rv: TTAAAGTTCTTCGATAGCATCTT |
Encodes the same protein sequence as GenBank accession KF879911, needs to confirm with Sanger sequencing | Qi et al. (2014) |
| Phosphorus efficiency | GmHAD1 | Tolerance to low-phosphorus stress | HC 2 |
qP10-2 Fw: CATTTATCATGTGAAAGCAG Rv: AACCAAAACAATCCAAGTAT |
Cai et al. (2018d) | |
| Low temperature | qCTR-A2 | High seed yielding ability at low temperature | Toyoharuka | Sat_181, Sat_162, Sat_233 | qCTR-A2 is located near Sat_162 | Ikeda et al. (2009) |
| Low temperature | GmIRCHS | Prevent low temperature induced seed coat discoloration | Toyoharuka |
Toyoharuka specific Fw: GAGTTTGAAAAATGTATTCTTTCTCTTCC Rv: GTATCGCAGATTCCTCCTGC Toyomusume specific Fw: GCAAACCAAATCAAGTAAGAGCG Rv: CCCATTCCTTGATTGCCTTA |
GmIRCHS is a candidate gene for the I allele at the I locus. | Ohnishi et al. (2011) |
| Biotic stress tolerance | ||||||
| Phytophthora sojae | RpsHC18 | Phytophthora sojae resistance | Huachun 18 |
BARCSOYSSR_03_0254 (0.3 cM) and BARCSOYSSR_03_0272 (0.7 cM) |
Zhong et al. (2018a) | |
| Phytophthora root rot |
Glyma.02g245700 Glyma.02g245800 Glyma.02g246300 |
Phytophthora root rot resistance | Zaoshu18 |
ZCSSR33 Fw: TAGTTGATAGCACCTGGGGACA Rv: TTCTCAGTCTCAAATGCC ZCSSR46 Fw: AAAGGGAGAAGCAAGTAAT Rv: TCGCAAACAGTAAACACG |
Zhong et al. (2018b) | |
| Phytophthora sojae | Rps1d | PSR resistance | PI 103091 | Satt152-Sat_186 | Rps1d is located at 5.7 cM from Sat_186 | Sugimoto et al. (2008) |
| Soybean cyst nematode | rhg1s, rhg1g | SCN resistance to race 1 and 3 | PI84751(rhg1s), Gedenshirazu (rhg1g) | Satt309 and Sat_210 | Both rhg1s and Rhg4 are required for SCN resistance to race 1. | Kato et al. (2017), Suzuki et al. (2011) |
| Soybean cyst nematode | Rhg4 | SCN resistance to race 1 | PI84751 | Satt632, Sat_162 and Sat_157 | Both rhg1s and Rhg4 are required for SCN resistance to race 1. | Kato et al. (2017), Suzuki et al. (2011) |
| Soybean mosaic virus |
Glyma.13g194700 and Glyma.13g195100 |
Resistance to Soybean mosaic virus SC20 | Qihuang-1 |
gm-ssr_13-14 Fw: ACAAAACGACAATTTTACCCA Rv: CGACCGGATACTGTAAAAGA gm-indel_13-12 Fw: GGTGAGTTTGGTCGATAATGT Rv: AGACTCGCGTAAGGTGTTCT |
Expected amplicon size of gm-ssr_13-14: 181 bp Expected amplicon size of gm-indel_13-12: 484 bp |
Karthikeyan et al. (2018) |
| Soybean mosaic virus | Rsv3 | Resistance to Soybean mosaic virus strain C and D | Harosoy, Fukuibuki, Dewamusume, Suzuyutaka |
BARCSOYSSR_14_1414, BARCSOYSSR_14_1416, BARCSOYSSR_14_1392, BARCSOYSSR_14_1403, M3SattM Fw: GATTGAAGGGTCACCAATCG, Rv: CGCTATCATCCAATGACCAA |
Kato et al. (2016) | |
| Peanut stunt virus | Rpsv1 | Resistance to two PSV isolates, PSV-K and PSV-T | Fukuyutaka, Harosoy, Hyuga | Satt435-Sat_244 | Saruta et al. (2011) | |
| Common cutworm | CCW-1 | Antibiosis resistance to common cutworm | Himeshirazu |
Satt220–Satt175 CCW1_15 Fw: GGGAAACAGGGCAAGGAAC Rv: CAAGTGTTGTCTCGAATGGAAA |
Komatsu et al. (2005) | |
| Common cutworm | CCW-2 | Antibiosis resistance to common cutworm | Himeshirazu | Satt567–Satt463 | Komatsu et al. (2005) | |
| Common cutworm | qRslx1 | Antixenosis resistance to common cutworm | Himeshirazu |
CCW1_15 Fw: GGGAAACAGGGCAAGGAAC Rv: CAAGTGTTGTCTCGAATGGAAA |
QTL for antixenosis resistance, qRslx1, is close to CCW-1. | Oki et al. (2018) |
| Common cutworm | qRslx2 | Antixenosis resistance to common cutworm | Himeshirazu | Sat_218 | Oki et al. (2018) | |
| Nutritional values | ||||||
| Seed oil content | qOIL_8_1 | Increased oil content | Jidou 12 |
Satt177 Satt341 |
Yan et al. (2019) | |
| P34 allergen content | Low P34 | Low P34 content | PI567476, PI603579A |
Fw: CAAACTGTCATTCCCTGTTGG Rv: AAGAAGCAACACAAGGAAACC |
Size of low P34-content allele larger, migrates more slowly on the gel | Jeong et al. (2013) |
| Morphological traits | ||||||
| Seed Coat color | I | Non-pigmented seed coat | All accessions with non-pigmented seed coat |
PCR marker: Inversion junction Fw: GGCCTGTTGTAGTGGAAAATTC Rv: CCTGGATACAGGTACGTTTTACT |
An inversion in the I locus corresponds to the non-pigmented seed coat phenotype | Xie et al. (2019) |
| Plant height | GmDW1 | Dwarf | Zp661 |
SNP08-1 Fw: TGCACCAAAACCAGCTCAAT Rv: AGGATCAGAAGGCTTGGGAC |
Li et al. (2018b) | |
| Growth Habit | Dt1 | Indeterminate growth | Moshidou Gong 503 |
CAPS marker: TFL1b-pro Fw: CCATGCTTAATCGGCATCACT Rv: GGTGGTGGCATAGTTTAATT dCAPS marker: TFL1b-ex4 Fw: GGCTGCTGTCTACTTCAATGTCTAG Rv: GCCACATGTGAAGATCAACTTCCA |
Digestion of TFL1b-pro amplification product of Dt1 allele with NdeI gives 217 and 193-bp fragments Digestion of TLF1b-ex4 amplification product of Dt1 allele with XbaI gives 155- and 21-bp fragments |
Liu et al. (2010a) |
| dt1 | Determinate growth | Misuzudaizu | ||||
| Internode distance | qSI13-1 | Short internode, lodging resistance | Y2 | Sat_375–Satt657 | Oki et al. (2018) | |
| Lodging | qLS19-1 | Lodging tolerance | Toyoharuka | Sat_099 | Yamaguchi et al. (2014) | |
| Seed Shattering | pdh1 | Shattering resistance | Hayahikari, Harosoy, Wasekogane, Karlyutaka, Jack, Yukihomare |
Dir_CAPS Fw: GCCCTCGTTGTGTTCTTCAT Rv: GCGTTGCTTCCGTTGTAGAT Dir_SNP_ARMS Outer forward: CTCTTGGCATAGCTAGGGAAAAGCACTA Outer reverse: GAAAACCACTATGTTTCCGAAGTGGAAC Inner forward: GAAGAGGCCACAACATGCACCATACT Inner reverse: TGTCGTGGAAGAAGAGGACTAAGTGTTT |
SR: can be digested with NheI SS: cannot be digested with NheI Common band: 262 bp SR: 169 bp (SNP: T) SS: 146 bp (SNP: A) SS = shattering sensitive |
Funatsuki et al. (2014) |
| Isoflavone content | qIF5-1 | Isoflavone-related traits | Wayao | Bin799–800 on chromosome 05 | Cai et al. (2018c) | |
| Developmental stages | ||||||
| Flowering time | E11 | Flowering time and maturity | Minsoy (PI27890) and Archer (PI54687) | Wang et al. (2019) | ||
| Flowering time | qFT12.1 | Delay flowering time | Wild soybean JWS156-1 |
BARCSOYSSR_12_0220 and BARCSOYSSR_12_0368 Satt192 |
Liu et al. (2018a) | |
| Flowering time in tropical region | j (HT1 m)/ j-4 | Delay flowering time in inductive short-day | Huaxia 3, etc |
J‐Indel‐spanning ‐Fw GGTTTTGTGATGTGATCGGAGCC J‐Indel‐spanning‐Rv AGCCACATTTGTCTGGTGCTGATTTCC |
A T>A indel results in a truncated GmELF3 gene product | Yue et al. (2017) |
| Flowering time in tropical region | j | Delay flowering time in inductive short-day |
PI 159925, BR121, etc |
J‐620F (haplotype analysis) CTATTGTGTGAGTGAGATTGATTTGGAT J‐5190R (haplotype analysis) CTACTCTTTCGGGTAAAGCAATTACTAC |
Detection of mutations on GmELF3 | Lu et al. (2017), Yue et al. (2017) |
| Flowering time | e1-fs, e1-as, e1-nl, e1-re | Flowering time and maturity | Sakamoto wase (e1-fs), Harosoy and Williams 82 (e1-as), Toyosuzu and Toyomusume (e1-nl), Gokuwase-Kamishunbetsu (e1-re) |
The e1-as and e1-fs are discriminated as shorter digested fragments from other alleles by the TaqαI and HinfI digestion of PCR products generated with specific primers G33snpTaqcutF TCAGATGAAAGGGAGCAGTGTCAAAAGAAGT G33snpTaqcutR1 TCCGATCTCATCACCTTTCC The e1-re and e1-nl alleles can be distinguished by amplification of a shorter fragment and no amplified product from other alleles using mixture of three primers E1M0535_Fw CCGTTTGATTGGTTTTTGGT E1P0305_Rv CCCTTCAGTTTCTGCAGCTC e1re_0188Rv GAGAAGACAAACAATTCGAG |
Tsubokura et al. (2014), Xia et al. (2012) | |
| Flowering time | e1-like-b (e1lb) | Incandescent long daylength insensitivity | Zeika, Yubileinaya, Sonata |
dCAPS Fw: GTGTAAACACTCAAAGTCCTT Rv: CGTCTTCTTGATCTTCCAACG PCR fragment of the loss-of-function e1lb allele is digested into two fragments by restriction enzyme of HpyCH4IV |
Glyma.04G143300 is considered as the candidate gene of e1lb | Zhu et al. (2019) |
| Flowering time | e2-ns | Flowering time and maturity | Enrei, Harosoy, Toyomusume, many others |
SoyGI_dCAP_Dra_Fw GAAGCCCATCAGAGGCATGTCTTATT SoyGI_dCAP_Dra_Rv GAGGCAGAGCCAAAGCCTAT PCR fragment of e2-ns can be digested with DraI. |
Tsubokura et al. (2014), Watanabe et al. (2011) | |
| Flowering time | e3-Mo, e3-tr | Flowering time and maturity | Moshidou Gong 503 (e3-Mo), Enrei (e3-tr) |
The e3-tr with a smaller PCR fragment can be distinguished from the other alleles using primer mixture of E3_08557Fw: TGGAGGGTATTGGATGATGC E3_09908Rv: CTAAGTCCGCCTCTGGTTTCAG E3Ha_1000Rv: CGGTCAAGAGCCAACATGAG e3tr_0716Rv: GTCCTATACAATTCTTTACGACG Further, the e3-Mo allele was distinguished from others by the MseI digestion of the PCR products using the primers E3_08094Fw: TTGCATGAAGTTTTGGTTGC E3_08417Rv: CAACTGAACTGAAGACCCACAA |
Tsubokura et al. (2014), Watanabe et al. (2009) | |
| Flowering time | e4-SORE-1 | Flowering time and maturity | Hayahikari, Yukihomare, Kariyutaka |
The e4-SORE-1 allele can be distinguished from E4 by allele-specific primer pairs PhyA2-Fw: AGACGTAGTGCTAGGGCTAT PhyA2-Rv/E4: GCATCTCGCATCACCAGATCA PhyA2-Rv/e4: GCTCATCCCTTCGAATTCAG |
Liu et al. (2008), Tsubokura et al. (2014) | |
| Flowering and maturity | e9 | Late flowering time and maturity | TK780, Shensei |
Interval between Satt215 and Satt431 M5-indel-30600002- Fw: TGAAGCATTGTCCCCTGTTTCTA Rv: GGTATTTGTCATCATGGCATCCA M7-indel-30845759 Fw: AGGATGAAAAGAGAAGATGTTG Rv: AGATTTTCTTGATAGAATAGTAACG. |
GmFT2a (Glyma16 g26660) or GmFT2b (Glyma16g26690) are considered as the candidate genes of E9 | Kong et al. (2014) |
| Flowering time | qDTF-J | Early flowering | Sakamoto wase, Otome wase |
Interval between SSR-J5 and SSR-FT3a SSR-J5 Fw: CAATATGACTGGAGGCTCATGA Rv: CCTAAGTAGGCCTACCAAT SSR-FT3a Fw: AAGCAGTGTGACTCAGTGAA Rv: GGACATCAGGATCCACCAT |
GmFT5a (Glyma.16G044100) is considered as the candidate gene | Takeshima et al. (2016) |
| Maturity | qdfm1 | Short reproductive period | Ippon-Sangoh | Interval between Sat_128 and Satt583, Satt519 is closest | Komatsu et al. (2011) | |
| Maturity | qdffgm1 | Short reproductive period | Peking |
Interval between SSR markers s008000014-2 and T001111280 m, s008000014-2 is closest. s008000014-2 Fw: TACAACTTTGATGTTCCCATCTT Rv: AAGCTCCTTCCTCTCAAGTTGTT T001111280 m Fw: GGTGCTCTTCCTCACATTAGAGA Rv: TAGGTTGCAAGTATAGCGTGTTT |
Watanabe et al. (2018) | |
aOnly major cultivars used in the reference study will be listed
bFw, forward primer; Rv, reverse primer; primer sequences are read from 5’ to 3’
China has its own system of classifying soybean mosaic virus (SMV), which designates the 22 SMV strains in China as SC1–SC22. Among them, SC20 is the most widespread in the five provinces in southern China, affecting soybean cultivation in that region. A SMV resistance QTL was fine-mapped to a 79-kb region on chromosome 13 of Qihuang-1, flanked by a new SSR marker and a new INDEL marker (Table 2) (Karthikeyan et al. 2018). Two TIR-NB-LRR protein-encoding genes that are highly polymorphic between the two parents in the mapping population are identified within this QTL, and they are highly likely to be the candidates of the SC20 resistance gene (Karthikeyan et al. 2018).
To introduce the resistance against Micosphaera diffusa Cooke & Peck, which causes powdery mildew disease (PMD), to Chinese soybean cultivars, researchers have made use of the PMD-resistant soybean cultivar BRSMG68 (B13) from Brazil (Jiang et al. 2018). The PMD resistance locus was mapped to a 188-kb region on chromosome 16 of B13 that contains 17 disease resistance genes accounting for 78% of the phenotypic differences within the mapping population (Jiang et al. 2018).
Alterations in plant architecture have played an important part during the Green Revolution to improve grain yield. Through EMS mutagenesis, a dwarf mutant (dw) was found in the Chinese soybean cultivar Zhongpin 661 background (Li et al. 2018b). The causal mutations were found on a gene encoding the ent-kaurene synthase, which disrupted the biosynthesis of gibberellins (Li et al. 2018b). Although the dwarf allele showed its potential for breeding new cultivars with high harvest index (Table 2), the actual effect of the mutation on yield has not yet been fully assessed.
Soybean cultivation has expanded beyond its natural habitat since domestication. Since soybean production is highly sensitive to photoperiod, efforts have been spent on studying the flowering and maturation time of soybean to maintain the yield at different latitudes. E1–E4 are the well-characterized major loci controlling the flowering and maturation times of soybean (Jiang et al. 2014; Miladinovic et al. 2018; Tsubokura et al. 2014). In a recent study, using two genetic populations with fixed genotypes in the E1–E4 loci, researchers were able to identify two other loci in soybean contributing to flowering and maturation, specifically in chromosome 4 of the Dongnong 50 × Wm82 RIL population and in chromosome 6 of the Suinong 14 × Enrei RIL population, respectively (Kong et al. 2018). However, these two QTLs are possibly linked with the other E loci, making the dissection of them difficult.
When grown in the tropical region, temperate soybean cultivars tend to flower before accumulating a reasonable amount of biomass, thus reducing the yield. Therefore, those soybean cultivars possessing the recessive long-juvenile (LJ) trait which allows them to stay vegetative for longer under inductive short-day conditions would be desirable for that region. The J locus controlling the long-juvenile trait was mapped to chromosome 4 on the soybean genome (Table 2) (Lu et al. 2017; Yue et al. 2017). Recessive j alleles were found to be actually GmELF3 carrying either frameshift or missense mutations that either abolish or diminish the functionality of the encoded protein (Lu et al. 2017; Yue et al. 2017). Manipulation of FT homologs in soybean provides another way to generate germplasm with longer growth period and higher yield in tropical region. Liu et al. (2018b) overexpressed GmFT1a, a flowering inhibitory gene, and created materials with delayed flowering and maturity and increased biomass and yield in short-day condition (Liu et al. 2018b). Cai et al. (2019) employed the CRISPR/Cas9 system to specifically knock out the soybean flowering-promoting genes GmFT2a and GmFT5a and the ft2a ft5a double mutant flowered about 31 days later than that wild type and produced more pods and seeds (Cai et al. 2019). Three Chinese soybean cultivars Longhuang #1-3 were developed by screening for the presence of the functinonal GmCHX1 gene (Qi et al. 2014) in drought tolerant accessions (Table 2), resulting in highly adaptive soybeans for improving agricultural productivity on marginal lands.
Pod distribution on the soybean plant also affects the harvest index and is influenced by a number of minor-effect loci. Using 7-year worth of data, a study identified 11 major QTLs and 90 epistatic pairs controlling the first pod height (Jiang et al. 2018). Another study identified 47 QTLs associated with the distribution of pods with different numbers of seeds at the upper, middle, and lower sections of the soybean plant (Ning et al. 2018). It appears that pod distribution is governed by a large number of QTLs that are difficult to be introduced using simple marker-assisted selection. Instead, genomic selection would be a better way of selecting supreme offsprings with a desirable pod distribution pattern.
Two Chinese cultivars with contrasting oil and protein contents, Linhefenqingdou and Meng 8206, were used to build an RIL population for QTL mapping (Karikari et al. 2019). With data from six different environmental conditions, 44 main-effect QTLs for protein and oil contents were identified in an environment-dependent manner (Karikari et al. 2019). Fifteen of the QTLs were novel, while 20 had R2 > 10%. Among them, qPro-7-1 was a new QTL on chromosome 7 that was consistently detected in three individual environmental conditions and in the combined environment, accounting for at least 13.6% of the total variations (Karikari et al. 2019). qOil-8-3, qOil-10-2, and qOil-10-4 were detected in at least two individual environmental conditions and in the combined environment, explaining 6.2–30.6% of the total variations (Karikari et al. 2019). There were significant additive interactions among seven of these loci, as well as interactions between themselves and the environmental conditions, while epistasis was observed in three loci pairs (Karikari et al. 2019).
Phosphorus (P) is a limiting macronutrient that the soybean can only obtain from soil deposition and fertilizer. Therefore, increasing P-efficiency in soybean would improve the adaptability of soybean to low-P environments. QTL mapping for the P-efficiency locus was done using an RI population originated from HC 2 and Wayao (Cai et al. 2018d). HC 2 was bred from the Chinese cultivar Guizao1 and the Brazilian cultivar CONFIANGA with high P-efficiency, while Wayao is a Chinese landrace with low P-efficiency. Fifteen major QTLs were identified, with qP10-2 at chromosome 10 explaining the highest percentage of variations (13.98%) (Cai et al. 2018d). This QTL contains an acid phosphatase-encoding gene, GmHAD1. Ectopic expression and overexpression of GmHAD1 in Arabidopsis and soybean hairy root were both shown to improve P-efficiency (Cai et al. 2018d).
To enhance the isoflavone contents in soybean seeds, a QTL mapping study identified 108 loci in a biparental population (Cai et al. 2018c). Fifteen of these QTLs were consistently found in different environmental conditions, with each explaining at least 1.8% of the variations (Cai et al. 2018c). Among them, qIF5-1, spanning a 611-kb region on chromosome 5, could explain 6.37–59.95% of the observed variations in the acetyldaidzin, daidzin, genistin, daidzein, glycitin, malonyldaidzin, malonylglycitin, malonylgenistin, genistein, and total isoflavone contents in the mapping population (Cai et al. 2018c). Although the causal gene of this QTL is still unknown, this QTL could be introduced into cultivars to improve isoflavone contents using MAS.
Progress on soybean molecular breeding in Japan
In Japan, soybeans are now cultivated in paddy fields in place of rice to prevent the overproduction of rice, according to a government policy. As a result, many problems related to lowland conditions have caused widely fluctuating and low yields in Japanese soybean production. Resistance genes to the widespread disease caused by Phytophthora sojae and Calonectria crotalaria, as well as seed-flooding tolerance at the germinating stage, have become very important for achieving higher yields. In addition, due to the widespread mechanization of farm operations, improvement in plant adaptation to high plant density and in the resistance against pod shattering during machine harvesting is desired goals. To this end, four cultivars with pod dehiscence resistance, ‘Sachiyutaka A1 gou’ (Hajika et al. 2016), ‘Fukuyutaka A1 gou’ (Yamada et al. 2017), ‘Enreinosora’ (Yamada et al. 2017), and ‘Kotoyutaka A1 gou’ (MAFF, Japan), have been developed by MAS through repeated backcrossing of progenies with the pod-shattering-resistant pdh1 mutant originating from ‘Hayahikari.’ Introduction of pdh1 into other leading Japanese soybean cultivars by MAS is underway.
The soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is one of the most damaging pests of soybean. SCN races 1, 3, and 5 have been reported in Japan. Soybean mosaic virus (SMV), soybean dwarf virus (SbDV), southern bean mosaic virus (SBMV), and peanut stunt virus (PSV) are also considered to be serious pathogens affecting seed quality. In southwestern Japan, significant damage to soybean plants is caused by the common cutworm (CCW, Spodoptera litura Fabricius). Therefore, breeding resistance into the crop against these pests and pathogens is essential for improving soybean yield.
There are two main SCN resistance resources for breeding in Japan. The Japanese cultivar ‘Gedenshirazu’ (PI 561360) has the SCN resistance QTLs Rhg4/rhg1g/rhg2g, while the Korean cultivar LG-59 (PI84751) has Rhg4/rhg1s/rhg2s. Rhg4, rhg1s, and rhg2g or rhg2s are necessary for race-1 resistance, and either rhg1s or rhg1g is necessary for race-3 resistance. rhg1g of ‘Gedenshirazu’ and rhg1s of PI84751 are allelic on chromosome 18, while rhg2g of ‘Gedenshirazu’ and rhg2s of PI84751 are non-allelic but are located close to each other on the same chromosome (Suzuki et al. 2011). SMV in Japan is classified into five major strains, A, B, C, D, and E (Takahashi et al. 1980). Many Japanese varieties are resistant to the A and B strains on account of existence of Rsv1, while resistance to the C and D strains has been limited. Rsv3 derived from ‘Harosoy’ on chromosome 14 has been confirmed to confer resistance to the Japanese SMV strains C and D by MAS using markers linked to Rsv3 (Kato et al. 2016). Rps1d and Rps1k for Phytophthora sojae resistance (PSR) are the most effective resistance genes among 14 Rps genes in Japan by using 109 P. sojae isolates from 14 regions (Moriwaki 2010). Sugimoto et al. (2008) mapped Rps1d to chromosome 3 using progenies from a cross between ‘Tanbakuro’ and ‘PI 103091’ to determine selectable SSR markers (Sugimoto et al. 2008). PSV resistance is traced to a single dominant gene, Rpsv1, near Satt435 on chromosome 7 by using RILs with PSV resistance from a cross between ‘Hyuga’ and ‘Enrei’ (Saruta et al. 2011). Two QTLs for antibiosis resistance to CCW, CCW-1, and CCW-2, both on chromosome 7, have been identified using progenies from a cross between ‘Fukuyutaka’ and ‘Himeshirazu’ (Komatsu et al. 2005). Two QTLs, qRslx1 and qRslx2, controlling antixenosis resistance have been identified from the same population (Oki et al. 2011).
By integrating the above information, the resistance genes against SCN, SMV, and SbDV can be incorporated into one target cultivar by high-resolution PCR fragment analyses using a florescent sequence machine (Kato et al. 2017). There has been a certain degree of success in this endeavor. For example, ‘Yukihomare R’ with resistance to SCN races 1 and 3 was developed by MAS for rhg1s and Rhg4 from PI 84751 and repeated backcrossing with ‘Yukihomare’, a leading cultivar in Hokkaido (Suzuki et al. 2017). In contrast, ‘Suzumaru R’ with resistance to SCN races 1 and 3 was developed by MAS for Rhg4 and rhg1s derived from PI 84751 and rhg2g from ‘Gendenshirazu’ through repeated backcrossing with ‘Suzumaru’, a leading cultivar for natto in Hokkaido (Kurosaki et al. 2017). ‘Fukuminori’ is developed by the introgression of two QTLs for antibiosis resistance, CCW-1 and CCW-2, into ‘Fukuyutaka’ by MAS with repeated backcrossing (Oki et al. 2011). Nearly all the characteristics of ‘Fukuminori’ are the same as those of ‘Fukuyutaka’ except for a slightly smaller seed size and the resistance to CCW. The SMV-resistant ‘Hyoukeikuro4 gou’ (MAFF, Japan) and PSR-resistant ‘Hyoukeikuro5 gou’ (MAFF, Japan) were developed by MAS and repeatedly backcrossed with a large black-seeded cultivar ‘Hyoukeikuro3.’
The combination of alleles at maturity-related loci is very important to develop a new cultivar with early harvesting for rotations with different crops or to adjust maturity of a new variety which can adapt to environments at different latitudes retaining the same seed quality with the similar genetic background. Among ten major classical genes, E1–E9 and J, E1 (Xia et al. 2012), E2 (Watanabe et al. 2011), E3 (Watanabe et al. 2009), E4 (Liu et al. 2008), E9 (Kong et al. 2014), and J (Lu et al. 2017) have been isolated. In addition, responsible genes for E1-like-b (Zhu et al. 2019) and qDTF-J (Takeshima et al. 2016) have been reported. Two QTLs, qDfm1 (Komatsu et al. 2011) and qDFFGm11 (Watanabe et al. 2018), have been identified as QTL controlling post-flowering period on chromosome 11. Allelic variation of E1, E2, E3, and E4 genes in the major soybean germplasm has been examined by using allele-specific DNA markers (Tsubokura et al. 2014).
Chilling tolerance in northern Japan, drought and high-temperature tolerance in southwestern Japan, and waterlogging tolerance throughout the whole country are also important for Japanese soybean. The cold (CD) tolerance QTL was mapped to chromosome 8 near the GmIRCHS gene by using two sets of RILs between a CD-tolerant ‘Toyoharuka’ and CD-susceptible cultivars (Ohnishi et al. 2011). The markers developed are currently used in MAS in Hokkaido. The QTL for the high seed yield trait at low temperature of ‘Toyoharuka’ was mapped near Sat_162 on chromosome 8 by using the RILs of ‘Toyoharuka’ × ‘Toyomusume’ (Ikeda et al. 2009). By using these same RILs, a research group found that the lodging tolerance of ‘Toyoharuka’ is controlled by a major QTL, qLS19-1, that is not associated with maturity or growth habit on chromosome 19 (Yamaguchi et al. 2014). Another QTL on chromosome 13, qSI13-1, the determinant of short internodal length and reduced plant height of the cultivar, ‘Y2’ (Oki et al. 2018), is being used to control plant height to prevent lodging.
Progress on soybean molecular breeding in the Republic of Korea
Officially, soybean breeding started in Korea with the release of the first landrace, Jangdanbaekmok in 1913. During the past 100 years, more than 178 soybean cultivars have been developed through hybridization-based breeding (87%) and registered with the two Korean national institutes, the RDA-Genebank Information Center (http://www.genebank.go.kr) and the Korea Seed and Variety Service (http://www.seed.go.kr). Most of the cultivars have been registered during the last 30 years, suggesting that the availability of genetic and genomic information has accelerated the soybean breeding efforts in Korea (Kim et al. 2011). Efforts of soybean breeding in the Republic of Korea have been dedicated to the improvement in nutritional values, disease and stress resistance, yield, and nitrogen fixation.
Soybean yield usually consists of three major components: the number of pods per plant, the number of seeds per pod, and seed weight. It has been found that the more branches the plant has, the higher will be the number of pods and thus the yield. One out of five branch number-associated quantitative trait nucleotides (QTNs) in the Korean soybean collection was found to overlap with a major branch number QTL, qBR6-1, which contains the gene BRANCHED1 (BRC1; Glyma.06g210600). Specifically, one missense mutation and two SNPs upstream of BRC1 were linked to branch numbers in 59 soybean accessions. BRC1 encodes the transcription factors TEOSINTE-BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTORS (TCP) and functions as a regulatory repressor of branching (Shim et al. 2019). Additionally, it has been proposed that the number of seeds per pod is directly linked to narrow leaflets in soybean. The mapping of the BC3F2 population from a cross between two cultivars, Sowon (narrow leaflets and high number of seeds per pod) and V94-5152 (broad leaflets and low number of seeds per pod), showed that Sowon carries a recessive allele of the ln gene on chromosome 20 (Jeong et al. 2012). The transition from broad (Ln) to narrow leaflet (ln) is associated with an amino acid substitution in the EAR motif encoded by a gene designated as Gm-JAGGED1 (Glyma20g25000), which is homologous to the Arabidopsis JAGGED (JAG) that regulates lateral organ development and its variant exerts a pleiotropic effect on fruit patterning (Jeong et al. 2012). Later studies by the same group suggested that Glyma.08g281900 could be the Lf1-encoding gene that controls lateral organ development. It was concluded that this gene possibly exerts a pleiotropic effect on the number of seeds per pod (Jeong et al. 2017), making it a good target for breeding.
While the soybean seed is nutritious, it also contains several anti-nutritional factors that are harmful to both humans and livestock. For example, the seed protein, P34, is a potential allergen which causes complications in soybean-sensitive patients. To develop lines with low P34, PI567476 was crossed with the Korean cultivar, Hwanggeum, and the resulting F2 and F3 populations were studied for the identification of molecular markers for the three copies of the gene encoding P34, i.e., Glyma08g12270, Glyma08g12280, and Glyma05g29130. As a result, a molecular marker (Table 2) and a polyclonal antibody were developed successfully for the selection of low P34-expressing lines (Jeong et al. 2013).
The beany flavor of the soybean seed is due to the seed lipoxygenase isozymes, LOX1-3, with LOX2 being the biggest contributor to the beany flavor, which is sometimes undesirable. Jinpumkong 2, a Korean soybean cultivar, lacks all three lipoxygenases (having the null alleles, lx1, lx2, and lx3). Using the RILs of a cross between Pureunkong 9 and Jinpumkong 2, a 175-bp fragment was identified within the Lx2 gene on chromosome 13 that is retained in G. soja but has become rare in the G. max. A single-nucleotide-amplified polymorphism (SNAP) marker was subsequently made available for soybean breeders to facilitate the identification of cultivars/lines lacking LOX2 (Shin et al. 2008, 2012).
To improve the oxidative stability and quality of soybean oil, breeding programs have mainly focused on reducing the saturated fatty acid and linolenic acid contents and increasing oleic acid in the oil. Hence, delta-12 fatty acid desaturase 2 (FAD2), which converts oleic acid (18:1) to linolenic acid (18:2), becomes the target for modification via molecular breeding. There are two FAD2 loci, FAD2-1A (Glyma10g42470) and FAD2-1B (Glyma20g24530), in the soybean genome. To develop high-oleic-acid lines (> 80% of total oil content vs. an average of 20-50% among existing soybean cultivars), efforts were made to combine a recessive mutant allele at the FAD2-1A locus (Glyma10g42470) and a recessive mutant allele at the FAD2-1B locus (Glyma20g24530) using molecular marker assays (Pham et al. 2011). In a later study, a non-synonymous SNP on FAD2-1A (S117 N) from 17D, an EMS mutant of Wm82, and another one on FAD2-1B (P137R), a mutant allele from PI 283327, were identified to be highly associated with the high-oleic-acid phenotype (Kulkarni et al. 2018). Similarly, an EMS mutant, PE1690, derived from the cultivar, Pungsannamul, was found to be low in linolenic acid. The phenotype was the result of a single-base mutation (W128*) in the GmFAD3A gene on the locus Glyma14g37350, which rendered the desaturase enzyme non-functional (Kim et al. 2015). A derived cleaved amplified polymorphic sequence (dCAPs) assay specific for this artificial polymorphism was thus developed for future breeding activities (Kim et al. 2015).
Soybean contains significant amounts of bioactive secondary metabolites, particularly soyasaponins. Studies in Korea reported natural variations in saponin contents in wild soybeans and identified different group A acetylsaponin-deficient mutants (Krishnamurthy et al. 2014a, b, c; Panneerselvam et al. 2013). Apart from saponins, other compounds such as soy isoflavones, polyphenols, flavonoids, proanthocyanidins, and phenolic acids have also been analyzed in large-scale wild soybean collections in Korea as well as in wild soybeans from other natural habitats. These studies identified unique germplasm accessions with higher amounts of these beneficial/desirable secondary metabolites that will be an important resource for future breeding (Ha et al. 2018; Nawaz et al. 2018; Tsukamoto et al. 2018). In additional to beneficial metabolites, trace harmful metabolites such as raffinose are also found in soybean. A novel allele of the putative soybean raffinose synthase gene, RS2, was discovered in PI200508, that was associated with low raffinose and stachyose contents. New Korean soybean cultivars, Daewon, Cheongja, and Danmiput, containing low levels of raffinose and stachyose were subsequently developed based on a specific marker assay for the PI200508 RS2 allele that allows for the direct selection of the low-raffinose and stachyose phenotype (Yang et al. 2014).
As mentioned before, the genetic control of flowering time is very important in photoperiod-sensitive soybean. Both wild (IT182932) and cultivated (Wm82) soybean genomes were blasted against Arabidopsis genes related to photoperiod-dependent flowering time and 118 genes were singled out based on functional DNA variations and insights into duplicated regions (Kim et al. 2012). Using two RIL populations, Jinpumkong 2 × SS2-2 (J × S) and Iksannamulkong × SS2-2 (I × S), QTLs related to days to flowering (DF) (Liu et al. 2010b) and days to maturity (Liu et al. 2011) were identified. In total, 18 QTLs for DF and days to maturity along with six yield-related traits were isolated from these two RIL populations (Liu et al. 2011). Whole-genome resequencing has also been used to identify genes governing DF in an early-flowering soybean cultivar, Josaengserori (mutant from Seoritae). Among 30 DF-related genes detected using SNPs, Glyma02g33040, Glyma06g22650, Glyma10g36600, Glyma13g01290, Glyma14g10530, Glyma16g01980, Glyma17g11040, Glyma18g53690, and Glyma20g29300 contained non-synonymous substitutions between Josaengserori and Seoritae. Among these genes, it was discovered that the changes in Glyma10g36600 (GI), Glya02g33040 (AGL18), Glyma17g11040 (TOC1), and Glyma14g10530 (ELF3) in Josaengserori affected the expression of GmFT2a and resulted in early flowering (Lee et al. 2016).
Since legume root nodules contain symbiotic nitrogen-fixing rhizobia, being able to increase nodule number could potentially increase biological nitrogen fixation. The cultivar SS2-2, having increased number of nodules, is an EMS mutant of a Korean soybean cultivar Sinpaldalkong 2. The causal mutation was found to be a nonsense mutation of GmNARK. Using this discovery, a PCR-based SNAP marker was developed for the identification of supernodulating soybean plants (Kim et al. 2005, Table 2). Genetic mapping has helped identify two nodule-specific genes, GmPGN (a polygalacturonase-encoding gene) and GmEKN (a short nodule-specific gene). The former was located near a known QTL conferring resistance to soybean cyst nematode (SCN) on the molecular linkage group (MLG) B1 and the latter on MLG A2 (Jeong et al. 2006). Studies on nodule development in other countries reported that three gene loci, rj1, rj5, and rj6, control the initial nodule development in soybean. Fine genetic and physical mapping of the non-nodulating locus, rj1, suggested that this simple recessive allele was created by a single-base-pair deletion from a spontaneous mutation, resulting in a premature stop codon and a non-functional NFR1α (nod factor receptor) (Lee et al. 2011b).
Cloning of a cluster of NBS-LRR resistance gene candidates from MLG F of the virus-resistant soybean line, PI96983, showed that this cluster contains multiple genes which interact to produce customized responses to different SMV strains. In order to improve SMV resistance in soybean cultivars, efforts have been made to identify potential resistance genes. Three independent R gene loci, Rsv1, Rsv3, and Rsv4, have been reported in different soybean cultivars to confer resistance to SMV strains G1–G7 (Kim et al. 2016) and are associated with a cluster of genes encoding NB-LRR proteins. Specifically, one candidate gene (Glyma14g38533) has been singled out in the Rsv3 cluster as a possible causal gene (Redekar et al. 2016). These three SMV resistance loci confer variable levels of resistance in soybean. However, Rsv4 in particular provides novel resources for map-based cloning and genetic improvement in soybean against SMV because it confers durable type resistance. The BC3F2 population (309 individuals from a cross between Sowon and V94-5152) were used to identify four SNPs perfectly associated with SMV resistance. Interestingly, haplotype analysis suggested that the Rsv4 locus in G. max was recently introgressed from wild soybean. Therefore, if we want to find variations in Rsv4 or the other two Rsv loci in order to fully understand the resistance mechanism of soybean against SMV, we must look to the wild soybean germplasms (Ilut et al. 2016).
Progress has also been made in the discovery of genes/loci associated with the resistance against other pests and pathogens. A novel QTL (Raso 2) for the Korean biotype foxglove aphid (Aulacorthum solani) resistance in soybean has been identified in the wild Japanese accession, PI366121, using GoldenGate SNP microarray (Lee et al. 2015). Phomopsis seed decay (PSD) caused by Phomosis longicolla results in major damage to seed quality and leads to yield loss. Two QTLs for resistance to PSD have been identified in an RIL population derived from a cross between the PSD-resistant cultivar, Taekwangkong, and the PSD-susceptible line SS2-2, namely PSD-6-1 and PSD-10-2 at the intervals Satt100–Satt460 and Sat_038-Satt243 on chromosomes 6 and 10, respectively. These newly identified QTLs will help improve soybean resistance to P. longicolla (Sun et al. 2013).
Perspectives
According to the statistics published by the Food and Agriculture Organization of the United Nations, the per-hectare yield of soybean in the USA is 1.5–2 times those in Asian countries. This suggests that there is still room for the yields of Asian soybeans to improve, despite reaching a plateau in recent years. In addition to achieving higher yields, agriculture also faces challenges due to climate change. To produce high-quality soybean using methods that are acceptable to the consumer, traditional and advanced breeding techniques making use of mined genetic resources from germplasm collections are essential. The distribution and maintenance of these resources usually involve high costs and are influenced by national policies. China, Japan, and Korea, as the origins of soybean domestication, are the guardians of an appreciable amount of wild soybean resources. It would be of tremendous benefits to the rest of the world for these governments to facilitate the sharing of the materials across national boundaries for sustained soybean crop improvement.
Genome-wide association analysis, genomic selection, and genome editing will become essential tools for assessing favorable alleles among germplasms and combining them to develop new cultivars. Nowadays, high-throughput genome sequencing is no longer the limiting factor for population genomics. In addition, high-throughput phenotyping tools such as unmanned aerial vehicles with multiple sensors and advanced digital image processing techniques are being used to evaluate traits which previously could only be estimated using labor-intensive and destructive methods. Large-scale evaluations of such traits as spatial and temporal changes in canopy biomass, branching, lodging, light capture, and photosynthesis, which are highly correlated with productivity, are expected to provide new insights to guide molecular breeding.
Nevertheless, specific potentially adaptive alleles and genes may not already exist in nature even with the availability of large germplasm collections. New genetic resources could be created de novo by chemical/radiation mutagenesis and genome editing. The drawback for random mutagenesis by chemical or radiation is that the mutated alleles may cause pleiotropic effects and require detailed characterization, and the residual background mutations in the mutants will take time to be completely removed. However, these mutants can be useful as the foundation for genome editing, along with alleles from wild accessions. Nonetheless, current protocols for genome editing still rely largely on modifying existing elements in the genome rather than the creation of completely new genes.
Author contribution statement
H-ML coordinated the writing. GZ, TH, GC, HN, and H-ML conceptualized the review. BJ, AK, and GC wrote the sections specifically related to China, Japan, and Korea, respectively. M-WL, ZW, and F-LW integrated the information and wrote the first draft. M-WL and H-ML wrote the final version.
Acknowledgements
We thank Dr. Masatsugu Hashiguchi and Dr. Fukuhiro Yamasaki for their kind assistance in preparing the statistics for the genetic resources in National BioResource Project and the statistics of NARO Genebank, respectively. Ms. Jee Yan Chu copyedited the manuscript.
Funding
This work was supported by Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M‐403/16), and Lo Kwee‐Seong Biomedical Research Fund awarded to H-ML, and the Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute (YFJGJS1.0).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Man-Wah Li, Email: limanwah@cuhk.edu.hk.
Zhili Wang, Email: wangzhili0804@gmail.com.
Bingjun Jiang, Email: jiangbingjun@caas.cn.
Akito Kaga, Email: kaga@affrc.go.jp.
Fuk-Ling Wong, Email: lingwong@cuhk.edu.hk.
Guohong Zhang, Email: zhangguohong223@163.com.
Tianfu Han, Email: hantianfu@caas.cn.
Gyuhwa Chung, Email: chung@chonnam.ac.kr.
Henry Nguyen, Email: nguyenhenry@missouri.edu.
Hon-Ming Lam, Email: honming@cuhk.edu.hk.
References
- Akond M, Yuan JZ, Liu SM, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Kassem MA. Detection of QTL underlying seed quality components in soybean [Glycine max (L.) Merr.] Can J Plant Sci. 2018;98:881–888. [Google Scholar]
- Bao AL, Chen HF, Chen LM, Chen SL, Hao QN, Guo W, Qiu DZ, Shan ZH, Yang ZL, Yuan SL, Zhang CJ, Zhang XJ, Liu BH, Kong FJ, Li X, Zhou XA, Tran LSP, Cao D. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019;19:131. doi: 10.1186/s12870-019-1746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai YP, Chen L, Liu XJ, Sun S, Wu CX, Jiang BJ, Han TF, Hou WS. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE. 2015;10:e0136064. doi: 10.1371/journal.pone.0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J. 2018;16:176–185. doi: 10.1111/pbi.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai YP, Chen L, Sun S, Wu CX, Yao WW, Jiang BJ, Han TF, Hou WS. CRISPR/Cas9-mediated deletion of large genomic fragments in soybean. Int J Mol Sci. 2018;19:3835. doi: 10.3390/ijms19123835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai ZD, Cheng YB, Ma ZW, Liu XG, Ma QB, Xia QJ, Zhang GY, Mu YH, Nian H. Fine-mapping of QTLs for individual and total isoflavone content in soybean (Glycine max L.) using a high-density genetic map. Theor Appl Genet. 2018;131:555–568. doi: 10.1007/s00122-017-3018-x. [DOI] [PubMed] [Google Scholar]
- Cai ZD, Cheng YB, Xian PQ, Ma QB, Wen K, Xia QJ, Zhang GY, Nian H. Acid phosphatase gene GmHAD1 linked to low phosphorus tolerance in soybean, through fine mapping. Theor Appl Genet. 2018;131:1715–1728. doi: 10.1007/s00122-018-3109-3. [DOI] [PubMed] [Google Scholar]
- Cai YP, Wang LW, Chen L, Wu TT, Liu LP, Sun S, Wu CX, Yao WW, Jiang BJ, Yuan S, Han TF, Hou WS. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J. 2019 doi: 10.1111/pbi.13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calyxt Inc. First commercial sale of Calyxt high oleic soybean oil. Minneapolis-St. Paul: Calyxt Inc.; 2019. [Google Scholar]
- Carter TE, Nelson RL, Sneller CH, Cui Z. Genetic diversity in soybean. In: Boerma HR, Specht JE, editors. Soybeans: improvement, production, and uses. Madison: American Society of Agronomy; 2004. [Google Scholar]
- Chinese Academy of Agricultural Sciences (2016) Highly adaptive and high yield new soybean cultivar—Zhonghuang 13 (in Chinese). http://www.caas.cn/zt/2016nygzky/hgsew/266697.html. Accessed 15 July 2019
- Chung WH, Jeong N, Kim J, Lee WK, Lee YG, Lee SH, Yoon W, Kim JH, Choi IY, Choi HK, Moon JK, Kim N, Jeong SC. Population structure and domestication revealed by high-depth resequencing of Korean cultivated and wild soybean genomes. DNA Res. 2014;21:153–167. doi: 10.1093/dnares/dst047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–196. [Google Scholar]
- Crossa J, Perez-Rodriguez P, Cuevas J, Montesinos-Lopez O, Jarquin D, de los Campos G, Burgueno J, Gonzalez-Camacho JM, Perez-Elizalde S, Beyene Y, Dreisigacker S, Singh R, Zhang XC, Gowda M, Roorkiwal M, Rutkoski J, Varshney RK. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 2017;22:961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Du HY, Zeng XR, Zhao M, Cui XP, Wang Q, Yang H, Cheng H, Yu DY. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2016;217:90–97. doi: 10.1016/j.jbiotec.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, Sayama T, Ishimoto M, Fujino K. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci USA. 2014;111:17797–17802. doi: 10.1073/pnas.1417282111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan RX, Qu Y, Guo Y, Yu LL, Liu Y, Jiang JH, Chen JG, Ren YL, Liu GY, Tian L, Jin LG, Liu ZX, Hong HL, Chang RZ, Gilliham M, Qiu LJ. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014;80:937–950. doi: 10.1111/tpj.12695. [DOI] [PubMed] [Google Scholar]
- Ha J, Kim M, Kim MY, Lee T, Yoon MY, Lee J, Lee YH, Kang YG, Park JS, Lee JH, Lee SH. Transcriptomic variation in proanthocyanidin biosynthesis pathway genes in soybean (Glycine spp.) J Sci Food Agric. 2018;98:2138–2146. doi: 10.1002/jsfa.8698. [DOI] [PubMed] [Google Scholar]
- Hajika M, Funatsuki H, Yamada T, Takahashi K, Hishinuma A, Hirata K, Oki N, Yamada N, Komaki K, Matsunaga R. Development of a new pod dehiscence-resistant soybean cultivar ‘Sachiyutaka A1 gou’. Bull NARO Inst Crop Sci. 2016;16:1–34. [Google Scholar]
- Hymowitz T. On the domestication of the soybean. Econ Bot. 1970;24:408–421. [Google Scholar]
- Hyten DL, Song QJ, Zhu YL, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Ohnishi S, Senda M, Miyoshi T, Ishimoto M, Kitamura K, Funatsuki H. A novel major quantitative trait locus controlling seed development at low temperature in soybean (Glycine max) Theor Appl Genet. 2009;118:1477–1488. doi: 10.1007/s00122-009-0996-3. [DOI] [PubMed] [Google Scholar]
- Ilut DC, Lipka AE, Jeong N, Bae DN, Kim DH, Kim JH, Redekar N, Yang K, Park W, Kang ST, Kim N, Moon JK, Maroof MAS, Gore MA, Jeong SC. Identification of haplotypes at the Rsv4 genomic region in soybean associated with durable resistance to soybean mosaic virus. Theor Appl Genet. 2016;129:453–468. doi: 10.1007/s00122-015-2640-8. [DOI] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15:16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SC, Yang K, Park JY, Han KS, Yu S, Hwang TY, Hur CG, Kim SH, Park PB, Kim HM, Park YI, Liu JR. Structure, expression, and mapping of two nodule-specific genes identified by mining public soybean EST databases. Gene. 2006;383:71–80. doi: 10.1016/j.gene.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Jeong N, Suh SJ, Kim MH, Lee S, Moon JK, Kim HS, Jeong SC. Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell. 2012;24:4807–4818. doi: 10.1105/tpc.112.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K-H, Choi M-S, Lee S-K, Seo M-J, Hwang T-Y, Yun H-T, Kim H-S, Kim J-T, Kwon Y-U, Kim Y-H. Development of low Gly m Bd 30K (P34) allergen breeding lines using molecular marker in soybean [Glycine max (L.) Merr.] Plant Breed Biotechnol. 2013;1:298–306. [Google Scholar]
- Jeong SC, Kim JH, Bae DN. Genetic analysis of the Lf1 gene that controls leaflet number in soybean. Theor Appl Genet. 2017;130:1685–1692. doi: 10.1007/s00122-017-2918-0. [DOI] [PubMed] [Google Scholar]
- Jeong SC, Moon JK, Park SK, Kim MS, Lee K, Lee SR, Jeong N, Choi MS, Kim N, Kang ST, Park E. Genetic diversity patterns and domestication origin of soybean. Theor Appl Genet. 2019;132:1179–1193. doi: 10.1007/s00122-018-3271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BJ, Nan HY, Gao YF, Tang LL, Yue YL, Lu SJ, Ma LM, Cao D, Sun S, Wang JL, Wu CX, Yuan XH, Hou WS, Kong FJ, Han TF, Liu BH. Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS ONE. 2014;9:e106042. doi: 10.1371/journal.pone.0106042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HW, Li YY, Qin HT, Li YL, Qi HD, Li CD, Wang NN, Li RC, Zhao YY, Huang SY, Yu JY, Wang XY, Zhu RS, Liu CY, Hu ZB, Qi ZM, Xin DW, Wu XX, Chen QS. Identification of major QTLs associated with first pod height and candidate gene mining in soybean. Front Plant Sci. 2018;9:1280. doi: 10.3389/fpls.2018.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A, Shimizu T, Watanabe S, Tsubokura Y, Katayose Y, Harada K, Vaughan DA, Tomooka N. Evaluation of soybean germplasm conserved in NIAS genebank and development of mini core collections. Breed Sci. 2011;61:566–592. doi: 10.1270/jsbbs.61.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazashi Y, Hirose A, Takahashi I, Mikami M, Endo M, Hirose S, Toki S, Kaga A, Naito K, Ishimoto M, Abe J, Yamada T. Simultaneous site-directed mutagenesis of duplicated loci in soybean using a single guide RNA. Plant Cell Rep. 2018;37:553–563. doi: 10.1007/s00299-018-2251-3. [DOI] [PubMed] [Google Scholar]
- Kang BC, Yun JY, Kim ST, Shin Y, Ryu J, Choi M, Woo JW, Kim JS. Precision genome engineering through adenine base editing in plants. Nat Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- Karikari B, Li SG, Bhat JA, Cao YC, Kong JJ, Yang JY, Gai JY, Zhao TJ. Genome-wide detection of major and epistatic effect QTLs for seed protein and oil content in soybean under multiple environments using high-density bin map. Int J Mol Sci. 2019;20:979. doi: 10.3390/ijms20040979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A, Li K, Li C, Yin JL, Li N, Yang YH, Song YP, Ren R, Zhi HJ, Gai JY. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor Appl Genet. 2018;131:461–476. doi: 10.1007/s00122-017-3014-1. [DOI] [PubMed] [Google Scholar]
- Katayose Y, Kanamori H, Shimomura M, Ohyanagi H, Ikawa H, Minami H, Shibata M, Ito T, Kurita K, Ito K, Tsubokura Y, Kaga A, Wu JZ, Matsumoto T, Harada K, Sasaki T. DaizuBase, an integrated soybean genome database including BAC-based physical maps. Breed Sci. 2011;61:661–664. doi: 10.1270/jsbbs.61.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Takada Y, Shimamura S, Hirata K, Sayama T, Taguchi-Shiobara F, Ishimoto M, Kikuchi A, Nishio T. Transfer of the Rsv3 locus from ‘Harosoy’ for resistance to soybean mosaic virus strains C and D in Japan. Breed Sci. 2016;66:319–327. doi: 10.1270/jsbbs.66.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Sayama T, Takada Y, Yumoto S, Ishimoto M, Shimada S, Hirata K, Kikuchi A (2017) Breeding of soybean lines conferred multiple disease and pest resistance by marker-assisted selection with a high-resolution PCR fragment analysis system. Bulletin of the NARO, Agricultural Research for Tohoku Region, pp 89–106
- Kelliher T, Starr D, Su XJ, Tang GZ, Chen ZY, Carter J, Wittich PE, Dong SJ, Green J, Burch E, McCuiston J, Gu WN, Sun YJ, Strebe T, Roberts J, Bate NJ, Que QD. One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol. 2019;37:287–292. doi: 10.1038/s41587-019-0038-x. [DOI] [PubMed] [Google Scholar]
- Kim MY, Van K, Lestari P, Moon JK, Lee SH. SNP identification and SNAP marker development for a GmNARK gene controlling supernodulation in soybean. Theor Appl Genet. 2005;110:1003–1010. doi: 10.1007/s00122-004-1887-2. [DOI] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma J, Kim WY, Kim BC, Park S, Lee KA, Kim DH, Kim KH, Shin JH, Jang YE, Do Kim K, Liu WX, Chaisan T, Kang YJ, Lee YH, Kim KH, Moon JK, Schmutz J, Jackson SA, Bhak J, Lee SH. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Van K, Kang YJ, Kim KH, Lee SH. Tracing soybean domestication history: from nucleotide to genome. Breed Sci. 2011;61:445–452. doi: 10.1270/jsbbs.61.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Shin JH, Kang YJ, Shim SR, Lee SH. Divergence of flowering genes in soybean. J Biosciences. 2012;37:857–870. doi: 10.1007/s12038-012-9252-0. [DOI] [PubMed] [Google Scholar]
- Kim M, Song JT, Bilyeu KD, Lee JD. A new low linolenic acid allele of GmFAD3A gene in soybean PE1690. Mol Breed. 2015;35:155. [Google Scholar]
- Kim HJ, Kim M-J, Pak JH, Im HH, Lee DH, Kim K-H, Lee J-H, Kim D-H, Choi HK, Jung HW, Chung Y-S. RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnol Rep. 2016;10:257–267. [Google Scholar]
- Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Okuda S, Takahashi M, Matsunaga R, Nakazawa Y. QTL mapping of antibiosis resistance to common cutworm (Spodoptera litura Fabricius) in soybean. Crop Sci. 2005;45:2044–2048. [Google Scholar]
- Komatsu K, Hwang TY, Takahashi M, Sayama T, Funatsuki H, Oki N, Ishimoto M. Identification of QTL controlling post-flowering period in soybean. Breed Sci. 2011;61:646–652. doi: 10.1270/jsbbs.61.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong FJ, Nan HY, Cao D, Li Y, Wu FF, Wang JL, Lu SJ, Yuan XH, Cober ER, Abe J, Liu BH. A new dominant gene E9 Conditions early flowering and maturity in soybean. Crop Sci. 2014;54:2529–2535. [Google Scholar]
- Kong LP, Lu SJ, Wang YP, Fang C, Wang FF, Nan HY, Su T, Li SC, Zhang FG, Li XM, Zhao XH, Yuan XH, Liu BH, Kong FJ. Quantitative trait locus mapping of flowering time and maturity in soybean using next-generation sequencing-based analysis. Front Plant Sci. 2018;9:995. doi: 10.3389/fpls.2018.00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P, Lee JM, Tsukamoto C, Takahashi Y, Singh RJ, Lee JD, Chung G. Evaluation of genetic structure of Korean wild soybean (Glycine soja) based on saponin allele polymorphism. Genet Resour Crop Evolut. 2014;61:1121–1130. [Google Scholar]
- Krishnamurthy P, Tsukamoto C, Singh RJ, Lee JD, Kim HS, Yang SH, Chung G. The Sg-6 saponins, new components in wild soybean (Glycine soja Sieb. and Zucc.): polymorphism, geographical distribution and inheritance. Euphytica. 2014;198:413–424. [Google Scholar]
- Krishnamurthy P, Tsukamoto C, Takahashi Y, Hongo Y, Singh RJ, Lee JD, Chung G. Comparison of saponin composition and content in wild soybean (Glycine soja Sieb. and Zucc.) before and after germination. Biosci Biotechnol Biochem. 2014;78:1988–1996. doi: 10.1080/09168451.2014.946389. [DOI] [PubMed] [Google Scholar]
- Kulkarni KP, Patil G, Valliyodan B, Vuong TD, Shannon JG, Nguyen HT, Lee JD. Comparative genome analysis to identify SNPs associated with high oleic acid and elevated protein content in soybean. Genome. 2018;61:217–222. doi: 10.1139/gen-2017-0158. [DOI] [PubMed] [Google Scholar]
- Kurosaki H, Fujita S, Ohnishi S, Kosaka F, Tanaka Y, Takeuchi T, Yamashita Y, Kiguchi T, Hiura S. A new soybean variety ‘Suzumaru R’. Bull Hokkaido Res Org Agric Exp Stn. 2017;101:1–13. [Google Scholar]
- Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL, Li MW, He WM, Qin N, Wang B, Li J, Jian M, Wang JA, Shao GH, Wang J, Sun SSM, Zhang GY. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053-U1041. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018;93:377–386. doi: 10.1111/tpj.13782. [DOI] [PubMed] [Google Scholar]
- Lee GA, Crawford GW, Liu L, Sasaki Y, Chen XX. Archaeological soybean (Glycine max) in East Asia: does size matter? PLoS ONE. 2011;6:e26720. doi: 10.1371/journal.pone.0026720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WK, Jeong N, Indrasumunar A, Gresshoff PM, Jeong S-C. Glycine max non-nodulation locus rj1: a recombinogenic region encompassing a SNP in a lysine motif receptor-like kinase (GmNFR1α) Theor Appl Genet. 2011;122:875–884. doi: 10.1007/s00122-010-1493-4. [DOI] [PubMed] [Google Scholar]
- Lee WK, Kim N, Kim J, Moon JK, Jeong N, Choi IY, Kim SC, Chung WH, Kim HS, Lee SH, Jeong SC. Dynamic genetic features of chromosomes revealed by comparison of soybean genetic and sequence-based physical maps. Theor Appl Genet. 2013;126:1103–1119. doi: 10.1007/s00122-012-2039-8. [DOI] [PubMed] [Google Scholar]
- Lee JS, Yoo MH, Jung JK, Bilyeu KD, Lee JD, Kang S. Detection of novel QTLs for foxglove aphid resistance in soybean. Theor Appl Genet. 2015;128:1481–1488. doi: 10.1007/s00122-015-2519-8. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Kim DS, Kim J-B, Jo S-H, Kang S-Y, Choi H-I, Ha B-K. Identification of candidate genes for an early-maturing soybean mutant by genome resequencing analysis. Mol Genet Genomics. 2016;291:1561–1571. doi: 10.1007/s00438-016-1183-2. [DOI] [PubMed] [Google Scholar]
- Li YH, Zhao SC, Ma JX, Li D, Yan L, Li J, Qi XT, Guo XS, Zhang L, He WM, Chang RZ, Liang QS, Guo Y, Ye C, Wang XB, Tao Y, Guan RX, Wang JY, Liu YL, Jin LG, Zhang XQ, Liu ZX, Zhang LJ, Chen J, Wang KJ, Nielsen R, Li RQ, Chen PY, Li WB, Reif JC, Purugganan M, Wang J, Zhang MC, Wang J, Qiu LJ. Molecular footprints of domestication and improvement in soybean revealed by whole genome re-sequencing. BMC Genom. 2013;14:579. doi: 10.1186/1471-2164-14-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Zhou GY, Ma JX, Jiang WK, Jin LG, Zhang ZH, Guo Y, Zhang JB, Sui Y, Zheng LT, Zhang SS, Zuo QY, Shi XH, Li YF, Zhang WK, Hu YY, Kong GY, Hong HL, Tan B, Song J, Liu ZX, Wang YS, Ruan H, Yeung CKL, Liu J, Wang HL, Zhang LJ, Guan RX, Wang KJ, Li WB, Chen SY, Chang RZ, Jiang Z, Jackson SA, Li RQ, Qiu LJ. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- Li C, Zong Y, Wang YP, Jin S, Zhang DB, Song QN, Zhang R, Gao CX. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZF, Guo Y, Ou L, Hong HL, Wang J, Liu ZX, Guo BF, Zhang LJ, Qiu LJ. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor Appl Genet. 2018;131:1001–1016. doi: 10.1007/s00122-017-3044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics. 2008;180:995–1007. doi: 10.1534/genetics.108.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BH, Watanabe S, Uchiyama T, Kong FJ, Kanazawa A, Xia ZJ, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J. The soybean stem growth habit gene Dt1 Is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010;153:198–210. doi: 10.1104/pp.109.150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kim MY, Van K, Sun S, Lee S-H. Identification of population-specific QTLs for flowering in soybean. J Crop Sci Biotechnol. 2010;13:213–218. [Google Scholar]
- Liu W, Kim MY, Van K, Lee Y-H, Li H, Liu X, Lee S-H. QTL identification of yield-related traits and their association with flowering and maturity in soybean. J Crop Sci Biotechnol. 2011;14:65–70. [Google Scholar]
- Liu DQ, Yan YL, Fujita Y, Xu DH. A major QTL (qFT12.1) allele from wild soybean delays flowering time. Mol Breed. 2018;38:45. [Google Scholar]
- Liu W, Jiang BJ, Ma LM, Zhang SW, Zhai H, Xu X, Hou WS, Xia ZJ, Wu CX, Sun S, Wu TT, Chen L, Han TF. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018;217:1335–1345. doi: 10.1111/nph.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gunapati S, Mihelich NT, Stec AO, Michno J-M, Stupar RM. Genome editing in soybean with CRISPR/Cas9. In: Qi Y, editor. Plant genome editing with CRISPR systems: methods and protocols. New York: Springer; 2019. pp. 217–234. [DOI] [PubMed] [Google Scholar]
- Lu SJ, Zhao XH, Hu YL, Liu SL, Nan HY, Li XM, Fang C, Cao D, Shi XY, Kong LP, Su T, Zhang FG, Li SC, Wang Z, Yuan XH, Cober ER, Weller JL, Liu BH, Hou XL, Tian ZX, Kong FJ. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet. 2017;49:773–779. doi: 10.1038/ng.3819. [DOI] [PubMed] [Google Scholar]
- Michno JM, Wang XB, Liu JQ, Curtin SJ, Kono TJY, Stupar RM. CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. Gm Crops Food. 2015;6:243–252. doi: 10.1080/21645698.2015.1106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miladinovic J, Ceran M, Dordevic V, Balesevic-Tubic S, Petrovic K, Dukic V, Miladinovic D. Allelic variation and distribution of the major maturity genes in different soybean collections. Front Plant Sci. 2018;9:1286. doi: 10.3389/fpls.2018.01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki J. Aiming at the construction of the race distinction system of Phytophthora sojae. Plant Prot. 2010;64:508–510. [Google Scholar]
- Nagatoshi Y, Fujita Y. Accelerating soybean breeding in a CO2-supplemented growth chamber. Plant Cell Physiol. 2019;60:77–84. doi: 10.1093/pcp/pcy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz MA, Golokhvast KS, Rehman HM, Tsukamoto C, Kim HS, Yang SH, Chung G. Soyisoflavone diversity in wild soybeans (Glycine soja Sieb. & Zucc.) from the main centres of diversity. Biochem Syst Ecol. 2018;77:16–21. [Google Scholar]
- Ning HL, Yuan JQ, Dong QZ, Li WB, Xuel H, Wang YS, Tian Y, Li WX. Identification of QTLs related to the vertical distribution and seed-set of pod number in soybean [Glycine max (L.) Merri] PLoS ONE. 2018;13:e0195830. doi: 10.1371/journal.pone.0195830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso-Tanaka E, Shimizu T, Hajika M, Kaga A, Ishimoto M. Highly multiplexed AmpliSeq technology identifies novel variation of flowering time-related genes in soybean (Glycine max) DNA Res. 2019;26:243–260. doi: 10.1093/dnares/dsz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S, Funatsuki H, Kasai A, Kurauchi T, Yamaguchi N, Takeuchi T, Yamazaki H, Kurosaki H, Shirai S, Miyoshi T, Horita H, Senda M. Variation of GmIRCHS (Glycine max inverted-repeat CHS pseudogene) is related to tolerance of low temperature-induced seed coat discoloration in yellow soybean. Theor Appl Genet. 2011;122:633–642. doi: 10.1007/s00122-010-1475-6. [DOI] [PubMed] [Google Scholar]
- Oki N, Komatsu K, Sayama T, Ishimoto M, Takahashi M, Takahashi M. Genetic analysis of antixenosis resistance to the common cutworm (Spodoptera litura Fabricius) and its relationship with pubescence characteristics in soybean (Glycine max (L.) Merr.) Breed Sci. 2011;61:608–617. doi: 10.1270/jsbbs.61.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki N, Sayama T, Ishimoto M, Yokota I, Kaga A, Takahashi M, Takahashi M. Quantitative trait loci associated with short inter-node length in soybean. Breed Sci. 2018;68:554–560. doi: 10.1270/jsbbs.18087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam K, Tsukamoto C, Honda N, Kikuchi A, Lee JD, Yang SH, Chung G. Saponin polymorphism in the Korean wild soybean (Glycine soja Sieb. and Zucc.) Plant Breed. 2013;132:121–126. [Google Scholar]
- Pham AT, Lee JD, Shannon JG, Bilyeu KD. A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor Appl Genet. 2011;123:793–802. doi: 10.1007/s00122-011-1627-3. [DOI] [PubMed] [Google Scholar]
- Qi XP, Li MW, Xie M, Liu X, Ni M, Shao GH, Song C, Yim AKY, Tao Y, Wong FL, Isobe S, Wong CF, Wong KS, Xu CY, Li CQ, Wang Y, Guan R, Sun FM, Fan GY, Xiao ZX, Zhou F, Phang TH, Liu X, Tong SW, Chan TF, Yiu SM, Tabata S, Wang J, Xu X, Lam HM. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun. 2014;5:4340. doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Wang Y, Wu SL, Wang YY, Ye CY, Bai XF, Li ZF, Yan CH, Wang WD, Wang ZQ, Shu QY, Xie JH, Lee SH, Fan LJ. Genome re-sequencing of semi-wild soybean reveals a complex soja population structure and deep introgression. PLoS ONE. 2014;9:e108479. doi: 10.1371/journal.pone.0108479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech EL, Vianna GR, Aragão FJL. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protoc. 2008;3:410. doi: 10.1038/nprot.2008.9. [DOI] [PubMed] [Google Scholar]
- Redekar NR, Clevinger EM, Laskar MA, Biyashev RM, Ashfield T, Jensen RV, Jeong SC, Tolin SA, Saghai Maroof MA. Candidate gene sequence analyses toward identifying -Type Resistance to. Plant Genome. 2016 doi: 10.3835/plantgenome2015.09.0088. [DOI] [PubMed] [Google Scholar]
- Saruta M, Takada Y, Kikuchi A, Yamada T, Komatsu K, Sayama T, Ishimoto M, Okabe A. Screening and genetic analysis of resistance to peanut stunt virus in soybean: identification of the putative Rpsv1 resistance gene. Breed Sci. 2011;61:625–630. doi: 10.1270/jsbbs.61.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Sedivy EJ, Wu FQ, Hanzawa Y. Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 2017;214:539–553. doi: 10.1111/nph.14418. [DOI] [PubMed] [Google Scholar]
- Shen YT, Liu J, Geng HY, Zhang JX, Liu YC, Zhang HK, Xing SL, Du JC, Ma SS, Tian ZX. De novo assembly of a Chinese soybean genome. Sci China Life Sci. 2018;61:871–884. doi: 10.1007/s11427-018-9360-0. [DOI] [PubMed] [Google Scholar]
- Shim S, Ha J, Kim MY, Choi MS, Kang ST, Jeong SC, Moon JK, Lee SH. GmBRC1 is a candidate gene for branching in soybean (Glycine max (L) Merrill) Int J Mol Sci. 2019;20:135. doi: 10.3390/ijms20010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura M, Kanamori H, Komatsu S, Namiki N, Mukai Y, Kurita K, Kamatsuki K, Ikawa H, Yano R, Ishimoto M, Kaga A, Katayose Y. The Glycine max cv. Enrei genome for improvement of Japanese soybean cultivars. Int J Genomics. 2015;2015:358127. doi: 10.1155/2015/358127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Van K, Kim DH, Do Kim K, Jang YE, Choi BS, Kim MY, Lee SH. The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 2008;8:133. doi: 10.1186/1471-2229-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Van K, Kim KD, Lee YH, Jun TH, Lee SH. Molecular sequence variations of the lipoxygenase-2 gene in soybean. Theor Appl Genet. 2012;124:613–622. doi: 10.1007/s00122-011-1733-2. [DOI] [PubMed] [Google Scholar]
- Shurtleff W, Aoyagi A. History of soybeans and soyfoods in Japan, and in Japanese cookbooks and restaurants outside Japan. Lafayette: Soyinfo Center; 2014. [Google Scholar]
- Splitter J. The latest gene-edited food is a soybean oil that comes with zero trans fats. New York: Forbes; 2019. [Google Scholar]
- Sugimoto T, Yoshida S, Watanabe K, Ano M, Kanto T, Maekawa K, Irie K. Identification of SSR markers linked to the Phytophthora resistance gene Rps1-d in soybean. Plant Breed. 2008;127:154–159. [Google Scholar]
- Sun S, Kim MY, Van K, Lee YW, Li BD, Lee SH. QTLs for resistance to Phomopsis seed decay are associated with days to maturity in soybean (Glycine max) Theor Appl Genet. 2013;126:2029–2038. doi: 10.1007/s00122-013-2115-8. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Hu Z, Chen R, Jiang QY, Song GH, Zhang H, Xi YJ. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep-UK. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C, Tanaka Y, Takeuchi T, Yumoto S, Shirai S. Genetic relationships of soybean cyst nematode resistance originated in Gedenshirazu and PI84751 on Rhg1 and Rhg4 loci. Breed Sci. 2011;61:602–607. doi: 10.1270/jsbbs.61.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C, Miyoshi T, Shirai S, Yumoto S, Tanaka Y, Hagihara S, Yamaguchi N, Kurosaki H, Yamazaki H, Ohnishi S. A new soybean variety ‘Yukihomare R’ introduced resistance for soybean cyst nematode race1 into ‘Yukihomare’ by marker assisted selection. Bull Hokkaido Res Organ Agric Exp Stat. 2017;101:33–47. [Google Scholar]
- Takahashi K, Tanaka T, Iida W, Tsuda Y. Studies on virus diseases and causal viruses of soybean in Japan. Bull Tohoku Natl Agric Exp Station. 1980;62:1–130. [Google Scholar]
- Takeshima R, Hayashi T, Zhu JH, Zhao C, Xu ML, Yamaguchi N, Sayama T, Ishimoto M, Kong LP, Shi XY, Liu BH, Tian ZX, Yamada T, Kong FJ, Abe J. A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J Exp Bot. 2016;67:5247–5258. doi: 10.1093/jxb/erw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubokura Y, Watanabe S, Xia ZJ, Kanamori H, Yamagata H, Kaga A, Katayose Y, Abe J, Ishimoto M, Harada K. Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann Bot-London. 2014;113:429–441. doi: 10.1093/aob/mct269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Kaga A, Anai T, Shimizu T, Sayama T, Takagi K, Machita K, Watanabe S, Nishimura M, Yamada N, Mori S, Sasaki H, Kanamori H, Katayose Y, Ishimoto M. Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genom. 2015;16:1014. doi: 10.1186/s12864-015-2079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto C, Nawaz MA, Kurosaka A, Le B, Lee JD, Son E, Yang SH, Kurt C, Baloch FS, Chung G. Isoflavone profile diversity in Korean wild soybeans (Glycine soja Sieb. & Zucc.) Turk J Agric For. 2018;42:248–261. [Google Scholar]
- Vaughn JN, Li ZL. Genomic signatures of north american soybean improvement inform diversity enrichment strategies and clarify the impact of hybridization. G3-Genes Genom Genet. 2016;6:2693–2705. doi: 10.1534/g3.116.029215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FF, Nan HY, Chen LY, Fang C, Zhang HY, Su T, Li SC, Cheng Q, Dong LD, Liu BH, Kong FJ, Lu SJ. A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol Breed. 2019;39:70. [Google Scholar]
- Watanabe S, Hideshima R, Xia ZJ, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics. 2009;182:1251–1262. doi: 10.1534/genetics.108.098772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Xia ZJ, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics. 2011;188:395-U260. doi: 10.1534/genetics.110.125062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Shimizu T, Machita K, Tsubokura Y, Xia ZJ, Yamada T, Hajika M, Ishimoto M, Katayose Y, Harada K, Kaga A. Development of a high-density linkage map and chromosome segment substitution lines for Japanese soybean cultivar Enrei. DNA Res. 2018;25:123–136. doi: 10.1093/dnares/dsx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F. On-the-spot harvesting ceremony of 100-million-mu of Zhonghuang 13 and the new cultivar field demonstration of Huang-Huai-Hai soybean breeding project were held. Beijing: Institute of Crop Sciences, Chinese Academy of Agricultural Sciences; 2018. [Google Scholar]
- Wolfgang G, An Y-QA. Genetic separation of southern and northern soybean breeding programs in North America and their associated allelic variation at four maturity loci. Mol Breed. 2017;37:8. doi: 10.1007/s11032-016-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZJ, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lu SX, Wu HY, Tabata S, Harada K. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA. 2012;109:E2155–E2164. doi: 10.1073/pnas.1117982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chung CYL, Li MW, Wong FL, Wang X, Liu AL, Wang ZL, Leung AKY, Wong TH, Tong SW, Xiao ZX, Fan KJ, Ng MS, Qi XP, Yang LF, Deng TQ, He LJ, Chen L, Fu AS, Ding Q, He JX, Chung G, Isobe S, Tanabata T, Valliyodan B, Nguyen HT, Cannon SB, Foyer CH, Chan TF, Lam HM. A reference-grade wild soybean genome. Nat Commun. 2019;10:1216. doi: 10.1038/s41467-019-09142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DH, Abe J, Gai JY, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor Appl Genet. 2002;105:645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- Yamada T, Makita H, Funatsuki H, Takahashi K, Hirata K, Hishinuma A, Tanaka J. Causal analysis of yield-increase by introgression of shattering resistance gene pdh1 in soybean. Jpn J Crop Sci. 2017;86:251–257. [Google Scholar]
- Yamaguchi N, Sayama T, Yamazaki H, Miyoshi T, Ishimoto M, Funatsuki H. Quantitative trait loci associated with lodging tolerance in soybean cultivar ‘Toyoharuka’. Breed Sci. 2014;64:300–308. doi: 10.1270/jsbbs.64.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Yan L, Di R, Wu CJ, Liu Q, Wei Y, Hou WH, Zhao QS, Liu BQ, Yang CY, Song QJ, Zhang MC. Haplotype analysis of a major and stable QTL underlying soybean (Glycine max) seed oil content reveals footprint of artificial selection. Mol Breed. 2019;39:57. [Google Scholar]
- Yang K, Ko JM, Ha TJ, Lee YH, Baek IY, Yang TJ, Nou IS. Development of molecular markers for low raffinose and stachyose in Korean soybean cultivars. Plant Breed Biotechnol. 2014;2:151–157. [Google Scholar]
- Yue YL, Liu NX, Jiang BJ, Li M, Wang HJ, Jiang Z, Pan HT, Xia QJ, Ma QB, Han TF, Nian H. A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol Plant. 2017;10:656–658. doi: 10.1016/j.molp.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Wang XZ, Lu YM, Bhusal SJ, Song QJ, Cregan PB, Yen Y, Brown M, Jiang GL. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol Plant. 2018;11:460–472. doi: 10.1016/j.molp.2017.12.016. [DOI] [PubMed] [Google Scholar]
- Zhao X, Meng ZG, Wang Y, Chen WJ, Sun CJ, Cui B, Cui JH, Yu ML, Zeng ZH, Guo SD, Luo D, Cheng JQ, Zhang R, Cui HX. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat Plants. 2017;3:956–964. doi: 10.1038/s41477-017-0063-z. [DOI] [PubMed] [Google Scholar]
- Zhong C, Sun S, Li Y, Duan C, Zhu Z. Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor Appl Genet. 2018;131:525–538. doi: 10.1007/s00122-017-3016-z. [DOI] [PubMed] [Google Scholar]
- Zhong C, Sun SL, Yao LL, Ding JJ, Duan CX, Zhu ZD. Fine mapping and identification of a novel Phytophthora root rot resistance locus RpsZS18 on chromosome 2 in soybean. Front Plant Sci. 2018;9:44. doi: 10.3389/fpls.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZK, Jiang Y, Wang Z, Gou ZH, Lyu J, Li WY, Yu YJ, Shu LP, Zhao YJ, Ma YM, Fang C, Shen YT, Liu TF, Li CC, Li Q, Wu M, Wang M, Wu YS, Dong Y, Wan WT, Wang X, Ding ZL, Gao YD, Xiang H, Zhu BG, Lee SH, Wang W, Tian ZX. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015;33:408-U125. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]
- Zhu JH, Takeshima R, Harigai K, Xu ML, Kong FJ, Liu BH, Kanazawa A, Yamada T, Abe J. Loss of function of the E1-Like-b gene associates with early flowering under long-day conditions in soybean. Front Plant Sci. 2019;9:1867. doi: 10.3389/fpls.2018.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]