Abstract
Diaporthe species (Sordariomycetes, Diaporthales) are often reported as important plant pathogens, saprobes and endophytes on a wide range of plant hosts. In this study, Diaporthe specimens were collected from symptomatic twigs and branches at the Huoditang Forest Farm in Shaanxi Province, China. Identification was done using a combination of morphology and comparison of DNA sequence data of the nuclear ribosomal internal transcribed spacer (ITS), calmodulin (cal), histone H3 (his3), partial translation elongation factor-1α (tef1) and β-tubulin (tub2) gene regions. Three new Diaporthe species are proposed: D. albosinensis, D. coryli and D. shaanxiensis. All species are illustrated and their morphology and phylogenetic relationships with other Diaporthe species are discussed.
Keywords: Diaporthaceae , Dieback, DNA phylogeny, Systematics, Taxonomy
Introduction
Diaporthe species (Sordariomycetes, Diaporthales) are associated with a wide range of plant hosts as pathogens, endophytes or saprobes of crops, ornamentals and forest trees (Murali et al. 2006, Rossman et al. 2007, Garcia-Reyne et al. 2011, Gomes et al. 2013, Udayanga et al. 2015, Dissanayake et al. 2017, Guarnaccia and Crous 2017, 2018, Wijayawardene et al. 2017, Yang et al. 2017a, b, 2018, Fan et al. 2018, Guarnaccia et al. 2018). The sexual morph of Diaporthe is characterised by immersed ascomata and an erumpent pseudostroma with elongated perithecial necks. Asci are unitunicate, clavate to cylindrical. Ascospores are fusoid, ellipsoid to cylindrical, hyaline, biseriate to uniseriate in the ascus, sometimes with appendages (Udayanga et al. 2011). The asexual morph is characterised by ostiolate conidiomata, with cylindrical phialides producing three types of hyaline, aseptate conidia (Udayanga et al. 2011, Gomes et al. 2013).
Species identification in Diaporthe has traditionally been based on host association, morphology and culture characteristics (Mostert et al. 2001, Santos and Phillips 2009, Udayanga et al. 2011), resulting in the description of over 200 species (Hyde et al. 2020). Multiple species of Diaporthe can colonise a single host and one species can be associated with different hosts (Santos and Phillips 2009, Diogo et al. 2010, Santos et al. 2011, Gomes et al. 2013). In addition, considerable within-species variability of phenotypic characters has been reported (Rehner and Uecker 1994, Mostert et al. 2001, Udayanga et al. 2011). Thus, a polyphasic taxonomic approach, based on multi-locus DNA data, morphology and ecology, has been increasingly employed for species boundaries in the genus Diaporthe (Gomes et al. 2013, Huang et al. 2013, 2015, Udayanga et al. 2014a, b, 2015, Fan et al. 2015, Du et al. 2016, Gao et al. 2016, 2017, Guarnaccia and Crous 2017, Guarnaccia et al. 2018, Long et al. 2019).
Huoditang is located in the middle part of the southern slope of the Qinling Mountains at 33°18'~33°28'N, 108°21'~108°29'E. It belongs to the transitional zone of the northern subtropical and warm temperate zone in China. The terrain is complex and the climate is changeable (Zhang and Cao 2007). The plant communities are complex and, as a result, species diversity of fungi in the forest area is high (Zhang and Cao 2007). During trips to collect forest pathogens causing dieback in Shaanxi Province, cankered branches with typical Diaporthe fruiting bodies were investigated and sampled. The aim of the present study was to identify these fungi, based on modern polyphasic taxonomic concepts.
Materials and methods
Isolates
Fresh specimens of Diaporthe were collected from symptomatic twigs or branches in Shaanxi Province (Table 1). Isolates were obtained by removing a mucoid spore mass from conidiomata and spreading the suspension on the surface of 1.8% potato dextrose agar (PDA) in a 9 cm diam. Petri dish. Petri dishes were incubated at 25 °C until spores germinated. Single germinating conidia were transferred on to new PDA plates, which were kept at 25 °C in the dark. Specimens are deposited in the Museum of the Beijing Forestry University (BJFC). Axenic cultures are maintained in the China Forestry Culture Collection Centre (CFCC).
Table 1.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaporthe.
| Species | Isolate | Host | Location | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | cal | his3 | tef1 | tub2 | ||||
| D. acericola | MFLUCC 17-0956 | Acer negundo | Italy | KY964224 | KY964137 | NA | KY964180 | KY964074 |
| D. acerigena | CFCC 52554 | Acer tataricum | China | MH121489 | MH121413 | MH121449 | MH121531 | NA |
| D. albosinensis | CFCC 53066 | Betula albosinensis | China | MK432659 | MK442979 | MK443004 | MK578133 | MK578059 |
| CFCC 53067 | Betula albosinensis | China | MK432660 | MK442980 | MK443005 | MK578134 | MK578060 | |
| D. alnea | CBS 146.46 | Alnus sp. | Netherlands | KC343008 | KC343250 | KC343492 | KC343734 | KC343976 |
| D. ambigua | CBS 114015 | Pyrus communis | South Africa | KC343010 | KC343252 | KC343494 | KC343736 | KC343978 |
| D. anacardii | CBS 720.97 | Anacardium occidentale | East Africa | KC343024 | KC343266 | KC343508 | KC343750 | KC343992 |
| D. angelicae | CBS 111592 | Heracleum sphondylium | Austria | KC343027 | KC343269 | KC343511 | KC343753 | KC343995 |
| D. apiculatum | CGMCC 3.17533 | Camellia sinensis | China | KP267896 | NA | NA | KP267970 | KP293476 |
| D. aquatica | IFRDCC 3051 | Aquatic habitat | China | JQ797437 | NA | NA | NA | NA |
| D. arctii | CBS 139280 | Arctium lappa | Austria | KJ590736 | KJ612133 | KJ659218 | KJ590776 | KJ610891 |
| D. aseana | MFLUCC 12-0299a | Unknown dead leaf | Thailand | KT459414 | KT459464 | NA | KT459448 | KT459432 |
| D. asheicola | CBS 136967 | Vaccinium ashei | Chile | KJ160562 | KJ160542 | NA | KJ160594 | KJ160518 |
| D. baccae | CBS 136972 | Vaccinium corymbosum | Italy | KJ160565 | NA | MF418264 | KJ160597 | NA |
| D. beilharziae | BRIP 54792 | Indigofera australis | Australia | JX862529 | NA | NA | JX862535 | KF170921 |
| D. benedicti | BPI 893190 | Salix sp. | USA | KM669929 | KM669862 | NA | KM669785 | NA |
| D. betulae | CFCC 50469 | Betula platyphylla | China | KT732950 | KT732997 | KT732999 | KT733016 | KT733020 |
| D. betulina | CFCC 52560 | Betula albo-sinensis | China | MH121495 | MH121419 | MH121455 | MH121537 | MH121577 |
| D. bicincta | CBS 121004 | Juglans sp. | USA | KC343134 | KC343376 | KC343618 | KC343860 | KC344102 |
| D. caryae | CFCC 52563 | Carya illinoensis | China | MH121498 | MH121422 | MH121458 | MH121540 | MH121580 |
| D. cassines | CPC 21916 | Cassine peragua | South Africa | KF777155 | NA | NA | KF777244 | NA |
| D. celeris | CPC 28262 | Vitis vinifera | Czech Republic | MG281017 | MG281712 | MG281363 | MG281538 | MG281190 |
| D. cercidis | CFCC 52565 | Cercis chinensis | China | MH121500 | MH121424 | MH121460 | MH121542 | MH121582 |
| D. chamaeropis | CBS 454.81 | Chamaerops humilis | Greece | KC343048 | KC343290 | KC343532 | KC343774 | KC344016 |
| D. charlesworthii | BRIP 54884m | Rapistrum rugostrum | Australia | KJ197288 | NA | NA | KJ197250 | KJ197268 |
| D. chensiensis | CFCC 52567 | Abies chensiensis | China | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| D. cichorii | MFLUCC 17-1023 | Cichorium intybus | Italy | KY964220 | KY964133 | NA | KY964176 | KY964104 |
| D. cinnamomi | CFCC 52569 | Cinnamomum sp. | China | MH121504 | NA | MH121464 | MH121546 | MH121586 |
| D. citriasiana | CGMCC 3.15224 | Citrus unshiu | China | JQ954645 | KC357491 | KJ490515 | JQ954663 | KC357459 |
| D. citrichinensis | CGMCC 3.15225 | Citrus sp. | China | JQ954648 | KC357494 | NA | JQ954666 | NA |
| D. compactum | CGMCC 3.17536 | Camellia sinensis | China | KP267854 | NA | KP293508 | KP267928 | KP293434 |
| D. conica | CFCC 52571 | Alangium chinense | China | MH121506 | MH121428 | MH121466 | MH121548 | MH121588 |
| D. coryli | CFCC 53083 | Corylus mandshurica | China | MK432661 | MK442981 | MK443006 | MK578135 | MK578061 |
| CFCC 53084 | Corylus mandshurica | China | MK432662 | MK442982 | MK443007 | MK578136 | MK578062 | |
| D. cucurbitae | CBS 136.25 | Arctium sp. | Unknown | KC343031 | KC343273 | KC343515 | KC343757 | KC343999 |
| D. cuppatea | CBS 117499 | Aspalathus linearis | South Africa | KC343057 | KC343299 | KC343541 | KC343783 | KC344025 |
| D. cynaroidis | CBS 122676 | Protea cynaroides | South Africa | KC343058 | KC343300 | KC343542 | KC343784 | KC344026 |
| D. cytosporella | FAU461 | Citrus limon | Italy | KC843307 | KC843141 | NA | KC843116 | KC843221 |
| D. discoidispora | ZJUD89 | Citrus unshiu | China | KJ490624 | NA | KJ490566 | KJ490503 | KJ490445 |
| D. dorycnii | MFLUCC 17-1015 | Dorycnium hirsutum | Italy | KY964215 | NA | NA | KY964171 | KY964099 |
| D. elaeagni-glabrae | CGMCC 3.18287 | Elaeagnus glabra | China | KX986779 | KX999281 | KX999251 | KX999171 | KX999212 |
| D. endophytica | CBS 133811 | Schinus terebinthifolius | Brazil | KC343065 | KC343307 | KC343549 | KC343791 | KC343065 |
| D. eres | AR5193 | Ulmus sp. | Germany | KJ210529 | KJ434999 | KJ420850 | KJ210550 | KJ420799 |
| D. eucalyptorum | CBS 132525 | Eucalyptus sp. | Australia | NR120157 | NA | NA | NA | NA |
| D. foeniculacea | CBS 123208 | Foeniculum vulgare | Portugal | KC343104 | KC343346 | KC343588 | KC343830 | KC344072 |
| D. fraxini-angustifoliae | BRIP 54781 | Fraxinus angustifolia | Australia | JX862528 | NA | NA | JX862534 | KF170920 |
| D. fraxinicola | CFCC 52582 | Fraxinus chinensis | China | MH121517 | MH121435 | NA | MH121559 | NA |
| D. fructicola | MAFF 246408 | Passiflora edulis × P. edulis f. flavicarpa | Japan | LC342734 | LC342738 | LC342737 | LC342735 | LC342736 |
| D. fusicola | CGMCC 3.17087 | Lithocarpus glabra | China | KF576281 | KF576233 | NA | KF576256 | KF576305 |
| D. garethjonesii | MFLUCC 12-0542a | Unknown dead leaf | Thailand | KT459423 | KT459470 | NA | KT459457 | KT459441 |
| D. guangxiensis | JZB320094 | Vitis vinifera | China | MK335772 | MK736727 | NA | MK523566 | MK500168 |
| D. helicis | AR5211 | Hedera helix | France | KJ210538 | KJ435043 | KJ420875 | KJ210559 | KJ420828 |
| D. heterophyllae | CBS 143769 | Acacia heterohpylla | France | MG600222 | MG600218 | MG600220 | MG600224 | MG600226 |
| D. hubeiensis | JZB320123 | Vitis vinifera | China | MK335809 | MK500235 | NA | MK523570 | MK500148 |
| D. incompleta | CGMCC 3.18288 | Camellia sinensis | China | KX986794 | KX999289 | KX999265 | KX999186 | KX999226 |
| D. inconspicua | CBS 133813 | Maytenus ilicifolia | Brazil | KC343123 | KC343365 | KC343607 | KC343849 | KC344091 |
| D. infecunda | CBS 133812 | Schinus terebinthifolius | Brazil | KC343126 | KC343368 | KC343610 | KC343852 | KC344094 |
| D. juglandicola | CFCC 51134 | Juglans mandshurica | China | KU985101 | KX024616 | KX024622 | KX024628 | KX024634 |
| D. kadsurae | CFCC 52586 | Kadsura longipedunculata | China | MH121521 | MH121439 | MH121479 | MH121563 | MH121600 |
| D. litchicola | BRIP 54900 | Litchi chinensis | Australia | JX862533 | NA | NA | JX862539 | KF170925 |
| D. lusitanicae | CBS 123212 | Foeniculum vulgare | Portugal | KC343136 | KC343378 | KC343620 | KC343862 | KC344104 |
| D. masirevicii | BRIP 57892a | Helianthus annuus | Australia | KJ197277 | NA | NA | KJ197239 | KJ197257 |
| D. middletonii | BRIP 54884e | Rapistrum rugostrum | Australia | KJ197286 | NA | NA | KJ197248 | KJ197266 |
| D. millettiae | GUCC9167 | Millettia reticulata | China | MK398674 | MK502086 | NA | MK480609 | MK502089 |
| D. miriciae | BRIP 54736j | Helianthus annuus | Australia | KJ197282 | NA | NA | KJ197244 | KJ197262 |
| D. musigena | CBS 129519 | Musa sp. | Australia | KC343143 | KC343385 | KC343627 | KC343869 | KC344111 |
| D. neilliae | CBS 144.27 | Spiraea sp. | USA | KC343144 | KC343386 | KC343628 | KC343870 | KC344112 |
| D. neoarctii | CBS 109490 | Ambrosia trifida | USA | KC343145 | KC343387 | KC343629 | KC343871 | KC344113 |
| D. nothofagi | BRIP 54801 | Nothofagus cunninghamii | Australia | JX862530 | NA | NA | JX862536 | KF170922 |
| D. novem | CBS 127270 | Glycine max | Croatia | KC343155 | KC343397 | KC343640 | KC343881 | KC344123 |
| D. oraccinii | CGMCC 3.17531 | Camellia sinensis | China | KP267863 | NA | KP293517 | KP267937 | KP293443 |
| D. ovalispora | ICMP20659 | Citrus limon | China | KJ490628 | NA | KJ490570 | KJ490507 | KJ490449 |
| D. ovoicicola | CGMCC 3.17093 | Citrus sp. | China | KF576265 | KF576223 | NA | KF576240 | KF576289 |
| D. osmanthi | GUCC9165 | Osmanthus fragrans | China | MK398675 | MK502087 | NA | MK480610 | MK502090 |
| D. padina | CFCC 52590 | Padus racemosa | China | MH121525 | MH121443 | MH121483 | MH121567 | MH121604 |
| D. pandanicola | MFLU 18-0006 | Pandanus sp. | Thailand | MG646974 | NA | NA | NA | MG646930 |
| D. pascoei | BRIP 54847 | Persea americana | Australia | JX862532 | NA | NA | JX862538 | KF170924 |
| D. passifloricola | CBS 141329 | Passiflora foetida | Malaysia | KX228292 | NA | KX228367 | NA | KX228387 |
| D. perseae | CBS 151.73 | Persea gratissima | Netherlands | KC343173 | KC343415 | KC343657 | KC343899 | KC344141 |
| D. pescicola | MFLUCC 16-0105 | Prunus persica | China | KU557555 | KU557603 | NA | KU557623 | KU557579 |
| D. phaseolorum | AR4203 | Phaseolus vulgaris | USA | KJ590738 | NA | KJ659220 | NA | KP004507 |
| D. podocarpi-macrophylli | CGMCC 3.18281 | Podocarpus macrophyllus | China | KX986774 | KX999278 | KX999246 | KX999167 | KX999207 |
| D. pseudomangiferae | CBS 101339 | Mangifera indica | Dominican Republic | KC343181 | KC343423 | KC343665 | KC343907 | KC344149 |
| D. pseudophoenicicola | CBS 462.69 | Phoenix dactylifera | Spain | KC343184 | KC343426 | KC343668 | KC343910 | KC344152 |
| D. psoraleae-pinnatae | CBS 136413 | Psoralea pinnata | South Africa | KF777159 | NA | NA | NA | KF777252 |
| D. pulla | CBS 338.89 | Hedera helix | Yugoslavia | KC343152 | KC343394 | KC343636 | KC343878 | KC344120 |
| D. racemosae | CBS 143770 | Euclea racemosa | South Africa | MG600223 | MG600219 | MG600221 | MG600225 | MG600227 |
| D. ravennica | MFLUCC 15-0479 | Tamarix sp. | Italy | KU900335 | NA | NA | KX365197 | KX432254 |
| D. rhusicola | CBS 129528 | Rhus pendulina | South Africa | JF951146 | KC843124 | NA | KC843100 | KC843205 |
| D. rosae | MFLU 17-1550 | Rosa sp. | Thailand | MG828894 | NA | NA | NA | MG843878 |
| D. rosicola | MFLU 17-0646 | Rosa sp. | UK | MG828895 | NA | NA | MG829270 | MG843877 |
| D. rudis | AR3422 | Laburnum anagyroides | Austria | KC843331 | KC843146 | NA | KC843090 | KC843177 |
| D. sackstonii | BRIP 54669b | Helianthus annuus | Australia | KJ197287 | NA | NA | KJ197249 | KJ197267 |
| D. salicicola | BRIP 54825 | Salix purpurea | Australia | JX862531 | NA | NA | JX862537 | JX862531 |
| D. sambucusii | CFCC 51986 | Sambucus williamsii | China | KY852495 | KY852499 | KY852503 | KY852507 | KY852511 |
| D. schini | CBS 133181 | Schinus terebinthifolius | Brazil | KC343191 | KC343433 | KC343675 | KC343917 | KC344159 |
| D. schoeni | MFLU 15-1279 | Schoenus nigricans | Italy | KY964226 | KY964139 | NA | KY964182 | KY964109 |
| D. sennicola | CFCC 51634 | Senna bicapsularis | China | KY203722 | KY228873 | KY228879 | KY228883 | KY228889 |
| D. serafiniae | BRIP 55665a | Helianthus annuus | Australia | KJ197274 | NA | NA | KJ197236 | KJ197254 |
| D. shaanxiensis | CFCC 53106 | on branches of liana | China | MK432654 | MK442976 | MK443001 | MK578130 | NA |
| CFCC 53107 | on branches of liana | China | MK432655 | MK442977 | MK443002 | MK578131 | NA | |
| D. siamensis | MFLUCC 10-573a | Dasymaschalon sp. | Thailand | JQ619879 | NA | NA | JX275393 | JX275429 |
| D. sojae | FAU635 | Glycine max | USA | KJ590719 | KJ612116 | KJ659208 | KJ590762 | KJ610875 |
| D. sterilis | CBS 136969 | Vaccinium corymbosum | Italy | KJ160579 | KJ160548 | MF418350 | KJ160611 | KJ160528 |
| D. stictica | CBS 370.54 | Buxus sampervirens | Italy | KC343212 | KC343454 | KC343696 | KC343938 | KC344180 |
| D. subclavata | ICMP20663 | Citrus unshiu | China | KJ490587 | NA | KJ490529 | KJ490466 | KJ490408 |
| D. subcylindrospora | MFLU 17-1195 | Salix sp. | China | MG746629 | NA | NA | MG746630 | MG746631 |
| D. subellipicola | MFLU 17-1197 | on dead wood | China | MG746632 | NA | NA | MG746633 | MG746634 |
| D. subordinaria | CBS 464.90 | Plantago lanceolata | New Zealand | KC343214 | KC343456 | KC343698 | KC343940 | KC344182 |
| D. tectonendophytica | MFLUCC 13-0471 | Tectona grandis | China | KU712439 | KU749354 | NA | KU749367 | KU749354 |
| D. tectonigena | MFLUCC 12-0767 | Tectona grandis | China | KU712429 | KU749358 | NA | KU749371 | KU743976 |
| D. terebinthifolii | CBS 133180 | Schinus terebinthifolius | Brazil | KC343216 | KC343458 | KC343700 | KC343942 | KC344184 |
| D. ternstroemia | CGMCC 3.15183 | Ternstroemia gymnanthera | China | KC153098 | NA | NA | KC153089 | NA |
| D. thunbergii | MFLUCC 10-576a | Thunbergia laurifolia | Thailand | JQ619893 | JX197440 | NA | JX275409 | JX275449 |
| D. tibetensis | CFCC 51999 | Juglandis regia | China | MF279843 | MF279888 | MF279828 | MF279858 | MF279873 |
| D. ueckerae | FAU656 | Cucumis melo | USA | KJ590726 | KJ612122 | KJ659215 | KJ590747 | KJ610881 |
| D. ukurunduensis | CFCC 52592 | Acer ukurunduense | China | MH121527 | MH121445 | MH121485 | MH121569 | NA |
| D. unshiuensis | CFCC 52594 | Carya illinoensis | China | MH121529 | MH121447 | MH121487 | MH121571 | MH121606 |
| D. vaccinii | CBS 160.32 | Oxycoccus macrocarpos | USA | KC343228 | KC343470 | KC343712 | KC343954 | KC344196 |
| D. velutina | CGMCC 3.18286 | Neolitsea sp. | China | KX986790 | NA | KX999261 | KX999182 | KX999223 |
| D. viniferae | JZB320071 | Vitis vinifera | China | MK341551 | MK500107 | NA | MK500119 | MK500112 |
| D. xishuangbanica | CGMCC 3.18282 | Camellia sinensis | China | KX986783 | NA | KX999255 | KX999175 | KX999216 |
| D. yunnanensis | CGMCC 3.18289 | Coffea sp. | China | KX986796 | KX999290 | KX999267 | KX999188 | KX999228 |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
Newly sequenced material is indicated in bold type. NA, not applicable.
Morphological analysis
Morphological observations of the asexual morph in the natural environment were based on features of the fruiting bodies produced on infected plant tissues and micromorphology, supplemented by cultural characteristics. Conidiomata from tree barks were sectioned by hand, using a double-edged blade and structures were observed under a dissecting microscope. The gross morphology of fruiting bodies was recorded using a Leica stereomicroscope (M205 FA). Fungal structures were mounted in clear lactic acid and micromorphological characteristics were examined at 1000× magnification using a Leica compound microscope (DM 2500) with differential interference contrast (DIC) optics. Thirty measurements of each structure were determined for each collection. Colony characters and pigment production on PDA were noted after 10 d. Colony colours were described according to Rayner (1970).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from colonies grown on cellophane-covered PDA, using the CTAB [cetyltrimethylammonium bromide] method (Doyle and Doyle 1990). PCR amplifications of phylogenetic markers were done using the same primer pairs and conditions as in Yang et al. (2018). PCR products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyzer with a BigDye Terminater Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Phylogenetic analyses
The quality of our amplified nucleotide sequences was checked and combined by SeqMan v.7.1.0 and reference sequences were retrieved from the National Center for Biotechnology Information (NCBI), based on recent publications on the genus Diaporthe (Guarnaccia et al. 2018, Yang et al. 2018, Long et al. 2019). Sequences were aligned using MAFFT v. 7.310 (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh and Standley 2016) and manually corrected using Bioedit 7.0.9.0 (Hall 1999). The best-fit nucleotide substitution models for each gene were selected using jModelTest v. 2.1.7 (Darriba et al. 2012) under the Akaike Information Criterion.
Phylogenetic analyses of the combined gene regions were performed using Maximum-Likelihood (ML) and Bayesian Inference (BI) methods. ML was conducted using PhyML v. 3.0 (Guindon et al. 2010), with 1000 bootstrap replicates. BI was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0b4 (Ronquist and Huelsenbeck 2003). Two MCMC chains, started from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed with FigTree v.1.3.1 (Rambaut and Drummond 2010) and processed by Adobe Illustrator CS5. Alignment and trees were deposited in TreeBASE (submission ID: S25522). The nucleotide sequence data of the new taxa have been deposited in GenBank (Table 1).
Results
Phylogenetic analyses
The five-gene sequence dataset (ITS, cal, his3, tef1 and tub2) was analysed to infer the interspecific relationships within Diaporthe. The dataset consisted of 124 sequences including the outgroup, Diaporthella corylina (culture CBS 121124). A total of 2555 characters including gaps (505 for ITS, 513 for cal, 528 for his3, 475 for tef1 and 522 for tub2) were included in the phylogenetic analysis. The best nucleotide substitution model for ITS, his3 and tub2 was TrN+I+G, while HKY+I+G was selected for both cal and tef1. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Fig. 1). Isolates from Shaanxi Province formed three individual clades representing three undescribed species.
Figure 1.
Phylogram of Diaporthe resulting from a maximum likelihood analysis based on combined ITS, cal, his3, tef1 and tub2. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.90). The tree is rooted with Diaporthella corylina. Isolates in current study are in blue. “-” indicates ML BS < 50% or BI PP < 0.90.
Taxonomy
Diaporthe albosinensis
C.M. Tian & Q. Yang sp. nov.
27FA9968-ECF3-56FB-9BA6-7478C45CE54B
829518
Figure 2.
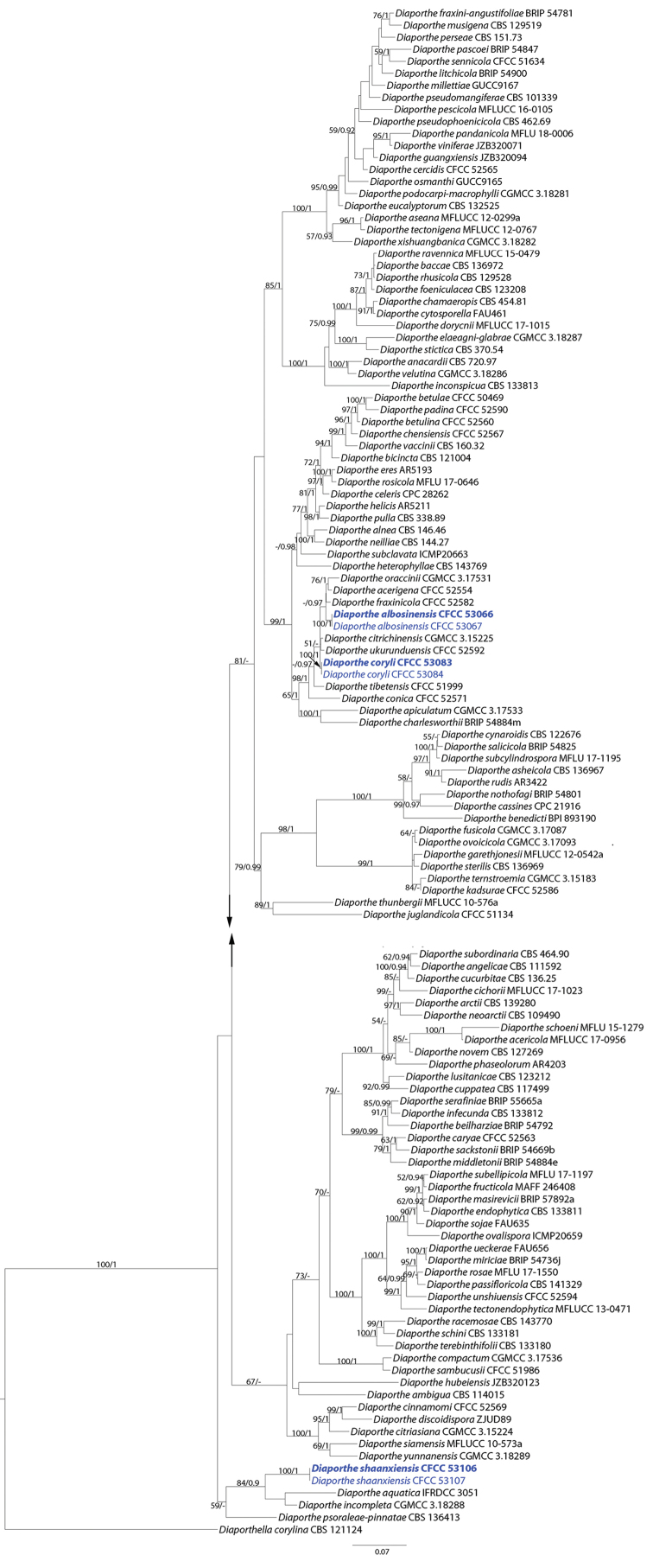
Diaporthe albosinensis on Betula albosinensis (BJFC-S1670). A Habit of conidiomata in wood B transverse section of conidiomata C longitudinal section through conidiomata D conidiogenous cells attached with beta conidia E conidiogenous cells attached with alpha conidia F beta conidia. Scale bars: 200 μm (B–C); 20 μm (D, F); 10 μm (E).
Diagnosis.
Distinguished from D. fraxinicola in having shorter conidiophores and longer beta conidia.
Etymology.
Named after the host plant, Betula albosinensis, from which the holotype was collected.
Description.
Conidiomata pycnidial, conical, immersed in bark, solitary to aggregated, erumpent through the bark surface, with a solitary undivided locule. Ectostromatic disc yellowish to brown, one ostiole per disc. Ostiole medium black, up to the level of disc. Locule undivided, (280–)290–375(–380) μm diam. Conidiophores (6–)8.5–13(–14.5) × (1.5–)2–2.5 μm, hyaline, cylindrical, smooth, phialidic, unbranched, straight or slightly curved. Alpha conidia hyaline, aseptate, fusiform, 0–1-guttulate, (7–)8–10(–11) × 2.5–3 μm. Beta conidia hyaline, aseptate, filiform, straight or slightly curved, eguttulate, base subtruncate, tapering towards one apex, (24–)25.5–30(–32) × 1–1.5 µm.
Culture characters.
Cultures incubated on PDA at 25 °C in the dark. Colony originally flat with white felted aerial mycelium, becoming light brown due to pigment formation, conidiomata irregularly distributed over agar surface, with yellowish conidial drops exuding from the ostioles.
Specimens examined.
China. Shaanxi Province: Ningshan County, Huoditang Forest Farm, 33°28'25"N, 108°29'39"E, on branches of Betula albosinensis, 10 July 2018, N. Jiang (holotype BJFC-S1670; ex-type living culture: CFCC 53066; living culture: CFCC 53067).
Notes.
Two isolates, representing D. albosinensis, are retrieved in a well-supported clade (ML BS/BPP=100/1) and appear most closely related to D. fraxinicola (Fig. 1). Diaporthe albosinensis can be distinguished from D. fraxinicola, based on tef1 and tub2 loci (3/335 in tef1 and 19/429 in tub2). Morphologically, D. albosinensis differs from D. fraxinicola in having shorter conidiophores (8.5–13 vs. 10.5–17.5 μm) and longer beta conidia (25.5–30 vs. 19–29.5 μm) (Yang et al. 2018).
Diaporthe coryli
C.M. Tian & Q. Yang sp. nov.
0790CB77-EE63-5681-AD57-D83CB470F50D
829520
Figure 3.
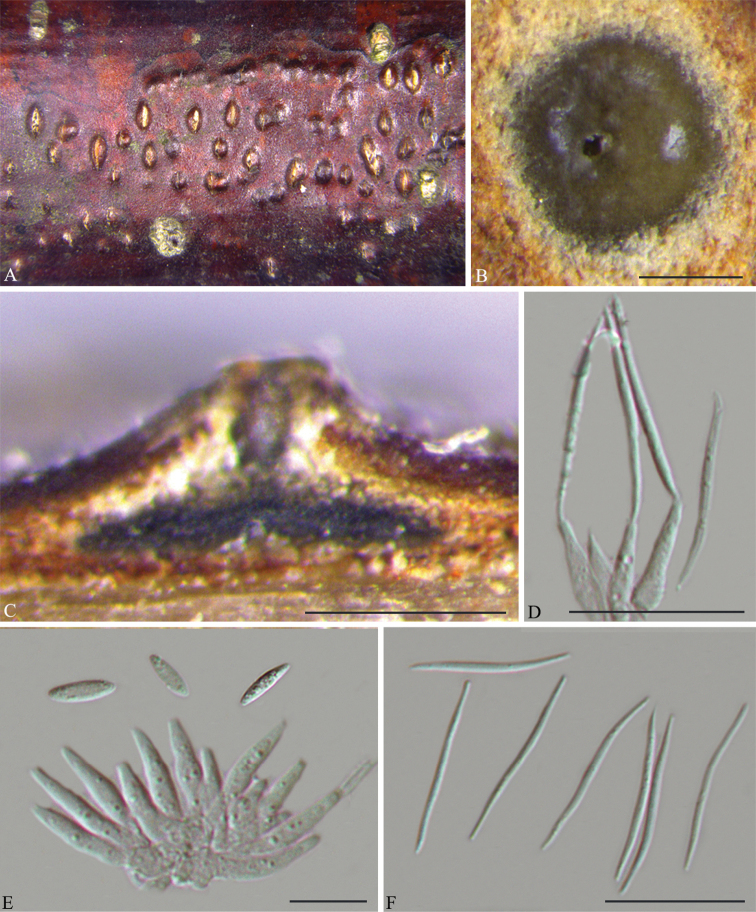
Diaporthe coryli on Corylus mandshurica (BJFC-S1671). A, B Habit of conidiomata in wood C transverse section of conidiomata D longitudinal section through conidiomata E conidiogenous cells attached with alpha conidia F alpha conidia. Scale bars: 500 μm (B–D); 10 μm (E); 20 μm (F).
Diagnosis.
Distinguished from D. ukurunduensis and D. citrichinensis in having larger alpha conidia.
Etymology.
Named after the genus of the host plant from which the holotype was collected, Corylus.
Description.
Conidiomata pycnidial, conical to spherical, immersed in the host bark, erumpent from surface of host branches, scattered, 950–1200 × 420–650 μm diam., covered by orange discharged conidial masses at maturity, usually conspicuous. Ectostromatic disc inconspicuous. Central column beneath the disc more or less conical, bright yellow. Conidiophores reduced to conidiogenous cells. Conidiogenous cells cylindrical, hyaline, smooth, unbranched, tapering towards the apex, (8.5–)10–12(–13) × (2–)2.5–3 μm. Alpha conidia hyaline, aseptate, fusiform, multiguttulate, rarely 2-guttulate, (10.5–)11.5–13(–13.5) × 3–3.5 μm. Beta conidia not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in the dark. Colony flat, felty with thick texture at the marginal area, with thin texture in the centre, producing beige pigment after 7–10 d. Aerial mycelium white, dense, conidiomata distributed in the centre, with translucent conidial drops exuding from the ostioles.
Specimens examined.
CHINA. Shaanxi Province: Ningshan County, Huoditang Forest Farm, 33°28'26"N, 108°29'40"E, on branches of Corylus mandshurica, 10 July 2018, N. Jiang (holotype BJFC-S1671; ex-type living culture: CFCC 53083); 33°28'26"N, 108°29'38"E, on branches of Corylus mandshurica, 10 July 2018, N. Jiang (paratype BJFC-S1672; living culture: CFCC 53084).
Notes.
We generated sequences for two isolates of D. coryli, CFCC 53083 and CFCC 53084. This new species is phylogenetically most closely related to D. ukurunduensis and D. citrichinensis (Fig. 1). Diaporthe coryli can be distinguished from D. ukurunduensis, based on ITS, his3 and tef1 loci (8/467 in ITS, 1/460 in his3 and 1/336 in tef1); and from D. citrichinensis based on tef1 and tub2 loci (4/335 in tef1 and 25/428 in tub2). Morphologically, D. coryli can be distinguished from both D. ukurunduensis (11.5–13 × 3–3.5 vs. 5–6 × 2–3 μm) and D. citrichinensis (11.5–13 × 3–3.5 vs. 5.5–9 × 1.5–2.5 μm) in having larger alpha conidia (Huang et al. 2013, Gao et al. 2016).
Diaporthe shaanxiensis
C.M. Tian & Q. Yang sp. nov.
1B8F6C9B-59D3-5D30-B416-6175363932FD
829527
Figure 4.
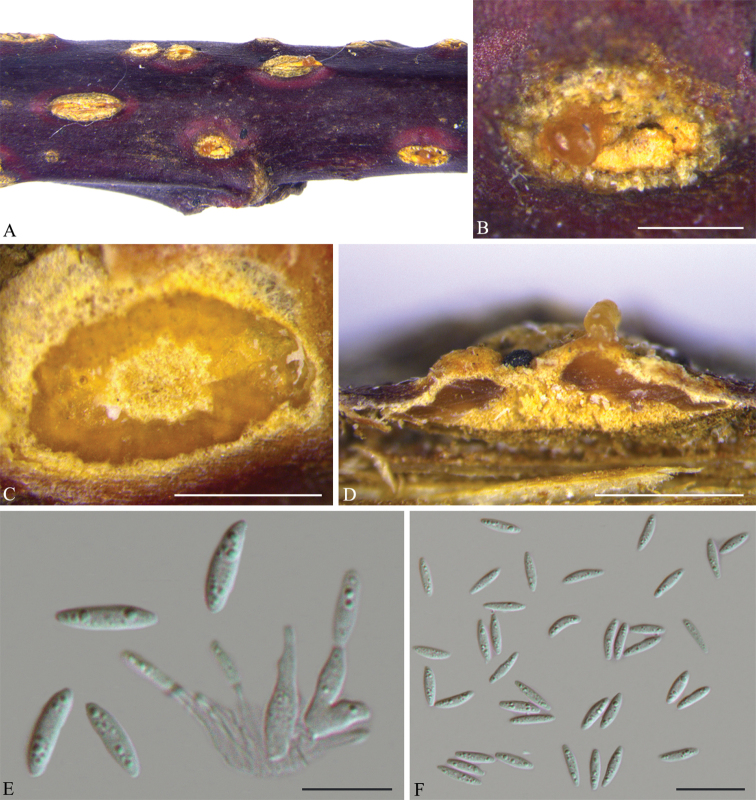
Diaporthe shaanxiensis on liana (BJFC-S1674). A, B Habit of conidiomata on twig C transverse section through conidiomata D longitudinal section through conidiomata E conidiogenous cells attached with beta conidia F beta conidia. Scale bars: 200 μm (B–D); 10 μm (E, F).
Diagnosis.
Distinguished from D. aquatica and D. incompleta in having longer beta conidia.
Etymology.
Named after Province Shaanxi, where the holotype was collected.
Description.
Conidiomata pycnidial, immersed in bark, scattered, erumpent through the bark surface, discoid, with a solitary undivided locule. Ectostromatic disc yellowish to pale brown, one ostiole per disc, usually conspicuous, (485–)500–687(–695) μm diam. Locule circular, undivided, (500–)526–765(–792) μm diam. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, cylindrical, unbranched, slightly curved, tapering towards the apex, (12.5–)14.5–17(–18) × 1–1.5(–2) μm. Alpha conidia not observed. Beta conidia hyaline, aseptate, filiform, straight to curved, eguttulate, (35.5–)37–47.5(–50) × 1 µm.
Culture characters.
Cultures incubated on PDA at 25 °C in the dark. Colony originally flat with white fluffy aerial mycelium, becoming pale brown with pigment, with visible solitary conidiomata at maturity.
Specimens examined.
CHINA. Shaanxi Province: Ningshan County, Huoditang Forest Farm, 33°28'25"N, 108°29'39"E, on branch of liana, 10 July 2018, N. Jiang (holotype BJFC-S1674; ex-type living culture: CFCC 53106); 33°28'24"N, 108°29'38"E, on branch of liana, 10 July 2018, N. Jiang (Paratype BJFC-S1675; living culture: CFCC 53107).
Notes.
In the combined tree, D. shaanxiensis is a distinct clade with maximum support and it appears to be most closely related to D. aquatica and D. incompleta (Fig. 1). Diaporthe shaanxiensis can be distinguished from D. aquatica by a 17 nt difference in the ITS region. For D. aquatica, only ITS sequences are available in NCBI GenBank (Hu et al. 2012). The new species can be distinguished from D. incompleta, based on ITS, cal, his3 and tef1 (24/454 in ITS, 14/443 in cal, 66/468 in his3 and 24/311 in tef1). Morphologically, D. shaanxiensis differs from both D. aquatica (37–47.5 vs. 9–12.5 µm) and D. incompleta (37–47.5 vs. 19–44 µm) in having longer beta conidia (Gao et al. 2016, 2017).
Discussion
In this study, an investigation of forest pathogens from Huoditang in Shaanxi Province was carried out and Diaporthe canker was observed as a common disease. Identification of our collections was conducted, based on isolates from fruiting bodies using five combined loci (ITS, cal, his3, tef1 and tub2), as well as morphological characters. Three new Diaporthe species were described. These are D. albosinensis sp. nov., D. coryli sp. nov. and D. shaanxiensis sp. nov.
Diaporthe albosinensis is associated with Betula albosinensis. Thus far, six Diaporthe species have been reported from Betula. These are D. alleghaniensis, D. betulae, D. betulicola, D. betulina, D. eres and D. melanocarpa (Kobayashi 1970, Gomes et al. 2013, Du et al. 2016, Yang et al. 2018). Morphologically, D. albosinensis differs from D. betulae (600–1250 μm), D. betulicola (700–1300 μm) and D. betulina (670–905 μm) in having smaller locules (Du et al. 2016, Yang et al. 2018); and from D. alleghaniensis (5–8 × 1.5–2 μm) and D. eres (6.5–8.5 × 3–4 μm) in having larger alpha conidia (Arnold 1967, Anagnostakis 2007, Gomes et al. 2013). In addition, our phylogenetic reconstruction of a five-locus dataset adds support for the new species, although no sequence data are currently available for D. alleghaniensis, D. betulicola and D. melanocarpa (Fig. 1). Interestingly, D. melanocarpa is found on different plant hosts; it was described from Pyrus melanocarpa in London and then recorded from Amelanchier, Betula and Cornus (Dearness 1926, Wehmeyer 1933, Kobayashi 1970). Diaporthe coryli is characterised by the ostiole with orange discharged conidial masses and a yellow central column (Fig. 3). Diaporthe shaanxiensis was found on branches of liana with an obvious ostiole per disc and characterised by hyaline, filiform beta conidia. Alpha conidia were found neither in the natural environment nor in culture for this species.
Species delimitation of Diaporthe has improved considerably by using a combination of morphological, cultural, phytopathological and molecular phylogenetic analyses (Udayanga et al. 2014a, b, 2015, Fan et al. 2015, Gao et al. 2017, Guarnaccia and Crous 2017, Hyde et al. 2017, 2020, Guarnaccia et al. 2018, Yang et al. 2018, Long et al. 2019). As a result, many Diaporthe canker diseases and new species have been discovered and reported from all over the world and also in China. The descriptions and molecular data of Diaporthe species represent an important resource for plant pathologists, plant quarantine officials and taxonomists.
Supplementary Material
Acknowledgements
This study is financed by the Research Foundation of Education Bureau of Hunan Province, China (Project No.: 19B608) and the introduction of talent research start-up fund project of CSUFT (Project No.: 2019YJ025). We are grateful to Chungen Piao, Minwei Guo (China Forestry Culture Collection Center, Chinese Academy of Forestry, Beijing) and reviewers Lu Quan and Jadson Bezerra.
Citation
Yang Q, Jiang N, Tian C-M (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. https://doi.org/10.3897/mycokeys.67.49483
Funding Statement
the Research Foundation of Education Bureau of Hunan Province, China (Project No.: 19B608) and the introduction of talent research start-up fund project of CSUFT (Project No.: 2019YJ025).
References
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- Dearness J. (1926) New and noteworthy fungi. Mycologia 18: 236–255. 10.1080/00275514.1926.12020515 [DOI] [Google Scholar]
- Diogo E, Santos JM, Phillips AJ. (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. 10.1007/s13225-010-0057-x [DOI] [Google Scholar]
- Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH. (2017) The current status of species in Diaporthe. Mycosphere 8: 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. 10.2307/2419362 [DOI] [Google Scholar]
- Du Z, Fan XL, Hyde KD, Yang Q, Liang YM, Tian CM. (2016) Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. in China. Phytotaxa 269: 90–102. 10.11646/phytotaxa.269.2.2 [DOI] [Google Scholar]
- Fan XL, Hyde KD, Udayanga D, Wu XY, Tian CM. (2015) Diaporthe rostrata, a novel ascomycete from Juglans mandshurica associated with walnut dieback. Mycological Progress 14: 1–8. 10.1007/s11557-015-1104-5 [DOI] [Google Scholar]
- Fan XL, Yang Q, Bezerra JDP, Alvarez LV, Tian CM. (2018) Diaporthe from walnut tree (Juglans regia) in China, with insight of Diaporthe eres complex. Mycological Progress 1–13. 10.1007/s11557-018-1395-4 [DOI]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8: 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyne A, López-Medrano F, Morales JM, Esteban CG, Martín I, Eraña I, Meije Y, Lalueza A, Alastruey‐Izquierdo A, Rodríguez‐Tudela JL, Aguado JM. (2011) Cutaneous infection by Phomopsis longicolla in a renal transplant recipient from Guinea: first report of human infection by this fungus. Transplant Infectious Disease 13: 204–207. 10.1111/j.1399-3062.2010.00570.x [DOI] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2018) Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathologia Mediterranea 57(2).
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40: 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hu DM, Cai L, Hyde KD. (2012) Three new ascomycetes from freshwater in China. Mycologia 104: 1478–1489. 10.3852/11-430 [DOI] [PubMed] [Google Scholar]
- Huang F, Hou X, Dewdney MM, Fu Y, Chen G, Hyde KD, Li HY. (2013) Diaporthe species occurring on Citrus in China. Fungal Diversity 61: 237–250. 10.1007/s13225-013-0245-6 [DOI] [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei DP, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li JF, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang SK, Thiyagaraja V, Lu YZ, Jayawardena RS, Dong W, Yang EF, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Lúcio R, Alvarenga M, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu JC, Sheng J. (2020) Fungal diversity notes 115–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. (1970) Taxonomic studies of Japanese Diaporthaceae with special reference to their life-histories. Bulletin of the Government Forest Experiment Station 226: 1–242. [Google Scholar]
- Long H, Zhang Q, Hao YY, Shao XQ, Wei XX, Hyde KD, Wang Y, Zhao DG. (2019) Diaporthe species in south-western China. MycoKeys 57: 113. 10.3897/mycokeys.57.35448 [DOI] [PMC free article] [PubMed]
- Mostert L, Crous PW, Kang JC, Phillips AJ. (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167. 10.1080/00275514.2001.12061286 [DOI] [Google Scholar]
- Murali TS, Suryanarayanan TS, Geeta R. (2006) Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52: 673–680. 10.1139/w06-020 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. (2010) FigTree v.1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK.
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew, UK.
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandečić K, Ćosić J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27: 9–19. 10.3767/003158511X603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014b) Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Diversity 67: 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014a) Species limits in Diaporthe: molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 32: 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lűcking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castanéda-Ruiz RF, Bahkali AH, Pang KL, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro JS, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel MA, Takamatsu S, Bensch K, Silva NI, De Kesel A, Karunarathna A, Boonmee S, Pfister DH, Lu YZ, Luo ZL, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng XY, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera PH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma XY, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC. (2017) Notes for genera: Ascomycota. Fungal Diversity 86: 1–594. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017a) Diaporthe species occurring on Senna bicapsularis in southern China, with descriptions of two new species. Phytotaxa 302: 145–155. 10.11646/phytotaxa.302.2.4 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017b) Diaporthe juglandicola sp. nov. (Diaporthales, Ascomycetes), evidenced by morphological characters and phylogenetic analysis. Mycosphere 8: 817–826. 10.5943/mycosphere/8/5/3 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CX, Cao ZM. (2007) Primary analysis of macrofungi flora of Huoditang Mts. Journal of Yunnan Agricultural University 22: 345–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.