Abstract
Alzheimer’s disease (AD) is a chronic and progressive neurological brain disorder. AD pathophysiology is mainly represented by formation of neuritic plaques and neurofibrillary tangles (NFTs). Neuritic plaques are made up of amyloid beta (Aβ) peptides, which play a central role in AD pathogenesis. In AD brain, Aβ peptide accumulates due to overproduction, insufficient clearance and defective proteolytic degradation. The degradation and cleavage mechanism of Aβ peptides by several human enzymes have been discussed previously. In the mean time, numerous experimental and bioinformatics reports indicated the significance of microbial enzymes having potential to degrade Aβ peptides. Thus, there is a need to shift the focus toward the substrate specificity and structure–function relationship of Aβ peptide-degrading microbial enzymes. Hence, in this review, we discussed in vitro and in silico studies of microbial enzymes viz. cysteine protease and zinc metallopeptidases having ability to degrade Aβ peptides. In silico study showed that cysteine protease can cleave Aβ peptide between Lys16–Cys17; similarly, several other enzymes also showed capability to degrade Aβ peptide at different sites. Thus, this review paves the way to explore the role of microbial enzymes in Aβ peptide degradation and to design new lead compounds for AD treatment.
Keywords: Alzheimer’s disease, Amyloid β-peptide, Aβ-degrading enzymes, Molecular modeling
Introduction
Alzheimer's disease (AD) is a neurological disorder of brain. It has been reported in the year 2000 that nearly 25 million people were affected worldwide by AD and this number would reach up to 63 and 114 millions by 2030 and 2050, respectively (Loncarevic et al. 2005). Because of this, AD is a serious problem in terms of medical and financial views to the society. There are different hypotheses in AD, including cholinergic, amyloid cascade, tau, oxidative imbalance and mild cognitive impairment (Schneider et al. 2011; Love et al. 2009; Ballard et al. 2011; Hardy and Selkoe 2002; Glenner and Wong 1984). The hypotheses, concepts and theories regarding AD have been discussed in earlier review (Barage and Sonawane 2015). In the cholinergic hypothesis, shortage of neurotransmitters like acetylcholine and butyrylcholine have been identified (Schneider et al. 2011). Therefore, inhibition of enzymes such as acetylcholinesterase (AChE), butyrylcholinesterase (BChE) which are responsible to break down acetylcholine and butyrylcholine has been considered as a standard approach to minimize symptomatic effects of AD (Schneider et al. 2011). The synthesis and molecular docking studies revealed potential of 2-substituted benzimidazoles as an acetyl cholinesterase inhibitor (AChE) (Sonawane et al. 2018).
Another hypothesis is the amyloid cascade, in which amyloid plaques are formed by accumulated Aβ peptides in AD patient’s brains (Reitz 2012; Barage et al. 2015). The amyloid plaque containing Aβ peptides is the pathological hallmark of AD (Hardy and Selkoe 2002; Glenner and Wong 1984). The Aβ1–42 peptides are main constituents of amyloid plaques in AD (Gouras et al. 1998; Masters et al. 1985; Tanzi and Bartman 2005). The Aβ peptide is produced after the sequential cleavage of amyloid precursor protein (APP) by beta- and gamma-secretases in the amyloidogenic pathway. The inhibition of these secretases becomes another approach in AD treatment (Love et al. 2009; Ballard et al. 2011). The steady-state concentration of Aβ peptide is tightly controlled by Aβ-degrading proteolytic enzymes and perivascular drainage (Weller et al. 2002; Nalivaeva et al. 2008; Hawkes et al. 2011). It has been showed that the arginine containing short peptide, RR-AFC can destabilize and inhibit the aggregation of Aβ protofibril by hydrogen bonding and hydrophobic interactions (Barale et al. 2019). There are various enzymes involved in Aβ peptide clearance such as insulin-degrading enzyme (IDE), neprilysin (NEP), endothelin-converting enzyme (ECE), angiotensin-converting enzyme (ACE), matrix metalloproteinases (MMPs) and plasmin (Iwata et al. 2001; Qiu et al. 1998; Tucker et al. 2000; Yin et al. 2006; Zou et al. 2007; Cimerman et al. 1999). Therefore, these enzymes can be targeted to reduce the Aβ peptide load in the AD brain. In addition, several in vitro and in silico studies reported the potential of microbial enzymes to degrade Aβ peptides (Hsu et al. 2009; Yoo et al. 2010; Dhanavade et al. 2013; Dhanavade and Sonawane 2014; Jalkute et al. 2015).
Earlier studies suggested that Nattokinase enzyme from Bacillus subtilis Natto (Hsu et al. 2009) and aminopeptidase from Streptomyces griseus KK565 can degrade Aβ peptides (Yoo et al. 2010). Similarly, experimental and some computational work revealed the role of microbial enzymes viz. aminopeptidase from Streptomyces griseus KK565, cysteine protease from Xanthomonas campestris and gelatinase from Enterococcus faecalis in Aβ peptides’ degradation (Yoo et al. 2010; Dhanavade et al. 2013; Dhanavade and Sonawane 2014; Jalkute et al. 2015). Thus, computational studies of microbial enzymes could be useful to understand Aβ peptide degradation in detail at the atomic level (Dhanavade et al. 2013; Dhanavade and Sonawane 2014; Jalkute et al. 2015). In this review article, we have summarized certain bacterial enzymes having potential to degrade Aβ peptides, which would be useful to design new therapeutic strategies to control Aβ peptides.
Microbial infections and Alzheimer's disease
Several microorganisms such as bacteria, viruses and yeasts have been suspected in AD brains and the oral infections were found to be responsible for the etiology of late onset of AD (LOAD) (Riviere et al. 2002; Poole et al. 2013) (Table 1). Numerous pathogens’ chronic periodontitis like P. gingivalis, T. forsythia, and T. denticola have been implicated in the development of several inflammatory diseases, out of which T. denticola represents a spirochete which mainly occurs in the AD brain (Riviere et al. 2002; Poole et al. 2013). The pathogenic bacteria like Chlamydophila pneumonia are likely to cause infection and cause pathogenesis in AD (Honjo et al. 2009; Maheshwari and Eslick 2015; Shima et al. 2010). Similarly, the role of bacterium such as H. pylori (monoinfection) has also been found in AD (Honjo et al. 2009). It was also observed that T. pallidum causes brain atrophy and deposition of Aβ in the atrophic form of general paresis (Miklossy 2011a, b). Also, the herpes simplex virus (HSV-1) is associated with amyloid-containing plaques and/or NFTs found to be involved in AD pathology (Harris and Harris 2018). Similarly, several studies have also been reported to reveal the association between Aβ accumulation in the brain and HSV infection (De Chiara et al. 2010; Itzhaki et al. 2008). Hence, understanding these microbial infections and pathogenesis could be an important task to control the adverse effects of AD.
Table 1.
The bacteria present in brain which can impact on AD
| Sr. no. | Name of bacteria which can influence in AD brain | References |
|---|---|---|
| 1 | Escherichia coli | Zhan et al. (2016) |
| 2 | Chlamydia pneumoniae | Balin et al. (2018) |
| 3 | Helicobacter pylori | Kountouras et al. (2009) |
| 4 | Lactobacillus fermentum | Wang et al. (2015) |
| 5 | Bacteroides fragilis | Lukiw (2016) |
| 6 | Klebsiella pneumonia | Bieler et al. (2005) |
| 7 | Mycobacterium tuberculosis | Alteri et al. (2007) |
| 8 | Salmonella typhimurium | Castelijn et al. (2012) |
| 9 | Salmonella enterica | Solomon et al. (2005) |
| 10 | Bacillus subtilis | Romero et al. (2014) |
| 11 | Pseudomonas | Dueholm et al. (2010) |
| 12 | Streptococcus mutans | Oli et al. (2012) |
| 13 | Candida albicans | Garcia et al. (2013) |
Gut-microbiota and Alzheimer's disease
Haran and co-workers have found that the microbiome of AD patients is associated with the disregulation of the anti-inflammatory p-glycoprotein pathway (Haran et al. 2019). Their conclusions are based on functional studies combined with machine learning approaches (Haran et al. 2019). It has been reported that the gut bacteria can produce amino acids like gamma-amino butyric acid (GABA), tryptophan and monoamines such as serotonin, histamine, dopamine, which play an important role in the brain (Briguglio et al. 2018; Lyte et al. 2018; Thomas et al. 2012; Wall et al. 2014). The relationship between gut microbiota and brain of the respective person has been discussed elegantly (Giau et al. 2018a, b). It has been showed that the commensal intestinal flora helps the host to produce necessary vitamins (Nishino et al. 2013). Borghammera and Beege hypothesized that the disease starts in the gut and then further spread to the brain (Borghammera and Berge 2019). The gut-related inflammation proves systematic inflammation, neuro-inflammation and endotoxemia, which has resulted into the development of neural disorders and psychiatric illness in later life (Sochocka et al. 2019). The amino acids produced by gut microbiota serves as a component of neurotransmitters influencing neurons in central nervous system (CNS) (Lyte et al. 2018). Pro-inflammatory cytokines and deposited Aβ peptides create positive feedback in the development of neurodegenerative diseases. Hence, there are considerable evidences which revealed that gut microbiota affect CNS, neuroinflammation and neurodegenerative disease such as AD (Sochocka et al. 2019). Hence, Giau and co-workers highlighted that cell signaling study might be useful to understand the role of different components produced by gut microbiota in AD (Giau et al. 2018a, b).
Human enzymes involved in Aβ peptide degradation and clearance
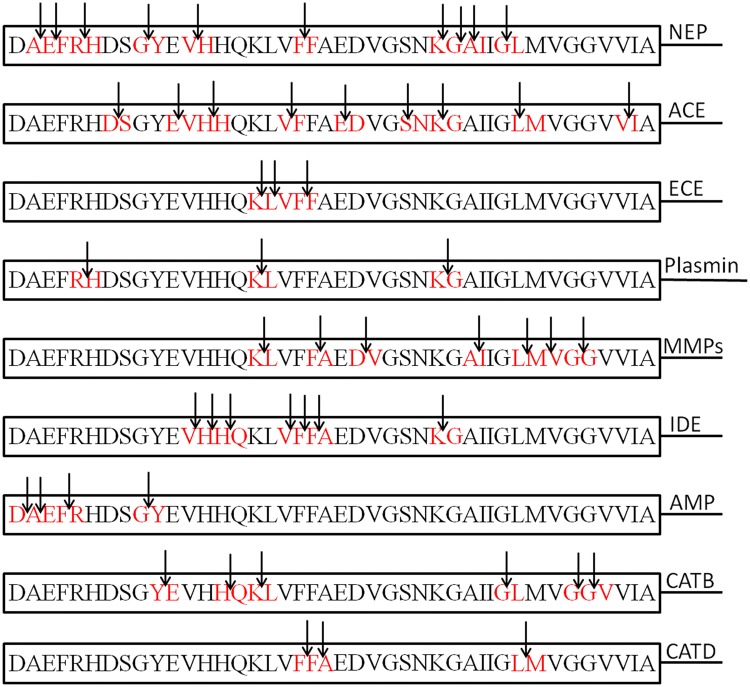
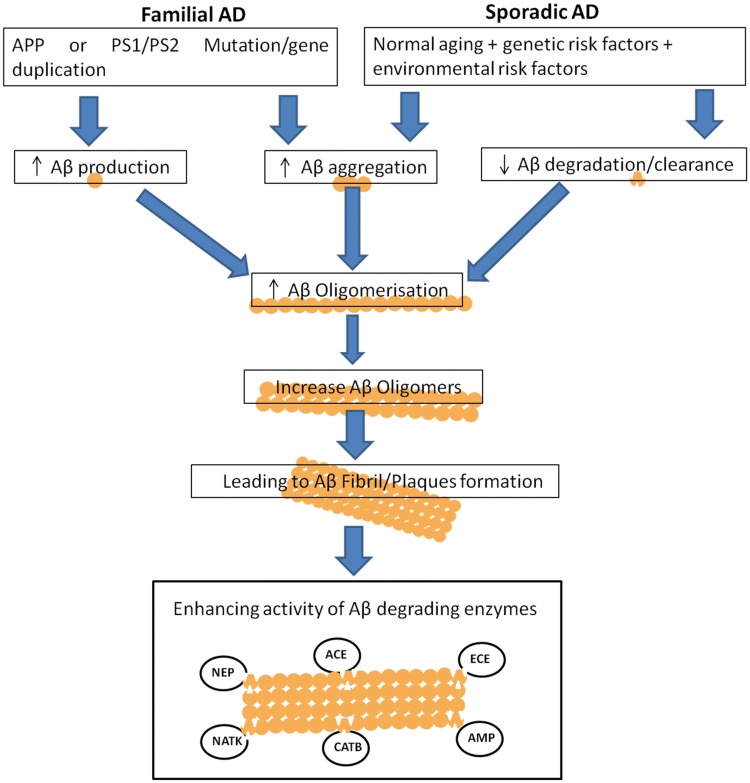
There are several drugs which can reduce Aβ peptides production, but they have been reported with certain side effects (Henley et al. 2009; Bateman et al. 2009). Some anti-dementia and memory-impairing drugs like nicotinic antagonist, mecamylamine, and the N-methyl-d-aspartic acid (NMDA) receptor antagonist, MK-801, have been described (Yuedea et al. 2007). Hence, research should also be focused on reducing the Aβ peptide levels by means of alternative approaches. In near future, enzymatic degradation can be mainly targeted to reduce the Aβ peptide levels and AD risk (Baranello et al. 2015). In human brain, there are many proteases or peptidases, which showed capability to degrade Aβ either in vitro or in vivo as can be seen in Fig. 1. These enzymes include neprilysin (NEP) (Iwata et al. 2001), endothelin-converting enzyme (ECE)-1 (Eckman et al. 2001), insulin-degrading enzyme (IDE) (Kurochkin and Goto 1994; McDermott and Gibson 1997; Qiu et al. 1998), angiotensin-converting enzyme (ACE) (Hu et al. 2001), uPA/tPA–plasmin system (Sasaki et al. 1988; Verde et al. 1988), cathepsin D (Yamada et al. 1995a, b; Hamazaki 1996), gelatinase A (Yamada et al. 1995a, b), gelatinase B (Backstrom et al. 1996), aminopeptidase A (Sevalle et al. 2009), cathepsin B (Mueller-Steiner et al. 2006), antibody light chain c23.5, hk14 (Rangan et al. 2003), and α2-macroglobulin complexes (Qiu et al. 1996). Many of these enzymes possess more than one cleavage sites in Aβ peptide (Fig. 1, Table 2). Therefore, up-regulation of these proteases in the brain could be a possible way to control accumulation and Aβ peptides’ aggregation, which could prove as an effective therapeutic strategy (Mueller-Steiner et al. 2006; Rangan et al. 2003; Qiu et al. 1996; Leissring et al. 2003). However, cross-reactivity of these enzymes could be another issue. Although these enzymes in human brain decrease the load of Aβ peptides, but the factors such as enzymatic loss through genetic mutations or non-genetic reasons like direct oxidative damage or enhanced production of inhibitors may result into abnormal Aβ catabolism (Wang et al. 2006). Because of these limitations, there is a need to discover new Aβ-degrading enzymes from other sources as well to study Aβ catabolism in detail at the molecular level. There are different enzymes from microbial sources having ability to degrade Aβ peptides. Further, these microbial enzymes can be studied to design novel drugs against AD. The enhanced activity of Aβ peptide-degrading enzymes could be a promising approach in AD treatment (Fig. 2).
Fig. 1.
Cleavage sites of amyloid beta (Aβ) peptide for human, microbial enzymes
Table 2.
The amyloid beta (Aβ) peptide degrading microbial, human enzymes and their cleavage sites
| Enzymes | Amyloid beta (Aβ) peptide cleavage site | References |
|---|---|---|
| Neprilysin (NEP) | A2–E3, E3–F4, R5–H6, G9–Y10, V12–H13, F19–F20, K28–G29, G29–A30, A30–I31 and G33–L34 | Howell et al. (1995); Nalivaeva et al. (2011) |
| Angiotensin-converting enzyme (ACE) | D7–S8, E11–V12, H13–H14, V18–F19, E22–D23, S26–N27, K28–G29, L34–M35 and V40–I41 | Zou et al. (2009); Hu et al. (2001) |
| Endothelin-converting enzyme (ECE) | K16–L17, L17–V18, F19–F20 | Eckman et al. (2001); Schulz et al. (2009); Sonawane and Barage (2015) |
| Plasmin | R5–H6, K16–L17, K28–G29 | Werb (1997); Ledesma et al. (2000) |
| Matrix metalloproteiases (MMPs) | K16–L17, F20–A21, D23–V24, A30–I31, L34–M35, M35–V36, G37–G38 | Carvalho et al. (1997); Yan et al. (2006) |
| Insulin degrading enzyme (IDE) | V12–H13, H13–H14, H14–Q15, V18–F19, F19–F20, F20–A21, K28–G29 | Farris et al. (2003) |
| Aminopeptidase (AMP) | D1–A2, A2–E3, F4–R5, G9–Y10 | Sevalle et al. (2009) |
| Cathepsin B (CATB) | Y10–E11, H14–Q15, K16–L17, G33–L34, G37–G38, G38–V39 | Mueller-Steiner et al. (2006) |
| Cathepsin D (CATD) | F19–F20, F20–A21, L34–M35 | Rogeberg et al. (2014); Sadik et al. (1999) |
Fig. 2.
Aβ peptide production and its clearance by enhance enzymatic activity (↑ increase, ↓ decrease)
Microbial enzymes having role in Aβ peptide degradation similar to human enzymes
Several in vitro studies have confirmed the Aβ peptide-degrading abilities of enzymes from microbial sources (Hsu et al. 2009; Yoo et al. 2010; Ningthoujama et al. 2019). Out of which, one report suggests that soluble Keratinase 1 reconstituted on neutral/cationic liposomes can degrade Aβ fibrils efficiently (Ningthoujama et al. 2019); an interesting report on small peptides having ability to inhibit oligomerization/aggregation of Aβ peptides and convert it into non-toxic oligomeric forms (Ribaric 2018). Such type of small peptides could be studied further for their potential role in AD treatment. There is one report in which authors pointed out strong connection between the general mitochondrial dysfunction and AD progression; they discussed how these two processes are interrelated (Giau et al. 2018a, b). Therefore, experimental and computational studies of Aβ peptide-degrading enzymes from microbial sources could be helpful to understand the Aβ peptide cleavage process in detail at molecular level. Sequence analysis, homology modeling, molecular docking and MD simulation studies of cysteine protease from Xantomonas campestris showed homology and Aβ peptide cleavage sites similar to human cathepsin B enzyme (Dhanavade et. al. 2013).
Matrix metalloproteinase (MMPs)
Matrix metalloproteinases (MMPs) are zinc and calcium-dependent endopeptidases produced by neuron and glial cells in humans (Yan et al. 2006). The experimental studies performed by Yan and coworkers confirmed that MMP-9 has ability to cleave Aβ peptide (Yan et al. 2006). The important ability of MMP9 over other Aβ-degrading enzymes is to degrade fibrillar Aβ peptides (Backstrom et al. 1996). The elevated activities of MMP-2 and MMP-9 were observed in the hippocampus tissue from AD patient’s brain. However, MMP-2, MMP-3 and MMP-9 expression has been found up-regulated in several brain tissues in response to Aβ stimulation (Deb et al. 1996; Miners et al. 2008; Gottschall et al. 1996). The MMP-2 is also known as gelatinase A or type IV collagenase (Roher et al. 1994). The studies done by Roher and coworker showed that MMP-2 cleaves peptide bonds of Aβ1–40 and Aβ1–42 in between different residues such as Ly16-Leu17, Leu34-Met35 and Met35-Val36 (Gottschall et al. 1996). Backstrom and co-workers determined that MMP-9 is present in the vicinity of extracellular amyloid plaques (Backstrom et al. 1996). Further, it has been confirmed that MMP-9 can also cleave aggregated Aβ fibrils (Gottschall et al. 1996). MMP-9 cleaves Aβ peptides predominantly in between Leu34-Met35 of membrane-spanning domain along with other numerous sites (Backstrom et al. 1996; Jung et al. 2003). The studies showed that MMP2 knockout has been resulted into increasing Aβ1–40 and Aβ1–42 levels in the soluble fraction of hippocampus and cortex (Yin et al. 2006). Also, the increased Aβ peptide levels in the brain were observed after infusion of a broad spectrum metalloproteinase inhibitor suggesting the importance of MMP-2 and other metalloproteinases in Aβ turnover to maintain the steady-state level (Yin et al. 2006).
Therefore, these metalloproteinase from human and other sources can be targeted to study the degradation of Aβ peptide. The X-ray crystal structure of MMP-9 (resolution—2.3 Å) has been solved by Rowsell and group members (Rowsell et al. 2002). The molecular modeling study depicted the potential of gelatinase from Enterococcus faecalis to degrade Aβ peptide (Jalkute et al. 2015). This study showed that the active site residue Glu328 of gelatinase from Enterococcus faecalis involved in catalytic mechanism by serving as a proton shuttle to cleave peptide bond in between Leu34-Meth35 of wild-type Aβ peptide (Jalkute et al. 2015). Thus, gelatinase from Eterococcus faecalis could also be a good target to study the exact cleavage mechanism of Aβ peptide degradation and to design new therapeutic approaches for the treatment of AD.
Cathepsin B (Catb)
The use of cysteine protease to degrade Aβ peptide in vivo has been shown pharmacologically (Frautschy et al. 1998). Further, in vivo study performed by Mueller-Steiner and co-workers also revealed the potential of CatB, a cysteine protease in degradation of Aβ peptide (Mueller-Steiner et al. 2006). Similarly, the role of CatB has also been shown in protein degradation, mainly Aβ peptides (Glabe 2001). However, in certain pathological conditions CatB is secreted by exocytosis (Cataldo and Nixon 1990; Cataldo et al. 1997). In AD, CatB has been found in the extracellular amyloid plaques (Maruyama et al. 2005). It is well known that CatB can also perform processing of APP and Aβ peptides (Cataldo and Nixon 1990; Cataldo et al. 1997). CatB cleaves Aβ peptides from carboxy terminus resulted into reduced Aβ1–42 peptides’ concentration (Cataldo et al. 1997). Although CatB can efficiently reduce the oligomeric Aβ1–42 levels in vitro, it is not known whether the over-expressed CatB also reduces toxic Aβ oligomers level in vivo. CatB can directly reduce the local monomeric and oligomeric Aβ1–42 peptide concentration through proteolytic cleavage (Cataldo et al. 1997). These studies showed that stimulating CatB activity might reduce Aβ peptides level, mainly Aβ1-42, resulted into the protection from AD-related deficits. The cystatin C is a cysteine protease inhibitor, which inhibits CatB activity (Cimerman et al. 1999; Turk et al. 1995). The increased CatB activity is achieved by reducing cystatin C levels (CysC, CST3) (Cimerman et al. 1999; Turk et al. 1995). Hence, from these studies it is confirmed that CatB degrades Aβ peptide and could be a possible target for AD treatment.
Bioinformatics study has been performed to find out microbial enzymes having ability to degrade Aβ peptide similar to human CatB (Dhanavade et al. 2013). Molecular docking study performed using AutoDock (Morris et al. 2009) showed that cysteine protease from Xanthomonas campestris possesses sequence similarity to human CatB (Dhanavade et al. 2013). The molecular docking and molecular dynamic simulation studies showed that the sulfhydryl hydrogen atom of Cys17 residue of cysteine protease from Xanthomonas campestris interacts with carboxylic oxygen of Lys16 of Aβ peptide, indicating the cleavage site (Dhanavade et al. 2013). In this study, three-dimensional fold of cysteine protease from Xanthomonas campestris found similar to crystal structure of human CatB (Dhanavade et al. 2013). This study may be helpful to understand Aβ peptide degradation and further, to design new therapeutic approaches for AD treatment (Dhanavade et al. 2013).
Aminopeptidase (Amp)
Aminopeptidases (AMP) (EC 3.4.11.-) are a heterogeneous group of exopeptidases, which catalyzes amino acid removal from the N-terminus of substrates such as proteins or peptides (Taylor et al. 1993a, b; Yao and Cohen 1999). These aminopeptidase enzymes are widely distributed in animal and plant kingdoms. Aminopeptidases are known to play important roles in activation, modulation, and degradation of bioactive peptides (Lendeckel et al. 2000; Stoltze et al. 2000; Hui 2007). The studies done by Sevalle and co-workers confirm that aminopeptidase A (APA) carries out Aβ peptide truncation at N-terminal (Sevalle et al. 2009). It has been noted that the N-terminal truncated Aβ peptide with first two residues deleted and glutamate at position 3 undergoes cyclization (Stoltze et al. 2000; Hui 2007; Saido 1996; Russo et al. 1997). The study performed by Medeiros and co-workers showed that aminopeptidase also plays an important role in tauopathies (Medeiros et al. 2011). Many neurodegenerative disorders are caused due to accumulation of hyperphosphorylated TAU proteins which can form neurofibrillary tangles (Wang et al. 2010; Ballatore et al. 2007). Therefore, the reduction of TAU helps to develop potential therapy for AD and other tauopathies (Bruden et al. 2009). It has been showed by Yoo and co-workers that aminopeptidase from Streptomyces griseus KK565 degrades Aβ peptide in vitro (Yoo et al. 2010). Therefore, such microbial enzymes may be targeted to understand the Aβ peptide degradation mechanism.
In silico experiments such as sequence analysis, homology modeling, molecular docking and molecular dynamics simulations have been found useful to understand the exact role of aminopeptidase from Streptomyces griseus KK565 in Aβ peptide degradation (Dhanavade and Sonawane 2014). The mutant Aβ peptide was compared with wild-type Aβ peptide, in which aminopeptidase enzyme showed more interactions with wild-type Aβ peptide than mutant Aβ peptide (Fede et al. 2009; Dhanavade and Sonawane 2014). Thus, these experimental and computational studies of aminopeptidase from Streptomyces griseus KK565 would be important to understand the Aβ peptide degradation process in detail at the molecular level.
Nattokinase
The experimental study performed by Hsu and coworkers suggest that Nattokinase enzyme from Bacillus subtilis Natto has potential to degrade Aβ peptide (Hsu et al. 2009). The bacterial strain Bacillus subtilis was isolated from fermented food Natto. Then, the Nattokinase enzyme was purified from this isolated Bacillus subtilis strain and further tested to know its ability to degrade Aβ peptide (Hsu et al. 2009). The nattokinase enzyme was found stable at 50 °C; which showed good activity at neutral pH. Therefore, Nattokinase enzyme could be a good target in the drug development studies. Further, exact Aβ peptide degradation mechanism by Nattokinase can be studied in detail at the atomic level using bioinformatics approaches.
Currently, AD treatment has been focused on cholinesterase inhibitors (minimizing symptomatic AD effects) and NMDA-receptor antagonists (Parsons et al. 2013). Many compounds have been tested to minimize the symptomatic effects of AD. The main concern is that only few of these compounds have been appeared in the drug development process or clinical testing by randomized clinical trials. Generally, negative results are reported for these clinical trials (Aprahamian et al. 2013). In last decade, most of the drugs were in phase 3 trials and all of them have failed to provide good clinical benefits, or were suspended because of having severe adverse side effects (Aprahamian et al. 2013). It has been demonstrated that the memantine nanoparticles are more effective than the free drug; hence, MEM–PEG–PLGA nanoparticles (NPs) could be an effective alternative for AD patients (Sanchez‑Lopez et al. 2018). Moreover, all drugs currently in clinical trials for AD treatment available in the database of Clinicaltrials.gov till February 12, 2019 have been reviewed thoroughly (Cummings et al. 2019). Therefore, there is a need to find new targets and approaches in AD treatment. Hence, above-discussed microbial enzymes having potential to degrade Aβ peptides could be an interesting topic for the development of new approaches in AD treatments.
Microbial enzymes and drug discovery in Alzheimer's disease
Earlier in vitro report revealed that stimulating the activity of Aβ-degrading enzymes using potent pharmacological agents could prevent the Aβ pathology (Kuruppua et al. 2016). It is well known that the pathways for Aβ synthesis and degradation have become crucial targets to develop effective treatment for AD. The recent report suggests that Aβ degradation is being used as a more effective strategy than preventing Aβ production (Sikanyika et al. 2019). Hence, Aβ-degrading enzymes are mainly targeted as an effective therapeutic agent in AD treatment (Sikanyika et al. 2019). One report suggests that people with coronary artery disease (CAD) are more prone to develop AD than those without CAD (De la Torre et al. 2004). It is believed that Aβ1–40 and APP are having an important role in atherothrombosis, vascular inflammation, vascular and cardiac aging; hence, Aβ1–40 can also be considered as a new biomarker for risk stratification in cardiovascular disease (Stakos et al. 2020).
It is quite difficult to isolate human enzymes from AD brain to study degradation mechanism of Aβ peptides. However, microbial enzymes having Aβ peptide-degrading ability can be easily isolated, identified from known microbial species and studied further (Hsu et al. 2009; Yoo et al. 2010; Dhanavade et al., 2013; Dhanavade and Sonawane, 2014; Jalkute et. al. 2015). Hence, these microbial enzymes having ability to degrade Aβ peptide can be further purified to fetch three-dimensional structural information, which could be useful to understand mechanism of Aβ peptide degradation. This experimental and structural information of microbial enzymes could be useful to find out effective drug molecules against the target. These drug molecules can be further tested on human enzymes having maximum sequence similarity with microbial enzymes.
Conclusion
In the current review, we discussed in vitro and in silico studies of microbial enzymes having potential to degrade Aβ peptides similar to human enzymes. These studies could be helpful to understand Aβ peptide degradation in detail at the atomic level. Such microbial enzymes having ability to degrade Aβ peptide at multiple sites may be targeted in the future drug-discovery process to design new therapeutic approaches for AD treatment. Thus, the present review would be helpful to get some useful information about the microbial enzymes involved in Aβ peptide degradation. Further, there is a need to find new enzymes having ability to degrade Aβ peptides from novel microbial sources, which could be useful to understand proper interactions between enzymes and Aβ peptide at molecular level.
Acknowledgements
KDS is thankful to University Grants Commission, New Delhi for providing financial support under UGC SAP DRS Phase-II programme sanctioned to Department of Biochemistry, Shivaji University, Kolhapur. Authors are thankful to Department of Science and Technology, New Delhi for providing fellowship as research assistance under DST-PURSE scheme.
Compliance with ethical standards
Conflict of interest
Authors have no conflict of interest.
References:s
- Aprahamian I, Stella F, Forlenza OV. New treatment strategies for Alzheimer’s disease: is there a hope? Indian J Med Res. 2013;138:449–460. [PMC free article] [PubMed] [Google Scholar]
- Backstrom JR, Lim GP, Cullen MJ, Tokes ZA. Matrix metalloproteinase-9 (MMP-9) is synthesized in neurons of the human hippocampus and is capable of degrading the amyloid-beta peptide (1–40) J Neurosci. 1996;16:7910–7919. doi: 10.1523/JNEUROSCI.16-24-07910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011;377:1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Barage SH, Sonawane KD. Amyloid cascade hypothesis: pathogenesis and therapeutic strategies in Alzheimer's disease. Neuropeptides. 2015;52:1–18. doi: 10.1016/j.npep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Barale S, Parulekar R, Fandilolu P, Dhanavade M, Sonawane K. Molecular insights into destabilization of Alzheimer’s Aβ protofibril by arginine containing short peptides: a molecular modeling approach. ACS Omega. 2019;4(1):892–903. [Google Scholar]
- Baranello R, Bharani K, Padmaraju V, Chopra N, Lahiri D, Greig N, Pappolla M, Sambamurti K. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer’s Disease. Curr Alzheimer Res. 2015;12(1):32–46. doi: 10.2174/1567205012666141218140953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R, Siemers E, Mawuenyega K, Wen G, Browning K, Sigurdson W, Yarasheski K, Friedrich S, Demattos R, May P, Paul S, Holtzman D. A gamma secretase inhibitor decreases amyloid-beta production in the central nervous system. Ann Neurol. 2009;66:48–54. doi: 10.1002/ana.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghammera P, Berge N. Brain-first versus gut-first Parkinson’s disease: a hypothesis. J Parkinson’s Dis. 2019;9:S281–S295. doi: 10.3233/JPD-191721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briguglio M, Dell’Osso B, Panzica G, Malgaroli A, Banfi G, Zanaboni Dina C, Galentino R, Porta M. Dietary neurotransmitters: a narrative review on current knowledge. Nutrients. 2018;10:591. doi: 10.3390/nu10050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunden K, Trojanowski J, Lee V. Advances in taufocused drug discovery for Alzheimer’s disease and related tauopathies. Nat Rev Drug Discov. 2009;8:783–793. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo A, Nixon R. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci USA. 1990;87:3861–3865. doi: 10.1073/pnas.87.10.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo A, Barnett J, Pieroni C, Nixon R. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased β-amyloidogenesis. J Neurosci. 1997;17:6142–6151. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimerman N, Prebanda M, Turk B, Popovic T, Dolenc I, Turk V. Interaction of cystatin C variants with papain and human cathepsins B, H and L. J Enzyme Inhib. 1999;14:167–174. doi: 10.3109/14756369909036552. [DOI] [PubMed] [Google Scholar]
- Cummings J, Lee G, Ritter A, Sabbagh M, Zhong K. Alzheimer's disease drug development pipeline: 2019. Alzheimers Dement (N Y) 2019;5:272–293. doi: 10.1016/j.trci.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara G, Marcocci M, Civitelli L, Argnani R, Piacentini R, Ripoli C, et al. APP processing induced by herpes simplex virus type 1 (HSV-1) yields several APP fragments in human and rat neuronal cells. PLoS ONE. 2010;5:e13989. doi: 10.1371/journal.pone.0013989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Torre J. Is Alzheimer’s disease a neurodegenerative or a vascular disorder? Data, dogma, and dialectics. Lancet Neurol. 2004;3:184–190. doi: 10.1016/S1474-4422(04)00683-0. [DOI] [PubMed] [Google Scholar]
- Deb S, Gottschall PE. Increased production of matrix metalloproteinases in enriched astrocyte and mixed hippocampal cultures treated with beta-amyloid peptides. J Neurochem. 1996;66:1641–1647. doi: 10.1046/j.1471-4159.1996.66041641.x. [DOI] [PubMed] [Google Scholar]
- Dhanavade M, Sonawane K. Insights into the molecular interactions between aminopeptidase and amyloid beta peptide using molecular modeling techniques. Amino Acids. 2014;46:1853–1866. doi: 10.1007/s00726-014-1740-0. [DOI] [PubMed] [Google Scholar]
- Dhanavade M, Jalkute C, Barage S, Sonawane K. Homology modeling, molecular docking and MD simulation studies to investigate role of cysteine protease from Xanthomonas campestris in degradation of Ab peptide. Comput Biol Med. 2013;43:2063–2070. doi: 10.1016/j.compbiomed.2013.09.021. [DOI] [PubMed] [Google Scholar]
- Eckman E, Reed D, Eckman C. Degradation of the Alzheimer’s amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276:24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli A, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantù L, Del Favero E, Levy E, Salmona M, Tagliavini F. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frautschy S, Horn D, Sigel J, Harris-White M, Mendoza J, Yang F, Saido T, Cole G. Protease inhibitor coinfusion with amyloid beta-protein results in enhanced deposition and toxicity in rat brain. J Neurosci. 1998;18:8311–8321. doi: 10.1523/JNEUROSCI.18-20-08311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giau V, An S, Hulme J. Mitochondrial therapeutic interventions in Alzheimer’s disease. J Neuro Sci. 2018 doi: 10.1016/j.jns.2018.09.033. [DOI] [PubMed] [Google Scholar]
- Giau V, Wu S, Jamerlan A, An S, Kim S, Hulme J. Gut microbiota and their neuroinflammatory implications in Alzheimer’s disease. Nutrients. 2018;10:1765–1782. doi: 10.3390/nu10111765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabe C. Intracellular mechanisms of amyloid accumulation and pathogenesis in Alzheimer’s disease. J Mol Neurosci. 2001;17:137–145. doi: 10.1385/JMN:17:2:137. [DOI] [PubMed] [Google Scholar]
- Glenner G, Wong C. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- Gottschall P. beta-Amyloid induction of gelatinase B secretion in cultured microglia: inhibition by dexamethasone and indomethacin. NeuroReport. 1996;7:3077–3080. doi: 10.1097/00001756-199611250-00057. [DOI] [PubMed] [Google Scholar]
- Gouras G, Xu H, Jovanovic J, Buxbaum J, Wang R, Greengard P, Relkin N, Gandy S. Generation and regulation of beta-amyloid peptide variants by neurons. J Neurochem. 1998;71:1920–1925. doi: 10.1046/j.1471-4159.1998.71051920.x. [DOI] [PubMed] [Google Scholar]
- Hamazaki H. Cathepsin D is involved in the clearance of Alzheimer’s beta-amyloid protein. FEBS Lett. 1996;396:139–142. doi: 10.1016/0014-5793(96)01087-3. [DOI] [PubMed] [Google Scholar]
- Haran J, Bhattarai S, Foley S, Dutta P, Ward D, Bucci V, McCormickb B. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. mbio. 2019;10:e00632–e719. doi: 10.1128/mBio.00632-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe D. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harris S, Harris E. Molecular mechanisms for herpes simplex virus type 1 pathogenesis in Alzheimer’s disease. Front Aging Neurosci. 2018;10:48–72. doi: 10.3389/fnagi.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes C, Hartig W, Kacza J, Schliebs R, Weller R, Nicoll J, Carare R. Perivascular drainage of solutes is impaired in the ageing mouse brain and in the presence of cerebral amyloid angiopathy. Acta Neuropathol. 2011;121:431–443. doi: 10.1007/s00401-011-0801-7. [DOI] [PubMed] [Google Scholar]
- Henley D, May P, Dean R, Siemers E. Development of semagacestat (LY450139), a functional gamma-secretase inhibitor, for the treatment of Alzheimer’s disease. Expert Opin Pharmacother. 2009;10:1657–1664. doi: 10.1517/14656560903044982. [DOI] [PubMed] [Google Scholar]
- Honjo K, van Reekum R, Verhoeff N. Alzheimer’s disease and infection: do infectious agents contribute to progression of Alzheimer’s disease? Alzheimers Dement. 2009;5:348–360. doi: 10.1016/j.jalz.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Hsu R, Lee K, Wang J, Lily Y, Lee L, Rita P, Chen Y. Amyloid-degrading ability of Nattokinase from Bacillus subtilis Natto. J Agric Food Chem. 2009;57:503–508. doi: 10.1021/jf803072r. [DOI] [PubMed] [Google Scholar]
- Hu J, Igarashi A, Kamata M, Nakagawa H. Angiotensin converting enzyme degrades Alzheimer amyloid beta-peptide (A beta); retards A beta aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem. 2001;276:47863–47868. doi: 10.1074/jbc.M104068200. [DOI] [PubMed] [Google Scholar]
- Hui K. Neuropeptidases. In: Lajtha A, Banik N, editors. Handbook of neurochemistry and molecular neurobiology: neural protein metabolism and function, vol 7. 3. Berlin: Springer; 2007. [Google Scholar]
- Itzhaki R, Cosby S, Wozniak M. Herpes simplex virus type 1 and Alzheimer’s disease: the autophagy connection. J Neurovirol. 2008;14:1–4. doi: 10.1080/13550280701802543. [DOI] [PubMed] [Google Scholar]
- Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, Gerard C, Hama E, Lee HJ, Saido TC. Metabolic regulation of brain Aβ by neprilysin. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- Jalkute C, Barage S, Dhanavade M, Sonawane K. Insight into molecular interactions of Aβ peptide and gelatinase from Enterococcus faecalis: a molecular modeling approach. RSC Adv. 2015;5:10488–10496. [Google Scholar]
- Jung S, Zhang W, Van Nostrand W. Pathogenic A beta induces the expression and activation of matrix metalloproteinase-2 in human cerebrovascular smooth muscle cells. J Neurochem. 2003;85:1208–1215. doi: 10.1046/j.1471-4159.2003.01745.x. [DOI] [PubMed] [Google Scholar]
- Kurochkin I, Goto S. Alzheimer’s beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345:33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- Kuruppua S, Rajapakseb N, Spicerd A, Parkingtonc H, Smith A. Stimulating the activity of amyloid-beta degrading enzymes: a novel approach for the therapeutic manipulation of amyloid-beta levels. J Alzheimers Dis. 2016;54:891–895. doi: 10.3233/JAD-160492. [DOI] [PubMed] [Google Scholar]
- Leissring M, Farris W, Chang A, Walsh D, Wu X, Sun X, Frosch M, Selkoe D. Enhanced proteolysis of β-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron. 2003;40:1087–1093. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- Lendeckel U, Arndt M, Frank K, Spiess A, Reinhold D, Ansorge S. Modulation of WNT-5A expression by actinonin: linkage of APN to the WNT-pathway? Adv Exp Med Biol. 2000;477:35–41. doi: 10.1007/0-306-46826-3_3. [DOI] [PubMed] [Google Scholar]
- Loncarevic N, Mehmedika-Sulic E, Alajibegovic A. The neurologist role in diagnostics and therapy of the Alzheimer’s disease. Med Arc. 2005;59(2):106–109. [PubMed] [Google Scholar]
- Love S, Miners S, Palmer J, Chalmers K, Kehoe P. Insights into the pathogenesis and pathogenicity of cerebral amyloid angiopathy. Front Biosci. 2009;14:4778–4792. doi: 10.2741/3567. [DOI] [PubMed] [Google Scholar]
- Lyte M, Villageliú D, Crooker B, Brown D. Symposium review: microbial endocrinology—why the integration of microbes, epithelial cells, and neurochemical signals in the digestive tract matters to ruminant health1. J Dairy Sci. 2018;101:5619–5628. doi: 10.3168/jds.2017-13589. [DOI] [PubMed] [Google Scholar]
- Maheshwari P, Eslick G. Bacterial infection and Alzheimer’s disease: a meta-analysis. J Alzheimers Dis. 2015;43:957–966. doi: 10.3233/JAD-140621. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Higuchi M, Takaki Y, Matsuba Y, Tanji H, Nemoto M, Tomita N, Matsui T, Iwata N, Mizukami H, Muramatsu S, Ozawa K, Saido TC, Arai H, Sasaki H. Cerebrospinal fluid neprilysin is reduced in prodromal Alzheimer’s disease. Ann Neurol. 2005;57:832–842. doi: 10.1002/ana.20494. [DOI] [PubMed] [Google Scholar]
- Masters C, Simms G, Weinman N, Multhaup G, McDonald B, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J, Gibson A. Degradation of Alzheimer’s beta-amyloid protein by human and rat brain peptidases: involvement of insulin-degrading enzyme. Neurochem Res. 1997;22:49–56. doi: 10.1023/a:1027325304203. [DOI] [PubMed] [Google Scholar]
- Medeiros R, Baglietto-Vargas D, LaFerla F. The Role of tau in Alzheimer's Disease and related disorders. CNS Neurosci Ther. 2011;17(5):514–524. doi: 10.1111/j.1755-5949.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklossy J. Alzheimer’s disease_a neurospirochetosis. Analysis of the evidence following Koch’s and Hill’s criteria. J Neuroinflammation. 2011;8:90. doi: 10.1186/1742-2094-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklossy J. Emerging roles of pathogens in Alzheimer disease. Expert Rev Mol Med. 2011;13:e30. doi: 10.1017/S1462399411002006. [DOI] [PubMed] [Google Scholar]
- Miners J, Baig S, Palmer J, Palmer L, Kehoe P, Love S. A beta-degrading enzymes in Alzheimer's disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G, Huey R, Lindstrom W, Sanner M, Belew R, Goodsell D, Olson A. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort J, Buttle D. Cathepsin B. Int J Biochem Cell Biol. 1997;29:715–720. doi: 10.1016/s1357-2725(96)00152-5. [DOI] [PubMed] [Google Scholar]
- Mueller-Steiner S, Zhou Y, Arai H, Roberson E, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, Gan L. Antiamyloidogenic and neuroprotective functions of cathepsinB: implications for Alzheimer's disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Nalivaeva N, Fisk L, Belyaev N, Turner A. Amyloid-degrading enzymes as therapeutic targets in Alzheimer’s disease. Curr Alzheimer Res. 2008;5:212–224. doi: 10.2174/156720508783954785. [DOI] [PubMed] [Google Scholar]
- Ningthoujama D, Mukherjeea S, Devia L, Singha E, Tamreihaoa K, Khunjamayuma R, Banerjeeb S, Mukhopadhyay D. In vitro degradation of β-amyloid fibrils by microbial keratinase. Alzheimer’s Dementia Transl Res Clin Interv. 2019;5:154–163. doi: 10.1016/j.trci.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino R, Mikami K, Takahashi H, Tomonaga S, Furuse M, Hiramoto T, Aiba Y, Koga Y, Sudo N. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterol Motil. 2013;25:521–528. doi: 10.1111/nmo.12110. [DOI] [PubMed] [Google Scholar]
- Parsons C, Danysz W, Dekundy A, Pulte I. Memantine and cholinesterase inhibitors: complementary mechanisms in the treatment of Alzheimer’s disease. Neurotox Res. 2013;24:358–369. doi: 10.1007/s12640-013-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole S, Singhrao S, Kesavalu L, Curtis M, Crean S. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J Alzheimers Dis. 2013;36:665–677. doi: 10.3233/JAD-121918. [DOI] [PubMed] [Google Scholar]
- Qiu W, Borth W, Ye Z, Haass C, Teplow D, Selkoe D. Degradation of amyloid beta-protein by a serine proteasealpha2-macroglobulin complex. J Biol Chem. 1996;271:8443–8451. doi: 10.1074/jbc.271.14.8443. [DOI] [PubMed] [Google Scholar]
- Qiu W, Walsh D, Ye Z, Vekrellis K, Zhang J, Podlisny M, Rosner M, Safavi A, Hersh L, Selkoe D. Insulin-degrading enzyme regulates extra-cellular levels of amyloid β-protein by degradation. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- Rangan S, Liu R, Brune D, Planque S, Paul S, Sierks M. Degradation of beta-amyloid by proteolytic antibody light chains. J Biochem. 2003;42:14328–14334. doi: 10.1021/bi035038d. [DOI] [PubMed] [Google Scholar]
- Reitz C. Alzheimer’s Disease and the amyloid cascade hypothesis: a critical review. Int J Alzheimer’s Dis. 2012 doi: 10.1155/2012/369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaric S. Peptides as potential therapeutics for Alzheimer’s Disease. Molecules. 2018;23:283–313. doi: 10.3390/molecules23020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere G, Riviere K, Smith K. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer’s disease. Oral Microbiol Immunol. 2002;17:113–118. doi: 10.1046/j.0902-0055.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- Roher A, Kasunic T, Woods A, Cotter R, Ball M, Fridman R. Proteolysis of A beta peptide from Alzheimer disease brain by gelatinase A. Biochem Biophys Res Commun. 1994;205:1755–1761. doi: 10.1006/bbrc.1994.2872. [DOI] [PubMed] [Google Scholar]
- Rowsell S, Hawtin P, Minshull C, Jepson H, Brockbank S, Barratt D, Slater A, McPheat W, Waterson D, Henney A, Pauptit R. Crystal structure of human MMP9 in complex with a reverse hydroxamate inhibitor. J Mol Biol. 2002;319(1):173–181. doi: 10.1016/S0022-2836(02)00262-0. [DOI] [PubMed] [Google Scholar]
- Russo C, Saido T, DeBusk L, Tabaton M, Gambetti P, Teller J. Heterogeneity of water-soluble b-peptide in Alzheimer’s disease and Down’s syndrome brains. FEBS Lett. 1997;409:411–416. doi: 10.1016/s0014-5793(97)00564-4. [DOI] [PubMed] [Google Scholar]
- Saido T, Yamao-Harigaya W, Iwatsubo T, Kawashima S. Amino-and carboxy-terminal heterogeneity of b-amyloid peptides deposited in human brain. Neurosci Lett. 1996;215:173–176. doi: 10.1016/0304-3940(96)12970-0. [DOI] [PubMed] [Google Scholar]
- Sánchez-López E, Ettcheto M, Egea M, Espina M, Cano A, Calpena A, Camins A, Carmona N, Silva A, Souto E, García M. Memantine loaded PLGA PEGylated nanoparticles for Alzheimer's disease: in vitro and in vivo characterization. J Nanobiotechnol. 2018;16(1):32. doi: 10.1186/s12951-018-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Saito Y, Hayashi M, Otsuka K, Niwa M. Nucleotide sequence of the tissue-type plasminogen activator cDNA from human fetal lung cells. Nucleic Acids Res. 1988;16(12):5692. doi: 10.1093/nar/16.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Insel P, Weiner M. Treatment with cholinesterase inhibitors and memantine of patients in the Alzheimer’s disease neuroimaging initiative treatment with ChEIs and Memantine in ADNI. Arch Neurol. 2011;68:58–66. doi: 10.1001/archneurol.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevalle J, Amoyel A, Robert P, Fournié-Zaluski M, Roques B, Checler F. Aminopeptidase A contributes to the N-terminal truncation of amyloid b-peptide. J Neurochem. 2009;109:248–256. doi: 10.1111/j.1471-4159.2009.05950.x. [DOI] [PubMed] [Google Scholar]
- Shima K, Kuhlenbäumer G, Rupp J. Chlamydia pneumonia infection and Alzheimer’s disease: a connection to remember? Med Microbiol Immunol. 2010;199:283–289. doi: 10.1007/s00430-010-0162-1. [DOI] [PubMed] [Google Scholar]
- Sikanyika N, Parkington H, Smith A, Kuruppu S. Powering amyloid beta degrading enzymes: a possible therapy for Alzheimer’s Disease. Neurochem Res. 2019 doi: 10.1007/s11064-019-02756-x. [DOI] [PubMed] [Google Scholar]
- Sochocka M, Donskow-Łysoniewska K, Diniz B, Kurpas D, Brzozowska E, Leszek J. The gut microbiome alterations and inflammation-driven pathogenesis of Alzheimer’s Disease—a critical review. Mol Neurobiol. 2019;56(3):1841–1851. doi: 10.1007/s12035-018-1188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakos D, Stamatelopoulos K, Bampatsias D, Sachse M, Zormpas E, Vlachogiannis N, Tual-Chalot S, Stellos K. The Alzheimer’s Disease amyloid-beta hypothesis in cardiovascular aging and disease. J Am Coll Cardiol. 2020;75(8):952–967. doi: 10.1016/j.jacc.2019.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson M, Hersh L, Kalbacher H, Stevanovic S, Rammensee H, Schild H. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- Tanzi R, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Taylor A. Aminopeptidases: structure and function. FASEB J. 1993;7:290–298. doi: 10.1096/fasebj.7.2.8440407. [DOI] [PubMed] [Google Scholar]
- Taylor A. Aminopeptidases: towards a mechanism of action. Trends Biochem Sci. 1993;18:167–171. [PubMed] [Google Scholar]
- Thomas C, Hong T, van Pijkeren J, Hemarajata P, Trinh D, Hu W, Britton R, Kalkum M, Versalovic J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE. 2012;7:e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker H, Kihiko M, Caldwell J, Wright S, Kawarabayashi T, Price D, Walker D, Scheff S, McGillis J, Rydel R, Estus S. The plasmin system is induced by and degrades amyloid-β aggregates. J Neurosci. 2000;20:3937–3946. doi: 10.1523/JNEUROSCI.20-11-03937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, Bieth J, Bjork I, Dolenc I, Turk D, Cimerman N, Kos J, Colic A, Stoka V, Turk V. Regulation of the activity of lysosomal cysteine proteinases by pH-induced inactivation and/or endogenous protein inhibitors, cystatins. Biol Chem Hoppe Seyler. 1995;376:225–230. doi: 10.1515/bchm3.1995.376.4.225. [DOI] [PubMed] [Google Scholar]
- Verde P, Boast S, Franze A, Robbiati F, Blasi F. An upstream enhancer and a negative element in the 5_flanking region of the human urokinase plasminogen activator gene. Nucleic Acids Res. 1988;16:10699–10716. doi: 10.1093/nar/16.22.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall R, Cryan J, Ross R, Fitzgerald G, Dinan T, Stanton C. Bacterial neuroactive compounds produced by psychobiotics. Adv Exp Med Biol. 2014;817:221–239. doi: 10.1007/978-1-4939-0897-4_10. [DOI] [PubMed] [Google Scholar]
- Wang D, Dickson D, Malter J. Beta-Amyloid degradation and Alzheimer's disease. J Biomed Biotechnol. 2006;2006:58406. doi: 10.1155/JBB/2006/58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Garg S, Mandelkow E, Mandelkow E. Proteolytic processing of tau. Biochem Soc Trans. 2010;38:955–961. doi: 10.1042/BST0380955. [DOI] [PubMed] [Google Scholar]
- Weller R, Yow H, Preston S, Mazanti I, Nicoll J. Cerebrovascular disease is a major factor in the failure of elimination of Ab from the aging human brain: implications for therapy of Alzheimer’s disease. Ann N Y Acad Sci. 2002;977:162–168. doi: 10.1111/j.1749-6632.2002.tb04812.x. [DOI] [PubMed] [Google Scholar]
- Yamada T, Kluve-Beckerman B, Liepnieks J, Benson M. In vitro degradation of serum amyloid A by cathepsin D and other acid proteases: possible protection against amyloid fibril formation. Scand J Immunol. 1995;41:570–574. doi: 10.1111/j.1365-3083.1995.tb03609.x. [DOI] [PubMed] [Google Scholar]
- Yamada T, Miyazaki K, Koshikawa N, Takahashi M, Akatsu H, Yamamoto T. Selective localization of gelatinase A, an enzyme degrading beta-amyloid protein, in white matter microglia and in Schwann cells. Acta Neuropathologica (Berl) 1995;89:199–203. doi: 10.1007/BF00309334. [DOI] [PubMed] [Google Scholar]
- Yan P, Hu X, Song H, Yin K, Bateman R, Cirrito J, Xiao Q, Hsu F, Turk J, Xu J, Hsu C, Holtzman D, Lee J. Matrix metalloproteinase-9 degrades amyloid-β fibrils in vitro and compact plaques in situ. J Biol Chem. 2006;281:24566–24574. doi: 10.1074/jbc.M602440200. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen R. Giant proteases: beyond the proteasome. Curr Biol. 1999;9:R551–R553. doi: 10.1016/s0960-9822(99)80352-2. [DOI] [PubMed] [Google Scholar]
- Yin K, Cirrito J, Yan P, Hu X, Xiao Q, Pan X, Bateman R, Song H, Hsu F, Turk J. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006;26:10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C, Ahn K, Park J, Kim M, Jo S. An aminopeptidase from Streptomyces sp. KK565 degrades beta amyloid monomers, oligomers and fibrils. FEBS Lett. 2010;584:4157–4162. doi: 10.1016/j.febslet.2010.08.048. [DOI] [PubMed] [Google Scholar]
- Yuedea C, Donga H, Csernansky J. Anti-dementia drugs and hippocampal-dependent memory in rodents. Behav Pharmacol. 2007;18(5–6):347–363. doi: 10.1097/FBP.0b013e3282da278d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Yamaguchi H, Akatsu H, Sakamoto T, Ko M, Mizoguchi K, Gong J, Yu W, Yamamoto T, Kosaka K. Angiotensin converting enzyme converts amyloid beta-protein1–42 (Aβ1–42) toAβ1–40, and its inhibition enhances brain Abeta deposition. J Neurosci. 2007;27:8628–8635. doi: 10.1523/JNEUROSCI.1549-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]