Abstract
Background
Arginine-vasopressin (AVP) is a neuropeptide and provides learning and memory modulation. The AVP (4–5) dipeptide corresponds to the N-terminal fragment of the major vasopressin metabolite AVP (4–9), has a neuroprotective effect and used in the treatment of Alzheimer’s and Parkinson’s disease.
Methods
The main objective of the present study is to evaluate the molecular mechanism of AVP (4–5) dipeptide and to develop and synthesize chitosan nanoparticle formulation using modified version of ionic gelation method, to increase drug effectiveness. For peptide loaded chitosan nanoparticles, the synthesized experiment medium was simulated for the first time by molecular dynamics method and used to determine the stability of the peptide, and the binding mechanism to protein (HSP70) was also investigated by molecular docking calculations. A potential pharmacologically features of the peptide was also characterized by ADME (Absorption, Distribution, Metabolism and Excretion) analysis. The characterization, in vitro release study, encapsulation efficiency and loading capacity of the peptide loaded chitosan nanoparticles (CS NPs) were performed by Dynamic Light Scattering (DLS), UV–vis absorption (UV), Scanning Electron Microscopy (SEM), Fourier transform infrared (FT-IR) spectroscopy techniques. Additionally, in vitro cytotoxicity of the peptide on human neuroblastoma cells (SH-SY5Y) was examined with XTT assay and the statistical analysis was evaluated.
Results
The results showed that; hydrodynamic size, zeta potential and polydispersity index (PdI) of the peptide-loaded CS NPs were 167.6 nm, +13.2 mV, and 0.211, respectively. In vitro release study of the peptide-loaded CS NPs showed that 17.23% of the AVP (4–5)-NH2 peptide was released in the first day, while 61.13% of AVP (4–5)-NH2 peptide was released in the end of the 10th day. The encapsulation efficiency and loading capacity were 99% and 10%, respectively. According to the obtained results from XTT assay, toxicity on SHSY-5Y cells in the concentration from 0.01 μg/μL to 30 μg/μL were evaluated and no toxicity was observed. Also, neuroprotective effect was showed against H2O2 treatment.
Conclusion
The experimental medium of peptide-loaded chitosan nanoparticles was created for the first time with in silico system and the stability of the peptide in this medium was carried out by molecular dynamics studies. The binding sites of the peptide with the HSP70 protein were determined by molecular docking analysis. The size and morphology of the prepared NPs capable of crossing the blood-brain barrier (BBB) were monitored using DLS and SEM analyses, and the encapsulation efficiency and loading capacity were successfully performed with UV Analysis. In vitro release studies and in vitro cytotoxicity analysis on SHSY-5Y cell lines of the peptide were conducted for the first time.
Grapical abstract
Electronic supplementary material
The online version of this article (10.1007/s40199-019-00325-9) contains supplementary material, which is available to authorized users.
Keywords: AVP (4–5), Hsp70, Drug delivery, Nanoparticle, Chitosan, Parkinson, MD
Background
Arginine-vasopressin (AVP), which acts like a growth factor, is a neuropeptide that plays important roles such as regulation of nephron recirculation, water contraction, cardiovascular functions, cognition, tolerance, adaptation and complex sexual or maternal behaviour in the body and modulating of learning and memory in human brain [1–3]. Degradation of the function or level of AVP hormone in the body with aging causes neurodegenerative diseases such as Alzheimer’s (AD) and especially, Parkinson’s disease (PD) [4]. Based on a clinical observation shows that the reduced AVP hormone secretion is also caused nocturia which is one of the commonest nonmotor symptoms in PD [5]. In an in vitro study which is conducted in 2006, it was predicted that the analog of arginine-vasopressin could be used in the treatment of (PD) [1]. The AVP (4–9) and AVP (4–8) are two different analogs of AVP which have the highest nootropic activity, however, have very low bioavailability. Due to this reason, the design of shorter dipeptides and tripeptide fragments of AVP, which can pass the blood-brain barrier (BBB) and the gastrointestinal tract with having high bioavailability, is of great importance to help to relieve of the symptoms of PD. On the other hand, AVP (4–5)-NH2 (pGlu-Asn-NH2) which is a short peptide chain is an important dipeptide that corresponds to N-terminal fragment of major vasopressin metabolite AVP (4–9), was synthesized with neuroprotective effect. In vitro studies which was carried out with mouse hippocampal neuronal cell line (HT-22) showed that when enough concentration of AVP (4–5)-NH2 dipeptide interacted with neurotrophins and especially, heat shock protein 70 (HSP70) which plays critical role in the cell growth and survival in many organisms, the content of endogenous neuroprotective effects increased [1]. It is also known that Hsp70 is one of the important components of the cellular network and folding catalysts [6] and inhibits the formation of α-synuclein fibrils and β-amyloid during PD and AD, respectively. Because of over-expression of Hsp70, molecular chaperones suppress the toxicity of aberrantly folded proteins that occur in AD and PD [2, 7, 8].
In recent year, pharmaceutical researchers focused on the encapsulation of the active compounds into the biocompatible polymers. Polymer encapsulated systems provide long term released of active compounds like peptides, proteins or genes [9]. Among the polymers, chitosan is safe and non-toxic material used for encapsulation studies [10]. Chitosan [(1, 4)-2-amino-2-deoxy-D-glucan] (CS), a derivative of the chitin which is included in the shell of crustacean and the cell wall of fungi, is frequently used in medical field due to its high biocompatibility and non-toxic effects [11, 12]. In recent years, some studies which were carried out with the nanoparticular forms of CS (CS NPs) showed that this organic material can be used for delivery of various neuroprotective peptides in the body [13]. One of these studies, levodopa (L-DOPA) which is used for treatment of symptoms of PD was encapsulated in CS NPs and their cellular effects were observed on PC12 cells [14]. According to the results, L-DOPA-loaded CS NPs increased the cell viability as nearly 85% while this rate was only as nearly 43% for L-DOPA. These results showed that CS NPs is not only a drug delivery agent but also it has neuroprotective effects. In this way, various neuroprotective drugs or peptides can be easily delivered and their effectiveness can be amplified with CS NPs. Therefore, the AVP (4–5)-NH2 dipeptide was encapsulated by using chitosan polymer for development of pharmaceutical formulation. The drug delivery system based on CS NPs was synthesized using modified version of ionic gelation method, to overcome low bioavailability and increase the peptide effectiveness. In addition, in vitro release profile of AVP (4–5)-NH2 loaded CS NPs was performed against phosphate-buffered saline (PBS) at pH 7.4. Furthermore, in vitro cytotoxicity and neuroprotective effect of peptide and CS NPs on human neuroblastoma cells (SH-SY5Y) was examined with a colorimetric method based on the tetrazolium salt (XTT).
In this study, the experimental medium of peptide-loaded chitosan nanoparticles was created for the first time with in silico system and the stability of the peptide in this medium was carried out by molecular dynamics studies. The active binding mechanism between AVP (4–5)-NH2 and HSP70 protein was investigated using molecular docking method and a potent pharmacologically features of peptide were also emerged by ADME (Absorption, Distribution, Metabolism and Excretion) profile. The characteristic wavenumbers of the AVP (4–5)-NH2 were observed in the experimental vibrational (IR and Raman) spectra together with the fundamental vibrational wavenumbers which were calculated at the level of DFT-RB3LYP / 6–31++G (d, p) basis set. Also, the assignments of the fundamental wavenumbers, which were calculated using the scaled quantum mechanical force field (SQM FF) method, were performed in accordance with the total energy distribution of the vibrational modes (TED).
Methods
Materials
AVP (4–5)-NH2 dipeptide was purchased from GL-Biochem Co. CS (Low Molecular Weight) (Cat No: 448869), and triphenylphosphate (TPP) (Cat No: 241288) were purchased from Sigma–Aldrich (USA). Acetic acid (Cat No: 1.00056.2500) was purchased from Merck Millipore (100%) (Darmstadt, Germany). DMEM-F12 Medium, Fetal Bovine Serum and Penicilum–Streptomycin were obtained from Gibco. XTT was obtained from Invitrogen. Ultrapure water from Millipore Milli-Q Gradient System was used to prepare the solutions for synthesis and cell culture experiments.
Instrumentation
The NPs were prepared with ultrasonic horn (Bandelin, Sonopuls) and centrifuge (Hettich, Germany). The absorption spectra were obtained from UV-Vis spectrometer (Shimadzu, Japan) for used in preparation of calibration curve of in vitro release study of AVP (4–5)-NH2 peptide. Hydrodynamic size, polydispersity index (PdI) and zeta potential values were obtained from Zeta-Sizer Nano ZS (Malvern Instruments, Malvern, UK). Scanning Electron Microscopy (SEM) (Zeiss Gemini 500) was used to observation of the morphology of AVP (4–5)-NH2 peptide-loaded CS NPs. Fourier-transform infrared (FT-IR) spectra were recorded using a Jasco 6300 FT-IR. In order to measure the cell viability, Microplate Photometer (Thermo Labsystems Multiskan Ascent 354) was used.
Molecular dynamics simulations
Firstly the optimized molecular geometries of the AVP (4–5)-NH2, chitosan, acetic acid, TPP (tripolyphosphate) molecules which are used at experimental step have been obtained by using the Gaussian16 software [15]. To obtain the parameters and topologies that belong to the molecules, some specific programs and tools were used [16–19]. After all topology and parameter files were generated for the peptide, (MD) simulation step was carried out. MD simulation was performed by GROMACS 2016.1 [20] program using AMBER99SB force field [21].
In the first system, AVP (4–5)-NH2 peptide was put in a cubic box that was filled with 751 water molecules using TIP4P water models [22] and then MD study was realized for comparing with other system. In the second system, AVP (4–5)-NH2 molecule was put in a cubic box including 30 chitosan molecules, 5 acetic acid and 5 TPP and then, the box was filled with 30,259 water molecules using TIP4P water models and added 12 Na+ ions to neutralize the net charge of the system. The energy minimization of systems was achieved using 50,000 steps of steepest descent algorithm. For equilibrations, the systems were stabilized at 310 K using V-rescale thermostat for 100 ps as NVT ensemble [23] and Parrinello-Rahman pressure-coupling with 1.0 bar pressure [24] for 100 ps as NPT ensemble. After the resulting ensembles, 5 ns MD simulations were run with 2 fs time step for each simulation. During simulations, LINCS algorithm [25] was used for constraining the bond length. The graphics were plotted using XMGRACE [26] and VMD was used for the visualization of the box [27].
Molecular docking and ADME analysis
Firstly, the protein used for the docking study was selected from PDB databank (PDB code: 1S3X) [28] and its homology model was obtained by the SWISS-MODEL [29] server for better results. Taking the molecular structure from the result of MD simulation, the preparation and calculations of molecular docking was done by Schrödinger Maestro software using the Glide SP (standard precision) module (Schrödinger Release 2017–4: Maestro, Schrödinger, LLC, New York, NY, 2017) [30–32].
Firstly, AVP (4–5)-NH2 as a ligand molecule was prepared for docking calculations by the LigPrep tool in the Maestro 11.4 version of the Schrödinger Software program using the OPLS3 force field [33]. The 32 stereoisomers were produced for the ligand after the ionization states at pH 7.0 ± 2.0 which were selected. The structure of selected receptor of Heat Shock 70 kDa protein 1 having a solubility of 1.84 Å was prepared by the Protein Preparation Wizard tool [34]. The polar hydrogens were added to the heavy atoms and all waters and ions in the structure were removed. The bond orders were assigned, charges were defined at pH 7.0 and the selected receptor was optimized using PROPKA [35]. The heavy atoms in the receptor were converged by preferring 0.3A° RMSD and the OPLS3 force field. The Grid box was defined to the receptor by centering the existing co-crystallized ligand using grid generation tool in Maestro 11.4 version. The ligand was docked into the receptor based on the grid using standard precision (SP) docking algorithm to rank the ligand with specific conformation of the receptor molecule [36]. Drug candidate molecules display favourable ADME parameters. The Qik-Prop module (Schrödinger Release 2017–4: QikProp, Schrödinger, LLC, New York, NY, 2017.) was used to determine the ADME profile of the drug candidate molecule.
FT-IR analysis
The FT-IR spectra for AVP (4–5)-NH2 loaded CS NPs were recorded on a Jasco FT/IR-6300 spectrometer with diamond ATR unit in the range 400–4000 cm−1 and 2 cm−1 resolution. The GRAMS/AI 7.02 (Thermo Electron Corporation) software package was used to provide the baseline adjustment, second derivative (Savitzky–Golay function with 2 polynomial and 9 points), and peak fitting which is used band component analysis procedure. The Raman spectrum of the sample was taken with a Jasco NRS-3100 micro-Raman spectrometer (1200 lines/mm grating and high sensitivity cooled CCD).
Preparation of AVP (4–5)-NH2 peptide loaded CS NPs
CS NPs were prepared using a simple ionic gelation process with some modifications as described elsewhere [37–40]. Optimization studies were conducted, and the effects of the concentration of chitosan, TPP and peptide to particles size, polydispersity index and zeta potential values were investigated. Finally, optimum preparation method was determined. Briefly, 2 mg of CS were dissolved in 10 mL of 0.33% of acetic acid solution under magnetic stirring at 55–60 °C for an hour. Then, the solution was sonicated another 1 h for completely dissolving of CS in the solution. Notably, dissolving of CS in aqueous medium is very difficult. For this reason, sonication process is essential. Finally, the pH of the solution was adjusted to 4.7 with aid of 0.1 M of NaOH. On the other hand, 10 mL of aqueous TPP solution (1.53 mM) was prepared and stored at +4 °C for further use. For synthesis of CS NPs, 3.3 mL of ice-cold TPP solution was injected to 10 mL of CS at 4 °C under magnetic stirring. After 10 min later, white turbidity was observed, and this indicated that CS NPs were successfully obtained. For the synthesis of AVP (4–5)-NH2 peptide-loaded CS NPs, same amounts and the method which is described above were used. However, to prepare of the peptide loaded CS NPs, 2 mg AVP (4–5)-NH2 peptide were dissolved in 1 mL of ethanol (EtOH) and mixed with 20 mL CS solution which include 0.33% acetic acid. After the mixing, 6.6 mL of TPP solution injected to this solution at 4 °C.
Dynamic light scattering (DLS) analysis
In order to determine the physicochemical properties of the NPs in the aqueous medium, DLS measurements were used. Every electrophoretic light scattering measurement was performed at 25 °C with 4.0 mW He-Ne laser operating at a wavelength of 633 nm. Hydrodynamic size, zeta potential, and PdI values were reported as the mean of at least five determinations.
Preparation of calibration curve of AVP (4–5)-NH2 peptide
The calibration curve was constructed with standard solutions of AVP (4–5)-NH2 peptide absorbance values were recorded at 217.2 nm. This curve was used for quantification for further encapsulation efficiency and loading capacity studies.
Encapsulation efficiency and loading capacity of AVP (4–5)-NH2 peptide loaded CS NPs
UV-Vis spectrometer was used to determine the encapsulation efficiency and loading capacity of AVP (4–5)-NH2 peptide.
The EE and LC were calculated as follows:
| 1 |
| 2 |
In vitro release of AVP (4–5)-NH2 peptide-loaded CS NPs
In vitro release study of the NPs was performed in PBS at pH 7.4 by dialysis method. Briefly, 1 mg/mL of AVP (4–5)-NH2 peptide-loaded CS NPs were sealed with a cellulose membrane (10 kDa cut-off value) in a dialysis capsule. The whole system was maintained at 37 °C and kept in a water bath shaker. During the release process, 1 mL of dialysis medium was used at a predetermined time point (0, 0.5, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 24, 48, 96, 120, and 240 h) while adding the same volume of fresh PBS. The released drug concentrations in each time points were determined as detailed in the following equation via UV–Vis spectrometer analysis.
| 3 |
Scanning electron microscopy (SEM) analysis
The morphology of AVP (4–5)-NH2 peptide-loaded CS NPs was analysed using SEM (Zeiss Supra 50 V). In order to increase the conductivity of the samples, 8.10−1 mbar / Pa vacuum was applied and 100 Å thickness gold-palladium (20–80%) plating was performed via Leica EM ACE600 device. The imaging was conducted using an accelerating voltage of 3 and 7 kV and with the 30 k times magnification.
Culture of SHSY-5Y cell line
SHSY-5Y (Neuroblastoma) cell line was used for cytotoxicity experiments (ATCC, Manassas, VA, USA). SHSY-5Y cells were seeded in 25 cm2 cell culture flasks in DMEM-F12 Medium (Gibco,11320074) supplemented with Fetal Bovine Serum 10% (FBS-Gibco, 10500064) and Penicillium–Streptomycin (Gibco,15140122) 0.5% from 10,000 unit/mL Penicillin −10 mg/mL Streptomycin). Cells were incubated at 37 °C in 5% CO2 until they reach 80% confluency. Then trypsinization was performed to remove cells from flask surface. After trypsinization, medium was added to the cells and centrifuged at 1000 rpm for 5 min. After centrifugation, the supernatant was removed, and cells were counted via a hemocytometry.
Treatment of AVP (4–5)-NH2, chitosan NPs, and AVP (4–5)-NH2 loaded chitosan NPs to SHSY-5Y cells
In vitro cytotoxicity assay was conducted on SHSY-5Y cell lines. The cells were seeded in 96 well flat bottom microplate with density of 1 × 104 cells for each well and incubated at 37 °C in 5% CO2 incubator for 24 h. Then, cells were treated with different dilutions (2, 8, 32, 48, and 64 μg/μL) of AVP (4–5)-NH2, Chitosan NPs, and AVP (4–5)-NH2 loaded Chitosan NPs. After 24 h of incubation, the medium was aspirated and 100 μL of 0.5 mg/mL 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2 H-tetrazolium-5-carboxanilide (XTT) solution (with 7.5 μg/mL phenazine methosulfate) in fresh medium were added to each well. The plate was incubated for 4 h at 37 °C in 5% CO2 incubator [41]. The absorbance at 450 nm were read by multiplate reader (Labline). Percentage of cell viability was calculated with this formula;
| 4 |
Evaluation of neuroprotective effect in vitro
Neuroprotective effects of AVP (4–5)-NH2, Chitosan NPs, and AVP (4–5)-NH2 loaded Chitosan NPs on SH-SY5Y cells were examined. Briefly, SH-SY5Y cells were seeded in 96-well plates and incubated 24 h at 37 °C in 5% CO2 incubator. After that 32 μg/μL AVP (4–5)-NH2, Chitosan NPs, and AVP (4–5)-NH2 loaded Chitosan NPs were separately added to the wells. After 24 h, medium was discarded, and cells were treated with 1,5 M of H2O2 for 30 min [1]. Medium was discarded again and XTT method were applied for assay cell viability as mentioned before. Non-treated cells were used as positive control and % cell viability calculated via Eq. 4.
Statistical analysis
Statistical comparisons were performed by unpaired Student’s t test assuming equal variance for cytotoxicity assay. Differences were considered as statistically significant at p ≤ 0.05. Data are the mean ± standard error (SE).
Results and discussion
Molecular dynamics results
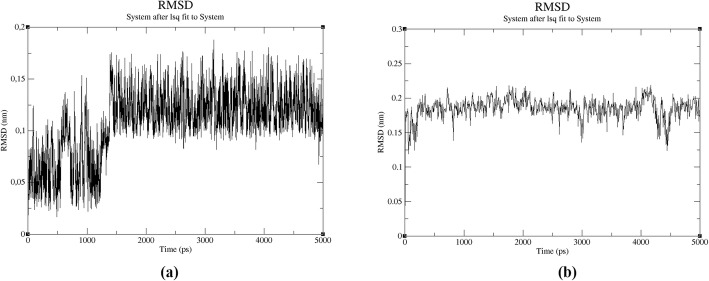
Firstly, all molecules were placed into the cubic box that was filled with 30,259 water molecules using TIP4P water model (Fig. 1). The net charge of the system was neutralized with counter ions. In this study, 12 Na+ ions were added to system for neutralization. For pH adjustment, NaOH were added to chitosan solution in the synthesis studies that were done using the ultra-pure water. According to this process, Na atoms were already available at preparation step of chitosan solution besides neutralization in the molecular dynamics study. After the system was completely constructed, the energy minimization was done using steepest descent algorithm. In energy minimization step, the potential energy of system was converged at 1120 steps with −1.5955162 × 106 kj/mol (Fig. S1). For equilibrations, NVT was achieved for 50,000 steps with a 2 fs time step at 310 K using the V-rescale thermostat that is a modified Berendsen thermostat and after NPT simulation at 1 bar result, the system’s average density was obtained as 1000.34 kg/m−3 in (Fig. S2). Lately, MD simulation of molecule with 2,500,000 steps was carried out for 5 ns. According to MD simulation, the average total, kinetic and potential energies were obtained at −1.14566 × 106 kj/mol, 2.39020 × 105 kj/mol and − 1.38807 × 106 kj/mol, respectively (Fig. S3 and Fig. S4). When the system was compared with MD study of AVP (4–5)-NH2 peptide in aqueous medium, it was observed that potential and total energies decreased while kinetic energy increased. The range of RMSD that belong to AVP (4–5)-NH2 peptide in the system was seen under 0.2 nm changed from 0.0004880 to 0.1893775 for 5 ns while the RMSD range of the peptide in the aqueous medium ranged from 0.0005046 to 0.1024015. According to these results, it was determined that the structure of AVP (4–5)-NH2 did not change significantly (Fig. 2). These RMSD values were important for verifying the stability of the structure. The MD study was given the preliminary information about stability of the peptide that exposed to the effects of other molecules in the synthesis of peptide loaded nanoparticles. Although molecular dynamics studies of various AVP derivatives have been performed in the literature [42, 43], there are no MD studies that simulate synthesized peptide loaded chitosan nanoparticles used in the experimental medium.
Fig. 1.
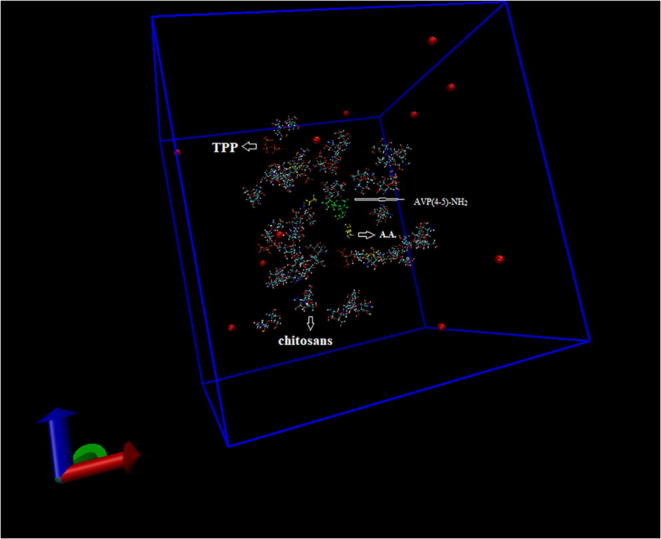
The initial confirmation of the system and counter ions in a cubic box
Fig. 2.
RMSD values of AVP(4–5)-NH2 peptide in aqueous medium (a) and in the system (b)
Molecular docking and ADME results
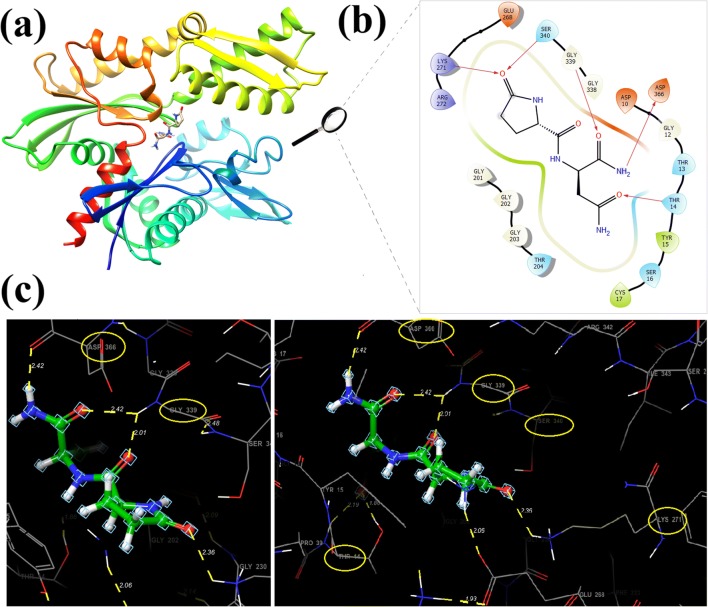
Molecular interaction and docking studies are very important for computer aided drug design and are used to better rationalize the action and prediction of the binding modes of drug candidate molecules. After the protein having 382 sequence length interacted with ligand, the energy of the docking score for AVP (4–5)-NH2 peptide obtained as −8.205 kcal/mol (Table 1 and Fig. 3). The binding regions of the Hsp70 kDa protein 1 for AVP (4–5)-NH2 were shown with hydrophobic interactions of TYR15, CYS17 and polar residues of THR14, THR13, THR204, SER16, SER340. The hydrogen bonds that linked to the ligand to the receptor which were observed on the binding regions are THR14 (1.86 Å and 2.19 Å), LYS271 (2.36 Å), GLY339 (2.42 Å and 2.01 Å) and ASP366 (2.42 Å), respectively (Fig. 3a., 3b., 3c.). The electrostatic potential map surfaces of the ligand and receptor protein were also constituted to define the regions that were electron-rich and electron-poor (Fig. S5). Moreover, favourable ADME properties which explain the pharmacological properties of a molecule that are necessary for being a drug candidate obtained using Qikprop tool of the Maestro software package and tabulated in Table 2. Lipinski’s five rules are used to determine whether the molecule having a pharmacological or biological activity, which has chemical and physical properties that make it an active drug. Lipinski’s rule generally refers to values that are multiples of 5, and generally an active drug must accomplish the following criteria [44, 45]. According to these rules, the molecular weight (mol_MW) of the drug candidate molecule should be less than 500 Da. The calculated molecular weight was 242.234 g/mol and is among the recommended values (recommended from 725 to 130). For a molecule of drug candidate, an octanol-water partition coefficient (log P) should not be greater than 5 [46]. Especially, logP coefficient, as one of estimative factors for BBB (Blood-Brain Barrier) [47], is crucial for the peptide having neuroprotective properties. The value of octanol-water partition coefficient that belongs to peptide was −4.023.
Table 1.
The conformation and docking score energies of AVP(4–5)-NH2
| Ligand | Energy(kcal/mol) | Docking Score(kcal/mol) |
|---|---|---|
| 1 | 3.340 | −8.205 |
| 2 | −1.918 | −8.204 |
| 3 | 4.229 | −8.182 |
| 4 | −1.255 | −7.098 |
Fig. 3.
The binding poses between that active site of Hsp70 Receptor and AVP(4–5)-NH2 peptide (a), 2D ligand interaction of AVP(4–5)NH2 peptide in the active side of the protein (b), the binding interaction of AVP(4–5)-NH2 peptide with THR14 (1.86 Å and 2.19 Å), LYS271 (2.36 Å), GLY339 (2.42 Å AND 2.01 Å) and ASP366 (2.42 Å) residues of protein, respectively
Table 2.
Docking score and calculated ADME properties of AVP(4–5)-NH2 peptide
| Property | Value | Recommended |
|---|---|---|
| Docking score (kcal/mol) | −8.205 | |
| Polar surface area PSA (A°2) | 180.528 | (7.0 / 200.0) |
| Molecular weight MW (g/mol) | 242.234 | (130.0 / 725.0) |
| Solute as Donor-Hydrogen Bonds | 4.250 | (0.0/ 6.0) |
| Solute as Acceptor-Hydrogen Bonds | 8.250 | (2.0/ 20.0) |
| Solute Ionization Potential (eV) | 9.382 | (7.9/ 10.5) |
| Solute Electron Affinity (eV) | −0.740 | (−0.9/ 1.7) |
| Polarizability (Angstroms^3) | 21.554 M | (13.0 / 70.0) |
| QP log P for hexadecane/gas | 8.503 M | (4.0 / 18.0) |
| QP log P for octanol/gas | 20.896 M | (8.0 / 35.0) |
| QP log P for water/gas | 25.811 M | (4.0 / 45.0) |
| QP log P for octanol/water | −4.023 | (−2.0 / 6.5) |
| QP log S for aqueous solubility | 2.000 | (−6.5 / 0.5) |
| QP log S - conformation independent | 1.220 | (−6.5 / 0.5) |
| QP log K hsa Serum Protein Binding | −2.059 | (−1.5 / 1.5) |
| QP log BB for brain/blood | −2.516 | (−3.0 / 1.2) |
| No. of Primary Metabolites | 6 | (1.0 / 8.0) |
| Predicted CNS Activity (−- to ++) | – | |
| HERG K+ Channel Blockage: log IC50 | 1.823 | (concern below −5) |
| Apparent Caco-2 Permeability (nm/s) | 0 | (<25 poor. >500 great) |
| Apparent MDCK Permeability (nm/s) | 3 | (<25 poor. >500 great) |
| QP log Kp for skin permeability | −6.820 | (Kp in cm/h) |
| Jm. max transdermal transport rate | 3.665 | (micrograms/cm^2-h) |
| Lipinski Rule of 5 Violations | 1 | (maximum is 4) |
| Jorgensen Rule of 3 Violations | 1 | (maximum is 3) |
| % Human Oral Absorption in GI (+ − 20%) | 0 | (<25% is poor) |
According to these rules, the drug candidate molecule must have 5 hydrogen bond donors (total nitrogen-hydrogen and oxygen-hydrogen bond). The sum of the NH and OH bonds gives the number of hydrogen bond donors and the sum of the N and O atoms in the structure gives the number of hydrogen bond acceptors. In this molecule; there are four oxygen atoms which formed at least one hydrogen bond with THR14, LYS271, SER340 and GLY339, two NH groups and two NH2 active groups which formed one hydrogen bond with ASP366 that make hydrogen bonds with the protein and shown in Fig. 3c. Also calculated value for solute as donor-hydrogen bonds was 4.250 (recommended from 0 to 6). In addition, up to 10 hydrogen bond receptors (all nitrogen or oxygen atoms) must be present in the molecule and the measured value of the molecule according to ADME is (8.250) again among the recommended values (from 2.0/ to 20.0). In addition to these rules, the polar surface area (PSA) and the number of routable bonds is two of the defining characteristics of the active drug determination and selection process. The active drug should have 10 or less routable bonds and the polar surface area should not be greater than 140 Ǻ2 [48]. For AVP (4–5)-NH2 peptide, the number of rotatable bonds is 8 and the polar surface area is measured as 180,528 Ǻ2 (Table 2). BBB permits diffusion of hydrophobic molecules and small polar molecules, while restricts the dissolution of bacteria and large or hydrophilic molecules in the blood into the cerebrospinal fluid (CSF) [49]. Neuroprotective drugs targeting the central nervous system must first cross BBB [50]. The corresponding value calculated by ADME is −2.516 recommended value range (−3.0 / 1.2). Additionally, solute electron affinity, solute ionization potential (IP (eV)) were also taken from the result of Qikprop analysis as −0.740 eV (standard limits from −0.9 to 1.7), 9.382 eV (standard limits from 7.9 to 10.5), respectively. It is presented in some oral prodrugs such as dirithromycin, tacrolimus, saquinavir, ceftidoren, efonidipine which do not have the expected molecular weight and PSA values [51]. In this study, AVP (4–5)-NH2 peptide having some drug-like properties was showed that have acceptable for molecular weight and BBB value for drug candidate.
FT-IR analysis results
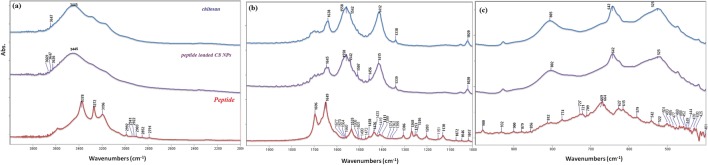
The typical absorption bands of CS are given in Table 3. The broad absorption peak at 3445 cm−1 and the prominent peaks at 1638 and 1558 cm−1 correspond to the O-H stretching and the C=O stretching for amide I and N-H bending for amide II vibrations, respectively and are marked in Fig. 4. The bands at 1412 and 1338 cm−1 were assigned to C-C-H bending. The C-N and C-O stretching vibrations were also seen at 1319 cm−1 and 1052; 1020 cm−1 for CS. The bands which are originated from TPP (P=O) (O-P-O) were also observed at 1126 and 929 cm−1. The observed and assigned peaks for the polymer are compatible with the literature [38, 52–57].
Table 3.
The FT-IR spectrum of Chitosan polymer, AVP(4–5)-NH2 peptide and AVP(4–5)-NH2 peptide-loaded CS NPs
| Chitosan (CS) | AVP(4–5)NH2 | NPs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FTIR | ATR | FTIR | FTIR | AVP(4–5)NH2 CS NPs | ||||||||
| References | This Study | |||||||||||
| Assignment of Chitosan | [51] | [52] | [53] | [54] | [55] | [56] | [38] | This Study | Assignment of PgluAsn NH2 | νexp. | νcal. | νexp. |
| O-H stretching | 3500–3300 | 3367 | 3800–3000 |
3290; 3285 |
3750–3000 | 3500–3200 | 3445; 3293 | O-H stretching | – | – | 3445; 3292 | |
| NH stretching | – | NH stretching | 3378;3272 | 3372;3320 | ||||||||
| C-H stretching | – | 2927 | 2867 | 2867 | 2920;2875 | 2865 | – | C-H stretching | 3196;2992 | 3001;2991 | 3180 | |
| CO stretching; NH2 bending | 1650;1600 | 1637;1559 | 1656;1554 | 1568 | 1645;1574 | 1650;1542 | 1635; 1530 | 1638; 15581542 | CO stretching (amide-I) | 1697* | 1680 | 1696* |
| H-N-H bending | 1648* | 1591 | 1646* | |||||||||
| C-N-H bending(amide-II) | 1534** | 1524 | 1534** | |||||||||
| C-C-Hbending | 1426;1375 | 1412; 1338 | H-C-H bending | 1458;1436 | 1472;1438 | 1474;1456 | ||||||
| C-N stretching+ νs(-CH3) | 1317 | 1322 | – | 1313 | – | – | 1319 | C-C-H bending | 1306;1268 1246; 1203 1130 | 1312;1263 s1247;1199 1139 |
1339;1320; 1124 |
|
| C-O stretching; O-C-H bending | 1261 | – | C-CPGLUstretching; | 980;900 | 994;900 | -;902 | ||||||
| COC asym. Stretching | 1150 | 1154 | – | 1152;1148 | 1150; | – | 1153 | – | C-CASNstretching; | 932;879 | 923;870 | 928 |
| C-O-C;C-O-H (CO stretching) | – | 1081 | 1107 | 1081; 1060 | 1080;1065;1025 | - 1027 | 1083; 1031 | - 1052; 1020 | C-C-N-H torsion | 669;615; 501 | 691;611; 495 | 642;626 519;473 |
| Saccharide structure | – | 897 | – | 892 | 890 | – | – | |||||
| TPP (P=O) (O-P-O) | 1170 | – | – | 1126;875 | - 912 | 1126; 929 | ||||||
Fig. 4.
The FT-IR spectrum of Chitosan polymer, AVP(4–5)-NH2 peptide and AVP(4–5)-NH2 peptide-loaded CS NPs in the range of 4000 cm−1 -2000 cm−1(a), 2000 cm−1 -1000 cm−1(b), 1000 cm−1 -400 cm−1 (c)
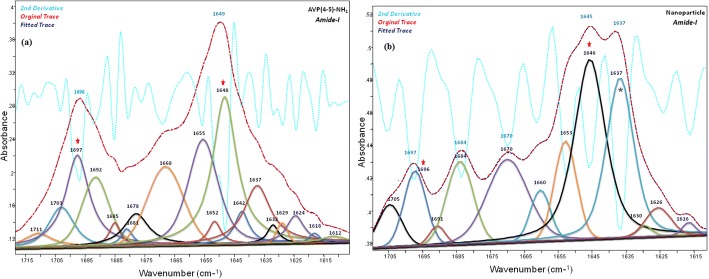
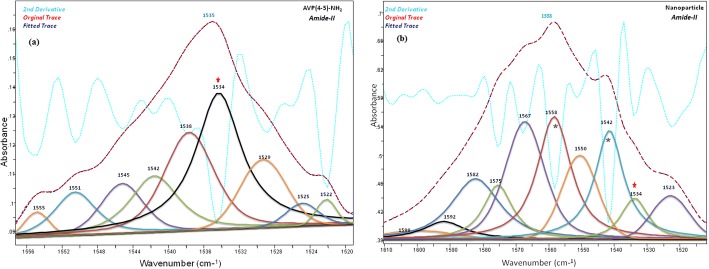
In present study, the characteristic wavenumbers of the AVP (4–5)-NH2 peptide, observed in the experimental vibrational (IR and Raman) spectra are given in Table 4, together with the fundamental vibrational wavenumbers, calculated at the level of DFT-RB3LYP / 6–31++G(d,p) basis set. The assignment of the Vibrational wavenumber was performed in accordance with the total energy distribution of the vibrational modes (TED), that was calculated using the scaled quantum mechanical force field (SQM FF) method [58], with the aid of scaled vibrational wavenumbers using dual scaling factors [59]. The major IR peaks at 3378 cm−1; 3272 cm−1 and 3196 cm−1; 2992 cm−1 indicate N-H and C-H stretching in FT-IR spectrum, respectively. The primary amide vibrations were appointed to 1696 cm−1(IR) and 1535 cm−1(IR) which characterized to amide-I and amide-II for AVP (4–5)-NH2 peptide. In the present study, the C=O stretching in peptide group (amide-I) which was calculated and assigned by TED contributions (77% C=O stretching, 5% C-N stretching, 3% C-N-H bending) and observed in the 1715–1610 cm−1 and 1705–1610 cm−1 intervals for AVP(4–5)-NH2 peptide and nanoparticle spectra, respectively.
Table 4.
Experimental (IR, Raman) and calculated wavenumbers (cm−1), and the TED of the vibrational modes of the AVP(4–5)NH2
| Assign. | IR | Raman | DFT-RB3LYP 6-31G++(d,p) | DFT-RB3LYP 6-31G(d,p) | DFT-RB3LYP 6-31G(d) | †TED-MONOMER DFT-RB3LYP 6-31G++(d,p) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| υexp | υexp | υcal | *υscaled | IRint. | υcal | *υscaled | IR int. | υcal | *υscaled | IR int. | |||
| 1 | υNasymm. | 3736 | 3568 | 45 | 3745 | 3576 | 40 | 3719 | 3552 | 36 | υNH (100) | ||
| 2 | υNHasymm. | 3682 | 3516 | 103 | 3685 | 3519 | 89 | 3662 | 3497 | 80 | υNH (100) | ||
| 3 | υNHpglu | 3615 | 3453 | 30 | 3627 | 3464 | 28 | 3606 | 3444 | 25 | υNH (99) | ||
| 4 | υNHsymm. | 3595 | 3433 | 55 | 3603 | 3441 | 53 | 3592 | 3430 | 51 | υNH (99) | ||
| 5 | υNH peptide | 3378 s | 3374 s | 3531 | 3372 | 114 | 3510 | 3352 | 144 | 3502 | 3345 | 134 | υNH (99) |
| 6 | υNH symm. | 3272 m | 3271 s | 3477 | 3320 | 181 | 3470 | 3313 | 188 | 3469 | 3312 | 179 | υNH (99) |
| 7 | υCH asymm-pgl | 3196 m | 3168 s | 3143 | 3001 | 14 | 3146 | 3004 | 15 | 3148 | 3006 | 17 | υCH (99) |
| 8 | υCH asymm-asn | 2992vw | 2996vs | 3132 | 2991 | 9 | 3135 | 2994 | 5 | 3136 | 2995 | 6 | υCH (100) |
| 9 | υCH asymm-pgl | 2978vs | 3130 | 2989 | 5 | 3131 | 2990 | 11 | 3132 | 2991 | 12 | υCH (97) | |
| 10 | υCH symm-asn | 3082 | 2944 | 4 | 3083 | 2944 | 5 | 3088 | 2949 | 5 | υCH (97) | ||
| 11 | υCH symm-pgl | 3079 | 2941 | 12 | 3081 | 2942 | 11 | 3086 | 2947 | 12 | υCH (97) | ||
| 12 | υCH symm-pgl | 2933vs | 3063 | 2925 | 12 | 3064 | 2926 | 13 | 3066 | 2928 | 14 | υCH (99) | |
| 13 | υCH-pglu cα | 3042 | 2906 | 24 | 3039 | 2903 | 30 | 3045 | 2908 | 22 | υCH (100) | ||
| 14 | υCH-asn cα | 3036 | 2900 | 10 | 3037 | 2901 | 12 | 3044 | 2907 | 21 | υCH (98) | ||
| 15 | υCOpglu | 1806 | 1724 | 531 | 1845 | 1762 | 433 | 1846 | 1763 | 433 | υOC(83) + δCCN(3) | ||
| 16 | υCO | 1766 | 1725 | 390 | 1803 | 1722 | 315 | 1806 | 1724 | 312 | υNC (5) + υOC(78) | ||
| 17 | υCO-asn | 1745 | 1704 | 415 | 1769 | 1710 | 334 | 1772 | 1713 | 336 | υNC (7) + υOC(80) | ||
| 18 | υCOpeptide (amide-I) | 1696s | 1687 m | 1720 | 1680 | 191 | 1748 | 1690 | 163 | 1751 | 1693 | 158 | υNC (5) + υOC(77) + δ CNH (3) |
| 19 | δHNHscis-asn. | 1649vs | 1604 m | 1629 | 1591 | 58 | 1634 | 1580 | 180 | 1653 | 1598 | 171 | υNC (5) + δHNH(57) + δCNH(28) |
| 20 | δHNHscis. | 1626 | 1589 | 254 | 1629 | 1575 | 138 | 1648 | 1594 | 136 | υNC (3) + δHNH(57) + δCNH(27) | ||
| 21 | δCNH peptide (amide-II) | 1535s | 1560 | 1524 | 344 | 1566 | 1515 | 373 | 1574 | 1522 | 364 | υCC (4) + υCN(30) + δOCN(3) + δCNH(51) | |
| 22 | δHCH scis-pglu | 1458vw | 1507 | 1472 | 6 | 1513 | 1463 | 5 | 1528 | 1477 | 5 | δHCH(27) + ΓHCCC(20) + ΓHCCH(28) + ΓNCCH(10) | |
| 23 | δHCH scis-asn | 1441 m | 1474 | 1440 | 22 | 1480 | 1431 | 2 | 1496 | 1447 | 2 | δCCH(8) + δHCH(31) + ΓHCCN(14) + ΓHCCC(8) + ΓHCCH(12) + ΓHCCO(14) | |
| 24 | δHCH scis-pglu | 1436v | 1472 | 1438 | 1 | 1476 | 1427 | 22 | 1493 | 1443 | 21 | δCCH(15) + δHCH(30) + ΓHCCN(18) + ΓHCCO(18) + ΓHCCH(3) | |
| 25 | δHCC-pglu | 1423 | 1390 | 43 | 1425 | 1378 | 73 | 1432 | 1385 | 67 | υCC(15) + υCN(9) + δHCC(18) + ΓHCCN(4) + ΓHCCC(3) + ΓHCC(4) | ||
| 26 | δCNH asn | 1411v | 1417 | 1384 | 58 | 1424 | 1377 | 40 | 1432 | 1385 | 37 | υNC (16) + υOC(4) + δCNH (42) + ΓHCNH(4) | |
| 27 | υCN | 1391 | 1359 | 136 | 1398 | 1351 | 107 | 1404 | 1357 | 118 | υCC (11) + υCN(21) + δCNH (15) + δCCH (5) + δ OCC (3) + δ OCN (4) + δ HNH (4) + ΓHCCC(4) + ΓHCCH(10) | ||
| 28 | δCCH pglu | 1340w | 1374 | 1343 | 15 | 1376 | 1330 | 13 | 1384 | 1338 | 11 | υNC (5) + δCCH(19) + ΓHCCC(11) + ΓNCCH(5) + ΓHCCH(12) | |
| 29 | δCNH | 1369 | 1337 | 6 | 1372 | 1327 | 7 | 1381 | 1335 | 6 | υNC (10) + δCNH (11) + δCCH(6) + ΓHCCH(5) + ΓCCCH(4) + ΓOCCH(7) + ΓNCCH(9) | ||
| 30 | δCCHpglu | 1306vw | 1343 | 1312 | 17 | 1351 | 1307 | 19 | 1360 | 1315 | 20 | υNC (3) + δCNH (5) + δCCH(21) + ΓCCCH(9) + ΓHCCH(26) + ΓOCCH(4) + ΓNCCH(4) | |
| 31 | δCCHpglu | 1326 | 1296 | 50 | 1328 | 1284 | 27 | 1337 | 1293 | 8 | υNC (6) + υCC (8) + δCCH(22) + ΓHCCH(11) + ΓHCNH(6) + ΓHCNC(7) + ΓHCCC(4) | ||
| 32 | υNC+δCCHasn | 1325 | 1294 | 40 | 1326 | 1283 | 63 | 1335 | 1291 | 81 | υNC (24) + δCCH(17) + δOCN(3) + δHCN(8) + ΓHCNH(3) + ΓHCNC(4) + ΓHCCH(6) | ||
| 33 | δCNHpeptide | 1282w | 1314 | 1283 | 21 | 1319 | 1275 | 16 | 1325 | 1282 | 17 | υNC (5) + δHCN(3) + δCCH(25) + ΓHCCC(4) + ΓNCCH(4) + ΓHCCH(3) | |
| 34 | δCCHwagg-pgl | 1268 m | 1253w | 1293 | 1263 | 19 | 1296 | 1253 | 21 | 1305 | 1262 | 24 | υCC (8) + υCN(13) + δCNH(6) + δCCH(25) |
| 35 | δCCHwagg.-asn | 1246vw | 1276 | 1247 | 68 | 1279 | 1237 | 66 | 1288 | 1246 | 67 | υCC (3) + υCN(11) + δCNH(7) + δCCH(30) + ΓHCCH(4) + ΓHCCN(3) + ΓOCCH(3) | |
| 36 | υCNpglu | 1262 | 1233 | 129 | 1262 | 1220 | 105 | 1269 | 1227 | 114 | υCC (6) + υCN(23) + δCNH (4) + δ OCN (4) + δCCH (18) + δ OCC (5) + ΓHCNH(3) + ΓOCCH(4) + ΓNCCH(3) | ||
| 37 | δCCHtwist.-pgl | 1203v | 1200w | 1227 | 1199 | 15 | 1227 | 1186 | 19 | 1234 | 1193 | 21 | υCC (8) + δCNH (4) + δCCH (33) + ΓNCCH(9) + ΓOCCH(3) + ΓHCCH(5) + ΓCCCH(5) |
| 38 | δCCH twist-asn | 1216 | 1188 | 18 | 1218 | 1178 | 13 | 1228 | 1187 | 15 | δCNH(7) + δCCH(38) + ΓNCCH(5) + ΓHCNH(3) + ΓHCCH(4) + ΓOCCH(4) | ||
| 39 | δCCH twist-pgl | 1130v | 1137vs | 1166 | 1139 | 27 | 1168 | 1130 | 29 | 1176 | 1137 | 32 | υCC (5) + δCCH(39) + ΓNCCH(3) + ΓCCCH(16) |
| 40 | υNC+δCNH | 1144 | 1117 | 2 | 1155 | 1117 | 2 | 1162 | 1124 | 2 | υNC(25) + υCC(20) + υOC(5) + δCNH(15) + δNCC(3) + δCCH (3) | ||
| 41 | δCNH+υNC | 1127 | 1101 | 3 | 1134 | 1097 | 1 | 1139 | 1101 | 1 | υNC (29) + υCC (10) + υOC (7) + δCNH(41) | ||
| 42 | υNCpglu | 1111 | 1085 | 12 | 1114 | 1077 | 12 | 1118 | 1081 | 10 | υNC (40) + υCC (13) + δCNC(4) + δCNH(8) | ||
| 43 | δCNH | 1072v | 1106 | 1081 | 6 | 1110 | 1073 | 4 | 1115 | 1078 | 6 | υNC (25) + υCC (8) + δCNC(3) + δCNH(39) | |
| 44 | υNC+δCNH | 1046v | 1057 | 1033 | 7 | 1062 | 1027 | 7 | 1069 | 1034 | 9 | υNC (14) + υCC (13) + δCNC(3) + δCNH(12) + δCCH(10) | |
| 45 | ρ(CH2)pglu | 1017v | 1014 m | 1047 | 1023 | 6 | 1052 | 1017 | 6 | 1058 | 1024 | 6 |
υNC(4) + δCCH(10) + ΓCCCH(3) + ΓNCCC(4) + ΓHCCH(27)+ ΓCCCC(6) + ΓCCCH(3) |
| 46 | υCCpglu | 980 m | 987w | 1017 | 994 | 5 | 1018 | 984 | 6 | 1020 | 987 | 6 | υNC (10) + υCC (48) |
| 47 | υCCpglu | 989 | 966 | 1 | 992 | 959 | 1 | 995 | 962 | 1 | υNC (4) + υCC (59) | ||
| 48 | υCCasn. | 932v | 947 m | 944 | 923 | 4 | 945 | 914 | 7 | 949 | 918 | 7 | υCC (26) |
| 49 | υCCring –asiym | 900v | 903w | 922 | 900 | 8 | 924 | 894 | 6 | 927 | 897 | 6 | υNC (16) + υCC (21) + δCCH(8) + δCCC(6) |
| 50 | υCCasn | 879v | 881 m | 890 | 870 | 4 | 892 | 863 | 4 | 895 | 866 | 4 | υNC (8) + υCC (8) + δCCH(5) + δCCC(3) + ΓOCCC(5) + ΓNCCH(12) |
| 51 |
υCC ring breathing |
856vw | 883 | 863 | 2 | 885 | 856 | 1 | 889 | 859 | 2 | υNC (6) + υCC (26) + δOCN(4) + ΓOCCH(4) | |
| 52 | υCC-asn | 861 | 842 | 0 | 861 | 832 | 1 | 863 | 835 | 1 | υNC (9) + υCC (55) + δCNH(9) | ||
| 53 | υCC-ring | 812vw | 826w | 833 | 814 | 10 | 833 | 806 | 11 | 836 | 809 | 12 | υCC (27) + δCCH(3) + ΓOCCH(13) + ΓNCCH(7) |
| 54 | ΓHNCO | 774v | 785 | 767 | 4 | 791 | 765 | 5 | 792 | 766 | 6 | υCC (11) + δOCN(8) + ΓHNCO(24) | |
| 55 | ΓHNCO | 760 | 742 | 5 | 761 | 736 | 8 | 762 | 737 | 7 | δCNH(4) + δCNC(4) + δOCC(7) + δOCN(3) + ΓNCCC(3) + ΓHNCO(15) | ||
| 56 | υCC | 727v | 727 | 710 | 2 | 733 | 708 | 3 | 734 | 710 | 4 |
υCC (12) + δCCN(4) + ΓOCCC(7) + ΓOCCH(3) + ΓNCCN(6)+ ΓNCCH(4) + ΓHNCO(7) + ΓONCO(9) |
|
| 57 | ΓNCCH | 709v | 718 | 701 | 13 | 719 | 696 | 21 | 721 | 697 | 23 | δCCN(4) + δOCN(5) + ΓNCCH(24) + ΓHNCO(21) | |
| 58 | ΓCCNH | 669v | 682 m | 707 | 691 | 19 | 712 | 688 | 14 | 713 | 690 | 16 | υCC (3) + υCN (5) + δOCC(4) + δOCN(5) + ΓNCCH(23) + ΓHNCO(11) |
| 59 | ΓHNCO | 664v | 671 | 656 | 28 | 679 | 656 | 46 | 682 | 659 | 55 | δOCC(4) + δCCN(10) + ΓOCCH(8) + ΓNCCH(4) + ΓHNCO(39) | |
| 60 | ΓOCNH | 627v | 643 | 629 | 50 | 657 | 635 | 48 | 659 | 637 | 44 |
υCN (5) + δOCC(14) + δCCC(4) + δOCN(16) + ΓOCCH(3)+ ΓOCNH(13) + ΓNCCH(3) |
|
| 61 | ΓCCNH | 615vw | 625 | 611 | 10 | 631 | 610 | 6 | 633 | 612 | 5 | δOCN(9) + δOCC(4) + ΓOCNH(14) + ΓHCNH(5) + ΓCCNH(27) | |
| 62 | ΓHNCC | 579vw | 611 | 597 | 19 | 619 | 599 | 38 | 621 | 600 | 42 | υCC (4) + δOCC(8) + δCCC(8) + δCCN(4) + ΓHCCH(4) + ΓHNCC(6) + ΓHNCO(5) | |
| 63 | δOCC | 572 | 559 | 55 | 581 | 562 | 51 | 583 | 564 | 56 | υCC (6) + δOCC(24) + δOCN(10) + ΓHNCC(8) + ΓHNCO(4) | ||
| 64 | ΓHNCC | 542v | 567 | 554 | 48 | 568 | 549 | 52 | 570 | 551 | 48 |
δCCN(6) + δOCC(4) + δCCC(4) + ΓHNCO(12) + ΓHNCC(20)+ ΓCCCH(4) |
|
| 65 | δCCN | 522vw | 546w | 552 | 540 | 29 | 555 | 536 | 33 | 555 | 537 | 35 | δCCN(28) + δCNH(3) + δOCN(33) |
| 66 | ΓCCNH | 501vw | 498w | 507 | 495 | 71 | 509 | 493 | 51 | 514 | 497 | 115 | ΓHNCC(48) + ΓCCCO(3) + ΓHCNH(4) + ΓHNCO(10) |
| 67 | ΓCCNH | 487 | 475 | 69 | 505 | 488 | 265 | 509 | 492 | 212 | ΓOCCH(15) + ΓNCCC(5) + ΓCCNH(52) + ΓHNCO(3) + ΓHCCC(3) | ||
| 68 | δOCC | 484 | 473 | 161 | 488 | 472 | 9 | 489 | 473 | 9 | υCC (9) + δOCC(25) + δOCN(16) + δCNC(5) + ΓCCNH(3) + ΓOCCN(4) | ||
| 69 | ΓHNCO | 481 | 470 | 22 | 481 | 465 | 7 | 481 | 465 | 7 | ΓCCNH(44) + ΓHNCO(43) | ||
| 70 | δOCC | 421 m | 409 | 400 | 6 | 411 | 398 | 8 | 412 | 399 | 8 | υCC (15) + υCN (12) + δOCN(10) + δOCC(25) + δNCC(7) | |
| 71 | ΓHNCC | 388 | 379 | 0 | 386 | 374 | 0 | 388 | 375 | 1 | υCC (10) + δNCC(16) + δOCN(4) + ΓCCNC(14) + ΓHCCN(24) | ||
| 72 | δNCC | 370 | 362 | 3 | 369 | 357 | 3 | 369 | 356 | 3 | υCC (10) + δOCC(14) + δOCN(9) + δNCC(56) | ||
| 73 | δNCC | 299w | 308 | 301 | 18 | 308 | 298 | 11 | 307 | 297 | 11 | υCN (4) + δOCN(5) + δOCC(4) + δNCC(33) + ΓHCCH(3) + ΓOCCC(5) + ΓNCCC(4) | |
| 74 | ΓCCNH | 294 | 287 | 170 | 272 | 263 | 125 | 269 | 260 | 10 | υCC (4) + δNCC(5) + δOCC(3) + ΓCCNH(29) + ΓHNCO(28) | ||
| 75 | ΓCCNH | 258w | 264 | 258 | 1 | 267 | 259 | 63 | 259 | 250 | 188 | υCC (9) + δNCC(6) + δOCC(5) + δCCC(3) + ΓCCNH(25) + ΓHNCO(21) | |
| 76 | δCCC | 246 | 240 | 19 | 248 | 239 | 19 | 247 | 239 | 23 |
δNCC(17) + δOCC(6) + δCCC(19) + ΓCCNH(8) + ΓCNCO(4)+ ΓCNCC(9) |
||
| 77 | δCCC | 205 | 201 | 25 | 212 | 205 | 26 | 213 | 206 | 26 |
δCCC(15) + δNCC(9) + ΓCNCC(7) + ΓCCNH(12) + ΓOCCN(6)+ ΓOCCC(3) |
||
| 78 | ΓCNCC | 176 | 172 | 5 | 180 | 174 | 4 | 181 | 175 | 3 | δCCC(19) + δNCC(7) + ΓCNCC(23) + ΓNCCN(3) + ΓCCNH(8) | ||
| 79 | δNCC | 154 | 150 | 8 | 154 | 149 | 5 | 153 | 148 | 5 |
δNCC(26) + δOCN(4) + δCCC(4) + ΓOCCH(9) + ΓCCCO(8)+ ΓNCCC(16) + ΓOCNC(7) |
||
| 80 | ΓHCCN | 134 | 130 | 2 | 143 | 139 | 4 | 144 | 139 | 4 |
δNCC(8) + ΓCNCC(14) + ΓHCCN(21) + ΓOCCC(9) + ΓOCCC(9)+ ΓCCCH(18) + ΓCCCC(7) |
||
| 81 | ΓNCCC | 105 | 103 | 3 | 111 | 107 | 3 | 111 | 108 | 3 |
δCCC(17) + ΓOCCN(5) + ΓOCCC(6) + ΓOCCH(4) + ΓNCCN(5)+ ΓNCCC(22) + ΓNCCH(13) + ΓHCCH(4) + ΓHCCC(3) |
||
| 82 | ΓHCCC | 72 | 70 | 10 | 71 | 69 | 6 | 72 | 69 | 7 | ΓNCCC(22) + ΓHCCC(34) + ΓCCCC(22) + ΓNCCN(3) | ||
| 83 | ΓHCCC | 69 | 67 | 0 | 70 | 67 | 1 | 70 | 68 | 0 | δNCC(18) + ΓOCCN(4) + ΓNCCC(15) + ΓHCCC(27) + ΓCCCC(16) + ΓNCCH(5) + ΓNCCN(4) | ||
| 84 | ΓNCCN | 49 | 48 | 1 | 51 | 49 | 2 | 52 | 50 | 1 |
ΓNCCN(18) + ΓNCCC(12) + ΓOCCN(16) + ΓOCCH(12)+ ΓOCCC(10) + ΓNCCH(11) |
||
| 85 | ΓNCCC | 41 | 40 | 4 | 48 | 47 | 6 | 50 | 48 | 6 |
δNCC(4) + ΓCCCC(4) + ΓCCCH(6) + ΓOCCC(4) + ΓNCCN(12)+ ΓNCCC(40) + ΓNCCH(20) + ΓOCCN(7) |
||
| 86 | ΓNCCC | 40 | 39 | 7 | 42 | 41 | 1 | 43 | 41 | 2 | ΓNCCC(34) + ΓOCCN(19) + ΓOCCC(8) + ΓHCNH(10) + ΓCCNH(10) | ||
| 87 | ΓNCCC | 32 | 32 | 1 | 32 | 31 | 2 | 32 | 31 | 2 | ΓOCCC(26) + ΓOCCH(20) + ΓNCCC(29) + ΓNCCH(26) | ||
The band component analyses of the amide-I and amide II regions of the IR spectra of the peptide and peptide loaded CS NPs were shown in Figs. 5 and 6, respectively. The C=O, amide-I and -II bands of the AVP(4–5)-NH2 peptide were clearly shown in the IR spectrum of CS NPs, indicating the presence of the peptide in CS NPs. Due to the encapsulation of the AVP (4–5)-NH2 peptide in CS NPs, slight wavelength and intensity changes occurred in both CS and peptide bands. Alterations of the amide I and II bands of the peptide can also be resulted due to the conformational alteration of the peptide after encapsulation. The peak observed at 1535 cm−1, in the experimental FT-IR spectrum (see Fig. 6), was calculated at 1524 cm−1 and was assigned to the amide-II band of the peptide group by the help of TED analysis results (δCNH(51) + υCC (4) + υCN(30) + δOCN(3)). The main H-N-H scissor vibration for ASN residue was assigned to 1649 cm−1 and the H-C-H scissors vibrations for PGLU and ASN moiety were assigned to 1458 cm−1 and 1436 cm−1 respectively, by the aid of TED (see Table 4).
Fig. 5.
The band component analysis of 1715–1610 cm−1region of the Infrared spectrum of AVP (4-5)-NH2 peptide (a) and AVP (4-5)-NH2 peptide-loaded CS NPs (b). Chitosan bands are marked (*)
Fig. 6.
The band component analysis of 1556–1520 cm−1region of the Infrared spectrum of AVP (4-5)-NH2 peptide (a) and AVP (4-5)-NH2 peptide-loaded CS NPs (b). Chitosan bands are marked (*)
The wagging and twisting vibrations for PGLU moiety were observed at 1268 cm−1 and 1203 cm−1; 1130 cm−1, whereas the wagging vibration for ASN moiety was observed at 1246 cm−1. The band at 1306 cm−1 was denominated to C-C-H bending for PGLU. The peaks at 980 cm−1, and 900 cm−1 corresponded to C-C stretching of PGLU, while the peaks at 932 cm−1and 879 cm−1 complied with C-C stretching of ASN moiety. The C-C-N-H torsion vibrations were also identified at 669, 615 and 501 cm−1.
DLS results
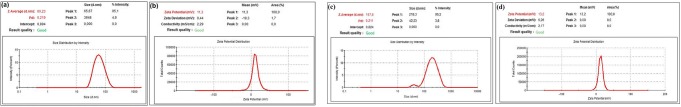
Among the different nanomaterial characterization techniques, DLS is the one of crucial and useful method to evaluate particle size, size distribution, and the zeta potential of nanomaterials in solution [60]. In this way, we can understand how to act of our synthetic drug transport agent in the liquid and learn about the hydrodynamic size. In Fig. 7a, b, the average hydrodynamic size, zeta potential and polydispersity index (PdI) values of blank CS NPs are given as 60.23 nm, +11.3 mV and 0.219, respectively. It was found that the AVP (4–5)-NH2 peptide-loaded CS NPs have a narrow size distribution with 0.211 and 167.6 nm average particle size as given in Fig. 7c. On the other hand, As seen from the Fig. 7d, the zeta potential value of AVP (4–5)-NH2 peptide-loaded CS NPs are +13.2 mV. The results indicated that both blank CS NPs and AVP (4–5)-NH2 peptide-loaded CS NPs are stable in the liquid because they are more resistant to aggregation due to their high zeta potential.
Fig. 7.
Dynamic light scattering results. Size (a) and Zeta-potential (b) graphics of blank CS NPs and Size (c) and Zeta-potential (d) graphics of AVP (4–5)-NH2 peptide-loaded CS NPs
These results coincide with other previous studies. One of these studies, optimization and evaluation of doxorubicin (DOX) loaded CS NPs were investigated [61]. The result of this study, zeta potential of CS NPs was found as +8.35 mV and it was reported that there was not any agglomeration in solution of CS NPs. On the other hand, in our study, zeta potential values of blank CS NPs and AVP (4–5)-NH2 peptide-loaded CS NPs were nearly 1.3 or 1.5 fold higher than +8.35 mV, respectively and therefore, we can say that both NPs have higher stability in the liquid medium. The effects of chitosan concentration, TPP concentration and peptide concentration to particle size, polydispersity index and zeta potential values were also investigated and tabulated in Table S1, Table S2 and Table S3, respectively. According to the conditions tested, there is a relationship between increased particle size and increased concentrations of chitosan and TPP, as well as the pH of the solution is effective on the stability of the nanoparticle system [62, 63].
Encapsulation efficiency and loading capacity results
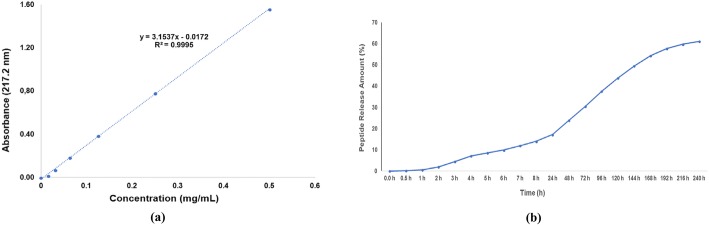
The encapsulation and loading efficiency are the two crucial indices for the evaluation of nanocarrier systems. The encapsulation value indicated that the AVP (4–5)-NH2 peptide was entrapped and AVP (4–5)-NH2 peptide-loaded CS NPs were obtained successfully. The calibration curve (Fig. 8a) of the AVP (4–5)-NH2 peptide was prepared to calculate the encapsulation efficiency of the AVP (4–5)-NH2 peptide-loaded CS NPs. The encapsulation efficiency of the AVP (4–5)-NH2 peptide was calculated via Eq. 1, and it was found as 99%. This result indicated that, almost all peptide was encapsulated in CS NPs. A previous study which was used of CS NPs and the other protein structures showed that molecular weights of protein structures is a key parameter for a higher encapsulation efficiency [64], and it is known that encapsulation efficiency can be increased with decreased of molecular weight. For this reason, our encapsulation efficiency result was found as very higher because of AVP (4–5)-NH2 has a small molecular weight. On the other hand, loading capacity of the AVP (4–5)-NH2 peptide-loaded CS NPs was calculated as 10%, from Eq. 2. This means that 10% of CS NPs’ weight consists of the AVP (4–5)-NH2 peptide and each 1 mg roughly contains 0.10 mg of the peptide.
Fig. 8.
Standard curve of AVP(4–5)-NH2 peptide (a) and in vitro release profil of AVP(4–5)-NH2 peptide-loaded chitosan NPs (b)
In vitro release profile
Figure 8b shows the in vitro release profile of AVP (4–5)-NH2 peptide-loaded CS NPs. Because of the degradation of the CS polymer, the slow diffusion of AVP (4–5)-NH2 peptide from the polymer matrix was obtained. It is also known that CS NPs have the advantage of slow and controlled drug release, which improves drug solubility and stability, enhances efficacy, and reduces toxicity [65]. The results showed that, in first 24 h 17.23% of the peptide was released and 61.13% of AVP (4–5)-NH2 peptide was also released at the end of the 240th hour. The in vitro release of catechin from CS NPs was found at 32% within 24 h [66]. According to the results of [66], the entrapment efficiency of the particles was calculated 60% and the in vitro release was found to be 32% over 24 h. However, in our study, the encapsulation efficiency of the AVP (4–5)-NH2 peptide was found as 99% and the release rate was found only 17.23% in the first 24 h and we reached about 60% release rate in 10 days.
The stability and integrity of AVP (4–5)-NH2 peptide-loaded CS NPs were checked by time of in vitro release study, and the dynamic light scattering and scanning electron microscope analysis were performed after 12nd and 24th hours of the in vitro release study. The DLS results were given in Fig. S6, and it was understood that the average particle size of the AVP (4–5)-NH2 peptide-loaded CS NPs increased significantly, and its zeta potential value decreased. After the 12 h in vitro release study, it was found that AVP (4–5)-NH2 peptide-loaded CS NPs had wide size distribution with 0.822, and 679.6 nm average particle size as given Fig. S6a. As seen from the Fig. S6b, the zeta potential value of AVP (4–5)-NH2 peptide-loaded CS NPs were − 4.80 mV. Scanning electron microscope was also used to investigate the stability and integrity of the AVP (4–5)-NH2 peptide-loaded CS NPs. SEM images of AVP (4–5)-NH2 peptide-loaded CS NPs after 12nd hours (during in vitro release study) were given in the Fig. S7. According to the SEM images obtained from different perspectives, it was found that the AVP (4–5)-NH2 peptide-loaded CS NPs were started to loose their spherical morphology. Besides, the DLS results for 24th hours were given in Fig. S8, and it was understood that the average particle size of the AVP (4–5)-NH2 peptide-loaded CS NPs increased, and its the zeta potential value decreased. It was found that AVP (4–5)-NH2 peptide-loaded CS NPs had wide size distribution with 0.281, and 751 nm average particle size as given Fig. S8a. As seen from the Fig. S8b, the zeta potential value of AVP (4–5)-NH2 peptide-loaded CS NPs were − 8.65 mV. SEM images of AVP (4–5)-NH2 peptide-loaded CS NPs after 24th hours (during in vitro release study) were given in the Fig. S9. Similarly, according to the SEM images, it was found that the AVP (4–5)-NH2 peptide-loaded CS NPs lost more their spherical morphology. When the 12nd and 24th hour results of in vitro release study were compared, it was seen that PdI value decreased at the end of 24 h. However, in the end of the first 12nd hours PdI value was larger and the particle sizes were in two different distributions as approximately 100 nm and 679 nm. The decrease of the PdI value can be explained by increases size of the chitosan nanoparticles in the in vitro release medium and reaching 751 nm size of most of the nanoparticles after 24 h. Besides, PdI value of nanoparticles was decreased, and only one peak was observed for average particle size distribution. Moreover, zeta potential values of the AVP (4–5)-NH2 peptide-loaded CS NPs were obtained negatively charged. It was thought that it was caused by negatively charged TPP with the degradation of the NPs. Because zeta potential values of nanoparticles increased more by the 24th hours of the in vitro release study. It was found that the AVP (4–5)-NH2 peptide presented release profile characterized by sustained release (Fig. 8b). In the first 12 h of in vitro release study, initial release was amounted as approximately 15% of loaded peptide. This was caused by peptide which was adsorbed or weakly bound to the surface area of the CS NPs. With the 24th hours, caused by cleavage of polymer chains, release was obtained (Fig. S9).
SEM result
SEM was used to investigate the morphology of the AVP (4–5)-NH2 peptide-loaded CS NPs. In Fig. 9, according to the SEM images, the AVP (4–5)-NH2 peptide-loaded CS NPs were spherical particles with a solid dense structure.
Fig. 9.

SEM images of AVP(4–5)-NH2 peptide-loaded CS NPs
Cytotoxicity experiments
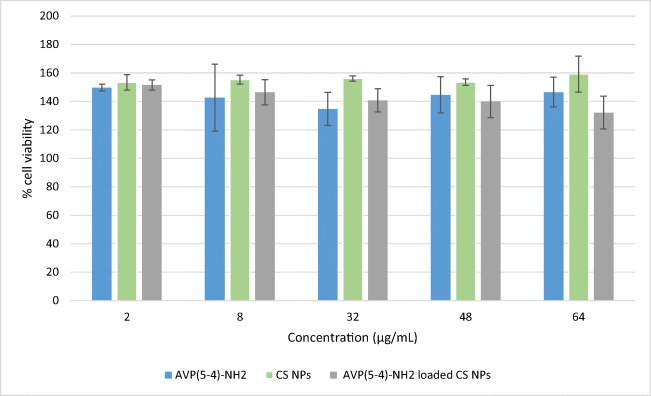
The neuroprotective effect of AVP (4–5)-NH2, CS NPs, and AVP (4–5)-NH2 peptide-loaded CS NPs was performed on SH-SY5Y cells, because SH-SY5Y cells were generally used for neuroprotective assays. XTT method were applied for assay cell viability [67, 68]. Firstly, the effect dipeptide and nanoparticle formulations on growth of SH-SY5Y cells were examined and results shown in Fig. 10. As seen in Fig. 10, none of the examined formulations has toxic effect on cells. On the contrary, the formulations increased cell viability compared to the control (p < 0.05). This showed that AVP (4–5)-NH2, Chitosan nanoparticles (with or without AVP(4–5)-NH2) have neuroproliferative effect on these cells. Because AVP (4–5)-NH2 dipeptide is a neuroprotective peptide and chitosan is a non-toxic and biocompatible polymer used for the polymeric nanoparticle structure, the obtained experimental results are consistent with the related literature results [1, 69].
Fig. 10.
Effect of AVP(4–5)-NH2, CS NPs, and AVP(4–5)-NH2 peptide-loaded CS NPs on cell viability of SH-SY5Y cells in vitro (Error bars indicate standard deviation)
Results of neuroprotective effect in vitro
In order to assay neuroprotective effect, it is necessary to treat with a damaging agent. For this purpose, cells exposed to AVP (4–5)-NH2 peptide, CS NPs, and AVP (4–5)-NH2 peptide-loaded CS NPs were co-treated with H2O2. As seen in Table 5, pretreatment with 32 μg/μL of AVP (4–5)-NH2 peptide and CS NPs (with or without AVP (4–5)-NH2), protected cell viability from undergoing H2O2−induced cell death. The evaluation of AVP (4–5)-NH2 peptide and CS NPs in the current study indicated that they all have neuroprotective effect on SH-SY5Y neuroblastoma cell line (p < 0,05). There are some studies on the neuroprotective effect of chitosan in the literature [70, 71] and our findings seem to be consistent with the literature. Besides, AVP (4–5)-NH2 is a peptide that is prominent with its neuroprotective effect in previous studies. In one of these studies, it has been shown that AVP (4–5)-NH2 peptide in the concentration of 10−5 - 10−7 M has a neuroprotective effect against the effect of H2O2 on cell cultures [1]. However, HT-22 cell line was used as the cell line in their study and the effect of AVP (4–5)-NH2 alone was investigated. Since there is no studies in literature on the encapsulation of AVP (4–5)-NH2 peptide with CS NPs, for the first time in literature, the neuroprotective efficacy of chitosan-loaded AVP (4–5)-NH2 was examined comparatively in this study, and it was shown that the combined effect of chitosan and AVP (4–5)-NH2 peptide was much higher than AVP (4–5)-NH2 peptide alone.
Table 5.
Neuroprotective effect of AVP(4–5)-NH2 peptide, CS NPs, and AVP(4–5)-NH2 peptide-loaded CS NPs on SH-SY5Y cells against H2O2 induced cell death (± sign indicates standard deviation)
| Control- H2O2 | AVP(5–4)-NH2 | Chitosan NPs | AVP(5–4)-NH2 loaded Chitosan NPs |
|---|---|---|---|
| 53,4 ± 2,2 | 73,2 ± 4,6 | 70,9 ± 3,3 | 78,3 ± 7,1 |
Conclusion
In the modelling part of our study; the experimental medium of AVP (4–5)-NH2 peptide-loaded CS were created for the first time with in silico system and the stability of the peptide in this medium was carried out by molecular dynamics studies. To evaluate the molecular mechanism of AVP (4–5)-NH2 peptide with HSP70 protein, molecular docking calculations were investigated. The active binding mechanism between peptide and protein were described using theoretical calculation methods and a potent pharmacologically features of peptide was also unveiled by ADME profile. The characteristic wavenumbers of the AVP (4–5)-NH2 peptide were observed using IR and Raman spectra and the fundamental vibrational wavenumbers of the peptide were calculated at DFT-RB3LYP /6–31++G (d, p) basis set. Also, the assignments of the fundamental wavenumbers were assigned in accordance with the total energy distribution of the vibrational modes (TED) analysis.
In the experimental part of our study; AVP (4–5)-NH2 peptide-loaded CS NPs was prepared by a simple and mild method of ionic gelation method to overcome low bioavailability and to increase drug effectiveness. By using dynamic light scattering (DLS), Fourier-Transform Infrared (FT-IR) spectroscopy, UV-Vis Spectrometer characterization methods, the structure of the nanoparticles were revealed. The morphology of the prepared nanoparticles was observed from SEM, and the nanoparticles were spherical form. Based on the in vitro controlled release study against PBS at pH 7.4, in first day 17.23% of the peptide was released, but 61.13% of AVP (4–5)-NH2 peptide was released at the end of the 10th day. Neuroblastoma cells were particularly selected for the reason of AVP (4-5)-NH2 peptide has neuroprotective effect and used in the treatment of Alzheimer’s and Parkinson’s disease. There is limited information about cytotoxicity of AVP (4–5)-NH2 peptide, so we studied the cytotoxicity of AVP (4–5)-NH2 peptide on neuroblastoma cells. CS NPs with or without AVP (4–5)-NH2 peptide was firstly obtained in this study and their cytotoxicity were also examined. The results of cytotoxicity experiments showed that there is no cytotoxicity of AVP (4–5)-NH2 peptide or CS NPs with the concentrations we studied. In fact, they have neuroproliferative effect on SH-SY5Y cells. Their neuroprotective effect was examined with co-treatment with H2O2 and consistent with the literature, AVP (4–5)-NH2 peptide was shown as neuroprotective effect in vitro. There is no studies in literature on the encapsulation of AVP (4–5)-NH2 peptide with CS NPs, for the first time in literature, the neuroprotective efficacy of chitosan-loaded AVP (4–5)-NH2 was examined comparatively in this study, and it was shown that the combined effect of chitosan and AVP (4–5)-NH2 was much higher than AVP (4–5)-NH2 alone.
Electronic supplementary material
(PNG 302 kb)
(PNG 271 kb)
(PNG 1059 kb)
(PNG 978 kb)
(PNG 7524 kb)
(PNG 1162 kb)
(PNG 1627 kb)
(PNG 1038 kb)
(PNG 1938 kb)
(DOCX 13 kb)
(DOCX 12 kb)
(DOCX 12 kb)
Acknowledgements
Authors are also very thankful to Rita Podzuna for allowing using the docking program with Schrödinger’s Small-Molecule Drug Discovery Suite. In this study, the infrastructure of Applied Nanotechnology and Antibody Production Laboratory established with TUBITAK support (project numbers: 115S132 and 117S097) was used. Authors would thank to TUBITAK for their support.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Abbreviations
- AVP
Arginine-vaspressin
- CS NPs
Chitosan nanoparticles
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- NGF
Nerve growth factor
- HSP
Heat shock proteins
- MD
Molecular Dynamics
- NVT
Number of particles, Volume, and Temperature
- NPT
Number of particles, Pressure, and Temperature
- Rg
Gyration
- RMSD
Root mean square deviation
- VMD
Visual Molecular Dynamics
- ADME
Absorption, Distribution, Metabolism and Excretion
- TED
Total energy distribution
- PdI
Polydispersity index
- FT-IR
Fourier Transform Infrared
- SEM
Scanning Electron Microscopy
- DLS
Dynamic Light Scattering
- EE
Encapsulation Efficiency
- LC
Loading Capacity
- PBS
Phosphate Buffered Saline
- XTT
sodium 3,3′-[1(phenylamino)carbonyl]-3,4-tetrazolium]-3is(4-methoxy-6-nitro) benzene sulfonic acid hydrate
- DMSO
Dimethyl Sulfoxide
Authors’ contributions
SG: Participated in the design of the study, carried out the FTIR, band component analysis study, molecular docking, and molecular dynamic simulation and drafted the manuscript. YBK and TZ: Participated in the design of the experimental study, (synthesize and characterize nanoparticles) drafted the manuscript. RK: Participated in the design of the experimental study (cytotoxicity studies) drafted the manuscript. BB and YK: Participated in the design of the molecular docking and molecular dynamic simulation. AO and SA: Responsible for the study design and gave final approval of the version to be published. All authors read and approved the final manuscript and provide financial and administrative support.
Funding
This study was supported by the Research funds of Istanbul University [ONAP-2423].
Compliance with ethical standards
Conflict of interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zenina TA, Gudasheva TA, Bukreyev YS, Seredenin SB. Neuroprotective effect of dipeptide AVP (4-5)-NH2 is associated with nerve growth factor and heat shock protein HSP70. Bull Exp Biol Med. 2007;144(4):543–545. doi: 10.1007/s10517-007-0373-7. [DOI] [PubMed] [Google Scholar]
- 2.Evans CG, Wisén S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta (1-42) aggregation in vitro. J Biol Chem. 2006. [DOI] [PubMed]
- 3.Kreek MJ, Zhou Y, Levran O. Functions of arginine vasopressin and its receptors: importance of human molecular genetics studies in bidirectional translational research. Biol Psychiatry. 2011;70(6):502. doi: 10.1016/j.biopsych.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolkowitz OM, Rothschild AJ. Psychoneuroendocrinology: the scientific basis of clinical practice. American psychiatric pub. 2008.
- 5.Batla A, Phé V, De Min L, Panicker JN. Nocturia in Parkinson's disease: why does it occur and how to manage? Movement Disorders Clinical Practice. 2016;3(5):443–451. doi: 10.1002/mdc3.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witt SN. Hsp70 molecular chaperones and Parkinson's disease. Biopolymers: Original Research on Biomolecules. 2010;93(3):218–228. doi: 10.1002/bip.21302. [DOI] [PubMed] [Google Scholar]
- 8.Lu RC, Tan MS, Wang H, Xie AM, Yu JT, Tan L. Heat shock protein 70 in Alzheimer’s disease. Biomed Res Int. 2014. [DOI] [PMC free article] [PubMed]
- 9.Calzoni E, Cesaretti A, Polchi A, Di Michele A, Tancini B, Emiliani C. Biocompatible polymer nanoparticles for drug delivery applications in cancer and neurodegenerative disorder therapies. J Funct Biomater. 2019;10(1). [DOI] [PMC free article] [PubMed]
- 10.Zhao LM, Shi LE, Zhang ZL, Chen JM, Shi DD, Yang J, Tang ZX. Preparation and application of chitosan nanoparticles and nanofibers. Braz J Chem Eng. 2011;28(3):353–362. [Google Scholar]
- 11.Ghadi A, Mahjoub S, Tabandeh F, Talebnia F. Synthesis and optimization of chitosan nanoparticles: potential applications in nanomedicine and biomedical engineering. Caspian journal of internal medicine. 2014;5(3):156–161. [PMC free article] [PubMed] [Google Scholar]
- 12.Landriscina A, Rosen J, Friedman AJ. Biodegradable chitosan nanoparticles in drug delivery for infectious disease. Nanomedicine. 2015;10(10):1609–1619. doi: 10.2217/nnm.15.7. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Chi N, Tang X. Preparation of estradiol chitosan nanoparticles for improving nasal absorption and brain targeting. Eur J Pharm Biopharm. 2008;70(3):735–740. doi: 10.1016/j.ejpb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Sadigh-Eteghad S, Talebi M, Farhoudi M, Mahmoudi J, Reyhani B. Effects of levodopa loaded chitosan nanoparticles on cell viability and caspase-3 expression in PC12 neural like cells. Neurosciences. 2013;18(3):281–283. [PubMed] [Google Scholar]
- 15.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 16. Revision A. 2016;3.
- 16.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 18.da Silva AWS, Vranken WF. ACPYPE-Antechamber python parser interface. BMC research notes. 2012;5(1):367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balajee R, Rajan MD. Molecular docking and simulation studies of farnesyl trasnferase with the potential inhibitor theflavin. Journal of Applied Pharmaceutical Science. 2011;1(8):141. [Google Scholar]
- 20.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 21.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 23.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. The Journal of Chemical Physics. 2007;126(1). [DOI] [PubMed]
- 24.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. [Google Scholar]
- 25.Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 26.Turner PJ. XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR. 2005.
- 27.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 28.Sriram M, Osipiuk J, Freeman BC, Morimoto RI, Joachimiak A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure. 1997;5(3):403–414. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- 29.Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T. The SWISS-MODEL repository—new features and functionality. Nucleic Acids Res. 2016;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 31.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 32.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 33.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2015;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 34.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 35.Søndergaard CR, Olsson MH, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p K a values. J Chem Theory Comput. 2011;7(7):2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesan A, Rambabu M, Jayanthi S. Febin Prabhu Dass J. Pharmacophore feature prediction and molecular docking approach to identify novel anti-HCV protease inhibitors. J Cell Biochem. 2018;119(1):960–966. doi: 10.1002/jcb.26262. [DOI] [PubMed] [Google Scholar]
- 37.Divya K, Jisha MS. Chitosan nanoparticles preparation and applications. Environ Chem Lett. 2018;16(1):101–112. [Google Scholar]
- 38.Sullivan DJ, Cruz-Romero M, Collins T, Cummins E, Kerry JP, Morris MA. Synthesis of monodisperse chitosan nanoparticles. Food Hydrocoll. 2018;83:355–364. [Google Scholar]
- 39.Ahsan SM, Thomas M, Reddy KK, Sooraparaju SG, Asthana A, Bhatnagar I. Chitosan as biomaterial in drug delivery and tissue engineering. Int J Biol Macromol. 2018;110:97–109. doi: 10.1016/j.ijbiomac.2017.08.140. [DOI] [PubMed] [Google Scholar]
- 40.Kilinc YB, Akdeste ZM, Koc RC, Bagirova M, Allahverdiyev A. Synthesis and characterization of antigenic influenza a M2e protein peptide-poly (acrylic) acid bioconjugate and determination of toxicity in vitro. Bioengineered. 2014;5(6):357–362. doi: 10.4161/21655979.2014.969131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erci F, Cakir-Koc R, Isildak I. Green synthesis of silver nanoparticles using Thymbra spicata L. var. spicata (zahter) aqueous leaf extract and evaluation of their morphology-dependent antibacterial and cytotoxic activity. Artificial Cells, Nanomedicine, and Biotechnology. 2018;46:150–158. doi: 10.1080/21691401.2017.1415917. [DOI] [PubMed] [Google Scholar]
- 42.Liwo A, Tempczyk A, Oldziej S, Shenderovich MD, Hruby VJ, Talluri S, et al. Exploration of the conformational space of oxytocin and arginine-vasopressin using the electrostatically driven Monte Carlo and molecular dynamics methods. Biopolymers. 1996;38(2):157–175. doi: 10.1002/(sici)1097-0282(199602)38:2<157::aid-bip3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt JM, Ohlenschläger O, Rüterjans H, Grzonka Z, Kojro E, Pavo I, Fahrenholz F. Conformation of [8-arginine] vasopressin and V1 antagonists in dimethyl sulfoxide solution derived from two-dimensional NMR spectroscopy and molecular dynamics simulation. Eur J Biochem. 1991;201(2):355–371. doi: 10.1111/j.1432-1033.1991.tb16293.x. [DOI] [PubMed] [Google Scholar]
- 44.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 45.Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Leo A, Hansch C, Elkins D. Partition coefficients and their uses. Chem Rev. 1971;71(6):525–616. [Google Scholar]
- 47.Tihanyi K, Vastag M (Eds.). Solubility, delivery and ADME problems of drugs and drug-candidates. Bentham Science Publishers. 2011.
- 48.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 49.Johansen A, Hansen HD, Svarer C, Lehel S, Leth-Petersen S, Kristensen JL, et al. The importance of small polar radiometabolites in molecular neuroimaging: a PET study with [11C] Cimbi-36 labeled in two positions. J Cereb Blood Flow Metab. 2018;38(4):659–668. doi: 10.1177/0271678X17746179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpenter TS, Kirshner DA, Lau EY, Wong SE, Nilmeier JP, Lightstone FC. A method to predict blood-brain barrier permeability of drug-like compounds using molecular dynamics simulations. Biophys J. 2014;107(3):630–641. doi: 10.1016/j.bpj.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dressman JB, Lennernas H. Oral drug absorption: prediction and assessment: CRC Press; 2000.
- 52.Mohammadpour Dounighi N, Eskandari R, Avadi MR, Zolfagharian H. Mir Mohammad Sadeghi a, Rezayat M. preparation and in vitro characterization of chitosan nanoparticles containing Mesobuthus eupeus scorpion venom as an antigen delivery system. Journal of Venomous Animals and Toxins Including Tropical Diseases. 2012;18(1):44–52. [Google Scholar]
- 53.Anicuta SG, Dobre L, Stroescu M, Jipa I. Fourier transform infrared (FTIR) spectroscopy for characterization of antimicrobial films containing chitosan. Analele Universită Ńii din Oradea Fascicula: Ecotoxicologie, Zootehnie şi Tehnologii de Industrie Alimentară. 2010:1234–40.
- 54.Liu CG, Desai KGH, Chen XG, Park HJ. Preparation and characterization of nanoparticles containing trypsin based on hydrophobically modified chitosan. J Agric Food Chem. 2005;53(5):1728–1733. doi: 10.1021/jf040304v. [DOI] [PubMed] [Google Scholar]
- 55.Negrea P, Caunii A, Sarac I, Butnariu M. The study of infrared spectrum of chitin and chitosan extract as potential sources of biomass. Digest Journal of Nanomaterials & Biostructures (DJNB). 2015;10(4).
- 56.Silva SM, Braga CR, Fook MV, Raposo CM, Carvalho LH, Canedo EL. Application of infrared spectroscopy to analysis of chitosan/clay nanocomposites. In Infrared Spectroscopy-Materials Science, Engineering and Technology. InTech. (2012).
- 57.Mazancová P, Némethová V, Treľová D, Kleščíková L, Lacík I, Rázga F. Dissociation of chitosan/tripolyphosphate complexes into separate components upon pH elevation. Carbohydr Polym. 2018;192:104–110. doi: 10.1016/j.carbpol.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 58.PQS version 3.1, Parallel Quantum Solutions, 2013 Green Acres Road, Suite A Fayetteville, Arkansas, 72703 USA.
- 59.Balci K, Akyuz S. A vibrational spectroscopic investigation on benzocaine molecule. Vib Spectrosc. 2008;48(2):215–228. [Google Scholar]
- 60.Murdock RC, Braydich-Stolle L, Schrand AM, Schlager JJ, Hussain SM. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol Sci. 2008;101(2):239–253. doi: 10.1093/toxsci/kfm240. [DOI] [PubMed] [Google Scholar]
- 61.Patil P, Bhoskar M. Optimization and evaluation of spray dried chitosan nanoparticles containing doxorubicin. Int J Curr Pharm Res. 2014;6(1):7–15. [Google Scholar]
- 62.Hu YL. Qi W, Han F, Shao JZ. Gao JQ Toxicity evaluation of biodegradable chitosan nanoparticles using a zebrafish embryo model International journal of nanomedicine. 2011;6:3351. doi: 10.2147/IJN.S25853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang ZX, Qian JQ, Shi LE. Preparation of chitosan nanoparticles as carrier for immobilized enzyme. Appl Biochem Biotechnol. 2007;136(1):77–96. doi: 10.1007/BF02685940. [DOI] [PubMed] [Google Scholar]
- 64.Jarudilokkul S, Tongthammachat A, Boonamnuayvittaya V. Preparation of chitosan nanoparticles for encapsulation and release of protein. Korean J Chem Eng. 2011;28(5):1247. [Google Scholar]
- 65.Wang JJ, Zeng ZW, Xiao RZ, Xie T, Zhou GL, Zhan XR, Wang SL. Recent advances of chitosan nanoparticles as drug carriers. Int J Nanomedicine. 2011;6:765. doi: 10.2147/IJN.S17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dudhani AR, Kosaraju SL. Bioadhesive chitosan nanoparticles: preparation and characterization. Carbohydr Polym. 2010;81(2):243–251. [Google Scholar]
- 67.Huyck L, Ampe C, Van Troys M. The XTT cell proliferation assay applied to cell layers embedded in three-dimensional matrix. Assay and drug development technologies. 2012;10(4):382–392. doi: 10.1089/adt.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.López-García J, Lehocký M, Humpolíček P, Sáha P. HaCaT keratinocytes response on antimicrobial atelocollagen substrates: extent of cytotoxicity, cell viability and proliferation. Journal of functional biomaterials. 2014;5(2):43–57. doi: 10.3390/jfb5020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao D, Yu S, Sun B, Gao S, Guo S, Zhao K. Biomedical applications of chitosan and its derivative nanoparticles. Polymers. 2018;10(4):462. doi: 10.3390/polym10040462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho Y, Shi R, Borgens RB. Chitosan nanoparticle-based neuronal membrane sealing and neuroprotection following acrolein-induced cell injury. J Biol Eng. 2010;4(1):2. doi: 10.1186/1754-1611-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pangestuti R, Kim SK. Neuroprotective properties of chitosan and its derivatives. Marine Drugs. 2010;8(7):2117–2128. doi: 10.3390/md8072117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 302 kb)
(PNG 271 kb)
(PNG 1059 kb)
(PNG 978 kb)
(PNG 7524 kb)
(PNG 1162 kb)
(PNG 1627 kb)
(PNG 1038 kb)
(PNG 1938 kb)
(DOCX 13 kb)
(DOCX 12 kb)
(DOCX 12 kb)
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.